Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal human cancers and shows resistance to any therapeutic strategy used. Here we tested small-molecule inhibitors targeting chromatin regulators as possible therapeutic agents in PDAC. We show that JQ1, an inhibitor of the bromodomain and extraterminal (BET) family of proteins, suppresses PDAC development in mice by inhibiting both MYC activity and inflammatory signals. The histone deacetylase (HDAC) inhibitor SAHA synergizes with JQ1 to augment cell death and more potently suppress advanced PDAC. Finally, using a CRISPR-Cas9–based method for gene editing directly in the mouse adult pancreas, we show that de-repression of p57 (also known as KIP2 or CDKN1C) upon combined BET and HDAC inhibition is required for the induction of combination therapy–induced cell death in PDAC. SAHA is approved for human use, and molecules similar to JQ1 are being tested in clinical trials. Thus, these studies identify a promising epigenetic-based therapeutic strategy that may be rapidly implemented in fatal human tumors.
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of human pancreatic cancer, representing more than 95% of all cases. Despite recent advances in surgical techniques and adjuvant therapy, survival has changed little in the last 20 years, with a 5-year survival rate hovering around 5%. Moreover, PDAC incidence has been increasing gradually to over 45,000 new cases in 2013 in the United States alone, where PDAC has been predicted to rapidly become the second most prevalent cancer killer1. Although some patients benefit from earlier diagnosis due to emerging imaging technologies (enabling the surgical removal of their tumors), even the most advanced chemotherapeutic regimens and virtually all targeted therapies have remained largely ineffective thus far (reviewed in refs. 2–5).
The most frequent oncogenic event in human PDAC is mutation of KRAS (occurring in >95% of cases), which results in Ras activation. Activation of Ras signaling is thought to be both an initiating event and a key driver of PDAC6. Although inhibitors of enzymes in the Ras pathway exist, clinical trials using these inhibitors have not shown meaningful effects in PDAC, in part because of dose-limited toxicities and the emergence of resistant disease5,7. Other genetic alterations frequently found in human PDAC include inactivation of TP53, CDKN2A (also known as p16) or SMAD4 (refs. 8–11). The functional roles of these alterations have been validated in mouse models of PDAC11–16, and the resulting mice constitute preclinical models in which to investigate the mechanisms of PDAC development and identify and test new therapeutic approaches17.
Recent next-generation sequencing efforts have revealed frequent alterations in genes regulating chromatin remodeling and modification in human tumors9,18, which has led to the idea that the proteins encoded by these genes may be used as therapeutic targets in cancer, including in PDAC (reviewed in refs. 19,20). Here we investigate the effect of targeting in PDAC one such family of chromatin regulators, the BET (bromodomain and extra-terminal) family of proteins, which recognize acetylated lysines on histones through their bromodomains (BRD) and control the transcription of oncogenic drivers such as MYC21–23.
RESULTS
BET inhibition suppresses pancreatic tumorigenesis
