Abstract
Bacteriocin producers normally possess dedicated immunity systems to protect themselves from their own bacteriocins. Lactococcus lactis strains LMG2081 and BGBM50 are known as lactococcin G producers. However, BGBM50 was sensitive to LMG2081, which indicated that LMG2081 might produce additional bacteriocins that are not present in BGBM50. Therefore, whole-genome sequencing of the two strains was performed, and a lantibiotic operon (called lctLMG) was identified in LMG2081 but not in BGBM50. The lctLMG operon contains six open reading frames; the first three genes, lmgA, lmgM, and lmgT, are involved in the biosynthesis and export of bacteriocin, while the other three genes, lmgF, lmgE, and lmgG, are involved in lantibiotic immunity. Mutational analysis confirmed that the lctLMG operon is responsible for the additional antimicrobial activity. Specifically, site-directed mutation within this operon rendered LMG2081 inactive toward BGBM50. Subsequent purification and electrospray ionization–time of flight mass spectrometric analysis confirmed that the lantibiotic bacteriocin called lacticin LMG is exported as a 25-amino-acid peptide. Lacticin LMG is highly similar to the lacticin 481 group. It is interesting that a bacteriocin producer produces two different classes of bacteriocins, whose operons are located in the chromosome and a plasmid.
INTRODUCTION
Many lactic acid bacteria (LAB) have the ability to produce ribosomally synthesized antimicrobial peptides or proteins, known as bacteriocins. These proteins are antimicrobials with different modes of action against closely related bacterial species, medically important pathogens, and bacteria involved in food spoilage (1). LAB frequently found in food constitute an important portion of our gut microbiota. Additionally, many LAB are generally recognized as safe (GRAS) for ingestion, which is one of the prerequisites for use as natural preservatives in foods and feeds and as antimicrobials in the treatment of infections. Bacteriocins produced by LAB have been studied intensively in recent years.
Bacteriocins produced by LAB are divided into two primary classes, i.e., lantibiotics (class I) and unmodified bacteriocins (class II) (2). Class I bacteriocins (lantibiotics) contain unusual amino acids such as lanthionine and dehydrated amino acids, as a result of posttranslational modifications such as dehydration of serine and threonine and formation of lanthionine bridges. Class II bacteriocins consist only of unmodified peptides or peptides with minor modifications (e.g., sulfide bridges and cyclization). Furthermore, class II bacteriocins are classified into four subclasses, i.e., pediocin-like bacteriocins (class IIa), two-peptide bacteriocins (class IIb), cyclic bacteriocins (class IIc), and linear non-pediocin-like bacteriocins (class IId) (3).
Thus far, over 95 different lantibiotics have been isolated from Gram-positive bacteria and characterized (4). According to the topology of their structures, lantibiotics have been classified as type A and type B (5). Type A lantibiotics are small (2 to 5 kDa) and can be elongated [subtype type A(I)] or contain a tail-and-ring region [subtype type A(II)] (6). For type A(I) lantibiotics, lanthionine and 3-methyllanthionine residues are formed by the action of two distinct enzymes (LanB and LanC) after the dehydration of serine and threonine; for type A(II) lantibiotics, in contrast, these residues are formed by the action of one enzyme (LanM) (7). Furthermore, for type A(I) lantibiotics, peptides are exported outside the cell and the leader peptide is removed by the exporter LanT and the protease LanP (4). For type A(II) lantibiotics, however, both processes (i.e., leader peptide cleavage and mature lantibiotic transport) are catalyzed by LanT (4), an exporter with an N-terminal protease domain. The immunity system of type A(II) lantibiotics is encoded and realized through the coexpression of three genes, namely, lanF, lanE, and lanG (8). This system of three genes ensures immunity by preventing lantibiotics from reaching the density necessary for pore formation, which is their mode of action. For some lantibiotics, such as nukacin ISK-1, the presence of two immunity systems (NukFEG and NukH) enhanced immunity, compared with the presence of only one system (9). The expression of nukFEG genes resulted in a greater degree of immunity than did nukH gene expression alone, which suggests that the NukFEG system plays a major role in immunity, while the NukH protein most likely functions as an accessory protein. Type B lantibiotics, such as mersacidin, cinnamycin, duramycin, and ancovenin, are small (approximately 2 kDa), are more globular and compact, and kill sensitive cells by interfering with cellular enzymatic reactions such as cell wall synthesis (10).
Nisin, a type A(I) lantibiotic, is one of the best known lantibiotics, with commercial applications in food processing and fermentation (11). Type A(II) lantibiotics, such as lacticin 481 and related lantibiotics, have demonstrated potential for use in food production. Previous results showed that lacticin 481, because it is active against LAB, can be used to speed cheese ripening by lysing starter cultures and increasing the amounts of intracellular enzymes (12–14). In addition, lacticin 481 could be used to inhibit the growth of nonstarter bacteria in cheese, which can cause major economic losses (15). Lacticin 481 and related lantibiotics do not show activity against pathogenic bacteria. In some cases, however, lacticin 481, in combination with high-pressure treatments, affected the survival of Listeria monocytogenes, Staphylococcus aureus, and Escherichia coli O157:H7 in raw milk cheeses (16–18).
