Abstract
Germ granules are the hallmark of all germ cells. These membrane-less, electron-dense structures were first observed over 100 years ago. Today, their role in regulating and processing transcripts critical for the establishment, maintenance and protection of germ cells is well-established and pathways outlining the biochemical mechanisms and physical properties associated with their biogenesis are emerging.
Keywords: Germ plasm, RNA granules, Germ granules, Nuage, Germ cells, RNA localization, RNA translation, Phase transition, Oskar, Vasa, Tudor, Nanos, Drosophila
In all sexually reproducing organisms, germ cells, the cells that generate the next generation, are marked by the inheritance of specialized RNA particles, collectively referred to as germ granules. In some organisms, like Drosophila, C. elegans, Xenopus or zebrafish, germ granule components are synthesized during oogenesis and assemble into granules that are passed on to the egg and embryo. As the embryo starts to develop, those cells that inherit the maternally derived germ granules will develop as germ cells. In other organisms, like mice or humans, that lack such maternally inherited germ cell determinants, germ cells are specified by cell-cell signaling and inductive events. Despite these differences, some of the same, conserved germ granule components are expressed in all germ cells irrespective of the mode of germ cell specification. This has led to the hypothesis that the organization and function of germ granules is intimately related to germ cell fate.
In contrast to evolutionary conserved, transcription-based mechanisms that control cell and tissue specification of somatic cell types, no ‘transcriptional master regulators of germ cell fate’ have been identified. Instead, the conserved components of germ cells include: structural, scaffolding proteins such as the Tudor class of proteins; RNA binding and processing proteins, such as the ATP dependent helicase Vasa and the Piwi class of Argonaute proteins that bind and processes small RNAs; and translational regulators, like the conserved Zn finger proteins Nanos and Dazl (Voronina et al., 2011). Furthermore, these components are assembled into larger RNP particles with strikingly similar electron-dense appearance suggesting that not just the molecules per se but their intracellular organization into large protein machines plays a critical role in all aspects of germ cell biology. Germ granules are present throughout the germ line cycle, where they are found in different cellular locations and have varied molecular composition (Voronina et al., 2011). In Drosophila, at least four different types of germ granules have been described and associated with distinct functions in germ line development (Fig 1): The Balbiani body (likely analogous to the mitochondrial cloud found in early frog oocytes) is found in the early, growing oocyte, where it is closely associated with mitochondria, Golgi, centrioles and ER rich fusome material (Cox and Spradling, 2003). It has been functionally linked to oocyte specification and initial microtubule organization. Nuage is associated with nuclear pores on the cytoplasmic side of the nurse cell nucleus and is the hub for the processing of small, piwi-interacting (pi) RNAs in defense of transposable elements (Pek et al., 2012). Sponge bodies are found in the nurse cells and the oocyte, they are large, ER rich structures that lack ribosomes but contain many components that are also found in P-bodies (Wilsch-Brauninger et al., 1997). Consistent with a role in RNA storage, sponge body architecture is highly dynamic depending on environmental conditions (Snee and Macdonald, 2009). Polar granules make up the germ plasm at the posterior pole of the mature egg and early embryo. They contain mRNA transcripts as well as piRNAs required to establish, maintain and protect the germ line of the next generation. Consistent with their role in primordial germ cell identity and function, polar granules are associated with ribosomes and mitochondria (Illmensee et al., 1976). Despite their morphological resemblance as membrane-less, RNA-rich granules and the identification of granule specific and shared components, it remains unclear how the structure of different granule types relates to their function in germ cell biology. Recent results from genetic and molecular analysis as well as structural and biophysical studies are beginning to shed new light on these issues.
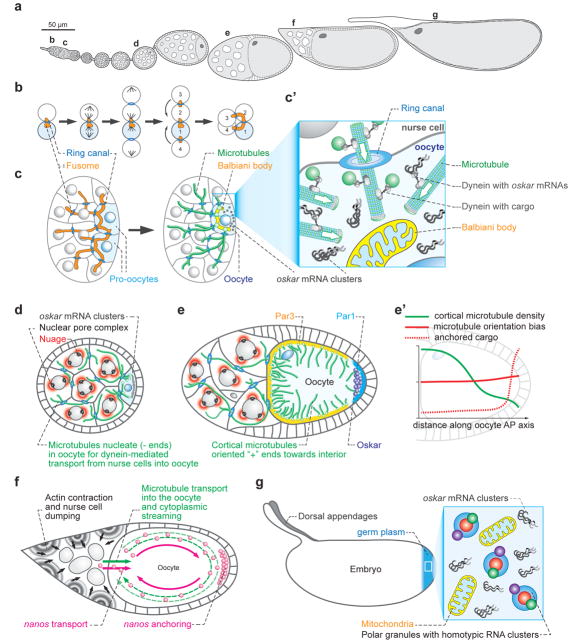
Figure 1.
Establishment of anterior-posterior polarity during Drosophila oogenesis
a. Ovariole with developing egg chambers. Each ovary contains 12–16 ovarioles. At the tip (left, b-c region) is the germarium, where 2–3 GSCs generate a cytoblast that matures into a egg chamber consisting of the oocyte and 15 nurse cells surrounded by somatic follicle cells (stage 1 egg chamber). Daily stem cell division produces a chain of maturing egg chambers that mature to the fully developed egg (right, g). Small letters on top refer to stages depicted in other figure panels.
b. Division pattern of the cystoblast, the GSC daughter in the germarium. After division cells remain connected via ring canals with fusome material traversing the cyst. The first two cells born are connected by four ring canals, the next two by three, the next four by two and the last 8 by one ring canal each. Note that the earlier born cells inherit proportionally more fusome material.
c. 16 cell cyst in germarium, all cells are connected by ring canals, and fusomes. The two cells with four ring canals “pro-oocytes” are positioned posteriorly within the cluster and enter into meiosis (left), only one of the two cells remains in meiosis and arrests in meiotic prophase (right). All other cells develop into polyploid nurse cells, which continue to be connected to the oocyte by ring canals. All centrosomes migrate into the oocyte and a microtubule network nucleate from the oocyte to the nurse cells. Dynein mediated transport brings cargo and RNAs from the nurse cell into the oocyte. Densely packed mitochondria and RNA form the Balbiani body (yellow in c and c’). c’ oocyte cytoplasm indicating microtubule polarity and RNA transport particles (after (Roth and Lynch, 2009)).
d. Stage 5 egg chamber. Nurse cells and oocyte are surrounded by somatic follicle cells. Continued dynein-mediated RNA transport from the nurse cells into oocyte. Microtubules are oriented to nucleate (- pole) from oocyte and extend into nurse cells. Nuage: RNA rich granules form around nurse cell nucleus often in close proximity of nuclear pore.
e. Stage 10A egg chamber. Mutually exclusive interactions between Par-1 (Blue) at the posterior pole and the Par-3/Bazooka (yellow) complex at the lateral and anterior cortex establish polarity within the oocyte. Microtubules nucleate from lateral and anterior cortex extending into the oocyte and are inhibited at the posterior pole by Par-1. Kinesin motors transport osk RNA to the posterior pole, where it is translated. Dynein-mediated transport continues to bring osk RNA from the nurse cells to the oocyte. Somatic follicle cells surround the egg chamber. e’ Summary of microtubule organization and osk RNA transport dynamics during stage 10A. Microtubule network is denser at the anterior than posterior of oocyte (green). A slight bias of kinesin-mediated microtubule transport (red line) leads to enrichment of osk RNA at posterior pole over time (red stippled) (after (Parton et al., 2011)).
f. Stage 10B/11, cytoplasmic streaming. The actin meshwork is disassembled in the oocyte and microtubules align in bundles (green). This leads to a streaming process where RNAs and proteins are mixed within the oocyte and additional RNAs/proteins are drawn into the oocyte (green arrow). Effector RNAs like nanos (red) become tethered at posterior pole.
g. Mature egg. Germplasm is assembled and polar granules of similar size to mitochondria are found in a precise region at the posterior pole. Effector RNAs are localized to the granules, Osk, Vasa, Tud and Aub proteins are essential part of the granules but osk RNA is not associated with polar granules (see Fig 3E).
