Abstract
Finger millet is an important cereal crop in eastern Africa and southern India with excellent grain storage quality and unique ability to thrive in extreme environmental conditions. Since negligible attention has been paid to improving this crop to date, the current study used Next Generation Sequencing (NGS) technologies to develop both Simple Sequence Repeat (SSR) and Single Nucleotide Polymorphism (SNP) markers. Genomic DNA from cultivated finger millet genotypes KNE755 and KNE796 was sequenced using both Roche 454 and Illumina technologies. Non-organelle sequencing reads were assembled into 207 Mbp representing approximately 13% of the finger millet genome. We identified 10,327 SSRs and 23,285 non-homeologous SNPs and tested 101 of each for polymorphism across a diverse set of wild and cultivated finger millet germplasm. For the 49 polymorphic SSRs, the mean polymorphism information content (PIC) was 0.42, ranging from 0.16 to 0.77. We also validated 92 SNP markers, 80 of which were polymorphic with a mean PIC of 0.29 across 30 wild and 59 cultivated accessions. Seventy-six of the 80 SNPs were polymorphic across 30 wild germplasm with a mean PIC of 0.30 while only 22 of the SNP markers showed polymorphism among the 59 cultivated accessions with an average PIC value of 0.15. Genetic diversity analysis using the polymorphic SNP markers revealed two major clusters; one of wild and another of cultivated accessions. Detailed STRUCTURE analysis confirmed this grouping pattern and further revealed 2 sub-populations within wild E. coracana subsp. africana. Both STRUCTURE and genetic diversity analysis assisted with the correct identification of the new germplasm collections. These polymorphic SSR and SNP markers are a significant addition to the existing 82 published SSRs, especially with regard to the previously reported low polymorphism levels in finger millet. Our results also reveal an unexploited finger millet genetic resource that can be included in the regional breeding programs in order to efficiently optimize productivity.
Introduction
Cultivated finger millet (Eleusine coracana subsp. coracana) is an annual grass that is widely cultivated as a staple food in eastern Africa and south Asia. It is tetraploid (2n = 4x = 36) and belongs to the Poaceae family and Chloridoideae sub-family [1]. Its origins can be traced to the highlands of Uganda and Ethiopia where it was likely domesticated around 5000 years ago from its wild progenitor E. coracana subsp. africana. Other wild species include E. kigeziensis, E. floccifolia, E. intermedia, E. tristachya, E. jaegeri and E. indica. Finger millet is a nutritious cereal high in protein, methionine and other essential amino acids. The small seeds can be stored for years without damage, making it an important food reserve in times of famine. The grain is used for bread, porridge, beer, soup and pudding. In countries where it is grown, it is commonly referred to as Wimbi (Swahili), Bulo (Uganda), Tellebun (Sudan) and Ragi (India) [2,3].
Despite finger millet’s importance as a subsistence crop, little attention has focused on improving production, probably because millets in general have been considered of little economic importance compared to maize, wheat and rice [4]. As a result, finger millet still lacks the required basic genomic resources for efficient breeding and remains one of the few cultivated cereal crops lacking a high-density genetic linkage map. There are currently <100 informative SSR markers available for finger millet [5,6,7], and only one study reported the identification of SNPs for this cereal using genotyping-by-sequencing (GBS) [8]. Nevertheless, the increasing affordability and access to Next Generation Sequencing (NGS) approaches facilitate the development of the genomic tools required to study complex traits and map quantitative trait loci (QTLs) of interest [9,10] in any crop, including finger millet.
Genetic markers have revolutionized crop improvement through the detection of DNA polymorphisms for precise, efficient and cost effective germplasm characterization and management. Such markers that have been used in finger millet include Random Amplified Polymorphic DNA (RAPD) [11,12], Inter Simple Sequence Repeats (ISSRs) [12], Random Fragment Length Polymorphisms (RFLPs), Amplified Fragment Length Polymorphisms (AFLPs), SSRs [5,6] and SNPs [8]. SSRs have become the markers of choice over the past decade for many crops including potato [13], rice [14] and wheat [15]. They remain the most commonly used marker for molecular analysis in finger millet and were among the markers used to construct the first genetic linkage map of finger millet [5]. More recently, Expressed Sequence Tags (EST)- derived SSRs have been developed for finger millet [16,17] although only a small percentage showed significant polymorphism across the accessions tested.
Cloning and hybridization-based SSR libraries and Sanger sequencing, which were used to develop the first 45 SSR markers for finger millet [5] can now be substituted with NGS, which generates larger numbers of sequences faster and cheaper. Roche 454 (Life Sciences) and Illumina platforms generate and process hundreds of thousands to millions of DNA templates in parallel resulting in low running costs per base of generated sequence and gigabase scale throughput [18], allowing the identification of large numbers of both SSR and SNP markers relatively cheaply. SSRs are PCR-based [19], highly polymorphic, hypervariable, co-dominant, reproducible, multi-allelic and distributed throughout the genome [20]. They can therefore be applied to finger millet improvement in genome-wide screens for variation and trait association, fingerprinting, genetic diversity analysis and genotyping [21,22].
On the other hand, SNPs have become the markers of choice for crop genotyping due to their abundance with up to 1 SNP per 140 bp being observed in rice [23]. They are co-dominant, bi-allelic, highly polymorphic, reproducible [24] and can be automated for high throughput genotyping. For these reasons, SNP markers are frequently used for genotyping large numbers of individuals for genomics-assisted breeding and genetic diversity applications. As an allotetraploid (AA and BB sub-genomes) with high levels of inbreeding, SNP discovery in finger millet can be challenging due to low polymorphism levels and high numbers of homeologous SNPs, which occur as a result of polymorphism between the AA and BB sub-genomes of the same individual. Using relevant filtering tools and stringent mapping parameters [25], SNP identification has been successful in several other polyploid crops including wheat [26], cotton [27], oats [28] and groundnut [29] and therefore can be successfully applied in finger millet. The current study capitalized on the power of NGS to develop additional SSRs as well as new SNP markers for finger millet using Roche 454 and Illumina sequencing.
Materials and Methods
Plant material
Finger millet genotypes KNE755 and KNE796 were used to generate sequence data for SSR and SNP marker development. Ten diverse finger millet genotypes (Table 1) that were used previously to assess polymorphism levels of published SSRs [5] were used to validate SSR markers in this study. Additional 89 genotypes (Table 1) were used to validate SNP markers. All the cultivated genotypes were obtained from the ICRISAT gene bank, the Tanzanian gene bank and the Gene Bank of Kenya. Maseno University, Kenya and Mikocheni Agricultural Research Institute (MARI), Tanzania kindly provided wild accessions. This study did not involve any endangered or protected species.
Table 1. Accessions of the genus Eleusine used for validating SSR and SNP markers.
| Species | Accession name | Codea | Origin | Purpose in the study |
|---|---|---|---|---|
| E. coracana ssp. coracana | KNE796 | 31 | Kenya | Generation of markers, SNP validation |
| KNE755 | 56 | Kenya | Generation of markers, SNP validation | |
| GBK-044047A | N/A | Kenya | SSR marker validation | |
| GBK-000414A | N/A | Kenya | SSR marker validation | |
| GBK-011135A | N/A | Kenya | SSR marker validation | |
| Sansamula | N/A | Tanzania | SSR marker validation | |
| Namakonta | N/A | Tanzania | SSR marker validation | |
| Ebega | N/A | Uganda | SSR marker validation | |
| Bulo | N/A | Uganda | SSR marker validation | |
| Emorumoru | N/A | Uganda | SSR marker validation | |
| IE2572 | N/A | Minicore collection | SSR marker validation | |
| IE2957 | N/A | Minicore collection | SSR marker validation | |
| GBK033383 | 32 | Kenya | SNP marker validation | |
| GBK033384 | 33 | Kenya | SNP marker validation | |
| GBK0333446 | 34 | Kenya | SNP marker validation | |
| GBK0333407A | 35 | Kenya | SNP marker validation | |
| GBK0333408A | 36 | Kenya | SNP marker validation | |
| GBK0333445A | 37 | Kenya | SNP marker validation | |
| GBK0333449A | 38 | Kenya | SNP marker validation | |
| GBK0333452A | 39 | Kenya | SNP marker validation | |
| GBK0333454A | 40 | Kenya | SNP marker validation | |
| GBK0333455A | 41 | Kenya | SNP marker validation | |
| GBK0333456A | 42 | Kenya | SNP marker validation | |
| GBK0333457A | 43 | Kenya | SNP marker validation | |
| GBK0333458A | 44 | Kenya | SNP marker validation | |
| GBK0333459A | 45 | Kenya | SNP marker validation | |
| GBK0333460A | 46 | Kenya | SNP marker validation | |
| GBK033373A | 47 | Kenya | SNP marker validation | |
| GBK033376A | 48 | Kenya | SNP marker validation | |
| GBK033377A | 49 | Kenya | SNP marker validation | |
| GBK033378A | 50 | Kenya | SNP marker validation | |
| GBK033379A | 51 | Kenya | SNP marker validation | |
| GBK033380A | 52 | Kenya | SNP marker validation | |
| GBK033381A | 53 | Kenya | SNP marker validation | |
| GBK033382A | 54 | Kenya | SNP marker validation | |
| IMULA | 55 | Uganda | SNP marker validation | |
| P224 | 57 | Uganda | SNP marker validation | |
| U15 | 81 | Uganda | SNP marker validation | |
| TZA128 | 58 | Tanzania | SNP marker validation | |
| TZA132 | 59 | Tanzania | SNP marker validation | |
| TZA137 | 60 | Tanzania | SNP marker validation | |