Lactococcus lactis strains LMG2081 and BGBM50 are known as producers of lactococcin G, a class IIb bacteriocin (19, 20). In cross-immunity tests, it was observed that strain LMG2081 inhibited the growth of strain BGBM50. To determine the differences between these two strains, whole-genome sequencing of strains LMG2081 and BGBM50 was performed. Genome analysis showed that strain LMG2081 contains an operon for the synthesis of a novel bacteriocin, lacticin LMG, belonging to the type A(II) class of lantibiotics. Purification and electrospray ionization–time of flight (ESI-TOF) mass spectrometric analysis showed that lacticin LMG is exported as a 25-amino-acid peptide.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. L. lactis LMG2081 was isolated from the European pear (Pyrus communis). Lactococcal strains were grown at 30°C in M17 medium (Merck GmbH, Darmstadt, Germany) supplemented with d-glucose (0.5% [wt/vol]) (GM17), unless otherwise indicated. For the cloning and propagation of constructs, Escherichia coli DH5α and EC101 were used; they were grown aerobically at 37°C in Luria-Bertani (LB) broth (21). Agar plates were prepared by adding 1.5% (wt/vol) agar (Torlak, Belgrade, Serbia) to the liquid medium. Lactococcal transformants were selected on GM17 plates containing 10 μg/ml erythromycin (Sigma-Aldrich Chemie GmbH, Munich, Germany), while E. coli transformants were selected on LB plates containing 300 μg/ml erythromycin or 100 μg/ml ampicillin, depending on the plasmids used. For blue/white screening of colonies carrying vectors with cloned fragments, 5-bromo-4-chloro-3-indolyl-d-galactoside (X-Gal) (Fermentas, Vilnius, Lithuania) was added to LB plates at a final concentration of 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Lactococcus lactis strains | ||
| LMG2081 | Bacteriocin (lactococcin G and lacticin LMG) producer | 19 |
| BGBM50 | Lactococcin G producer | 37 |
| BGMN1-596 | Plasmid-free derivate of L. lactis subsp. lactis BGMN1-5 | 35 |
| LMG2081/pG+host9lcnG | LcnG−, LctLMG+, LctLMGr | This work |
| LMG2081/pG+host9lctLMG | LcnG+, LctLMG−, LcnGr | This work |
| Escherichia coli strains | ||
| DH5α | λ− Ф80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 38 |
| EC101 | JM101 containing repA gene of pWV01 in chromosome | 39 |
| DH5α/pGEM-T EasylcnG | This work | |
| DH5α/pGEM-T EasylctLMG | This work | |
| DH5α/pGEM-T EasyLL/LD | This work | |
| EC101/pG+host9lcnG | This work | |
| EC101/pG+host9lctLMG | This work | |
| Plasmids | ||
| pG+host9 | Emr; thermosensitive vector | 40 |
| pG+host9lcnG | pG+host9 carrying fragment of lcnG gene | This work |
| pG+host9lctLMG | pG+host9 carrying fragment of lmgM gene | This work |
| pGEM-T Easy | 3,015 bp; Ampr; PCR cloning vector | Promega |
| pGEM-T EasylcnG | This work | |
| pGEM-T EasylctLMG | pGEM-T Easy carrying 814 bp of lmgM gene | This work |
| ppGEM-T EasyLL/LD | This work |
Bacteriocin activity.
The bacteriocin activity of tested strains and mutants was analyzed by an agar well diffusion test (22). Bacteriocin extracts were obtained after centrifugation (15,871 × g for 10 min) of 16-h-old cultures and subsequent filtration through filters with 0.45-μm pores (Sarstedt, Numbrecht, Germany). Lactococcus lactis subsp. lactis BGMN1-596 was used as an indicator strain. Positive signals of bacteriocin activity presented as clear zones of inhibition around the wells.
Whole-genome sequencing.
DNA from strain LMG2081 was sequenced at the Norwegian Sequencing Centre (Oslo, Norway) using an Illumina MiSeq instrument, according to the manufacturer's recommendations. Approximately 1.5 million paired-end reads of 2 by 250 bp were obtained. Using ABySS, a preliminary assembly of the LMG2081 genome was obtained, with 100 contigs covering 2,548,962 bp.
Inverse PCR.
The inverse PCR procedure was used to determine neighboring contigs. The plasmid DNA of strain LMG2081 was digested with EcoRI and then self-ligated to yield circularized fragments. The ligation mixture was used as the template for PCR with the specific primers LL and LD (Table 2). The PCR fragments obtained were purified and cloned into a commercial pGEM-T Easy vector to yield pGEM-T EasyLL/LD, using E. coli DH5α. The resulting products were purified and sequenced.
TABLE 2.
Sequences of specific primers used in this study
| Primer name | Sequence of primer | Gene | Source or reference |
|---|---|---|---|
| LcnG-Fw | 5′-GAAAGAATTATCAGAAAAAG-3′ | Lactococcin G gene | 41 |
| LcnG-Rev | 5′-CCACTTATCTTTATTTCCCTCT-3′ | Lactococcin G gene | 41 |
| LctLMG-Fw | 5′-TGCAGAAGTGGTTACGG-3′ | lmgM gene (position, bp 1095–1111) | This work |
| LctLMG-Rev | 5′-GGTTGAATAAGCAGGAG-3′ | lmgM gene (position, bp 1892–1908) | This work |
| LL | 5′-CCATTTTGCTAGTACAGTC-3′ | This work | |
| LD | 5′-GAACATAGTATGCAAAGGGG-3′ | This work |
Construction of mutants.