In this review, I will focus on the germ plasm and germ granule biology of one species, Drosophila melanogaster. A ‘pioneer’ for the study of germ plasm, as germ cells of Drosophila have been studied for over a century. Early morphological studies have been complemented by functional and molecular analysis and can now be connected to the striking structural properties of the granules.
This essay summarizes some of the original findings that led to the identification of germ plasm components (see also excellent reviews on the topic written by Tony Mahowald one of the early leaders of germ plasm analysis (Mahowald, 2001), and more recently by (Gao and Arkov, 2013) and will highlight new observations that have shaped our cellular, molecular and structural understanding of the process of germ granule biogenesis under the control of the germ plasm determinant Oskar.
Genetic analysis of genes required for germ plasm assembly
The first mutants in germ plasm assembly were identified because of their abdominal segmentation defects in screens for maternal effect genes required for embryonic pattern formation (Boswell and Mahowald, 1985; Lehmann and Nusslein-Volhard, 1986; Schupbach and Wieschaus, 1989). Subsequently, it was found that such mutant embryos also lack germ plasm and germ cells. The genetics agreed well with transplantation experiments that showed that germ plasm when transplanted to an ectopic site is not only able to induce germ cell formation but can also generate a second abdomen at the new location (Illmensee and Mahowald, 1974; Sander, 1975). Thus information about germ cells and posterior embryonic patterning are contained in the germ plasm. Maternal effect genes with a similar role in embryonic abdomen formation were collectively referred to as ‘posterior group genes’. Most posterior group genes affect germ plasm and germ cell formation as well as abdominal development. Only, two posterior group genes, nanos and pumilio specifically affect abdominal development by repressing the translation of maternal hunchback in the future abdominal region, thereby allowing the abdomen to form (Barker et al., 1992; Irish et al., 1989; Lehmann and Nusslein-Volhard, 1991; Struhl et al., 1992). While nos and pum are not directly involved in germ cell formation, they do play an important role in primordial germ cell specification and development (Asaoka-Taguchi et al., 1999; Kobayashi et al., 1996).
Although phenotypically the posterior group genes are very similar, molecular and genetic analysis revealed a key role for oskar (osk) in germ plasm organization. First, the pattern of osk RNA distribution during oogenesis foreshadows events that lead to germ plasm biogenesis (Ephrussi et al., 1991; Kim-Ha et al., 1991). Second, mutations in Oskar protein affect the enrichment of other posterior group RNAs and/or proteins at the posterior pole. Finally, mislocalization of osk RNA to the anterior pole and expression of Oskar protein at this ectopic location is sufficient to instruct germ plasm assembly, leading to the formation of ectopic germ cells and a second abdomen (Ephrussi and Lehmann, 1992). The later finding informed the design of genetic epistasis experiments that distinguished between those genes regulating osk (upstream genes) and those genes that depend on osk for their posterior localization and function (downstream genes) (Fig 3). The ‘upstream’ group includes genes required for the establishment of oocyte polarity, genes involved in the processing and localization of osk RNA, and genes that control the translation and stability of Oskar protein. The ‘downstream’ group includes genes that act together with Oskar in germ plasm assembly and also “effector genes” whose products are not involved in germ plasm assembly but are localized to the germ plasm and have diverse functions in primordial germ cell formation, germ cell specification and migration as well as abdominal patterning (Fig 2, 3).
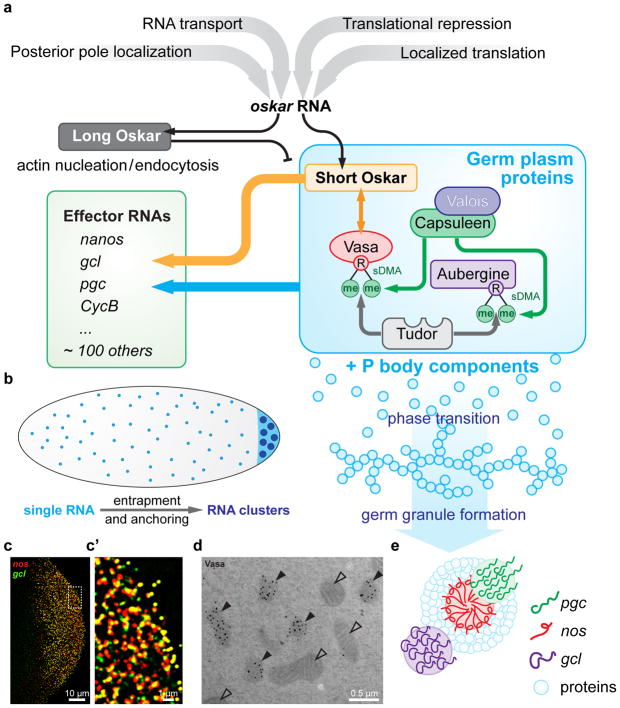
Figure 3.
Germ plasm assembly
a. Multiple controls described in Fig 1 and 2 feed into the localization and translation of osk. Osk protein recruits other components of the germplasm such as Vasa, Tudor and Aubergine. Symmetric dimethylation of Arginine (sDMA) in Vasa and Aubergine by the methyltransferase Capsuleen allow binding to Tudor domain proteins (Kirino et al., 2010a; Kirino et al., 2010b; Liu et al., 2010; Webster et al., 2015). This together with liquid to gel phase transitions could provide a possible basis for the dense assembly of polar granules.
b. Model for phase transition that occurs at posterior pole when concentration of germ plasm protein and RNAs become high (Brangwynne, 2013).
c. SIM image of nos and gcl RNA hybridization using Stellaris© RNA in situ hybridization probes. C’ higher magnification to reveal particle overlap.
d. Germ plasm with polar granules marked by Vasa (filled arrow) and mitochondria (open arrow).
e. Organization of homotypic RNA clusters within polar granules according to (Trcek et al., 2015).
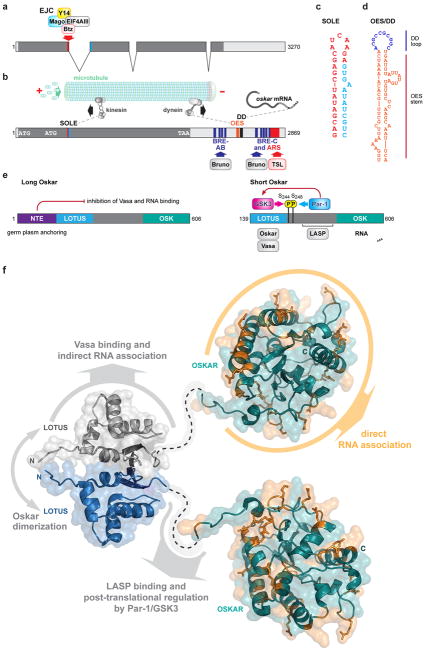
Figure 2.
