Abstract
Growing experimental evidences suggest the existence of direct relationships between the surface chemistry of nanomaterials and their biological effects. Herein, we have employed computational approaches to design a set of biologically active Carbon Nanotubes (CNTs) with controlled protein binding and cytotoxicity. Quantitative Structure-Activity Relationships (QSAR) models were built and validated using a dataset of 83 surface-modified CNTs. A subset of a combinatorial virtual library of 240,000 ligands potentially attachable to CNTs was selected to include molecules that were within the chemical similarity threshold with respect to the modeling set compounds. QSAR models were then employed to virtually screen this subset and prioritize CNTs for chemical synthesis and biological evaluation. Ten putatively active and ten putatively inactive CNTs decorated with the ligands prioritized by virtual screening for either protein binding or cytotoxicity assays were synthesized and tested. We found that all 10 putatively inactive and 7 out of 10 putatively active CNTs were confirmed in the protein binding assay, whereas all 10 putatively inactive and 6 out of 10 putatively active CNTs were confirmed in the cytotoxicity assay. This proof-of-concept study shows that computational models can be employed to guide the design of surface-modified nanomaterials with the desired biological and safety profiles.
Keywords: QSAR, cheminformatics, nanotoxicity, virtual screening, carbon nanotubes
Introduction
Nanotechnology is emerging as a multi-purpose platform in many industrial applications (Demirdjian, 2006; Jones, 2009; Luther, 2004) resulting in various marketed products (Sips, 2009) based on manufactured nanoparticles (MNPs). An increasing amount of evidence (Baalousha & Lead, 2013) suggests that nanomaterials may cause detrimental effects in living organisms (Bai et al., 2010; Maynard et al., 2006; Murray et al., 2012; Nel et al., 2006; Qu et al., 2009; Service, 2004; Zhang et al., 2014). As interactions between MNPs and biological systems are extremely complex (Nel et al., 2009), both manufacturers and regulators are in need for new methods to design MNPs with controlled and safe bioprofiles (Kumar et al., 2014; Sips, 2009). Experimental in vivo toxicological studies (usually LD50 assessment, but also MNPs biodistribution, serum binding chemistry, or tissue histopathology after exposure to MNPs) are time- and resource-consuming, making it unrealistic to test all marketed consumer products incorporating MNPs (Savolainen et al., 2010). Extrapolating in vitro measurements to in vivo outcomes (Monteiro-Riviere et al., 2013) is even more challenging. Meanwhile, computer simulation approaches have been widely explored and exploited in chemical toxicology (e.g., Tox21 project (Austin, 2009)) especially for organic compounds (Arts et al., 2008; European Parliament, 2010; Hartung, 2010) to quickly assess their potential safety concerns (e.g., carcinogenicity (Zhu et al., 2008a), aquatic toxicity (Zhu et al., 2008b), liver toxicity (Fourches et al., 2010a; Rodgers et al., 2010), etc.) with reasonable accuracy. Similarly, it would be highly desirable to develop computer-based approaches to (i) predict the biological (including toxicological) effects of MNPs solely from their physical, geometrical, and chemical properties (Carbó-Dorca & Besalú, 2011), and (ii) guide experimental investigations by focusing costly toxicological studies on a small number of selected or rationally designed MNPs. The need to develop such approaches has been well articulated in recent scientific literature (Burello & Worth, 2011; Kar et al., 2014; Luan et al., 2014; Puzyn et al., 2009; Singh & Gupta, 2014; Winkler et al., 2012).
In an attempt to address these challenges, our group has recently pioneered a QSAR modeling approach (Fourches et al., 2011, 2010c) to develop computational models capable of predicting biological activities of MNPs from their composition and structural properties only. In our previous studies, we showed that computational models could help forecasting various biological effects such as protein binding or cellular uptake for diverse sets of MNPs with a reasonable prediction accuracy evaluated by the means of external validation using MNPs that were not employed for model development. Similar retrospective studies have been reported by our colleagues (Gajewicz et al., 2014; Kar et al., 2014; Luan et al., 2014; Puzyn et al., 2011; Singh & Gupta, 2014; Toropova et al., 2014) for different sets of MNPs. However, so far there has not been any published report where molecular modeling was employed prospectively as part of an experimental and iterative design of MNPs with the desired biological activity.
Since MNPs have very large surface area, surface chemistry is thought to play a key role in controlling their biological activities (Mu et al., 2014). One of our research groups published a promising study (Zhou et al., 2008) showing the possibility of perturbing both biocompatibility and toxicity of MNPs that have been synthesized using a combinatorial chemistry approach (Su & Yan, 2010). In order to discover biologically benign nanotubes without a priori knowledge of related targets or potential mechanisms, we have exposed the biological targets of interest to multi-walled carbon nanotubes (CNTs) with diverse surface modifiers and very different molecular and physicochemical properties obtained through combinatorial nanotube library synthesis (Yan, 2011; Q. Zhang et al., 2010). Then we demonstrated the existence of relationships between the surface chemistry of CNTs covalently decorated with small organic ligands and their biological profiles assessed by a panel of protein binding and cytotoxicity assays. Better understanding and accurate prediction of biological effects caused by CNTs are of critical importance for ensuring their safe use in various medical applications with a therapeutic potential (e.g., drug carriers) (Kaiser et al., 2009; Y. Zhang et al., 2010). To the best of our knowledge, there is no published report on the prospective use of any computational approach for designing and prioritizing novel CNTs with the desired bioactivity and safety profiles.
Herein, we have modeled the structure-activity relationships for a set of 83 CNTs tested in six different biological or toxicological in vitro assays (Zhou et al., 2008). Using cheminformatics methods to characterize and cluster CNTs using structural descriptors of their surface-modifying molecules, we have uncovered chemical features of the modifiers that correlate with a particular biological activity. These initial results confirmed the direct relationships between surface chemistry and CNT bioprofiles observed previously (Zhou et al., 2008). Furthermore, we have developed several QSAR models with external prediction power as high as 75% and 77% balanced accuracy for protein binding and acute toxicity endpoints, respectively. In combination with similarity search, these models were used to screen in silico a virtual library of 240,000 ligands considered as potentially attachable to the surface of a CNT. A series of hits were selected for experimental validation, and the results showed a good agreement between predicted and measured bioprofiles: all rationally prioritized, synthesized, and tested CNTs predicted as inactive were confirmed experimentally in both protein binding and cytotoxicity assays, whereas six out of ten rationally prioritized CNTs predicted as toxic were also confirmed as were seven out of ten surface-modified CNTs predicted as protein binders. Thus, statistical QSAR models were used to rationally prioritize and design a series of surface modified CNTs that were experimentally confirmed with the expected biological and toxicological properties.
Materials and methods
CNT dataset and assays
We considered a library of 83 multi-walled CNTs, each one being decorated with small organic molecules through combinatorial and chemical conjugation. The initial series of surface molecules used to decorate CNTs were selected to have the most diverse molecular and physicochemical properties, such as solubility, hydrophobicity, stereochemical, and topological properties on the basis of computation. They were chosen from 806 combinatorial conjugations (31 amines and 26 acylators), for which molecular properties were calculated by Accord and Pipeline Pilot from Accelrys. The entire selection workflow was published previously (Zhou et al., 2008). In this work, we used 83 multiwalled carbon nanotubes with a controlled size distribution: diameter of 40±10 nm and length of 250±120 nm. Functional group modifications are made at a density of five modifications per 10 nm2 on the basis of a loading of 0.4 mmol/g and a surface area of 500 m2/g (from experimental measurements). Then every CNT was biologically characterized using six short-term in vitro assays: protein binding to bovine serum albumin (BSA), carbonic anhydrase (CA), chymotrypsin (CT), and hemoglobin (HB), as well as acute toxicity and immune toxicity properties. Briefly, the binding of CNTs to proteins were studied by steady-state fluorescence spectroscopy (acquired on a Hitachi F-4500 spectrofluorometer). Fluorescence quenching indicates differential protein binding affinity profile for the two groups of designed CNTs. Carbonic anhydrase (50 µg/mL) was excited at 280 nm and emission spectra (300–400 nm) were recorded. Spectra were measured before and immediately after addition of CNT (15 µg/mL). Ratio of F0 (initial) to F (modified) at 340 nm was used to evaluate protein binding affinity: F0/F indicates the extent of protein fluorescence quenching by 15 µg/ml of CNT as a result of CNT binding to the protein; F0 is the protein fluorescence intensity before CNT binding and F is the fluorescence intensity after CNT binding. The higher F0/F value represents stronger protein binding. Acute toxicity of CNTs was investigated by WST-1 assays, and the survival percentages after exposure of macrophages to CNTs were recorded. The WST-1 assay is commonly used to evaluate cell proliferation and cytotoxicity. It was used to test cell viability by measuring cellular dehydrogenase activity in the mitochondrion. Cytotoxicity is also the consequence of CNTs’ binding with protein receptors on cell surface. For the whole set of 83 decorated CNTs, the average of cell survival variations was found to be as low as 3.6% at (200 µg/mL) and 3.8% at (50 µg/mL). For immune toxicity endpoint, the secretion of nitric oxide (NO) by macrophages was measured after the exposure to CNTs. More details about the protocols and other experimental details can be found in the supplementary materials and in (Zhou et al., 2008). The chemical structures and in vitro bioprofiles of the 83 CNTs are also available in the Supplementary Material.
