ABSTRACT
Cryptococcus neoformans is a major opportunistic fungal pathogen that causes fatal meningoencephalitis in immunocompromised individuals and is responsible for a large proportion of AIDS-related deaths. The fungal cell wall is an essential organelle which undergoes constant modification during various stages of growth and is critical for fungal pathogenesis. One critical component of the fungal cell wall is chitin, which in C. neoformans is predominantly deacetylated to chitosan. We previously reported that three chitin deacetylase (CDA) genes have to be deleted to generate a chitosan-deficient C. neoformans strain. This cda1Δ2Δ3Δ strain was avirulent in mice, as it was rapidly cleared from the lungs of infected mice. Here, we report that clearance of the cda1Δ2Δ3Δ strain was associated with sharply spiked concentrations of proinflammatory molecules that are known to be critical mediators of the orchestration of a protective Th1-type adaptive immune response. This was followed by the selective enrichment of the Th1-type T cell population in the cda1Δ2Δ3Δ strain-infected mouse lung. Importantly, this response resulted in the development of robust protective immunity to a subsequent lethal challenge with a virulent wild-type C. neoformans strain. Moreover, protective immunity was also induced in mice vaccinated with heat-killed cda1Δ2Δ3Δ cells and was effective in multiple mouse strains. The results presented here provide a strong framework to develop the cda1Δ2Δ3Δ strain as a potential vaccine candidate for C. neoformans infection.
IMPORTANCE
The most commonly used anticryptococcal therapies include amphotericin B, 5-fluorocytosine, and fluconazole alone or in combination. Major drawbacks of these treatment options are their limited efficacy, poor availability in limited resource areas, and potential toxicity. The development of antifungal vaccines and immune-based therapeutic interventions is promising and an attractive alternative to chemotherapeutics. Currently, there are no fungal vaccines in clinical use. This is the first report of a C. neoformans deletion strain with an avirulent phenotype in mice exhibiting protective immunity when used as a vaccine after heat inactivation, although other strains that overexpress fungal or murine proteins have recently been shown to induce a protective response. The data presented here demonstrate the potential for developing the avirulent cda1Δ2Δ3Δ strain into a vaccine-based therapy to treat C. neoformans infection.
INTRODUCTION
Cryptococcal meningitis is the most frequent result of Cryptococcus neoformans infection of the central nervous system, observed mainly in patients with AIDS. Worldwide, it has been estimated that cryptococcal meningitis accounts for more than 1 million cases with about 625,000 deaths annually (1). Even though infections due to C. neoformans are more common, Cryptococcus gattii is emerging as an important fungal pathogen with significant virulence, wide-spread environmental prevalence, and the ability to cause infections even in immunocompetent individuals (2, 3). The anticryptococcal treatment regimen of choice consists of a combination of amphotericin B and 5-fluorocytosine. Unfortunately, this combination can have substantial toxicity and is not available in much of the developing world, where most cases are seen (4, 5). In some regions, fluconazole is widely used as an alternate to amphotericin B. However, it is not as effective, and there are reports of strains of C. neoformans that have developed resistance to these drugs (6). Although echinocandins are effective for treating other fungal infections, they are ineffective against C. neoformans infections. Therefore, there is an urgent need for the development of safe and effective treatment strategies against cryptococcal infections. The development of vaccine-based immunotherapeutics is an attractive alternative for controlling cryptococcal infections.
Upon entering the host, Cryptococcus is initially challenged by the complement system and the phagocytic activity of different innate immune cells. Innate defense is specifically triggered by the recognition of the pathogen by pattern recognition receptors (PRRs) on the surface of immune cells. C. neoformans is able to modulate host immune responses through a combination of its polysaccharide capsule- and cell wall-associated mannans, mannoproteins, glucans, and chitin. The adaptive immune response against Cryptococcus includes both antibody- and cell-mediated responses. Effective cross talk between the innate and adaptive arms of the immune system is critical for the defense against the pathogen and the resolution of the fungal infection (7–9). It is well established that cell-mediated immunity (CMI) plays a critical role in anticryptococcal defense, as is evident from the higher prevalence of cryptococcal infections in immunocompromised patients (1). This is recapitulated in animal models of cryptococcosis, where either immunodeficient transgenic mice or mice that are depleted of CD4+ and/or CD8+ T cells succumb to cryptococcal infection more rapidly than immunocompetent mice (10, 11). To further support the importance of an adaptive response, several studies have demonstrated a role for humoral immunity in contributing to host protection against experimental cryptococcal infections (12). Antigens demonstrated to induce partial protective immunity include glucuronoxylomannan (GXM), which is a component of the cryptococcal capsule, peptide mimotopes of GXM, complex mixtures of cell surface mannoproteins, and melanin (13–16). Moreover, fungal β-glucan particles have been exploited as an adjuvant and, also, as a vaccine delivery system due to their ability to stimulate dendritic cells to secrete cytokines that mediate beneficial host immune responses and to cause robust stimulation of T cells in murine vaccine models (17, 18).
Evidence that a strain of C. neoformans could confer complete protective immunity came from studies of a C. neoformans H99 strain that was engineered to express and secrete murine gamma interferon (IFN-γ). In this case, a protective response was mediated primarily by a Th1-type T cell immune response, without the contribution of a B-cell mediated processes (19–21). Vaccination of mice with IFN-γ-expressing C. neoformans (strain H99γ) resulted in significant increases in the levels of Th1-type proinflammatory cytokines and chemokines in the lung, with concomitant decreases in the levels of cytokines that are mediators of Th2-type anti-inflammatory activities. This polarized Th1 immunogenic response was associated with clearance of the H99γ cells used for vaccination and, in turn, induced protective immunity against a lethal pulmonary infection with wild-type C. neoformans (19). While the H99γ strain described in the above-mentioned study exploited an inflammatory cytokine of the host to induce protective immunity, a strain of C. neoformans in which the SLG1 gene was deleted was recently reported to induce protective immunity in mice both under immunocompetent and experimentally induced immunocompromised conditions (22). SGL1 encodes a sterylglucosidase that when deleted results in the accumulation of sterylglucosides, which in turn modulates immune responses (23). Therefore, protective immunity developed during infection with an sgl1Δ strain is shown to be triggered by the C. neoformans-derived sterylglucosides (22).
The cryptococcal cell wall is essential for survival, and its components are critical for mediating host-pathogen interactions. The modified fungal cell wall architecture in a mutant strain may provoke a different host immune response than its wild-type counterpart during infection. This was recently demonstrated by a C. neoformans strain engineered to overexpress Znf2, a master regulator of hyphal development. The A/J mouse strain vaccinated with the Znf2-overexpressing strain showed an altered host immune response that resulted in the induction of protective immunity, even when the strain was heat killed (HK) (24).
The C. neoformans cell wall differs from those of other pathogenic fungi by the presence of a polysaccharide capsule. In addition, we have previously discovered that, unlike other yeasts, the chitin fiber in the cell wall of C. neoformans predominantly exists in its deacetylated chitosan form under laboratory growth conditions and in the infected mouse lung (25). We found that deletion of three C. neoformans chitin deacetylase genes, CDA1, CDA2, and CDA3, was sufficient to render the yeast cells devoid of chitosan. The cda1Δ2Δ3Δ strain was avirulent in an inhalation infection model in mice and was rapidly cleared from the lungs of infected mice (25). The inability of the mutant yeast cells to deacetylate chitin results in compromised cell wall integrity, as revealed by their increased sensitivity to cell wall stressors and altered melanin phenotype (26). Therefore, we hypothesized that chitosan-deficient yeast cells trigger an altered host immune response compared to the host response to the wild type, which in turn is responsible for their efficient eradication from the host lung. In the present study, we extend our observation of rapid mutant fungal clearance and show that the clearance of the chitosan-deficient cda1Δ2Δ3Δ strain was associated with a strong induction of the proinflammatory immune response which was able to specifically attract Th1-type adaptive T cells to the site of infection. We further demonstrate that pulmonary vaccination with the chitosan-deficient strain was able to stimulate a robust protective immunity to a subsequent lethal challenge with a virulent wild-type C. neoformans strain in multiple mouse strains. We also report here that heat-killed cells of the cda1Δ2Δ3Δ strain were able to induce effective protective immunity to a pulmonary challenge with wild-type C. neoformans. The observation that viability of the mutant strain is not required to elicit protective immunity indicates a means to an even-safer vaccine.
RESULTS
Chitosan-deficient cda1Δ2Δ3Δ C. neoformans strain was rapidly cleared from the infected mouse lung.
We previously reported that the chitosan-deficient cda1Δ2Δ3Δ strain was avirulent in CBA/J mice when an inhalation mode of infection was used. This avirulent phenotype was associated with the rapid clearance of the mutant strain from the infected lung when 105 yeast cells were used for infection (25). To characterize the host factors potentially responsible for rapidly clearing the cda1Δ2Δ3Δ mutant, we first determined whether the ability of the host to eradicate fungal cells depends on the number of yeast cells used for inoculation. Therefore, we infected CBA/J mice with 105, 106, and 107 CFU of the wild-type, KN99α, and cda1Δ2Δ3Δ mutant strains. At 1, 3, and 7 days postinfection (p.i.), lungs were excised from the infected mice and the fungal burdens were determined. Mice infected with 105 CFU of the cda1Δ2Δ3Δ strain completely eliminated live fungal cells by day 3 p.i. (Fig. 1). Mice inoculated with 106 CFU of the cda1Δ2Δ3Δ mutant had cleared more than 99% of the initial inoculum by day 1 p.i., with complete removal by day 7 p.i. (Fig. 1). Mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant, however, took significantly longer to clear the yeast cells. At day 1 p.i., 2 × 106 CFU were recovered, and at day 7 p.i., there were still ~5,000 CFU recovered in the lungs (Fig. 1). In a separate experiment, we performed fungal burden analysis beyond 7 days p.i. for the 107 CFU-infected group and found no detectable CFU on day 10 p.i., indicating that all live chitosan-deficient cells were completely eliminated from the host lung (data not shown). The fungal burden in the lungs of the mice infected with the wild-type KN99α strain continued to proliferate as expected (Fig. 1).
FIG 1 .
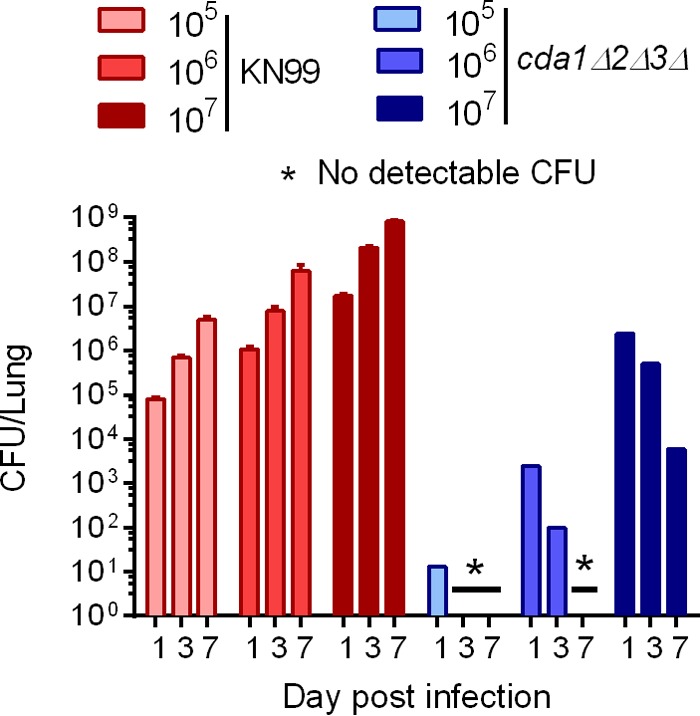
The time required for the host to completely eradicate chitosan-deficient cda1Δ2Δ3Δ cells in the lung depends on the size of the yeast inoculum. CBA/J mice were inoculated with various doses (equivalent to 105, 106, or 107 CFU) of either strain KN99α or cda1Δ2Δ3Δ mutant cells through intranasal inoculation. At 1, 3, and 7 days p.i., lungs were harvested, homogenized, and serially diluted for CFU enumeration by plating on YPD. Fungal burden is expressed as CFU per lung. The detection limit of the assay was <10 CFU/lung. Error bars indicate standard errors of the means for three mice per treatment group.
