ABSTRACT
African trypanosomes, except Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense, which cause human African trypanosomiasis, are lysed by the human serum protein apolipoprotein L1 (ApoL1). These two subspecies can resist human ApoL1 because they express the serum resistance proteins T. b. gambiense glycoprotein (TgsGP) and serum resistance-associated protein (SRA), respectively. Whereas in T. b. rhodesiense, SRA is necessary and sufficient to inhibit ApoL1, in T. b. gambiense, TgsGP cannot protect against high ApoL1 uptake, so different additional mechanisms contribute to limit this uptake. Here we report a complex interplay between trypanosomes and an ApoL1 variant, revealing important insights into innate human immunity against these parasites. Using whole-genome sequencing, we characterized an atypical T. b. gambiense infection in a patient in Ghana. We show that the infecting trypanosome has diverged from the classical T. b. gambiense strains and lacks the TgsGP defense mechanism against human serum. By sequencing the ApoL1 gene of the patient and subsequent in vitro mutagenesis experiments, we demonstrate that a homozygous missense substitution (N264K) in the membrane-addressing domain of this ApoL1 variant knocks down the trypanolytic activity, allowing the trypanosome to avoid ApoL1-mediated immunity.
IMPORTANCE
Most African trypanosomes are lysed by the ApoL1 protein in human serum. Only the subspecies Trypanosoma b. gambiense and T. b. rhodesiense can resist lysis by ApoL1 because they express specific serum resistance proteins. We here report a complex interplay between trypanosomes and an ApoL1 variant characterized by a homozygous missense substitution (N264K) in the domain that we hypothesize interacts with the endolysosomal membranes of trypanosomes. The N264K substitution knocks down the lytic activity of ApoL1 against T. b. gambiense strains lacking the TgsGP defense mechanism and against T. b. rhodesiense if N264K is accompanied by additional substitutions in the SRA-interacting domain. Our data suggest that populations with high frequencies of the homozygous N264K ApoL1 variant may be at increased risk of contracting human African trypanosomiasis.
INTRODUCTION
The human innate immunity is crucial to resist infection with African trypanosomes. Only the subspecies Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense can resist lysis by most human sera and thus cause human African trypanosomiasis (HAT) in western/central and eastern/southern Africa, respectively. T. b. gambiense has been historically divided into two groups (1). T. b. gambiense group 1 (T. b. gambiense 1) is a homogenous group of isolates, constitutively resistant to human serum and responsible for the majority of HAT cases. T. b. gambiense 2 is a rather ambiguous group of strains which were isolated from patients in the 1970s but do not form a monophyletic group (2) and show varied levels of resistance to human serum (3). T. b. gambiense 1 and T. b. rhodesiense are pathogenic to humans because they developed resistance mechanisms against apolipoprotein L1 (ApoL1), the trypanolytic component in human serum (4).
ApoL1 is contained within two serum complexes, trypanolytic factors 1 and 2 (TLF1 and -2) (5, 6). Both TLF1 and TLF2 are able to lyse other trypanosomes, such as Trypanosoma brucei brucei and Trypanosoma evansi, which cause nagana and surra in animals, respectively, so these trypanosomes are not infectious to humans. TLF1 can be efficiently taken up by trypanosomes through binding to the haptoglobin-hemoglobin (HpHb) receptor (6, 7). However, in normal human serum, the HpHb receptor is saturated with serum HpHb, and ApoL1 enters the trypanosome only via TLF2, independently of the HpHb receptor (6, 7). The exact mechanism behind TLF2 uptake by trypanosomes is still unknown. The trypanolytic activity of ApoL1 results from ionic pore formation in endolysosomal membranes, mediated by the combination of contiguous pore-forming and membrane-addressing domains (8). T. b. gambiense 1 and T. b. rhodesiense resist ApoL1 through the activities of distinct resistance proteins. T. b. gambiense expresses the T. b. gambiense-specific glycoprotein (TgsGP), which is necessary but not sufficient for full resistance, and T. b. rhodesiense expresses the serum resistance-associated protein (SRA) (9, 10). While TgsGP prevents ApoL1 pore-forming activity by stiffening the target endolysosomal membranes, SRA inhibits ApoL1 by direct interaction with the C-terminal domain of this protein (4, 9, 11). The TgsGP gene is physically linked to a truncation of a gene homologous to yeast AUT1 (ATG3), a gene involved in internal vesicular formation (12). T. b. gambiense is heterozygous for AUT1, with the full and truncated alleles located in independent chromosome II homologues (12). In hypohaptoglobinemic patients, receptor-mediated uptake of TLF1 is efficient due to the absence of the competing HpHb ligand for the HpHb receptor and can overcome the TgsGP-mediated resistance of T. b. gambiense 1 (9). Therefore, this parasite is characterized by an L210S substitution in the HpHb receptor, which inhibits TLF1 uptake and allows T. b. gambiense 1 infection in regions with a high frequency of hypohaptoglobinemia (6, 9). Two variant ApoL1 alleles (G1 and G2) are frequently observed in West Africans, presumably because these variants resist inhibition by SRA and can thus lyse T. b. rhodesiense (13). The G1 ApoL1 genotype is characterized by the coding single nucleotide polymorphisms (SNPs) rs73885319 (S342G) and rs60910145 (I384M) in the SRA-interacting domain, and the G2 ApoL1 genotype is characterized by the allele for indel rs71785313 (N388del and Y389del) in the same domain (13). None of 75 human sera with different combinations of G1 and G2 genotypes were able to lyse T. b. gambiense in vitro, as was also observed with recombinant G1 or G2 ApoL1 (13). Both TgsGP and SRA are noncanonical variant surface glycoproteins (VSG), which are routed to the lysosome of the trypanosome, in contrast to functional native VSGs. Native VSGs cover the trypanosome plasma membrane as a dense layer of homodimers and help to evade the host’s immune system through continuous variation of surface-exposed antigenic loops (14).
