Abstract
Arboviruses (arthropod borne viruses) have life cycles that include both vertebrate and invertebrate hosts with substantial differences in vector and host specificity between different viruses. Most arboviruses utilize RNA for their genetic material and are completely dependent on host tRNAs for their translation, suggesting that virus codon usage could be a target for selection. In the current study we analyzed the relative synonymous codon usage (RSCU) patterns of 26 arboviruses together with 25 vectors and hosts, including 8 vertebrates and 17 invertebrates. We used hierarchical cluster analysis (HCA) and principal component analysis (PCA) to identify trends in codon usage. HCA demonstrated that the RSCU of arboviruses reflects that of their natural hosts, but not that of dead-end hosts. Of the two major components identified by PCA, the first accounted for 62.1% of the total variance, and among the 59 codons analyzed in this study, the leucine codon CTG had the highest correlation with the first principal component, however isoleucine had the highest correlation during amino acid analysis. Nucleotide and dinucleotide composition were the variables that explained most of the total codon usage variance. The results suggest that the main factors driving the evolution of codon usage in arboviruses is based on the nucleotide and dinucleotide composition present in the host. Comparing codon usage of arboviruses and potential vector hosts can help identifying potential vectors for emerging arboviruses.
Introduction
Rather than a taxonomic classification, arboviruses are a broad group of viruses transmitted biologically by hematophagous (blood-feeding) arthropod vectors (e.g. mosquitoes, ticks, biting flies) to vertebrate hosts [1]. This type of transmission cycle involves virus-host interactions with both invertebrate and vertebrate hosts. Arboviruses not only affect humans but also other animal species and some are transmitted and many cause disease in both humans and animals (zoonotic diseases). Understanding this three-component transmission cycle, is of great importance as recently arboviral infections have been seen with increasing frequency and magnitude for both old and newly emerging arboviruses (e.g. Zika virus). For example, dengue virus affects over 400 million people a year (http://www.cdc.gov/dengue/). Recent examples of emerging arboviral activity in human populations include the 2003 West Nile fever outbreak[2], and the Chikungunya virus, which reached the Western hemisphere sparking over 1.5 million new clinical cases [3]. A recent non-human arboviral infection; Schmallenburg virus was identified in Germany in 2011 has induced thousands of cases in eight European countries [4].
Arboviruses consist mostly of RNA virus with the exception of African swine fever virus a dsDNA virus[1], the RNA viruses that are classified as arboviruses are comprised of many different taxa, having in common only that they are viruses that infect vertebrate hosts but are transmitted by arthropod vectors. Current biological information regarding the infectious cycles of arboviruses shows substantial differences in vectors, hosts and transmission modes. For example, West Nile virus (Flaviviridae) is maintained and amplified in nature within an enzootic transmission cycle among birds and Culex mosquitoes, with outbreaks caused by tangential or spillover transmission to equids and humans, which may develop terminal neuroinvasive disease. However, these are considered dead-end hosts because they do not develop high enough viremia adequate for mosquito infection [1]. In some cases, the role of the vertebrate in the life cycle of the virus may be minimal. For example, many phleboviruses seem to be maintained in their vectors, phlebotomine sandflies, by vertical (transovarial) transmission, which may allow persistence during periods when susceptible vertebrate hosts are not available [5].
Viruses are intracellular pathogens that have to exploit and co-evolve with host molecular mechanisms to prosper in a cellular environment [6]. Most amino acids are encoded by 2–6 different synonymous codons due to the redundancy of the genetic code. Codon usage bias refers to the phenomenon that some synonymous codons are used more often than others and how this preference varies within and among species [7]. There are two non-mutually exclusive models that propose mechanisms to account for codon usage bias in viruses. The transitional model assumes that codon usage is under selection because RNA viruses are completely dependent of host tRNAs [8] and the bias results from viruses matching the codon usage of their hosts [9]. In addition, other factors such as mononucleotide and dinucleotide composition are likely to influence codon usage in RNA viruses. The second model proposes that mutational pressures and the probability of fixation for different mutations determine codon usage bias [10]. Evolution can sometimes favor viruses that match host codon usage to promote speed of replication, as is the case of poliovirus [11, 12, 13] and influenza A virus [14].
In order to investigate possible patterns of coevolution between arboviruses and their vertebrate and invertebrate hosts, we analyzed the base composition and codon usage bias of different arboviruses and their respective vertebrate and invertebrate host. Our results suggest that codon usage patterns among different arbovirus are consistent with the codon usage of their respective natural hosts, with the mimicking of nucleotide and dinucleotide compositions being the main factors that explain these patterns. Correlations between these factors may have practical application for the characterization of emerging arboviruses and the identification of their hosts and vectors.
Materials and Methods
Viral Dataset
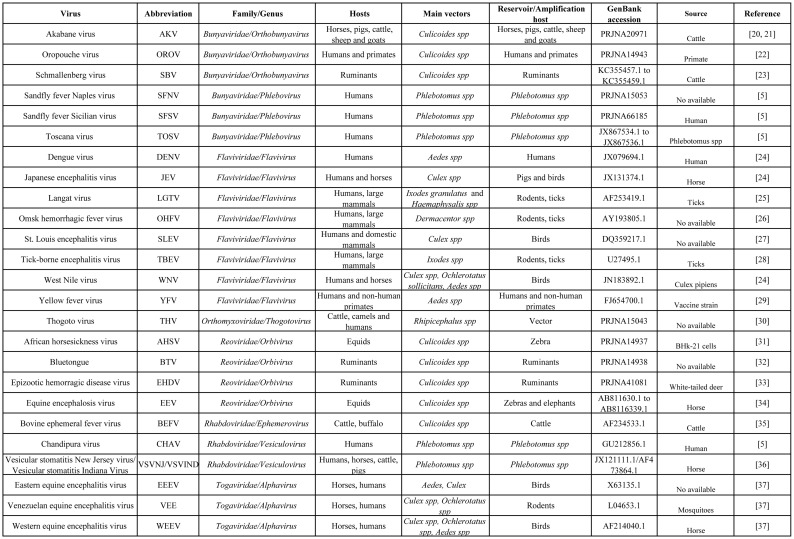
All complete viral genome sequences included in this study were downloaded from the National Center for Biotechnology Information [15]. These viral genomes were randomly chosen in order to obtain one representative member from each of the different viral families included in this study. Detailed information about the viruses used in this study is presented in Fig 1.
Fig 1. General information about viral species used in this study.
