ABSTRACT
A major challenge facing bacterial intestinal pathogens is competition for nutrient sources with the host microbiota. Vibrio cholerae is an intestinal pathogen that causes cholera, which affects millions each year; however, our knowledge of its nutritional requirements in the intestinal milieu is limited. In this study, we demonstrated that V. cholerae can grow efficiently on intestinal mucus and its component sialic acids and that a tripartite ATP-independent periplasmic SiaPQM strain, transporter-deficient mutant NC1777, was attenuated for colonization using a streptomycin-pretreated adult mouse model. In in vivo competition assays, NC1777 was significantly outcompeted for up to 3 days postinfection. NC1777 was also significantly outcompeted in in vitro competition assays in M9 minimal medium supplemented with intestinal mucus, indicating that sialic acid uptake is essential for fitness. Phylogenetic analyses demonstrated that the ability to utilize sialic acid was distributed among 452 bacterial species from eight phyla. The majority of species belonged to four phyla, Actinobacteria (members of Actinobacillus, Corynebacterium, Mycoplasma, and Streptomyces), Bacteroidetes (mainly Bacteroides, Capnocytophaga, and Prevotella), Firmicutes (members of Streptococcus, Staphylococcus, Clostridium, and Lactobacillus), and Proteobacteria (including Escherichia, Shigella, Salmonella, Citrobacter, Haemophilus, Klebsiella, Pasteurella, Photobacterium, Vibrio, and Yersinia species), mostly commensals and/or pathogens. Overall, our data demonstrate that the ability to take up host-derived sugars and sialic acid specifically allows V. cholerae a competitive advantage in intestinal colonization and that this is a trait that is sporadic in its occurrence and phylogenetic distribution and ancestral in some genera but horizontally acquired in others.
IMPORTANCE
Sialic acids are nine carbon amino sugars that are abundant on all mucous surfaces. The deadly human pathogen Vibrio cholerae contains the genes required for scavenging, transport, and catabolism of sialic acid. We determined that the V. cholerae SiaPQM transporter is essential for sialic acid transport and that this trait allows the bacterium to outcompete noncatabolizers in vivo. We also showed that the ability to take up and catabolize sialic acid is prevalent among both commensals and pathogens that colonize the oral cavity and the respiratory, intestinal, and urogenital tracts. Phylogenetic analysis determined that the sialic acid catabolism phenotype is ancestral in some genera such as Yersinia, Streptococcus, and Staphylococcus and is acquired by horizontal gene transfer in others such as Vibrio, Aeromonas, and Klebsiella. The data demonstrate that this trait has evolved multiple times in different lineages, indicating the importance of specialized metabolism to niche expansion.
INTRODUCTION
Vibrio cholerae is the causative agent of cholera, the deadly diarrheal disease that affects millions each year. To date, there have been seven cholera pandemics, the latest of which, the 7th pandemic, began in 1961 (1–3). Pandemic cholera-causing isolates of V. cholerae belong to the O1 serogroup, which is divided into two biotypes: the classical biotype and the El Tor biotype. The current 7th pandemic is of the El Tor biotype, and the classical biotype strains are believed to have caused the first six pandemics of cholera but have now disappeared from the regions of cholera endemicity (1, 3). Additionally, in the early 1990s, a new serogroup, O139, emerged as a leading cause of cholera in what was believed at the time to be the beginning of the 8th pandemic; however, within a few years, the El Tor biotype reemerged as the predominant cause of cholera (1, 3, 4). Interestingly, O139 isolates lack the sialic acid catabolism (SAC) gene cluster (5). Others have speculated that one explanation of why the El Tor biotype was able to dominate the classical biotype is that it was a consequence of enhanced metabolic fitness (6, 7). They proposed that the ability to produce a neutral fermentation end product rather than an acid by-product from metabolized sugars improves fitness in a range of environments.
With nearly 100 trillion commensal bacteria colonizing the intestine, being highly efficient at scavenging for nutrients becomes essential for survival (8–10). Enhanced metabolic fitness is especially important for gastroenteritis-causing intestinal pathogens, as these pathogens compete for nutrients with the already established microbiota (11–15). The gastrointestinal (GI) tract is lined with a mucus layer serving as a barrier between epithelial cells and bacteria as well as protecting the epithelium from digestive enzymes found in the lumen (16). The main components of mucus are mucins, which comprise a family of heavily glycosylated proteins that are secreted from the epithelial tissue and form the gel-like mucus layer that covers the cells (16). The glycosylation of mucin is of particular importance for the gut microbiota, as many commensal as well as pathogenic species are able to cleave sugars from mucin in order to use them as nutrients (17, 18). Some of the main sugars that are typically found in mucus include ribose, mannose, hexuronates, galactose, fucose, arabinose, N-acetylglucosamine (GlcNAc), and the sialic acid N-acetylneuraminic acid (Neu5Ac) (19, 20). Sialic acid is of particular interest because it is the most common terminal carbohydrate present on N- and O-linked glycans, which are predominant in the mucus-covered oral, lung, intestinal, and vaginal surfaces. Additionally, there have been over 50 structural sialic acid derivatives that been characterized, the most abundant of which is Neu5Ac.
Cholera-causing strains of V. cholerae carry genes necessary for transport (siaPQM) and catabolism (nanA, nanEK) of sialic acid as well as a gene encoding a scavenging enzyme, sialidase (nanH), which can cleave the sialic acid residues from the glycan, allowing them to be taken up into the bacterial cell (5). These genes are contained within the 57-kb Vibrio pathogenicity island 2 (VPI-2) region that integrates at a tRNA-serine locus (5). The canonical VPI-2 region is present only in V. cholerae O1 serogroup strains; however, the majority of the region has been deleted from O139 serogroup isolates (5, 21). Previously, we have shown that V. cholerae N16961, an O1 serogroup El Tor isolate, can utilize Neu5Ac as a sole carbon source and that a mutant lacking the sialic acid aldolase enzyme N-acetylneuraminic acid lyase (NanA) is defective in colonization of an infant mouse model compared to wild-type V. cholerae (22). Additionally, it was demonstrated that SiaPQM, the tripartite ATP-independent periplasmic (TRAP) transporter within VPI-2, is solely responsible for the transport of the Neu5Ac sialic acid (23). Four diverse families of sialic acid transporters have been shown to be genetically linked with SAC genes among bacteria: the TRAP transporter system, the type present in most Vibrio species; the major facilitator superfamily (MFS)-type NanT transporters; an ATP-binding cassette (ABC)-type transporter; and the sodium solute symporter (SSS)-type transporters (24–28).
In this study, we investigated the ability of V. cholerae to grow in mouse intestinal mucus as well as in individual sugar components of mucus as sole carbon sources. We investigated the ability of V. cholerae to utilize sialic acid derivatives N-glycolylneuraminic acid (Neu5Gc) and 2-keto-3-deoxy-d-glycero-d-galacto-nononic acid (KDN) as sole carbon sources. We determined the significance of sialic acid transport in vivo using a streptomycin (Sm)-pretreated adult mouse model of colonization as well as competition persistence assays between the wild-type strain and NC1777, a SiaPQM transporter-deficient mutant strain. The results of the in vivo study as well as of in vitro competition assays suggest that sialic acids are important nutrients that V. cholerae scavenges for during host colonization. We determined the distribution of the ability to take up and catabolize sialic acid and mapped the different types of sialic acid transporters onto the phylogeny of the NanA protein among bacteria to demonstrate that this phenotype evolved multiple times.
RESULTS AND DISCUSSION
Vibrio cholerae utilizes intestinal mucus and its components as sole carbon sources.