Chemical Descriptors
In our previous proof-of-concept study (Fourches et al., 2010c), we hypothesized that relative biological activity of surface-modified nanoparticles with the same core correlates with structural features of organic molecules attached to the surface. This approach allows the use of classical molecular descriptors for characterizing surface modifiers. Thus, each CNT has been represented by a single copy of its surface modifying organic molecules encoded as SMILES strings. Following the guidelines published by us recently (Fourches et al., 2010b), structural curation procedures were carried out to obtain standardized two-dimensional representations of surface modifiers. The latter were then represented by molecular descriptors computed with Dragon (Mauri et al., 2006) and MOE (Vilar et al., 2008) software including constitutional, functional group counts, atom-centered fragments, walk and path counts, information indices, topological and electrostatic indices. Descriptors with zero value or zero variance as well as one of each pair of highly inter-correlated descriptors (R2 ≥ 0.95) were removed leaving 180 (Dragon) and 158 (MOE) descriptors for further cheminformatics analysis. For clustering (Downs & Barnard, 2002; Liu et al., 2008; Monti et al., 2003) compounds by chemical similarity, we employed the Sequential Agglomerative Hierarchical Non-overlapping (SAHN, classical Johnson’s algorithm) method implemented in the ISIDA/Cluster program (Varnek et al., 2008). When using this combination of descriptors and clustering algorithm, the clusters were very stable and ensured to contain compounds with structural similarity. To validate the clusters, we analyzed the content of each cluster by studying the structural variations of molecules and that stage enabled the discovery of clusters containing CNT surface modifiers having consistent experimental responses in the considered protein binding assays.
QSAR Modeling
The QSAR modeling workflow can be divided into three major steps (Tropsha & Golbraikh, 2007; Tropsha, 2010): (i) data preparation/analysis (selection of compounds and descriptors), (ii) model building, and (iii) model validation/selection. Here we followed a 5-fold external cross-validation procedure: the full set of compounds with known experimental activity is randomly split into five modeling (80% of the full set) and external validation sets (remaining 20%). Models are built using the modeling set only and it is important to emphasize that the external set compounds are never taken into account to build and/or select models. Briefly, each modeling set is split into many training and test sets; then models are built using compounds of each training set and applied to test set compounds to assess their properties. The following statistical metrics were used to assess model accuracy:
| (1) |
| (2) |
| (3) |
| (4) |
Best models are identified and selected according to acceptable values of CCRtraining and CCRtest thresholds (≥0.7 in this study). Other statistical tests (e.g., Y-randomization) are applied to assess the overall robustness and predictivity of the selected models. Then, these models are applied to the external set compounds to predict their experimental properties. This procedure is repeated five times to ensure that every compound is present in exactly one external test set. External test sets are never used to derive, bias, or select the models, so that the entire procedure gives a fair estimation of the true predictivity of the models. Technical details concerning the machine-learning methods used in this study (k Nearest Neighbors – kNN (Zheng & Tropsha, 2000), Random Forest – RF (Breiman, 2001), and Support Vector Machines (Cristianini & Shawe-Taylor, 2000) - SVM) and definitions of the model applicability domain were published elsewhere (Fourches et al., 2010a; Tropsha & Golbraikh, 2007; Zhu et al., 2008b). For SVM, the Radial Basis Function (RBF) kernel has been used for the training of SVM models. For each fold (at each fold corresponds a pair of modeling set 80% and an external validation set 20%), a different number of models is selected (with gamma and cost parameters taking values in different ranges) according to both training and internal test sets. Despite the fact that the activity is binary in classification QSAR modeling, quantitative relationships between descriptors and the binary bioactivity are computed and optimized to achiev the best prediction performances. Importantly, the most predictive models (and details about their kernel parameters) are freely available online on our Chembench webportal (http://chembench.mml.unc.edu/). SVM/Dragon models for CA binding and acute toxicity are given as examples in Supplementary Tables 1 and 2.
Results
Clustering of CNTs chemical surface modifiers
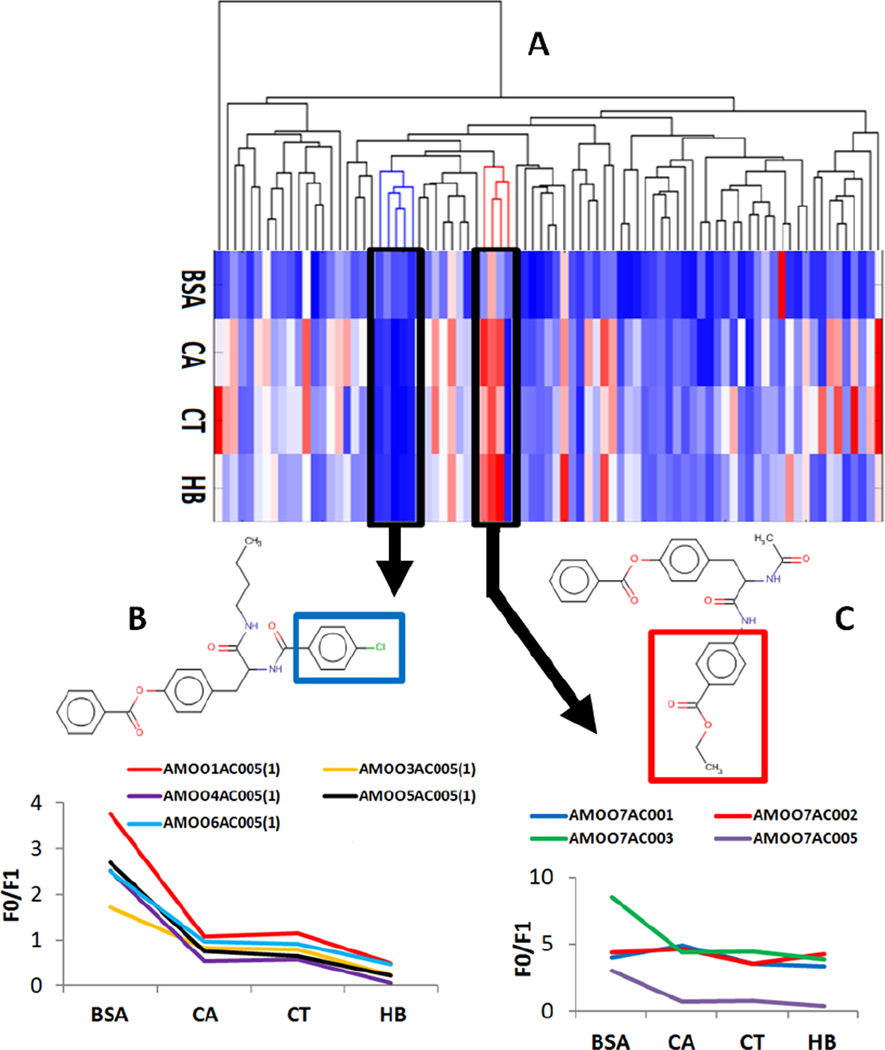
All 83 surface-decorated CNTs were clustered according to the similarity of the chemical structures of the modifiers characterized by Dragon molecular descriptors (see Methods section). Initially, we were particularly interested in identifying and analyzing clusters of CNTs associated to extreme bioprofiles (e.g., strong protein binders, highly toxic CNTs). Protein binding of nanomaterials has been recently shown to affect their cellular uptake (Monteiro-Riviere et al., 2013). Since bovin serum albumin (BSA) binding of CNTs correlated rather poorly with their binding to other proteins, we focused on identifying clusters of CNTs (Figure 1) with consistent binding characteristics towards carbonic anhydrase (CA), chymotrypsin (CT), and hemoglobin (HB). Two clusters of CNTs with strong or weak protein binding were identified:
a cluster with weak binders consisted of five CNTs with benzene chloride (or 4-chlorophenyl) as R1 subgroup. This observation was in agreement with the previous experimental finding that benzene chloride moiety is a key building block that leads to low protein binding affinity (Zhou et al., 2008);
a cluster with strong binders was comprised of four CNTs with high protein binding activity across three assays. All these four CNTs included ethyl benzoate as R2 substituent. Among them, the CNT with the weakest protein binding contained a benzene chloride subgroup as R1 substituent.