Oskar RNA and protein history
a. Structure of nascent osk RNA. Exon junction complex components bind to sequences flanking the first intron, which is critical for kinesin-mediated osk RNA transport.
b. Osk RNA transport particles are transported along microtubule by dynein (directed towards – end of microtubule) and kinesin (directed towards + end of microtubule) motors. The oocyte entry signal (d) regulates the dynein-mediated transport of osk RNA into the early oocyte. (c) The Spliced Oskar Localization Element (SOLE) forms as a consequence of first intron splicing, where 18nt from the first and 10 nt from the second exon merge into a stem loop strcuture. The DD palindromic sequence element (d) is located in the loop of the OES stem loop. Via RNA-RNA “kissing interaction” this element promotes RNA transport particle formation and hitchhiking of RNA lacking the SOLE element on those with the element. AB, C region are regions rich of Bruno regulatory elements (BRE), which repress RNA translation, and IMP binding elements, which promote translation and localization. Binding elements close to the most distal part of the 3′UTR are important for translational activation, via Adenine-rich sequences (ARS) that mediate polyadenylation.
e. Structure of Oskar isoforms. Left: Long Oskar is a 606 aminoacid protein, the N-terminal extension (NTE) is required for actin filament nucleation and enhanced endocytosis at the posterior pole. Somehow with NTE suppresses short-Oskar functions. Right: Short -Oskar start at the second Methionine 139. The Lotus domain dimerizes and interacts with Vasa (see also g), the C-terminal OSK domain resembles a SGNH hydrolase domain, however the enzymatic amino acids are missing. Instead the OSK domain is thought to interact with effector RNAs.
f. Structure diagram of Lotus and OSK domain. The Lotus domain forms a homodimer of two winged helix structures and interacts with Vasa, while the OSK domain forms a globular structure with basic and hydrophilic aminoacids exposed to the surface, which are thought to interact with RNA.
In the following, I will review how oocyte specification and establishment of oocyte polarity lead to the spatial restriction of osk RNA, how Oskar protein synthesis is regulated, and how Oskar together with other posterior group genes and effector RNAs assemble into a functional germ plasm that instructs the next generation.
Oocyte Specification and dynein-mediated nurse cell to oocyte RNA transport
Biogenesis of germ plasm is intimately linked to successive polarizing events leading from the asymmetric division of a germ line stem cell to a mature egg that harbors a prepattern of the embryonic axes (Fig 1A). In Drosophila, germ line stem cells (GSC) reside in a somatic niche, which supports GSC maintenance (Spradling et al., 1997). GSCs divide asymmetrically to self-renew and produce a differentiating daughter. This daughter undergoes 4 rounds of mitosis to produce a 16 cell cyst. During each division, the sisters do not complete cytokinesis and remain connected by cytoplasmic bridges, the ring canals (Fig 1B). Through the ring canals weaves the fusome, an endoplasmic reticulum-rich structure associated with cytoskeletal components and microtubules. As a consequence of this division strategy and an apparent asymmetry of fusome material a bias is created that orients the microtubule network toward one of the cells with four ring canals (Fig 1C) (Roth and Lynch, 2009). This cell remains in meiosis and becomes the oocyte. The other fifteen cells develop into polyploid nurse cells that feed RNA, protein and even mitochondria into the transcriptionally silent oocyte (Fig 1C’). As part of the Balbiani body a Microtubule Organizing Center (MTOC) forms in the oocyte. The MTOC nucleates microtubules that direct dynein motor mediated transport from the nurse cells to the oocyte (Fig 1D). Mutants in the dynein cargo adapters, Egalitarian and Bicaudal-D fail to specify an oocyte and all 16 cells take on equal, nurse cell fate (Mach and Lehmann, 1997; Ran et al., 1994). Large scale RNA in situ analysis shows that 17% of all germ line expressed genes are specifically enriched in the oocyte during stage 1–7 of oogenesis (Jambor et al., 2015). Among these are many RNAs involved in germ plasm assembly and function including osk (Ephrussi et al., 1991; Kim-Ha et al., 1991). The transport of osk RNA into the oocyte is mediated by a 67nt stem loop in the osk 3′UTR, the oocyte entry signal (OES), which shares functional similarity with the K10 oocyte localization signal (Fig 1D, Figure 2B, D) (Jambor et al., 2014; Serano and Cohen, 1995). Both signals mediate dynein-dependent, microtubule minus-end directed transport dependent on a short dsRNA stem. It is likely that these and RNAs with localization signals of similar structure are transported into the oocyte possibly direct association with Egalitarian (Dienstbier et al., 2009).
Oocyte polarization and kinesin-mediated transport of osk RNA within the oocyte
Beginning at stage 7, the MTOC is disassembled and replaced by a non-centromeric microtubule network that nucleates from the egg cortex (Fig 1E). This reorganization of the microtubule network is initiated by a signal sent by the posterior follicle cells (Gonzalez-Reyes et al., 1995; Roth et al., 1995). While the nature of this signal is still unknown, it is thought to trigger the targeted degradation of Par-6 and atypical Protein Kinase C complex at the posterior pole by Slmb, the SCF ubiquitin ligase (Morais-de-Sa et al., 2014). This allows recruitment of Par-1 kinase to the posterior pole, which excludes Par-3/Bazooka subdividing the oocyte cortex into an anterior lateral and posterior region (Huynh et al., 2001; Shulman et al., 2000). As a consequence, microtubules nucleate from the anterior and lateral cortex, with a higher microtubule concentration at the anterior, extending plus ends in all directions. While these microtubules mediate both kinesin (+ pole directed) and dynein mediated (-pole directed) transport, osk travels only by kinesin-dependent transport (Brendza et al., 2000; Parton et al., 2011; Zimyanin et al., 2008). Direct live observation of osk RNA transport particles and mathematical modeling suggest that microtubule + ends are oriented with a slight bias towards posterior and that, over time, this bias leads to the accumulation of osk RNA at the posterior pole (Fig 1E, F) (Khuc Trong et al., 2015; Zimyanin et al., 2008).
An isotropic actin meshwork maintains microtubule organization until stage 10B when this meshwork disassembles and microtubules align in parallel bundles close to the cortex of the oocyte and initiate minus-end directed, kinesin dependent ooplasmic streaming (Dahlgaard et al., 2007; Serbus et al., 2005). Ooplasmic streaming along the microtubule bundles mixes nurse cell and oocyte contents that are not yet anchored in the oocyte (Fig 1F). Active nurse cell to oocyte transport ceases and nurse cells ‘dump’ their contents into the oocyte as the actin cytoskeleton within the nurse cells constricts followed by nurse cell death (Figure 1G). While osk RNA localization at the posterior begins well before ooplasmic streaming and nurse cell dumping, green fluorescent protein (GFP) tagging of osk RNA with the MS2/MCP system allowed observation of the localization process live and revealed a 45% increase in osk RNA accumulation in the oocyte after stage 10 (Sinsimer et al., 2011; Snee et al., 2007). This suggests that osk RNA is continuously produced and transported into the oocyte even after the microtubule network-dependent, directed transport into and within the oocyte ceases. Late accumulation and sustained maintenance of osk RNA is mediated by the RNA-binding protein Rump and its associated factor Lost, the actin cytoskeleton, and requires Oskar protein translation (see below) (Fig 1G) (Babu et al., 2004; Sinsimer et al., 2011; Suyama et al., 2009).