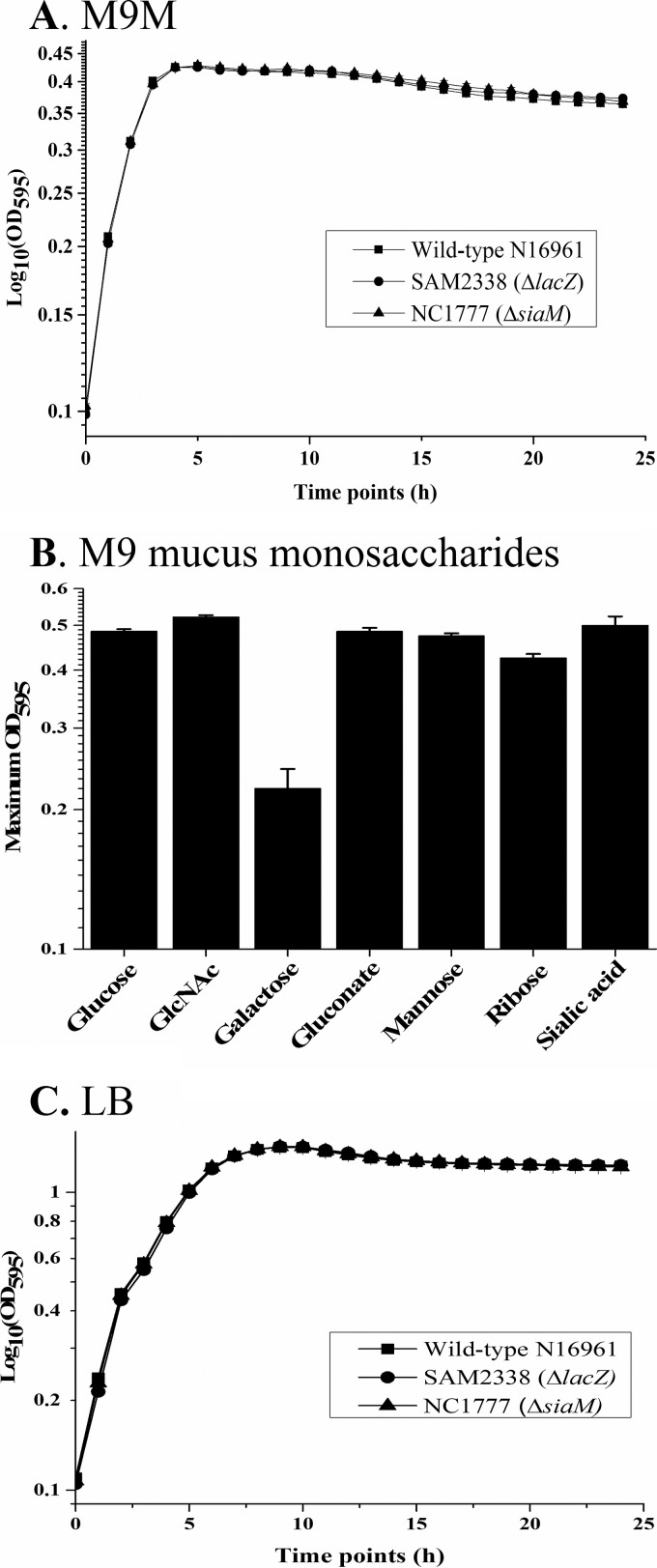
During colonization in the GI tract, one of the main sources of nutrients is mucus secreted from the epithelial cells lining the lumen (16). Here, we assessed the ability of V. cholerae N16961 to grow in M9 minimal medium supplemented with intestinal mucus (M9M) as the sole carbon source. We demonstrated that V. cholerae is able to grow in intestinal mucus, showing no observable lag phase (Fig. 1A). We next determined the growth patterns on the main monosaccharides that are commonly found as part of intestinal mucus: ribose, mannose, hexuronates (gluconate and glucuronate), galactose, fucose, arabinose, N-acetylglucosamine (GlcNAc), and Neu5Ac. We found that V. cholerae N16961 achieved comparable levels of biomass (maximum optical density [OD]) when grown on glucose, ribose, mannose, gluconate, GlcNAc, or Neu5Ac as the sole carbon source. Only moderate growth was shown with galactose as a sole carbon source (Fig. 1B).
FIG 1 .
Growth analysis of V. cholerae N16961 on mouse intestinal mucus and mucus sugars. (A) V. cholerae growth in M9 minimal medium supplemented with mucus (M9M). (B) Maximum density (OD595) of V. cholerae grown in M9 supplemented with glucose, N-acetylglucosamine (GlcNAc), galactose, gluconate, mannose, ribose, or Neu5Ac. (C) V. cholerae growth in LB media. Data are presented as averages of results of two biological replicates with three technical replicates. Error bars represent standard errors of the means (SEM).
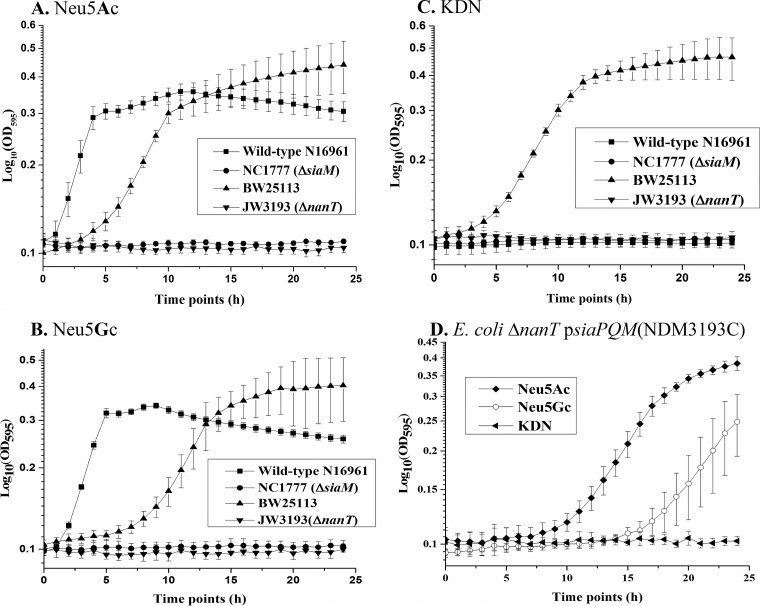
It is unknown whether V. cholerae can utilize additional sialic acids such as Neu5Gc, which is a major component of all mammalian mucins, with the exception of mucins of humans, or KDN, a recently identified sialic acid of lower-order vertebrates (29, 30). Our growth analysis showed that N16961 can utilize Neu5Ac and Neu5Gc but not KDN as a sole carbon source (Fig. 2A to C). To determine whether the SiaPQM TRAP transporter was required for Neu5Gc transport, we examined a SiaPQM-deficient strain, NC1777, for Neu5Gc uptake and found that the mutant did not grow on either Neu5Ac or Neu5Gc (Fig. 2A and B). Interestingly, the growth pattern of V. cholerae showed a shorter lag phase on Neu5Ac and Neu5Gc than that seen with Escherichia coli, which had an extended lag phase. This suggests that V. cholerae SiaPQM transports both of these substrates with greater efficiency than E. coli BW25113, which is consistent with TRAP transporters being high-affinity transporters for their respective solutes. To further examine the ability of Vibrio species to utilize KDN and to ensure that our phenotype was not strain specific, we examined the growth patterns of Vibrio vulnificus, a species distantly related to V. cholerae and containing an identical sialic acid transport and catabolism region (26, 31). Similarly to what we showed for V. cholerae, V. vulnificus CMCP6 can utilize Neu5Ac and Neu5Gc but not KDN as a sole carbon source (see Fig. S1 in the supplemental material). To determine whether SiaPQM was required for Neu5Gc transport, we examined a SiaPQM-deficient strain, JJK0731, for Neu5Gc uptake and also demonstrated that the mutant cannot grow on either Neu5Ac or Neu5Gc (see Fig. S1).
FIG 2 .
Growth analysis of V. cholerae N16961 with sialic acid derivatives as sole carbon sources. V. cholerae N16961, V. cholerae NC1777 (ΔsiaM), E. coli BW25113, and E. coli JW3193 (ΔnanT) in M9 Neu5Ac (A), M9 Neu5Gc (B), or M9 KDN (C) or E. coli JW3193 in M9 Neu5Ac, M9 Neu5Gc, or M9 KDN (D) were complemented with siaPQM and tested for restored growth on sialic acid derivatives. Growth curves represent results of at least two biological replicates performed in triplicate. Error bars represent SEM.
A recent study by Hopkins and colleagues demonstrated the ability of E. coli to catabolize both Neu5Gc and KDN (32). In their study, they found that the E. coli permease NanT was required for transport of these substrates. They also demonstrated that complementation of a NanT mutant with the SiaPQM transporter from Haemophilus influenzae also resulted in uptake of both Neu5Gc and KDN but that the affinity for KDN was 50-fold lower than for Neu5Ac (32). We complemented an E. coli nanT mutant with siaPQM, strain NDM3193C, and showed that in the E. coli background, both Neu5Ac and Neu5Gc were transported but not KDN, suggesting that SiaPQM is not a transporter for KDN (Fig. 2D). In addition, we also complemented the V. cholerae NC1777 mutant with siaPQM and showed that Neu5Ac and Neu5Gc were transported and catabolized in the complemented mutant (see Fig. S1 in the supplemental material).
The ability to transport sialic acid is essential for V. cholerae fitness in vivo.