Figure 1.
Clusters of CNTs; protein binders are shown in red and non-binders are in blue. (A) Dendrogram generated with non-supervised hierarchical clustering method; the heatmap represents the protein binding activities of the CNTs ordered according to the dendrogram. Red color stands for high activity while blue color for low activity. (B) The blue cluster represents five compounds incorporating common substructure (benzene chloride) highlighted in blue square. They behave similarly (low) activity in four protein binding assays. (C) The red cluster represents four compounds sharing a common substituent (ethyl benzoate, highlighted in the red square) causing similar (strong) protein binding responses as measured in four different assays.
Thus, the presence of benzene chloride on the CNT surface not only reduces the potential of CNTs to bind proteins, but also dominates the overall protein binding behavior of CNTs. Note that (i) the use of different molecular descriptors (e.g., fragment descriptors) would lead to different clusters, and (ii) these results reconfirm that CNTs with similar chemical surface modifiers elicit similar biological and toxicological responses
QSAR modeling of CNT protein binding and toxicity
Since all 83 CNTs had the same core structure, it is reasonable to hypothesize that the variation of their behavior in biological systems would be related, at least for the most part, to the structural modifications of the surface-attached organic ligands (as shown by the dramatic changes of biological properties highlighted in the previous section). Therefore, we approximated MNPs’ structures in the context of QSAR modeling by surface modifiers solely represented by their chemical descriptors, which enables a classical cheminformatics treatment of the system. This approach was successfully employed in our previous study (Fourches et al., 2011, 2010c). A similar approach was also used by (Puzyn et al., 2011), when properties of metal-oxide MNPs were approximated by quantum mechanical properties of the metal oxide molecules themselves. We aimed to establish quantitative links between computed physical, chemical, and structural properties of CNTs’ surface modifiers with experimentally measured biological or toxicological activities. Although the modeling was carried out for surface ligands, the experimentally measured biological activities relate to nanoconjugates (i.e., the whole decorated CNT), not the free ligands. The same assays done with free ligands would have given completely different outputs as confirmed by recent investigations. Indeed, in our previous research on CNT/Chymotrypsin binding and enzymatic inhibition (Zhang et al., 2009; Zhang et al., 2010), we found that four CNTs showed a typical feature for competitive inhibition. QSAR analysis found that a common building block dibutylamine group at the surface molecules of these CNTs helped to explain the structure-activity relationships of the CNT/ChT recognition. We further analyzed whether free dibutylamine molecules had any binding or inhibition of ChT. An interesting finding was that free dibutylamine molecules did not inhibit ChT’s enzymatic activity. Considering the large CNT/protein contact areas, this functional group, in combination with the nanotube and adjacent ligands, was likely crucial for targeting the catalytic site of ChT. In the present study, the QSAR models were trained, selected, and validated solely based on the bioactivities of the ligand-decorated nanomaterials, not free ligands. Nevertheless, we could use descriptors of surface modifying molecules only, not descriptors of entire nanoconjugates because QSAR models intrinsically correlate changes in chemical structure with changes in their bioactivities. Since the core structures are the same for all particles, we could ignore their (similar) contribution to bioactivity of different modified CNTs.
We have explored all six binary combinations of three machine learning techniques (kNN, RF and SVM) and two sets of chemical descriptors (calculated with Dragon or MOE) in an effort to develop QSAR models that would have the highest internal fitness and external predictive power (see Methods section for modeling workflow and computation details). Initial attempts to build QSAR models to compute the continuous response (e.g., actual percentage value of cell survival in cytotoxicity assay) failed due to poor-to-mediocre external predictivity irrespective of the combination of the modeling approach and the type of descriptors. This negative result can be explained by the small size of the dataset, the distribution of measured responses for protein binding and cytotoxicity assays as well as the experimental variability. Therefore, we switched to classification modeling by introducing an activity threshold such that the original set of CNTs was split into two balanced classes (labeled as 0 and 1, for low and high activity, respectively) with the intent to accurately predict the class membership of each CNT. QSAR models for each individual biological endpoint were constructed using our standard workflow (Tropsha, 2010). We were capable of developing statistically significant models for CA binding and acute cytotoxicity (measured by cellular metabolic activity in the WST-1 assay) (Supplementary Table 3). Here, the expression “statistically significant” refers to the comparison of the prediction accuracy obtained by actual models with that obtained with models developed with randomized response variable (Y-randomization procedure); the latter models were characterized by very low internal accuracy estimated by cross-validation and no external predictivity.
In the case of CA binding, the activity values ranged from 0.53 to 5.29 at CNT concentration of 15 µg/ml (see Method for assay information). Based on the distribution of CA binding activities, we chose a separation threshold at 2.0 to split the dataset in two classes of CNTs with balanced distribution of actives and inactives. Thus, CNTs with the CA binding activity greater than 2.0 were assigned an activity value of 1 (CA binder) and those with binding affinity under 2.0 were assigned the activity value of 0 (CA non-binder). As a result, there were 44 CA binders and 39 CA non-binders. A standard 5-fold external cross-validation procedure was carried out and the results showed that the cumulative Correct Classification Rate (CCR) ranged from 63% to 75% (Supplementary Table 3). Only models with CCR no less than 70% were considered to have acceptable predictive power. Therefore, only kNN-Dragon (75%) and RF-Dragon (73%) models were retained. Obviously, changing the separation threshold, for instance from 2.00 to 4.00, leads to an extremely unbalanced split of 8 CA strong binders and 75 CA non-binders for which no robust model can be built. Note that CNTs' protein binding measurements obtained in the CA, CT, and HB assays showed good pairwise correlation in the range of R2 = 0.58 – 0.65 (for instance, the correlation between CA and HB was found to be as high as R2 = 0.65), whereas BSA assay results were less correlated (R2 = 0.20 between BSA and CA). Therefore it is not surprising that modeling results obtained for CT and HB binding were similar to the ones obtained for CA.
Similarly to CA binding, the acute toxicity endpoint was modeled using the same workflow. The experimental survival percentage ranged from 2% to 68% at high CNT concentration of 200 µg/ml. One could note that even as such high CNT concentration, some specific surface modifiers can still lead to weak CNT-induced toxicity. From the analysis of cytotoxicity values following a normal distribution, CNTs around the median cell survival range [37–43%] were not included in the refined modeling set of 73 CNTs including 35 CNTs labeled as class 0 (“non-toxic”) and 38 CNTs labeled as class 1 (“toxic”). Frequently applied in QSAR modeling, this technique of discarding threshold was used to bias the selection of QSAR models for facilitating the discrimination between the most toxic CNTs and the less-toxic CNTs. Calculated statistics showed that cumulative CCR ranged from 63% to 77%; kNN-Dragon, RF-Dragon and SVM-Dragon afforded the best external prediction performances in the range of CCR = 70–77%. To ensure the robustness of selected QSAR models, we employed standard Y-randomization test and found that no statistically significant models (i.e., with CCR >0.7) for datasets with randomized response variables were retrieved.