Cis-acting sequences and RNA transport particles
Initially all data suggested that the 3′UTR of osk was both necessary and sufficient for posterior localization (Kim-Ha et al., 1993). However, with the identification of osk RNA null mutations, it became clear that additional elements outside the osk 3′UTR are needed for posterior localization. These additional elements were found in a ‘Spliced Oskar Localization Element’ (‘SOLE’), that forms when sequences proximal and distal to the 1st intron fuse during splicing and (Fig 2A, B) (Hachet and Ephrussi, 2004; Simon et al., 2015). The SOLE element (Fig 2C) acts in association with the Exon Junction Complex (EJC) components Mago Nashi, Y14, eIF4AIII, which associate with the 5′splice site in the nucleus of the nurse cells, where osk is transcribed (Ghosh et al., 2012; Hachet and Ephrussi, 2004). Barentz and the dsRNA binding protein Staufen join the complex, possibly via the widened grove formed by the A’ helix in SOLE in the nurse cell cytoplasm to form large transport particles (Macchi et al., 2003; Micklem et al., 2000; Simon et al., 2015; St Johnston et al., 1991; van Eeden et al., 2001) (Fig 2A, B). In these transport particles, osk RNA is thought to self-associate via palindromic sequences, located in the dimerization domain (DD), contained within the distal loop of the OES stem-loop element (Fig 2D) (Jambor et al., 2011). RNA-RNA hybrid formation between palindromic loops apparently enables “hitchhiking” of osk RNA lacking the SOLE element on RNA molecules with intact SOLE elements. This model provides an explanation why constructs containing only the osk 3′UTR localize properly when tested in the background of protein null mutations, which produce stable, spliced RNA (Kim-Ha et al., 1993; Munro et al., 2006). Additional elements besides the DD ‘kissing loop’ are found in other regions of the 3′UTR and play roles in osk RNA localization and anchoring (Kim-Ha et al., 1993).
Curiously, osk RNA travels in a dynein dependent manner into the oocyte after which it depends on kinesin motors to get to the posterior pole. Mutants in EJC components, as well as Tropomyosin and Staufen alter osk RNA transport bias in the oocyte toward the anterior but do not affect dynein-mediated transport into the oocyte (Erdelyi et al., 1995; Micklem et al., 2000; Zimyanin et al., 2008). Thus, splicing and EJC/Tropomysin/Staufen association with osk RNA defines kinesin motor preference of the transport particle. This type of transport seems only necessary within the oocyte and could indeed be a hindrance for efficient dynein-mediated transport from the nurse cells into the oocyte. How such a switch in motor preference is achieved remains to be determined. Since kinesin directed transport is only observed within the oocyte, it is possible that the ‘DD-kissing-loop’, which is located at the distal tip of the stem that mediates dynein-directed oocyte entry, could ‘passively’ pull RNA associated with EJC/SOLE transport complex into the oocyte. Here, abundant kinesin motors could switch the balance. Whether such regulation occurs and if so how other part of the osk 3′UTR known to be required for posterior localization interact with EJC loaded RNA molecules remains to be seen.
Translational control of osk RNA
During transport osk RNA is packaged into transport particles that contain EJC components, the RNA binding proteins Staufen and the translational repressor Bruno. Staufen contains five double stranded RNA binding domains, two of which have been implicated in microtubule dependent transport of osk RNA and in translational activation of osk RNA upon localization at the posterior pole (Micklem et al., 2000). Bruno is a RRM (RNA Recognition Motif) protein that binds to multiple regions in the 3′UTR of osk, termed Bruno response elements (BRE) (Kim-Ha et al., 1995; Snee et al., 2008). Bruno represses translation of osk during transport and prior to its localization to the posterior pole in a process that involves the 5′Cap-binding protein Cup (Nakamura et al., 2004; Wilhelm et al., 2003). Two mechanisms have been proposed to achieve osk translational repression. One mechanism is based on the interaction of Bruno with Cup. In this scenario initiation of translation is inhibited by Cup competing with the 5′UTR Cap-binding protein and translational mediator eIF4E (Kinkelin et al., 2012; Wilhelm et al., 2003). The other model proposes that Bruno, independently of Cup, packages oligomerized osk RNA molecules into large silencing particles (Chekulaeva et al., 2006). Both mechanisms may coexist, the first acting at the level of ribosome engagement, the second consistent with the large transport particles observed in vivo.
Precise control of osk translational activation is critical for germ plasm assembly. During transport, osk should not be translated prematurely as this can lead to patterning defects (Smith et al., 1992). Once osk RNA becomes localized, it is equally important that this translational repression is released, so sufficient quantities of Oskar protein can be synthesized for germ plasm biogenesis. Indeed, overexpression of the osk 3′UTR alone is sufficient to activate translation, suggesting that the concentration of repressors, such as Bruno, is limiting and may be titrated as concentration of osk RNA at the pole increases (Kanke and Macdonald, 2015; Smith et al., 1992). Interestingly, proteins involved in osk transport and translational repression such as Staufen and Bruno have also been implicated in translational activation (Kim et al., 2015; Micklem et al., 2000). One of the three regions with BRE binding sites (C-region) is required for osk translation. A role of Bruno in translational activation may depend on the Bruno phosphorylation state (Reveal et al., 2010; Yoshida et al., 2004). Such a scenario is reminiscent of the mechanism that was shown to release translational repression of actin RNA by phosphorylation of Zip Code Binding protein 1 (ZBP1) at the leading edge of mammalian tissue culture cells (Huttelmaier et al., 2005). Sequences with binding specificity similar to ZBP1, a class of conserved KH (K homology) domain proteins that also includes the insulin growth factor II mRNA–binding protein (IMP), as well as its Xenopus laevis homologue, Vg1 RNA and endoplasmic reticulum–associated protein (VERA)/Vg1 RNA-binding protein, have been identified in the osk 3′UTR and are required for osk transport and translation. While the Drosophila IMP protein binds to these sequences, it is apparently not required for their regulation (Munro et al., 2006). In general, trans-acting sequences mediating osk translation are found in the distal, 3′ end of the 3′UTR, close to the polyadenylation signal. Consistent, proteins involved in polyadenylation such as the cytoplasmic poly-A-binding protein Orb (Castagnetti and Ephrussi, 2003) and Poly-A-binding protein PABP, which binds to A rich sequences (ARS) are required for oskar translation (Fig. 2B) (Vazquez-Pianzola et al., 2011).
A non-coding RNA function for oskar
In addition to its role in Oskar protein synthesis, oskar RNA also has a role as a noncoding RNA (Jenny et al., 2006). This non-coding RNA function maps to the C-terminal end of the oskar 3′UTR and largely overlaps with Bruno and ARS binding sequences (Fig. 2B) (Kanke et al., 2015; Ryu and Macdonald, 2015). osk RNA null mutants show a strikingly different, much earlier oogenesis phenotype than Oskar protein null mutants that seems unrelated to germ plasm organization. In osk RNA null mutants, oogenesis is arrested early during oogenesis and prior to the stage when Oskar protein is first synthesized, compaction of the oocyte nucleus is defective. Consistent with osk RNA sequestering Bruno protein, the phenotype of the non-coding RNA is partially rescued by lowering Bruno levels (Kanke et al., 2015). These results argue against an instructive role of osk noncoding RNA in oogenesis. Rather, they favor the idea that binding sites in the osk 3′UTR sequester Bruno and possibly other osk RNA binding proteins and thereby prevent inappropriate binding of these factors to off-target sites in RNAs with roles in early oogenesis.