In this work, we report an ApoL1 variant in a patient in Ghana who was infected with an atypical T. b. gambiense strain lacking the TgsGP defense mechanism against ApoL1. The ApoL1 variant is characterized by a homozygous N264K missense substitution that knocks down its lytic activity against T. b. gambiense devoid of TgsGP.
RESULTS
Patient.
In 2013, a 54-year-old man presented at the Ghana Ports and Harbours Authority Hospital in Takoradi City, Ghana, with complaints of gastroenteritis, fever, and nausea. The patient was HIV positive but not on antiretroviral therapy at the time of admission. He was taking co-trimoxazole as prophylaxis for HIV-related opportunistic infections and artemisinin-based combination therapy against malaria. His hemoglobin level was 9.2 g/dl, white blood cell count was 9.3 × 109/liter (58% neutrophils, 42% lymphocytes), and platelet level was 259 × 109/liter. Giemsa-stained thin blood smears revealed an infection with trypanosomes, and he was treated with 10 injections of pentamidine at 4 mg/kg of body weight/day intramuscularly for 7 days. No lumbar puncture was performed. He responded well to the trypanocidal treatment, and no trypanosomes were microscopically observed in blood films prepared after treatment. These blood films did show the presence of malaria parasites. Because no HAT cases have been reported in Ghana in the last 10 years, the blood of the patient before treatment was collected on Whatman filter paper for further analysis. Ethical clearance for the study was obtained from the institutional ethics committee of the Institute of Tropical Medicine, Antwerp, Belgium.
Trypanosome genome sequencing and SNP analysis.
We extracted the DNA from the dried blood spot, and subsequent whole-genome sequencing generated 10,738,766 paired-end reads of 600 nucleotides (2 × 300 bp). Most (79.29%) of these reads mapped to the human genome and were discarded. Many (1,509,189) of the remaining reads could be mapped to the T. b. brucei TREU927 reference genome, with 10-fold average and 7-fold median coverage. Sixty-two percent of the sample genome (here referred to as the GHANA genome) with a minimum coverage of 5 was used for SNP calling. Quality-scored reads have been submitted to the NCBI Sequence Read Archive (accession number SRP072325). Following mapping of all sequence data, including publically available sequence data from T. b. gambiense types 1 and 2, T. evansi, and T. b. brucei, SAMtools mpileup identified 78,899 SNPs. A RAxML maximum-likelihood tree revealed a single monophyletic clade of T. b. gambiense 1 strains [DAL972, 1829(Aljo), and Fontem strain 10] (Fig. 1). The GHANA trypanosome was genetically more related to T. b. gambiense 1 than to any other trypanosome in this data set. The GHANA genome differed from the T. b. gambiense 1 strains Fontem strain 10, DAL972, and 1829(Alijo) by, respectively, 7,950, 8,233, and 8,050 SNPs and from the T. b. gambiense 2 strains TH126 and STIB386 by, respectively, 15,050 and 15,437 SNPs. To exclude the possibility that the GHANA trypanosome originated by recombination of T. b. gambiense 1 with another, related trypanosome, a NeighborNet Split network was generated (Fig. S1). Reticulation was limited to the center of the network, and no ambiguous phylogenetic signal related to T. b. gambiense 1 and GHANA was observed. In addition, the GHANA sample showed the least number of heterozygous SNPs compared to the other trypanosomes (see Table S2 in the supplemental material). These results suggest that the GHANA trypanosome is not a product of recombination. The gene for the T. b. gambiense 1 serum resistance TgsGP protein was completely absent from the GHANA genome. The absence of this gene was further confirmed by PCR analysis with TgsGP-internal primers (Fig. S2). Since the TgsGP gene is physically linked to a heterozygous truncated AUT1 allele, we have tested the GHANA DNA by PCR with a primer set that amplifies the full AUT1 allele and a primer set that amplifies the truncated AUT1 allele (Fig. S3). GHANA showed a homozygous intact AUT1 gene, as in T. b. brucei. However, the GHANA genome reads that mapped to the reference sequence of the HpHb receptor gene harbored the T. b. gambiense 1-specific codon that is responsible for the L210S substitution in the HpHb receptor. Because only one read mapped to this region, we further confirmed this substitution by Sanger sequencing of the complete GHANA HpHb receptor gene. GHANA has the z1/z1 alleles of the T. b. gambiense 1 DAL972 strain (Fig. S4A) described by Symula et al. (15) and has the codon for the L210S substitution in the HpHb receptor (Fig. S4B). This L210S substitution in the HpHb receptor decreases the binding and internalization of TLF1 in hypohaptoglobinemic-patient serum (9). Given that our patient was anemic and coinfected with malaria, the reduced uptake of TLF1 via the trypanosome’s mutated HpHb receptor partially explains the resistance of the trypanosome to human serum. However, it is known that ApoL1 can still enter the trypanosome via TLF2 and lyse trypanosomes that do not have the TgsGP or SRA resistance proteins against ApoL1 (9).
FIG 1 .