Twenty-six different viruses comprised of 6 different viral families that represent the most common fully sequenced arboviruses were used for this study.
Host Dataset
The host and vector codon usage and GC composition in coding regions were obtained from the Codon Usage Data Base [16], which includes sequences of complete protein-coding genes. The vertebrate dataset was composed of the following species: Cow (Bos taurus), horse (Equus caballus), chicken (Gallus gallus), human (Homo sapiens), turkey (Meleagris gallopavo), mouse (Mus musculus), chimpanzee (Pan troglodytes) and pig (Sus scrofa). The invertebrate dataset included insects and arachnids: mosquitoes in the family Culicidae (Aedes aegypti, Aedes albopictus, Culex nigripalpus, Culex pipens quinquefasciatus, Culex pipiens, Culex tritaeniorhynchus and Ochlerotatus sollicitans), sand flies in the family Psychodidae (Lutzomyia longipalpis, Phlebotomus argentipes, Phlebotomus ariasi, Phlebotomus duboscqi, Phlebotomus papatasi, Phlebotomus perniciosus), midges in the family Ceratopogonidae (Culicoides sonorensis), as well as the tick species Ixodes rinicus and Rhipicephalus microplus of the family Ixodidae.
Compositional bias measures
For each virus included in this study, codon usage, overall GC-content, and the GC1, GC2, and GC3 (GC content at the first, second and third codon position, respectively) were calculated using the CAIcal software [17].
Dinucleotide odds ratio is defined as the quotient of the probability of finding a dinucleotide in a given sequence divided by the product of the probabilities of finding each nucleotide that forms the pair in the same sequence, calculated as shown in the following equation: Pxy = (fxy)/(fxfy). Where fx and fy denote the frequency of mononucleotides x and y in a given sequence and fxy denotes the frequency of dinucleotide xy in the same sequence. In the case of organisms with double-stranded genomic material, the frequency of each dinucleotide must be calculated in a symmetric manner, also considering the complementary strand as described in the following equation: Pxy = 2(fxy +fzw) / (fx +fy) (fz + fw). Where x and y denote two dinucleotides, and z and w denote the two complementary nucleotides of y and x, respectively [18].
Dinucleotide odds ratios for all viral sequences used in this study were calculated using single-stranded odds ratios; the only exception being viruses from the Reoviridae family, which contain a double RNA strain in their genome. In this case, we used the same method applied to the hosts and calculated the dinucleotide odds ratio using the symmetric odds ratio.
Amino acid composition calculation
Average amino acid compositions among different host proteins were obtained directly from the Codon Usage Data Base, which uses coding sequence composition for to determine Codon Usage for individual species. For the viruses, the amino acid composition was inferred considering the protein coding region for the viruses under study. Calculations were conducted in the ExPASy Bioinformatics Resource Portal using the software ProtoParm [16].
Relative Synonymous Codon Usage (RSCU)
RSCU is a common measure used to estimate codon bias for all codons that code for any amino acid with a degeneracy greater than one (i.e. all except methionine and tryptophan). It is defined as the observed frequency of the codon j in a sequence x divided by the expected frequency E if all synonymous codons for the amino acid coded by j were equally frequent. Calculations were conducted using the following equation: RSCUj(x) = (fjx /Ejx,) Where fjx is the observed frequency of codon j in the genome x and Ejx is the expected frequency of the codon j. Expected values are calculated by counting the total number of synonymous codons for a given amino acid in the sequence divided by the number of existing codons that code for it. RSCU values larger than 1.0 indicate that a given synonymous codon is favored over the rest; RSCU values less than 1.0 indicate a disfavored codon; and RSCU values of 1.0 indicate no preference [19].
Two-way hierarchical clustering analysis (TWHCA)
Two-way hierarchical clustering analysis is a statistical method that classifies objects into groups (clusters) according to similarities between them, and is used to identify a subset in one dimension that is useful for clustering the other dimension [20]. We organized all organisms (viruses, hosts and vectors), in a matrix of N x M dimensions, with N being the number of species and M the number of degenerate codons, represented by their RSCU values. Monocodonic amino acids (tryptophan and methionine) and stop codons (UAA, UAG and UGA) were excluded to generate a final multivariate data set of 59 codons for each organism. The original dataset with all RSCU values is included in S1 Table. For the cluster analysis, the values for each codon were scaled and centered by subtracting the mean and dividing by the standard deviation. With the standardized values, cluster analysis using Ward’s minimum variance method [21] in combination with initial Manhattan (city-block) distance measures was conducted in the R statistical environment. Ward’s method joins clusters based on minimizing the within-group sum of squares and tends to produce compact clusters. The results of the cluster analysis are presented as dendrograms, and the order of clusters is used to reorder the rows and columns of a heat map showing the RSCU values.
To assess the uncertainty in the first dimension of the hierarchical cluster analysis, approximately unbiased (AU) p-values were calculated by a multiscale bootstrap procedure [22]. The AU values are expressed as percentages and are superimposed over the corresponding dendrograms. For a cluster with a given AU p-value, the null hypothesis of non-existence of the cluster is rejected at a significance level of (100-AU)/100; i.e., for high AU values, it can be assumed that these clusters do actually exist in the original data, and are not merely caused by sampling error.
Principal component analysis (PCA)
PCA is an orthogonal linear transformation that converts the original data set into a new coordinate system, in which the greatest variance represented by any projection of the data (the first principal component; PC) comes to lie on the first coordinate and the second greatest variance on the second PC [23]. This method is frequently used to analyze multivariate data sets. Reliable components in the analysis where retained based on the eigenvalues (the variance of the principal components), applying the criteria of the eigenvalues greater than one rule [24].
Correlations between different variables and the main principal components were examined by correlation analysis and analysis of variance (ANOVA) using the statistical software JMP 11.