Mutants for the bacteriocins lactococcin G and lacticin LMG were constructed using the plasmid pG+host9 (carrying segments of the targeted genes), by insertion into operons. The thermosensitive erythromycin-resistant plasmid pG+host9 was used to disrupt the lcnG and lmgM genes (24). PCR fragments of 378 bp and 814 bp, containing segments of the lcnG and lmgM genes, respectively, were cloned into a pGEM-T Easy vector (Promega, Madison, WI, USA) to yield pGEM-T EasylcnG and pGEM-T EasylctLMG, respectively. Then, fragments were subcloned into a pG+host9 vector using the EcoRI restriction enzyme, resulting in the constructs pG+host9lcnG and pG+host9lctLMG. These plasmid constructs were introduced into L. lactis LMG2081 by electroporation. To enable integration of the constructs, the transformants obtained at 28°C were streaked onto GM17 agar plates with erythromycin (10 μg/ml) and incubated at 37°C for 48 h. Transformants containing the plasmids pG+host9lcnG and pG+host9lctLMG integrated in the lcnG and lmgM genes, respectively, were confirmed by their Bac− phenotype, sensitivity to bacteriocin, and pulsed-field gel electrophoresis (PFGE) and hybridization analysis results. Southern blot hybridization was performed with a digoxigenin (DIG) DNA labeling and detection kit (Roche Diagnostics GmbH, Germany), following the manufacturer's protocol, using an 814-bp fragment from lmgM as a biotin-labeled probe.
Molecular methods.
For clonal confirmation, PFGE was used (25). For the isolation of total DNA from lactococci, a modified version (with the addition of lysozyme treatment) of the method described by Hopwood et al. (26) was used, while the isolation of plasmid DNA from lactococci was performed by the miniprep method described by O'Sullivan and Klaenhammer (23). Plasmid DNA from E. coli was isolated by using a QIAprep spin miniprep kit (Qiagen, Hilden, Germany), according to the manufacturer's protocol. For DNA ligation, T4 DNA ligase (Agilent Technologies, Santa Clara, CA, USA) was used, according to the manufacturer's instructions. Plasmid constructs were inserted in lactococci by electroporation using an Eporator (Eppendorf, Hamburg, Germany) (27). For the amplification of DNA fragments by PCR using a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA, USA), KapaTaq DNA polymerase (KapaBiosystem, Inc., Boston, MA, USA) was used. A QIAquick gel extraction kit was used for the purification of DNA fragments from agarose gels, according to the manufacturer's recommendations (Qiagen, Hilden, Germany). For the purification of PCR products, a QIAquick PCR purification kit (Qiagen) was used, according to the supplier's protocol. For the cloning of PCR products, the pGEM-T Easy (Promega) vector was used. The purified PCR products obtained were sequenced by the Macrogen Sequencing Service (Macrogen Europe, Amsterdam, The Netherlands) and analyzed by using BLAST. The sets of specific primers used in this study are listed in Table 2. The functions of the proteins encoded by the lctLMG operon were attributed on the basis of homology with proteins determined by BLAST comparisons with the Entrez protein database (Table 3). Sequence comparisons of LmgA and representative lantibiotics forming the lacticin 481 group were performed using Clustal Omega software (http://www.ebi.ac.uk/Tools/msa/clustalo).
TABLE 3.
Results of BLAST comparison of proteins encoded by lctLMG operon with Entrez protein database
| ORF | No. of amino acids (position)a | Protein with greatest homology (accession no.) | Predicted domain(s) or superfamily encoded by ORF | Organism | Amino acid identity (%) |
|---|---|---|---|---|---|
| lmgA | 49 (bp 118–267) | Lantibiotic nukacin (WP_037580734.1) | Type A lantibiotic (pfam04604) | Streptococcus equi | 69 |
| lmgA | 49 (bp 118–267) | Lantibiotic lacticin 481 (WP_032489363.1) | Type A lantibiotic (pfam04604) | Lactococcus lactis | 73 |
| lmgM | 884 (bp 757–3411) | Lacticin 481/lactococcin biosynthesis protein LcnDR2 (WP_032489360.1) | Lantibiotic-modifying enzyme (COG4403), cyclases involved in biosynthesis of class II lantibiotics (cd04792) | Lactococcus lactis | 45 |
| lmgT | 696 (bp 3420–5510) | Lacticin 481/lactococcin DR transport/processing ATP-binding protein LcnDR3 (WP_032489362.1) | Peptidase C39 family (pfam03412) | Lactococcus lactis | 53 |
| lmgF | 303 (bp 5587–6498) | Bacitracin transport ATP-binding protein BcrA (CIT27467.1) | ATP-binding cassette domain of bacitracin-resistance transporter (cd03268) | Streptococcus pneumoniae | 65 |
| lmgE | 250 (bp 6498–7250) | NukE (AKQ51582.1) | ABC-2 family transporter protein (pfam12730), lantibiotic protection ABC transporter permease subunit (TIGR03733) | Staphylococcus epidermidis | 48 |
| lmgG | 245 (bp 7247–7984) | NukG (AKQ51591.1) | Predicted lantibiotic-exporting membrane permease, EfiE/EfiG/ABC2 family (COG4200) | Staphylococcus epidermidis | 49 |
The number of amino acids and the position of the lctLMG operon from the beginning of the sequence are given for each ORF.
Purification and mass spectrometric analysis of bacteriocin lacticin LMG.