| TZA138 | 61 | Tanzania | SNP marker validation | |
| TZA141 | 62 | Tanzania | SNP marker validation | |
| TZA1628 | 63 | Tanzania | SNP marker validation | |
| TZA1629 | 64 | Tanzania | SNP marker validation | |
| TZA1632 | 65 | Tanzania | SNP marker validation | |
| TZA1633 | 66 | Tanzania | SNP marker validation | |
| TZA1634 | 67 | Tanzania | SNP marker validation | |
| TZA1636 | 68 | Tanzania | SNP marker validation | |
| TZA1637 | 69 | Tanzania | SNP marker validation | |
| TZA1638 | 70 | Tanzania | SNP marker validation | |
| TZA1640 | 71 | Tanzania | SNP marker validation | |
| TZA1655 | 72 | Tanzania | SNP marker validation | |
| TZA1656 | 73 | Tanzania | SNP marker validation | |
| TZA1658 | 74 | Tanzania | SNP marker validation | |
| TZA1659 | 75 | Tanzania | SNP marker validation | |
| TZA1661 | 76 | Tanzania | SNP marker validation | |
| TZA1662 | 77 | Tanzania | SNP marker validation | |
| TZA1663 | 78 | Tanzania | SNP marker validation | |
| TZA1665 | 79 | Tanzania | SNP marker validation | |
| TZA1666 | 80 | Tanzania | SNP marker validation | |
| MS19 | 82 | Kenya | SNP marker validation | |
| MS17 | 83 | Kenya | SNP marker validation | |
| LEN24 | 88 | Ethiopia | SNP marker validation | |
| MS21 | 84 | Kenya | SNP marker validation | |
| EDL34 | 85 | Tanzania | SNP marker validation | |
| MS18 | 86 | Kenya | SNP marker validation | |
| EDL25 | 89 | Tanzania | SNP marker validation | |
| UG10 | 87 | Uganda | SNP marker validation | |
| Wild accessions, true species not confirmed | EDL30 | 1 | Tanzania | SNP marker validation |
| EDL15 | 2 | Tanzania | SNP marker validation | |
| MS9 | 3 | Kenya | SNP marker validation | |
| MS13 | 4 | Kenya | SNP marker validation | |
| UG19 | 5 | Uganda | SNP marker validation | |
| MS5 | 6 | Kenya | SNP marker validation | |
| MS4 | 7 | Kenya | SNP marker validation | |
| UG1 | 8 | Uganda | SNP marker validation | |
| UG18 | 9 | Uganda | SNP marker validation | |
| MSN10 | 10 | Kenya | SNP marker validation | |
| MS8 | 11 | Kenya | SNP marker validation | |
| AAU-ELU-48 | 12 | Ethiopia | SNP marker validation | |
| UG9 | 13 | Uganda | SNP marker validation | |
| UG11 | 14 | Uganda | SNP marker validation | |
| UG20 | 15 | Uganda | SNP marker validation | |
| MS3 | 16 | Kenya | SNP marker validation | |
| MS6 | 17 | Kenya | SNP marker validation | |
| MS7 | 18 | Kenya | SNP marker validation | |
| MS11 | 19 | Kenya | SNP marker validation | |
| MS12 | 20 | Kenya | SNP marker validation | |
| MS15 | 21 | Kenya | SNP marker validation | |
| EDL9 | 23 | Tanzania | SNP marker validation | |
| EDL16 | 24 | Tanzania | SNP marker validation | |
| LEN7 | 25 | Ethiopia | SNP marker validation | |
| LESK10 | 26 | Ethiopia | SNP marker validation | |
| MD48 | 22 | Kenya | SNP marker validation | |
| UG3 | 27 | Uganda | SNP marker validation | |
| UG8 | 28 | Uganda | SNP marker validation | |
| MS16 | 29 | Kenya | SNP marker validation | |
| EDL10 | 30 | Tanzania | SNP marker validation |
aThis is the code used in STRUCTURE outputs. Genotypes that were not included in the STRUCTURE analysis are represented with N/A.
Library preparation and sequencing
For Roche 454 sequencing, leaves of each genotype (KNE755 and KNE796) were sampled 2–3 weeks after planting, dried using silica gel and sent to Ecogenics (Schlieren, Switzerland) for DNA extraction, SSR enrichment and sequencing (Roche 454/FLX). For Illumina sequencing, DNA was extracted from two weeks old seedlings of KNE755 and KNE796 and sent to Georgia Genomics Facility at the University of Georgia (USA). A 1-μg portion of each DNA sample was fragmented using Covaris (Covaris Inc., MA, USA) ultrasonication. A second DNA portion of 5μg of each sample was digested using PstI methylation sensitive restriction endonuclease for 1 hour at 37°C in order to enrich for genic regions. After end-repair of both Covaris-sheared and enzyme digested DNA, sequencing libraries were prepared following the TruSeq protocol (Illumina, San Diego, USA) and sequenced on an Illumina Hi-Seq 2000.
Processing of Illumina reads for SNP and SSR marker identification
Fastq-mcf [30] was used to remove adaptors and trim for quality. Finger millet chloroplast and mitochondrial sequences were removed by mapping trimmed reads to the rice reference chloroplast and mitochondrial genomes downloaded from GOBASE [31] using Bowtie2 [32]. De novo assembly of all non-organelle sequences was done using Velvet software [33] to create a reference file. Only reference contigs with at least 200 bp were maintained for marker identification and functional analysis. SSR motifs with a maximum of 4-nucleotide repeats were identified from the reference file using the software GMATo [34] with a minimum repeat value of 5. We specifically searched for only di-, tri-, and tetra-nucleotide repeats due to their abundance in plant genomes [35–37] and excluded other nucleotide repeats because of the high error rates [38] and less informative nature of mono-nucleotide repeats [39,40] and the low abundance of penta- and hexa-nucleotide repeats in monocots [41,42].
For SNP identification, BWA software [43] was used to map the Illumina reads from each genotype to the reference file. Generating reference sequences of each genotype and mapping back reads to the reference identified homeologous SNPs and their frequency in each genotype. SAMtools [44] was used to view and sort the mapped reads. Duplicate reads were removed from respective alignment sequences using Picard-tools 1.94 (http://picard.sourceforge.net) before running FreeBayes [45] to identify genetic variants. The raw SNPs obtained were filtered using VCFtools [46] based on a quality score of 30, maximum allele number of 2 and a minimum coverage of 3. Homeologous SNPs identified from each genotype were eliminated using VCFtools [46]. The raw data was submitted to the NCBI Sequence Read Archive (accession number SRP073162).
Functional analysis
Reference sequence contigs that were at least 200bp long were masked for repetitive elements using RepeatMasker (http://www.repeatmasker.org/) and aligned using blastx [47] against rice genes retrieved from the UniProt database (http://www.uniprot.org/) as well as the non-redundant plant protein database retrieved from the Genbank (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/plant/plant.1.protein.faa.gz) setting an e-value cutoff of 1e-5 and minimum similarity of 80%. A non-redundant protein list generated from the two databases was compiled and submitted to PANTHER [48] (http://pantherdb.org/) classification system for Gene Ontology (GO) term annotation (molecular function).
SSR and SNP marker validation
Sequence assembly of Roche 454 data and identification of SSRs was done by Ecogenics (Switzerland). Primers were designed for 101 of the identified SSRs using the following parameters: primer length between 18–23 with an optimum of 21 bp, PCR products of 100 to 300 bp, primer TM between 58°- 64°C with an optimum of 60°C and GC content from 45–70%. All forward primers contained an M13-tag (5’- CACGACGTTGTAAAACGAC—3’) on the 5’ end that was fluorescently labelled to allow detection of amplification products [49]. SSR marker validation was performed across 10 selected genotypes as described by De Villiers et al. [7].
One hundred and one SNP markers were selected randomly and submitted for Competitive allele-specific PCR (KASPar) genotyping at LGC Genomics (http://www.lgcgenomics.com/genotyping/kasp_technical_resources/), UK. The data generated was viewed graphically as cluster plots using SNPviewer V2 (www.lgcgenomics.com). For SNP validation, two weeks-old seedlings of the 89 genotypes (Table 1) were sampled by placing 1 cm long leaf pieces in strip tubes supplied by LGC Genomics (www.lgcgroup.com). The tubes were sealed in a plastic bag with desiccant and immediately shipped to LGC Genomics (Herts, United Kingdom) for genotyping.
Phylogenetic and population structure analysis
PowerMarker v.3.25 [50] was used to compute PIC and total numbers of alleles. Polymorphism information content (PIC) was calculated using the method of Botstein et al. [51] as below;
Where, pi and pj are the frequencies of alleles i and j, respectively
The UPGMA based clustering was computed using TASSEL [52] and rooted using MD48, which belongs to the species E. kigeziensis. The genetic structure of finger millet accessions was determined using the admixture model with correlated allele frequencies based on the Monte Carlo Markov Chain (MCMC) algorithm implemented in STRUCTURE 2.3.3 software [53]. The admixture model assumed that the genome of each individual resulted from the mixture of K ancestral populations. The estimated proportions of each individual’s genotype originating from each of the K ancestral populations (q) was calculated for K ranging from 1 to 10 with 10 runs for each K value. For each run, a burn-in period of 10000 and MCMC replications of 100000 was used. The optimum K value was calculated using STRUCTURE HARVESTER [54], which computed the log likelihood of the data [LnP(D)] in the STRUCTURE output and an ad hoc statistic Δk based on the rate of change in LnP(D) between successive k [55]. Results from each replicate run were combined using the CLUMPP software [56].
Results
Sequence assembly
Table 2 provides a summary of reads generated and assembled for each genotype. Reads mapping to the rice organelle genomes were excluded from further analysis. The non-organelle finger millet reads from KNE755 (1,778,492) were assembled into 906,426 nodes/contigs consisting of 34,469,967 bp while those from KNE796 (5,706,821) were assembled into 5,552,610 nodes/contigs consisting of 167,333,449 bp (Table 2). All nuclear sequences from KNE755 and KNE796 were assembled into a reference fasta file containing 6,810,971 nodes/contigs spanning 207,197,804 bp. Assuming a genome size of 1.593 Gb [57], the assembled reads generated from both KNE755 and KNE796 genotypes represented about 13% of the finger millet genome. Contigs that were at least 200 bp long were retrieved from the reference file and used for SNP and SSR marker identification.