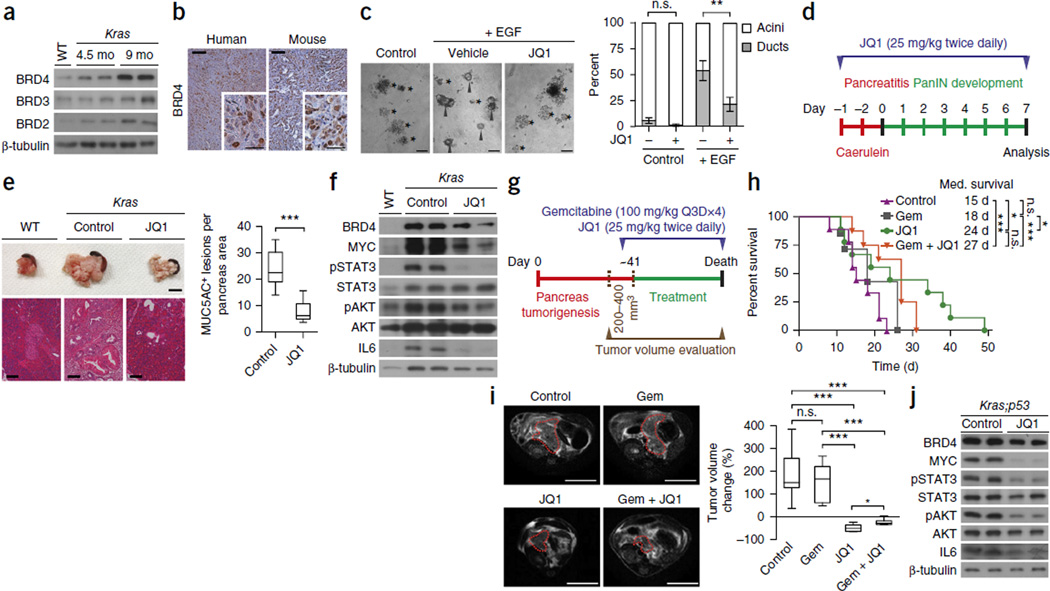
First we examined the expression of BET family proteins in PDAC. We detected expression of BRD2, BRD3, and BRD4 in preneoplastic lesions and frank tumors in the Ptf1a+/Cre;Kras+/LSL-G12D mouse model of endogenous PDAC (hereafter referred to as Kras)12, in human tumors and in human cell lines (Fig. 1a,b and Supplementary Fig. 1a,b). On the basis of these observations, we examined the effects of treating primary mouse pancreatic acinar explants in culture with a small-molecule inhibitor of BET proteins ((+)-JQ-1, hereafter JQ1; ref. 21). JQ1 treatment blocked acinar-to-ductal metaplasia (ADM), a key step in PDAC initiation24 observed in response to constant EGF stimulation, which mimics Ras activation (Fig. 1c). This repro-gramming block correlated with decreased c-MYC (hereafter MYC) levels in mouse acinar explants and the pancreas of Kras mice in vivo (Supplementary Fig. 2a,b). JQ1 treatment blocked pancreatic cell proliferation and the development of pancreatic intraepithelial neoplastic lesions (PanINs) in a mouse model of PDAC co-triggered by oncogenic K-Ras and caerulein-induced inflammation25 (Fig. 1d,e and Supplementary Fig. 2a–c). Immunoblot analysis showed decreased activation of the pro-survival kinase AKT in pancreata from JQ1-treated mice; we also observed downregulation of the activity of inflammatory regulators such as STAT3 and IL6 in pancreata extracts upon JQ1 treatment, correlating with tumor inhibition (Fig. 1f and Supplementary Fig. 2b,c). These data suggest that JQ1 treatment may have chemopreventive effects in PDAC.
Figure 1.
BET protein inhibition suppresses PDAC growth and improves survival in a PDAC mouse model. (a) Immunoblot analysis with the indicated antibodies on tumor lysates from wild-type pancreas and from pancreas of Ptf1a+/Cre;Kras+/LSL-G12D (Kras) mutant mice at 4.5 and 9 months of age (two biological replicates). β-tubulin serves as a loading control. (b) Immunohistochemical analysis of BRD4 expression (brown signal with hematoxylin purple counterstain) on sections from mouse and human PDAC tumors (representative of 12 independent samples). Scale bars, 100 µm, insets 50 µm. (c) Representative images of wild-type mouse acinar clusters (asterisks) undergoing acinar-to-ductal metaplasia (ADM) forming ducts (arrowheads) ex vivo in response to co-culture with EGF or vehicle control for 3 d. Scale bars, 100 µm. Quantification of acinar and ductal clusters on day 3 of culture (right panel), (four independent biological replicates with three technical replicates each). **P < 0.01; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (d) Schematic of the caerulein pancreatitis–induced precancerous (PanINs) lesion formation protocol used for JQ1 treatment of Kras mice. (e) Representative examples of six pancreata images (scale bars, 1 cm) and hematoxylin and eosin (HE) staining (scale bars, 100 µm) (left panel). Quantification of MUC5AC-positive lesions in caerulein-treated pancreata from Kras control (vehicle) (n = 6) and JQ1 treated (n = 6) mice (right panel). ***P < 0.001 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (f) Immunoblots with the indicated antibodies of pancreatic tissue lysates from wild-type (WT) and Kras mutant control (vehicle) and JQ1-treated mice. (g) Treatment schedule for administration