Clearance of the cda1Δ2Δ 3Δ mutant is associated with robust stimulation of cytokines that are critical for inducing protective immunity.
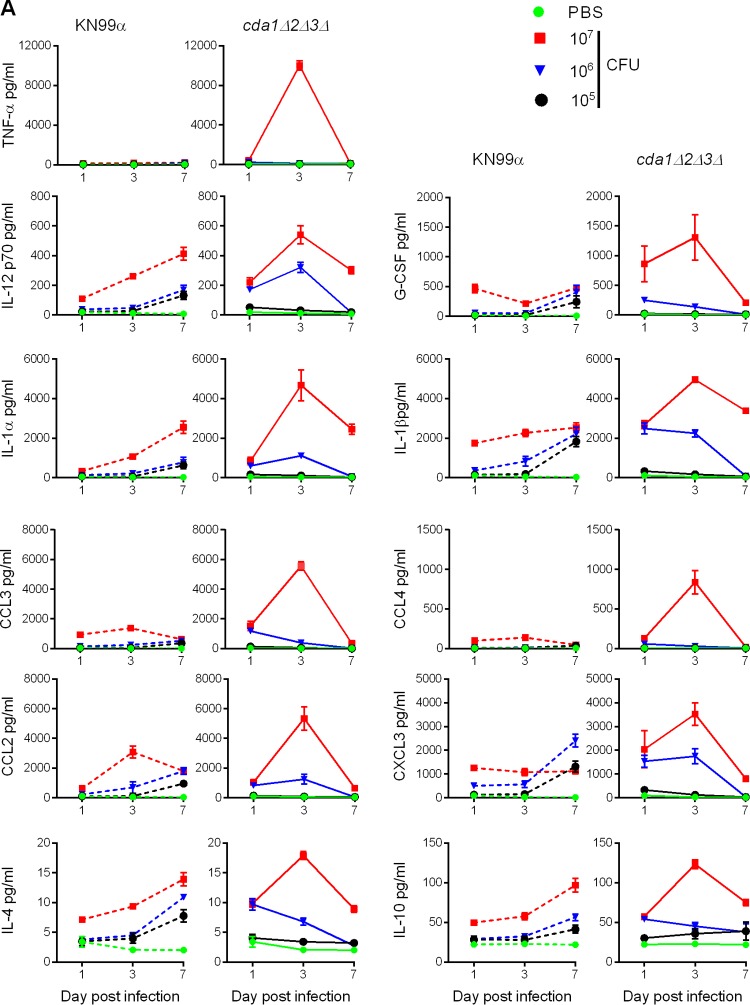
To gain insight into the potential role of the host immune response in clearing the mutant fungal cells, we analyzed the concentrations of cytokines in lung homogenates at 1, 3, and 7 days p.i. by employing the Bio-Plex pro mouse cytokine assay (Bio-Rad Laboratories). We inoculated CBA/J mice with 105, 106, and 107 CFU, as for the clearance studies. Overall, we found two major factors affecting the type and magnitude of the host cytokine response (Fig. 2; see also Table S1 in the supplemental material). First, the number of cells being introduced into the lung had a major influence on the magnitude of the host cytokine immune response: 105 CFU gave minimal induction, while 107 CFU elicited maximum induction. This trend was found for both the wild type and the cda1Δ2Δ3Δ strain (Fig. 2A). Second, the absence of chitosan in the cell wall of the cda1Δ2Δ3Δ mutant affected the timing and magnitude of the induction of various cytokines in the infected lungs. One of the characteristic features of the immune response triggered by cda1Δ2Δ3Δ mutant infection was that the levels of various cytokines peaked on day 3 p.i., before declining sharply on day 7 p.i. to around day 1 p.i. levels (Fig. 2A). However, in mice infected with wild-type KN99α, the concentrations of many cytokines gradually increased as the infection progressed (Fig. 2A).
FIG 2 .
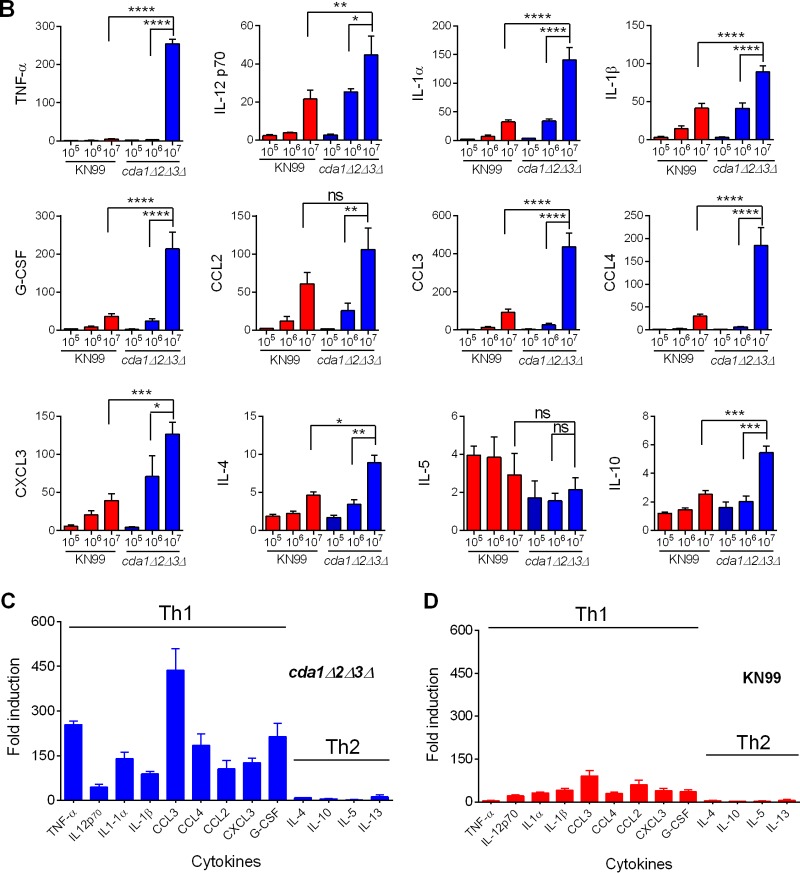
The kinetic profiles of mouse pulmonary cytokines were dependent on the inoculum load of C. neoformans cells and the presence or absence of chitosan in the cell wall. (A) Representative cytokine profiles in the lungs at various times after infecting the mice with various doses (equivalent to 105, 106, or 107 CFU) of either KN99α or cda1Δ2Δ3Δ cells. Pulmonary cytokines that showed significantly different levels at day 3 p.i. in mice infected with 107 CFU of KN99α or the cda1Δ2Δ3Δ mutant, in two independent experiments with three mice per treatment group, are displayed. Error bars indicate standard errors of the means. (B) Data from panel A are expressed as fold induction in protein levels of cytokines on day 3 p.i. in the lungs of the mice infected with various doses of either the cda1Δ2Δ3Δ mutant or KN99α compared to their levels in the PBS-inoculated control mouse lungs (n = 3). ****, P < 0.0001 by two-way ANOVA and Bonferroni’s multiple comparisons test. For IL2p70, **, P = 0.0053, and *, P = 0.0193; for CCL2, **, P = 0.002; for CXCL3, ***, P = 0.0006, and *, P = 0.026; for IL-4, **, P = 0.0027, and *, P = 0.05; and for IL-10, ***, P = 0.0009. All data are presented as mean values ± standard errors of the means (SEM). (C, D) Selective high-level induction of Th1-associated cytokines was observed only in cda1Δ2Δ3Δ-infected mouse lungs (C), while their upregulation was not as robust in KN99α-infected mouse lungs (D). The intensity of upregulation of Th2-associated cytokines was not as dramatic as that of the Th1 cytokines. The cytokine levels for each group were normalized to the results for the PBS control group.
We did not find statistically significant differences between the levels of cytokines in the lungs of mice infected with 105 CFU of strain KN99α and the cda1Δ2Δ3Δ mutant (Fig. 2A and B; see also Table S1 in the supplemental material). However, with higher inocula, significant differences in the concentrations of cytokines were observed between KN99α- and cda1Δ2Δ3Δ mutant-infected lungs. With an inoculum of 106 CFU, 14 cytokines (interleukin 1α [IL-1α], IL-1β, IL-12p70, tumor necrosis factor alpha [TNF-α], CCL3, CCL4, CCL2, IFN-γ, IL-2, IL-3, IL-4, IL-5, IL-10, and CCL5) showed significantly different levels in lung lysates of mice infected with KN99α and cda1Δ2Δ3Δ mutant cells on day 3 p.i. (see Table S1 in the supplemental material). When an inoculum of 107 CFU was used, the levels of 11 of the 23 cytokines tested were significantly different in the lungs of mice infected with the wild type and the corresponding cda1Δ2Δ3Δ mutant on day 3 p.i. (Fig. 2A). Importantly, the magnitude of cytokine induction triggered by 107 CFU of the cda1Δ2Δ3Δ mutant was distinct and dramatically higher than for all other infections (Fig. 2B).
We determined the fold induction of cytokines at day 3 p.i. in various C. neoformans-infected groups compared to the induction in the PBS-inoculated control group (Fig. 2B). Then, we compared the cytokine levels induced by 107 CFU of the cda1Δ2Δ3Δ mutant to the levels induced by 107 CFU of the wild type and 106 CFU of the cda1Δ2Δ3Δ mutant. We consistently observed a larger induction of Th1-associated cytokines in mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant than in mice infected with the wild type or the lower inoculum of the cda1Δ2Δ3Δ strain. TNF-α, an important early-response cytokine, increased about 250-fold in the lungs of mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant, in contrast to 2- to 4-fold increases seen in mice infected with either 107 CFU of KN99α or 106 CFU of cda1Δ2Δ3Δ cells (Fig. 2B). Similarly, in mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant, cytokines IL-12, granulocyte colony-stimulating factor (G-CSF), IL-1α, and IL-1β increased about 45-, 215-, 140-, and 90-fold, respectively (Fig. 2B). IL-12p70 is an important proinflammatory cytokine that is known to promote the maturation of naive T cells to helper Th1 cells through the induction of IFN-γ (27). While both the KN99α and cda1Δ2Δ3Δ strains induced the upregulation of IL-12p70 in an inoculum size-dependent manner, its regulation was tightly controlled during infection with the cda1Δ2Δ3Δ mutant compared to its persistently higher levels in the KN99α-infected murine lung in the later days of infection (Fig. 2A). G-CSF specifically boosts the proliferation and maturation of neutrophils (28). The contribution of IL-1 in resisting fungal infections has been demonstrated using neutralizing antibodies to IL-1β and using IL-1 receptor-deficient (IL-1R−/−) mice (29).