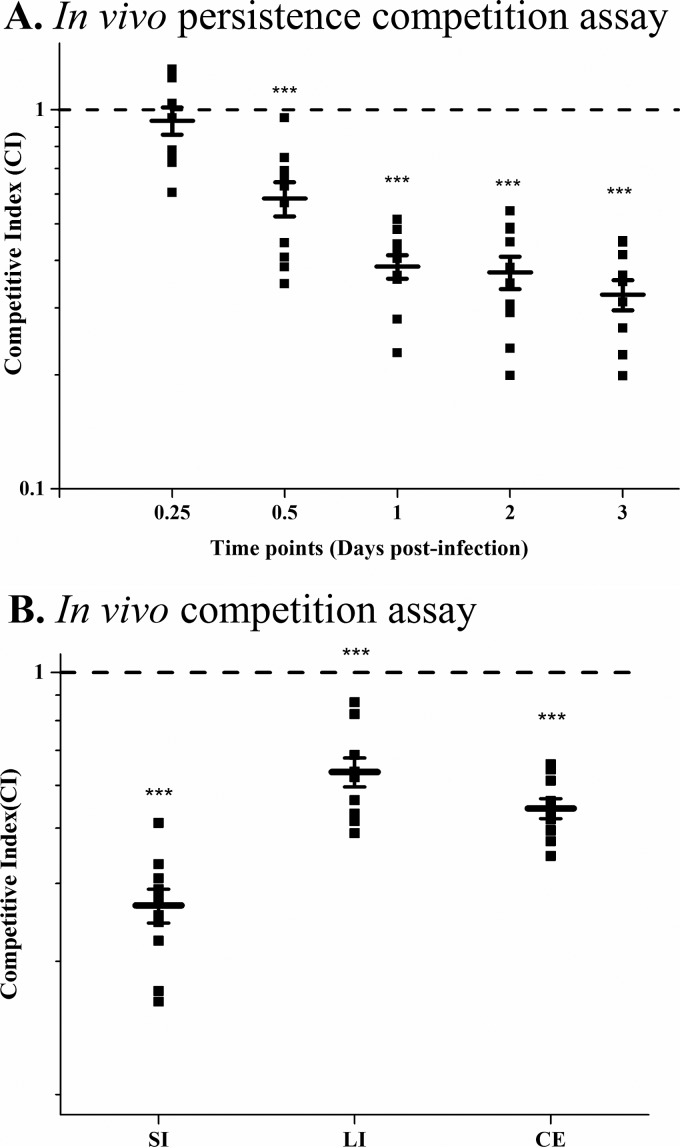
To address whether the ability to take up sialic acids is a significant attribute of V. cholerae in vivo, we performed in vivo competition and persistence assays using a V. cholerae streptomycin-pretreated adult mouse model of colonization (33). Conventionally reared mice contain diverse microbiotas of more than 500 species predominantly from two phyla, Firmicutes and Bacteroidetes, that prevent infection by many enteric pathogens (reviewed in reference 12). Pretreatment of mice with streptomycin to allow pathogen colonization has been utilized to study the mechanisms of pathogenesis as well as the host response to these pathogens in many studies (34–42). The antibiotic treatment does not change the overall number of bacteria but favors expansion of Proteobacteria at the expense of Bacteroidetes, with the total number of Firmicutes remaining the same (12, 43). Several studies have used this model to examine the role of metabolism, and some have suggested that the initial colonization of a pathogen is dependent on nonlimiting nutrients that are available due to the removal of facultative anaerobes; however, during persistence in the gut, the pathogen must compete for the limiting nutrients (13–15, 44–50). For these reasons, the streptomycin-treated mouse model was used to investigate the competition between wild-type V. cholerae and a knock-out strain that cannot transport sialic acid. To accomplish this, we compared NC1777 (ΔsiaM) and a β-galactosidase N16961 mutant strain, SAM2338 (ΔlacZ), which allows a blue/white colony screen. First, to confirm that NC1777 and SAM2338 grow similarly to wild-type N16961, we determined growth in LB and M9M at 37°C overnight and found no difference in growth patterns between the two strains (Fig. 1). In addition, we performed in vitro competition assays between SAM2338 and N16961 grown overnight in LB at 37°C and found no difference in the CFU counts between the strains (data not shown). Streptomycin-pretreated mice were inoculated with a 1:1 ratio mix of equal amounts of SAM2338 and NC1777, resulting in a final concentration of 1 × 1010 CFU/ml. Following inoculation, at 6, 12, 24, 48, and 72 h postinfection, fecal pellets were collected from each of the mice, the CFU were enumerated, and the competitive index (CI) was calculated. First, we compared SAM2338 and N16961 in coculture in vivo and confirmed that, similarly to the in vitro assays, there was no competition between the strains, with a CI not significantly different from 1 (data not shown). Then, we examined a coculture of SAM2338 and NC1777 (ΔsiaM) and determined that the ΔsiaM strain had a CI close to 1 after 6 h postinfection. However, at 12 h postinfection, the ΔsiaM strain was outcompeted 1.7-fold (P < 0.001) by SAM2338 (mean CI of 0.58) (Fig. 3A). This trend continued at time points 24 h, 48 h, and 72 h, with the ΔsiaM strain significantly (P < 0.001) outcompeted, with mean CIs of 0.39, 0.37, and 0.33, respectively (Fig. 3A). These data indicate that at the early stages of colonization, sialic acid may not be a limiting nutrient; however, as colonization persists, sialic acid represents a carbon source that strains must compete for and that contributes to fitness. On day 3 of the colonization persistence assay, mice were sacrificed, and the gastrointestinal tracts were removed, sectioned into the small intestine, cecum, and large intestine, homogenized in phosphate-buffered saline (PBS), serially diluted, and plated on LB X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). It was determined that the ΔsiaM strain was significantly outcompeted in the small intestine, cecum, and large intestine by SAM2338, corroborating the fecal pellet data (Fig. 3B). SAM2338 showed the greatest fitness advantage over the ΔsiaM strain in the small intestine, with a CI of 0.30, followed by the cecum (CI = 0.49) and, finally, the large intestine (CI = 0.59) (Fig. 3B).
FIG 3 .
Vibrio cholerae in vivo competition assays. In vivo competition assays were performed using streptomycin-pretreated mice which were orally inoculated with an equal amounts of SAM2338 (ΔlacZ) and NC1777 (ΔsiaM). (A) Competitive index data were determined by homogenizing fecal pellets collected at 6, 12, 24, 48, and 72 h postinoculation followed by plating on LB X-Gal for a blue/white screen. The competitive index is calculated as follows: CI = ratio out(NC1777 / SAM2338)/ratio in(NC1777/SAM2338). (B) Upon completion of the in vivo persistence assay (72 h), mice were sacrificed, gastrointestinal tracts were removed and sectioned into small intestine (SI), large intestine (LI), and cecum (CE), and CFU values were calculated. In vivo assays were performed with two biological replicates and n = 10 mice. Error bars represent SEM. Statistics were calculated using a one-sample t test with the means compared to the hypothetical CI of 1. Asterisks represent the following: ***, P < 0.01.
Loss of SiaPQM is detrimental in vitro.