Application of QSAR models for in silico screening of a virtual library of CNT surface modifiers
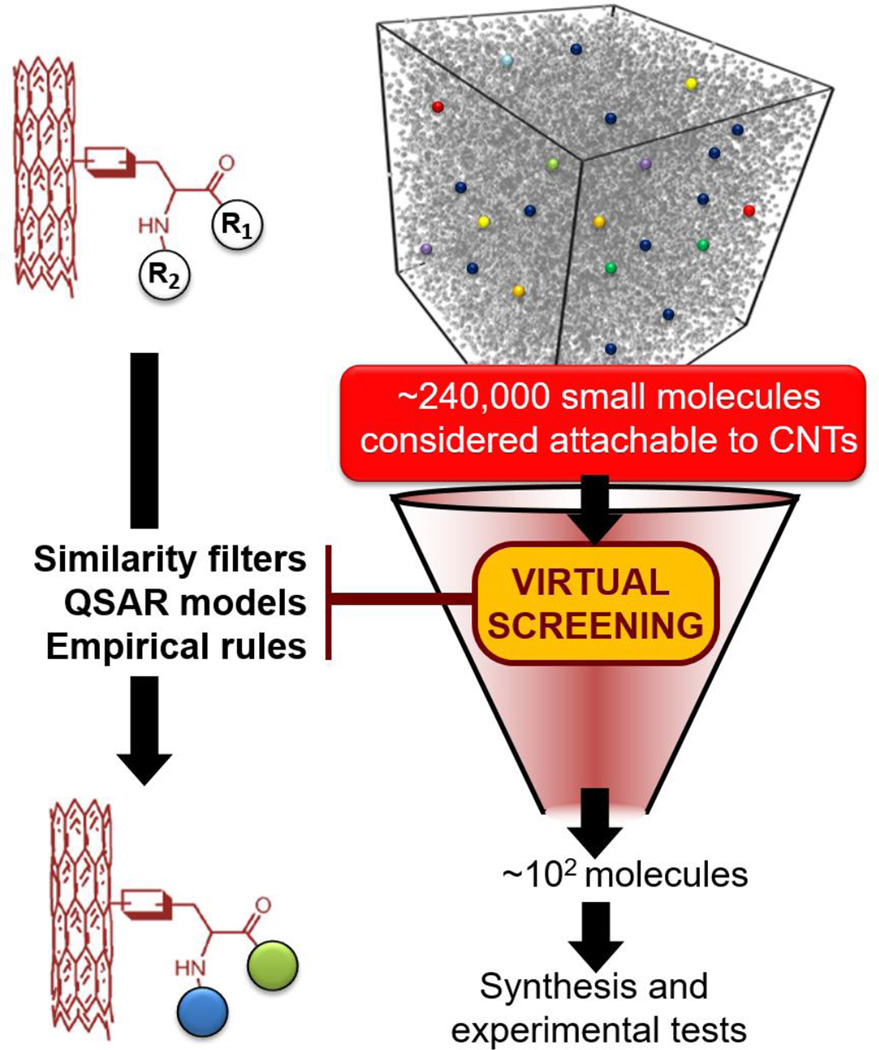
The most important objective of this study was the prioritization of CNTs for experimental testing and their confirmation to demonstrate how computational screening incorporating similarity search and QSAR models can be used to facilitate the design of novel CNTs with the desired biological properties. To this end, we virtually screen an external compound collection consisting of 240,000 small molecules considered to be synthetically feasible and potentially attachable to the surface of CNTs. Figure 2 illustrates the workflow we implemented and followed.
Figure 2. Workflow for virtual screening of 240 000 small molecules attachable to CNTs.
Construction of a virtual MWNT combinatorial library
We used the same scaffold as in the modeling set of CNTs with two sites of diversification. The amines and acylators were selected as modifying building blocks based on the following criteria: (i) diverse amines and acylators were selected to get the maximum surface diversity based on their estimated molecular properties such as hydrophobicity (MlogP), solubility, hydrogen-bonding, and stereochemical properties calculated by Accord and Pipeline Pilot from Accelrys and (ii) both amines and acylators had to be commercially available. We finally selected 400 Acylators and 600 Amines to build a virtual library containing 240,000 compounds being attachable to CNT surfaces.
Similarity based filtering of surface modifiers
The similarity concept underlies many rational molecular design approaches; i.e., it is expected that similar chemical structures will have similar physical-chemical properties and thus cause similar biological responses. The first step in our virtual screening procedure aimed to identify among the 240K compounds of the virtual library, those molecules that were not too dissimilar from the ones already tested as surface modifiers in our modeling set of 83 CNTs. The dissimilarity between compounds was computed using molecular descriptors (Dragon) and Euclidean metric between molecules represented as vectors in high-dimensional descriptor space. If the pairwise Euclidean distances between a compound from the library and any of the modeling set compounds exceeded a pre-defined distance threshold (here, a very strict z = 0.5 threshold, see reference (Tropsha & Golbraikh, 2007)), this compound was excluded. Finally, only 462 compounds from the library of 240K molecules remained after the similarity-based filtering. It is important to note that setting up the distance threshold is directly related to the final number of compounds one wants to select in regard to the feasible number of compounds to be experimentally synthesized and tested. For instance, increasing our distance threshold to z =1 would have enabled the selection of several thousands of library compounds.
Selection of Virtual Hits Using QSAR-based Consensus Predictions
The 462 compounds retrieved from similarity search were evaluated using QSAR models developed for two endpoints, i.e., CA binding and acute toxicity. Models’ applicability domains (Tropsha & Golbraikh, 2007) were used to confirm that surface modifiers were similar enough from the modeling set compounds. Models predicted 352 ligand-decorated CNTs as binders, whereas only 110 compounds were predicted as non-binders. For the toxicity prediction, 189 out of 462 CNTs were predicted to be non-toxic, and the remaining 273 compounds were predicted as potential toxicants. Final hit selections were made based on consensus scores (computed as the averaged predictions between individual QSAR models), and overall confidence coefficients (based on molecule similarity z to the modeling set and the standard deviation of predictions between the individual QSAR models). Finally, ten predicted CA binders and ten predicted CA non-binders as well as ten predicted toxic CNTs and ten predicted non-toxic CNTs were recommended for experimental testing regarding their protein binding and cytotoxicity, respectively. Importantly, these compounds were selected based on (i) the highest prediction confidence from QSAR models, (ii) their structural diversity compared to the surface modifiers already present in the initial set of 83 CNTs, and (iii) their overall synthetic feasibility.