Oskar protein structure and function
Once osk RNA reaches the posterior pole it is translated into two protein isoforms: Long Oskar, which is translated at the first Methionine codon and encodes a protein of 606 amino acids (Fig 2E); and short-Oskar which is initiated at the second Methionine codon and encodes a protein of 467 amino acids (Fig 2F) (Markussen et al., 1995; Rongo et al., 1995). Despite one being a N-terminal extension of the other, the two proteins have distinct localization patterns and strikingly different functions. Long Oskar localizes to the oocyte and embryo posterior cortex and is associated with endosomes, while short Oskar is an integral part of the polar granules (Tanaka and Nakamura, 2008; Vanzo et al., 2007). Long Oskar has been implicated in the formation of an extended actin meshwork at the posterior pole and implicated in increased endocytic activity (Tanaka et al., 2011). While interaction partners specific to the N-terminal extension of Long-Oskar have not been identified, loss of long-Oskar causes germ plasm to be less tightly associated with the posterior pole (Rongo et al., 1997; Vanzo and Ephrussi, 2002). This and the fact that long-Oskar can act in trans to increase the localization efficiency of short Oskar suggest that enhanced actin nucleation and endocytosis somehow contribute to the tethering of germ plasm. In support Lasp, an actin binding protein, which interacts with a region of Oskar shared by long and short isoforms, affects oskar RNA localization (Suyama et al., 2009). An Oskar-dependent actin network may also attract additional Par-1 to the posterior pole, which, as part of a positive feedback loop, stabilizes the microtubule network that transports more oskar RNA to the posterior (Doerflinger et al., 2006; Zimyanin et al., 2007). Despite these roles of long-Oskar in stabilizing osk RNA and protein localization its function is dispensable for embryonic patterning and germ cell formation, although fewer form in long-Oskar mutants.
Short Oskar, in contrast, is absolutely required for germ plasm assembly, germ cell formation and normal posterior patterning (Markussen et al., 1995; Rongo et al., 1995; Vanzo and Ephrussi, 2002). Indeed, Oskar short expressed ectopically at the anterior pole is capable of inducing pole cell formation and abdominal patterning at the new location independent of posteriorly produced Oskar (Tanaka and Nakamura, 2008). A number of interaction partners for Short-Oskar have been identified. These include Vasa, an ATP dependent helicase, Valois, a methyltransferase associated WD-repeat protein, and Lasp (Anne, 2010; Breitwieser et al., 1996; Jeske et al., 2015; Suyama et al., 2009). Recently, two studies provided insight into the function of Oskar by determining the structure of two domains in Oskar (Fig 2F): a domain at the very N-terminus of short-Oskar (139–240), which was initially identified by bioinformatics and termed LOTUS domain due to its presence in Limkain, Oskar and Tudor containing proteins 5 and 7; and the C-terminal “OSK” domain, which resembles a SGNH hydrolase or lipase domain found in mammalian platelet-activating factor acetylhydrolase (PAF-AH) and GDSL-like lipases (Jeske et al., 2015; Yang et al., 2015). The studies found that the LOTUS domain forms a winged-helix-turn-helix structure with three-to-four alpha helices (depending on the size of the fragment used for crystallization) as wings emerging from two antiparallel beta strands. In contrast to other winged-helix structures, the Osk-N-terminal LOTUS domain homodimerizes masking the helices that in other winged-helix proteins bind DNA. The Oskar-LOTUS domain binds to the RNA helicase Vasa an important component of germ plasm. The C-terminal ‘OSK’ domain forms a globular structure composed of central beta sheets surrounded by alpha helices with extended loops. While the structure of this domain resembles lipase-fold structures, three of the four residues required for enzymatic activity are missing, making it highly unlikely that the OSK domain functions as a hydrolase (Jeske et al., 2015; Yang et al., 2015). Instead, the structure reveals a number of basic, positively charged residues at its surface suggesting chemical property more typical for RNA binding proteins. Indeed, in vitro experiments suggest that this domain can bind to RNA and demonstrate specific interaction with the osk and nos 3′UTRs that can be disrupted by mutations in positively charged amino acids in the LOTUS domain (Yang et al., 2015). In support, these key residues are in proximity to genetically identified point mutations in Osk that strongly affect posterior pattern and germ plasm assembly. Furthermore, in vivo pull-down experiments from embryo extracts after UV-crosslinking, which stabilizes RNA-protein interactions, revealed significant interactions between Osk and nos, pgc and to a lesser extent gcl RNA in vivo (Jeske et al., 2015). All three RNAs are known to be localized to the germ plasm.
Assembly of germ plasm
Germ plasm assembly initiated by Oskar requires the function of Vasa and Tudor. While homologs of these two proteins are widely conserved as universal germ cell markers, oskar is a novel gene that evolved within the order of insects (Ewen-Campen et al., 2012). Like osk, females mutant for vasa or tudor produce embryos that lack germ cells and fail to form an abdomen (Boswell and Mahowald, 1985; Dehghani and Lasko, 2015). Vasa encodes a DEAD-box RNA helicase and has been proposed to be involved in translational activation of RNA localized to the posterior pole (Lasko and Ashburner, 1988). However, while Vasa binds the translation factor eIF5E, a direct role of Vasa in binding to or regulating the translation of germ plasm localized RNAs is missing (Johnstone and Lasko, 2004). In addition to its role in germ plasm assembly Vasa also plays an important role in piRNA biogenesis, likely as part of the piRNA processing machinery that is assembled in the nuage of the nurse cells (for reviews on piRNA processing see: (Malone and Hannon, 2009; Pek et al., 2012; Siomi et al., 2011)). Tudor is the founding member of the Tudor domain family of proteins (Golumbeski et al., 1991). Tudor encodes a large protein of 2515 amino acids that contains 11 copies of the Tudor domain motif (Arkov et al., 2006). Structural analysis of a single Tudor domain revealed that the core Tud barrel-like structure, consisting of four β strands that forms an aromatic cage, is extended N-and C-terminal helices, folded into an oligonucleotide binding (OB) fold, commonly called the extended Tudor domain. The ~ 120 aminoacid eTud domain is predicted to be the common structural motif found in germ line Tudor proteins. Tudor domain 11 specifically recognizes symmetrically dimethylated Arginines (sDMA) within the Piwi protein Aubergine (Liu et al., 2010). Point mutations in Tudor that abolish sDMA peptide binding or mutations of the corresponding arginines in Aubergine abolish Aubergine localization to the germ plasm (Liu et al., 2010; Webster et al., 2015). With its many domains it is likely that Tudor acts as a scaffold for germ plasm assembly by interacting with sDMA modified target proteins (Figure 3A) (Ren et al., 2014; Thomson and Lasko, 2004). sDMA modification of target proteins could be achieved specifically at the posterior pole through the localization of the sDMA methyltransferase Capsuleen and its associated protein Valois to the germ plasm, possibly via direct interaction of Valois with Oskar (Anne et al., 2007; Gonsalvez et al., 2006). Another example is Pyruvate kinase (Pyk), which contains sDMAs modification, localizes to polar granules and interacts with Tudor (Thomson et al., 2008). Interestingly, not only Pyk but several enzymes in the glycolytic pathway localize specifically to the germ plasm and affect germ cell formation (Gao et al., 2015). It has therefore been suggested that Tudor may recruit glycolytic enzymes to the germ plasm to provide a local source of ATP for helicases like Vasa. Taken together, sDMA modification-mediated binding of proteins to the Tudor scaffold could lead to a robust network assembly of proteins in the germ plasm (Fig 3A, B).