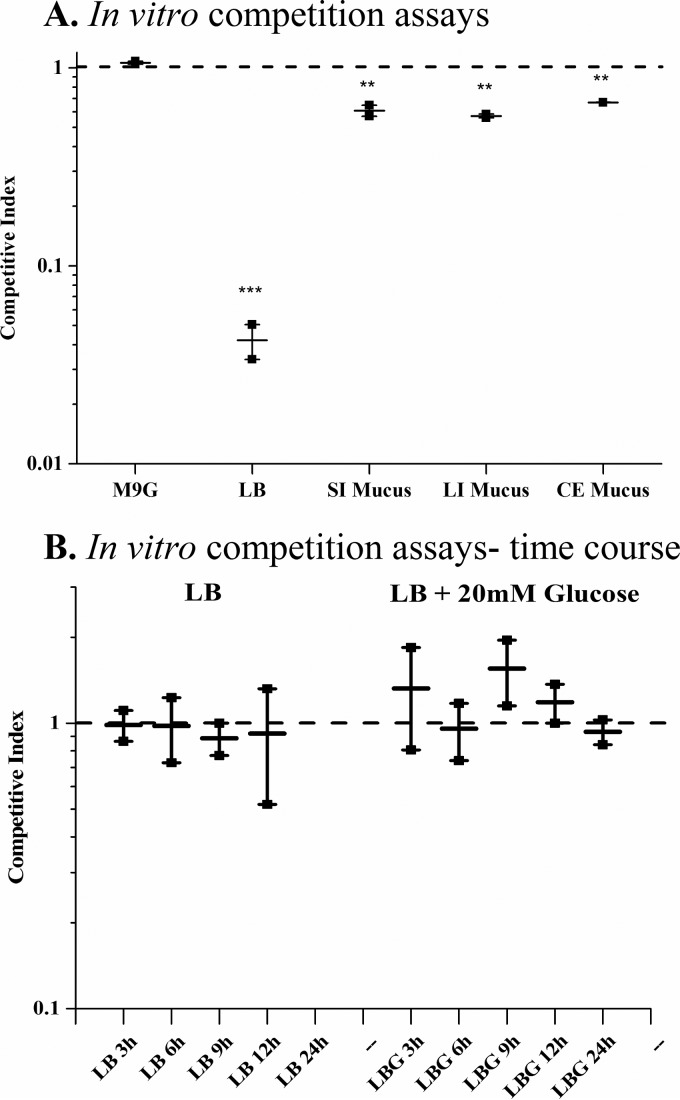
An in vitro competition assay was performed using the same inoculum mix for the in vivo assays containing equal amounts of NC1777 and SAM2338. The strains were competed against each other in either LB or M9 plus glucose (M9G) (Fig. 4A). The coinoculated cultures were incubated for 24 h, serially diluted, and plated on LB X-Gal. The CI in M9G was 1.06, indicating that neither strain exhibits any growth advantage over the other in this medium; however; in LB, the CI was 0.04 (Fig. 4A). Thus, the siaM mutant has a significant defect in competition assays in LB. In order to further investigate the significant defect of the siaM mutant cocultured in LB, a time course competition assay was performed in LB and LB supplemented with 20 mM glucose. The CI was calculated at 3, 6, 9, 12, and 24 h postcoinoculation. In LB, at up to 24 h, there was no significant competition between SAM2338 and NC1777; however, at the 24-h time point, the siaM mutant was again significantly outcompeted by SAM2338 with a CI that was below the limit of detection of 8.1 × 10−8 CFU/ml (Fig. 4B). When glucose was supplied to the LB media, there was no competition between the strains over the course of the assay (Fig. 4B).
FIG 4 .
In vitro competition assays. Results of assays of in vitro competition between V. cholerae strain SAM2338 (ΔlacZ) and the sialic acid transporter mutant NC1777 (ΔsiaM) are presented. (A) Competition assays were performed in M9G, LB, or M9 supplemented with small-intestine (SI), large-intestine (LI), or cecum mucus. (B) Time course in vitro competition assays were performed at 3, 6, 9, 12, and 24 h postinoculation in either LB or LB supplemented with glucose (0.4% [wt/vol]). In vitro data represent averages of results of two biological replicates, and error bars represent SEM. Statistics were calculated using an unpaired Student’s t test with a 95% confidence interval. All samples were compared to M9G to determine statistical significance. Asterisks represent the following: **, P < 0.05.
To investigate additional substrates that could require SiaPQM for transport, we examined growth of N16961 and NC1777 on 190 different carbon sources, 62 of which were utilized by both strains under the conditions examined (see Fig. S2 in the supplemental material). There were six substrates in the presence of which NC1777 either showed no growth (Neu5Ac) or reached a lower OD than N16961 (d-glucosamine, l-proline, l-lactic acid, methyl pyruvate, and α-keto-valeric acid). With the exception of d-glucosamine, all of these substrates are structurally similar, being carboxylic acids or carboxylic acid derivatives. Additionally, previous studies have shown evidence of TRAP substrate binding proteins able to bind each of these compounds except d-glucosamine (51, 52). The main components of LB are tryptone and yeast extract; the former is composed of peptides of casein, which contain a high number of proline residues and sialic acid. Our data suggest that SiaPQM may play a secondary role in the transport of additional substrates, which may explain the significant defect in in vitro competitions assays in LB.
In an attempt to more closely mimic an in vivo nutrient environment, strains were competed against one another in mouse mucus isolated from the small intestine, large intestine, and cecum. In vitro competition assays in M9M were performed as previously described. The cultures were incubated for 24 h, serially diluted, and plated on LB X-Gal plates. The CI values for the small intestine, large intestine, and cecum were 0.61, 0.58, and 0.67, respectively (Fig. 4A). These results showed that the sialic acid transporter mutant was less fit in each of the three mucus samples and that the ability to transport and catabolize sialic acids is essential for the optimal fitness of choleragenic V. cholerae in mucosal environments.
Distribution and phylogenetic analysis of N-acetylneuraminic acid lyase (NanA) among bacteria.
Since we and others have shown how significant sialic acid utilization is in vivo among a number of pathogens (22, 26, 28), we next determined the prevalence of sialic acid utilization genes in all available completed bacterial genomes. We identified 452 bacterial species with the potential to utilize sialic acid as a carbon source from ~7,400 species representing 23 major bacterial groups, nearly a 10-fold increase from our previous study (26). These 452 species were comprised of primarily commensal and pathogen species and encompassed eight phyla: Actinobacteria, with 83 representatives; Bacteroidetes, with 70 representatives; Firmicutes, with 134 representatives; Fusobacteria; with 9 representatives; Planctomycetes, with 6 representatives; Proteobacteria, with 142 representatives; Spirochaetes, with 5 representatives; and Verrucomicrobia, with 3 representatives. Of the 300 species representing Crenarchaeota, Cyanobacteria, Deinococcus-Thermus, and Euryarchaeota from environmental samples, none contained NanA; similarly, of the 188 representatives of the Bacillus genus, predominantly soil organisms, only one species had the ability to utilize sialic acid. Within some species, SAC is present in all strains; in other species, it is present in only a subset of strains. The 452 species are comprised of Gram-positive and Gram-negative species, and a few species from divergent phyla are within each of these major groups. These data demonstrate that SAC is phylogenetically widespread but is sporadic in its occurrence within phyla, families, genera, and even species. The data suggest that the ability to utilize sialic acid as a carbon source was acquired by a select group of species that are colonizers.
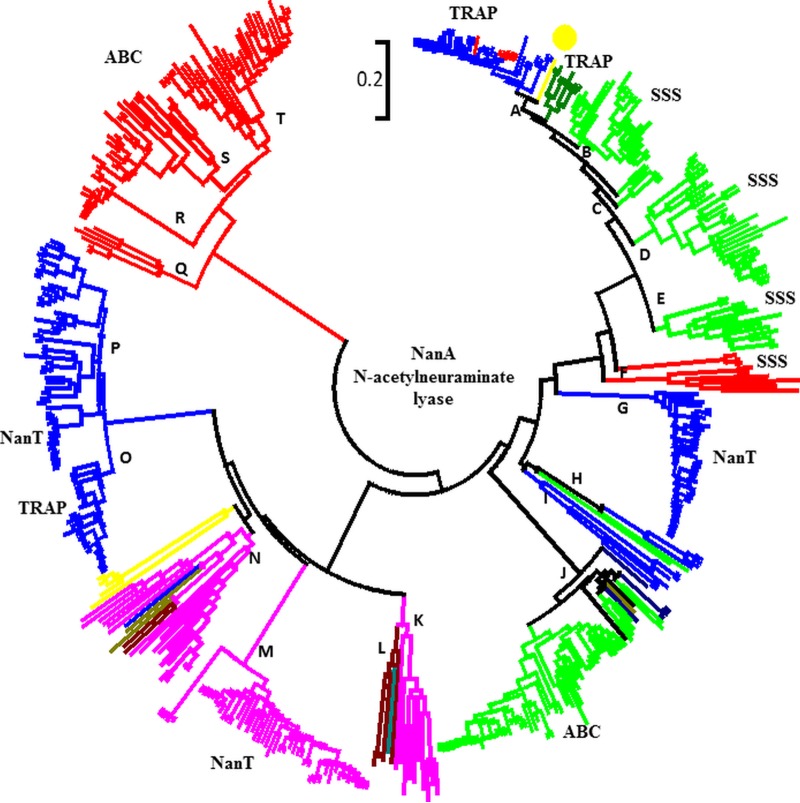
In order to examine the evolutionary history of this trait more closely, we constructed a phylogenetic tree of NanA among the 452 species and examined the branching patterns among these species (Fig. 5). In addition, we mapped the different types of transporters associated with the sialic acid catabolism genes from each species onto this tree. For ease of analysis, we have divided the tree into 20 major clusters designated A to T on the tree (Fig. 5). What is immediately apparent from this analysis is that representatives of each of the eight major phyla do not cluster together, indicating different evolutionary histories of NanA among the groups (Fig. 5).