of JQ1, gemcitabine (Gem) or vehicle. Ptf1a+/Cre; Kras+/LSL-G12D;Trp53loxP/loxP (Kras;p53) mutant mice undergoing gemcitabine monotherapy also received vehicle. (h) Kaplan-Meier survival curves of Kras;p53 mutant mice from enrollment time in control (vehicle) (n = 9, median survival = 15 d), gemcitabine (Gem; n = 7, median survival = 18 d), JQ1 (n = 9 median survival = 24 d) and combined Gem + JQ1 (n = 8, median survival = 27 d) treatment groups. *P < 0.05; ***P < 0.001; n.s., not significant by log-rank test for significance. (i) Left, representative MRI scan at endpoint measurement of tumor size in Kras;p53 mice. Scale bars, 1 cm. Red dotted lines indicate tumor area. Right, tumor volume quantification at endpoint based on MRI scan (detailed procedure description in Online Methods and Supplementary Fig. 3) of mice in the treatment groups: control, n = 9; Gem, n = 5; JQ1, n = 6; Gem + JQ1, n = 6. *P < 0.05; ***P < 0.001; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (j) Immunoblot analysis of MYC, phospho-STAT3 (pSTAT3), phospho-AKT (pAKT) and IL6 levels in tumor biopsies collected from control and JQ1-treated Kras;p53 mutant mice (multiple independent mouse tumors were obtained and analyzed, two independent and representative samples are shown).
Treatment of human PDAC cell lines with JQ1 also led to lower MYC levels, induction of cell death, and decreased tumorigenic features in culture (Supplementary Fig. 1c – e; see also ref. 26). To further investigate the effects of JQ1 treatment on tumor maintenance in vivo, we used a highly aggressive mouse model of PDAC (Ptf1a+/Cre;Kras+/LSL-G12D;Trp53loxP/loxP mice, hereafter referred to as Kras;p53), in which tumors develop within the first 6 weeks of life11. Short-term treatment (5 d) with JQ1 after tumors had formed resulted in reduced MYC expression and proliferation and increased apoptosis in this model (Supplementary Fig. 2d – g). This led us to investigate the long-term consequences of JQ1 treatment in established PDAC, both as a single agent and in combination with standard-of-care chemotherapy (gemcitabine), in a preclinical therapy trial setting7,27,28. Kras;p53 mutant mice were screened weekly for cancer development by MRI29 and were enrolled in the study upon development of a tumor volume of approximately 200–400 mm3 (Fig. 1g and Supplementary Fig. 3). As reported before in various mouse models29–31, gemcitabine monotherapy had no significant effect on tumor composition, tumor volume or mouse survival in this context (Fig. 1h – j), similar to what is seen in humans with PDAC. In contrast, JQ1 alone or in combination with gemcitabine led to a significant reduction in tumor volume, an increase in apoptosis and a decrease in proliferation, as well as decreased activity in pro-survival and pro-proliferative pathways (Supplementary Fig. 4). Inflammatory signals such as STAT3 and IL6 (Fig. 1j) and expression of the tumor-associated stroma marker smooth muscle actin and collagen (Supplementary Fig. 4) were also reduced upon JQ1 treatment. Notably, JQ1 treatment had only a modest effect on survival on its own and did not significantly synergize with chemotherapy under these conditions (Fig. 1h). Nevertheless, these experiments and recent reports32,33 support an acute therapeutic effect of JQ1 treatment in established PDAC tumors in vivo.