Experimental validation of the selected hits
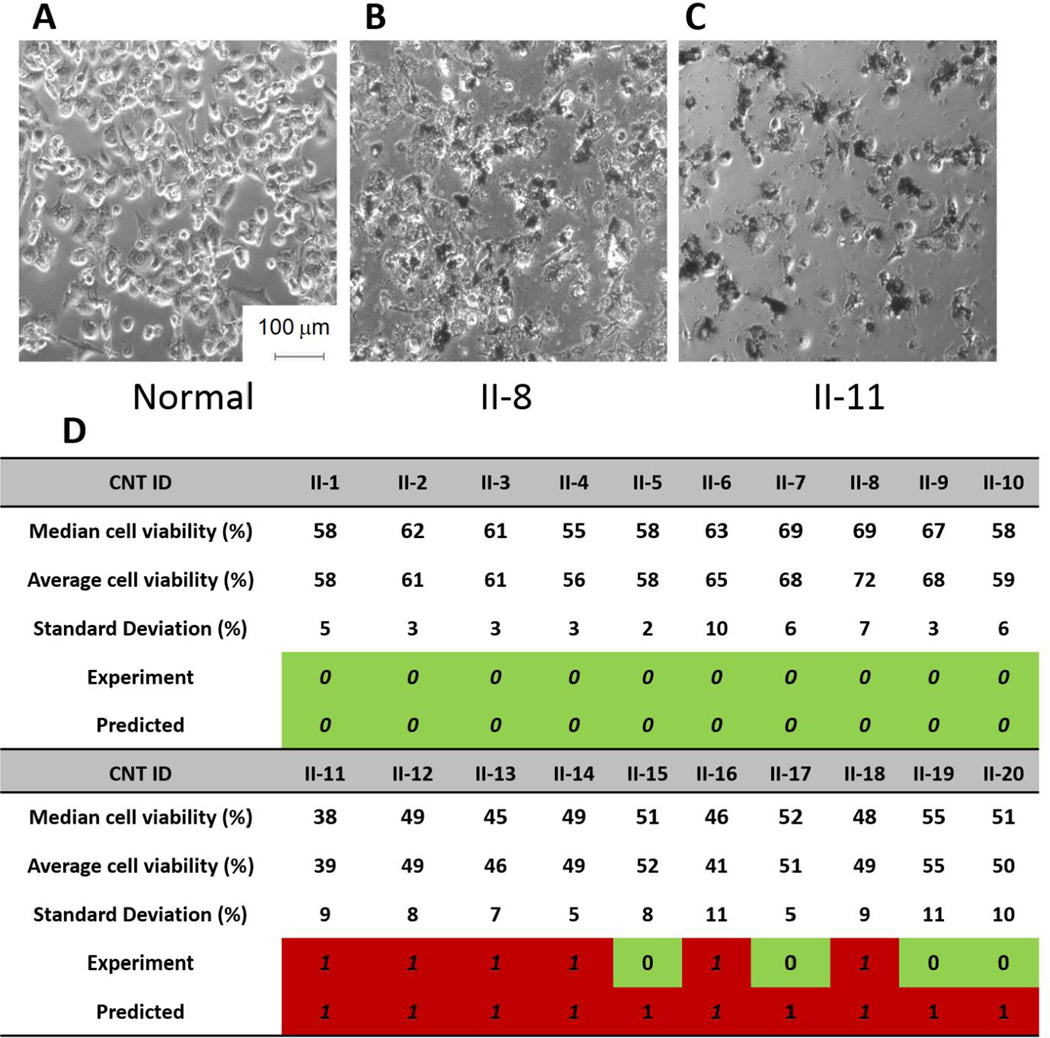
CNTs decorated by computer-prioritized molecules were synthesized, purified, and characterized according to our established procedures (Zhou et al., 2008). Their chemical structures are given in Supplementary Table 4. These CNTs were experimentally tested in CA binding and cytotoxicity assays. For the cytotoxicity endpoint (Figure 3), 10 toxic and 10 non-toxic hits as predicted by QSAR models were tested in THP-1 derived macrophage by measuring cellular metabolic activity (WST-1 method). Similarly, 10 QSAR-predicted CA binders and 10 non-binder CNTs (Table 1) were tested in a protein binding assay using fluorescence spectroscopy (Mu et al., 2008). Overall, for cytotoxicity endpoint, all 10 CNTs predicted to be non-toxic were confirmed as non-toxic, whereas 6 out of 10 CNTs predicted to be toxic were verified as being toxic (implying prediction accuracy of 80%, Figure 3). All QSAR-predicted non-toxic CNTs were characterized by higher cell survival percentages compared with those of predicted toxic CNTs. The statistical test (one sided student’s t-test, p<0.0001) showed that average cell viability of non-toxic hits was significantly higher than that of toxic hits. Given that the modeling set is biased towards cytotoxic CNTs, it is interesting to realize that our QSAR models were indeed able to identify surface modifiers leading to relatively low cytotoxicity such as II-7, II-8, and II-9 with cell survival percentages around 70%. Note the examination of replicate measurements for these compounds confirms the low cytotoxic CNTs with cell viability rates as high as 78.7% for II-6 and 82.4% for II-8 (Supplementary Figure 1). These results are very encouraging in the context of developing non-toxic surface-decorated CNTs. On the contrary, II-11 and II-16 were found to be severe toxicants with cell survival rates as low as 28.9% and 24.4% confirming the predictions by our QSAR models. Nevertheless, despite their statistical significance, the results obtained for the compounds predicted to be cytotoxic were less clear due to experimental variability. For CA binding endpoint, all 10 CNTs predicted as binders were validated as binders, whereas 7 out of 10 CNTs predicted to be non-binders were confirmed as non-binders (overall prediction accuracy as high as 85%, Table 1).
Figure 3. Cytotoxicity of novel CNTs prioritized for testing based on QSAR predictions.
Cell viability of THP-1 macrophage cells in the presence of CNTs (200 µg/mL, 24h). THP-1 macrophage cell microscopy images incubated (A) without CNT for 24 h, (B) with 200 µg/mL II-8 CNT, and (C) with 200 µg/mL II-11 CNT. (D) Summary of experimental validation results for cytotoxicity of selected hits. CNTs are labeled as “0” (non-toxic) if their associated cell viability is greater than 50% and “1” (toxic) if their associated cell viability is smaller than 50%.
Table 1. Carbonic anhydrase binding measured for novel CNTs prioritized for testing based on QSAR predictions.
Summary of experimental validation results for selected CA binders and non-binders. CNTs are labeled as “0” (non-binder) if their CA bindings are smaller than 2.00 and “1” (binder) if their CA bindings are greater than 2.00. Overall prediction accuracy is 85% (17/20).
| CNT ID | II-21 | II-22 | II-23 | II-24 | II-25 | II-26 | II-27 | II-28 | II-29 | II-30 |
|---|---|---|---|---|---|---|---|---|---|---|
| Average protein binding (F0/F1) |
1.77 | 1.78 | 1.87 | 1.76 | 1.82 | 2.30 | 1.74 | 2.00 | 1.67 | 2.33 |
| Standard Deviation | 0.05 | 0.06 | 0.02 | 0.03 | 0.02 | 0.02 | 0.02 | 0.01 | 0.06 | 0.02 |
| Experiment | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 |
| Predicted | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CNT ID | II-31 | II-32 | II-33 | II-34 | II-35 | II-36 | II-37 | II-38 | II-39 | II-9 |
| Average protein binding (F0/F1) |
3.40 | 2.28 | 2.04 | 2.22 | 2.17 | 2.95 | 2.08 | 2.25 | 2.65 | 2.24 |
| Standard Deviation | 0.03 | 0.05 | 0.08 | 0.01 | 0.04 | 0.00 | 0.02 | 0.06 | 0.05 | 0.11 |
| Experiment | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Predicted | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
Discussion
This proof-of-concept study demonstrates the applicability of classical molecular modeling approaches that are well-established in the field of drug discovery to an unusual class of substances exemplified by decorated CNTs. Most importantly, we show that a virtual screening protocol can reasonably prioritize a small number of CNTs for experimental investigations calling for broader application of computational approaches to enable “safe-by-design” development of MNP-based materials.
QSAR-prioritized descriptors of the surface modifiers
The preliminary analysis of individual descriptors involved in the best QSAR models was conducted. For CA binding models, several descriptors were found to take much higher values in binders: for instance, MATS2m (Moran autocorrelation of lag 2 weighted by mass) values were found to be twice as high in CA binders compared to CA non-binders; the H-051 descriptor (number of hydrogens attached to alpha carbons) was also more elevated in CA binders. Meanwhile, other descriptors such as nCl (number of chlorine groups) and several electrotopological indices (maxDN, GATS2m, and JGI2) were significantly more elevated in CA non-binders. One could note that all CNTs from the modeling set with 4-chlorophenyl groups (directly encoded by nCl descriptor) in position R2 were found to be CA non-binders. For the WST model, several descriptors were more elevated in toxic modifiers such as nCp (number of terminal sp3 carbons) and BAC (Balaban centric index). Importantly, the selected descriptor nRNH2 (primary amines) highlights the fact that all primary amines were found to be toxic, whereas the descriptor nPyrrolidines (number of pyrrolidine substructure) helped revealing that 8 out of 10 CNTs incorporating a pyrrolidine group were found to be non-toxic. The aforementioned descriptors could potentially be refined as structural alerts in the context of a given class of surface modifiers. However, our models take into account the actual number of structural fragments (e.g., number of Cl, number of pyrrolidine) to forecast the bioactivity, not only the absence/presence as standard alerts would do.