Table 2. A summary of sequencing reads generated for each genotype and the resulting assemblies.
| Genotype | Roche 454 Raw Reads | Illumina Raw Reads | Nuclear Sequences (Reads) | Assembled Nuclear Sequences (bp) | Genome Coverage |
|---|---|---|---|---|---|
| KNE755 | 5,774 | 4,804,190 | 1,778,492 | 34,469,967 | ~2% |
| KNE796 | 5,266 | 13,007,430 | 5,706,821 | 167,333,449 | ~10% |
| Combined Assembly | 207,197,804 | ~13% | |||
Homeologous SNPs
KNE755 was more abundant in homeologous SNPs with a frequency of 1/657 bp compared to a frequency of 1/956 bp in KNE796. The most abundant homeologous SNP in both genotypes was CT/AG (~62%) while CG was the rarest SNP at about 7.5%. The Ts/Tv ratios of the homeologous SNPs were comparable across the two genotypes but slightly higher in KNE755 (1.8) than in KNE796 (1.5).
SSRs and Non-homeologous SNP mining
We identified 10,327 SSRs (di-, tri- and tetra-nucleotide repeats) (S1 Table) and 23,285 non-homeologous SNPs (S2 Table) from 77 Mbp compared to 38 SSRs and 1415 SNPs from the 1.3 Mbp putative genic regions. Table 3 shows a summary of SSRs and SNPs identified from different regions and the estimated frequencies of the markers across the two genotypes (KNE755 and KNE796). The most abundant SSRs were di-nucleotide repeats (80%), followed by tri-nucleotide (18.3%) and tetra-nucleotide repeats (1.2%) (Table 3). Of the di-nucleotide repeats, AG/CT was the most prevalent (39%) while CAA/TTG were the most prevalent (~9%) tri-nucleotide repeats. Within the putative genic regions, AT di-nucleotide repeats were the most abundant. The overall Ts/Tv SNP ratio was 1.8 compared to a ratio of 2 within the putative genic regions.
Table 3. A summary of identified SSR and SNP markers and their frequency across genotypes KNE796 and KNE755.
| Marker Type | Type | Total Identified | Frequency |
|---|---|---|---|
| SSR | ALL | 10,327 | 1 per 7.5 Kb |
| di-nucleotide | 8,308 | 1 per 9.3 kb | |
| Tri-nucleotide | 1,895 | 1 per 40 kb | |
| Tetra-nucleotide | 124 | 1 per 623 Kb | |
| Putative Genic SSRs | ALL | 38 | 1 per 2 Mb |
| di-nucleotide SSR | 27 | 1 per 2.9 Mb | |
| Tri-nucleotide | 11 | 1 per 7 Mb | |
| Tetra-nucleotide | - | ||
| SNPs | ALL | 23,285 | 1 per 3.3 kb |
| Transition | 14962 | 1 per 5.1 Kb | |
| Transversion | 8323 | 1 per 9.3 Kb | |
| Putative Genic SNPs | ALL | 1,415 | 1 per 54.6 Kb |
| Transition | 952 | 1 per 81 Kb | |
| Transversion | 463 | 1 per 167 kb |
Functional annotation
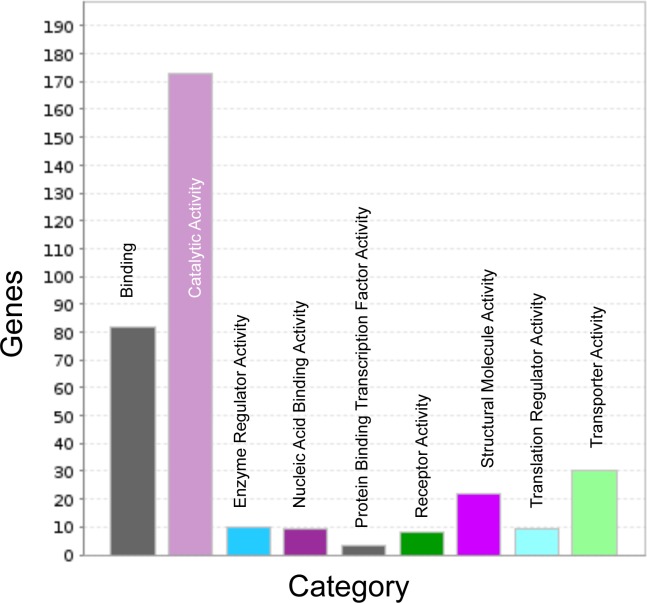
Searches against the plant protein and rice gene databases revealed 6,821 (1,340,261 bp) and 6707 (1,328,367 bp) sequences respectively containing putative genes. While 9,175 and 11,632 of the reference contigs contained SSRs and SNPs respectively, only 36 and 564 of the putative genic sequences contained SNPs and SSRs respectively. Out of the 5,094 rice genes that showed sequence similarity with 6,707 finger millet sequences, 4,240 GO terms were retrieved for biological processes, 2,835 related to molecular functions and 1,147 to cellular components. For the 564 SNP-containing putative genic sequences, 519, 346 and 146 GO terms for biological processes, molecular functions and cellular components could be assigned respectively. Fig 1 shows the breakdown of different categories of the 346 GO terms for molecular functions as revealed by PANTHER. Genes with catalytic activity were over-represented (50%) followed by genes involved in binding (24%). Protein binding transcription factor activity was the least represented (1%).
Fig 1. Molecular function categories.
The distribution of SNP-containing putative genes that were assigned GO terms in PANTHER (http://pantherdb.org/). Catalytic activity category was over-represented.
SSR and SNP validation using diverse finger millet genotypes
Of the 101 SSRs tested (S3 Table), 49 were polymorphic, 10 were monomorphic and 42 did not amplify products that could be scored unambiguously. Among the polymorphic markers, the PIC ranged from 0.16 to 0.77 with an average of 0.42 (Table 4). SSR loci ICECP54, ICECP47 and ICECP89 were the most polymorphic and revealed at least 5 alleles across the 10 genotypes. The rest of the markers revealed between 2–4 alleles with an average of 3 alleles per locus (Table 4).
Table 4. Characteristics of polymorphic SSRs after validation across 10 genotypes.
| Marker | Major Allele Frequency | Allele Number | Gene Diversity | PIC |
|---|---|---|---|---|
| ICECP54 | 0.30 | 6 | 0.80 | 0.77 |
| ICECP47 | 0.29 | 5 | 0.78 | 0.74 |
| ICECP89 | 0.35 | 5 | 0.72 | 0.67 |
| ICECP50 | 0.45 | 4 | 0.68 | 0.62 |
| ICECP58 | 0.50 | 4 | 0.66 | 0.61 |
| ICECP84 | 0.45 | 4 | 0.67 | 0.60 |
| ICECP5 | 0.50 | 4 | 0.64 | 0.57 |
| ICECP96 | 0.44 | 3 | 0.64 | 0.57 |
| ICECP3 | 0.50 | 3 | 0.61 | 0.54 |
| ICECP95 | 0.60 | 4 | 0.58 | 0.54 |
| ICECP4 | 0.55 | 3 | 0.60 | 0.53 |
| ICECP68 | 0.60 | 3 | 0.56 | 0.50 |
| ICECP73 | 0.50 | 3 | 0.58 | 0.49 |
| ICECP53 | 0.60 | 3 | 0.54 | 0.47 |
| ICECP63 | 0.60 | 3 | 0.54 | 0.47 |
| ICECP64 | 0.60 | 3 | 0.54 | 0.47 |
| ICECP90 | 0.60 | 3 | 0.54 | 0.47 |
| ICECP61 | 0.70 | 4 | 0.48 | 0.45 |
| ICECP62 | 0.70 | 4 | 0.48 | 0.45 |
| ICECP37 | 0.70 | 3 | 0.46 | 0.41 |
| ICECP69 | 0.70 | 3 | 0.46 | 0.41 |
| ICECP66 | 0.75 | 4 | 0.42 | 0.39 |
| ICECP11 | 0.50 | 2 | 0.50 | 0.38 |
| ICECP67 | 0.50 | 2 | 0.50 | 0.38 |
| ICECP70 | 0.50 | 2 | 0.50 | 0.38 |
| ICECP71 | 0.50 | 2 | 0.50 | 0.38 |
| ICECP97 | 0.50 | 2 | 0.50 | 0.38 |
| ICECP46 | 0.57 | 2 | 0.49 | 0.37 |
| ICECP40 | 0.60 | 2 | 0.48 | 0.36 |
| ICECP85 | 0.60 | 2 | 0.48 | 0.36 |
| ICECP98 | 0.60 | 2 | 0.48 | 0.36 |
| ICECP99 | 0.60 | 2 | 0.48 | 0.36 |
| ICECP42 | 0.63 | 2 | 0.47 | 0.36 |
| ICECP44 | 0.67 | 2 | 0.44 | 0.35 |
| ICECP59 | 0.67 | 2 | 0.44 | 0.35 |
| ICECP82 | 0.67 | 2 | 0.44 | 0.35 |
| ICECP92 | 0.67 | 2 | 0.44 | 0.35 |
| ICECP93 | 0.78 | 3 | 0.37 | 0.34 |
| ICECP56 | 0.70 | 2 | 0.42 | 0.33 |
| ICECP52 | 0.71 | 2 | 0.41 | 0.32 |
| ICECP72 | 0.80 | 3 | 0.34 | 0.31 |
| ICECP80 | 0.80 | 3 | 0.34 | 0.31 |
| ICECP101 | 0.80 | 3 | 0.34 | 0.31 |
| ICECP1 | 0.75 | 2 | 0.38 | 0.30 |
| ICECP43 | 0.80 | 2 | 0.32 | 0.27 |
| ICECP48 | 0.89 | 2 | 0.20 | 0.18 |
| ICECP57 | 0.90 | 2 | 0.18 | 0.16 |
| ICECP81 | 0.90 | 2 | 0.18 | 0.16 |
| ICECP91 | 0.90 | 2 | 0.18 | 0.16 |
| Mean | 0.62 | 3 | 0.48 | 0.42 |
We developed 8,740 KASPar assays from 92 SNP regions across 93 finger millet accessions (S4 Table). The assays produced 8,099 identified allele calls, 640 unidentified allele calls and 1 bad call. The mean number of calls made per SNP was 87 with an allele call rate of 93%. Four genotypes that revealed > 80% missed calls and 12 SNP assays that revealed more than 90% unidentified allele calls as well as those that were monomorphic were excluded from further analysis. This resulted in 80 polymorphic markers (Table 5) tested across 89 genotypes (Table 1). The PIC ranged from 0.01 to 0.38 with a mean of 0.29 while heterozygosity ranged from 0 to 0.989 with a mean of 0.534 (Table 5). The most polymorphic markers were ICECSNT26 and ICECSNT94.