Results
Hosts are influencing codon usage in arboviruses
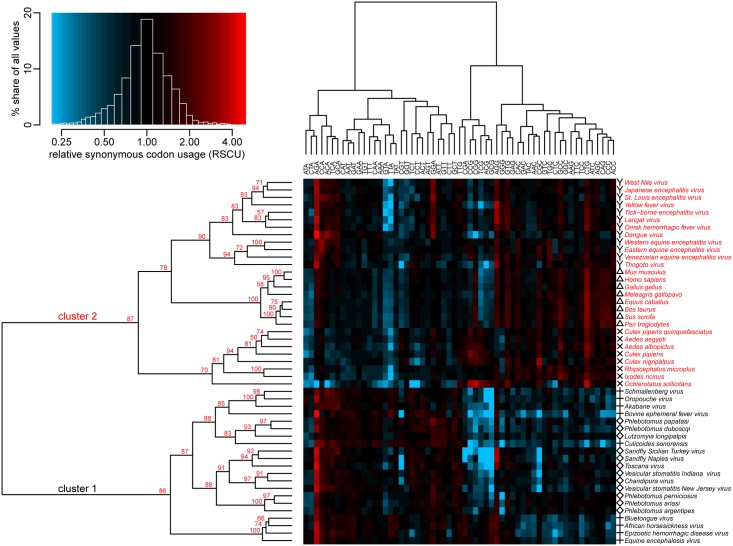
TWHCA was utilized to investigate intra-species differences in codon usage and evaluate similarities among species. In the first dimension of the analysis, there was a clear division of all species, viral and eukaryotic, into two main clusters labeled as cluster one and cluster two (Fig 2). This cluster division was further supported by the multivariate bootstrap analysis, in which unbiased p-values (AU) of 88 and 86 were obtained for cluster one and two, respectively.
Fig 2. Influence of host on codon usage in arboviruses.
The two-way hierarchical cluster analysis shows a correlation of the RSCU between viruses and their respective hosts. In the first dimension, viral and invertebrate species were split into two main clusters, while vertebrate species clustered together, in the second dimension, there was a contrasting pattern between species based on their preference for A/T or G/C-end codons. The cluster analysis was done with centered and scaled RSCU values, but cell colors in the heat map represent the original values. High RSCU values are shown in red and low values in blue; values around 1 are shown in black. The AU p-values from a multiscale bootstrap analysis (n = 10000) are overlaid over the first-dimension dendrogram.
In the first dimension of these analyses, cluster one included all viruses from the genera Vesiculovirus, Ephemerovirus, Orthobunyavirus, Phlebovirus and Orbivirus, together with their main invertebrate vectors, which belong to the families Psychodidae and Ceratopogonidae. Within cluster one, viruses associated more closely with their respective vectors than with other related viruses. For example, the Culicoides-transmitted viruses, particularly AKV, OROV, SBV and BEFV, grouped closely with Culicoides sonorensis, while SFNV, SFSV, TOSV, VSNJV, and VSIV grouped closely with members of the Psychodidae family (their associated sand fly vectors).
Cluster two included the genera Alphavirus, Thogotovirus and Flavivirus, which were similar in their codon usage to both vertebrates (mammals and birds) and invertebrates of the families Culicidae and Ixodidae. Unlike the species in cluster one, the distinctive codon usage patterns of species in cluster two placed them into subgroups retracing their phylogenetic origin rather than the biological interaction between viral species and their respective vector or host.
Patterns of codon usage between arboviruses
Analyzing the second dimension of the TWHCA, it is possible to distinguish two main clusters based on their preference for codons with A/T or G/C endings. Although the viruses were split into two different clusters, all of them shared a common pattern of codon usage correlated with the usage of codons TCA, AGA, ACA, GCA, AGG, TTG and CCA, for which the average RSCU for all viral groups was higher than one. With the exception of the orbiviruses, the remaining viral groups avoided the usage of certain codons containing CpG dinucleotides, such as TCG, CCG, ACG, GCG and CGA, where the average RSCU was lower than one. The same pattern of low usage of codons containing CpG was displayed only by the vertebrate group and the invertebrates associated with cluster one. Invertebrates associated with cluster two had a high preference for the usage of these codons.
A second pattern of usage reflected the preferences among viruses and their respective hosts. In the case of viruses associated with cluster one, similar preferences of usage between them and their hosts were seen for codons TTT, TTA, TAT, ATA, CAT, AAT, GAT, GGT, AAT, TCT, CCT, ACT and GCT. For viruses in cluster two, these similarities were seen for codons CTG, ACC, ATC, AGC and CAC.
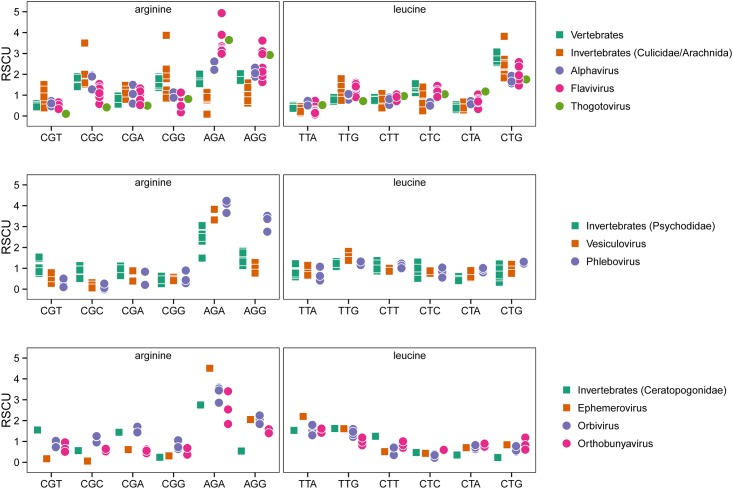
Interestingly, the RSCU values of all 59 codons analyzed in this study were fairly disparate across the two main clusters. Based on their RSCU values, two of the most relevant codons identified by this analysis were CTG and AGA. CTG, one of the six codons used to encode leucine, had the highest RSCU difference between the two main clusters (1.54±0.45), with average values of 0.85±0.34 for species contained in cluster one and 2.39±0.57 for species contained in cluster two. Among the six available codons for the amino acid leucine, the CTG codon had the highest RSCU within the coding regions analyzed in vertebrates (2.82±0.19) and invertebrates of the Culicidae family (2.67±0.62). Flaviviruses, alphaviruses and thogotovirus displayed the same predilection for the CTG codon, contrasting with viruses in the genera Vesiculovirus and Phlebovirus usually associated with invertebrates of the Psychodidae family, as well as with ephemeroviruses, orbiviruses and orthobunyaviruses commonly associated with invertebrates of the Ceratopogonide family (Fig 3). Contrastingly, AGA, one of the six codons used to encode arginine was by far the codon with the highest RSCU (3.33 ±0.77) among viral populations, independently of their host preference, suggesting that a common evolutionary pattern among may be favoring the high usage of this codon.
Fig 3. Frequencies of relative synonymous codon usage for encoding the amino acids leucine and arginine.