Structural diversity of newly-synthesized CNTs
Although the initial set of 83 ligands is certainly limited in terms of chemical diversity, the overall dataset of surface modified CNTs is not built using highly congeneric compounds only. Importantly, measured bioactivities for these CNTs spread over the whole range of protein binding (no binding for AMOO2AC005 to very strong binding for MWNT-Tyr-Fmoc) and cytotoxicity (very low for AMOO6AC003 to very high for AMOO5AC006). This significant distribution of bioactivity is predicated on the underlying chemical diversity of tested substances.
The use of rather conservative similarity threshold for the virtual screening only kept a small fraction of compounds in the diverse 240K compound library that were relatively similar to the training set molecules. Nevertheless, hits identified by the screening protocol still showed some unique chemical features. Let’s consider the example of 4-trifluoromethylphenyl group present at the R2 position in six out of 10 toxic CNTs: this group was not present in the modeling set, whereas its closest analog being the 3-trifluoromethylphenyl group was indeed present in 10 CNTs from the modeling set but always in position R1 (never in R2). We can also cite the example of the 4-nitro-3-(trifluoromethyl)phenyl group present in three CNTs selected by QSAR models, (II-11, II-12 and II-13); this group is not found in any of the modeling set modifiers. Also note that cyclopentyl and cycloheptyl groups were present in four non-toxic CNTs selected by the models but were not present in the modeling set structures at all.
Solely using potential structural alerts (e.g., presence of cyclohexyl group in R1) would not allow a reliable ranking of hits in the virtual screening library. Indeed, cyclohexyl group (AM003 in R1) is present in 10 CNTs of the modeling set: 6 are CA-binders, 4 CA-nonbinders, 6 toxic, 4 non-toxic. Phenyl group (AM004 in R1) is present in 10 CNTs of the modeling set: 4 CA-binders, 6 CA-nonbinders, 6 toxic, 4 non-toxic. Benzyl group (AM005 in R1) is present in 10 CNTs of the modeling set: 5 CA-binders, 5 CA-nonbinders, 5 toxic, 5 non-toxic. Therefore only the use of screening protocol involving similarity search and QSAR-based prioritization could both enable the accurate classification of CNTs and the rational design of novel substances with the desired bioactivity profiles. Moreover, the screening of 240,000 molecules is a task that is unlikely to be accomplished effectively manually. We plan to expand the chemical diversity in the future series of decorated CNTs.
Limitations of this study
In this work, CNTs only differ by the types of small organic compounds that decorate their surface. Experiments showed that the biological properties of these decorated CNTs dramatically vary from one organic modifier to another. Given that their cores are the same, it is logical to expect that certain chemical features (encoded by molecular descriptors) of the surface modifiers do influence biological activities of decorated CNTs. Molecular descriptors were exclusively computed for modifiers to characterize their molecular connectivity, size, partial charges, bulkiness, that could contribute to CNTs bioactivity. QSAR models using these descriptor values were shown to achieve a reasonable predictive power justifying their utility to prioritize the modifiers to be use for the synthesis of novel CNTs.
We should underline again that the choice of activity classification thresholds greatly influences the modeling performances and especially the mispredictions of CNTs near the threshold borders (e.g., 2.0 for the CA binding assay). Choosing a different threshold would affect the balance between active and inactive compounds making it impossible to develop any model. In the case of CA, using a threshold of 2.0 (the activity ranging from 0.53 to 5.29) discriminates 44 binders versus 39 non-binders. If the threshold is changed to 1.8, 49 binders and 34 non-binders are defined. If the threshold is changed to 2.2, 40 binders and 43 non-binders are defined. We do observe that the activity values follow a normal distribution around the threshold (no bimodal distribution here). Indeed, we could have moved the threshold numerous times and rebuild models to optimize the prediction performances and identify the best threshold; given the above analysis, relatively few compounds would have changed their class assignment for as long as we keep the dataset balanced and therefore the model statistics would stay the same (e.g., a threshold of 2.1 leads to identical QSAR models). Similarly, the threshold used for cytotoxicity is also critical since the modeling set is biased toward cytotoxic CNTs (only 6 CNTs from the modeling set are associated with a cellular survival rate higher than 60% at 200 µg/mL). However, given that we have built binary models using an ensemble QSAR approach where individual models make binary predictions (toxic or “1”, non-toxic or “0”), these individual predictions are averaged to output a predicted value between “0” and “1” so our choice of overall threshold is valid.
This study was strictly related to evaluating and predicting toxicity for surface modified CNTs tested in the reported in vitro assays. The issue of in vitro to in vivo extrapolation was obviously beyond the scope of this work. We shall also stress that due to study design, our observations and conclusions are strictly limited to modified CNTs. Graphene oxides, polycyclic aromatic hydrocarbons and CNTs can theoretically be decorated with the same small molecules on their surface. Although the intensity and diversity of toxicity effects reported in the literature (cf. the excellent review by (Jastrzebska et al., 2012)) resonates with the ones reported for CNTs, there is no indication that graphene/PAHs decorated with the same surface modifiers would induce the same biological effects as those of CNTs. Our results do not suggest that our models could be applied to any other types of surface-modified nanomaterials since their toxicity or any measured property is likely governed by different structural features.
It is also interesting to note that some other research groups (Shao et al., 2013; Singh & Gupta, 2014) recently attempted and succeeded to model 29 CNTs (subset of the 83 CNTs from this work) but used the indirect CNT rank (ranging from 1 to 83) as the modeling endpoint (not the actual, measured activity as done in this study).
Limitations of the approach
The approach used in this study is not without important limitations due to the assumptions that are inherent to this technique of surface characterization. Indeed, QSAR modeling is not been used to fully analyze and understand the complexity of all nano-bio interactions (Nel et al., 2009) and features that are involved in defining the bioactivity of the CNTs. As illustrated by our results, we used this approach to explain the differences in reported bioactivity in terms of the differences in the structure of the surface modifiers. Our experimental results show that the collection of 83 CNTs is characterized by a quite significant diversity of biological responses that cannot be explained by minor variations in the size and other properties of the same CNT core common to all substances. Thus, we explain the variability of biological response by the variance in the structures of the surface modifiers and identify structural features and SAR trends that are responsible for this variability. Importantly, cytotoxicity is also the consequence of the binding of CNTs with protein receptors on cell surface. We should also underline that: (i) if the molecular descriptors were completely inadequate to characterize CNT surfaces, then the prediction power of the QSAR models built using these descriptors would be close to random and this is not the case (as confirmed by Y-randomization results); (ii) as of today, there is no other available approach capable of characterizing 80+ CNTs with computationally derived parameters and using them to build predictive models and prospectively guide the design of novel nanomaterials with the desired properties; (iii) our approach was validated by accurately predicting both active and inactive CNTs that were confirmed experimentally.
Perspectives
The unfortunate truth is that records on biologically tested MNPs are extremely scarce. Our pioneering study of a dataset of 100+ surface-modified MNPs with the same metal core tested for their cancer cell uptake (Fourches et al., 2010c) has been repeated in the literature at least five times (e.g., (Chau and Yap, 2012; Ghorbanzadeh et al., 2012; Chandana Epa et al., 2012; Toropov et al., 2013)) with similar if not poorer statistical characteristics for the reported models. This unfortunate situation continues to severely limit the opportunities to develop and validate additional QSAR models; even a dataset of 80+ CNTs is relatively small to try meaningful sensitivity and descriptor analyses given the need to set aside a significant fraction of compounds for external model validation. Additional obvious pitfalls possibly affecting model accuracy is, by necessity, a simplified representation of surface-modified CNTs that does not take into account the location, density, and other structural aspects of these substances related to modifier-core interaction. We are also exploring additional approaches to improve model predictivity, e.g., by using layers of classification models (binary and multi-class) with different activity thresholds and testing other types of chemical descriptors. Nevertheless, similar to our previous studies (Fourches et al., 2010c), we have managed as a proof-of-concept to develop validated models and showed that with all the above limitations they could guide experimental synthesis. QSAR models could aid in the rational design of surface-modified MNPs and this study may encourage more experimental, large scale investigations in nanomaterial science, which will in return promote further development of the nascent modeling field for nanomaterials.