Effector RNA localization
More than 100 different, maternally synthesized mRNAs have been reported by in situ hybridization to be localized to the germ plasm (Jambor et al., 2015; Lecuyer et al., 2007). For only a few of these have their function in germ cell biology been demonstrated. The best-studied effector RNA is nanos. Nanos encodes a Zinc-finger protein that together with the RNA-binding protein Pumilio acts as a translational repressor (Curtis et al., 1997). Targets of Nos/Pum mediated translational repression include the maternally provided hunchback (hb) RNAs (Sonoda and Wharton, 1999). Elimination of hb RNA from the posterior domain is necessary for normal abdomen formation (Irish et al., 1989; Struhl, 1989). Nos and Pum also affect early germ cell development, one of the target for this function is CyclinB RNA, which is localized to the germ plasm but translationally repressed due to Nos and Pum function (Hayashi et al., 2004; Kadyrova et al., 2007). PGCs do not divide until this translational block is relieved at the end of embryogenesis. Other targets include the cell death-promoting gene hid and yet unidentified targets that mediate defects in germ cell migration, PGC survival and PGC chromatin remodeling (Sato et al., 2007). Other germ granule localized RNAs whose functions are known include germ cell-less, which encodes a BTB protein that is required for the formation of PGCs (Cinalli and Lehmann, 2013; Jongens et al., 1992), and polar granule component, which encodes a small protein that inhibits transcription in early germ cells by preventing the recruitment P-TEFb kinase complex to chromatin. (Hanyu-Nakamura et al., 2008; Martinho et al., 2004; Nakamura et al., 1996)
The localization efficiency for effector RNAs is low and estimated at 2.5–4% depending on the RNA studied (Bergsten and Gavis, 1999; Little et al., 2015; Trcek et al., 2015). A more detailed understanding of how posteriorly localized RNAs reach the germ plasm has come from in vivo studies using GFP and the MS2/MCP RNA labeling system. These studies revealed that nanos RNAs reaches the germ plasm during the late stage of oogenesis starting during cytoplasmic streaming and continuing throughout nurse cell dumping (Forrest and Gavis, 2003; Ganguly et al., 2012). Once at the posterior pole, persistent microtubule dependent trafficking keeps RNAs associated with the germ plasm (Sinsimer et al., 2013). These observations have led to a model by which RNAs destined for posterior localization have an affinity for the posterior germplasm and are trapped by this specialized cytoplasm (Fig. 3).
The localization patterns of other RNAs, such as CycB, gcl and pgc seem to follow similar patterns (Little et al., 2015; Trcek et al., 2015). In all cases studied, the 3′UTR was sufficient to recapitulate the posterior RNA localization pattern (Rangan et al., 2009). Single molecule analysis of nos, cyclinB, pgc and gcl RNA has revealed that RNAs first arrive in the germ plasm as single molecules and then organize into larger RNA particles associated with polar granules (Fig 1A, 3B) (Little et al., 2015; Trcek et al., 2015). Structural illumination microscopy, a super resolution method, further showed that RNAs form homotypic clusters within polar granules and that these RNA clusters have unique positions within the granule, with nos and cycB RNA more towards the center of the granule and pgc and gcl tend to be more peripheral (Fig 3C, C’, E) (Trcek et al., 2015). While large osk RNA clusters are found in the germ plasm, these do not associate with polar granules (Fig 1G) (Little et al., 2015; Trcek et al., 2015; Vanzo et al., 2007). It is presently unclear how this organization is achieved as protein components of the germ granule, such as Vasa, Oskar, Tudor and Aubergine show uniform distribution throughout the granule (Fig 3D) (Little et al., 2015; Trcek et al., 2015). One intriguing possibility is that RNAs self-associate once they are localized. Indeed, the concentration of effector RNAs in the germ plasm is about 8–12 fold higher than their concentration in the rest of the embryo (Little et al., 2015; Trcek et al., 2015). This increase in concentration depends on the germ plasm proteins. RNA binding proteins have been shown to undergo phase transitions from a soluble to viscous state dependent on their concentration and association with RNA (Brangwynne, 2013; Lin et al., 2015; Zhang et al., 2015). Thus, RNAs may be trapped by germ plasm aggregates, which create specific, homotypic RNA clusters due to RNA-protein as well as RNA-RNA recognition (Fig 3B). Such an organization could provide an efficient mechanism to organize a large number of localized RNAs in an RNA-specific manner.
Like osk, other germ plasm RNAs are also translationally silent during transport (Gavis and Lehmann, 1994). 3′UTR sequences from different effector RNAs fused to GFP reporter transcripts showed that the respective 3′UTRs are sufficient to recapitulate the posterior RNA localization pattern, to mediate translational of unlocalized RNA and translationly de-repress upon localization (Rangan et al., 2011). For nos RNA, translational repression is achieved through cis-acting sequences in the nos 3′UTR and binding of the hnRNP protein Glorund during oogenesis and the SAM domain protein Smaug together with Argonaute 1 during embryogenesis (Andrews et al., 2011; Crucs et al., 2000; Gavis, 1995; Kalifa et al., 2006; Pinder and Smibert, 2013; Smibert et al., 1996). The onset of translation is strikingly different depending on the effector RNA: nos RNA is translated as soon as it becomes localized to the posterior pole, gcl is translated as nuclei reach the embryo cortex, pgc is translated when germ cells form and cycB is not translated until the end of embryogenesis (Clark et al., 2000; Dalby and Glover, 1993; Hanyu-Nakamura et al., 2008; Jongens et al., 1994). Curiously, the position of a specific RNA cluster with respect to the germ granule seems not to correlate with the timing of translational de-repression. Indeed, nos RNA, which is located in the center of the granule is translated first, while pgc and gcl, located more peripherally are translated later (Trcek et al., 2015). This is in contrast with the organization of RNA in sponge bodies, another large RNA-protein granule found at the anterior of oocytes. Here translational activity is correlated with position, as translationally active gurken RNA is located more peripherally, and quiescent bicoid RNA is positioned in the center of the sponge body (Delanoue et al., 2007). This type of regulation has also been proposed for stress granules and P bodies. While polar granules share RNA decay and translational regulators with sponge bodies, stress granules and P bodies, the access of specific RNAs to RNA regulators cannot be controlled by position within the granule alone. Thus, it remains unclear how RNA clusters are organized with respect to each other, how they control their position within the germ plasm and what controls the timing of their translation.
Summary and Outlook
Regulation of osk exemplifies how the nuclear history of an RNA and cell polarity control RNA localization and subcellular translation. Osk protein has multiple roles in the assembly and anchoring of germ plasm components at the egg cortex. In contrast to Vasa, Tudor and Nanos, Osk is not a conserved components of germ plasm. Interestingly in other species, seemingly unrelated proteins, like Bucky ball in fish and the Pgl proteins in C. elegans seem to play similar roles in the initial steps of germ granule assembly (Bontems et al., 2009; Hanazawa et al., 2011). With the emerging concept that phase transitions and proteins aggregates play an important role in RNA granule assembly and biology, these functional similarities may be grounded in common biophysical properties.
Electron-dense granules of the germ line share many components that are also common to other cytoplasmic, membrane-less granules like P bodies and stress granules. However, certain proteins are specific identifiers for granule classes. While Osk is critical for the assembly of germ plasm, it is not needed or present in the nuage, which requires Vasa and a new set of Tudor proteins as well as the Argonaute proteins Ago3 and Aubergine. Similarly, sponge bodies rely on the Exuperantia protein (Wilsch-Brauninger et al., 1997). While all germ line granules are involved in the regulation of RNA, the processes controlled are strikingly different. Nuage is required for the processing of small RNAs involved in the recognition and defense against transposable elements and sponge bodies have a major role in the localization of RNA and possibly secretion of signaling factors. Germ plasm regulates the translation of RNAs destined for the germ line of the next generation. How protein composition is determined and whether RNAs are sorted between these compartments and whether there is direct exchange between specific components like Vasa, Tudor or Aubergine, is presently unclear. The Tudor class of proteins may play a critical role here whereby individual Tudor domains recognize and bind proteins depending on their modification.
Phase transition from a single molecule, liquid composition throughout the cell to an Oskar-dependent increase in concentration of specific germ plasm proteins and effector RNAs creates a steep transition between aggregated germ plasm at the posterior pole and the rest of the embryo. This transition may facilitate homotypic RNA cluster formation but how within these clusters micro-regulating domains are established remains to be determined.
Acknowledgments
My special thanks go to Dr. Alexey Soshnev for generating the figures. I thank members of my lab for discussion and particulalry Drs. Thomas Hurd and Tatjana Trcek for comments on the manuscript and images in Figure 3. Paul Wasserman’s his encouragement and patience were critical in completing this review. Due to space limitation, I apologize for neglecting to cite all pertinent literature. Work in our lab on germ cells is supported by NIH R01/R37HD41900 and RL is a HHMI investigator.