Table 5. A list of polymorphic SNP markers and their characteristics after validation across 89 Eleusine accessions.
| SNP Locus | MAF | Gene Diversity | Heterozygosity | PIC (All) | PIC (Wild) | PIC (Cultivated) |
|---|---|---|---|---|---|---|
| ICECSNT2 | 0.70 | 0.42 | 0.057 | 0.33 | 0.18 | N/A |
| ICECSNT3 | 0.98 | 0.03 | 0.034 | 0.03 | 0.09 | N/A |
| ICECSNT4 | 0.76 | 0.37 | 0.077 | 0.30 | 0.23 | 0.02 |
| ICECSNT5 | 0.54 | 0.50 | 0.857 | 0.37 | 0.37 | 0.37 |
| ICECSNT6 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT8 | 0.51 | 0.50 | 0.955 | 0.37 | 0.37 | 0.37 |
| ICECSNT9 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT11 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT12 | 0.85 | 0.25 | 0.092 | 0.22 | 0.37 | 0.03 |
| ICECSNT13 | 0.92 | 0.15 | 0.045 | 0.14 | 0.27 | N/A |
| ICECSNT14 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | 0.03 |
| ICECSNT15 | 0.97 | 0.07 | 0.000 | 0.06 | 0.16 | N/A |
| ICECSNT16 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT17 | 0.51 | 0.50 | 0.977 | 0.37 | 0.37 | N/A |
| ICECSNT18 | 0.70 | 0.42 | 0.081 | 0.33 | 0.22 | N/A |
| ICECSNT20 | 0.51 | 0.50 | 0.943 | 0.37 | 0.37 | N/A |
| ICECSNT22 | 0.75 | 0.38 | 0.082 | 0.31 | 0.29 | N/A |
| ICECSNT23 | 0.51 | 0.50 | 0.977 | 0.37 | 0.37 | N/A |
| ICECSNT24 | 0.74 | 0.38 | 0.060 | 0.31 | 0.21 | N/A |
| ICECSNT26 | 0.50 | 0.50 | 0.977 | 0.38 | 0.38 | N/A |
| ICECSNT27 | 0.52 | 0.50 | 0.954 | 0.37 | 0.37 | 0.37 |
| ICECSNT28 | 0.51 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT31 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT32 | 0.67 | 0.44 | 0.038 | 0.34 | 0.19 | 0.16 |
| ICECSNT33 | 0.99 | 0.01 | 0.011 | 0.01 | N/A | 0.02 |
| ICECSNT34 | 0.51 | 0.50 | 0.989 | 0.37 | 0.37 | N/A |
| ICECSNT35 | 0.58 | 0.49 | 0.000 | 0.37 | 0.18 | 0.27 |
| ICECSNT36 | 0.69 | 0.43 | 0.047 | 0.34 | 0.22 | 0.12 |
| ICECSNT38 | 0.96 | 0.08 | 0.011 | 0.07 | 0.17 | 0.02 |
| ICECSNT39 | 0.70 | 0.42 | 0.045 | 0.33 | 0.20 | N/A |
| ICECSNT40 | 0.99 | 0.02 | 0.000 | 0.02 | 0.06 | N/A |
| ICECSNT41 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT42 | 0.62 | 0.47 | 0.713 | 0.36 | 0.17 | N/A |
| ICECSNT43 | 0.52 | 0.50 | 0.964 | 0.37 | 0.37 | N/A |
| ICECSNT44 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT45 | 0.61 | 0.48 | 0.778 | 0.36 | 0.19 | N/A |
| ICECSNT46 | 0.99 | 0.01 | 0.012 | 0.01 | 0.03 | N/A |
| ICECSNT47 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT48 | 0.52 | 0.50 | 0.913 | 0.37 | 0.37 | 0.37 |
| ICECSNT49 | 0.88 | 0.21 | 0.171 | 0.19 | 0.36 | 0.02 |
| ICECSNT51 | 0.61 | 0.47 | 0.722 | 0.36 | 0.19 | N/A |
| ICECSNT52 | 0.89 | 0.20 | 0.150 | 0.18 | 0.37 | N/A |
| ICECSNT53 | 0.51 | 0.50 | 0.977 | 0.37 | 0.37 | N/A |
| ICECSNT54 | 0.97 | 0.07 | 0.000 | 0.06 | 0.16 | N/A |
| ICECSNT55 | 0.74 | 0.39 | 0.482 | 0.31 | 0.38 | 0.21 |
| ICECSNT56 | 0.54 | 0.50 | 0.871 | 0.37 | 0.36 | N/A |
| ICECSNT57 | 0.51 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT58 | 0.51 | 0.50 | 0.951 | 0.37 | 0.38 | 0.37 |
| ICECSNT59 | 0.96 | 0.07 | 0.000 | 0.07 | 0.17 | N/A |
| ICECSNT61 | 0.63 | 0.47 | 0.744 | 0.36 | 0.08 | N/A |
| ICECSNT62 | 0.70 | 0.42 | 0.000 | 0.33 | 0.12 | N/A |
| ICECSNT66 | 0.82 | 0.29 | 0.000 | 0.25 | 0.33 | N/A |
| ICECSNT67 | 0.97 | 0.07 | 0.000 | 0.06 | 0.17 | N/A |
| ICECSNT68 | 0.52 | 0.50 | 0.965 | 0.37 | 0.37 | N/A |
| ICECSNT69 | 0.98 | 0.05 | 0.000 | 0.04 | 0.12 | N/A |
| ICECSNT72 | 0.97 | 0.07 | 0.023 | 0.06 | N/A | 0.09 |
| ICECSNT73 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT76 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT77 | 0.72 | 0.40 | 0.069 | 0.32 | 0.24 | N/A |
| ICECSNT78 | 0.99 | 0.01 | 0.011 | 0.01 | N/A | 0.02 |
| ICECSNT79 | 0.94 | 0.11 | 0.000 | 0.11 | 0.26 | N/A |
| ICECSNT80 | 0.51 | 0.50 | 0.989 | 0.37 | 0.37 | N/A |
| ICECSNT81 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT82 | 0.52 | 0.50 | 0.965 | 0.37 | 0.37 | N/A |
| ICECSNT83 | 0.69 | 0.43 | 0.198 | 0.34 | 0.30 | 0.16 |
| ICECSNT84 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT85 | 0.67 | 0.44 | 0.074 | 0.35 | 0.20 | 0.11 |
| ICECSNT86 | 0.85 | 0.25 | 0.115 | 0.22 | 0.37 | N/A |
| ICECSNT88 | 0.55 | 0.49 | 0.894 | 0.37 | 0.34 | N/A |
| ICECSNT89 | 0.51 | 0.50 | 0.977 | 0.37 | 0.37 | N/A |
| ICECSNT90 | 0.75 | 0.38 | 0.061 | 0.30 | 0.22 | N/A |
| ICECSNT91 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT93 | 0.72 | 0.40 | 0.000 | 0.32 | 0.29 | 0.15 |
| ICECSNT94 | 0.50 | 0.50 | 0.977 | 0.38 | 0.38 | N/A |
| ICECSNT95 | 0.80 | 0.32 | 0.267 | 0.27 | 0.37 | 0.03 |
| ICECSNT96 | 0.80 | 0.32 | 0.179 | 0.27 | 0.35 | N/A |
| ICECSNT99 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSN99 | 0.99 | 0.01 | 0.011 | 0.01 | N/A | 0.02 |
| ICECSN100 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| ICECSNT100 | 0.52 | 0.50 | 0.966 | 0.37 | 0.37 | N/A |
| Mean | 0.67 | 0.38 | 0.524 | 0.29 | 0.30 | 0.15 |
MAF–Major Allele Frequency; PIC–Polymorphism Information Content
Markers that were monomorphic among wild and/or cultivated accessions are represented by “N/A” under the respective PIC columns
Genetic diversity analysis
Ninety five % (76 out of 80) of the SNP markers showed polymorphism across 30 wild accessions while only 27.5% (22 out of 80) were polymorphic across the 59 cultivated genotypes revealing low variability within the cultivated finger millet. However, the 4 SNP markers (ICECSN99, ICECSNT33, ICECSNT72, ICECSNT78) that were monomorphic across wild accessions all showed polymorphism among the cultivated accessions. These polymorphisms resulted either from a single genotype harboring a heterozygous allele (for ICECSN99, ICECSNT33, and ICECSNT78) or from a few segregating genotypes (in the case of ICECSNT72). KNE796, which was also one of the genotypes used to generate the sequencing data, uniquely harbored the only heterozygous alleles for loci ICECSN99 and ICECSNT78 while P224, a popular high yielding variety, harbored the only heterozygous allele for locus ICECSNT33 and one of the 2 alternative homozygous alleles of locus ICECSNT72. The average PIC of the 80 SNP markers tested across 89 diverse genotypes was 0.29 (Table 5). The wild accessions revealed significantly higher polymorphism levels with 76 polymorphic markers and a mean PIC of 0.30. Only 22 SNP markers were polymorphic across the cultivated accessions, with an average PIC of 0.15 (Table 5).