BET inhibition blocks MYC and inflammation
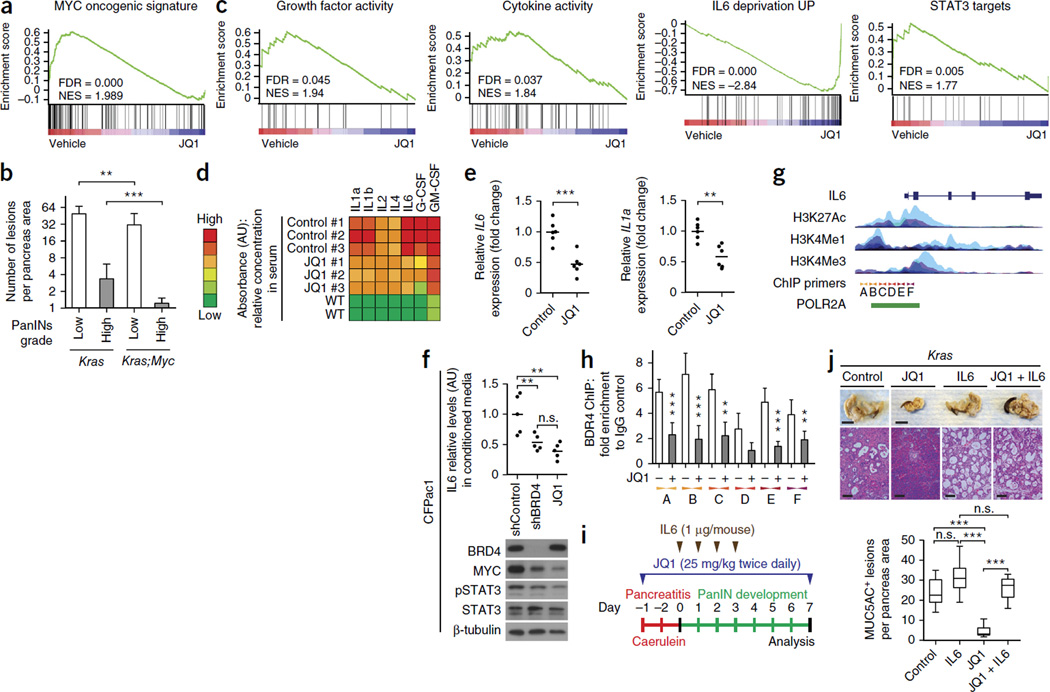
To determine how JQ1 treatment blocks PDAC growth, we performed gene set enrichment analysis (GSEA) on gene expression profiles from JQ1-treated and control Kras;p53 mouse primary PDAC cells. This analysis identified a substantial downregulation of MYC targets in JQ1 treated cells (Fig. 2a). The MYC family of oncogenes has been implicated in the pathogenesis of most types of human tumors, including PDAC34, and we confirmed amplification and increased expression of MYC in human and mouse PDAC (Supplementary Fig. 5a–d). To investigate a role for MYC in PDAC development, we crossed Kras mice to Myc conditional mutant mice. Unlike Kras mice, which showed the expected formation of preneoplastic lesions, mice with concomitant oncogenic Kras activation and Myc deletion at the time of cancer initiation showed attenuated development of preneoplastic lesions of both low and higher grades (Fig. 2b and Supplementary Fig. 5e,f). These data indicate that the inhibitory effects of JQ1 treatment on PDAC cells may in part be mediated by decreased MYC levels; however, BRD family proteins may also control the activity of other drivers of PDAC development.
Figure 2.
MYC and inflammation are key tumorigenic drivers of PDAC that are inhibited by JQ1 treatment and BET protein inhibition. (a) Gene set enrichment analysis (GSEA) in JQ1-treated compared to vehicle-treated Kras;p53 mouse primary PDAC cells. (b) Quantification of spontaneous PanIN lesions formed in 6-month-old Kras (n = 8) and Kras;Myc (n = 8) mutant mice. The grade of lesions is indicated. **P < 0.01; ***P < 0.001 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (c) GSEA from data sets comparing vehicle- to JQ1-treated Kras;p53 mouse primary PDAC cells (see also Supplementary Fig. 5g). (d) Relative serum concentration of inflammatory cytokines in the serum of Kras;p53 mutant mice treated with vehicle control or JQ1 (n = 3 for each experimental group) and wild-type (WT) animals, (e) Quantitative RT-PCR analysis of IL6 and IL1a mRNA expression in patient-derived PDAC xenografts following treatment with JQ1 or vehicle control (see Fig. 4e) (six biological replicates for each experimental condition). **P < 0.01; ***P < 0.001 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (f) Effects of BRD4 inhibition via shRNA or JQ1 treatment on IL6 levels in the conditioned medium of human PDAC CFPac1 cells and for MYC and STAT3 as assessed by immunoblot. **P < 0.01; ***P < 0.001; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (g) Schematic representation of the human IL6 promoter (UCSC Gene Browser) with integrated epigenetic regulation marks and RNA polymerase II binding (POL2RA, green bar) (ENCODE). Localization of primers used for chromatin immunoprecipitation analysis is indicated by A–F. (h) Chromatin immunoprecipitation analysis of BRD4 at the IL6 promoter in CFPac1 cells treated with vehicle control (−) or JQ1 (+). The data are plotted relative to the values obtained with IgG control antibodies. **P < 0.01; ***P < 0.001 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (i) Schematic of the caerulein pancreatitis– induced preneoplastic (PanIN) lesion formation protocol for the rescue experiment with exogenous IL6 injection upon JQ1 treatment. (j) Top, representative pancreata images (of n = 5 each, scale bars, 1 cm) and HE staining (scale bars, 100 µm) in Kras mutant mice in the four experimental conditions indicated (IHC and immunoblot biopsy analysis shown in Supplementary Fig. 7a,b). Bottom, quantification of MUC5AC-positive lesions in caerulein-treated pancreata from each experimental group (the control group was injected with vehicle control) (n = 5 each; IHC staining on Supplementary Fig. 7a). ***P < 0.001; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m.