FIG 5 .
NanA phylogeny. A phylogenetic tree of NanA is shown. The tree was constructed using the neighbor-joining tree method and the p-distance model of evolutionary distances as parameters and MEGA6 (75–77). Letters indicate major bacterial phyla present in the corresponding NanA clade. Colors indicate the major phylogenetic groups that contain nanA. Red, Actinobacteria; purple, Bacteroidetes; lime green, Firmicutes; dark green, Fusobacteria; dark red, Planctomycetes; blue, Proteobacteria; yellow, eukaryotes; gray, Spirochaetes.
For example, species from the phylum Proteobacteria that contain a NanA homologue are found in 6 highly divergent clusters designated A, G, H, I, O, and P, indicating that this trait has evolved several times in this phylum (Fig. 5). Cluster A is a tight group of closely related NanA alleles from the family Pasteurellaceae represented by Aggregatibacter, Haemophilus, and Pasturella species, and all encode a TRAP-type sialic acid transporter, with the exception of H. ducreyi, which contains an ABC sialic acid transporter (53) (see Fig. S3A in the supplemental material). Sialic acid transport and catabolism was demonstrated in H. influenzae, H. ducreyi, and P. multocida (24, 53–57). Nested within cluster A are species of the phylum Actinobacteria, which suggests that NanA in these species may have been acquired from Proteobacteria (see Fig. S3A). Branching separately within cluster A are 8 Fusobacteria representatives, which all also have a sialic acid TRAP transporter system.
The second Proteobacteria group is cluster G and is comprised of Enterobacteriaceae species, E. coli, Shigella spp., Salmonella spp., Citrobacter spp., and Klebsiella spp., all of which form a tight group with short branch lengths and contain species that are known pathogens, and many have been shown to utilize sialic acid as a carbon source (see Fig. S3B in the supplemental material) (25, 26, 58–60).
We identified 31 species of Aliivibrio, Photobacterium, and Vibrio that can putatively transport and catabolize sialic acid. Three species, Vibrio nigripulchritudo, V. sinaloensis, and V. orientalis, contained two copies of NanA which share <30% amino acid identity. One NanA copy clustered with all NanA proteins from all other Vibrio species (cluster O), whereas the second copy branched in a distant lineage with members of the Enterobacteriaceae (cluster I) (see Fig. S3C in the supplemental material). Cluster O is comprised solely of 23 Vibrio species, and all contain a TRAP transporter (see Fig. S3D in the supplemental material). Cluster P is comprised mainly of Aliivibrio, Photobacterium, Enterobacteria, Psychromonas, Pseudoalteromonas, and Yersinia species. Subgroup P1 is comprised of all 12 Yersinia species, and NanA is present in all strains of each species, indicating that this is an ancestral trait within the genus. Subgroup P2 contains Plesiomonas shigelloides, Aeromonas hydrophila, and A. veronii isolates, human and fish pathogens. Five A. hydrophila strains from among the 31 sequenced and 2 A. veronii strains from among 12 sequenced contained NanA. The gene orders of the A. hydrophila and A. veronii SAC clusters differed, and both integrase and transposase genes were associated with both regions, suggesting that the SAC cluster represents a horizontally acquired trait in this genus. Indeed, in A. hydrophila strain ML09-119, the sialic acid region is on a genomic island that also contains the genes for fucose utilization, indicating the acquisition of a metabolic island. It is of interest that A. hydrophila ML09-119 represents an invasive species from Asia that caused serious disease epidemics in United States catfish farms. The island region is absent from indigenous strains, suggesting that acquisition by the Asian strain may be one of the factors that make them so deadly. These species branched with NanA from three Photobacterium species, and all contained an SSS-type transporter. Subgroup P4 represented mainly Pseudoalteromonas species, and P5 contained Klebsiella, Edwardsiella, and Proteus species (see Fig. S3D). In Klebsiella oxytoca, a second divergent copy of NanA is found within cluster P4. Our growth curve analysis of Aeromonas hydrophila, Enterobacter aerogenes, Klebsiella oxytoca, and Providencia rettgeri on Neu5Ac demonstrated that all four species can utilize sialic acid as a sole carbon source (see Fig. S4).
The phylum Firmicutes is represented by 5 divergent clusters, and either an SSS transporter (clusters B, C, D, and E) or an ABC transporter (cluster J) is associated with the SAC genes in these groups (Fig. 5). Cluster B contains 13 Clostridium species, and within cluster C is a sole representative of the genus Bacillus, B. aquimaris, along with 3 Planococcus species, all of which contain a TRAP transporter, which is highly unusual for Gram-positive bacteria (see Fig. S5A in the supplemental material). Cluster D represents all Staphylococcus species, and most Lactobacillus species are present in cluster E (see Fig. S5B). NanA appears to be present in all sequenced representatives of Staphylococcus, indicating that SAC represents an ancestral trait. Both Staphylococcus aureus and Lactobacillus plantarum have been shown to utilize sialic acid (61, 62). Cluster J, which is related to clusters B, D, and E only distantly, contains 29 Streptococcus species (see Fig. S5C).
The phylum Bacteroidetes is represented by 72 species, which are found on three divergent clusters, K, N, and M, on the NanA tree (Fig. 5; also Fig. S6 in the supplemental material). Cluster K is a highly divergent group with long-branch lengths and is comprised of members of the phyla Planctomycetes and Bacteroidetes. Branching divergently from this group in cluster L are representatives of the phylum Verrucomicrobia (see Fig. S6A in the supplemental material). Cluster M consists of closely related NanA alleles from Bacteroides, Prevotella, and Tannerella species, many of which are human gut commensals, and all contain a NanT-type transporter (see Fig. S6B). Tannerella forsythiae and Bacteroides fragilis were shown to utilize sialic acid as a sole carbon source (63, 64).
The phylum Actinobacteria is represented by 83 species, the majority of which are present on the most divergent branches of the NanA tree, clusters Q, R, S, and T (Fig. 5; also Fig. S7 in the supplemental material). These cluster present mainly Corynebacterium, Arthrobacter, and Bifidobacterium species; nested within cluster S was a NanA from Garderella vaginalis, which suggests that it may have been acquired from a Bifidobacterium (see Fig. S7A in the supplemental material). All members of clusters Q, R, S, and T had an ABC type putative sialic acid transporter. Experimental evidence for sialic acid utilization was shown for Gardnerella vaginalis, Corynebacterium diphtheriae, C. glutamicum, and Bifidobacterium breve (65–68).
Conclusions.
The adaptability to new nutritional sources is crucial for a species to switch between different environments. Adaptation to host environments requires the ability to switch from nutrient-poor to nutrient-rich and from free-living to host-associated conditions, including competing with the resident microbiota for both space and nutrients. Some of the major nutrient capabilities of V. cholerae are mucus associated, and pathogenic V. cholerae bacteria specifically scavenge and utilize sialic acids to allow niche expansion. Similarly, a recent study has proposed that the ability of Salmonella enterica and Clostridium difficile to colonize antibiotic-treated mice is due to the disruption of intestinal microbiota that allows these species to scavenge and utilize host sialic acids (69). It is of interest that, although the members of the TRAP family of transporters have a high affinity for sialic acid, their distribution is limited, being confined mainly to the genus Vibrio and the family Pasteurellaceae. Among other Gram-negative bacteria, NanT is the predominant sialic acid transporter, whereas the predominant types in Gram-positive bacteria are the SSS and ABC transporters. As the transporter distribution and phylogenetic tree suggest, sialic acid catabolism has evolved multiple times and is an ancestral trait in some species and newly acquired in others.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. All V. cholerae strains were grown aerobically with aeration (225 rpm) at 37°C in Luria Bertani (LB) broth (Fisher Scientific, Waltham, MA) with a 1% NaCl concentration. For growth analysis on intestinal mucus, mucus sugars, and sialic acid derivatives as sole carbon sources, M9 minimal medium was supplemented with 0.02 mM MgSO4 and 0.1 mM CaCl2. M9 minimal medium supplemented with intestinal mucus (M9M) (30 µg/ml) or glucose (M9G) was used or N-acetylglucosamine, d-ribose, d-mannose, d-gluconate, or d-galactose (all 10 mM), or Neu5Ac (3 mM), 2-keto-3-deoxy-d-glycero-d-galacto-nononic acid (KDN) (4 mM), or N-glycolylneuraminic acid (Neu5Gc) (3 mM) (Sigma-Aldrich) was added to M9 to serve as the carbon source. Antibiotics were used at the following concentrations: streptomycin (Sm) at 200 µg/ml, ampicillin (Amp) at 100 µg/ml, chloramphenicol (Cm) at 25 µg/ml, and kanamycin at 50 µg/ml.