For the purification and mass spectrometric analysis of the bacteriocin lacticin LMG, a L. lactis LMG2081 lcnG mutant was grown at 30°C for 16 h in 500 ml of GM17 broth, to the early stationary phase. The removal of cells was performed by centrifugation (4,500 × g for 30 min at 4°C). The bacteriocin present in the supernatant was subjected to precipitation with ammonium sulfate at 40% (wt/vol) saturation, with stirring, for 3 h at 4°C. The precipitate was collected by centrifugation (10,000 × g for 30 min at 4°C) and then dissolved in 5 ml of Milli-Q water containing 0.1% trifluoroacetic acid. The bacteriocin was purified using reverse-phase high-performance liquid chromatography (HPLC). Reverse-phase chromatography of the bacteriocin sample was performed using an Äkta Purifier 10 system (GE Healthcare, Uppsala, Sweden) with a Discovery BIO Wide Pore C5 column (10 cm by 4.6 mm; particle size, 5 μm; Supelco, Bellefonte, PA, USA). The protein was eluted using an acetonitrile gradient (0 to 90% with 0.1% trifluoroacetic acid for 10 column volumes). The chromatography was monitored by measuring absorbance at 215 nm. The protein fractions obtained were dried under a stream of nitrogen, dissolved in the same volume of Milli-Q water, and tested for antibacterial activity.
Mass spectrometric analysis.
Mass analysis of the isolated bacteriocin was carried out using a mass spectrometer coupled with HPLC. The sample was injected onto a reverse-phase C18 column (RRHT column; 4.6 by 50 mm; particle size, 1.8 μm) coupled with a Zorbax Eclipse XDB-C18 column installed in a 1200 series HPLC system (Agilent Technologies). The sample components were separated using an acetonitrile gradient (5 to 95% with 0.2% formic acid for 10 min and then 95% for 5 min). The mass spectrometer, a 6210 TOF liquid chromatography-mass spectrometry (LC-MS) system (G1969A; Agilent Technologies, Santa Clara, CA, USA), was run in positive ESI mode with a capillary voltage of 4,000 V, a fragmentor voltage of 200 V, and a mass range of m/z 100 to 3,200. Agilent MassHunter Workstation software and Analyst QS were used for data processing.
Nucleotide sequence accession number.
The DNA sequence presented in this article has been deposited in the European Nucleotide Archive (ENA) and GenBank databases, under accession no. LN879392.
RESULTS
Genome sequencing results.
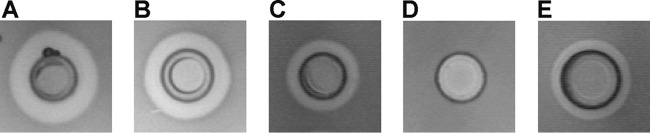
In a bacteriocin assay, we observed that the lactococcin G producer L. lactis LMG2081 killed not only the L. lactis subsp. lactis BGMN1-596 indicator strain but also L. lactis subsp. lactis BGBM50, another lactococcin G producer. This indicated that LMG2081 produces one or more bacteriocins that are not present in BGBM50 (Fig. 1A and B).
FIG 1.
Analysis of the bacteriocin activity of L. lactis LMG2081 and its mutants. (A) LMG2081 against BGMN1-596. (B) LMG2081 against BGBM50 (lactococcin G producer). (C) LMG2081/pG+host9lcnG (ΔlcnG) against BGMN1-596. (D) LMG2081/pG+host9lctLMG (ΔlcnLMG) against BGBM50. (E) LMG2081/pG+host9lctLMG against BGMN1-596.
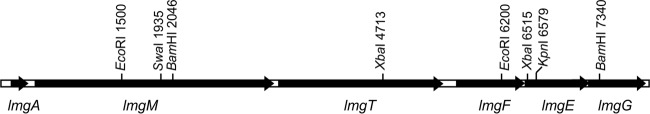
To search for additional bacteriocins, we performed genome sequencing of LMG2081. An incomplete bacteriocin-like operon was identified at the end of one contig. By primer walking, the entire operon was revealed in an 8-kb fragment. The operon, called lctLMG, contains six open reading frames (ORFs), named lmgA, lmgM, lmgT, lmgF, lmgE, and lmgG (Fig. 2; Table 3). In silico analysis of the surrounding sequences of lctLMG revealed a downstream ORF encoding a Pin-related site recombinase/DNA invertase with the greatest amino acid identity (83%) with the Pin-related site recombinase/DNA invertase from L. lactis (GenBank accession no. WP_046782557.1). The flanking DNA sequence from the upstream region is currently unavailable. The lmgA gene appears to be a structural gene encoding a 49-amino-acid precursor peptide that is expected to be cleaved between alanine and glycine following posttranslational modifications, to form a 25-residue mature peptide. We derived a consensus sequence of lacticin 481-like bacteriocins based on invariant residues in a protein sequence comparison of representative lantibiotics forming the lacticin 481 group and LmgA (Fig. 3). The N-terminal extremities, or leader peptides, of the peptides analyzed were variable in length and sequence but contained three invariant residues. The propeptides of the lantibiotics compared were more conserved than the leader peptides, and we found 12 invariant residues in the propeptides. The C termini of these propeptides corresponded to invariant regions of four amino acids (TCCS). In addition to invariant residues, we found four conserved residues in the leader peptides and seven conserved residues in the propeptides (Fig. 3).
FIG 2.
Linear gene map of the region carrying the 8,000-bp lctLMG operon in L. lactis LMG2981. The positions of relevant restriction sites are indicated. Arrows, size and orientation of predicted ORFs.
FIG 3.