Among the chemokines analyzed, macrophage inflammatory proteins CCL3 and CCL4, both of which are C-C chemokines, rose by 435- and 185-fold, respectively, in the lungs of mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant, while such a robust induction was not observed in the lungs of other mouse groups (infected with the wild type or a lower inoculum of the cda1Δ2Δ3Δ mutant) (Fig. 2B). We also saw statistically significant upregulation in the level of CXCL3, a CXCL-1–type chemokine that is important for attracting neutrophils to the site of infection, in the lungs of mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant (Fig. 2B). All the above-mentioned cytokines have been demonstrated to create an environment conducive for the development of a protective Th1-type adaptive immune response (30). In contrast, even though we observed statistically significant differences in the induction levels of IL-4 and IL-10 in mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant and mice infected with 107 CFU of the wild type, the magnitudes of upregulation for infection with 107 CFU of the cda1Δ2Δ3Δ mutant were small (5- to 10-fold) compared to the levels seen for the Th1-specific inflammatory molecules mentioned above (Fig. 2B and C). IL-4 and IL-10 are cytokines with known roles in the Th2-type immune response that are also known to facilitate fungal pathogenesis (31, 32).
Overall, we found that infection by and subsequent clearance of the cda1Δ2Δ3Δ strain triggered a host immune response characterized by significant, although temporally limited stimulation of cytokines known to be important for orchestrating a protective Th1-type environment in the lung (Fig. 2C). This Th1 cytokine enrichment in the lungs of mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant was in conjunction with lower levels of induction of the Th2-type cytokines IL-4, IL-5, IL-10, and IL-13. This suggested to us that the chitosan-deficient cda1Δ2Δ3Δ C. neoformans cells may be capable of skewing the host immune response in favor of a Th1-type adaptive response.
Pulmonary inflammatory response triggered by infection with 107 CFU of the cda1Δ2Δ3Δ mutant was followed by a specific increase in the recruitment of Th1 CD4+ T cells in the infected mouse lung.
We conducted a pulmonary leukocyte analysis to study the recruitment of adaptive immune cells in response to infection. We infected mice with 107 CFU of either KN99α or the cda1Δ2Δ3Δ mutant, and at days 1, 3, 7, 10, and 21 p.i., excised the lungs and prepared single-cell suspensions for analysis by multicolor flow cytometry. Since mice infected with KN99α did not survive beyond 19 days, they were not available for leukocyte analysis on day 21 p.i. The total number of leukocytes (i.e., CD45+) in the lungs was significantly higher for mice infected with the cda1Δ2Δ3Δ mutant than for either KN99α-infected or PBS control mice on day 3 p.i. (P < 0.05), which reached similar levels at the later stages of infection (Fig. 3A). However, the number of leukocytes in the KN99α-infected mice was not significantly higher than for the PBS control group on day 3. Similarly, there were significant increases in the numbers of neutrophils on days 1 (P = 0.0034) and 3 (P < 0.01) p.i. in the lungs of the mice infected with cda1Δ2Δ3Δ mutant compared to the numbers in KN99α-infected mice (Fig. 3B). However, no statistical differences were found between the total numbers of macrophages, dendritic cells, eosinophils, total CD4+ and CD8+ T cells, Th17 cells, and Treg cells for KN99α and cda1Δ2Δ3Δ mutant infection at each time point (data not shown). The frequencies of CD4+ T cells with a Th1 phenotype (CD45+ CD3+ CD4+ CD183+ CD196−) were significantly higher in the lungs of mice infected with cda1Δ2Δ3Δ cells (P < 0.05) than in KN99α-infected lungs on day 10 p.i. (Fig. 3C). In contrast, the number of CD4+ Th2 cells (CD45+ CD3+ CD4+ CD183− CD196−) was significantly lower in cda1Δ2Δ3Δ mutant-infected lungs than in wild type-infected lungs (P < 0.05) on day 10 p.i. (Fig. 3D). Furthermore, significant populations of Th1 cells were found to express CD44 on day 10 p.i. only in the lungs of cda1Δ2Δ3Δ mutant-infected mice and not in the lungs of either KN99α-infected mice (P < 0.05) or PBS control mice (P = 0.0005) (Fig. 3E). High expression of CD44 receptor is associated with the expansion of Th1 cells into antigen-experienced memory cells (33). Taken together, the abundance of Th1-type T cells that were also CD44-expressing memory cells in the cda1Δ2Δ3Δ mutant-infected mouse lungs indicated that a host immune response triggered during the clearance of cda1Δ2Δ3Δ cells might induce protective immunity to subsequent C. neoformans infection.
FIG 3 .
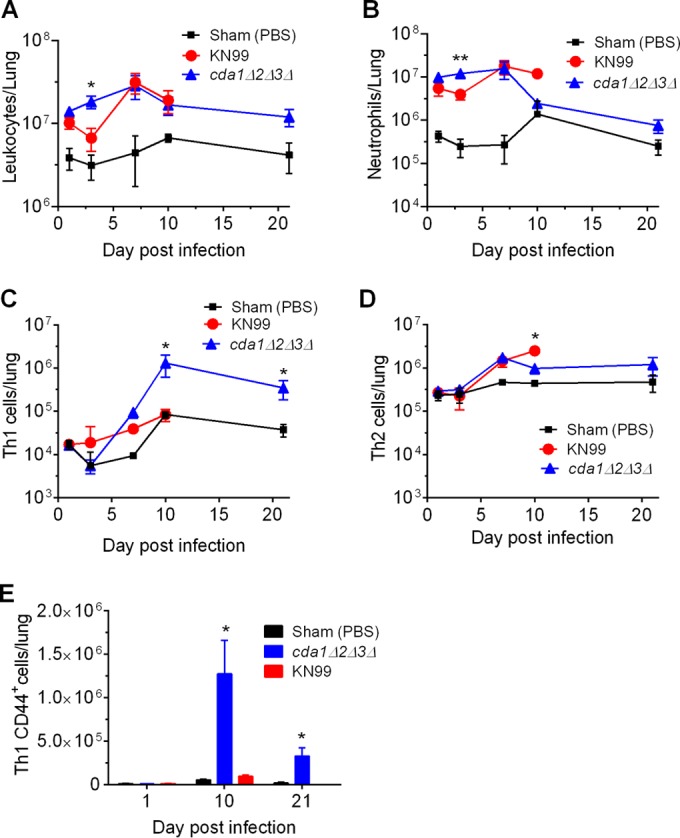
Pulmonary leukocyte analysis revealed increased recruitment of Th1-type CD4+ T cells in the lungs of mice infected with the chitosan-deficient cda1Δ2Δ3Δ C. neoformans cells. (A, B) Mice inoculated with 107 CFU of the cda1Δ2Δ3Δ strain exhibited significant enrichment of total leukocytes (A) and neutrophils (B) in the lungs on day 3 p.i. compared to their levels in the lungs of mice inoculated with either 107 CFU of KN99α or 50 µl of PBS. (C) The populations of CD4+ Th1 cells on days 10 and 21 p.i. were significantly higher in the cda1Δ2Δ3Δ mutant-infected mouse lungs than in the lungs of either PBS- or KN99α-inoculated animals. (D) The frequencies of Th2-type CD4+ leukocytes on day 10 p.i. were significantly higher in the KN99α-infected animals than in either PBS- or cda1Δ2Δ3Δ-inoculated animals. (E) Th1 cells induced during infection with cda1Δ2Δ3Δ cells showed increased expression of CD44 on their surface on day 10 p.i., and this was maintained during immune homeostasis (day 21 p.i.). The gating strategy for total leukocytes, neutrophils, Th1 and Th2 cells, and CD44 expression on Th1 cells per whole lung was based upon these leukocyte populations being CD45+, CD45+ CD24+ CD11c− Siglec F-LY6G+, CD45+ CD3+ CD4+ CD183+ CD196−, CD45+ CD3+ CD4+ CD183− CD196−, and CD45+ CD3+ CD4+ CD183+ CD196− CD44+, respectively. The total number of leukocytes was calculated by multiplying the frequency of the leukocyte by the total number of cells determined from the single-cell mouse lung suspensions. Data are presented as the mean results ± standard deviations for three mice per group from one biological experiment. Means were compared among groups within each day using one-way ANOVA followed by the Bonferroni multiple-comparison test. *, P < 0.05; **, P < 0.01.
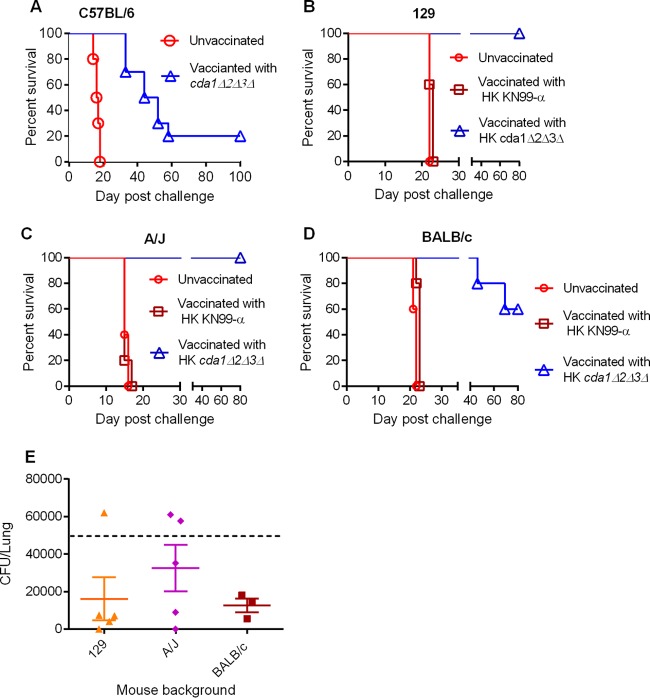
Immunizing mice with cda1Δ2Δ3Δ cells conferred complete protection to subsequent challenge with highly virulent wild-type C. neoformans cells.
To evaluate whether the pulmonary immune response elicited by the cda1Δ2Δ3Δ mutant and the subsequent increased recruitment of Th1-type CD4+ T cells is able to trigger protective immunity, we first infected CBA/J mice with 107 CFU of cda1Δ2Δ3Δ cells in PBS through intranasal inoculation. Mice inoculated with PBS served as a control group. Mice were allowed 40 days to resolve the infection and then were challenged intranasally with 105 CFU of the wild-type KN99α strain. Mice preinfected (immunized) with the cda1Δ2Δ3Δ strain were completely protected, while the PBS control group mice exhibited 100% mortality by day 19 postchallenge (Fig. 4). On day 80 p.i., the study was ended and the lung and brain homogenates of mice were plated for enumerating fungal CFU. We found that the fungal burden was restricted to the lung and was approximately equal to the challenge inoculum of 105 CFU (data not shown).
FIG 4 .