RSCU for the different codons to encode the amino acids leucine and arginine, were calculated and compared between different groups of viruses and their respective hosts and for each virus and host. Each dot in the graphic represents a single virus or host. Different families of hosts and viruses were represented in different colors. In the case of leucine, Flaviviruses, alphaviruses and thogotoviruses displayed the same high predilection for the CTG codon as their natural hosts, using this codon as a first option to encode leucine, while in case of arginine, AGA was the codon with the highest RSCU among viral populations independently of their host preferences.
Factors influencing codon usage in arboviruses
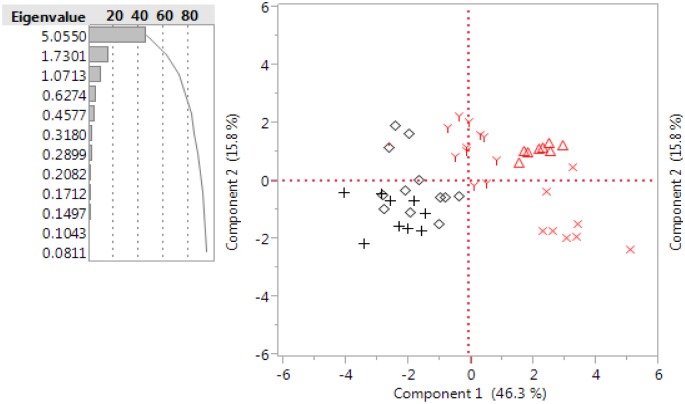
In order to gain a more thorough understanding of which factors, might be relevant in the choice of codon usage in arboviruses and their respective vectors and hosts, we conducted principal component analysis (PCA). The results showed that the two principal components were able to explain 62.1% of the total variation among the 59 RSCU indices. The first principal component with an eigenvalue of 5.05 accounted for 46.3% of the total variance, while the second principal component with an eigenvalue of 1.73 contributed with 15.8% of the differences. Viral species were clustered in two separate groups, each containing their respective vectors or hosts, confirming the prior observations in the TWHCA and providing further support to the hypothesis that host or vector codon usage are major determinants of virus codon usage (Fig 4).
Fig 4. Two main principal components explain 62.1% of the total variation among RSCU indices.
The principal component analysis was conducted using RSCU values corresponding to 59 codons of all eukaryotic and viral species used in this study. Based on eigenvalues of 5.05 and 1.73 the first principal component one accounts for the 46.3% of the total variance, while the second principal component for the 15.8%. In black are represented the following species: (+) Orthobunyavirus, Ephemerovirus, Orbivirus and Ceratopogonide. (◊) Vesiculovirus, Phlebovirus and Psychodidae. In red are represented the following species: (Y) Flavivirus, Alphavirus and Thogotoviridae. (Δ) Vertebrates. (X) Culicidae and Arachnida.
Relevant codons associated with the two principal components
Correlation analysis was utilized in order to identify which codons correlated best with the two first principal components. There were significant positive and negative correlations (p<0.05) between the first principal component and several codons, associated with the two main clusters of the TWHCA (Fig 3. The highest positive correlation was with CTG (r = 0.94), followed by ACC (r = 0.87), GGC (r = 0.81), CAC (r = 0.81), CAG (r = 0.77), CGG (r = 0.77), GTG (r = 0.77) and negative correlations were found for AAA (r = -0.9), AAT (r = -0.87), ATT (r = -0.86), CAA (r = -0.84), TTA (r = -0.82), TAT (r = -0.82) and TCA (r = -0.78). In addition, we found significant negative and positive correlations (p<0.05) with the second principal component for codons TCG (r = 0.75), CCG (r = 0.66), ACG (r = 0.52), TTC (r = 0.52), GAT (r = 0.46), CGT (r = 0.44), AGG (r = -0.66), AGA (r = -0.64), ACA (r = -0.63), GGG (r = -0.62), GCA (r = -0.55) CTA (r = -0.45).
Nucleotide and dinucleotide compositions are the main factors influencing codon usage in arbovirus
Nucleotide composition
To determine the influence of nucleotide, dinucleotide and amino acid compositions on the variability of codon usage, these parameters were calculated for each species and ANOVA was used to examine correlation with the two main principal components. The results indicated that differences in GC nucleotide composition were by far the best correlated variable with the first principal component. Significant associations (p<0.0001) were found for GC3 (R2 = 0.87), GC2 (R2 = 0.74) and GC1 (R2 = 0.61), suggesting that GC3 is the main factor influencing the variability associated with the first principal component. In general, vertebrates, invertebrates in the family Culicidae or class Arachnida, as well as viruses from the genera Alphavirus, Flavivirus and Thogotovirus have the highest total percentage of GC3, accounting for more than 50% of the total mononucleotide content. In contrast, invertebrates in the families Psychodidae and Ceratopogonidae and viruses in the genera Vesiculovirus, Ephemerovirus, Orthobunyavirus, Phlebovirus and Orbivirus have a total GC3 content below 50%. These differences correlate strongly (p<0.0001) (R2 = 0.85) with the usage of the CTG codon among species in this study.