Alignment of the deduced peptide sequence for lacticin LMG and homologous type A(II) lantibiotics, including nukacin ISK-1 from Staphylococcus warneri (GenBank accession no. Q9KWM4), lacticin 481 from Lactococcus lactis (GenBank accession no. WP_032489363), variacin from Kocuria varians (GenBank accession no. CAA63706), butyrivibriocin OR79 from Butyrivibrio fibrisolvens (GenBank accession no. AAC19355), lacticin LmgA from Lactococcus lactis (GenBank accession no. CUH82811.1), and salivaricin G32 from Streptococcus salivarius (GenBank accession no. AFB74480). Underlined amino acids, leader peptide; x, undefined residues. Gray shading and asterisks indicate identical amino acids, periods indicate amino acids belonging to similar groups, and colons indicate amino acids belonging to the same group.
In the region downstream of the lmgA gene, an ORF was found and designated lmgM (2,654 bp). The product of lmgM is likely a modification enzyme involved in the biosynthesis of the lantibiotic lacticin LMG (Table 3) (28, 29). Moreover, lmgT (2,090 bp) presumably codes for the ABC-containing maturation and secretion (AMS) protein, which cleaves off the leader peptide and secretes the mature lantibiotic (Table 3) (30, 31). At the end of the operon, three genes, i.e., lmgF (911 bp), lmgE (752 bp), and lmgG (737 bp), likely code for proteins similar to the ABC transporter and provide immunity by ejecting the lantibiotic from the membrane (Table 3). Similar immunity systems are present for the lantibiotics lacticin 481, mutacin II, and streptococcin SA-FF22; in each case, a system of three genes, lanF, lanE, and lanG, provides immunity against lantibiotics (32). In the case of the lantibiotic nukacin ISK-1, the presence of two immunity systems, NukFEG and NukH, provides a much higher level of immunity than each system alone (9).
Construction of mutant strains.
To check the functionality of bacteriocin operons in strain LMG2081, insertional mutagenesis of structural genes for the bacteriocins lactococcin G and lacticin LMG was performed separately. Plasmid constructs (pG+host9lcnG and pG+host9lctLMG) were integrated into the corresponding genes lcnG and lmgM, yielding two different mutants, i.e., LMG2081/pG+host9lcnG (minus lactococcin G) and LMG2081/pG+host9lctLMG (minus lacticin LMG), respectively. Transformants of L. lactis LMG2081 were grown at 37°C for 48 h and were analyzed by Bac− phenotype analysis, PFGE, and hybridization. The mutants with an integrated pG+host9lcnG plasmid construct in the lcnG gene showed lower activity against the L. lactis subsp. lactis BGMN1-596 indicator strain than did the wild-type (WT) LMG2081 strain (Fig. 1A and C). The second group of mutants, which had an integrated pG+host9lctLMG plasmid construct in the lmgM gene, lost inhibition activity against L. lactis subsp. lactis BGBM50 (lactococcin G producer) (Fig. 1D) and showed significantly lower activity against BGBMN1-596 (Fig. 1E). The high antimicrobial activity of strain LMG2081 is the result of the additive effects of the bacteriocins lactococcin G and lacticin LMG. The bacteriocin activity of the constructed mutants confirmed that strain LMG2081 contains two functional bacteriocin operons. The antimicrobial activities of the WT strain and both mutants were tested against various pathogenic or nonpathogenic bacteria. The WT strain and mutants producing lacticin LMG or lactococcin G showed activity only against lactococci.
Localization of lacticin LMG operon.
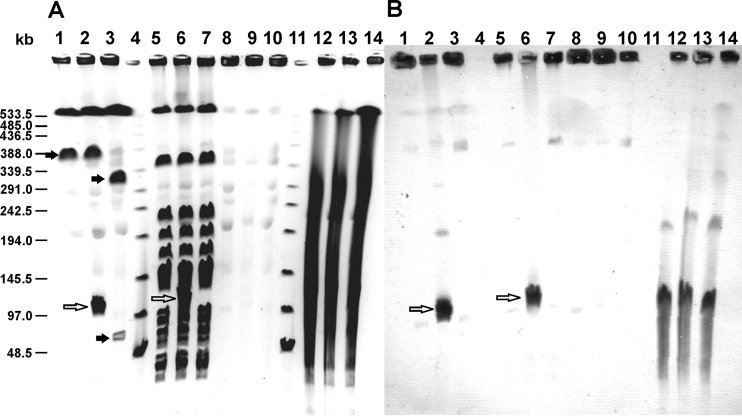
PFGE together with Southern blot hybridization was used for localization of the lacticin LMG and lactococcin G operons in the L. lactis LMG2081 strain. For that purpose, total DNA of the WT strain and the two mutants (with a disrupted lacticin LMG operon [LMG2081/pG+host9lctLMG] or lactococcin G operon [LMG2081/pG+host9lcnG]) that had been digested with NotI or SmaI, not digested, or treated with S1 nuclease was used. Bacteriocin mutants of strain LMG2081 were made by integration of a pG+host9 plasmid carrying NotI and SmaI sites, which are introduced into given operons by plasmid integration. The results of PFGE-hybridization experiments confirmed that the lctLMG operon is located on a plasmid of approximately 115 kb that does not possess NotI and SmaI restriction sites (Fig. 4A and B, lanes 2 and 6). In addition, when nondigested DNA and S1 nuclease-treated DNA from LMG2081 and the mutants were analyzed by PFGE and hybridization, signals from the lacticin LMG (lmgM) probe were obtained only with extrachromosomal bands (Fig. 4B, lanes 12 to 14) that are positioned on the gel in the region of approximately 115 kb, very similar to the findings obtained with linearized plasmids. In contrast, the lactococcin G operon is chromosomally located on a NotI fragment of 380 kb. Integration of pG+host9 into the lactococcin G operon split this fragment into fragments of 310 and 70 kb (Fig. 4A, lane 3).