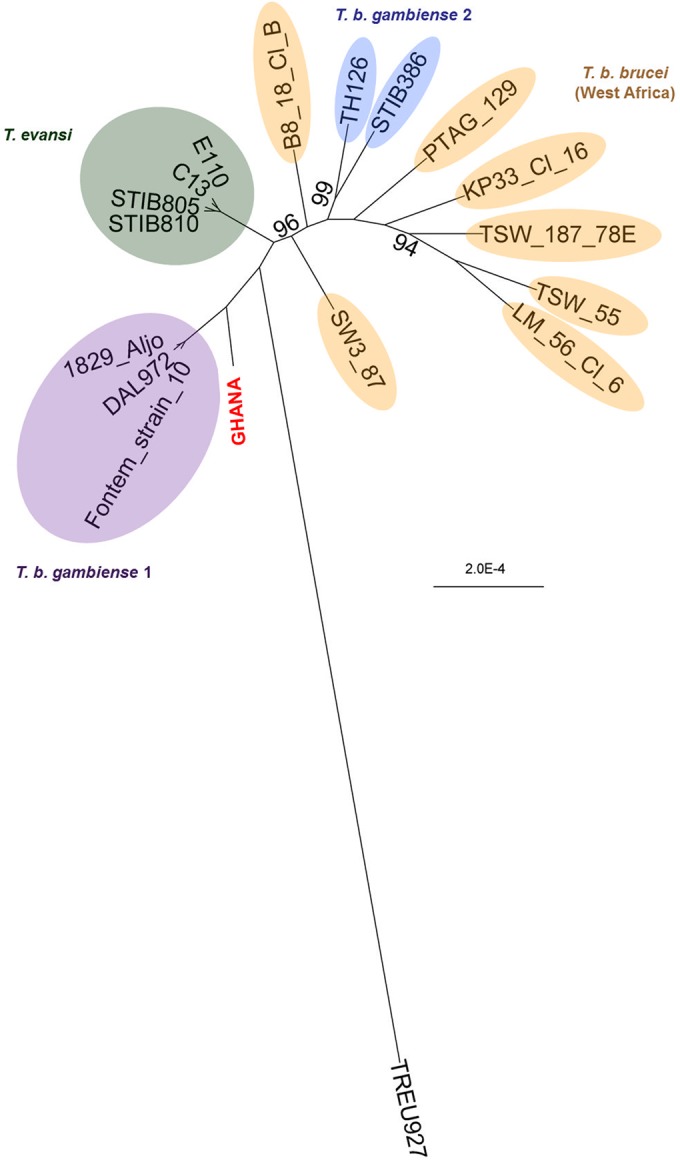
SNP analysis of the trypanosome in the patient and reference strains. Shown is a bootstrapped RAxML maximum-likelihood tree based on 78,899 SNP loci identified in the patient’s trypanosome (GHANA) and on publically available sequence data from three T. b. gambiense 1 strains, two T. b. gambiense 2 strains, five T. evansi strains, seven West African T. b. brucei strains, and the East African T. b. brucei reference strain TREU927. Bootstrap values were calculated on the basis of 1,000 replicates and are presented in the figure if they were below 100%.
ApoL1 genotyping and in vitro mutagenesis experiments.
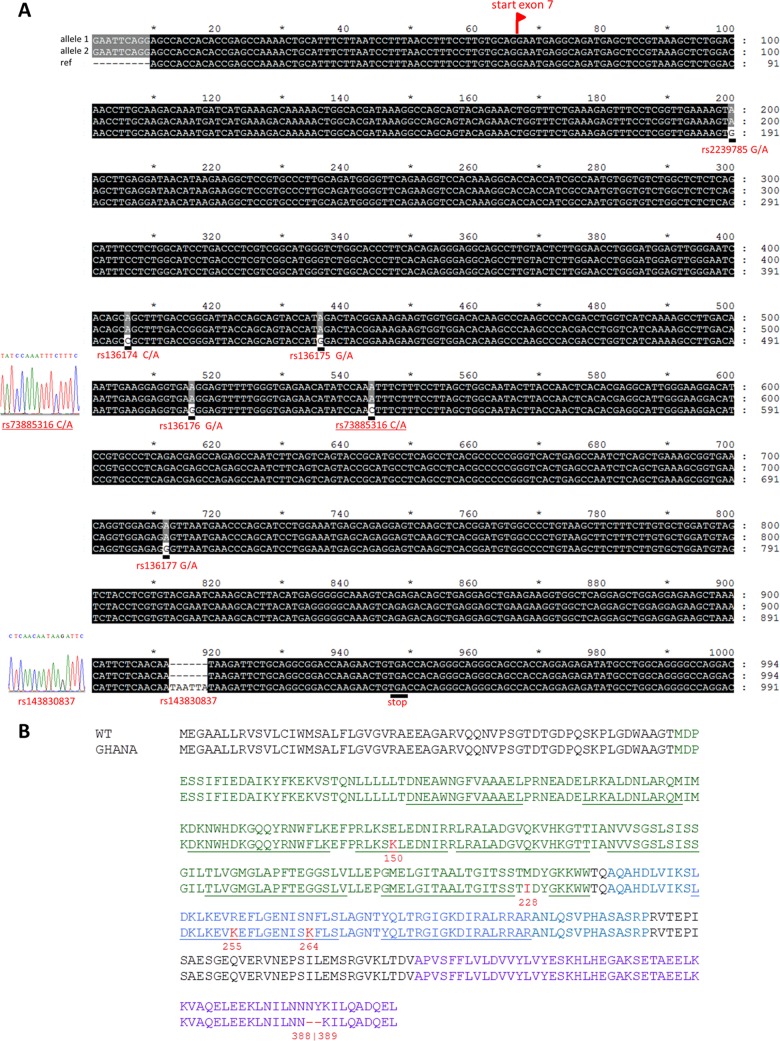
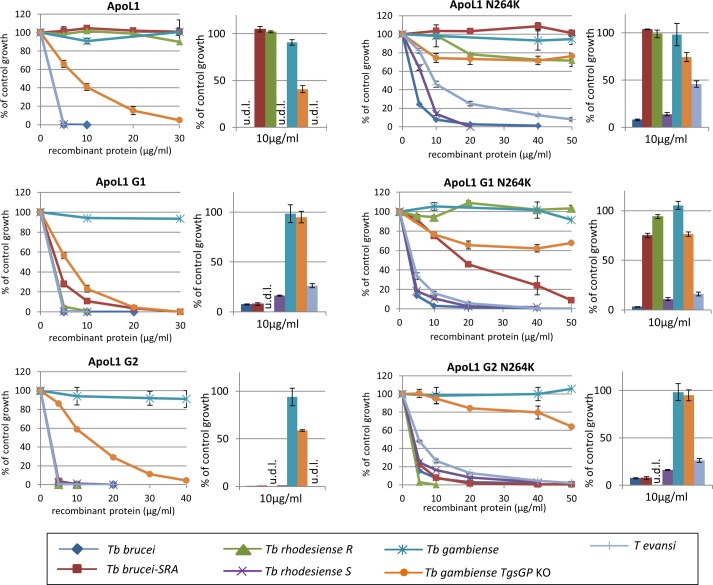
Sanger sequencing of exons 3 to 7 of the ApoL1 gene in the patient’s genome revealed a G2 ApoL1 genotype with homozygous rs143830837 alleles (N388del and Y389del) in the SRA-interacting domain of ApoL1 (Fig. 2). The ApoL1 gene showed additional substitutions in the pore-forming domain, rs2239785 G/A (E150K) and rs136174 C/A (M228I), and in the membrane-addressing domain, rs136176 G/A (R255K) and rs73885316 C/A (N264K) (Fig. 2). The E150K, M228I, and R255K variants are actually present in the ApoL1 wild-type (WT) version used in our laboratory since the discovery of the trypanolytic potential of ApoL1 (4), and these variants do not affect ApoL1 activity. M228I and R255K are coded by the ApoL1 G3 genotype (16), and the trypanolytic activity of ApoL1 G3 was confirmed using plasma samples (17). Since previous experiments have indicated that mutations in the membrane-addressing domain can be deleterious to trypanolytic activity (8), we decided to check the activity of the N264K variant. Therefore, we experimentally introduced the N264K substitution in WT, G1, and G2 ApoL1s by site-directed mutagenesis and expressed these ApoL1 variants as recombinant proteins (Fig. S5). The N264K variants of WT, G1, and G2 ApoL1 variants knocked down the lytic activity against T. b. gambiense 1 TgsGP knockout (KO) parasites (Fig. 3). Our mutagenesis experiments also showed that the lytic activity of G1 ApoL1 against serum-resistant T. b. rhodesiense was completely destroyed by the N264K substitution. The same trend was observed with T. b. brucei ribSRA. This is a T. b. brucei strain in which SRA was genetically introduced and which thus ectopically expresses SRA (10).
FIG 2 .
(A) Nucleotide alignment of ApoL1 exon 7 of the patient (alleles 1 and 2) and the wild-type ApoL1 gene (ref; GenBank accession number NM_003661.3); (B) amino acid alignment of WT ApoL1 and ApoL1 of the patient (GHANA). Allele variants in panel A are indicated with the dbSNP codes, and Sanger sequencing chromatograms are included for alleles rs73885316 C/A and rs14380837. The green, blue, and purple sequences in panel B represent, respectively, the pore-forming domain, the membrane-addressing domain, and the SRA-interacting domain of ApoL1 (8). Underlined sequences represent alpha helixes. These helices are defined in reference 8; in the pore-forming domain, they were defined by homology modeling using the pore-forming domain of colicin A as the template. Amino acid substitutions or deletions, as well as the amino acid positions, are indicated in red.
FIG 3 .
Trypanolytic activities of WT, G1, and G2 ApoL1s and the N264K variants. Shown is the growth of T. b. brucei, T. b. brucei ectopically expressing SRA (ribSRA), human-serum-resistant (R) and sensitive (S) clones of T. b. rhodesiense, T. b. gambiense 1, the T. b. gambiense 1 TgsGP knockout (KO), and T. evansi in complete growth medium supplemented with various concentrations of recombinant WT ApoL1 (A), ApoL1 G1 (B), and ApoL1 G2 (C), as well as the N264K variants ApoL1 N264K (D), ApoL1 G1 N264K (E), and ApoL1 G2 N264K (F). Histograms for the 7 strains in the presence of 10 µg/ml recombinant ApoL1 protein are shown for each ApoL1 variant. The physiological concentration of ApoL1 in human blood is approximately 10 µg/ml. Completely lysed trypanosomes in the histograms are indicated by “u.d.l.” (under the detection limit).