Inflammatory mediators including STAT3 and IL6 drive pancreatic carcinogenesis35–38. Although MYC is known to promote tumorigenesis by controlling the tumor microenvironment39, we did not observe significant changes in pSTAT3 levels, inflammatory molecules or tumor stromal response in the absence of MYC (Supplementary Fig. 5e and data not shown). In contrast, we observed a marked reduction in IL6 and phospho-STAT3 (pSTAT3) levels in Kras;p53 mutant mice treated with JQ1 (Fig. 1j and Supplementary Fig. 4a). Furthermore, GSEA of JQ1-treated and control tumors revealed that signatures for growth factors, cytokines and immune-related processes were significantly decreased in JQ1-treated mice (Fig. 2c and Supplementary Fig. 5g–i). On the basis of these observations, we analyzed the serum of wild-type, control and JQ1-treated mice and found lower levels of inflammatory cytokines, including IL1a and IL6, in the serum of JQ1-treated animals (Fig. 2d). IL1a and IL6 both promote pancreatic tumorigenesis35,36,40,41. Reduced levels of IL1A and IL6 mRNA were also noted in xenografts derived from humans with PDAC (patient-derived xenografts, PDXs) treated with JQ1 (Fig. 2e). In addition, we observed a correlation between the expression levels of BET family members, MYC and a set of cytokines in human PDAC, further indicating that JQ1 treatment broadly affects inflammatory molecules (Supplementary Fig. 5j). Like JQ1 treatment, shRNA-mediated knockdown of BRD4 led to the downregulation of IL6 and pSTAT3 in human CFPac1 PDAC cells in culture, supporting a cell-intrinsic role for BET proteins in the regulation of the expression of inflammatory mediators in PDAC cells (Fig. 2f). Furthermore, chromatin immunoprecipitation assays in CFPac1 cells showed direct BRD4 association to the IL6 promoter, which was disrupted by JQ1 pretreatment (Fig. 2g,h). Finally, the protective effect of JQ1 treatment on tumor development could be reversed by exogenous injections of IL6, which led to reactivation of STAT3 phosphorylation and accelerated tumorigenesis (Fig. 2i,j and Supplementary Fig. 6). Collectively, these observations indicate that, in addition to MYC, inflammatory signals represent a second oncogenic driver that is downstream of BET family proteins in pancreatic tumorigenesis.