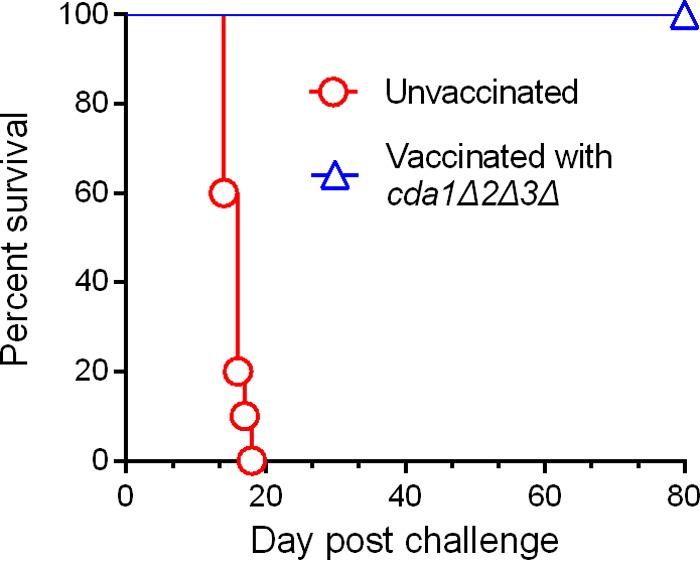
Vaccination of CBA/J mice with 107 CFU of live cda1Δ2Δ3Δ cells conferred robust protective immunity to subsequent infection with wild-type KN99α C. neoformans cells. Mice were inoculated with 107 CFU of cda1Δ2Δ3Δ cells through inhalation. PBS-inoculated mice served as control. Animals were left for 40 days to resolve the infection. Subsequently, both groups of mice were challenged with 105 CFU of KN99α cells. Virulence was recorded as mortality of mice. Mice that lost 25% of starting body weight were considered to be moribund and were sacrificed. The percentage of mice that survived was plotted against the day p.i. Each survival curve is the average of three independent experiments that had five mice per experimental group.
Heat-killed cda1Δ2Δ3Δ cells are equally effective in inducing strong protective immunity to C. neoformans infection.
To determine whether any biological activity present in the cda1Δ2Δ3Δ strain is specifically required for inducing protective immunity in mice, we wanted to test the feasibility of using its heat-killed (HK) cells as a vaccine. We determined that incubation of either KN99α or cda1Δ2Δ3Δ cells at 70°C for 15 min resulted in the complete loss of their viability, and we proceeded to inoculate mice intranasally with 107 CFU of HK cells. At 40 days p.i., mice were challenged with 105 CFU of the wild-type strain KN99α. We observed that mice inoculated with either PBS or HK KN99α succumbed to the KN99α challenge infection by day 19 (Fig. 5A). However, the mice vaccinated with HK cda1Δ2Δ3Δ cells were completely protected throughout the period of the experiment (80 days) (Fig. 5A). Similar to the results for vaccination with live cda1Δ2Δ3Δ cells, the fungal burden in the surviving mice was restricted to the lungs, with no dissemination to brain, spleen, or kidneys (data not shown). When 50,000 CFU of KN99α was used for challenging mice, we found on average an 85% decrease in the lung CFU on day 70 after the challenge infection, suggesting that HK cda1Δ2Δ3Δ mutant-induced protective immunity has the capacity to contain and partly clear an otherwise lethal infection. We also observed in a separate experiment that mice vaccinated with HK cda1Δ2Δ3Δ cells and later challenged with 50,000 CFU of KN99α cells survived for 140 days postchallenge, at which time the experiment was terminated (data not shown). Surviving mice from both these experiments showed various levels of fungal clearance, with some mice showing no fungal burden in the lung (Fig. 5B). In all mice, the fungal burden was restricted to the lungs. In the experiment that was ended on day 70, mice had a median fungal burden in the lungs of about 8,000 CFU/lung (Fig. 5B). The median fungal burden for the experiment that was ended on day 140 was around 5,500 CFU/lung (Fig. 5B).
FIG 5 .
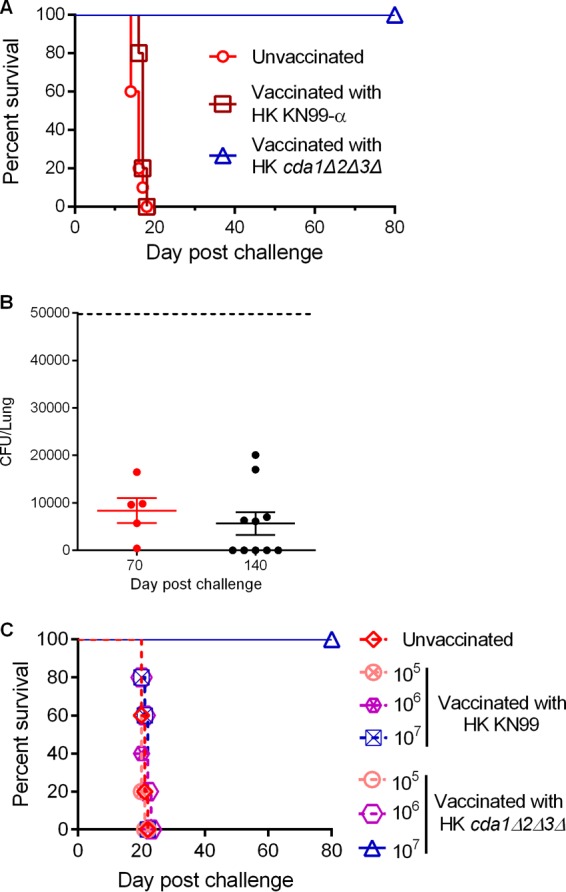
Heat-killed (HK) cda1Δ2Δ3Δ cells of C. neoformans induced strong protective immunity to a subsequent challenge with wild-type KN99α infection. (A) Mice were immunized with 107 CFU of HK cells of either the wild-type KN99α or the cda1Δ2Δ3Δ strain. Control mice were inoculated with PBS. After 40 days, mice were challenged with 105 CFU of wild-type KN99α cells. Survival of the animals was recorded as described above. The data shown are the average results from four experiments with five mice per experimental group. (B) Pulmonary fungal burden analysis of the mice that were vaccinated initially with 107 HK cda1Δ2Δ3Δ cells and later challenged with 50,000 virulent KN99α cells. At 70 days p.i., lungs were harvested and fungal CFU were enumerated. (C) Mice were vaccinated with various doses (105, 106, or 107 CFU) of HK cells of either the wild-type KN99α or the cda1Δ2Δ3Δ strain. After 40 days, they were challenged with 50,000 wild-type KN99α cells. Survival of the mice was monitored as described above.
Based upon our discovery that animals infected with 107 CFU of cda1Δ2Δ3Δ cells responded with a vigorous induction of proinflammatory cytokines in the lungs, while the cytokine response was less dramatic in the mice infected with either 105 or 106 CFU, we wondered whether vaccination with lower doses (105 or 106 CFU) of heat-killed cda1Δ2Δ3Δ cells could induce protective immunity. To test this, we vaccinated mice with doses of 105, 106, and 107 CFU of HK cells of either wild-type KN99α or the cda1Δ2Δ3Δ mutant. We found that mice vaccinated with 107 HK cda1Δ2Δ3Δ cells were the only group that was able to mount a robust protective response to a subsequent lethal challenge with KN99α (Fig. 5C). Mice vaccinated with lower numbers of cda1Δ2Δ3Δ cells succumbed to infection similarly to those in the unvaccinated group, with a median survival time of between 20 and 22 days postchallenge (Fig. 5C). These data further support the requirement of a minimum dose of antigen to induce an effective early innate response for successful priming of an adaptive immune response.
Vaccination of mice with the C. neoformans cda1Δ2Δ3Δ strain confers significant cross protection against subsequent pulmonary challenge with a lethal dose of C. gattii.
Even though C. neoformans and C. gattii are closely related species, they differ significantly in their natural habitats, in the clinical manifestations of the disease they cause, in the specificity toward the organs they target in the murine infection model, in their carbon and nitrogen assimilation pathways, and finally, in the types of host immune response they provoke (34, 35). The differences in the immune responses they induce suggest that there may be differences in the organization of antigens on the cell surface of these two species. Therefore, we tested whether vaccination of mice with the cda1Δ2Δ3Δ mutant generated in a C. neoformans background would protect them from a subsequent challenge with C. gattii. Using the vaccination and infection protocol described above, we challenged mice vaccinated with HK cells of the C. neoformans cda1Δ2Δ3Δ mutant with 105 CFU of either C. gattii strain R265 or C. gattii strain WM276. Mice vaccinated with PBS served as a sham vaccination control. We observed that control mice challenged with either C. gattii R265 or C. gattii WM276 had mean survival times of 18 and 17 days, respectively (Fig. 6). On the other hand, mice that were vaccinated with the C. neoformans cda1Δ2Δ3Δ strain exhibited significantly delayed mortality, with mean survival times of 33 and 30 days for C. gattii R265 and C. gattii WM276, respectively. This demonstrated that some cross protection developed with the C. neoformans cda1Δ2Δ3Δ vaccine.
FIG 6 .
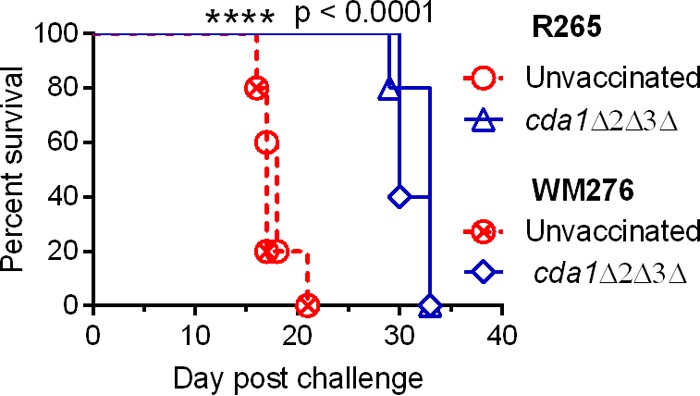
Vaccination of mice with HK cells of cda1Δ2Δ3Δ C. neoformans conferred partial protection to subsequent challenge with a lethal dose of a wild-type strain of C. gattii. A group of 10 mice (CBA/J) was subjected to initial vaccination with 107 HK cda1Δ2Δ3Δ cells and later challenged with 105 CFU of either C. gattii R265 or C. gattii WM276. Survival of the mice was monitored as described above. PBS-inoculated mice served as control.
Heat-killed cda1Δ2Δ3Δ cell-induced protective immunity to C. neoformans infection is effective in multiple strains of mice.
Previous work by several laboratories demonstrated that mice of different genetic backgrounds exhibit various levels of sensitivity to cryptococcal infection (36–38). Therefore, we chose to test the effectiveness of the cda1Δ2Δ3Δ mutant vaccination in C57BL/6, 129, A/J, and BALB/c inbred mice. We vaccinated C57BL/6 mice with 107 CFU of live cda1Δ2Δ3Δ cells and, after 40 days, challenged them with 105 CFU of KN99α. Vaccination of C57BL/6 mice with the cda1Δ2Δ3Δ strain conferred a partial but significant level of protection, with a mean survival time of 48 days compared to 16.5 days in the unvaccinated control group (Fig. 7A).
FIG 7 .