Dinucleotide composition
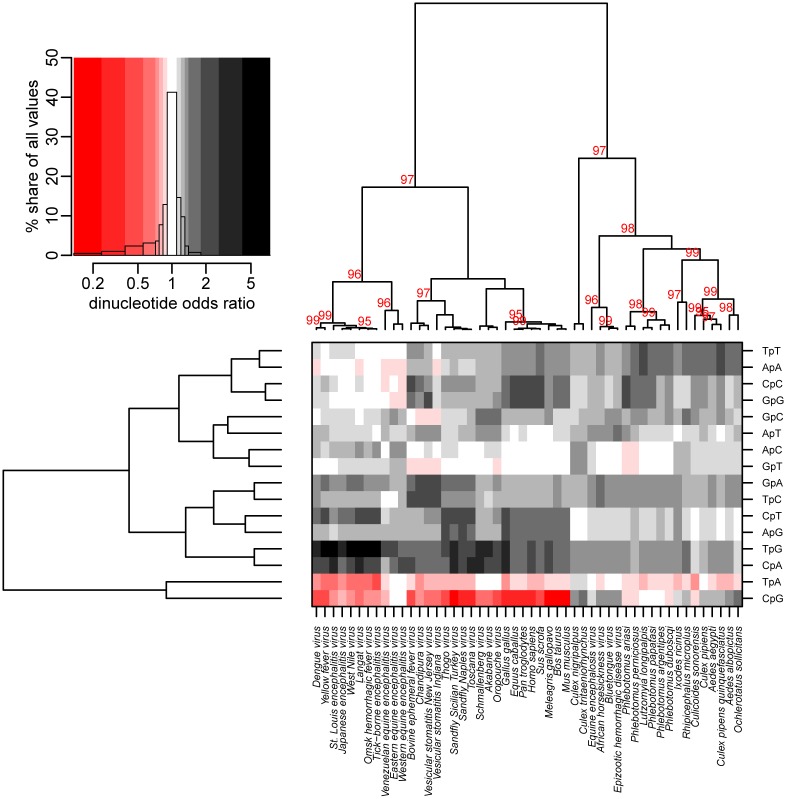
Dinucleotide composition was the variable that was most significantly associated with the second principal component. There were significant correlations (p<0.0001) with dinucleotides CpT (R2 = 0.87), ApG (R2 = 0.64), CpG (R2 = 0.63), CpA (R2 = 0.66) and TpG (R2 = 0.65). Comparing patterns of dinucleotide usage among viruses, vertebrates and invertebrates hosts, using TWHCA, we found that the hosts were assigned to separate clusters. With the exception of orbiviruses, which clustered with invertebrates, the arboviruses grouped together with the vertebrate group and presented a typical dinucleotide pattern with significant underrepresentation of dinucleotides CpG and TpA and overrepresentation of dinucleotides CpA, and TpG, (Fig 5). These results suggest that viruses acquire these dinucleotide patterns as a consequence of their replication in the vertebrate host and these patterns shape codon usage similarities between viruses associated with different clusters (cluster one vs. cluster two) in the TWHCA based on RSCU values.
Fig 5. Dinucleotide odds ratios in most arboviruses resemble the vertebrate hosts.
Hierarchical cluster analysis was conducted using the dinucleotide composition of all species included in this study. AU values of 97 (n = 10000) support the existence of two main clusters with either invertebrates or vertebrates. With the exception of orbiviruses, which clustered with invertebrates, the arboviruses grouped together with the vertebrates.
Amino acid composition
Comparing amino acid frequencies by the Tukey-Kramer test, leucine (Leu) with an average of 9±1% (p<0.05) was predominant among the proteins of both viruses and hosts (S1 Fig). However, isoleucine (Ile) had the best association with the first principal component (p<0.0001, R2 = 0.53). Differences in Ile composition correlated best with the general CG composition (p<0.0001, R2 = 0.73). Interestingly, it has been demonstrated previously that a strong bias in the nucleotide composition can also affect the average amino acid composition of the encoded proteins, suggesting that AT-rich coding sequences would encode proteins with excess of FYMINK amino acids (phenylalanine, tyrosine, methionine, isoleucine, asparagine, and lysine), whereas GC-rich sequences would produce proteins with high levels of GARP amino acids (glycine, alanine, arginine, and proline) [25]. The influence of GC content on amino acid composition was probed using ANOVA to look for associations between GC content and the GARP/FYMINK ratio. The results demonstrated a statistically significant linear correlation (p < 0.0001) between that ratio and GC (R2 = 0.61), GC1 (R2 = 0.67), GC2 (R2 = 0.77) and GC3 (R2 = 0.38). Thus, GC composition is not only influencing 46.2% of the RSCU variance but also might be influencing the differences in amino acid composition between different species. However, because our analysis was only done using the coding region of arboviruses, we cannot rule out the possibility that amino acid composition is driving base composition.
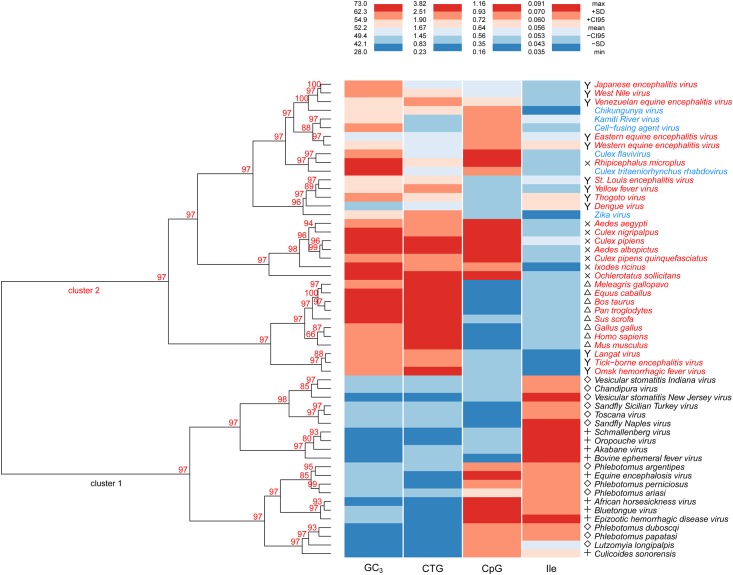
Biological markers to infer host preferences
Based on the differences in codon usage, nucleotide, dinucleotide and amino acid compositions among different arboviruses and their vertebrates and invertebrates hosts, it is possible to propose relevant biological markers that might be useful for the characterization of future emerging arboviruses. For this propose, using the values of GC3, CTG, CpG, and Ile, it was conducted a new TWHCA in order to confirm the previous cluster association obtained using RSCU values. Additionally, the potential use of this analysis was demonstrated adding six new viruses. Four viruses were chosen based on their ability to replicate solely in invertebrate cell culture and were isolated from insect cell lines or from field-collected mosquitoes. Specifically viruses in this category and their sources were: cell-fusing agent virus (flavivirus), isolated form Aedes aegypti [26], Kamiti River virus (flavivirus), isolated from Aedes macintoshi [27], Culex flavivirus (flavivirus) isolated from Cluex pipiens [28] and Culex tritaeniorhynchus rhabdovirus isolated from Culex tritaeniorhynchus [29]. Additionally, two different reemerging arboviruses were chosen: Chikungunya virus (Alphavirus) and Zika virus, both transmitted by Aedes mosquitoes [30,31]. Using just these four biological markers, a biologically-relevant cluster of viruses and their invertebrate host was obtained, statistically supported by high AU values (Fig 6). The only exceptions were LGTV, OHFV and TBEV (transmitted by ticks) that appear more associated with the vertebrate cluster. Interestingly, although phylogenetically Culex tritaeniorhynchus rhabdovirus, is more related to the rest of the rest of rhabdoviruses used in this study (BEFV, CHAV, VSVNJ and VSVIND), it grouped in the opposite cluster clearly influenced by its host preference, associated with viruses transmitted by mosquitoes of the Culicidae family.