References
- Andrews S, Snowflack DR, Clark IE, Gavis ER. RNA. 2011;17:967–77. doi: 10.1261/rna.2478611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne J. PLoS One. 2010;5:e14362. doi: 10.1371/journal.pone.0014362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM. Development. 2007;134:137–46. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- Arkov AL, Wang JY, Ramos A, Lehmann R. Development. 2006;133:4053–62. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Nat Cell Biol. 1999;1:431–7. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- Babu K, Cai Y, Bahri S, Yang X, Chia W. Genes Dev. 2004;18:138–43. doi: 10.1101/gad.282604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DD, Wang C, Moore J, Dickinson LK, Lehmann R. Genes Dev. 1992;6:2312–26. doi: 10.1101/gad.6.12a.2312. [DOI] [PubMed] [Google Scholar]
- Bergsten SE, Gavis ER. Development. 1999;126:659–69. doi: 10.1242/dev.126.4.659. [DOI] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, Dosch R. Curr Biol. 2009;19:414–22. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Boswell RE, Mahowald AP. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP. J Cell Biol. 2013;203:875–81. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitwieser W, Markussen FH, Horstmann H, Ephrussi A. Genes Dev. 1996;10:2179–88. doi: 10.1101/gad.10.17.2179. [DOI] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Duffy JB, Saxton WM. Science. 2000;289:2120–2. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnetti S, Ephrussi A. Development. 2003;130:835–43. doi: 10.1242/dev.00309. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. Cell. 2006;124:521–33. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Cinalli RM, Lehmann R. Nat Cell Biol. 2013;15:839–45. doi: 10.1038/ncb2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Wyckoff D, Gavis ER. Curr Biol. 2000;10:1311–4. doi: 10.1016/s0960-9822(00)00754-5. [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. Development. 2003;130:1579–90. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- Crucs S, Chatterjee S, Gavis ER. Mol Cell. 2000;5:457–67. doi: 10.1016/s1097-2765(00)80440-2. [DOI] [PubMed] [Google Scholar]
- Curtis D, Treiber DK, Tao F, Zamore PD, Williamson JR, Lehmann R. EMBO J. 1997;16:834–43. doi: 10.1093/emboj/16.4.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Dev Cell. 2007;13:539–53. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby B, Glover DM. EMBO J. 1993;12:1219–27. doi: 10.1002/j.1460-2075.1993.tb05763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani M, Lasko P. Biol Open. 2015;4:450–62. doi: 10.1242/bio.201410579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Dev Cell. 2007;13:523–38. doi: 10.1016/j.devcel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Dienstbier M, Boehl F, Li X, Bullock SL. Genes Dev. 2009;23:1546–58. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger H, Benton R, Torres IL, Zwart MF, St Johnston D. Curr Biol. 2006;16:1090–5. doi: 10.1016/j.cub.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R. Nature. 1992;358:387–92. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Erdelyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A. Nature. 1995;377:524–7. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Srouji JR, Schwager EE, Extavour CG. Curr Biol. 2012;22:2278–83. doi: 10.1016/j.cub.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER. Curr Biol. 2003;13:1159–68. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Ganguly S, Williams LS, Palacios IM, Goldstein RE. Proc Natl Acad Sci U S A. 2012;109:15109–14. doi: 10.1073/pnas.1203575109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Arkov AL. Mol Reprod Dev. 2013;80:610–23. doi: 10.1002/mrd.22115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Thomson TC, Creed TM, Tu S, Loganathan SN, Jackson CA, McCluskey P, Lin Y, Collier SE, Weng Z, Lasko P, Ohi MD, Arkov AL. EMBO Rep. 2015;16:379–86. doi: 10.15252/embr.201439694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis ER. Curr Biol. 1995;5:1252–4. doi: 10.1016/s0960-9822(95)00250-8. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R. Nature. 1994;369:315–8. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Marchand V, Gaspar I, Ephrussi A. Nat Struct Mol Biol. 2012;19:441–9. doi: 10.1038/nsmb.2257. [DOI] [PubMed] [Google Scholar]
- Golumbeski GS, Bardsley A, Tax F, Boswell RE. Genes Dev. 1991;5:2060–70. doi: 10.1101/gad.5.11.2060. [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG. Curr Biol. 2006;16:1077–89. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Elliott H, St Johnston D. Nature. 1995;375:654–8. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Nature. 2004;428:959–63. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Hanazawa M, Yonetani M, Sugimoto A. J Cell Biol. 2011;192:929–37. doi: 10.1083/jcb.201010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Nature. 2008;451:730–3. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, Kobayashi S. Proc Natl Acad Sci U S A. 2004;101:10338–42. doi: 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Nature. 2005;438:512–5. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Huynh JR, Petronczki M, Knoblich JA, St Johnston D. Curr Biol. 2001;11:901–6. doi: 10.1016/s0960-9822(01)00244-5. [DOI] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP. Proc Natl Acad Sci U S A. 1974;71:1016–20. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP, Loomis MR. Dev Biol. 1976;49:40–65. doi: 10.1016/0012-1606(76)90257-8. [DOI] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. Nature. 1989;338:646–8. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- Jambor H, Brunel C, Ephrussi A. RNA. 2011;17:2049–57. doi: 10.1261/rna.2686411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Mueller S, Bullock SL, Ephrussi A. RNA. 2014;20:429–39. doi: 10.1261/rna.041566.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. Elife. 2015:4. doi: 10.7554/eLife.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Hachet O, Zavorszky P, Cyrklaff A, Weston MD, Johnston DS, Erdelyi M, Ephrussi A. Development. 2006;133:2827–33. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- Jeske M, Bordi M, Glatt S, Muller S, Rybin V, Muller CW, Ephrussi A. Cell Rep. 2015;12:587–98. doi: 10.1016/j.celrep.2015.06.055. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Development. 2004;131:4167–78. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- Jongens TA, Ackerman LD, Swedlow JR, Jan LY, Jan YN. Genes Dev. 1994;8:2123–36. doi: 10.1101/gad.8.18.2123. [DOI] [PubMed] [Google Scholar]
- Jongens TA, Hay B, Jan LY, Jan YN. Cell. 1992;70:569–84. doi: 10.1016/0092-8674(92)90427-e. [DOI] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP. Development. 2007;134:1519–27. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER. Dev Cell. 2006;10:291–301. doi: 10.1016/j.devcel.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Kanke M, Jambor H, Reich J, Marches B, Gstir R, Ryu YH, Ephrussi A, Macdonald PM. RNA. 2015;21:1096–109. doi: 10.1261/rna.048298.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M, Macdonald PM. PLoS One. 2015;10:e0125849. doi: 10.1371/journal.pone.0125849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuc Trong P, Doerflinger H, Dunkel J, St Johnston D, Goldstein RE. Elife. 2015:4. doi: 10.7554/eLife.06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Pai CI, Sato K, Person MD, Nakamura A, Macdonald PM. PLoS Genet. 2015;11:e1004992. doi: 10.1371/journal.pgen.1004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. Cell. 1995;81:403–12. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Webster PJ, Smith JL, Macdonald PM. Development. 1993;119:169–78. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- Kinkelin K, Veith K, Grunwald M, Bono F. RNA. 2012;18:1624–34. doi: 10.1261/rna.033639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z. J Biol Chem. 2010a;285:8148–54. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z. RNA. 2010b;16:70–8. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T. Nature. 