Bacterial growth assays.
Strains (diluted 1:100) were grown aerobically in LB broth at 37°C overnight and incubated for 3 h to reach mid-exponential-phase growth, and cells were pelleted by centrifugation, washed with M9 media, and resuspended in M9 media. The cultures were then diluted 1:40 in M9 media supplemented with each carbon source and incubated at 37°C with intermittent shaking for 24 h. The optical density at 595 nm (OD595) was taken hourly using a Tecan Sunrise microplate reader (Tecan Group, Ltd.) and Magellan plate reader software. High-throughput screens of carbon utilization abilities were performed using Biolog PM1 and PM2A plates as described above. Each experiment was performed with at least two biological replicates.
Complementation of the E. coli sialic acid transporter mutant.
Genomic DNA from V. cholerae N16961 was used as a template for PCR amplification of siaPQM (VC1777-VC1779) transporter genes using primer pair VcsiaPQMF (TCTAGATGCTAAGCCATCAACATCTG) and VcsiaPQMR (GAGCTCTCAACTACTGTCCGTATCCG), which included XbaI and SacI restriction enzyme sites (underlined), respectively. The product was subcloned into pJET1.2 and transformed into E. coli DH5α. Recombinant pJET1.2 plasmids were ligated into pBAD33 and were used to transform into E. coli JW3193, resulting in strain NDM3193C, which was used for the functional complementation assays described below. Strain NDM3193C was examined for growth in M9 media supplemented with arabinose (0.05% [wt/vol]) and with Neu5Ac, Neu5Gc, or KDN at 37°C. This construct was also used to complement NC1777 and demonstrate function complementation. Each strain and condition was tested using at least two biological replicates and in triplicate.
Mucus extraction.
The intestinal mucus used in in vitro competition assays and growth analyses was isolated from mouse gastrointestinal tracts as previously described (70, 71). Briefly, mice gastrointestinal tracts were collected and the small intestine, cecum, and large intestine were first flushed with PBS followed by gentle scraping of the walls of the intestine to remove the mucus. The mucus was processed by homogenizing 200 mg of mucus in 5 ml of sterile PBS with streptomycin and centrifuged at 500 × g to pellet out tissue and fecal material. The protein content of the mucus was determined using a Bradford assay. Competition assays and growth analyses were performed in M9 media supplemented with mucus (final concentration of 30 µg/ml of protein) (M9M).
In vivo and in vitro competition assays.
All experiments involving mice were approved by the University of Delaware Institutional Animal Care and Use Committee. Male C57BL/6 mice between 6 and 10 weeks of age were treated with streptomycin and inoculated as described previously (33, 70, 72). In short, the mice (n = 10) were fasted for 4 h before being orogastrically administered streptomycin and were then resupplied with food and water, supplemented with streptomycin (2.5 g/500 ml). At 20 h after antibiotic treatment, the mice were again fasted for 4 h and orogastrically inoculated with 100 µl of bacterial suspension in PBS with a final concentration of 1 × 1010 CFU/ml. Vibrio cholerae strain SAM2338 (ΔlacZ) was used as a surrogate wild-type strain in competition experiments as it allows white/blue colony selection with the NC1777 strain (ΔsiaM) when grown on X-Gal LB plates.
For in vitro competition assays, an aliquot of the in vivo inoculum was used to inoculate M9G, M9M, or LB. Cultures were incubated at 37°C for 24 h, serially diluted and plated on LB X-Gal plates, and then incubated overnight at 37°C. CFU were counted as either blue (N16961 or NC1777) or white (SAM2338), and the ratio between the strains was determined. The competitive index (CI) was determined as follows: CI = ratio out(NC1777 / SAM2338)/ratio in(NC1777 / SAM2338). A CI of <1 indicated that the wild-type strain outcompeted the mutant strain; a CI of >1 indicated the mutant strain outcompeted the wild-type strain. For the in vivo persistence competition assay, fecal pellets were collected (2 to 4 per animal) at daily time intervals from coinfected mice. The fecal pellets were weighed and then mechanically homogenized in 2 ml of sterile PBS, serially diluted and plated on LB Sm X-Gal agar, and incubated at 37°C overnight. The ratio of NC1777 colonies to SAM2338 colonies was termed the “ratio out” at each time point. After the last time point of the persistence assay, the mice were sacrificed and the entire gastrointestinal tract was harvested from each mouse and separated into samples of the small intestine, cecum, and large intestine. Each sample was homogenized in 4 ml of PBS, serially diluted in PBS, plated on LB X-Gal plates, and incubated overnight at 37°C, and a blue/white colony screen was performed as described above.
Phylogenetic analysis.
We queried using pBLAST with NanA of V. cholerae N16961 (VC1776) and Staphylococcus aureus N315 (SA0304) as seeds, and all completed and draft bacterial genomes (~7,400 genomes) were published on the National Center for Biotechnology Information (NCBI) website (73). In addition to a putative NanA, candidate species were determined by the presence of homologues to a sialic acid transporter, a putative nanE or nanK gene, or a gene encoding sialidase in proximity to the NanA-encoding gene on the genome. Sequences were aligned using ClustalW (see Table S2 in the supplemental material) (74). Aligned sequences were manually checked, and phylogenetic analysis was conducted with MEGA6 using the neighbor-joining (NJ) tree-building method and p distance and Poisson correction models for estimating evolutionary distances (75, 76).
SUPPLEMENTAL MATERIAL
Growth analysis of V. vulnificus CMCP6 with sialic acid derivatives as sole carbon sources. Data represent results of growth analysis of V. vulnificus CMCP6 (black circles), JJK0731 (ΔsiaM) (a sialic acid transporter mutant) (black downward-facing triangles), V. cholerae N16961 (black squares), NC1777 (ΔsiaM) (black diamonds), and E. coli BW25113 (black upward-facing triangles) in M9 minimal media supplemented with N-acetylneuraminic acid (Neu5Ac) (A), N-glycolylneuraminic acid (Neu5Gc) (B), or 2-keto-3-deoxy-d-glycero-d-galacto-nononic acid (KDN) (C) as a sole carbon source. (D) Complementation of NC1777 with siaPQM. All sialic acid derivatives were used at a concentration of 1 mg/ml. Growth curves represent results of at least two biological replicates performed in triplicate. Error bars represent SEM. Download
High-throughput screen of carbon utilization abilities of V. cholerae pathogenic isolates. Vibrio cholerae strains N16961 (El Tor) (black bars) and NC1777 (ΔsiaM) (gray bars) were screened for growth on 190 different carbon sources using Biolog PM1 and PM2A plates. The area under the curve (AUC) was determined for each condition tested. The growth was normalized by subtracting the AUC value corresponding to the well containing no carbon source from the AUC value corresponding to each well with a carbon source. Data represent the averages of results of two biological replicates for each strain for each of the PM plates. Download
NanA phylogeny subtrees A, G, H, I, O, and P. (A) Subtree containing representative species from cluster A. (B) Subtree containing representative species from cluster G. (C) Subtree containing representative species from clusters H and I. (D) Subtree containing representative species from clusters O and P. Download
Growth analysis of various species on M9 supplemented with glucose or N-acetylneuraminic acid as a sole carbon source. Sialic acid was used at a concentration of 10 mM. Growth curves represent results of at least two biological replicates performed in triplicate. Error bars represent SEM. Download
NanA phylogeny subtrees B, D, E, F, and J. (A) Subtree containing representative species from cluster B. (B) Subtree containing representative species from clusters D, E, and F. (C) Subtree containing representative species from cluster J. Download
NanA phylogeny subtrees K to N. (A) Subtree containing representative species from clusters K and L. (B) Subtree containing representative species from cluster M. (C) Subtree containing representative species from cluster N. Download
NanA phylogeny subtrees Q to T. (A) Subtree containing representative species from clusters Q, R, and S. (B) Subtree containing representative species from cluster T. Download
Bacterial strains and plasmids used in this study.
ACKNOWLEDGMENTS
We gratefully acknowledge members of the Boyd Group for reviewing and giving constructive feedback on the manuscript.
This research was supported in part by a National Science Foundation CAREER award, DEB-0844409, to E.F.B. N.D.M. and J.-B.L. were supported in part by the Chemistry Biology Interface graduate program at the University of Delaware.