Fig 6. Inferring arbovirus host preferences.
A two way hierarchical cluster analysis (Euclidean distance, Ward’s D clustering) was conducted using some of the relevant markers identified in this study (GC3, CTG, CpG, Ile). The cluster relationship was congruent with the current biological information regarding to the different infectious cycles of these viruses. The AU p-values from a multiscale bootstrap analysis (n = 10000) are overlaid over the dendrogram Viruses with blue labels represent new viruses included to assess the reliability of this analysis.
Discussion
In the investigation described here we sought to understand possible selective factors associated with the evolution of codon usage in natural populations of arboviruses. For this purpose we used a combination of different bioinformatic tools to analyze the complete genomes of 26 arboviral species from six different viral families.
Vertebrates and invertebrates could influence codon usage in distinct arboviruses lineages
Subsequently we used HCA to test the hypothesis that specific virus-host interactions play a role in arboviral RSCUs. Among hosts, vertebrates displayed the lowest variation in codon usage bias and all species were found in a specific sub-cluster. In contrast there was substantial variation among the invertebrates, which appeared split into the two main clusters. These patterns are consistent with a previous study in which the vertebrates demonstrated lower variation of codon usage bias compared with the average of other groups of eukaryotes as well as prokaryotes [32]. The authors of that study suggested that there is a negative correlation between codon usage bias diversity and genomic complexity.
In contrast to vertebrates, insects are considered the most diverse organisms in the history of eukaryotic life, with a projected number of recognized species close to one million. [33]. The insect order Diptera (which includes Culicidae, Psychodidae and Ceratopogonide familes) is the most species-rich, anatomically varied and ecologically innovative group, making up 10–15% of known animal species [34]. A previous study using 22 insect species of order Diptera (Culicidae and Drosophila) and Hymenoptera showed the contrasting patterns of codon frequencies between these two orders [35]. In the current study, we have demonstrated that there is high codon usage diversity associated with the Diptera order. This is interesting, because the high diversity of codon usage identified in the invertebrate group was consistent with the pattern shown by arboviruses, which grouped into the two main clusters, suggesting that invertebrates rather than vertebrates might be responsible for differences in codon usage between arboviruses. This is consistent with the fact that the evolutionary origin of most of the viral families included in this study is within insects [36] However, it is possible that the differences in clustering between vertebrates and the two main clusters of invertebrates could be influenced by shared ancestry between the invertebrates in each cluster, and the clustering in the virus families could be due to a common ancestor within the viruses studied.
Although all viral species included in this study have been recovered from vertebrates in nature, invertebrates can be involved in completing the infectious cycle for these viruses in nature. For example, a study conducted in Barkehji-Senegal from 1990 to 1995, used insect surveillance that determined CHAV and WNV were only isolated from their biological vectors (phlebotomine sand flies and Aedes species, respectively) [37]. Because other invertebrates were commonly found in the same area, and no CHAV or WNV were found in other invertebrates, it suggests that the main phase of restriction of arboviral replication could be the invertebrate host. In VSV, Phlebotomine sand flies are the only vector in the absence of clinical cases, to be confirmed biologically to host VSV. However during epidemics VSV has been isolated from midges (Culicoides spp and black flies) and mosquitoes (Aedes spp),[38]. Different in vivo and in vitro experiments conducted using VSIV, DENV and VEEV have also suggested the preponderance of the insect phase over the mammalian phase in evolutionary terms, suggesting that this may be a common feature of arbovirus evolution [16,17,39,40].
Codon usage could reflect the natural transmission cycle in arboviruses
Cluster analysis of codon usage appeared to be consistent with biological studies of host and vector competence during natural transmission cycles. For example, in viruses belonging to the Alphavirus, Thogothovirus and Flavivirus genera, codon usage bias appeared related to both the vertebrate and the invertebrate host, while for viruses in the Vesiculovirus, Ephemerovirus, Orthobunyavirus, Phlebovirus and Orbivirus genera, codon usage bias remained clearly distinct from that of vertebrates and clustered just with their invertebrate host. In the first group of viruses, vertebrates play an important role in maintenance of the virus in nature and vertebrate reservoirs produce transient, high-titer viremias that allow transmission of the virus to feeding mosquitoes or ticks. Additional vertebrates, like humans or horses, can be infected but are considered dead-end hosts because of their lack of ability to produce sufficient viremia. In some cases, amplifier hosts, like birds infected with SLEV or with WNV, may develop high viremia without producing any symptoms or adverse effects on their health, thus increasing the possibility of transmission [41,42]. Conversely, in the second group viruses, that were recovered from vertebrate infections in nature, codon usage appears different than the codon usage of vertebrates. Previous research done in phleboviruses and VSV has produced inconclusive results trying to identify a vertebrate reservoir for the long-term persistence suggesting the possibility that long-term persistence is maintained solely by their respective vector. [43–45]. Finally, experimental work carried out with BTV indicates that once the virus infects a Culicoides vector, the midge will be able to infect a vertebrate host between 10–14 days after infection, corresponding to the period of time between the ingestion and the replication of the virus in the salivary gland. [46].
Matching nucleotide and dinucleotide host compositions could be a mechanism influencing codon usage in arboviruses
The principal component analysis was used in order to identify potential factors influencing codon usage bias in arboviruses. Our results were consistent with previous research conducted in flaviviruses that determined hosts ware an important factor in shaping nucleotide motifs in flaviviruses [18]. Our results suggest that differences in nucleotide and dinucleotide compositions also influenced patterns of codon usage in distinct arboviruses. Experimental evidence in human immunodeficiency virus (HIV) showed that the biased nucleotide composition of HIV RNA is detected in human cells and induces a stronger immune response as compared to HIV RNA that had human optimized codon usage bias[47]. Studies conducted in polioviruses have also shown that differences in the nucleotide composition even at silent sites can determine the mutational robustness, evolutionary capacity and virulence, all factors that facilitate replication and spread within the dynamic host environment [13].
In the case of dinucleotide composition, selection favoring CpG depletion in RNA viruses has been associated with decreasing the innate immune response imposed by the vertebrate host [48]. This response may be mediated by the intracellular Pattern Recognition Receptor (PRR) Toll-like receptor 9 (TLR9), which recognizes in vertebrates CpG unmethylated DNA as a sign of infection [49]. Some experimental work conducted in echovirus (Picornaviridae family), altering the frequencies of CpG dinucleotides in the viral composition of this virus, showed that those viruses with increased frequencies of CpG had impaired replication kinetics and higher physical particule/infectious particle ratios as well as higher expression of mRNA for tumor necrosis factor and interferon beta genes compared with the wild type virus. Interestingly, the mutants with CpG dinucleotide depletion, showed enhanced replication and outcompeted wild-type virus during co-infections [50]. This work suggested that some viruses could have evolved with decreased CpG to avoid detection with TLR9, and thus avoid recognition by the immune system.