1996;380:708–11. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. Nature. 1988;335:611–7. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Cell. 2007;131:174–87. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. Cell. 1986;47:141–52. doi: 10.1016/0092-8674(86)90375-2. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. Development. 1991;112:679–91. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Mol Cell. 2015;60:208–19. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Sinsimer KS, Lee JJ, Wieschaus EF, Gavis ER. Nat Cell Biol. 2015;17:558–68. doi: 10.1038/ncb3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. Genes Dev. 2010;24:1876–81. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P, Kroening S, Palacios IM, Baldassa S, Grunewald B, Ambrosino C, Goetze B, Lupas A, St Johnston D, Kiebler M. J Neurosci. 2003;23:5778–88. doi: 10.1523/JNEUROSCI.23-13-05778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach JM, Lehmann R. Genes Dev. 1997;11:423–35. doi: 10.1101/gad.11.4.423. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. Int Rev Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markussen FH, Michon AM, Breitwieser W, Ephrussi A. Development. 1995;121:3723–32. doi: 10.1242/dev.121.11.3723. [DOI] [PubMed] [Google Scholar]
- Martinho RG, Kunwar PS, Casanova J, Lehmann R. Curr Biol. 2004;14:159–65. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- Micklem DR, Adams J, Grunert S, St Johnston D. EMBO J. 2000;19:1366–77. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais-de-Sa E, Mukherjee A, Lowe N, St Johnston D. Development. 2014;141:2984–92. doi: 10.1242/dev.109827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro TP, Kwon S, Schnapp BJ, St Johnston D. J Cell Biol. 2006;172:577–88. doi: 10.1083/jcb.200510044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Science. 1996;274:2075–9. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Parton RM, Hamilton RS, Ball G, Yang L, Cullen CF, Lu W, Ohkura H, Davis I. J Cell Biol. 2011;194:121–35. doi: 10.1083/jcb.201103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Patil VS, Kai T. Dev Growth Differ. 2012;54:66–77. doi: 10.1111/j.1440-169x.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- Pinder BD, Smibert CA. EMBO Rep. 2013;14:80–6. doi: 10.1038/embor.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran B, Bopp R, Suter B. Development. 1994;120:1233–42. doi: 10.1242/dev.120.5.1233. [DOI] [PubMed] [Google Scholar]
- Rangan P, DeGennaro M, Jaime-Bustamante K, Coux RX, Martinho RG, Lehmann R. Curr Biol. 2009;19:72–7. doi: 10.1016/j.cub.2008.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. Curr Biol. 2011;21:1373–9. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Liu H, Wang W, Wang M, Yang N, Dong YH, Gong W, Lehmann R, Xu RM. Cell Res. 2014;24:1146–9. doi: 10.1038/cr.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveal B, Yan N, Snee MJ, Pai CI, Gim Y, Macdonald PM. Dev Cell. 2010;18:496–502. doi: 10.1016/j.devcel.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Broihier HT, Moore L, Van Doren M, Forbes A, Lehmann R. Cold Spring Harb Symp Quant Biol. 1997;62:1–11. [PubMed] [Google Scholar]
- Rongo C, Gavis ER, Lehmann R. Development. 1995;121:2737–46. doi: 10.1242/dev.121.9.2737. [DOI] [PubMed] [Google Scholar]
- Roth S, Lynch JA. Cold Spring Harb Perspect Biol. 2009;1:a001891. doi: 10.1101/cshperspect.a001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schupbach T. Cell. 1995;81:967–78. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Ryu YH, Macdonald PM. Dev Biol. 2015 doi: 10.1016/j.ydbio.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander K. Ciba Found Symp. 1975;0:241–63. doi: 10.1002/9780470720110.ch12. [DOI] [PubMed] [Google Scholar]
- Sato K, Hayashi Y, Ninomiya Y, Shigenobu S, Arita K, Mukai M, Kobayashi S. Proc Natl Acad Sci U S A. 2007;104:7455–60. doi: 10.1073/pnas.0610052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Genetics. 1989;121:101–17. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serano TL, Cohen RS. Development. 1995;121:3809–18. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- Serbus LR, Cha BJ, Theurkauf WE, Saxton WM. Development. 2005;132:3743–52. doi: 10.1242/dev.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, Benton R, St Johnston D. Cell. 2000;101:377–88. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Simon B, Masiewicz P, Ephrussi A, Carlomagno T. RNA. 2015;21:1444–53. doi: 10.1261/rna.049601.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsimer KS, Jain RA, Chatterjee S, Gavis ER. Development. 2011;138:3431–40. doi: 10.1242/dev.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsimer KS, Lee JJ, Thiberge SY, Gavis ER. Cell Rep. 2013;5:1169–77. doi: 10.1016/j.celrep.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM. Genes Dev. 1996;10:2600–9. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Smith JL, Wilson JE, Macdonald PM. Cell. 1992;70:849–59. doi: 10.1016/0092-8674(92)90318-7. [DOI] [PubMed] [Google Scholar]
- Snee M, Benz D, Jen J, Macdonald PM. RNA Biol. 2008;5:1–9. [PubMed] [Google Scholar]
- Snee MJ, Harrison D, Yan N, Macdonald PM. Differentiation. 2007;75:246–55. doi: 10.1111/j.1432-0436.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM. Dev Dyn. 2009;238:918–30. doi: 10.1002/dvdy.21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Genes Dev. 1999;13:2704–12. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, de Cuevas M, Drummond-Barbosa D, Keyes L, Lilly M, Pepling M, Xie T. Cold Spring Harbor symposia on quantitative biology. 1997;62:25–34. [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Struhl G. Nature. 1989;338:741–4. doi: 10.1038/338741a0. [DOI] [PubMed] [Google Scholar]
- Struhl G, Johnston P, Lawrence PA. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- Suyama R, Jenny A, Curado S, Pellis-van Berkel W, Ephrussi A. Development. 2009;136:95–105. doi: 10.1242/dev.027698. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kato Y, Matsuda K, Hanyu-Nakamura K, Nakamura A. Development. 2011;138:2523–32. doi: 10.1242/dev.062208. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nakamura A. Development. 2008;135:1107–17. doi: 10.1242/dev.017293. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Genesis. 2004;40:164–70. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Mech Dev. 2008;125:865–73. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. Nat Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St Johnston D. J Cell Biol. 2001;154:511–23. doi: 10.1083/jcb.200105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo N, Oprins A, Xanthakis D, Ephrussi A, Rabouille C. Dev Cell. 2007;12:543–55. doi: 10.1016/j.devcel.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Vanzo NF, Ephrussi A. Development. 2002;129:3705–14. doi: 10.1242/dev.129.15.3705. [DOI] [PubMed] [Google Scholar]
- Vazquez-Pianzola P, Urlaub H, Suter B. Dev Biol. 2011;357:404–18. doi: 10.1016/j.ydbio.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster A, Li S, Hur JK, Wachsmuth M, Bois JS, Perkins EM, Patel DJ, Aravin AA. Mol Cell. 2015;59:564–75. doi: 10.1016/j.molcel.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. J Cell Biol. 2003;163:1197–204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsch-Brauninger M, Schwarz H, Nusslein-Volhard C. J Cell Biol. 1997;139:817–29. doi: 10.1083/jcb.139.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Yu Z, Hu M, Wang M, Lehmann R, Xu RM. Proc Natl Acad Sci U S A. 2015;112:11541–6. doi: 10.1073/pnas.1515568112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Muller HA, Wodarz A, Ephrussi A. Development. 2004;131:1401–10. doi: 10.1242/dev.01034. [DOI] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. Mol Cell. 2015;60:220–30. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin V, Lowe N, St Johnston D. Curr Biol. 2007;17:353–9. doi: 10.1016/j.cub.2006.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. Cell. 2008;134:843–53. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]