Conclusions
This study presents the computer-aided rational design of nanomaterials, in which a complete development cycle comprising the initial chemical and biological data generation, QSAR model building, and QSAR-based virtual screening followed by computational hit identification and experimental validation, was realized. We have demonstrated that QSAR models built and selected following rigorous guidelines (Tropsha, 2010) can be used to (i) evaluate the activity profiles of surface-decorated nanomaterials, (ii) virtually screen large chemical libraries of MNP-attachable ligands, (iii) select and identify chemicals based on predicted bioprofiles, and overall (iv) help designing safer MNPs. The reasonable agreement between computer-selected hits and their experimentally assessed biological profiles validates both the rationale and the overall efficacy of our modeling methods and workflow. The results of this study suggest that QSAR models can be successfully employed for rational discovery of novel nanoparticles with the desired properties (i.e., reduced toxicity and low protein binding) and more generally, for the design of MNPs with controlled biological and safety profiles.
Supplementary Material
Acknowledgments
DF and AT gratefully acknowledge the support from a Semiconductor Research Corporation (SRC)/Sematech grant, NIH (grants R01-GM66940 and R01-GM096967), and EPA (grant RD832720). This work was also supported by the Natural Science Foundation of China (21137002 to BY) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB14030401 to BY). DF thank the NCSU Chancellor’s Faculty Excellence Program. We also gratefully acknowledge the excellent contributions from reviewers that helped us improve our manuscript.
Footnotes
Declaration of Interest
The authors declare no competing financial interest. The authors alone are responsible for the content and writing of the paper.
References
- Arts JHE, Muijser H, Jonker D, Van De Sandt JJM, Bos PMJ, Feron VJ. Inhalation toxicity studies: OECD guidelines in relation to REACH and scientific developments. Exp Toxicol Pathol Off J Gesellschaft fur Toxikologische Pathol. 2008;60:125–133. doi: 10.1016/j.etp.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Austin CP. The Tox21 Collaboration on Predictive Toxicology The best of times, the worst of times. Toxicology. 2009 [Google Scholar]
- Baalousha M, Lead JR. Nanoparticle dispersity in toxicology. Nat Nanotechnol. 2013;8:308–309. doi: 10.1038/nnano.2013.78. [DOI] [PubMed] [Google Scholar]
- Bai Y, Zhang Y, Zhang J, Mu Q, Zhang W, Butch ER, Snyder SE, Yan B. Repeated administrations of carbon nanotubes in male mice cause reversible testis damage without affecting fertility. Nat Nanotechnol. 2010;5:683–689. doi: 10.1038/nnano.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Burello E, Worth A. Computational nanotoxicology: Predicting toxicity of nanoparticles. Nat Nanotechnol. 2011;6:138–139. doi: 10.1038/nnano.2011.27. [DOI] [PubMed] [Google Scholar]
- Carbó-Dorca R, Besalú E. Construction of coherent nano quantitative structure-properties relationships (nano-QSPR) models and catastrophe theory. SAR QSAR Environ Res. 2011;22:661–665. doi: 10.1080/1062936X.2011.623319. [DOI] [PubMed] [Google Scholar]
- Chandana Epa V, Burden FR, Tassa C, Weissleder R, Shaw S. Modeling Biological Activities of Nanoparticles. Nano Lett. 2012;12:5808–5812. doi: 10.1021/nl303144k. [DOI] [PubMed] [Google Scholar]
- Chau YT, Yap CW. Quantitative Nanostructure-Activity Relationship modelling of nanoparticles. RSC Adv. 2012;2:8489–8496. [Google Scholar]
- Cristianini N, Shawe-Taylor J. An introduction to Support Vector Machines, History. Cambridge University Press; 2000. [Google Scholar]
- Demirdjian ZS. Nanotechnology: The New Frontier for Business and Industry. J Am Acad Bus Cambridge. 2006;8:I–II. [Google Scholar]
- Downs GM, Barnard JM. Clustering Methods and Their Uses in Computational Chemistry. Rev Comput Chem. 2002;18:1–40. [Google Scholar]
- European Parliament EC. [Accessed 7/1/2014];REACH regulation. 2010 Available at http://ec.europa.eu/enterprise/sectors/chemicals/reach/index_en.htm.
- Fourches D, Barnes JC, Day NC, Bradley P, Reed JZ, Tropsha A. Cheminformatics analysis of assertions mined from literature that describe drug-induced liver injury in different species. Chem Res Toxicol. 2010a;23:171–183. doi: 10.1021/tx900326k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D, Muratov E, Tropsha A. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model. 2010b;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D, Pu D, Tassa C, Weissleder R, Shaw SY, Mumper RJ, Tropsha A. Quantitative nanostructure-activity relationship modeling. ACS Nano. 2010c;4:5703–5712. doi: 10.1021/nn1013484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D, Pu D, Tropsha A. Exploring quantitative nanostructure-activity relationships (QNAR) modeling as a tool for predicting biological effects of manufactured nanoparticles. Comb Chem High Throughput Screen. 2011;14:217–225. doi: 10.2174/138620711794728743. [DOI] [PubMed] [Google Scholar]
- Gajewicz A, Schaeublin N, Rasulev B, Hussain S, Leszczynska D, Puzyn T, Leszczynski J. Towards understanding mechanisms governing cytotoxicity of metal oxides nanoparticles: Hints from nano-QSAR studies. Nanotoxicology. 2014:1–13. doi: 10.3109/17435390.2014.930195. [DOI] [PubMed] [Google Scholar]
- Ghorbanzadeh M, Fatemi MH, Karimpour M. Modeling the Cellular Uptake of Magnetofluorescent Nanoparticles in Pancreatic Cancer Cells: A Quantitative Structure Activity Relationship Study. Ind Eng Chem Res. 2012;51:10712–10718. [Google Scholar]
- Hartung T. Evidence-based toxicology - the toolbox of validation for the 21st century? ALTEX Altern zu Tierexperimenten. 2010;27:253–263. doi: 10.14573/altex.2010.4.253. [DOI] [PubMed] [Google Scholar]
- Jastrzębska AM, Kurtycz P, Olszyna AR. Recent advances in graphene family materials toxicity investigations. J Nanopart Res. 2012;14:1320–1328. doi: 10.1007/s11051-012-1320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. Nanotechnology, energy and markets. Nat Nanotechnol. 2009;4:75. doi: 10.1038/nnano.2008.420. [DOI] [PubMed] [Google Scholar]
- Kaiser J-P, Krug HF, Wick P. Nanomaterial cell interactions: how do carbon nanotubes affect cell physiology? Nanomedicine. 2009;4:57–63. doi: 10.2217/17435889.4.1.57. [DOI] [PubMed] [Google Scholar]
- Kar S, Gajewicz A, Puzyn T, Roy K. Nano-quantitative structure-activity relationship modeling using easily computable and interpretable descriptors for uptake of magnetofluorescent engineered nanoparticles in pancreatic cancer cells. Toxicol In Vitro. 2014;28:600–606. doi: 10.1016/j.tiv.2013.12.018. [DOI] [PubMed] [Google Scholar]
- Kumar A, Kumar P, Anandan A, Fernandes TF, Ayoko GA, Biskos G. Engineered Nanomaterials: Knowledge Gaps in Fate, Exposure, Toxicity, and Future Directions. J Nanomater. 2014;2014:1–16. [Google Scholar]
- Liu Y, Hayes DN, Nobel A, Marron JS. Statistical Significance of Clustering for High-Dimension, Low-Sample Size Data. J Am Stat Assoc. 2008;103:1281–1293. [Google Scholar]
- Luan F, Kleandrova VV, González-Díaz H, Ruso JM, Melo A, Speck-Planche A, Cordeiro MNDS. Computer-aided nanotoxicology: assessing cytotoxicity of nanoparticles under diverse experimental conditions by using a novel QSTR-perturbation approach. Nanoscale. 2014;6:10623–10630. doi: 10.1039/c4nr01285b. [DOI] [PubMed] [Google Scholar]
- Luther W. Industrial application of nanomaterials - chances and risks, Technology analysis. [Accessed 7/1/2014];Future Technologies Division of VDI Technologiezentrum, Dusseldorf (Germany) 2004 Available at http://www.innovationsbegleitung.de/11.pdf.