Funding Statement
This research was supported in part by a National Science Foundation CAREER award DEB-0844409 to EFB
Footnotes
Citation McDonald ND, Lubin J-B, Chowdhury N, Boyd EF. 2016. Host-derived sialic acids are an important nutrient source required for optimal bacterial fitness in vivo. mBio 7(2):e02237-15. doi:10.1128/mBio.02237-15.
REFERENCES
- 1.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Balakrish Nair G, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 2.Faruque SM, Sack DA, Sack RB, Colwell RR, Takeda Y, Nair GB. 2003. Emergence and evolution of Vibrio cholerae O139. Proc Natl Acad Sci U S A 100:1304–1309. doi: 10.1073/pnas.0337468100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sack DA, Sack RB, Nair GB, Siddique AK. 2004. Cholera. Lancet 363:223–233. doi: 10.1016/S0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 4.Waldor MK, Mekalanos JJ. 1994. Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype. J Infect Dis 170:278–283. doi: 10.1093/infdis/170.2.278. [DOI] [PubMed] [Google Scholar]
- 5.Jermyn WS, Boyd EF. 2002. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology 148:3681–3693. doi: 10.1099/00221287-148-11-3681. [DOI] [PubMed] [Google Scholar]
- 6.Kovacikova G, Lin W, Skorupski K. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator APhA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol Microbiol 57:420–433. doi: 10.1111/j.1365-2958.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- 7.Yoon SS, Mekalanos JJ. 2006. 2,3-Butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect Immun 74:6547–6556. doi: 10.1128/IAI.00695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley RE, Knight R, Gordon JI. 2007. The human microbiome: eliminating the biomedical/environmental dichotomy in microbial ecology. Environ Microbiol 9:3–4. doi: 10.1111/j.1462-2920.2006.01222_3.x. [DOI] [PubMed] [Google Scholar]
- 10.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freter R. 1962. In vivo and in vitro antagonism of intestinal bacteria against Shigella flexneri. II. The inhibitory mechanism. J Infect Dis 110:38–46. doi: 10.1093/infdis/110.1.38. [DOI] [PubMed] [Google Scholar]
- 12.Stecher B, Hardt WD. 2011. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol 14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. 2009. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun 77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertin Y, Chaucheyras-Durand F, Robbe-Masselot C, Durand A, de la Foye A, Harel J, Cohen PS, Conway T, Forano E, Martin C. 2013. Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environ Microbiol 15:610–622. doi: 10.1111/1462-2920.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corfield AP, Carroll D, Myerscough N, Probert CS. 2001. Mucins in the gastrointestinal tract in health and disease. Front Biosci 6:D1321–D1357. doi: 10.2741/Corfield. [DOI] [PubMed] [Google Scholar]
- 17.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. 2013. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23:1038–1046. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manzi AE, Norgard-Sumnicht K, Argade S, Marth JD, van Halbeek H, Varki A. 2000. Exploring the glycan repertoire of genetically modified mice by isolation and profiling of the major glycan classes and nano-NMR analysis of glycan mixtures. Glycobiology 10:669–689. doi: 10.1093/glycob/10.7.669. [DOI] [PubMed] [Google Scholar]
- 20.Bansil R, Turner BS. 2006. Mucin structure, aggregation, physiological functions and biomedical applications Curr Opin Colloid Interface Sci 11:164–170. doi: 10.1016/j.cocis.2005.11.001. [DOI] [Google Scholar]
- 21.Jermyn WS, Boyd EF. 2005. Molecular evolution of Vibrio pathogenicity island-2 (VPI-2): mosaic structure among Vibrio cholerae and Vibrio mimicus natural isolates. Microbiology 151:311–322. doi: 10.1099/mic.0.27621-0. [DOI] [PubMed] [Google Scholar]
- 22.Almagro-Moreno S, Boyd EF. 2009. Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect Immun 77:3807–3816. doi: 10.1128/IAI.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury N, Norris J, McAlister E, Lau SY, Thomas GH, Boyd EF. 2012. The VC1777-VC1779 proteins are members of a sialic acid-specific subfamily of TRAP transporters (SiaPQM) and constitute the sole route of sialic acid uptake in the human pathogen Vibrio cholerae. Microbiology 158:2158–2167. doi: 10.1099/mic.0.059659-0. [DOI] [PubMed] [Google Scholar]
- 24.Vimr E, Lichtensteiger C, Steenbergen S. 2000. Sialic acid metabolism’s dual function in Haemophilus influenzae. Mol Microbiol 36:1113–1123. doi: 10.1046/j.1365-2958.2000.01925.x. [DOI] [PubMed] [Google Scholar]
- 25.Severi E, Hood DW, Thomas GH. 2007. Sialic acid utilization by bacterial pathogens. Microbiology 153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 26.Almagro-Moreno S, Boyd EF. 2009. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol 9:118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer M, Zhang QY, Hubbard RE, Thomas GH. 2010. Caught in a TRAP: substrate-binding proteins in secondary transport. Trends Microbiol 18:471–478. doi: 10.1016/j.tim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Haines-Menges BL, Whitaker WB, Lubin JB, Boyd EF. 2015. Host sialic acids: a delicacy for the pathogen with discerning taste. Microbiol Spectr 3(4). doi: 10.1128/microbiolspec.MBP-0005-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varki A. 2001. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol 33(Suppl):54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, Wright KL, Muchmore EA, Nelson DL, Warren ST, Varki A. 1998. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci U S A 95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lubin JB, Kingston JJ, Chowdhury N, Boyd EF. 2012. Sialic acid catabolism and transport gene clusters are lineage specific in Vibrio vulnificus. Appl Environ Microbiol 78:3407–3415. doi: 10.1128/AEM.07395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopkins AP, Hawkhead JA, Thomas GH. 2013. Transport and catabolism of the sialic acids N-glycolylneuraminic acid and 3-keto-3-deoxy-d-glycero-d-galactonononic acid by Escherichia coli K-12. FEMS Microbiol Lett 347:14–22. doi: 10.1111/1574-6968.12213. [DOI] [PubMed] [Google Scholar]
- 33.Nygren E, Li BL, Holmgren J, Attridge SR. 2009. Establishment of an adult mouse model for direct evaluation of the efficacy of vaccines against Vibrio cholerae. Infect Immun 77:3475–3484. doi: 10.1128/IAI.01197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohnhoff M, Miller CP. 1962. Enhanced susceptibility to salmonella infection in streptomycin-treated mice. J Infect Dis 111:117–127. doi: 10.1093/infdis/111.2.117. [DOI] [PubMed] [Google Scholar]
- 35.Miller CP, Bohnhoff M. 1963. Changes in the mouse’s enteric microflora associated with enhanced susceptibility to salmonella infection following streptomycin treatment. J Infect Dis 113:59–66. doi: 10.1093/infdis/113.1.59. [DOI] [PubMed] [Google Scholar]
- 36.Miller CP, Bohnhoff M, Rifkind D. 1956–1957. The effect of an antibiotic on the susceptibility of the mouse’s intestinal tract to salmonella infection. Trans Am Clin Climatol Assoc 68:51–55; discussion 55–58. [PMC free article] [PubMed] [Google Scholar]
- 37.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myhal ML, Laux DC, Cohen PS. 1982. Relative colonizing abilities of human fecal and K 12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur J Clin Microbiol 1:186–192. doi: 10.1007/BF02019621. [DOI] [PubMed] [Google Scholar]
- 39.Wadolkowski EA, Laux DC, Cohen PS. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of adhesion to mucosal receptors. Infect Immun 56:1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCormick BA, Laux DC, Cohen PS. 1990. Neither motility nor chemotaxis plays a role in the ability of Escherichia coli F-18 to colonize the streptomycin-treated mouse large intestine. Infect Immun 58:2957–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sweeney NJ, Klemm P, McCormick BA, Moller-Nielsen E, Utley M, Schembri MA, Laux DC, Cohen PS. 1996. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect Immun 64:3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sweeney NJ, Laux DC, Cohen PS. 1996. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect Immun 64:3504–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun 76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones SA, Chowdhury FZ, Fabich AJ, Anderson A, Schreiner DM, House AL, Autieri SM, Leatham MP, Lins JJ, Jorgensen M, Cohen PS, Conway T. 2007. Respiration of Escherichia coli in the mouse intestine. Infect Immun 75:4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leatham-Jensen MP, Frimodt-Møller J, Adediran J, Mokszycki ME, Banner ME, Caughron JE, Krogfelt KA, Conway T, Cohen PS. 2012. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect Immun 80:1716–1727. doi: 10.1128/IAI.06193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stern AM, Hay AJ, Liu Z, Desland FA, Zhang J, Zhong Z, Zhu J. 2012. The NorR regulon is critical for Vibrio cholerae resistance to nitric oxide and sustained colonization of the intestines. mBio 3(2):e00013-12. doi: 10.1128/mBio.