DISCUSSION
We successfully sequenced the genome of a trypanosome in a dried blood spot from a HAT patient in Ghana. Using SNP and targeted gene analysis, we showed that the trypanosome is an atypical T. b. gambiense strain that lacks TgsGP for its defense against trypanolytic ApoL1 in human serum but has the T. b. gambiense 1-specific L210S substitution in its HpHb receptor. Given that the patient was anemic, reduced uptake of TLF1 by the trypanosome due to the mutated HpHb receptor may partially explain the resistance to human serum. However, ApoL1 can still enter the trypanosome via TLF2 and lyse trypanosomes that do not have the TgsGP serum resistance protein. We have shown that the N264K substitution in the membrane-addressing domain of the ApoL1 protein of the patient knocks down the trypanolytic activity against this atypical T. b. gambiense strain devoid of TgsGP. Interestingly, the intact AUT1 gene in GHANA indicates that this strain has never harbored the TgsGP gene and that GHANA diverged from the common ancestor before the T. b. gambiense type 1 strains. Because GHANA had the mutation for the L210S substitution in its HpHb receptor, this also indicates that the T. b. gambiense-specific HpHb mutation existed before TgsGP was introduced and before the monophyletic human pathogen T. b. gambiense type 1 emerged.
ApoL1 is targeted to membranes by its membrane-addressing domain (8). After ApoL1 is inserted into the lysosomal membrane, its pore-forming domain creates membrane pores that lead to trypanosome death (18). The N264K substitution in the membrane-addressing domain of G2 ApoL1 of the patient thus explains why he was infected with the atypical T. b. gambiense strain devoid of TgsGP. In contrast, G2 ApoL1 N264K still efficiently lysed T. b. brucei, T. evansi, and T. b. rhodesiense. When we introduced the N264K substitution in G1 and G2 ApoL1 proteins, both variants could lyse T. b. brucei and T. evansi. The observation that N264K affects only the lytic activity of ApoL1 against T. b. gambiense devoid of TgsGP and not T. b. brucei and T. evansi is probably due to the lower inherent sensitivity of the T. b. gambiense TgsGP KO to ApoL1, caused by a decreased ApoL1 uptake in hypohaptoglobinemic serum and an accelerated degradation of the internalized ApoL1 (9). Interestingly, the N264K substitution also completely knocked down the lytic activity of G1 ApoL1 against T. b. rhodesiense, although N264K is located in the membrane-addressing domain of ApoL1 and not in the domain that binds to the T. b. rhodesiense resistance protein SRA. However, the N264K G2 ApoL1 could still lyse T. b. rhodesiense. G1 and G2 ApoL1 proteins have different mutations in the SRA-binding domain ablating the serum resistance of T. b. rhodesiense, the G1 variant being weaker than the G2 variant in lysing T. b. rhodesiense, presumably due to higher residual binding to SRA (13). Therefore, the higher susceptibility to N264K knockdown for the G1 variant in T. b. rhodesiense may result from the increased probability of interaction with SRA following a lowered ability to access the target membrane.
The G2 ApoL1 gene of the patient was characterized by a homozygote missense mutation (rs73885316 C/A) causing an N264K substitution in the membrane-addressing domain of ApoL1 and a homozygote 6-bp deletion (rs143830837) causing the N388del and Y389del deletions in the SRA-interacting domain of ApoL1. According to the 1000 Genomes Project, 31 individuals were heterozygous for the rs73885316 C/A allele, of which 8 individuals were heterozygous for the rs143830837 deletion; none were homozygous for rs73885316 C/A or rs143830837. This suggests that both mutations probably arose independently on different haplotypic backgrounds, given that they are only 368 bp apart. Therefore, the observation that the patient is homozygous for both rs73885316 (N264K) and rs143830837 (N388del and Y389del) is remarkable, as it could have happened only through convergent evolution (allowing the same mutations to arise in different haplotypes) or through recombination (allowing the mutations to be merged on the same haplotype). Ko et al. reported that the allele for the N264K ApoL1 variant is frequently observed (34%) in the Hadza population in Tanzania (16). Following the Hardy-Weinberg equilibrium, we expect 11.5% homozygous N264K ApoL1 variants in the Hadza population. Given that the Hadza live in an area where T. b. rhodesiense is endemic, our data thus suggest that about 11% of the people may be at increased risk of contracting HAT, since the N264K substitution knocks down the protective effect of G1 ApoL1 against T. b. rhodesiense. The G1 genotype is characterized by an S342G (rs73885319) and an I384M (rs71785313) substitution in the SRA-interacting domain of ApoL1, and both alleles are present in the Hadza population with a 5% frequency (16).
In conclusion, we report that the missense N264K substitution in the membrane-addressing domain of WT, G1, and G2 ApoL1s knocks down the lytic activity against T. b. gambiense strains devoid of the TgsGP serum resistance mechanism. Interestingly, the N264K substitution also knocks down the lytic activity of G1 ApoL1 against T. b. rhodesiense. Trypanosomes are ancient parasites that have existed together with human populations on the African continent through evolution. Our findings illustrate the sensitive interplay between ApoL1 genotypes and susceptibility to trypanosome infection (19).
MATERIALS AND METHODS
Ethics.
Ethical clearance for the study was obtained from the Institutional Review Board (IRB) of the Institute of Tropical Medicine, Antwerp, Belgium (approval number IRB/AB/ac/108). No informed consent of the study participant could be given because the patient died before the results of the molecular laboratory tests were available. The IRB gave approval to publish our findings without informed consent, as the patient is anonymized in the report.
Genome sequencing and SNP analysis.