Synergistic effects of JQ1 and the HDAC inhibitor SAHA
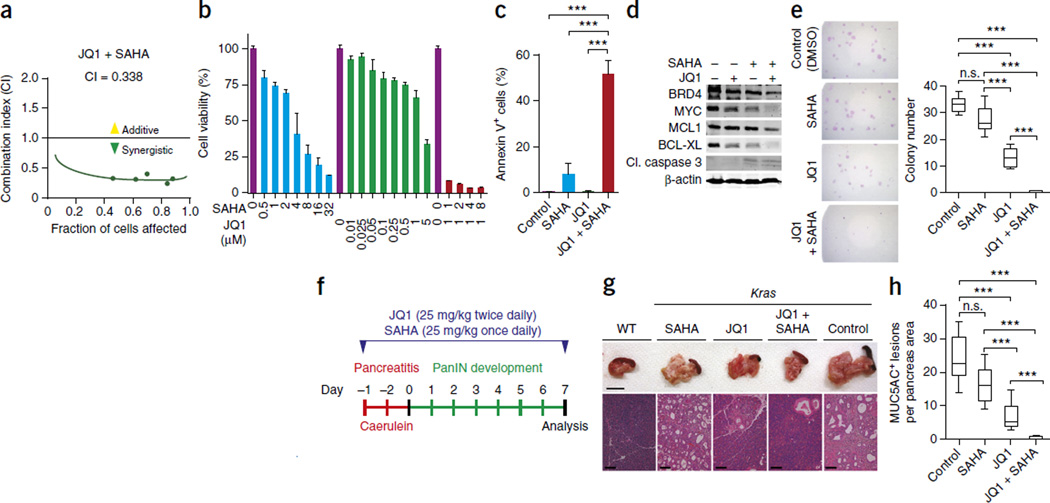
JQ1-treated mice eventually succumbed to PDAC (Fig. 1h). This led us to investigate additional agents that could be combined with JQ1 to more durably suppress PDAC. We selected eight drugs that target key Ras-dependent oncogenic or CDKN2A- or TP53-dependent pathways or epigenetic regulators, and established a cell culture assay to screen for effective combination therapies. Inhibition of histone deacetylases (HDACs) with the small molecule SAHA (vorinostat) had a synergistic effect with BET protein inhibition in human CFPac1 cells, whereas other drug combinations were less effective (Fig. 3a and Supplementary Fig. 7). Co-treatment with JQ1 and SAHA led to a strong reduction in cell viability (Fig. 3b), which correlated with marked induction of apoptosis as measured by annexin V staining, caspase 3 cleavage and reduction of expression of the anti-apoptotic proteins MCL1 and BCL-XL in several PDAC cell lines; in addition, JQ1 and SAHA treatment blocked the ability of human and mouse PDAC cells to form colonies at low density, a measure of decreased tumorigenic potential (Fig. 3b–e and Supplementary Fig. 8). JQ1 and SAHA also synergized to suppress the formation of MUC5AC-positive preneoplastic lesions in Kras mutant mice (Fig. 3f–h and Supplementary Fig. 9).
Figure 3.
Synergistic inhibitory effects of JQ1 and the HDAC inhibitor SAHA in PDAC. (a) Combination index (CI) calculation for JQ1 and SAHA. CFPac1 cell viability was measured after 72 h of drug treatment by the MTT survival assay. (b) Validation of the synergistic inhibitory effects of combined treatment with JQ1 and SAHA as measured by the MTT assay in human DanG PDAC cells after 72 h of treatment. Data are represented as mean ± s.e.m. (c) Quantification of apoptotic cell death by annexin V staining in DanG cells following treatment with JQ1 and SAHA (n = 3 independent replicates). ***P < 0.001 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (d) Immunoblot analysis of BRD4, MYC and apoptosis markers in DanG cells treated with JQ1 and SAHA. β-actin served as a loading control. (e) Representative pictures of colony-formation assays with DanG cells in response to JQ1 and SAHA treatment, with quantification at right (n = 3 for each experimental group). ***P < 0.001; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. The control group was treated with vehicle control (b,c–e). (f) Schematic of the caerulein pancreatitis–induced preneoplastic (PanIN) lesion formation protocol in Kras mutant mice for JQ1 and SAHA co-treatment or control (vehicle) experiment. (g) Representative pancreata images (from n = 5 for each experimental group). Scale bars, 1 cm. (h) Quantification of MUC5AC-positive lesions in caerulein-treated Kras mutant mice pancreata from each experimental group (n = 5 for each experimental group; IHC staining in Supplementary Fig. 9a,b). ***P < 0.001; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m.