Protective immunity induced by HK cda1Δ2Δ3Δ cells was effective in different inbred mouse strains. (A) A group of 10 C57BL/6 mice were vaccinated with 107 CFU of live cda1Δ2Δ3Δ cells and later challenged with 105 CFU of KN99α cells as described above. Survival was monitored, and the percentage of survival was plotted against the day postchallenge. (B to D) Groups of five mice each of strains 129 (B), A/J (C), and BALB/c (D) were vaccinated with 107 HK cda1Δ2Δ3Δ cells. At 40 days postvaccination, mice were challenged with 50,000 wild-type KN99α cells. Survival was monitored as described above. (E) At 80 days postchallenge, lungs from the surviving mice from the experiment whose results are shown in panels B, C, and D were harvested and subjected to lung fungal burden analysis.
We then vaccinated 129, A/J, and BALB/c mice with 107 HK cda1Δ2Δ3Δ cells. The A/J and 129 mice were protected similarly to the CBA/J mice (Fig. 7B and C). All vaccinated animals survived the challenge with the KN99α strain for the length of the experiment (80 days postchallenge). However, for BALB/c mice, the efficacy of the cda1Δ2Δ3Δ mutant vaccine was slightly decreased (Fig. 7D). Of the five mice used for the experiment, three survived the infection, while two succumbed, on day 46 and day 64 (Fig. 7D). All mice either inoculated with PBS or vaccinated with an HK preparation of KN99α cells succumbed by 23 days postchallenge. Among these control groups, the A/J mice were the most sensitive to KN99α challenge, with a median survival time of 15 days, while 129 and BALB/c mice showed median survival times of 22 and 23 days, respectively. We also observed significant but varied clearance of infection in vaccinated mice of different genetic backgrounds (Fig. 7E).
DISCUSSION
Our initial goal for the investigation was to understand the mechanism of the clearance of chitosan-deficient mutant strains of C. neoformans in the mouse lung. In our previous study, we reported that cells of C. neoformans chs3Δ, csr2Δ, and cda1Δ2Δ3Δ mutant strains were devoid of chitosan and displayed compromised cell wall integrity (26). Interestingly, while the chs3Δ and csr2Δ strains were temperature sensitive, the cda1Δ2Δ3Δ strain was able to grow at host body temperature. Therefore, we selected the cda1Δ2Δ3Δ strain to further understand the mechanism of its clearance from the mouse lung. Since 105 CFU of the cda1Δ2Δ3Δ mutant were cleared in 24 h, we wondered whether increasing the CFU of the inoculum would alter clearance (25). We found that using higher inocula delayed complete eradication (Fig. 1). Moreover, when 107 CFU of the cda1Δ2Δ3Δ mutant were used, we did recover 5,000 CFU of cells from the infected lung at 7 days p.i., showing that cda1Δ2Δ3Δ cells were able to survive in the host. It will be interesting to see whether those that survived for 7 days were the product of newly replicated fungal cells or persisted from the original inoculum without replication. Nevertheless, the complete clearance of cda1Δ2Δ3Δ cells from the mouse lung by day 10 p.i. suggested that host-specific mechanisms may be responsible for clearing the chitosan-deficient mutant cells, as opposed to the mere inability of the cda1Δ2Δ3Δ cells to survive or grow in the host lung.
The results from using increasing sizes of cda1Δ2Δ3Δ inocula for intranasal inoculation not only suggested the potential role of host factors in eradicating the mutant fungal cells but also implied that the intensity of the host immune response was dependent on the size of the initial infection. We chose to measure cytokine levels in lung homogenates as a measure of the host response. Infection with 105 CFU resulted in the detection of low levels of cytokines on different days p.i. This may be due to either the limitations in the sensitivity of the assay employed or the difficulty in choosing an ideal time point for cytokine analysis due to the dynamic nature of the inflammatory response induced. Moreover, at this size of inoculum, we did not find a statistically significant difference between the levels of cytokines in KN99α- and cda1Δ2Δ3Δ mutant-infected murine lungs. However, there were statistically significant differences between the amounts of various cytokines in the lung homogenates of mice infected with KN99α and with the cda1Δ2Δ3Δ mutant for mice inoculated with both 106 and 107 CFU, and the magnitude of their induction on day 3 p.i. was clearly different for the mice infected with 107 CFU.
TNF-α has been shown to be necessary for the development of Th1-mediated immunity and plays a pivotal role in clearing various fungal infections, including C. neoformans infection, in animal models (reference 39 and references therein). The production of TNF-α in the afferent phase (0 to 7 days p.i.) of the host immune response has been demonstrated to be critical for the development of protective immunity to C. neoformans infection in the CBA/J mouse model (40). Such a dependency of cda1Δ2Δ3Δ mutant-induced protective immunity on the early expression of TNF-α may be playing a role in our experiments. While infection with 105 and 106 CFU of cda1Δ2Δ3Δ cells did not elicit significant TNF-α induction, infection with 107 CFU of the cda1Δ2Δ3Δ mutant clearly produced a dramatic spike in its levels in the lung on day 3 p.i. Accordingly, vaccination with 105 and 106 CFU of the cda1Δ2Δ3Δ mutant did not confer any protection. However, robust protection was observed when 107 CFU of the cda1Δ2Δ3Δ mutant was used for vaccination. Therefore, vaccination experiments when performed in the presence of TNF-α-specific neutralizing antibodies may shed light on the essential nature of early TNF-α induction during vaccination, on the clearance of the cda1Δ2Δ3Δ strain, and on subsequent induction of effective protective immunity to C. neoformans infection. More importantly, the increased levels of TNF-α detected during infection with 107 cda1Δ2Δ3Δ cells did not persist after 3 days p.i. This might perhaps have prevented deleterious damage to the host lung due to an unregulated TNF-α-mediated inflammatory response. Several fungus-related virulence factors, such as polysaccharide capsule, melanin, and prostaglandinlike molecules, have been documented to inhibit the stimulation of TNF-α during wild-type C. neoformans infection (41–43). However, determination of cellular factors associated with the cda1Δ2Δ3Δ strain that resulted in the controlled expression of TNF-α production during infection will provide more insight into the regulation of TNF-α during C. neoformans infection. In any case, this effect of the cda1Δ2Δ3Δ strain in controlled regulation of TNF-α expression during vaccination may be an added benefit as far as the safety of the vaccine is concerned.
IL-12 is a heterodimeric cytokine that has been shown to be essential for the development of a protective Th1 response and resistance to C. neoformans infection, and its expression is under the influence of TNF-α (39, 44, 45). We found that the expression of IL-12p70 in both KN99α- and cda1Δ2Δ3Δ mutant-infected lungs followed the pattern of TNF-α expression (see Table S1 in the supplemental material). We did not detect significant differences between the levels of either IFN-γ or IL-2 in KN99α- and cda1Δ2Δ3Δ mutant-infected mouse lungs at the chosen time points. This may be due to the complex and dynamic nature of the regulation of cytokine expression, the limited number of time points chosen in our experiment for cytokine analysis, or the limitations in the sensitivity of the assay employed.
IL-1α and IL-1β both belong to the IL-1 family of proinflammatory cytokines that play a major role in the host’s defense against infections, not only by activating innate immune cells but also by triggering further release of proinflammatory cytokines that participate in the modulation of the host adaptive immune response. The critical role of IL-1β in controlling C. neoformans and other pathogenic fungal infections through the activation of the NLRP3 inflammasome has recently been documented (46–48). We do not know the source of IL-1β in the present study; however, dendritic cell-derived IL-1β has been shown to be important for effective priming of T cells (49). Moreover, when recombinant IL-β was used as an adjuvant together with a heat-killed formulation of a weak attenuated strain of Blastomyces dermatitidis, it enhanced the protective effect of the vaccine strain (50). Even though infecting mice with 107 CFU of KN99α caused the stimulation of both IL-1α and IL-1β, the levels were only comparable to those induced by infection with 106 CFU of the cda1Δ2Δ3Δ mutant, which was nonprotective. Therefore, the timing (3 days p.i.) and the effective concentration of both IL-1α and IL-1β induced by 107 CFU of the cda1Δ2Δ3Δ mutant may have contributed to the protective immunity.
CCL3 is a C-C chemokine that acts as a chemoattractant for leukocytes, is critical for the promotion of the Th1-type immune response, inhibits the expression of IL-4/IL-13, and plays an important role in preventing lung eosinophilia during C. neoformans infection (51, 52). Of all the cytokines tested, CCL3 showed a maximum intensity of induction in the cda1Δ2Δ3Δ mutant-infected mouse lung which was specific to mice infected with 107 CFU of cells, similar to the response seen for TNF-α. Consistent with this, we saw a significant increase in leukocyte migration to the lungs on day 3 p.i. in the 107 cda1Δ2Δ3Δ mutant-infected mouse lung compared to that in the KN99α-infected mouse lung. Therefore, CCL3 may be one of the important cytokines responsible for the migration of leukocytes to the lung. These results, along with the previously documented requirement of early induction of CCL3 during C. neoformans infection in promoting the Th1-type protective cellular response, suggest a role of CCL3 in the cda1Δ2Δ3Δ mutant-induced protective immunity (52).
CCL2 is a potent chemotactic factor for monocytes and dendritic cells, in addition to its role in regulating the migration and infiltration of memory T-lymphocytes and natural killer (NK) cells. The role of CCL2 in inducing Th1 protective immunity and fungal clearance during C. neoformans infection has been demonstrated by its involvement in recruiting macrophages, CD4+ T cells, and NK T cells to the site of infection (53, 54). Even though we observed increased levels of CCL2 in the lungs of mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant, significantly increased levels of CCL2 were also seen in mice infected with 107 CFU of strain KN99α. These results suggest a minimal role of CCL2 in cda1Δ2Δ3Δ mutant-induced protective immunity.
CXCL3 is a CXCL1 type of chemokine that is associated with neutrophil recruitment. The contribution of CXCL3 in inducing resistance to C. neoformans infection is not well characterized. However, we observed that this chemokine was significantly elevated in murine lungs infected with 107 CFU of the cda1Δ2Δ3Δ mutant. This is consistent with previously published results showing the expression pattern of CXCL3 during infection of the SJL/J mouse strain that is resistant to a moderately virulent strain of C. neoformans (strain 24067) (55). The higher concentration of CXCL3, followed by the increased number of neutrophils in the lungs of mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant, may indicate an important role of this chemokine in the clearance of the cda1Δ2Δ3Δ mutant and in the induction of adaptive immunity. In addition, the significant increase in the levels of G-CSF, only in the lungs of mice infected with 107 CFU of the cda1Δ2Δ3Δ mutant, might have promoted the proliferation and maturation of neutrophils, which is consistent with previously published results showing a beneficial role of recombinant G-CSF therapy during bacterial and opportunistic fungal infections (28). Furthermore, recombinant human G-CSF has been reported to enhance the ability of human effector cells to kill C. neoformans (56). Therefore, we speculate that this property of G-CSF may be conserved and may be partly responsible for the clearance of the cda1Δ2Δ3Δ mutant. Cytokines secreted upon the infiltration of potent neutrophils might have contributed to the eventual polarization of the adaptive immunity and the subsequent protection against wild-type C. neoformans challenge.
The selective induction of the above-mentioned proinflammatory cytokines during infection with 107 CFU of the cda1Δ2Δ3Δ mutant was associated with minimal changes in the concentrations of cytokines driving a Th2 response, such as IL-4, IL-5, IL-10, and IL-13 (Fig. 2C). A Th2-type T cell response exacerbates C. neoformans infection by allowing fungal proliferation (31, 57). Overall, our cytokine analysis suggested an overwhelming proinflammatory response that should direct Th1 skewing of the early host immune response. This was supported by the results of multicolor flow cytometry, which showed an increase of Th1-type cells in the infected lung on day 10 p.i. This immunological data prompted us to evaluate whether chitosan-deficient cda1Δ2Δ3Δ cells can induce protective immunity.