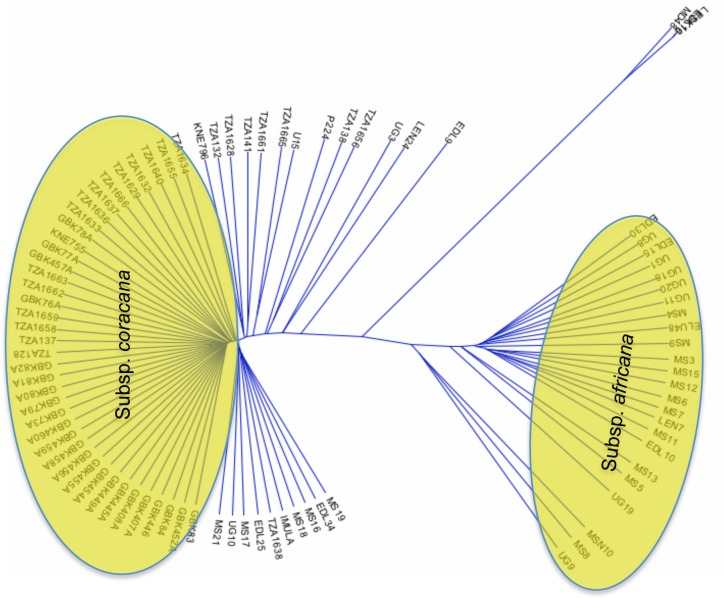
The dendrogram (Fig 2) generated showed two major clusters (highlighted in yellow) and an outgroup formed by three E. kigeziensis accessions (MD48, LESK10 and EDL16). Cluster I comprised mainly of cultivated E. coracana subsp. coracana accessions while the second cluster comprised of E. coracana subsp. africana accessions (24 accessions). Accessions MS19, MS17, MS21, EDL34, MS18, UG10, LEN24, MS16 and EDL25, which were previously classified as wild, grouped closer to the subsp. coracana accessions, suggesting they may have hybridized with cultivated accessions.
Fig 2. Clustering of 89 Eleusine spp using UPGMA.
A dendrogram showing clustering of both wild and cultivated accessions generated using UPGMA in TASSEL software. Two major clusters (subsp. coracana and subsp. africana) are shown in yellow and one out-group consisting of 3 accessions (MD48, LESK10 and EDL16). Accessions MS19, MS17, MS21, EDL34, MS18, UG10, LEN24, EDL25, EDL9, MS16 and UG3, which were previously morphologically classified under subsp. africana clustered closer to the subsp. coracana genotypes and are therefore likely to be hybrids between subsp. africana and coracana.
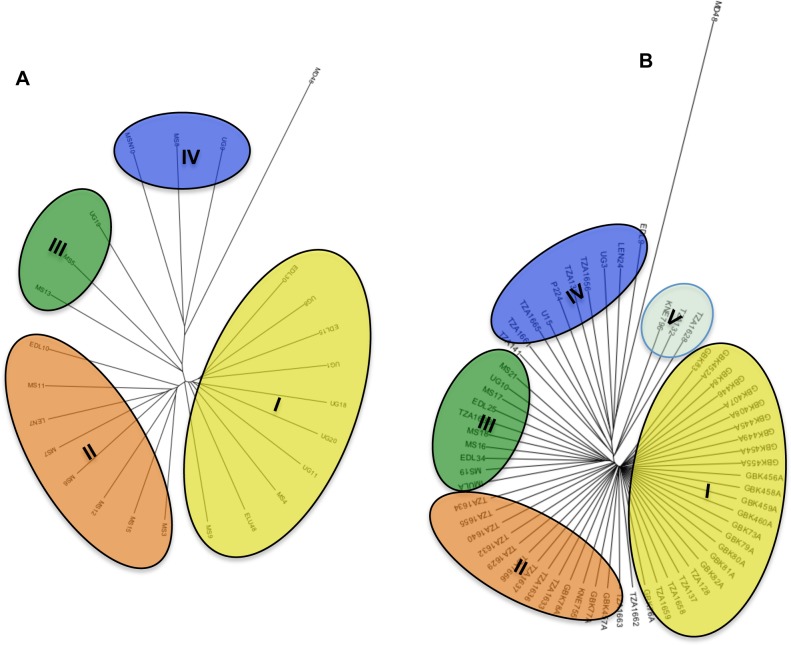
We wanted to understand the extent of diversity within each of the 2 clusters identified in Fig 2 so we selected all accessions from each cluster and independently analysed them using E. kigeziensis accession MD48 as an outgroup. We generated two dendrograms (Fig 3), the first one composed of all the 24 accessions of E. coracana subsp. africana (Fig 3A) and the second one comprising of 62 accessions that had clustered within or closer to the E. coracana subsp. coracana group (Fig 3B). The 2 dendrograms showed relatively similar clustering patterns (clusters I, II, III, IV) except in Fig 3B, in which 3 genotypes (KNE796, TZA132 and TZA1628) formed an additional cluster V.
Fig 3. Comparison of clustering within the different sub species of E. coracana.
Two dendrograms generated using genotypes that clustered under subsp. africana (A) and subsp. coracana cluster (B) in Fig 1 above, each rooted using MD48 (E. kigeziensis). In (A), 69 polymorphic markers were used to cluster 24 africana accessions while in (B), 71 polymorphic markers were used to cluster 62 accessions. Four major sub-clusters of africana are seen (I, II, III, IV), which correspond to similar sub-clusters in coracana (B). There is an additional cluster (V) in coracana (B), comprising only of high yielding improved accessions, which are likely to have been improved using Indian material.
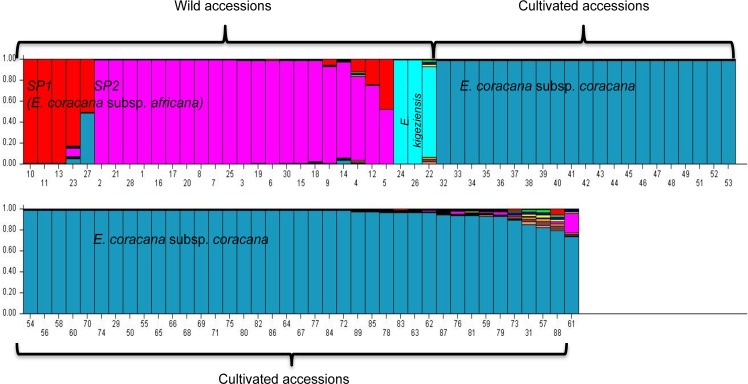
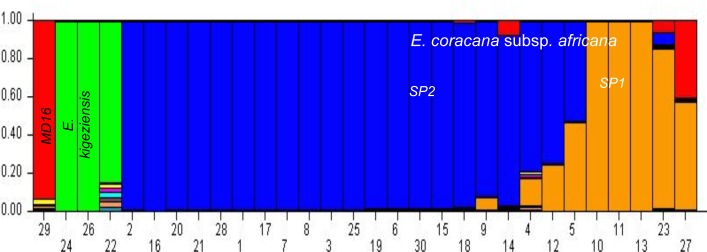
The 89 accessions were further classified into two sub-populations (delta K = 2) (S1 Fig) using STRUCTURE. The sub-populations generated were complementary to the UPGMA tree analysis (Fig 2) and could distinguish the wild accessions (24 of subsp. africana and 3 of E. kigeziensis) from the cultivated (Fig 4). We further confirmed that the 9 accessions (MS19, MS21, MS18, MS17, EDL34, EDL25, LEN24, UG10, MS16) that had been previously recorded as subsp. africana were actually subsp. coracana as they grouped together with the cultivated accessions (Fig 4). Although LEN24 and UG10 showed some degree of admixture, there were also other cultivated genotypes such as TZA138, P224 and KNE796 that equally showed some levels of admixture but have been classified as E. coracana subsp coracana. The 3 out-group accessions (MD48, LESK10 and EDL16) revealed using UPGMA maintained a distinct grouping from the E. coracana subsp. africana accessions (Fig 4) confirming that they belonged to the distinct species E. kigeziensis.
Fig 4. Population structure of 89 Eleusine accessions.
Output of STRUCTURE analysis done using 80 polymorphic markers across 89 Eleusine accessions. Each vertical line represents an individual accession. The corresponding accession codes are provided in Table 1. The y-axis displays % estimated membership of each individual cluster or population. Accessions numbered 1 to 30 are assumed to be “wild”. The rest of the accessions are cultivated and were either collected from farmers’ fields or gene banks. Four sub-populations can be seen here highlighted in red (E. coracana subsp. africana sub-population 1), purple (E. coracana subsp. africana sub-population 2), turquoise (E. kigeziensis) and sky blue (E. coracana subsp. coracana). Each color indicates the proportion of DNA segments for each individual, represented by a vertical bar, in each group.
We subsequently explored the sub-populations within the wild accessions (three from E. kigeziensis and 24 of E. coracana subsp. africana) and attempted to correctly identify the two accessions (EDL9 and UG3) that were previously classified as wild but which now clustered with the cultivated accessions under both UPGMA clustering (Fig 2) and STRUCTURE (Fig 4) grouping. An independent STRUCTURE analysis of the 29 accessions, including MS16 as check, detected a maximum delta K at K = 3. The 30 “wild” accessions were classified into 3 sub-populations–one in E. kigeziensis and two in subpopulations of subsp. africana. Cultivated accession MS16 formed a distinct cluster (Fig 5). EDL9 and UG3 showed mixed ancestry with cultivated species further confirming their clustering with cultivated accessions in Fig 2. The latter STRUCTURE analysis also revealed minimal admixture of the two E. kigeziensis accessions (LESK10 and EDL16) with the other accessions in comparison with MD48 (Fig 5). This is understandable since MD48 has been used in the breeding program for a long time while LESK10 and EDL16 are new collections.