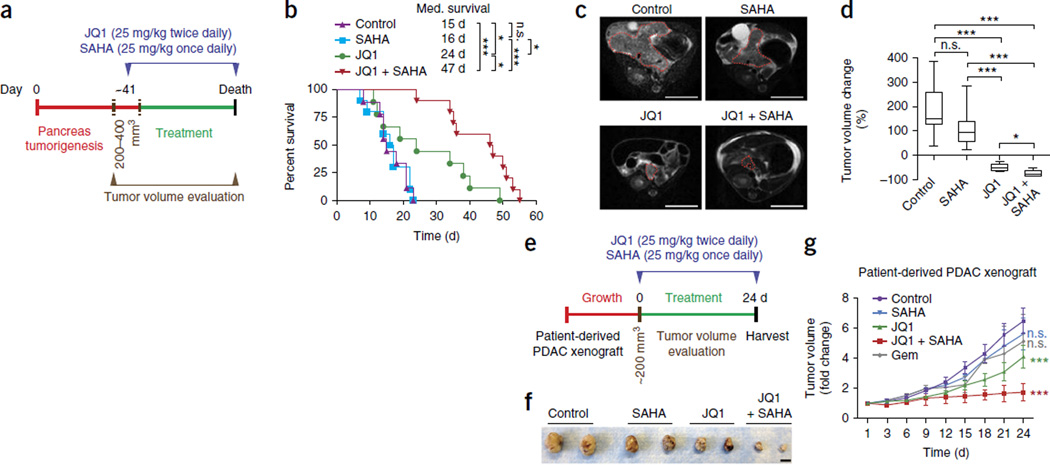
Based on these experiments on cancer initiation in vivo and tumor maintenance in vitro, we next evaluated the effect of combined JQ1+SAHA treatment on established PDAC in vivo. Kras;p53 mutant mice treated with JQ1+SAHA after development of a tumor volume of 200–400 mm3 (Supplementary Fig. 3) showed a greatly reduced tumor volume, more so than Kras;p53 mutant mice treated with JQ1 alone, and had a significant improvement in overall survival(Fig. 4a–d). Of note, 4 of 10 mice treated in the combination arm had to be euthanized because of neurological symptoms due to Cre expression and tumor development in the brain42 (data not shown); this is usually not an issue in this mouse model because these mice normally die much faster from PDAC. Five of the remaining mice treated with the combination therapy developed bile drainage problems and were morbid due to jaundice rather than high tumor burden; one mouse died of unknown cause. Importantly, mice treated with JQ1+SAHA showed no evidence of tumor relapse, a sign of inherent or acquired resistance, such as is typically seen with other therapies, nor did they develop metastasis (data not shown). Residual small tumor clusters had a moderately differentiated histology, with some poorly or undifferentiated areas with strongly depleted stroma. The combination therapy led to a significant cell cycle inhibition (P < 0.05) and a significant increase in apoptotic cell death (P < 0.001) compared to JQ1 treatment alone (Supplementary Fig. 10).
Figure 4.
Combined treatment with JQ1 and SAHA inhibits PDAC progression in vivo. (a) Treatment schedule for administration of JQ1 and SAHA. Kras;p53 mutant mice undergoing monotherapy also received placebo (vehicle) (IHC and immunoblot biopsy analysis in Supplementary Fig. 11a–f). (b) Kaplan-Meier survival curves of Kras;p53 mutant mice from enrollment time in control (vehicle) (n = 9, median survival = 15 d), SAHA (n = 9, median survival = 16 d), JQ1 (n = 9 median survival = 24 d, as in Fig. 1j) and combined JQ1 + SAHA (n = 10, median survival = 47 d) treatment groups. *P < 0.05; ***P < 0.001; n.s., not significant by log-rank test for significance. (c) Representative MRI scan showing measurement of tumor size in Kras;p53 mutant mice at time of animal morbidity. Red dotted lines indicate tumor area. Scale bars, 1 cm. (d) Tumor volume quantification based on MRI scan of Kras;p53 mutant mice in the treatment groups: control (vehicle), n = 9; SAHA, n = 9; JQ1, n = 5; JQ1 + SAHA, n = 5. *P < 0.05; ***P < 0.001; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (e) Treatment schedule for administration of JQ1 and SAHA in a PDX model. (f) Representative macroscopic picture of xenografts from control (vehicle), SAHA, JQ1 and combined JQ1 + SAHA treatment groups at the end of the experiment. Scale bar, 1 cm. (g) Tumor volume quantification of patient-derived PDAC xenografts in mice (n = 4 mice, two tumors per mouse for each treatment group). Mice undergoing monotherapy also received vehicle. ***P < 0.001; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m.