Unlike C. neoformans, C. gattii causes disseminated infections in immunocompetent and apparently healthy individuals. C. gattii strain R265, which was responsible for the Pacific Northwest outbreak, was found to be more virulent than C. gattii strain WM276, an environmental isolate from Australia, when tested for virulence in either C57BL/6 or A/JCr mice (58). Interestingly, we observed similar levels of virulence for these strains in the CBA/J mouse model both in the immunized and naive mouse groups. It is encouraging that the cda1Δ2Δ3Δ mutant engineered in C. neoformans generated significant cross protection to C. gattii infections. This is most likely due to the presence of common immunodominant proteins in the cell wall fraction of both C. neoformans and C. gattii (30, 59). However, the failure of the C. neoformans cda1Δ2Δ3Δ vaccine to confer complete protection against C. gattii infection emphasizes the underlying differences between the antigenic determinants in these two species of Cryptococcus. In addition, previous studies have reported that C. gattii infection causes inhibition of efficient neutrophil migration and pulmonary cytokine production and downregulation of the dendritic cell-mediated Th1 immune response (34, 58). Therefore, it is plausible that challenging cda1Δ2Δ3Δ strain-vaccinated mice with C. gattii might have decreased the intensity of the protective host inflammatory response, which in turn resulted in the less-effective cross protection observed in our experiments. The mechanisms of chitin and chitosan biosynthesis and its regulation in C. gattii have not been studied. It will be interesting to see whether vaccination with a cda1Δ2Δ3Δ strain engineered in C. gattii confers complete protection against C. gattii infections.
Several studies have demonstrated the influence of host genetic factors on the susceptibility to Cryptococcus infection (36, 37). CBA/J mice have been demonstrated to be resistant to C. neoformans strain 52 D by developing a strong Th1 adaptive immunity that results in the progressive clearance of the infection. In contrast, C57BL/6 mice respond with a skewed Th2 response and succumb to infection. In such infection experiments, BALB/c mice have exhibited an intermediate phenotype (57). These patterns are mirrored by the results of our vaccination and challenge experiments. CBA/J mice showed a strong resistance to infection after vaccination, with significant clearing, while the efficiency of protection was slightly reduced in BALB/c mice and appreciably decreased in C57BL/6 mice. Interestingly, all the surviving BALB/c mice showed significant clearance of the challenge inoculum. The vaccination of 129 and A/J strains with the cda1Δ2Δ3Δ mutant resulted in the generation of protective immunity similar to that in the CBA/J strain, with all mice showing clearance of the challenge inoculum to various degrees. Even though naive A/J mice were more sensitive to C. neoformans infection than 129 and BALB/c mice, they were able to mount a strong protection after vaccination.
Previous reports have shown that commercial and fungal chitin preparations elicit potent immune responses in the host (60–62). The type of immune response stimulated by a chitin preparation depended on its source, size, purity, and concentration in in vitro and in vivo studies (63, 64). Purified chitosan also stimulates immune responses, including inflammasome activation, which suggests a proinflammatory role for chitosan as well (65). However, the biological context of the material being presented may be crucial for how it is interpreted by the host. Additional pathogen-associated molecular patterns (PAMPs) present in the wild-type cell surface may be modulating a host response to whole Cryptococcus cells that is distinct from the response to purified chitosan.
We have previously shown that all of the chitin-derived fiber in the cda1Δ2Δ3Δ mutant and most of the chitin-derived material in chs3Δ and csr2Δ cells exists in its fully acetylated chitin form. These chitosan-deficient mutants exhibited sensitivity to cell wall stressors and a leaky melanin phenotype, suggesting a potentially disorganized cell wall architecture with possible exposure of underlying chitin and other immunogenic molecules on the cell surface which, in turn, may have resulted in the induction of a strong host immune response (26). It is possible that the chitin induces the protective immune response. The nature of the chitin fiber present in the cda1Δ2Δ3Δ, chs3Δ, and csr2Δ strains is not known, but all of the strains were cleared rapidly from the infected murine lung (25). In the case of the chs3Δ and csr2Δ strains, their clearance can almost surely be attributed in part to their temperature-sensitive phenotype. However, whether their clearance is followed by a concomitant polarization of the host immune response toward Th1 adaptive immunity is yet to be determined, which may provide more insight into the connection between chitin/cell wall structure and the host immune response. In wild-type cells, the presence of chitosan may mask the exposure of the chitin, resulting in a subdued host inflammatory response. It is possible that the host response is due to the exposure of chitin and not the quantity of chitin in the cell wall.
Various C. neoformans cell wall-associated proteins have been shown to induce a proinflammatory response that was followed by partial protection against C. neoformans infection when they were used as vaccines by themselves or in conjunction with yeast glucan particles (18, 59). Lack of chitosan in the cda1Δ2Δ3Δ strain may have resulted in the exposure of these antigenic cell wall proteins and other potential PAMPs which, in addition to the potent immunomodulatory property of chitin, may have contributed to the mechanisms of protective immunity. Comparison of the surface-exposed PAMPs on additional chitin or chitosan mutants or mutants with other mutations that affect the cell surface, such as Znf2 overexpression (which causes a morphogenesis defect) and Rim101 deletion (which causes cell wall defects), and the nature of the host immune responses may lead to a better understanding of the molecular signals that drive the protective response (24, 66).
In the absence of potent subunit vaccines that confer complete protection, the use of attenuated strains as potential vaccine candidates is one of the most promising approaches. The finding that HK cells of the cda1Δ2Δ3Δ mutant conferred protective immunity makes it attractive for development into a safe vaccine for clinical use.
MATERIALS AND METHODS
Mice.
All mice used were females between 6 and 8 weeks old. CBA/J, C57BL/6, 129, BALB/c, and A/J mice were from The Jackson Laboratory (Bar Harbor, ME). Mice were housed at the Washington University School of Medicine and handled according to guidelines approved by the Institutional Animal Care and Use Committee.
Cryptococcus strains.
C. neoformans strain KN99α and the cda1Δ2Δ3Δ (LBCN 632) strain derived from it were described previously (25). C. gattii strain R265 is a representative of the common VGIIa molecular subtype and is from the outbreak in British Columbia. C. gattii strain WM276 is a representative of subtype VGI and is an environmental isolate from Australia. Both of these strains were kindly provided by Joseph Heitman (Duke University Medical Center). Cells were recovered from −80°C stocks prior to use in each experiment. The strains were cultured in yeast extract-peptone-dextrose (YPD) medium.
Pulmonary infections.
Yeast strains were grown in 50 ml of YPD medium in a 250-ml Erlenmeyer flask (Corning Incorporated, Tewkesbury, MA) for 36 h at 30°C and at 300 rpm in a shaking incubator. Cells were harvested by centrifugation at 1,800 × g for 10 min, washed twice with phosphate-buffered saline (PBS) and resuspended in PBS at an appropriate cell density. Mice were infected intranasally as previously described (25). Briefly anesthetized mice were suspended by their incisors on a thread. A yeast inoculum of 50 µl containing the appropriate number of yeast cells was slowly dripped directly into the nares. Mice were left suspended for an additional 10 min and were closely monitored until they recovered from anesthesia. Mice were monitored daily and those showing signs of morbidity (weight loss or extension of the cerebral portion of the cranium) were sacrificed by CO2 asphyxiation.
Pulmonary fungal burden and cytokine analysis.
At various time points after infection, mouse organs were excised, weighed, and homogenized in 2 ml of PBS containing Complete protease inhibitor cocktail tablet (Roche Diagnostics) using a Pro200 homogenizer (Pro Scientific, Oxford, CT). Homogenates were serially diluted, plated on YPD agar, and incubated at 30°C. Yeast colonies were counted after 2 to 3 days of incubation. For pulmonary cytokine analysis, 0.05% Triton X-100 was added to a portion of the homogenized lung lysate and the sample was incubated for 2 h at 4°C with intermittent mixing. At the end of incubation, the lysate was centrifuged at 12,000 × g for 20 min. The supernatant was collected and spun again for 20 min at 12,000 × g. Aliquots of the supernatant were stored at −80°C. Cytokine analysis was performed using the Bio-Plex pro mouse cytokine 23-plex assay kit, following the protocol supplied with the kit (Bio-Rad Laboratories, Hercules, CA). Fifty microliters of lung sample was analyzed in each well.
Preparation of lung leukocyte single-cell suspensions.
Mice were sacrificed, and their left lungs were removed and added to homogenization C tubes containing proprietary catalysts for mechanical and enzymatic digestion (Miltenyi Biotec, Auburn, CA). This was followed by homogenization of lung tissue in the gentleMACS dissociator based upon the manufacturer’s instructions (Miltenyi Biotec). Homogenized tissue was then passed through the 70-µm-pore-size MACS SmartStrainer (Miltenyi Biotec) and centrifuged at 300 × g for 10 min to pellet cells. Erythrocytes were removed using ACK (ammonium-chloride-potassium) lysing buffer (Gibco, Grand Island, NY). Briefly, 1 ml of ACK buffer was used to suspend pelleted cells. After a maximum of 10 min, the ACK lysing buffer was diluted 1:10 with complete RPMI medium. Cells were then washed with cell staining buffer (CSB) containing final concentrations of 1 × PBS (Corning Cellgro, Manassas, VA), 2 mM EDTA (Amresco, Solon, OH), and fraction V BSA (Fisher Scientific, Pittsburgh, PA). Cells were then counted in a hemocytometer, using trypan blue staining to exclude dead cells.
Fluorochrome-conjugated antibodies for flow cytometry.
The following fluorochrome-conjugated mouse antibodies from BioLegend (San Diego, CA) were used: peridinin chlorophyll protein (PerCP)-Cy5.5-conjugated anti-CD45 antibody (Ab) (30-F11), phycoerythrin (PE)-conjugated anti-CD24 Ab (30-F1), allophycocyanin (APC)-conjugated anti-CD11c Ab (N418), PE-Cy7-conjugated anti-CD206 Ab (C068C2), and PE-Cy7-conjugated anti-CD Ly-6G Ab (1A8). The following BD Biosciences (San Jose, CA) fluorochrome-conjugated antibodies were also used: BUV737-conjugated anti-CD11b Ab (M1/70), BV605-conjugated anti-major histocompatibility complex class II (MHC-II) (M5/114.15.2), BV421-conjugated anti-CD80 Ab (16-10A1), Alexa Fluor 647-conjugated anti-Siglec F Ab (E50-2440), BV711-conjugated anti-CD3e Ab (145-2C11), Alexa Fluor 700-conjugated anti-CD4 Ab (RM4-5), BB515-conjugated anti-CD25 Ab (PC61), APC-H7-conjugated anti-CD8a Ab (53-6.7), BV605-conjugated anti-CD27 Ab (LG.3A10), PE-CF594-conjugated anti-CD28 Ab (37.51), BV421-conjugated anti-CD183 Ab (CXCR3-173), Alexa Fluor 647-conjugated anti-CD196 Ab (140706), BUV737-conjugated anti-CD44 Ab (IM7), and PE-conjugated anti-CD45RB Ab (16A).