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. 2013. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One 8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Bäumler AJ. 2013. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4(4):e00430-13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meador JP, Caldwell ME, Cohen PS, Conway T. 2014. Escherichia coli pathotypes occupy distinct niches in the mouse intestine. Infect Immun 82:1931–1938. doi: 10.1128/IAI.01435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schinner SA, Mokszycki ME, Adediran J, Leatham-Jensen M, Conway T, Cohen PS. 2015. Escherichia coli EDL933 Requires gluconeogenic nutrients to successfully colonize the intestines of streptomycin-treated mice pre-colonized with E. coli Nissle 1917. Infect Immun 83:1983–1991. doi: 10.1128/IAI.02943-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonin S, Arnoux P, Pierru B, Lavergne J, Alonso B, Sabaty M, Pignol D. 2007. Crystal structures of an extracytoplasmic solute receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding. BMC Struct Biol 7:11. doi: 10.1186/1472-6807-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulligan C, Leech AP, Kelly DJ, Thomas GH. 2012. The membrane proteins SiaQ and SiaM form an essential stoichiometric complex in the sialic acid tripartite ATP-independent periplasmic (TRAP) transporter SiaPQM (VC1777-1779) from Vibrio cholerae. J Biol Chem 287:3598–3608. doi: 10.1074/jbc.M111.281030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Post DM, Mungur R, Gibson BW, Munson RS Jr.. 2005. Identification of a novel sialic acid transporter in Haemophilus ducreyi. Infect Immun 73:6727–6735. doi: 10.1128/IAI.73.10.6727-6735.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steenbergen SM, Lichtensteiger CA, Caughlan R, Garfinkle J, Fuller TE, Vimr ER. 2005. Sialic acid metabolism and systemic Pasteurellosis. Infect Immun 73:1284–1294. doi: 10.1128/IAI.73.3.1284-1294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen S, Zaleski A, Johnston JW, Gibson BW, Apicella MA. 2005. Novel sialic acid transporter of Haemophilus influenzae. Infect Immun 73:5291–5300. doi: 10.1128/IAI.73.9.5291-5300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Severi E, Randle G, Kivlin P, Whitfield K, Young R, Moxon R, Kelly D, Hood D, Thomas GH. 2005. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol Microbiol 58:1173–1185. doi: 10.1111/j.1365-2958.2005.04901.x. [DOI] [PubMed] [Google Scholar]
- 57.Johnston JW, Zaleski A, Allen S, Mootz JM, Armbruster D, Gibson BW, Apicella MA, Munson RS Jr.. 2007. Regulation of sialic acid transport and catabolism in Haemophilus influenzae. Mol Microbiol 66:26–39. doi: 10.1111/j.1365-2958.2007.05890.x. [DOI] [PubMed] [Google Scholar]
- 58.Vimr ER, Troy FA. 1985. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J Bacteriol 164:845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Severi E, Hosie AH, Hawkhead JA, Thomas GH. 2010. Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiol Lett 304:47–54. doi: 10.1111/j.1574-6968.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- 60.Joseph S, Hariri S, Masood N, Forsythe S. 2013. Sialic acid utilization by Cronobacter sakazakii. Microb Inform Exp 3:3. doi: 10.1186/2042-5783-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Molenaar D, Bringel F, Schuren FH, de Vos WM, Siezen RJ, Kleerebezem M. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J Bacteriol 187:6119–6127. doi: 10.1128/JB.187.17.6119-6127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marco ML, Peters TH, Bongers RS, Molenaar D, van Hemert S, Sonnenburg JL, Gordon JI, Kleerebezem M. 2009. Lifestyle of Lactobacillus plantarum in the mouse caecum. Environ Microbiol 11:2747–2757. doi: 10.1111/j.1462-2920.2009.02001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roy S, Douglas CW, Stafford GP. 2010. A novel sialic acid utilization and uptake system in the periodontal pathogen Tannerella forsythiae. J Bacteriol 192:2285–2293. doi: 10.1128/JB.00079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brigham C, Caughlan R, Gallegos R, Dallas MB, Godoy VG, Malamy MH. 2009. Sialic acid (N-acetyl neuraminic acid or NANA) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine (manNAc) epimerase. J Bacteriol 191:3629–3638. doi: 10.1128/JB.00811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilbert NM, Lewis WG, Lewis AL. 2013. Clinical features of bacterial vaginosis in a murine model of vaginal infection with Gardnerella vaginalis. PLoS One 8:e59539. doi: 10.1371/journal.pone.0059539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lewis WG, Robinson LS, Gilbert NM, Perry JC, Lewis AL. 2013. Degradation, foraging, and depletion of mucus sialoglycans by the vagina-adapted Actinobacterium Gardnerella vaginalis. J Biol Chem 288:12067–12079. doi: 10.1074/jbc.M113.453654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arden SB, Chang WH, Barksdale L. 1972. Distribution of neuraminidase and n-acetylneuraminate lyase activities among corynebacteria, mycobacteria, and nocardias. J Bacteriol 112:1206–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitaker WB, Richards GP, Boyd EF. 2014. Loss of sigma factor RpoN increases intestinal colonization of Vibrio parahaemolyticus in an adult mouse model. Infect Immun 82:544–556. doi: 10.1128/IAI.01210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haines-Menges B, Whitaker WB, Boyd EF. 2014. Alternative sigma factor RpoE is important for Vibrio parahaemolyticus cell envelope stress response and intestinal colonization. Infect Immun 82:3667–3677. doi: 10.1128/IAI.01854-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whitaker WB, Parent MA, Boyd A, Richards GP, Boyd EF. 2012. The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect Immun 80:1834–1845. doi: 10.1128/IAI.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 77.Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166. In Bryson V, Vogel HJ (ed), Evolving Genes and Proteins. Academic Press, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth analysis of V. vulnificus CMCP6 with sialic acid derivatives as sole carbon sources. Data represent results of growth analysis of V. vulnificus CMCP6 (black circles), JJK0731 (ΔsiaM) (a sialic acid transporter mutant) (black downward-facing triangles), V. cholerae N16961 (black squares), NC1777 (ΔsiaM) (black diamonds), and E. coli BW25113 (black upward-facing triangles) in M9 minimal media supplemented with N-acetylneuraminic acid (Neu5Ac) (A), N-glycolylneuraminic acid (Neu5Gc) (B), or 2-keto-3-deoxy-d-glycero-d-galacto-nononic acid (KDN) (C) as a sole carbon source. (D) Complementation of NC1777 with siaPQM. All sialic acid derivatives were used at a concentration of 1 mg/ml. Growth curves represent results of at least two biological replicates performed in triplicate. Error bars represent SEM. Download
High-throughput screen of carbon utilization abilities of V. cholerae pathogenic isolates. Vibrio cholerae strains N16961 (El Tor) (black bars) and NC1777 (ΔsiaM) (gray bars) were screened for growth on 190 different carbon sources using Biolog PM1 and PM2A plates. The area under the curve (AUC) was determined for each condition tested. The growth was normalized by subtracting the AUC value corresponding to the well containing no carbon source from the AUC value corresponding to each well with a carbon source. Data represent the averages of results of two biological replicates for each strain for each of the PM plates. Download
NanA phylogeny subtrees A, G, H, I, O, and P. (A) Subtree containing representative species from cluster A. (B) Subtree containing representative species from cluster G. (C) Subtree containing representative species from clusters H and I. (D) Subtree containing representative species from clusters O and P. Download
Growth analysis of various species on M9 supplemented with glucose or N-acetylneuraminic acid as a sole carbon source. Sialic acid was used at a concentration of 10 mM. Growth curves represent results of at least two biological replicates performed in triplicate. Error bars represent SEM. Download
NanA phylogeny subtrees B, D, E, F, and J. (A) Subtree containing representative species from cluster B. (B) Subtree containing representative species from clusters D, E, and F. (C) Subtree containing representative species from cluster J. Download
NanA phylogeny subtrees K to N. (A) Subtree containing representative species from clusters K and L. (B) Subtree containing representative species from cluster M. (C) Subtree containing representative species from cluster N. Download
NanA phylogeny subtrees Q to T. (A) Subtree containing representative species from clusters Q, R, and S. (B) Subtree containing representative species from cluster T. Download
Bacterial strains and plasmids used in this study.