Fig 5. Population structure of 30 “wild” Eleusine accessions.
Output of STRUCTURE analysis done using 76 polymorphic SNP markers across 30 “wild” accessions only. Four sub-populations are shown in red, green, blue and orange. The only genotype appearing under red (MS16) has been confirmed in Fig 2 to be cultivated. The 3 genotypes highlighted in green are E. kigeziensis (Lesk10, EDL16 and MD48) while blue and orange represent subsp. africana subpopulations 2 and 1 respectively. The orange sub-population is composed of the same samples clustering in red under Fig 3. The corresponding accession codes are shown in Table 1.
Discussion
Abundance of molecular markers in the finger millet genome
We report here the simultaneous mining of SSR and SNP markers from cultivated finger millet (E. coracana subsp. coracana), for which there are currently very few polymorphic markers. As expected, SNP markers were more abundant (1/3.3 kbp) than SSRs (1/7.5 kbp) despite the stringent filtering criteria used to eliminate homeologous SNPs. Homeologous SNPs were much more abundant (at least 1/kbp) suggesting a recent polyploidization event within the cultivated species. Recently formed polyploids have not undergone extensive genetic or genomic changes, and therefore their genomes would be additive with respect to their parental species [58]. The higher levels of homeologous SNPs may also suggest independent segregation of the AA and BB sub-genomes. The allotetraploid origin of finger millet remains unresolved and the detection of homeologous loci in the nuclear genome can provide critical information to elucidate its evolutionary history. Future evolution studies should include E. indica (AA), which is believed to be the maternal genome donor [59], alongside the two supspecies of E. coracana.
Consistent with previous studies in finger millet [6], there were fewer markers within the putative genic regions compared to those from genomic regions. The Ts/Tv ratio was higher within putative genic regions (2.0) compared with genomic regions (1.8). AT di-nucleotide repeats were the most abundant SSR markers within the genic regions. These results are in agreement with findings for Petunia [60] and mungbean [61] but lower than what was reported for rice [62]. Transitions (A/G, C/T) are always more common than Transversions (A/C, A/T, G/C, G/T) as they provide easy tolerance from selection pressure [63] but may also be an indication of low levels of genetic divergence [61].
We focused on di-, tri- and tetra-nucleotide SSRs due to their abundance in plant genomes [37–39] and selected those that were at least 10 bp long to maximize the polymorphism levels. Mononucleotide repeats were excluded due to the higher error rates [40] and less informative nature compared to di- and tri-nucleotide repeats [41,42]. Penta- and hexa-nucleotide repeats were similarly excluded due to low abundance as reported in monocot genomes [43,44]. The frequency of the chosen three classes of repeats (di-, tri-, tetra-) was 1/7.5 kb in finger millet genomic sequences. This was higher than observed in other grasses including foxtail millet (69/Mb) [64] and Brachypodium (101/Mb) [44] but lower than in rice [65] even though more classes of nucleotide repeats (di-, tri-, tetra-, penta-, hexa-) were considered in the latter study. In comparison with dicots, the SSR frequency distribution in finger millet was lower than in cucumber [66] and citrus [67]. However, meaningful SSR frequency distribution comparisons across species can only be done if SSR mining criteria and algorithms are fully standardized across studies [68].
The distribution of SNPs across the two genotypes (1/3.3 kb or 303 SNPs/Mb) was comparable to those observed between two Japonica rice varieties (346.6 SNPs/Mb) [69] but much lower than those reported across Japonica and Indica genomes [70]. However, the rice study identified SNPs from two Japonica varieties that included a landrace (Omachi) and an improved line (Nipponbare), whilst the current study included only improved finger millet varieties (KNE755 and KNE796), suggesting that higher numbers of SNP markers could be identified if more diverse genotypes were included in the discovery process. Stringent SNP identification criteria would need to be maintained to eliminate homeologous SNPs and reduce the numbers of false positives that have been observed in other polyploid crops [71]. Future SNP discovery studies in finger millet should also include genotypes from other Eleusine species, which are likely to be exploited in future breeding programs.
Conversion of SNP markers into assays
KBiosciences allele-specific PCR (KASPar) technology was used to convert a random set of the identified SNPs into assays, as this system is flexible and was shown to work well with other polyploids [72,73]. Ninety-two of the 101 (91%) randomly selected SNP regions were successfully converted into assays, of which 80 (~80%) were polymorphic across across diverse Eleusine species. This conversion rate was quite compared to similar studies in hexaploid wheat (67%) [72] and polyploid Spartina pectinata (78.5%) [73], and demonstrated that KASPar-technology is suitable for quick validation of markers and low-throughput genotyping within and across different Eleusine species. However, for high-throughput SNP assays in the future, all available SNP genotyping platforms [74–76], as well as genotyping-by-sequencing (GBS) options [77,78] should be considered for the rapid analysis of high numbers of SNPs.
SNP/SSR allelic diversity
In most plants, di-nucleotide SSRs have been reported to be the most polymorphic [79], followed by tri-nucleotide [37] repeats. The 12 most polymorphic SSR markers (PIC ≥ 0.5) in the current study were di-nucleotide repeats with a minimum of 22 bp in length. Previous studies on SSR marker development in finger millet reported the same [7]. Although most of the SSRs identified in this study were not further tested across diverse finger millet germplasm, we recommend that future investigations intending to make use of SSRs in finger millet should focus on testing mainly di-nucleotide repeats with a minimum of 20 bp in length.
The allelic diversity of the SNP markers identified in this study was relatively low (mean PIC of 0.29) and remarkably different across wild (mean PIC = 0.30) and cultivated (mean PIC = 0.15) germplasm. Narrow genetic diversity had been observed in cultivated finger millet [17,80] due to high inbreeding levels. Compared with wild species in which 76 SNP markers showed polymorphism, there were only 22 polymorphic markers across the cultivated species. Similar low PIC values were reported for other polyploids including wheat [81]. However, it must be noted that unlike SSRs, SNPs are bi-allelic with a maximum PIC values of 0.5 [81]. Since this was one of the few SNP diversity analysis studies in finger millet, additional studies are recommended before detailed conclusions can be made. Nevertheless, the abundant diversity observed among the wild accessions should be used in breeding programs to broaden the genetic base of cultivated accessions.
Finger millet genetic diversity
The 80 polymorphic SNP markers developed in the current investigation successfully discriminated various Eleusine species and enabled correct classification of unknown genotypes. Most wild accessions used in the current study were new collections that were expected to be cross-compatible with cultivated species except MD48. The clustering pattern observed when all accessions were analysed together clearly distinguished between E. coracana subsp. coracana and that of subsp. africana confirming the distinctness of the two subspecies. We were also able to classify ten “unidentified” new collections (MS16, MS19, MS17, MS21, EDL34, MS18, UG10, LEN24, EDL25, EDL16, LESK10) correctly based on the phylogenetic and STRUCTURE analysis. Although MS16 was previously considered a wild accession belonging to E. coracana subsp. africana, our clustering confirmed it was a cultivated accession with mixed ancestry. Although Lesk10 and EDL16 were previously classified as E. coracana subsp. africana, their consistent clustering with MD48 (E. kigeziensis) left no doubt that these two genotypes belong to the same (E. kigeziensis) species. Due to the tight clustering between Lesk10 and EDL16, it was difficult to conclude whether the samples were contaminated or if the genotypes were indeed genetically very similar. More studies are needed to combine both morphological and genetic analysis to correctly distinguish these two genotypes (Lesk10 and EDL16) in the future.
Some degree of geographical clustering was also observed within the two major clusters. For example, within the africana subspecies, most of the accessions from Uganda (UG11, UG20, UG18, UG1) and western Kenya (at the border with Uganda) clustered together, while the Kenyan (names starting with GBK) and Tanzanian (names starting with TZA) cultivated accessions also clustered together. The deviation of some of the improved varieties from the major cluster may have been due to gene flow between subsp. africana and coracana, as well as the recent history of finger millet breeding in eastern Africa, which was influenced by Indian accessions. Some finger millet varieties released in Africa (such as P224) were bred through hybridization of African and Indian germplasm [18]. Previous studies including both African and Asian collections often resulted in distinct clustering of Asian accessions from African ones [17,82,83]. More surprising was the clustering of EDL9 and UG3 with the subsp. coracana accessions. Both EDL9 and UG3 were previously classified as belonging to subsp. africana using morphological features. Given the high levels of gene flow between africana and coracana, it is likely possible that these two genotypes have crossed with coracana and are therefore genetically more similar to subsp. coracana but morphologically similar to subsp. africana.
Population structure
Eastern Africa, being the center of diversity for finger millet, contains a large number of landraces as well as finger millet wild relatives. Using STRUCTURE software, we identified four distinct sup-populations when K = 10 (Fig 4) representing E. kigeziensis, two sub-populations of E. coracana subsp. africana and E. coracana subsp. coracana. Of the three genotypes (MD48, LESK10 and EDL16) that clustered under E. kigeziensis, it was only MD48 that displayed some degree of admixture. MD48 was originally collected from Uganda [80], but had been used in the breeding program at Maseno University (Kenya) for many years. MD48 may have received pollen from other cross-compatible accessions growing side-by-side, although LESK10 and/or EDL16, appeared to be pure and could therefore be used in future studies aiming to capture pure E. kigeziensis genomes. While earlier studies within E. coracana species reported two subpopulations of coracana (Asian and African subpopulations) [80], this is the first study suggesting two subpopulations within subsp. africana. The two subpopulations of africana observed did not reveal any geographical clustering pattern and therefore need to be investigated further to determine the basis for grouping.