FIG 4.
PFGE profiles of Lactococcus lactis LMG2081 and lacticin LMG and lactococcin G mutants (A) and Southern blot hybridization with a lmgM probe (B). Total DNA from Lactococcus lactis LMG2081 (lanes 1, 5, 8, and 12), LMG2081/pG+host9lctLMG (lanes 2, 6, 9, and 13), or LMG2081/pG+host9lcnG (lanes 3, 7, 10, and 14) was digested with NotI (lanes 1 to 3) or SmaI (lanes 5 to 7), not digested (lanes 8 to 10), or treated with S1 nuclease (lanes 12 to 14). Lanes 4 and 11, λ concatemers. White arrows, linearized plasmids carrying the lctLMG operon; black arrows, differences in chromosomal fragments between the WT strain and the lactococcin G mutant.
Mass spectrometry.
A novel active lacticin LMG was purified to homogeneity by ammonium sulfate precipitation and reverse-phase chromatography. The active fraction was spotted on GM17 soft agar that had been inoculated with L. lactis subsp. lactis BGBM50, for 24 h at 30°C (Fig. 5A). The molecular mass determined for the active fraction by ESI-TOF mass spectrometry was 2,759 Da (Fig. 5B). The mass analysis showed that the recorded molecular ions of the active fraction were m/z 2,760 ([M + H]+), 2,782 ([M + Na]+), and 2,798 ([M + K]+). Based on amino acid sequence analysis, cleavage of the leader peptide occurred between amino acids Ala23 and Gly24 to form a 25-residue excreted peptide with a calculated molecular mass of 2,831 Da. Considering the high degree of conservation of important amino acid residues between lacticin LMG and lacticin 481-type lantibiotics, it could be assumed that lacticin LMG underwent the same types of posttranslational modifications as its homologue lacticin 481. Lacticin 481-type lantibiotics contain threonine, serine, and cysteine, which tend to be converted to unusual amino acids such as dehydrobutyrine, lanthionine, and 3-methyllanthionine, followed by the elimination of one water molecule. Therefore, the positions of the thioether bridges were predicted to be between residues 7 and 12, 9 and 23, and 16 and 24, while the position of dehydrobutyrine was predicted to be at residue 22. According to the proposed posttranslational modifications, the calculated molecular mass of mature lacticin LMG was 2,759 Da, exactly the same as determined by mass spectrometry.
FIG 5.
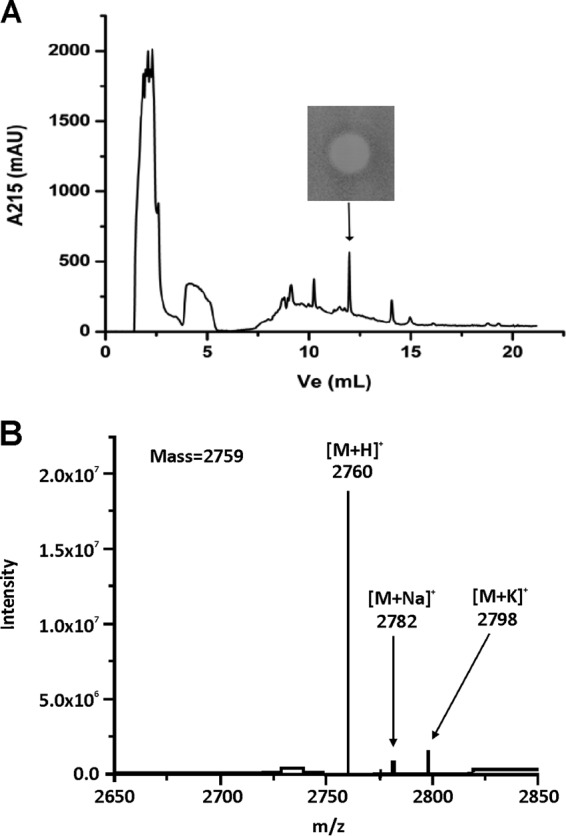
Bioassay for the production of lacticin LMG in L. lactis LMG2081 and mass spectrometric analysis. (A) Reverse-phase HPLC. The active fraction was spotted on GM17 soft agar that had been inoculated with the indicator strain L. lactis subsp. lactis BGBM50. (B) Calculated molecular mass of lacticin LMG, determined by ESI-TOF mass spectrometry.
DISCUSSION
In this study, we provide strong evidence that lacticin LMG is a novel lantibiotic produced by L. lactis strain LMG2081. The lctLMG operon showed organization similar to that of the other operons of the lacticin 481 group. Downstream of the operon, we identified an ORF encoding a Pin-related site recombinase/DNA invertase, indicating the end of the lctLMG operon. However, the sequence upstream of the lmgA gene is presently not available; therefore, whether there are additional genes involved in the synthesis or regulation of expression of lacticin LMG is not known. Analysis of the lctLMG operon sequence showed that lacticin LMG was synthesized as a 49-amino-acid precursor peptide, similar to lacticin 481-type lantibiotics. The amino acid sequences of the propeptide of lacticin LMG and the leader peptide show different levels of identity with those of lacticin 481 and other representative lacticin 481-like lantibiotics (Fig. 3). The propeptide of lacticin LMG shares 76% and 74% identity with lacticin 481 and nukacin ISK-1, respectively, while the leader sequence of lacticin LMG shares 76% identity with lacticin 481 and 50% identity with nukacin ISK-1. The mature lacticin LMG is shorter by two amino acids at its amino-terminal end and has six conservative amino acid substitutions in comparison with lacticin 481 and seven substitutions in comparison with nukacin ISK-1. Furthermore, in lacticin LMG, three cysteine residues, two threonine residues, and a Ser-9 residue involved in lanthionine ring formation were conserved, while Ser-18 was replaced by a Thr-16, also dehydrated. In the case of streptococcin SA-FF22, the formation of thioether bonds between Cys and dehydrated Thr-18 results in 3-methyllanthionine (Abu-S-Ala, in which Abu is an aminobutyric acid and Ala is alanine) (33). Consequently, streptococcin SA-F22 includes one lanthionine (Ala-S-Ala) and two 3-methyllanthionines instead of two lanthionines and one 3-methyllanthionine, but it has the same bridging pattern as lacticin 481 and nukacin ISK-1 (34). Based on these results, we assume that the same posttranslational modifications occur in the case of lacticin LMG.