Relevance of the CTG codon
Based on the contrasting usage preferences between the species used in this study, CTG was one of the most relevant codons. In the case of vertebrates, previous research showed a correlation between tRNA abundance and CTG preference in 12 out of 24 species [51]. Two species (Gallus gallus and Mus musculus) were also included in the current study. Interestingly, the mammalian CTG codon has evolved as an alternative start codon, contributing to the production of protein isoforms [52], a fact that might explain the high RSCU values for this codon in vertebrates. Only two insect species are available in the Genomic tRNA Database [53], one associated with the Culicidae family (Anopheles gambiae) and the other with the Drosophilidae family (Drosophila melanogaster). Both of these species demonstrated the same correlation between tRNA composition and the preference for CTG codon. This suggests the possibility that transitional selection (abundance of tRNA in a cell) might be one of the forces associated with the usage of this codon. However currently with only these two insect species available to evaluate the tRNA composition and the preference for the CTG codon its possible that other insect species may not have the same preference towards the CTG codon.
Viral hosts have been demonstrated to be an important factor in shaping nucleotide composition, amino acid composition and codon usage, it is possible as more sequenced arboviruses become available, and the codon usage data for more insect species is determined, that new codon bias factors could be identified. The main factors identified in this study, are a first step, and have shown promise to predict the host and vector species associated with newly identified viruses with unknown natural transmission cycles. However further analysis will be required to identify other codon bias factors that could be used to rationally predict the hosts for emerging arboviruses.
Supporting Information
Tukey-Kramer analysis conducted among vertebrate, invertebrate and virus groups to determinate the frequency of amino acids among the proteins analyzed in this study.
(TIF)
(XLSX)
Acknowledgments
The authors thank Elizabeth Ramirez-Medina and Steven J. Pauszek from Plum Island Animal Disease Center for their technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1.Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral research. 2010. February;85(2):328–45. 10.1016/j.antiviral.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuno G, Chang GJ. Biological transmission of arboviruses: reexamination of and new insights into components, mechanisms, and unique traits as well as their evolutionary trends. Clinical microbiology reviews. 2005. October;18(4):608–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broeckel R, Haese N, Messaoudi I, Streblow DN. Nonhuman Primate Models of Chikungunya Virus Infection and Disease (CHIKV NHP Model). Pathogens (Basel, Switzerland). 2015;4(3):662–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez M, Gache K, Touratier A, Perrin JB, Fediaevsky A, Collin E, et al. Spread and impact of the Schmallenberg virus epidemic in France in 2012–2013. BMC veterinary research. 2012;10:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Depaquit J, Grandadam M, Fouque F, Andry PE, Peyrefitte C. Arthropod-borne viruses transmitted by Phlebotomine sandflies in Europe: a review. Euro Surveill. 2010. March 11;15(10):19507 [PubMed] [Google Scholar]
- 6.Su MW, Lin HM, Yuan HS, Chu WC. Categorizing host-dependent RNA viruses by principal component analysis of their codon usage preferences. Journal of computational biology: a journal of computational molecular cell biology. 2009; 16(11):1539–47. [DOI] [PubMed] [Google Scholar]
- 7.Behura SK, Severson DW. Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes. Biological reviews of the Cambridge Philosophical Society. 2013. February;88(1):49–61. 10.1111/j.1469-185X.2012.00242.x [DOI] [PubMed] [Google Scholar]
- 8.Stedman KM, Kosmicki NR, Diemer GS. Codon usage frequency of RNA virus genomes from high-temperature acidic-environment metagenomes. Journal of virology. 2013. February;87(3):1919 10.1128/JVI.02610-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus research. 2003. March;92(1):1–7. [DOI] [PubMed] [Google Scholar]
- 10.Duret L. Evolution of synonymous codon usage in metazoans. Current opinion in genetics & development. 2002. December;12(6):640–9. [DOI] [PubMed] [Google Scholar]
- 11.Wimmer E, Mueller S, Tumpey TM, Taubenberger JK. Synthetic viruses: a new opportunity to understand and prevent viral disease. Nature biotechnology. 2009. December;27(12):1163–72. 10.1038/nbt.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorba J, Campagnoli R, De L, Kew O. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. Journal of virology. 2008. May;82(9):4429–40. 10.1128/JVI.02354-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauring AS, Acevedo A, Cooper SB, Andino R. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell host & microbe. 2012. November 15;12(5):623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller S, Coleman JR, Papamichail D, Ward CB, Nimnual A, Futcher B, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nature biotechnology. 2010. July;28(7):723–6. 10.1038/nbt.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic acids research. 2012. July;40(Web Server issue):W597–603. 10.1093/nar/gks400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Presloid JB, Ebendick-Corpus BE, Zarate S, Novella IS. Antagonistic pleiotropy involving promoter sequences in a virus. Journal of molecular biology. 2008. October 3;382(2):342–52. 10.1016/j.jmb.2008.06.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, Weaver SC. Arbovirus evolution in vivo is constrained by host alternation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(19):6970–5. 10.1073/pnas.0712130105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobo FP, Mota BE, Pena SD, Azevedo V, Macedo AM, Tauch A, et al. Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PloS one. 2009;4(7):e6282 10.1371/journal.pone.0006282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic acids research. 1986. July 11;14(13):5125–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mooi E, Sarsted M. Cluster analysis In: Springer –Verlag Berlin Heidelberg editions. A concise guide to market research; 2011. pp. 237–283. [Google Scholar]
- 21.Ward H. Hierarchical grouping to optimize and objective function. 1963. (58): 263–244. [Google Scholar]
- 22.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics (Oxford, England). 2006. June 15;22(12):1540–2. [DOI] [PubMed] [Google Scholar]