Flow cytometry analysis of leukocyte populations.
Briefly, cell surface immunofluorescence staining involved the addition of 0.015 to 0.15 µg of a fluorochrome-conjugated antibody mixture containing antibodies specific to various leukocyte subpopulations to 25 µl of an equal mixture of CSB (defined above) and BD Horizon Brilliant stain buffer (San Jose, CA) in the wells of a 96-well microtiter plate. This staining mixture was then used to resuspend 2.5 × 105 cells in triplicate. Cells were then incubated on ice for 30 min in the dark. This was followed by two washes with CSB, and then cells were fixed at 4°C overnight in 1% paraformaldehyde (PFA). The next day, cells were washed and resuspended in CSB and added to a 96-well microtiter plate for high-throughput flow cytometry sampling using a BD LSRFortessa X-20 (BD Biosciences). BD FACSDiva software was used to process samples on the flow cytometer, and FlowJo (FlowJo, LLC, Ashland, OR) was used for data analysis of the acquired samples to facilitate the identification of leukocyte populations. Thirty thousand cell events were recorded, and dead cells and debris were excluded based upon lower forward scatter (FSC) and side scatter (SSC) signals (i.e., smaller size and granularity). Leukocyte populations were identified by the following markers as previously described: eosinophils (CD45+ CD24+ Siglec F+), neutrophils (CD45+ CD24+ LY6G+), CD11b+ dendritic cells (DCs) (CD45+ CD11b+ CD11c+ CD24+ MHC-II+), CD11b− DCs (CD45+ CD11b− CD11c+ CD24+ MHC-II+), alveolar (CD45+ CD11b− CD11c+ CD24− MHC-II+) and interstitial (CD45+ CD11b+ CD11c+ CD24− MHC-II+) macrophages, Th1 CD4 T cells (CD45+ CD3+ CD4+ CD183+ CD196−), Th2 CD4 T cells (CD45+ CD3+ CD4+ CD183− CD196−), Th17 cells (CD45+ CD3+ CD4+ CD183− CD196+), regulatory T cells (Tregs) (CD45+ CD3+ CD4+ CD25+), CD8 cytotoxic T cells (CD45+ CD3+ CD8+ CD28+), and CD8 T suppressor cells (CD45+ CD3+ CD8+ CD28−) (34, 67, 68). The absolute number of each leukocyte population was determined by multiplying the frequency of the leukocyte by the number of cells determined from the single-cell suspensions and then multiplying by 2 to account for the whole lung.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism version 6.0 (GraphPad Software, Inc., San Diego, CA). Statistical significance for survival studies was assessed by log-rank test. Other group comparisons were performed by two-way analysis of variance (ANOVA) with Bonferroni’s multiple-comparison test.
SUPPLEMENTAL MATERIAL
Kinetic profiles of various cytokines in the lungs of the mice infected with various doses (equivalent to CFU of 105 or 106 or 107) of either the wild-type KN99α or the cda1Δ2Δ3Δ mutant strain, determined at 1, 3, and 7 days postinfection. Lungs were excised and homogenized as described in Materials and Methods, and 50 µl of the homogenate was used for the assay. Concentrations of the cytokines are expressed as pg/ml of the homogenate. Comparisons of the concentrations of the various cytokines were performed between the lungs of the mice infected with KN99α and with the corresponding cda1Δ2Δ3Δ strain. Data for the 107-CFU infection are representative of two biological experiments with three mice per each time point. Data for the 105- and 106-CFU infections are from one biological experiment with three animals per each time point. *, P < 0.05 compared to the corresponding wild-type or cda1Δ2Δ3Δ mutant strain-infected lung homogenate.
ACKNOWLEDGMENTS
We thank Maureen Donlin for editing the manuscript.
This work is supported by NIH grants RO1 AI072195 to J.K.L., RO1 AI125045 to J.K.L., S.M.L., and C.A.S., and RO1 AI25780 to S.M.L.
Footnotes
Citation Upadhya R, Lam WC, Maybruck B, Specht CA, Levitz SM, Lodge JK. 2016. Induction of protective immunity to cryptococcal infection in mice by a heat-killed, chitosan-deficient strain of Cryptococcus neoformans. mBio 7(3):e00547-16. doi:10.1128/mBio.00547-16.
REFERENCES
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Byrnes EJ III, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6:e00547-16. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrnes EJ III, Bartlett KH, Perfect JR, Heitman J. 2011. Cryptococcus gattii: an emerging fungal pathogen infecting humans and animals. Microbes Infect 13:895–907. doi: 10.1016/j.micinf.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson MD, Perfect JR. 2010. Use of antifungal combination therapy: agents, order, and timing. Curr Fungal Infect Rep 4:87–95. doi: 10.1007/s12281-010-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krysan DJ. 2015. Toward improved anti-cryptococcal drugs: novel molecules and repurposed drugs. Fungal Genet Biol 78:93–98. doi: 10.1016/j.fgb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Smith KD, Achan B, Huppler Hullsiek K, McDonald T, Okagaki LH, Akampurira A, Rhein JR, Meya DB, Boulware DR, Nielsen K, ASTRO-CM/COAT Team . 2015. Increased antifungal drug resistance in clinical isolates of Cryptococcus neoformans in Uganda. Antimicrob Agents Chemother 59:7197–7204. doi: 10.1128/AAC.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olszewski MA, Zhang Y, Huffnagle GB. 2010. Mechanisms of cryptococcal virulence and persistence. Future Microbiol 5:1269–1288. doi: 10.2217/fmb.10.93. [DOI] [PubMed] [Google Scholar]
- 8.Coelho C, Bocca AL, Casadevall A. 2014. The intracellular life of Cryptococcus neoformans. Annu Rev Pathol 9:219–238. doi: 10.1146/annurev-pathol-012513-104653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kronstad JW, Attarian R, Cadieux B, Choi J, D’Souza CA, Griffiths EJ, Geddes JM, Hu G, Jung WH, Kretschmer M, Saikia S, Wang J. 2011. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol 9:193–203. doi: 10.1038/nrmicro2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cauley LK, Murphy JW. 1979. Response of congenitally athymic (nude) and phenotypically normal mice to Cryptococcus neoformans infection. Infect Immun 23:644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huffnagle GB, Yates JL, Lipscomb MF. 1991. Immunity to a pulmonary Cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. J Exp Med 173:793–800. doi: 10.1084/jem.173.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta K, Pirofski LA. 2006. Towards a vaccine for Cryptococcus neoformans: principles and caveats. FEMS Yeast Res 6:525–536. doi: 10.1111/j.1567-1364.2006.00073.x. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigues ML, Travassos LR, Miranda KR, Franzen AJ, Rozental S, de Souza W, Alviano CS, Barreto-Bergter E. 2000. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect Immun 68:7049–7060. doi: 10.1128/IAI.68.12.7049-7060.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosas AL, Nosanchuk JD, Casadevall A. 2001. Passive immunization with melanin-binding monoclonal antibodies prolongs survival of mice with lethal Cryptococcus neoformans infection. Infect Immun 69:3410–3412. doi: 10.1128/IAI.69.5.3410-3412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nosanchuk JD, Rosas AL, Casadevall A. 1998. The antibody response to fungal melanin in mice. J Immunol 160:6026–6031. [PubMed] [Google Scholar]
- 16.Maitta RW, Datta K, Lees A, Belouski SS, Pirofski LA. 2004. Immunogenicity and efficacy of Cryptococcus neoformans capsular polysaccharide glucuronoxylomannan peptide mimotope-protein conjugates in human immunoglobulin transgenic mice. Infect Immun 72:196–208. doi: 10.1128/IAI.72.1.196-208.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitz SM, Huang H, Ostroff GR, Specht CA. 2015. Exploiting fungal cell wall components in vaccines. Semin Immunopathol 37:199–207. doi: 10.1007/s00281-014-0460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Specht CA, Lee CK, Huang H, Tipper DJ, Shen ZT, Lodge JK, Leszyk J, Ostroff GR, Levitz SM. 2015. Protection against experimental Cryptococcosis following vaccination with glucan particles containing Cryptococcus alkaline extracts. mBio 6:e00547-16. doi: 10.1128/mBio.01905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wormley FL Jr., Perfect JR, Steele C, Cox GM. 2007. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect Immun 75:1453–1462. doi: 10.1128/IAI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wozniak KL, Ravi S, Macias S, Young ML, Olszewski MA, Steele C, Wormley FL. 2009. Insights into the mechanisms of protective immunity against Cryptococcus neoformans infection using a mouse model of pulmonary cryptococcosis. PLoS One 4:e00547-16. doi: 10.1371/journal.pone.0006854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wozniak KL, Young ML, Wormley FL Jr.. 2011. Protective immunity against experimental pulmonary cryptococcosis in T cell-depleted mice. Clin Vaccine Immunol 18:717–723. doi: 10.1128/CVI.00036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rella A, Mor V, Farnoud AM, Singh A, Shamseddine AA, Ivanova E, Carpino N, Montagna MT, Luberto C, Del Poeta M. 2015. Role of sterylglucosidase 1 (Sgl1) on the pathogenicity of Cryptococcus neoformans: potential applications for vaccine development. Front Microbiol 6:836. doi: 10.3389/fmicb.2015.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe T, Ito T, Goda HM, Ishibashi Y, Miyamoto T, Ikeda K, Taguchi R, Okino N, Ito M. 2015. Sterylglucoside catabolism in Cryptococcus neoformans with endoglycoceramidase-related protein 2 (EGCrP2), the first steryl-beta-glucosidase identified in fungi. J Biol Chem 290:1005–1019. doi: 10.1074/jbc.M114.616300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai B, Wozniak KL, Masso-Silva J, Upadhyay S, Hole C, Rivera A, Wormley FL Jr., Lin X. 2015. Development of protective inflammation and cell-mediated immunity against Cryptococcus neoformans after exposure to hyphal mutants. mBio 6:e00547-16. doi: 10.1128/mBio.01433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker LG, Specht CA, Lodge JK. 2011. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot Cell 10:1264–1268. doi: 10.1128/EC.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker LG, Specht CA, Donlin MJ, Lodge JK. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell 6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cain JA, Deepe GS Jr.. 2000. Interleukin-12 neutralization alters lung inflammation and leukocyte expression of CD80, CD86, and major histocompatibility complex class II in mice infected with Histoplasma capsulatum. Infect Immun 68:2069–2076. doi: 10.1128/IAI.68.4.2069-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Root RK, Dale DC. 1999. Granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor: comparisons and potential for use in the treatment of infections in nonneutropenic patients. J Infect Dis 179(Suppl 2):S342–S352. doi: 10.1086/513857. [DOI] [PubMed] [Google Scholar]
- 29.Deepe GS Jr., McGuinness M. 2006. Interleukin-1 and host control of pulmonary histoplasmosis. J Infect Dis 194:855–864. doi: 10.1086/506946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaturvedi AK, Wormley FL Jr.. 2013. Cryptococcus antigens and immune responses: implications for a vaccine. Expert Rev Vaccines 12:1261–1272. doi: 10.1586/14760584.2013.840094. [DOI] [PubMed] [Google Scholar]