- Mauri A, Consonni V, Pavan M, Todeschini R. Dragon software: An easy approach to molecular descriptor calculations. Match Commun Math Comput Chem. 2006;56:237–248. [Google Scholar]
- Maynard AD, Aitken RJ, Butz T, Colvin V, Donaldson K, Oberdörster G, Philbert MA, Ryan J, Seaton A, Stone V, Tinkle SS, Tran L, Walker NJ, Warheit DB. Safe handling of nanotechnology. Nature. 2006;444:267–269. doi: 10.1038/444267a. [DOI] [PubMed] [Google Scholar]
- Monteiro-Riviere NA, Samberg ME, Oldenburg SJ, Riviere JE. Protein binding modulates the cellular uptake of silver nanoparticles into human cells: Implications for in vitro to in vivo extrapolations? Toxicol Lett. 2013;220:286–293. doi: 10.1016/j.toxlet.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J, Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Mach Learn. 2003;52:91–118. [Google Scholar]
- Mu Q, Jiang G, Chen L, Zhou H, Fourches D, Tropsha A, Yan B. Chemical Basis of Interactions Between Engineered Nanoparticles and Biological Systems. Chem Rev. 2014;114:7740–7781. doi: 10.1021/cr400295a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Q, Liu W, Xing Y, Zhou H, Li Z, Zhang Y, Ji L, Wang F, Si Z, Zhang B, Yan B. Protein Binding by Functionalized Multiwalled Carbon Nanotubes Is Governed by the Surface Chemistry of Both Parties and the Nanotube Diameter. J Phys Chem B. 2008;112:3300–3307. [Google Scholar]
- Murray AR, Kisin E, Inman A, Young S-H, Muhammed M, Burks T, Uheida A, Tkach A, Waltz M, Castranova V, Fadeel B, Kagan VE, Riviere JE, Monteiro-Riviere N, Shvedova AA. Oxidative Stress and Dermal Toxicity of Iron Oxide Nanoparticles In Vitro. Cell Biochem Biophys. 2012;67:461–476. doi: 10.1007/s12013-012-9367-9. [DOI] [PubMed] [Google Scholar]
- Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–677. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- Nel AE, Mädler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- Puzyn T, Leszczynska D, Leszczynski J. Toward the development of “nano-QSARs”: advances and challenges. Small. 2009;5:2494–2509. doi: 10.1002/smll.200900179. [DOI] [PubMed] [Google Scholar]
- Puzyn T, Rasulev B, Gajewicz A, Hu X, Dasari TP, Michalkova A, Hwang H-M, Toropov A, Leszczynska D, Leszczynski J. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat Nanotechnol. 2011;6:175–178. doi: 10.1038/nnano.2011.10. [DOI] [PubMed] [Google Scholar]
- Qu G, Bai Y, Zhang Y, Jia Q, Zhang W, Yan B. The effect of multiwalled carbon nanotube agglomeration on their accumulation in and damage to organs in mice. Carbon N Y. 2009;48:2060–2069. [Google Scholar]
- Rodgers AD, Zhu H, Fourches D, Rusyn I, Tropsha A. Modeling liver-related adverse effects of drugs using knearest neighbor quantitative structure-activity relationship method. Chem Res Toxicol. 2010;23:724–732. doi: 10.1021/tx900451r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen K, Pylkkänen L, Norppa H, Falck G, Lindberg H, Tuomi T, Vippola M, Alenius H, Hämeri K, Koivisto J. Nanotechnologies, engineered nanomaterials and occupational health and safety – A review. Saf Sci. 2010;48:957–963. [Google Scholar]
- Service RF. Nanotoxicology. Nanotechnology grows up. Science. 2004;304:1732–1734. doi: 10.1126/science.304.5678.1732. [DOI] [PubMed] [Google Scholar]
- Shao C-Y, Chen S-Z, Su B-H, Tseng YJ, Esposito EX, Hopfinger AJ. Dependence of QSAR models on the selection of trial descriptor sets: a demonstration using nanotoxicity endpoints of decorated nanotubes. J Chem Inf Model. 2013;53:142–158. doi: 10.1021/ci3005308. [DOI] [PubMed] [Google Scholar]
- Singh KP, Gupta S. Nano-QSAR modeling for predicting biological activity of diverse nanomaterials. RSC Adv. 2014;4:13215–13230. [Google Scholar]
- Sips M. Nanotechnology in perspective Risks to man and the environment. Public Health. 2009:138. [Google Scholar]
- Su G, Yan B. Nano-combinatorial chemistry strategy for nanotechnology research. J Comb Chem. 2010;12:215–221. doi: 10.1021/cc900193g. [DOI] [PubMed] [Google Scholar]
- Toropov AA, Toropova AP, Puzyn T, Benfenati E, Gini G, Leszczynska D, Leszczynski J. QSAR as a random event: modeling of nanoparticles uptake in PaCa2 cancer cells. Chemosphere. 2013;92:31–37. doi: 10.1016/j.chemosphere.2013.03.012. [DOI] [PubMed] [Google Scholar]
- Toropova AP, Toropov AA, Benfenati E, Korenstein R. QSAR model for cytotoxicity of SiO2 nanoparticles on human lung fibroblasts. J Nanoparticle Res. 2014;16:2282–2289. [Google Scholar]
- Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol Inform. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- Tropsha A, Golbraikh A. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr Pharm Des. 2007;13:3494–3504. doi: 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, Solov’ev V, Hoonakker F, Tetko IV, Marcou G. ISIDA - Platform for virtual screening based on fragment and pharmacophoric descriptors. Curr Comput Aided Drug Des. 2008;4:191–198. [Google Scholar]
- Vilar S, Cozza G, Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- Winkler DA, Mombelli E, Pietroiusti A, Tran L, Worth A, Fadeel B, McCall MJ. Applying quantitative structure-activity relationship approaches to nanotoxicology: Current status and future potential. Toxicology. 2012;313:15–23. doi: 10.1016/j.tox.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Yan B. Nano-combinatorial and catalyst screening technologies. Comb Chem high throughput Screen. 2011;14:146–154. doi: 10.2174/138620711794728734. [DOI] [PubMed] [Google Scholar]
- Zhang B, Xing Y, Li Z, Zhou H, Mu Q, Yan B. Functionalized carbon nanotubes specifically bind to alpha-chymotrypsin’s catalytic site and regulate its enzymatic function. Nano Lett. 2009;9:2280–2284. doi: 10.1021/nl900437n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhou H, Yan B. Reducing nanotube cytotoxicity using a nano-combinatorial library approach. Methods Mol Biol. 2010;625:95–107. doi: 10.1007/978-1-60761-579-8_9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bai Y, Jia J, Gao N, Li Y, Zhang R, Jiang G, Yan B. Perturbation of physiological systems by nanoparticles. Chem Soc Rev. 2014;43:3762–3809. doi: 10.1039/c3cs60338e. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bai Y, Yan B. Functionalized carbon nanotubes for potential medicinal applications. Drug Discov Today. 2010;15:428–435. doi: 10.1016/j.drudis.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Tropsha A. Novel variable selection quantitative structure--property relationship approach based on the k-nearest-neighbor principle. J Chem Inf Comput Sci. 2000;40:185–194. doi: 10.1021/ci980033m. [DOI] [PubMed] [Google Scholar]
- Zhou H, Mu Q, Gao N, Liu A, Xing Y, Gao S, Zhang Q, Qu G, Chen Y, Liu G, Zhang B, Yan B. A nano-combinatorial library strategy for the discovery of nanotubes with reduced protein-binding, cytotoxicity, and immune response. Nano Lett. 2008;8:859–865. doi: 10.1021/nl0730155. [DOI] [PubMed] [Google Scholar]
- Zhu H, Rusyn I, Richard A, Tropsha A. Use of cell viability assay data improves the prediction accuracy of conventional quantitative structure-activity relationship models of animal carcinogenicity. Environ Health Perspect. 2008a;116:506–513. doi: 10.1289/ehp.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Tropsha A, Fourches D, Varnek A, Papa E, Gramatica P, Oberg T, Dao P, Cherkasov A, Tetko IV. Combinatorial QSAR modeling of chemical toxicants tested against Tetrahymena pyriformis. J Chem Inf Model. 2008b;48:766–784. doi: 10.1021/ci700443v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.