- 23.Penny KI, Jolliffe IT. Multivariate outlier detection applied to multiply imputed laboratory data. Statistics in medicine. 1999. July 30;18(14):1879–95; dicussion 97. [DOI] [PubMed] [Google Scholar]
- 24.Gaskin CJ, Happell B. On exploratory factor analysis: a review of recent evidence, an assessment of current practice, and recommendations for future use. International journal of nursing studies. 2014;51(3):511–21. 10.1016/j.ijnurstu.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 25.Singer GA, Hickey DA. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Molecular biology and evolution. 2000;17(11):1581–8. [DOI] [PubMed] [Google Scholar]
- 26.Sang RC, Gichogo A, Gachoya J, Dunster MD, Ofula V, Hunt AR, et al. Isolation of a new flavivirus related to cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Archives of virology. 2003;148(6):1085–93. [DOI] [PubMed] [Google Scholar]
- 27.Crabtree MB, Sang RC, Stollar V, Dunster LM, Miller BR. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Archives of virology. 2003;148(6):1095–118. [DOI] [PubMed] [Google Scholar]
- 28.Hoshino K, Isawa H, Tsuda Y, Yano K, Sasaki T, Yuda M, et al. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359(2):405–14. [DOI] [PubMed] [Google Scholar]
- 29.Kuwata R, Isawa H, Hoshino K, Tsuda Y, Yanase T, Sasaki T, et al. RNA splicing in a new rhabdovirus from Culex mosquitoes. Journal of virology. 2011;85(13):6185–96. 10.1128/JVI.00040-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broeckel R, Haese N, Messaoudi I, Streblow DN. Nonhuman Primate Models of Chikungunya Virus Infection and Disease (CHIKV NHP Model). Pathogens. 2015;4(3):662–81. 10.3390/pathogens4030662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes EB. Zika virus outside Africa. Emerging infectious diseases. 2009;15(9):1347–50. 10.3201/eid1509.090442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biro JC. Does codon bias have an evolutionary origin? Theoretical biology & medical modelling. 2008;5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimaldi D, Engel M. Evolution of insects. 1 st. ed New York: University of Cambridge press; 2005. [Google Scholar]
- 34.Yeates DK, Wiegman BM, Courtney GW, Meir R, Pape T. Phylogeny and systematics of Diptera: two decades of progress and prospects. Zootaxa. 2007;. 1668:565–590. Available: http://www.mapress.com/zootaxa/2007f/zt01668p590.pdf. [Google Scholar]
- 35.Behura SK, Severson DW. Comparative analysis of codon usage bias and codon context patterns between dipteran and hymenopteran sequenced genomes. PloS one. 2012;7(8):e43111 10.1371/journal.pone.0043111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Blerkom LM. Role of viruses in human evolution. American journal of physical anthropology. 2003;Suppl 37:14–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traoré-Lamizana M, Fontenille D, Diallo M, Bâ Y, Zeller HG, Mondo M, et al. Arbovirus surveillance from 1990 to 1995 in the Barkedji area (Ferlo) of Senegal, a possible natural focus of Rift Valley fever virus. Entomology. 2001. July; 38(4):480–92. [DOI] [PubMed] [Google Scholar]
- 38.Maroli M, Feliciangeli MD, Bichaud L, Charrel RN, Gradoni L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Medical and veterinary entomology. 2013. June;27(2):123–47. 10.1111/j.1365-2915.2012.01034.x [DOI] [PubMed] [Google Scholar]
- 39.Zarate S, Novella IS. Vesicular stomatitis virus evolution during alternation between persistent infection in insect cells and acute infection in mammalian cells is dominated by the persistence phase. Journal of virology. 2004. November;78(22):12236–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasilakis N, Deardorff ER, Kenney JL, Rossi SL, Hanley KA, Weaver SC. Mosquitoes put the brake on arbovirus evolution: experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS pathogens. 2009;5(6):e1000467 10.1371/journal.ppat.1000467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell GL, Marfin AA, Lanciotti RS, Gubler DJ. West Nile virus. The Lancet Infectious diseases. 2002;2(9):519–29. [DOI] [PubMed] [Google Scholar]
- 42.Day JF. Predicting St. Louis encephalitis virus epidemics: lessons from recent, and not so recent, outbreaks. Annual review of entomology. 2001;46:111–38. [DOI] [PubMed] [Google Scholar]
- 43.Charrel RN, Bichaud L, de Lamballerie X. Emergence of Toscana virus in the mediterranean area. World journal of virology. 2012;1(5):135–41. 10.5501/wjv.v1.i5.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Comer JA, Stallknecht DE, Nettles VF. Incompetence of domestic pigs as amplifying hosts of vesicular stomatitis virus for Lutzomyia shannoni (Diptera: Psychodidae). Journal of medical entomology. 1995;32(5):741–4. [DOI] [PubMed] [Google Scholar]
- 45.Howerth EW, Stallknecht DE, Dorminy M, Pisell T, Clarke GR. Experimental vesicular stomatitis in swine: effects of route of inoculation and steroid treatment. Journal of veterinary diagnostic investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 1997;9(2):136–42. [DOI] [PubMed] [Google Scholar]
- 46.Mellor PS. Replication of arboviruses in insect vectors. Journal of comparative pathology. 2000;123(4):231–47. [DOI] [PubMed] [Google Scholar]
- 47.Vabret N, Bailly-Bechet M, Najburg V, Muller-Trutwin M, Verrier B, Tangy F. The biased nucleotide composition of HIV-1 triggers type I interferon response and correlates with subtype D increased pathogenicity. PloS one. 2012;7(4):e33502 10.1371/journal.pone.0033502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenbaum BD, Levine AJ, Bhanot G, Rabadan R. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS pathogens. 2008;4(6):e1000079 10.1371/journal.ppat.1000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agrawal S, Kandimalla ER. Modulation of Toll-like Receptor 9 Responses through Synthetic Immunostimulatory Motifs of DNA. Annals of the New York Academy of Sciences. 2003;1002:30–42. [DOI] [PubMed] [Google Scholar]
- 50.Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. eLife. 2014;3:e04531 10.7554/eLife.04531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doherty A, McInerney JO. Translational selection frequently overcomes genetic drift in shaping synonymous codon usage patterns in vertebrates. Molecular biology and evolution. 2013;30(10):2263–7. 10.1093/molbev/mst128 [DOI] [PubMed] [Google Scholar]
- 52.Gerashchenko MV, Su D, Gladyshev VN. CUG start codon generates thioredoxin/glutathione reductase isoforms in mouse testes. The Journal of biological chemistry. 2010;285(7):4595–602. 10.1074/jbc.M109.070532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research. 1997;25(5):955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tukey-Kramer analysis conducted among vertebrate, invertebrate and virus groups to determinate the frequency of amino acids among the proteins analyzed in this study.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.