- 31.Arora S, Olszewski MA, Tsang TM, McDonald RA, Toews GB, Huffnagle GB. 2011. Effect of cytokine interplay on macrophage polarization during chronic pulmonary infection with Cryptococcus neoformans. Infect Immun 79:1915–1926. doi: 10.1128/IAI.01270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murdock BJ, Teitz-Tennenbaum S, Chen GH, Dils AJ, Malachowski AN, Curtis JL, Olszewski MA, Osterholzer JJ. 2014. Early or late IL-10 blockade enhances Th1 and Th17 effector responses and promotes fungal clearance in mice with cryptococcal lung infection. J Immunol 193:4107–4116. doi: 10.4049/jimmunol.1400650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baaten BJ, Li CR, Deiro MF, Lin MM, Linton PJ, Bradley LM. 2010. CD44 regulates survival and memory development in Th1 cells. Immunity 32:104–115. doi: 10.1016/j.immuni.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angkasekwinai P, Sringkarin N, Supasorn O, Fungkrajai M, Wang YH, Chayakulkeeree M, Ngamskulrungroj P, Angkasekwinai N, Pattanapanyasat K. 2014. Cryptococcus gattii infection dampens Th1 and Th17 responses by attenuating dendritic cell function and pulmonary chemokine expression in the immunocompetent hosts. Infect Immun 82:3880–3890. doi: 10.1128/IAI.01773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngamskulrungroj P, Chang Y, Roh J, Kwon-Chung KJ. 2012. Differences in nitrogen metabolism between Cryptococcus neoformans and C. gattii, the two etiologic agents of cryptococcosis. PLoS One 7:e00547-16. doi: 10.1371/journal.pone.0034258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhodes JC, Wicker LS, Urba WJ. 1980. Genetic control of susceptibility to Cryptococcus neoformans in mice. Infect Immun 29:494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll SF, Lafferty EI, Flaczyk A, Fujiwara TM, Homer R, Morgan K, Loredo-Osti JC, Qureshi ST. 2012. Susceptibility to progressive Cryptococcus neoformans pulmonary infection is regulated by loci on mouse chromosomes 1 and 9. Infect Immun 80:4167–4176. doi: 10.1128/IAI.00417-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shourian M, Flaczyk A, Angers I, Mindt BC, Fritz JH, Qureshi ST. 2015. The Cnes2 locus on mouse chromosome 17 regulates host defense against cryptococcal infection through pleiotropic effects on host immunity. Infect Immun 83:4541–4554. doi: 10.1128/IAI.00697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herring AC, Falkowski NR, Chen GH, McDonald RA, Toews GB, Huffnagle GB. 2005. Transient neutralization of tumor necrosis factor alpha can produce a chronic fungal infection in an immunocompetent host: potential role of immature dendritic cells. Infect Immun 73:39–49. doi: 10.1128/IAI.73.1.39-49.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huffnagle GB, Toews GB, Burdick MD, Boyd MB, McAllister KS, McDonald RA, Kunkel SL, Strieter RM. 1996. Afferent phase production of TNF-alpha is required for the development of protective T cell immunity to Cryptococcus neoformans. J Immunol 157:4529–4536. [PubMed] [Google Scholar]
- 41.Huffnagle GB, Chen GH, Curtis JL, McDonald RA, Strieter RM, Toews GB. 1995. Down-regulation of the afferent phase of T cell-mediated pulmonary inflammation and immunity by a high melanin-producing strain of Cryptococcus neoformans. J Immunol 155:3507–3516. [PubMed] [Google Scholar]
- 42.Noverr MC, Phare SM, Toews GB, Coffey MJ, Huffnagle GB. 2001. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun 69:2957–2963. doi: 10.1128/IAI.69.5.2957-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vecchiarelli A, Retini C, Pietrella D, Monari C, Tascini C, Beccari T, Kozel TR. 1995. Downregulation by cryptococcal polysaccharide of tumor necrosis factor alpha and interleukin-1beta secretion from human monocytes. Infect Immun 63:2919–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decken K, Köhler G, Palmer-Lehmann K, Wunderlin A, Mattner F, Magram J, Gately MK, Alber G. 1998. Interleukin-12 is essential for a protective Th1 response in mice infected with Cryptococcus neoformans. Infect Immun 66:4994–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoag KA, Lipscomb MF, Izzo AA, Street NE. 1997. IL-12 and IFN-gamma are required for initiating the protective Th1 response to pulmonary cryptococcosis in resistant C.B-17 mice. Am J Respir Cell Mol Biol 17:733–739. doi: 10.1165/ajrcmb.17.6.2879. [DOI] [PubMed] [Google Scholar]
- 46.Netea MG, Simon A, van de Veerdonk F, Kullberg BJ, Van der Meer JW, Joosten LA. 2010. IL-1beta processing in host defense: beyond the inflammasomes. PLoS Pathog 6:e00547-16. doi: 10.1371/journal.ppat.1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo C, Chen M, Fa Z, Lu A, Fang W, Sun B, Chen C, Liao W, Meng G. 2014. Acapsular Cryptococcus neoformans activates the NLRP3 inflammasome. Microbes Infect 16:845–854. doi: 10.1016/j.micinf.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 48.Caffrey AK, Lehmann MM, Zickovich JM, Espinosa V, Shepardson KM, Watschke CP, Hilmer KM, Thammahong A, Barker BM, Rivera A, Cramer RA, Obar JJ. 2015. IL-1alpha signaling is critical for leukocyte recruitment after pulmonary Aspergillus fumigatus challenge. PLoS Pathog 11:e00547-16. doi: 10.1371/journal.ppat.1004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, Perfettini JL, Schlemmer F, Tasdemir E, Uhl M, Génin P, Civas A, Ryffel B, Kanellopoulos J, Tschopp J, André F, Lidereau R, McLaughlin NM, Haynes NM, Smyth MJ, Kroemer G, Zitvogel L. 2009. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med 15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 50.Wüthrich M, LeBert V, Galles K, Hu-Li J, Ben-Sasson SZ, Paul WE, Klein BS. 2013. Interleukin 1 enhances vaccine-induced antifungal T-helper 17 cells and resistance against Blastomyces dermatitidis infection. J Infect Dis 208:1175–1182. doi: 10.1093/infdis/jit283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doyle HA, Murphy JW. 1997. MIP-1alpha contributes to the anticryptococcal delayed-type hypersensitivity reaction and protection against Cryptococcus neoformans. J Leukoc Biol 61:147–155. [DOI] [PubMed] [Google Scholar]
- 52.Olszewski MA, Huffnagle GB, Traynor TR, McDonald RA, Cook DN, Toews GB. 2001. Regulatory effects of macrophage inflammatory protein 1alpha/CCL3 on the development of immunity to Cryptococcus neoformans depend on expression of early inflammatory cytokines. Infect Immun 69:6256–6263. doi: 10.1128/IAI.69.10.6256-6263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huffnagle GB, Strieter RM, Standiford TJ, McDonald RA, Burdick MD, Kunkel SL, Toews GB. 1995. The role of monocyte chemotactic protein-1 (MCP-1) in the recruitment of monocytes and CD4+ T cells during a pulmonary Cryptococcus neoformans infection. J Immunol 155:4790–4797. [PubMed] [Google Scholar]
- 54.Kawakami K, Kinjo Y, Uezu K, Yara S, Miyagi K, Koguchi Y, Nakayama T, Taniguchi M, Saito A. 2001. Monocyte chemoattractant protein-1-dependent increase of V alpha 14 NKT cells in lungs and their roles in Th1 response and host defense in cryptococcal infection. J Immunol 167:6525–6532. doi: 10.4049/jimmunol.167.11.6525. [DOI] [PubMed] [Google Scholar]
- 55.Guillot L, Carroll SF, Homer R, Qureshi ST. 2008. Enhanced innate immune responsiveness to pulmonary Cryptococcus neoformans infection is associated with resistance to progressive infection. Infect Immun 76:4745–4756. doi: 10.1128/IAI.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiller T, Farrokhshad K, Brummer E, Stevens DA. 2002. Effect of granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor on polymorphonuclear neutrophils, monocytes or monocyte-derived macrophages combined with voriconazole against Cryptococcus neoformans. Med Mycol 40:21–26. doi: 10.1080/mmy.40.1.21.26. [DOI] [PubMed] [Google Scholar]
- 57.Huffnagle GB, Boyd MB, Street NE, Lipscomb MF. 1998. IL-5 is required for eosinophil recruitment, crystal deposition, and mononuclear cell recruitment during a pulmonary Cryptococcus neoformans infection in genetically susceptible mice (C57BL/6). J Immunol 160:2393–2400. [PubMed] [Google Scholar]
- 58.Cheng PY, Sham A, Kronstad JW. 2009. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect Immun 77:4284–4294. doi: 10.1128/IAI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chaturvedi AK, Weintraub ST, Lopez-Ribot JL, Wormley FL Jr.. 2013. Identification and characterization of Cryptococcus neoformans protein fractions that induce protective immune responses. Proteomics 13:3429–3441. doi: 10.1002/pmic.201300213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reese TA, Liang HE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, Locksley RM. 2007. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447:92–96. doi: 10.1038/nature05746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagener J, Malireddi RK, Lenardon MD, Köberle M, Vautier S, MacCallum DM, Biedermann T, Schaller M, Netea MG, Kanneganti TD, Brown GD, Brown AJ, Gow NA. 2014. Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog 10:e00547-16. doi: 10.1371/journal.ppat.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiesner DL, Specht CA, Lee CK, Smith KD, Mukaremera L, Lee ST, Lee CG, Elias JA, Nielsen JN, Boulware DR, Bohjanen PR, Jenkins MK, Levitz SM, Nielsen K. 2015. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS Pathog 11:e00547-16. doi: 10.1371/journal.ppat.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez FJ. 2014. The effect of chitin size, shape, source and purification method on immune recognition. Molecules 19:4433–4451. doi: 10.3390/molecules19044433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Da Silva CA, Chalouni C, Williams A, Hartl D, Lee CG, Elias JA. 2009. Chitin is a size-dependent regulator of macrophage TNF and IL-10 production. J Immunol 182:3573–3582. doi: 10.4049/jimmunol.0802113. [DOI] [PubMed] [Google Scholar]
- 65.Bueter CL, Lee CK, Wang JP, Ostroff GR, Specht CA, Levitz SM. 2014. Spectrum and mechanisms of inflammasome activation by chitosan. J Immunol 192:5943–5951. doi: 10.4049/jimmunol.1301695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 6:e00547-16. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. 2013. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol 49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Groom JR, Luster AD. 2011. CXCR3 in T cell function. Exp Cell Res 317:620–631. doi: 10.1016/j.yexcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kinetic profiles of various cytokines in the lungs of the mice infected with various doses (equivalent to CFU of 105 or 106 or 107) of either the wild-type KN99α or the cda1Δ2Δ3Δ mutant strain, determined at 1, 3, and 7 days postinfection. Lungs were excised and homogenized as described in Materials and Methods, and 50 µl of the homogenate was used for the assay. Concentrations of the cytokines are expressed as pg/ml of the homogenate. Comparisons of the concentrations of the various cytokines were performed between the lungs of the mice infected with KN99α and with the corresponding cda1Δ2Δ3Δ strain. Data for the 107-CFU infection are representative of two biological experiments with three mice per each time point. Data for the 105- and 106-CFU infections are from one biological experiment with three animals per each time point. *, P < 0.05 compared to the corresponding wild-type or cda1Δ2Δ3Δ mutant strain-infected lung homogenate.