All E. coracana subsp. coracana accessions formed one distinct grouping and also enabled further identification of previously misidentified accessions, such as MS16. We were also able to confirm that both EDL9 and UG3 were admixtures and contained DNA from subsp. coracana. Clearly, UPGMA and morphological characterization alone are not enough for correct classification of accessions within a genus where gene flow is rampant. Other studies that included both Asian and African accessions in the past [80] have revealed two subpopulations of subsp. coracana that reflected the two geographies. Our study specifically included only eastern African collections of subsp coracana, and this may explain the low levels of genetic diversity and the observation of only one subpopulation of coracana.
This study illustrated the power of NGS to advance research in previously under-studied crops such as finger millet and demonstrated the immediate application of such resources in breeding programs. Although genic molecular markers are considered more useful due to their likely association with functional genes and wider application across related species, genomic markers are still extremely useful in finger millet due to their abundance and high polymorphism levels that would facilitate the immediate implementation of genomics-assisted breeding. No doubt, future marker development studies in finger millet would need to exploit the decreasing sequencing costs in order to generate higher numbers of genic markers as has been done in other polyploids [27,73]. While the narrow genetic diversity within cultivated finger millet can be immediately addressed through the exploitation of primary, secondary and tertiary genepools, such an effort will also require an extensive assembly of a well-characterized genetic resource alongside well-developed genomic resources. Finger millet breeders will therefore need to modernize their breeding tools in order to reduce the current tedious varietal development process, especially in Africa.
Supporting Information
(TIF)
(XLS)
(XLSX)
(DOCX)
(DOCX)
Data Availability
All sequences have been deposited to the NCBI database accession number SRP073162.
Funding Statement
The work published here was funded by Bio-Innovate (www.bioinnovate-africa.org) GRANT NUMBER: 01/2010 as well as the CGIAR Research Program on Dryland Cereals (CRP-DC). Bio-Innovate funds were used for germplasm collection, sequencing, marker development and genotyping. CRP-DC funds were used to pay cater for laboratory bench fees, germplasm maintenance, post-graduate student stipend and the publication costs. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Srinivasachary, Dida MM, Gale MD, Devos KM. Comparative analyses reveal high levels of conserved colinearity between the finger millet and rice genomes. Theor Appl Genet. 2007. August;115(4):489–99. [DOI] [PubMed] [Google Scholar]
- 2.Hilu KW, de Wet JMJ. Domestication of Eleusine coracana. Econ Bot. 1976. July;30(3):199–208. [Google Scholar]
- 3.Bheema Lingeswara Reddy IN, Srinivas Reddy D, Lakshmi Narasu M, Sivaramakrishnan S. Characterization of disease resistance gene homologues isolated from finger millet (Eleusine coracana L. Gaertn). Mol Breed. 2010. April 23;27(3):315–28. [Google Scholar]
- 4.Kothari S.L., Kumar S., Vishnoi R.K., Kothari O. and Watanabe KN. Application of biotechnology for improvement of millet crops: Review of progress and future prospects. Plant Biotechnol. 2005;22(2):81–8. [Google Scholar]
- 5.Dida MM, Srinivasachary, Ramakrishna S, Bennetzen JL, Gale MD, Devos KM. The genetic map of finger millet, Eleusine coracana. Theor Appl Genet. 2007;114:321 10.1007/s00122-006-0435-7. [DOI] [PubMed] [Google Scholar]
- 6.Arya L, Verma M, Gupta VK, Karihaloo JL. Development of EST-SSRs in finger millet (Eleusine coracana ssp coracana) and their transferability to pearl millet (Pennisetum glaucum). J Plant Biochem and Biotechnol. 2009;18(1):97–100. [Google Scholar]
- 7.De Villiers SM, Michael VN, Manyasa EO, Saiyiorri AN, Deshpande S. Compilation of an informative SSR set for genetic characterisation of East African finger millet (Eleusine coracana). Electron J Biotechnol. Elsevier B.V.; 2015;18(2):77–82. [Google Scholar]
- 8.Kumar A, Sharma D, Tiwari A, Jaiswal JP, Singh NK, Sood S. Genotyping-by-Sequencing analysis for determining population structure of finger millet germplasm of diverse origins. The Plant Genome. 2016; 9(2):1–15. 10.3835/plantgenome2015.07.0058 [DOI] [PubMed] [Google Scholar]
- 9.Varshney RK, Graner A, Sorrells ME. Genic SSR markers in plants: features and applications. Trends Biotechnol. 2005. January;23(1):48–55. [DOI] [PubMed] [Google Scholar]
- 10.Perez-de-Castro AM, Vilanova S, Canizares J, Pascual L, Blanca JM, Diez MJ, et al. Application of genomic tools in plant breeding. Curr Genomics. 2012;13(3):179–195. 10.2174/138920212800543084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bardakci F. Random Amplified Polymorphic DNA (RAPD) Markers. Turkish J Biol. 2000;25:185–96. [Google Scholar]
- 12.Gupta R, Verma K, Joshi DC, Yadav D, Singh M. Assessment of genetic relatedness among three varieties of finger millet with variable seed coat color using RAPD and ISSR Markers. Genet Eng Biotechnol J. 2010;2:1–9. [Google Scholar]
- 13.Coombs J, Frank L, Douches D. An applied fingerprinting system for cultivated potato using simple sequence repeats. Am J potato Res. 2004. March;81:243–50. [Google Scholar]
- 14.Rahman ML, Jiang W, Chu SH, Qiao Y, Ham T- H, Woo M- O, et al. High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor Appl Genet. 2009. November;119(7):1237–46. 10.1007/s00122-009-1125-z [DOI] [PubMed] [Google Scholar]
- 15.Hao C, Wang L, Ge H, Dong Y, Zhang X. Genetic diversity and linkage disequilibrium in Chinese bread wheat (Triticum aestivum L.) revealed by SSR markers. PLoS One. 2011. January;6(2):e17279 10.1371/journal.pone.0017279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obidiegwu ON. Development and genotyping potentials of EST-SSRs in finger millet (E. Coracana (L.) Gaertn.). Int J Genet Genomics. 2014;2(3):42. [Google Scholar]
- 17.Babu BK, Dinesh P, Agrawal PK, Sood S, Chandrashekara C, et al. Comparative Genomics and Association Mapping Approaches for Blast Resistant Genes in Finger Millet Using SSRs. PLoS ONE. 2014. November;9(6): e99182 10.1371/journal.pone.0099182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner D. Next-generation DNA sequencing technologies. Encycl Anal Chem. 2011. [Google Scholar]
- 19.Parida SK, Kalia SK, Kaul S, Dalal V, Hemaprabha G, Selvi A, et al. Informative genomic SSR markers for efficient genotyping applications in sugarcane. Theor Appl Genet. 2009. January;118(2):327–338. 10.1007/s00122-008-0902-4 [DOI] [PubMed] [Google Scholar]
- 20.Kalia RK, Rai MK, Kalia S, Singh R, Dhawan a. K. SSR markers: an overview of the recent progress in plants. Euphytica. 2010. November 9;177(3):309–34. [Google Scholar]
- 21.Anithakumari AM, Tang J, van Eck HJ, Visser RGF, Leunissen JM, Vosman B, et al. A pipeline for high throughput detection and mapping of SNPs from EST databases. Mol Breed. 2010. June;26(1):65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawson IK, Hedley PE, Guarino L, Jaenicke H. Does biotechnology have a role in the promotion of underutilised crops? Food Policy. 2009. August 1;34(4):319–28. [Google Scholar]
- 23.IRGSP. The map-based sequence of the rice genome. Nature. 2005. August 11;436(7052):793–800. [DOI] [PubMed] [Google Scholar]
- 24.Singhal D, Gupta P, Sharma P. In-silico single nucleotide polymorphisms (SNP) mining of Sorghum bicolor genome. African J Biotechnol. 2013;10(4):580–3. [Google Scholar]
- 25.Clevenger JP, Ozias-Akins P. SWEEP: A tool for filtering high-quality SNPs in polyploid crops. Genes Genomes Genetics. 2015. September;5:1797–1803. 10.1534/g3.115.019703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai K, Duran C, Berkman PJ, Lorenc MT, Stiller J, Manoli S, et al. Single nucleotide polymorphism discovery from wheat next-generation sequence data. Plant Biotechnol J. 2012. March 10;6:743–749. [DOI] [PubMed] [Google Scholar]
- 27.Hulse-Kemp AM, Ashrafi H, Stoffel K, Zheng X, Saski CA, Scheffler BE, et al. BAC-end sequence-based SNP mining in allotetraploid cotton (Gossypium) utilizing resequencing data, phylogenetic inferences, and perspectives for genetic Mapping. Genes Genomes Genetics. 2015. April;5:1095–1105. 10.1534/g3.115.017749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y- F, Poland JA, Wight CP, Jackson EW, Tinker NA. Using Genotyping-By-Sequencing (GBS) for genomic discovery in cultivated oat. PLoS ONE. 2014. July;9(7):e102448 10.1371/journal.pone.0102448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, Xia Y, Ren X, Chen Y, Huang L, Huang S, et al. Construction of a SNP-based genetic linkage map in cultivated peanut based on large scale marker development using next-generation double-digest restriction-site-associated DNA sequencing (ddRADseq). BMC Genomics. 2014. May;15:351 10.1186/1471-2164-15-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aronesty E. ea-utils: Command-line tools for processing biological sequencing data. 2011. Available from: http://code.google.com/p/ea-utils.