DNA from three 5-mm punches of dried blood of the patient was extracted from the filter paper using the QIAamp DNA microkit according to the manufacturer’s instructions (Qiagen, Germany). One nanogram of genomic DNA was subjected to Illumina’s Nextera XT library preparation, and the library was quantified with the KAPA library quantification kit (KAPA Biosystems, MA). The indexed library was sequenced at a final concentration of 33 pM in a MiSeq sequencer using a reagent v3 600-cycle cartridge (Illumina, CA). With Bowtie 2 under default settings (20), we first discarded the GHANA reads that mapped to human genome version 19 and then mapped the reads of GHANA and the publically available genome reads from the T. b. gambiense 1 strains DAL972 (21), Fontem strain 10, and 1829(Aljo) (22), the T. b. gambiense 2 strains TH126 and STIB386 (21), the T. evansi strains STIB810, E110, C13, KETRI2479, and STIB805 (23), and the West African T. b. brucei strains TSW187/78E, LM56clone6, TSW55, KP33clone16, PTAG129, B8/18cloneB, and SW3/87 (22) to the T. b. brucei TREU927 reference genome (24). The list of strains with country, host, and date of collection is presented in the supplemental material (see Table S1). Variants were called with SAMtools’ mpileup function and bcftools (25). The resulting vcf file was filtered to retain only SNPs with read depths of ≥5 in all strains, an SNP quality of ≥50, a mapping quality of ≥40, and no significant strand bias (P > 0.05). For each strain, SNPs were concatenated to a FASTA sequence in which each SNP was represented as 2 bases, one for each allele, and invariant sites were subsequently removed. RAxML v8.1.17 was used to construct a maximum-likelihood tree with 1,000 bootstraps using a GTR CAT substitution model and the Felsenstein correction for ascertainment bias inherent to SNP alignments (26). To check for possible recombination events, a NeighborNet Split network of the SNP alignment was generated in Splitstree4 (27). The presence and sequence of the HpHb receptor, TgsGP, and SRA genes in the genomes was checked by mapping the sequencing reads against the FASTA sequences of these genes (NCBI accession number JX143837 for the HpHb receptor gene, FN555988 for TgsGP, and Z37159 for SRA) using Bowtie2 and by visual inspection with the Integrated Genomics Viewer (IGV) (28).
TgsGP and HpHb receptor analysis.
The presence of the TgsGP gene in GHANA was verified by PCR amplification with TgsGP internal primers reported by Radwanska et al. (29). The presence of a truncated AUT1 allele was verified as described by Felu et al. (12) but with the primers AUT1-Fwd (5′ GTTGAGGATGAACAAACAAAGTTTGTATGAAGG 3′), AUT1-Full-Rev (5′ CATTTTTAAATGTCGATACCTGTCGACAAATCATATTCG 3′), and AUT1-Trunc-Rev (5′ ATATTCGTATATACACTCTCACAACACTCTCATATCATTATTATTATC 3′). The HpHb receptor gene was PCR amplified and Sanger sequenced as described elsewhere (9, 15). Successful DNA extraction from the dried blood on filter paper was verified by amplifying a part of the trypanosome’s 18S rRNA gene and the patient’s cytochrome b gene with primers described previously (30, 31).
ApoL1 genotyping and mutagenesis.
To characterize the patient’s ApoL1 protein, exons 3 to 7 of this gene were amplified using PCR as described previously (32). ApoL1 PCR products were cloned in pCR-Blunt vectors, and six to eight clones per exon were Sanger sequenced. Amino acid sequences of the exons were aligned against the ApoL1 reference sequence (NM_003661.3, NCBI). ApoL1 G1, ApoL1 G2, and the N264K variants were obtained by site-directed mutagenesis of WT ApoL1 cloned in the pStaby expression vector (Delphi Genetics). Recombinant ApoL1 variants were expressed in Escherichia coli in fusion with the His tag and affinity purified on nickel columns (11). Trypanosome strains used in the lysis experiment were T. b. gambiense 1 ELIANE (clone LiTat1.3), T. b. gambiense 1 TgsGP KO, T. b. brucei Lister 427, T. b. brucei ribSRA, T. b. rhodesiense Etat1.2 R (serum-resistant phenotype) and S (serum-sensitive phenotype), and T. evansi RoTat 1.2. The transgenic cell lines T. b. gambiense 1 TgsGP KO and T. b. brucei ribSRA were described earlier (9, 10). Bloodstream forms were diluted at 1 × 105 trypanosomes/ml (T. b. brucei and T. b. rhodesiense) or 2 × 105 trypanosomes/ml (T. b. gambiense 1 and T. evansi) in HMI (Hirumi Modified Escove [33]) medium containing 10% fetal calf serum (FCS) and 10% Serum Plus. Appropriate dilutions of the recombinant proteins were made in 20 mM acetic acid and added to the trypanosomes as 10% of the final volume. Lysis tests were performed in triplicate, and parasites were incubated overnight under standard culture conditions. Quantification was performed with the CellTiter Glo luminescence assay (Promega), and control growth conditions with addition of only acetic acid were used as the reference (100% growth).
SUPPLEMENTAL MATERIAL
Splitstree4 NeighborNet network based on 78,899 SNP loci identified in the patient’s trypanosome (GHANA) and in publically available sequence data from three T. b. gambiense 1 strains, two T. b. gambiense 2 strains, five T. evansi strains, seven West African T. b. brucei strains, and the East African T. b. brucei reference strain TREU927. Download
PCR analysis of the GHANA DNA extract with TgsGP-specific primers (A), Trypanozoon-specific primers (B), and vertebrate cytochrome b-specific primers (C). Download
PCR analysis of the GHANA DNA extract with primers that amplify the full AUT1 allele (A) and primers that amplify the truncated AUT1 allele (B). Download
DNA alignment of the HpHb receptor gene (A) and amino acid alignment of the HpHb receptor protein (B) of GHANA together with the known HpHb receptor sequences of T. b. gambiense 1 (y, z1, z2, and z3 alleles), T. b. gambiense 2 (n, c t1, t3, and u1 alleles), T. b. brucei (b1, c, d, e, f1, f2, g, h, I, j, k, l, m, n, o, p1, p2, q, r, s, t1, t2, u1, u2, u3, v, w, and x alleles), and T. evansi type A (u1 allele). The codon and the L210S substitution are indicated by the red box. Download
Coomassie blue stains of 400 ng recombinant ApoL1, ApoL1 N264K, ApoL1 G1, Apol1 G1 N264K, ApoL1 G2, and ApoL1 G2 N264K proteins. These recombinant proteins are all tagged with a 6-His sequence at the C terminus and are solubilized in 20 mM acetic acid. Download
Taxon, country, host, and year of isolation of the reference strains included in the genome-wide SNP analysis.