The proteolytic cleavage site of the leader peptide in the lacticin LMG precursor peptide is predicted to be between Ala and Gly, yielding a 25-amino-acid mature lantibiotic peptide. This is in contrast to all other known lantibiotic peptides, which contain 27 amino acid residues. Using this theory, the calculated molecular mass of the active lacticin LMG is 2,759 Da, which is in agreement with the molecular mass determined by ESI-TOF mass spectrometry (100% match). Although lacticin LMG shares a high level of identity with the previously described lacticins (lacticin 481 and nukacin ISK-1), this novel lantibiotic has specific features that make it unique, such as its molecular weight, amino acid substitutions, and operon structure. It is known that lactococci can synthesize more than one bacteriocin, such as in strains L. lactis subsp. cremoris 9B4 and L. lactis subsp. lactis BGMN1-5, which produce at least three different bacteriocins (35, 36) despite belonging to the same group (class II). In the present study, the lactococcal strain LMG2081 has been shown to produce two bacteriocins belonging to different groups, one in class I (lantibiotics; lacticin LMG) and the other in class IIb (lactococcin G). In addition, operons encoding lacticin LMG and lactococcin G are located on different genetic elements, i.e., on a plasmid and the chromosome, respectively.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Education, Science, and Technological Development, Republic of Serbia (grant 173019). The scientific work of Nemanja Mirkovic at the Norwegian University of Life Sciences was supported by a FEMS short-term fellowship.
REFERENCES
- 1.Nes IF. 2011. History, current knowledge, and future directions on bacteriocin research in lactic acid bacteria, p 3–12. In Drider D, Rebuffat S (ed), Prokaryotic antimicrobial peptides: from genes to applications. Springer Science & Business Media, New York, NY. [Google Scholar]
- 2.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 3.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins: a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. [DOI] [PubMed] [Google Scholar]
- 4.Dischinger J, Basi Chipalu S, Bierbaum G. 2014. Lantibiotics: promising candidates for future applications in health care. Int J Med Microbiol 304:51–62. doi: 10.1016/j.ijmm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Jung G. 1991. Nisin and novel lantibiotics, p 1–34. In Jung G, Sahl H-G (ed), Nisin and novel lantibiotics. ESCOM, Leiden, Netherlands. [Google Scholar]
- 6.Nagao J, Asaduzzaman SM, Aso Y, Okuda K-I, Nakayama J, Sonomoto K. 2006. Lantibiotics: insight and foresight for new paradigm. J Biosci Bioeng 102:139–149. doi: 10.1263/jbb.102.139. [DOI] [PubMed] [Google Scholar]
- 7.Gilmore MS, Segarra RA, Booth MC, Bogie CP, Hall LR, Clewell DB. 1994. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol 176:7335–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rincé A, Dufour A, Uguen P, Le Pennec JP, Haras D. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl Environ Microbiol 63:4252–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aso Y, Okuda K, Nagao J, Kanemasa Y, Thi Bich Phuong N, Koga H, Shioya K, Sashihara T, Nakayama J, Sonomoto K. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci Biotechnol Biochem 69:1403–1410. doi: 10.1271/bbb.69.1403. [DOI] [PubMed] [Google Scholar]
- 10.Riley MA, Wertz JE. 2002. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 11.Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. 1996. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 12.Garde S, Avila M, Gaya P, Medina M, Nuñez M. 2006. Proteolysis of Hispanico cheese manufactured using lacticin 481-producing Lactococcus lactis ssp. lactis INIA 639. J Dairy Sci 89:840–849. doi: 10.3168/jds.S0022-0302(06)72147-6. [DOI] [PubMed] [Google Scholar]
- 13.Garde S, Carbonell M, Fernández-García E, Medina M, Nuñez M. 2002. Volatile compounds in Hispánico cheese manufactured using a mesophilic starter, a thermophilic starter, and bacteriocin-producing Lactococcus lactis subsp. lactis INIA 415. J Agric Food Chem 50:6752–6757. doi: 10.1021/jf020577v. [DOI] [PubMed] [Google Scholar]
- 14.Garde S, Tomillo J, Gaya P, Medina M, Nuñez M. 2002. Proteolysis in Hispánico cheese manufactured using a mesophilic starter, a thermophilic starter, and bacteriocin-producing Lactococcus lactis subsp. lactis INIA 415 adjunct culture. J Agric Food Chem 50:3479–3485. doi: 10.1021/jf011291d. [DOI] [PubMed] [Google Scholar]
- 15.O'Sullivan L, Ross RP, Hill C. 2003. A lacticin 481-producing adjunct culture increases starter lysis while inhibiting nonstarter lactic acid bacteria proliferation during Cheddar cheese ripening. J Appl Microbiol 95:1235–1241. doi: 10.1046/j.1365-2672.2003.02086.x. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez E, Arques JL, Nuñez M, Gaya P, Medina M. 2005. Combined effect of high-pressure treatments and bacteriocin-producing lactic acid bacteria on inactivation of Escherichia coli O157:H7 in raw-milk cheese. Appl Environ Microbiol 71:3399–3404. doi: 10.1128/AEM.71.7.3399-3404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arqués JL, Rodríguez E, Gaya P, Medina M, Guamis B, Nuñez M. 2005. Inactivation of Staphylococcus aureus in raw milk cheese by combinations of high-pressure treatments and bacteriocin-producing lactic acid bacteria. J Appl Microbiol 98:254–260. doi: 10.1111/j.1365-2672.2004.02507.x. [DOI] [PubMed] [Google Scholar]