- 31.O'Brien EA, Zhang Y, Wang E, Marie V, Badejoko W, Lang BF, et al. GOBASE: an organelle genome database. Nucleic Acids Res. 2009;37:D946–950. 10.1093/nar/gkn819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009. January;10(3):R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008. May;18(5):821–9. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Lu P, Luo Z. GMATo: A novel tool for the identification and analysis of microsatellites in large genomes. Bioinformation. 2013. June;9(10):541–544. 10.6026/97320630009541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez L, Barreiro R, Fischer M, Koch MA. Mining microsatellite markers from public expressed sequence tags databases for the study of threatened plants. BMC Genomics. 2015;16:781 10.1186/s12864-015-2031-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merritt BJ, Culley TM, Avanesyan A, Stokes R, Brzyski J. An empirical review: characteristics of plant microsatellite markers that confer higher levels of genetic variation. Applications in Plant Sciences. 2015;3(8):1500025 10.3732/apps.1500025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Zhang G, Li X, Wang L, Yuan J, Deng C, Gao W. Genome-wide identification and validation of simple sequence repeats (SSRs) from Asparagus officinalis. Molecular and Cellular Probes. 2016. 10.1016/j.mcp.2016.03.003 [DOI] [PubMed] [Google Scholar]
- 38.Fungtammasan A, Ananda G, Hile SE, Su MS, Sun C, Harris R, et al. Accurate typing of short tandem repeats from genome-wide sequencing data and its applications. Genome Research. 2016. May;25:736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng SP, Li WG, Huang HS, Wang JY, Wu YT. Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breed. 2009;28:85–97. 10.1007/s11032-008-9216-0 [DOI] [Google Scholar]
- 40.Xiao J, Zhao J, Liu M, Liu P, Dai L, Zhao Z. Genome-wide characterization of Simple Sequence Repeat (SSR) loci in Chinese Jujube and Jujube SSR primer transferability. PLoS ONE. 2015. May;10(5):e0127812 10.1371/journal.pone.0127812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang ZJ, Peng ZS, Yang H. Identification of novel and useful EST-SSR markers from de novo transcriptome sequence of wheat (Triticum aestivum L.). Genetics and Molecular Research. 2016;15(1):gmr.15017509. doi 10.4238/gmr.15017509. [DOI] [PubMed] [Google Scholar]
- 42.Sonah H, Deshmukh RK, Sharma A, Singh VP, Gupta DK, Gacche RN, et al. Genome-wide distribution and organization of microsatellites in plants: An insight into marker development in Brachypodium. PLoS ONE. 2011. June;6(6):e21298 10.1371/journal.pone.0021298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009. July 15;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009. August 15;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv Preprint arXiv. 2012;1207.3907 [q-bio.GN].
- 46.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011. August 1;27(15):2156–8. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. [DOI] [PubMed] [Google Scholar]
- 48.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nature Protocols. 2013;8:1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 2000. February;18:233–234. [DOI] [PubMed] [Google Scholar]
- 50.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005. May 1;21(9):2128–2129. [DOI] [PubMed] [Google Scholar]
- 51.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 52.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. [DOI] [PubMed] [Google Scholar]
- 53.Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4(2):359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- 55.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. [DOI] [PubMed] [Google Scholar]
- 56.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. [DOI] [PubMed] [Google Scholar]
- 57.Solanke AU, Mithra SVA. Genetic and genomic resources in “finger millet—a climate resilient nutri-millet”. http://www.biotecharticles.com/Biotech-Research-Article/Genetic-and-Genomics-Resources-in-Finger-Millet-3098.html. 2013-12-12 03:20:15.
- 58.Baumel A, Ainouche M, Kalendar R, Schulman AH. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Molecular Biology and Evolution. 2002;19:1218–1227. [DOI] [PubMed] [Google Scholar]
- 59.Neves SS, Swire-Clark G, Hilu KW, Baird WV. Phylogeny of Eleusine (Poaceae: Chloridoideae) based on nuclear ITS and plastid trnT-trnF sequences. Mol Phylogenet Evol. 2005;35:395–419. [DOI] [PubMed] [Google Scholar]
- 60.Guo Y, Wiegert-Rininger KE, Vallejo VA, Barry CS, Warner RM. Transcriptome-enabled marker discovery and mapping of plastochron-related genes in Petunia spp. BMC Genomics. 2015;16:726 10.1186/s12864-015-1931-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yadav CB, Bhareti P, Muthamilarasan M, Mukherjee M, Khan Y, Rathi P, Prasad M. Genome-Wide SNP Identification and Characterization in Two Soybean Cultivars with Contrasting Mungbean Yellow Mosaic India Virus Disease Resistance Traits. PLoS ONE. 2015. April;4:e0123897 10.1371/journal.pone.0123897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mehra P, Pandey BK, Giri J. Genome-wide DNA polymorphisms in low Phosphate tolerant and sensitive rice genotypes. Scientific Reports. 2015. August;5:13090 10.1038/srep13090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wakeley J. The excess of transitions among nucleotide substitutions: new methods of estimating transition bias underscore its significance. Trends Ecol Evol. 1996;11:158–162. [DOI] [PubMed] [Google Scholar]
- 64.Pandey G, Misra G, Kumari K, Gupta S, Parida SK, Chattopadhyay D, et al. Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res. 2013. February;20:197–207. 10.1093/dnares/dst002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, Deng Y, Tan J, Hu S, Yu J, Xue Q. A Genome-wide microsatellite polymorphism database for the Indica and Japonica rice. DNA Res. 2007;14:37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J, Qu J, Hu K, Zhang L, Li J, Wu B, et al. Development of genomewide simple sequence repeat fingerprints and highly polymorphic markers in cucumbers based on next-generation sequence data. Plant Breeding. 2015. July;134:605–611. [Google Scholar]
- 67.Liu S-R, Li W-Y, Long D, Hu C-G, Zhang J-Z. Development and characterization of genomic and expressed SSRs in citrus by genome-wide analysis. PLoS ONE. 2013. October;8(10):e75149 10.1371/journal.pone.0075149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uncu AÖ, Gultekin V, Allmer J, Frary A, Doganlar S. Genomic Simple Sequence Repeat markers reveal patterns of genetic relatedness and diversity in sesame. The Plant Genome 8. 2015;8 10.3835/plantgenome2014.11.0087 [DOI] [PubMed] [Google Scholar]
- 69.Arai-Kichise Y, Shiwa Y, Nagasaki H, Ebana K, Yoshikawa H, Yano M, Wakasa K. Discovery of genome-wide DNA polymorphisms in a landrace cultivar of Japonica rice by whole-genome sequencing. Plant Cell Physiol. 2011;52(2): 274–282. 10.1093/pcp/pcr003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH. An SNP resource for rice genetics and breeding based on subspecies Indica and Japonica genome alignments. Genome Research. 2004;14:1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khera P, Upadhyaya HD, Pandey MK, Roorkiwal M, Sriswathi M, Janila P, et al. Single Nucleotide Polymorphism–based genetic diversity in the reference set of peanut (Arachis spp.) by developing and applying cost-effective Kompetitive Allele Specific Polymerase Chain Reaction genotyping assays. The Plant Genome. 2013;6(3): 10.3835/plantgenome2013.06.0019 [DOI] [Google Scholar]
- 72.Allen AM, Barker GLA, Berry ST, Coghill JA, Gwilliam R, Kirby S, et al. Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnology J. 2011;9:1086–1099. [DOI] [PubMed] [Google Scholar]
- 73.Graves H, Rayburn AL, Gonzalez-Hernandez JL, Nah G, Kim D-S, Lee DK. Validating DNA polymorphisms using KASP assay in Prairie cordgrass (Spartina pectinata Link) populations in the U.S. Front. Plant Sci. 2016. January;6:1271 10.3389/fpls.2015.01271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Syvänen A-C. Toward genome-wide SNP genotyping. Nature Genetics Supplement. 2005. June;37:S5–S10. [DOI] [PubMed] [Google Scholar]
- 75.Paux E, Sourdille P, Mackay I, Feuillet C. Sequence-based marker development in wheat: Advances and applications to breeding. Biotechnology Advances. 2012;30:1071–1088. 10.1016/j.biotechadv.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 76.Clevenger J, Chavarro C, Pearl SA, Ozias-Akins P, Jackson SA. Single nucleotide polymorphism identification in polyploids: A review, example, andrecommendations. Molecular Plant. 2015. June;8:831–846. 10.1016/j.molp.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 77.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple Genotyping-by-Sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6(5): e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD. Switchgrass genomic diversity, ploidy, and evolution: Novel insights from a network-based SNP discovery protocol. PLoS Genet. 2013;9(1):e1003215 10.1371/journal.pgen.1003215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang S, Tang C, Zhao Q, Li J, Yang L, Qie L, et al. Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in Foxtail millet [Setaria italica (L.) P. Beauv.]. BMC Genomics 2014, 15:78 10.1186/1471-2164-15-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dida MM, Wanyera N, Harrison Dunn ML, Bennetzen JL, Devos KM. Population structure and diversity in finger millet (Eleusine coracana) germplasm. Trop Plant Biol. 2008. June 18;1(2):131–41. [Google Scholar]
- 81.Chao S, Zhang W, Akhunov E, Sherman J, Ma Y, Luo M-C, Dubcovsky J. Analysis of gene-derived SNP marker polymorphism in US wheat (Triticum aestivum L.) cultivars. Mol Breeding. 2009;23:23–33. [Google Scholar]
- 82.Arya L, Verma M, Gupta VK, Seetharam A. Use of genomic and genic SSR markers for assessing genetic diversity and population structure in Indian and African finger millet (Eleusine coracana (L.) Gaertn.) germplasm. Plant Syst Evol. 2013;299:1395–401. [Google Scholar]
- 83.Ramakrishnan M, Ceasar SA, Duraipandiyan V, Al-Dhabi NA, Ignacimuthu S. Using molecular markers to assess the genetic diversity and population structure of finger millet (Eleusine coracana (L.) Gaertn.) from various geographical regions. Genet. Resour. Crop Evol. 2015. April;62(4). 10.1007/s10722-015-0255-1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLS)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All sequences have been deposited to the NCBI database accession number SRP073162.