Number of heterozygous SNPs within the 78,899 characterized SNP loci in the Trypanosoma genomes.
ACKNOWLEDGMENTS
We thank the Center of Medical Genetics at the University of Antwerp for hosting the NGS facility. The computational resources used for this work were provided by the VSC (Flemish Supercomputer Center) at the University of Antwerp. We thank Mark Sistrom and Adalgisa Caccone (Yale University) for providing T. evansi, T. b. brucei, and T. b. gambiense 2 genome reads. We thank the laboratory staff at the Ghana Ports and Harbours Authority Hospital for the diagnosis and management of the patient.
We have no conflicts of interest.
Footnotes
Citation Cuypers B, Lecordier L, Meehan CJ, Van den Broeck F, Imamura H, Büscher P, Dujardin J, Laukens K, Schnaufer A, Dewar C, Lewis M, Balmer O, Azurago T, Kyei-Faried S, Ohene S, Duah B, Homiah P, Mensah EK, Anleah F, Franco JR, Pays E, Deborggraeve S. 2016. Apolipoprotein L1 variant associated with increased susceptibility to trypanosome infection. mBio 7(2):e02198-15. doi:10.1128/mBio.02198-15.
REFERENCES
- 1.Gibson WC. 1986. Will the real Trypanosoma b. gambiense please stand up? Parasitol Today 2:255–257. doi: 10.1016/0169-4758(86)90011-6. [DOI] [PubMed] [Google Scholar]
- 2.Balmer O, Beadell JS, Gibson W, Caccone A. 2011. Phylogeography and taxonomy of Trypanosoma brucei. PLoS Negl Trop Dis 5:e961. doi: 10.1371/journal.pntd.0000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capewell P, Veitch NJ, Turner MR, Raper J, Berriman M, Hajduk SL, MacLeod A. 2011. Differences between Trypanosoma brucei gambiense groups 1 and 2 in their resistance to killing by trypanolytic factor 1. PLoS Negl Trop Dis 5:e1287. doi: 10.1371/journal.pntd.0001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, Pays A, Tebabi P, Van Xong H, Jacquet A, Moguilevsky N, Dieu M, Kane JP, De Baetselier P, Brasseur R, Pays E. 2003. Apolipoprotein l-I is the trypanosome lytic factor of human serum. Nature 422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 5.Bullard W, Kieft R, Capewell P, Veitch NJ, MacLeod A, Hajduk SL. 2012. Haptoglobin-hemoglobin receptor independent killing of African trypanosomes by human serum and trypanosome lytic factors. Virulence 3:72–76. doi: 10.4161/viru.3.1.18295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Pérez-Morga D. 2014. The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol 12:575–584. doi: 10.1038/nrmicro3298. [DOI] [PubMed] [Google Scholar]
- 7.Vanhollebeke B, De Muylder G, Nielsen MJ, Pays A, Tebabi P, Dieu M, Raes M, Moestrup SK, Pays E. 2008. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science 320:677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
- 8.Pérez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, Homblé F, Vanhamme L, Tebabi P, Pays A, Poelvoorde P, Jacquet A, Brasseur R, Pays E. 2005. Apolipoprotein l-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science 309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 9.Uzureau P, Uzureau S, Lecordier L, Fontaine F, Tebabi P, Homblé F, Grélard A, Zhendre V, Nolan DP, Lins L, Crowet JM, Pays A, Felu C, Poelvoorde P, Vanhollebeke B, Moestrup SK, Lyngsø J, Pedersen JS, Mottram JC, Dufourc EJ, Pérez-Morga D, Pays E. 2013. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature 501:430–434. doi: 10.1038/nature12516. [DOI] [PubMed] [Google Scholar]
- 10.Xong HV, Vanhamme L, Chamekh M, Chimfwembe CE, Van Den Abbeele J, Pays A, Van Meirvenne N, Hamers R, De Baetselier P, Pays E. 1998. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell 95:839–846. doi: 10.1016/S0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 11.Lecordier L, Vanhollebeke B, Poelvoorde P, Tebabi P, Paturiaux-Hanocq F, Andris F, Lins L, Pays E. 2009. C-terminal mutants of apolipoprotein l-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathog 5:e1000685. doi: 10.1371/journal.ppat.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felu C, Pasture J, Pays E, Pérez-Morga D. 2007. Diagnostic potential of a conserved genomic rearrangement in the Trypanosoma brucei gambiense-specific TgsGP locus. Am J Trop Med Hyg 76:922–929. [PubMed] [Google Scholar]
- 13.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. 2010. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borst P, Rudenko G. 1994. Antigenic variation in African trypanosomes. Science 264:1872–1873. doi: 10.1126/science.7516579. [DOI] [PubMed] [Google Scholar]