- 18.Arqués JL, Rodríguez E, Gaya P, Medina M, Nuñez M. 2005. Effect of combinations of high-pressure treatment and bacteriocin-producing lactic acid bacteria on the survival of Listeria monocytogenes in raw milk cheese. Int Dairy J 15:893–900. doi: 10.1016/j.idairyj.2004.07.020. [DOI] [Google Scholar]
- 19.Nissen-Meyer J, Holo H, Håvarstein LS, Sletten K, Nes IF. 1992. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol 174:5686–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nissen-Meyer J, Oppegård C, Rogne P, Haugen HS, Kristiansen PE. 2010. Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob Proteins 2:52–60. doi: 10.1007/s12602-009-9021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller HJ. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 22.Lozo J, Vukasinovic M, Strahinic I, Topisirovic L. 2004. Characterization and antimicrobial activity of bacteriocin 217 produced by natural isolate Lactobacillus paracasei subsp. paracasei BGBUK2-16. J Food Prot 67:2727–2734. [DOI] [PubMed] [Google Scholar]
- 23.O'Sullivan DJ, Klaenhammer TR. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol 59:2730–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. 1992. New thermosensitive plasmid for Gram-positive bacteria. J Bacteriol 174:5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojic M, Strahinic I, Topisirovic L. 2005. Proteinase PI and lactococcin A genes are located on the largest plasmid in Lactococcus lactis subsp. lactis bv diacetylactis S50. Can J Microbiol 51:305–314. doi: 10.1139/w05-009. [DOI] [PubMed] [Google Scholar]
- 26.Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate CM, Smith CP, Ward JM, Schrempf H. 1985. Genetic manipulation of Streptomyces: a laboratory manual. John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 27.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uguen P, Le Pennec JP, Dufour A. 2000. Lantibiotic biosynthesis: interactions between prelacticin 481 and its putative modification enzyme, LctM. J Bacteriol 182:5262–5266. doi: 10.1128/JB.182.18.5262-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. 2004. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science 303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 30.Uguen P, Hindré T, Didelot S, Marty C, Haras D, Le Pennec J-P, Vallée-Réhel K, Dufour A. 2005. Maturation by LctT is required for biosynthesis of full-length lantibiotic lacticin 481. Appl Environ Microbiol 71:562–565. doi: 10.1128/AEM.71.1.562-565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee C, Paul M, Xie L, van der Donk WA. 2005. Biosynthesis and mode of action of lantibiotics. Chem Rev 105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 32.Dufour A, Hindré T, Haras D, Le Pennec J-P. 2007. The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol Rev 31:134–167. doi: 10.1111/j.1574-6976.2006.00045.x. [DOI] [PubMed] [Google Scholar]
- 33.Jack RW, Carne A, Metzger J, Stefanović S, Sahl HG, Jung G, Tagg J. 1994. Elucidation of the structure of SA-FF22, a lanthionine-containing antibacterial peptide produced by Streptococcus pyogenes strain FF22. Eur J Biochem 220:455–462. doi: 10.1111/j.1432-1033.1994.tb18643.x. [DOI] [PubMed] [Google Scholar]
- 34.Asaduzzaman SM, Sonomoto K. 2009. Lantibiotics: diverse activities and unique modes of action. J Biosci Bioeng 107:475–487. doi: 10.1016/j.jbiosc.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Kojic M, Strahinic I, Fira D, Jovcic B, Topisirovic L. 2006. Plasmid content and bacteriocin production by five strains of Lactococcus lactis isolated from semi-hard homemade cheese. Can J Microbiol 52:1110–1120. doi: 10.1139/w06-072. [DOI] [PubMed] [Google Scholar]
- 36.van Belkum MJ, Hayema BJ, Jeeninga RE, Kok J, Venema G. 1991. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl Environ Microbiol 57:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirkovic N, Radulovic Z, Uzelac G, Lozo J, Obradovic D, Topisirovic L, Kojic M. 2015. Isolation and characterisation of bacteriocin and aggregation-promoting factor production in Lactococcus lactis ssp. lactis BGBM50 strain. Food Technol Biotechnol 53:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 39.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177:7011–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maguin E, Prévost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other Gram-positive bacteria. J Bacteriol 178:931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alegría A, Delgado S, Roces C, López B, Mayo B. 2010. Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. Int J Food Microbiol 143:61–66. doi: 10.1016/j.ijfoodmicro.2010.07.029. [DOI] [PubMed] [Google Scholar]