- 15.Symula RE, Beadell JS, Sistrom M, Agbebakun K, Balmer O, Gibson W, Aksoy S, Caccone A. 2012. Trypanosoma brucei gambiense group 1 is distinguished by a unique amino acid substitution in the HpHb receptor implicated in human serum resistance. PLoS Negl Trop Dis 6:e1728. doi: 10.1371/journal.pntd.0001728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko WY, Rajan P, Gomez F, Scheinfeldt L, An P, Winkler CA, Froment A, Nyambo TB, Omar SA, Wambebe C, Ranciaro A, Hirbo JB, Tishkoff SA. 2013. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am J Hum Genet 93:54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limou S, Nelson GW, Lecordier L, An P, O’huigin CS, David VA, Binns-Roemer EA, Guiblet WM, Oleksyk TK, Pays E, Kopp JB, Winkler CA. 2015. Sequencing rare and common APOL1 coding variants to determine kidney disease risk. Kidney Int 88:754–763. doi: 10.1038/ki.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanwalleghem G, Fontaine F, Lecordier L, Tebabi P, Klewe K, Nolan DP, Yamaryo-Botté Y, Botté C, Kremer A, Burkard GS, Rassow J, Roditi I, Pérez-Morga D, Pays E. 2015. Coupling of lysosomal and mitochondrial membrane permeabilization in trypanolysis by APOL1. Nat Commun 6:8078. doi: 10.1038/ncomms9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, Winkler CA, Kopp J, Rotimi C, Adeyemo A, Doumatey A, Ayodo G, Alper SL, Pollak MR, Friedman DJ, Raper J. 2014. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A 111:E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, Quail MA, Chukualim B, Capewell P, MacLeod A, Melville SE, Gibson W, Barry JD, Berriman M, Hertz-Fowler C. 2010. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African trypanosomiasis. PLoS Negl Trop Dis 4:e658. doi: 10.1371/journal.pntd.0000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sistrom M, Evans B, Bjornson R, Gibson W, Balmer O, Mäser P, Aksoy S, Caccone A. 2014. Comparative genomics reveals multiple genetic backgrounds of human pathogenicity in the Trypanosoma brucei complex. Genome Biol Evol 6:2811–2819. doi: 10.1093/gbe/evu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carnes J, Anupama A, Balmer O, Jackson A, Lewis M, Brown R, Cestari I, Desquesnes M, Gendrin C, Hertz-Fowler C, Imamura H, Ivens A, Koreny L, Lai DH, MacLeod A, McDermott SM, Merritt C, Monnerat S, Moon W, Myler P, Phan I, Ramasamy G, Sivam D, Lun ZR, Lukes J, Stuart K, Schnaufer A. 2015. Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl Trop Dis 9:e3404. doi: 10.1371/journal.pntd.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Böhme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, et al.. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 28.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radwanska M, Claes F, Magez S, Magnus E, Perez-Morga D, Pays E, Büscher P. 2002. Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am J Trop Med Hyg 67:289–295. [DOI] [PubMed] [Google Scholar]
- 30.Deborggraeve S, Lejon V, Ekangu RA, Mumba Ngoyi D, Pati Pyana P, Ilunga M, Mulunda JP, Büscher P. 2011. Diagnostic accuracy of PCR in gambiense sleeping sickness diagnosis, staging and post-treatment follow-up: a 2-year longitudinal study. PLoS Negl Trop Dis 5:e972. doi: 10.1371/journal.pntd.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC. 1989. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A 86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanhollebeke B, Truc P, Poelvoorde P, Pays A, Joshi PP, Katti R, Jannin JG, Pays E. 2006. Human Trypanosoma evansi infection linked to a lack of apolipoprotein L-I. N Engl J Med 355:2752–2756. doi: 10.1056/NEJMoa063265. [DOI] [PubMed] [Google Scholar]
- 33.Hirumi H, Hirumi K. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol 75:985–989. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Splitstree4 NeighborNet network based on 78,899 SNP loci identified in the patient’s trypanosome (GHANA) and in publically available sequence data from three T. b. gambiense 1 strains, two T. b. gambiense 2 strains, five T. evansi strains, seven West African T. b. brucei strains, and the East African T. b. brucei reference strain TREU927. Download
PCR analysis of the GHANA DNA extract with TgsGP-specific primers (A), Trypanozoon-specific primers (B), and vertebrate cytochrome b-specific primers (C). Download
PCR analysis of the GHANA DNA extract with primers that amplify the full AUT1 allele (A) and primers that amplify the truncated AUT1 allele (B). Download
DNA alignment of the HpHb receptor gene (A) and amino acid alignment of the HpHb receptor protein (B) of GHANA together with the known HpHb receptor sequences of T. b. gambiense 1 (y, z1, z2, and z3 alleles), T. b. gambiense 2 (n, c t1, t3, and u1 alleles), T. b. brucei (b1, c, d, e, f1, f2, g, h, I, j, k, l, m, n, o, p1, p2, q, r, s, t1, t2, u1, u2, u3, v, w, and x alleles), and T. evansi type A (u1 allele). The codon and the L210S substitution are indicated by the red box. Download
Coomassie blue stains of 400 ng recombinant ApoL1, ApoL1 N264K, ApoL1 G1, Apol1 G1 N264K, ApoL1 G2, and ApoL1 G2 N264K proteins. These recombinant proteins are all tagged with a 6-His sequence at the C terminus and are solubilized in 20 mM acetic acid. Download
Taxon, country, host, and year of isolation of the reference strains included in the genome-wide SNP analysis.
Number of heterozygous SNPs within the 78,899 characterized SNP loci in the Trypanosoma genomes.