A similar therapeutic effect was observed in a second highly aggressive mouse model of PDAC (Ptf1a+/Cre;Kras+/LSL-G12Dp16/p19loxP/loxP mutant mice, hereafter Kras;p16/19), in which tumors develop with a latency of 8 weeks43 (Supplementary Fig. 11a–d). Also, the combination therapy significantly inhibited the growth of two independent PDAC PDXs; this growth suppression was accompanied by reduced expression of MYC and pSTAT3 and reduced stromal response (Fig. 4e–g and Supplementary Fig. 11e–i).
Typically, increased cancer tumor burden is accompanied by the development of cachexia, but this weight loss was completely prevented by JQ1+SAHA treatment (Supplementary Fig. 12a,b). In addition, the histopathology and blood markers of the treated mice revealed no overt systemic toxicity due to the combination therapy (Supplementary Fig. 12c–h). Thus, the combined inhibition of BET family proteins and HDACs results in a potent and sustained antitumoral response.
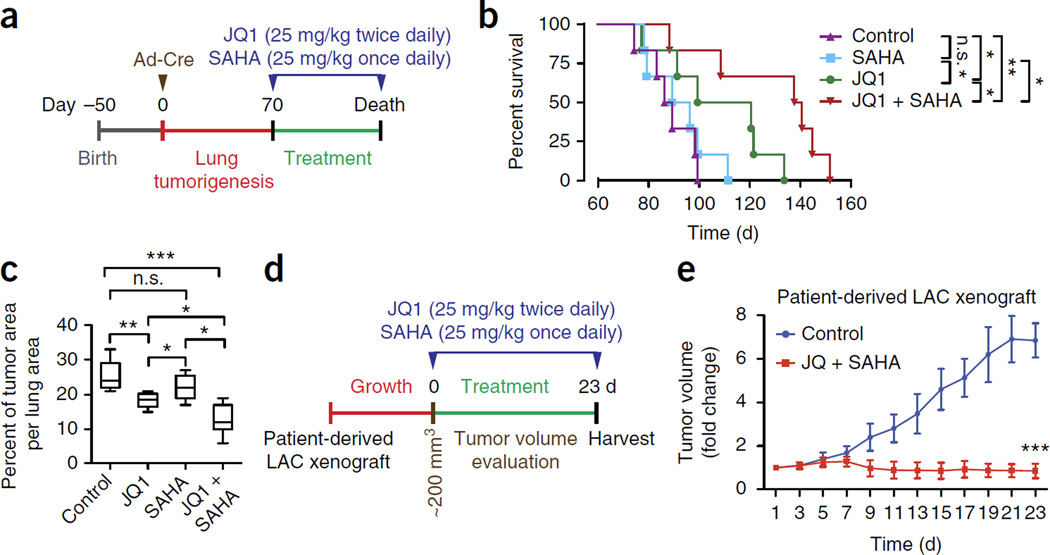
To investigate whether the JQ1+SAHA combination therapy could be effective in other tumor types, we performed another set of experiments in lung cancer cells. As in PDAC, MYC and BET family proteins, including BRD4, were expressed in human lung adenocarcinoma (LAC) (Supplementary Fig. 13a–c). In a well-established mouse model of LAC based on activation of KrasG12D and loss of p53 (ref. 44), treatment with JQ1 and SAHA 70 d after cancer initiation, when mice have developed tumors, significantly increased the survival of the mutant mice (Fig. 5a–c), associated with lower MYC levels, induction of cell death, and inhibition of proliferation (Supplementary Fig. 13c–g). Similarly, a lung cancer PDX showed significant growth inhibition upon JQ1 and SAHA treatment (Fig. 5d,e). Thus, the combination therapy seems to be effective against several types of transformed cells and tumor entities.
Figure 5.
JQ1 and SAHA synergistically suppress lung adenocarcinoma growth in vivo. (a) Treatment schedule for administration of JQ1 and SAHA. Kras;p53 mutant mice undergoing monotherapy also received placebo (vehicle). (b) Kaplan-Meier survival curves of Kras;p53 mutant mice from enrollment time in control (vehicle) (n = 6, median survival = 87.5 d), SAHA (n = 6, median survival = 92.5 d), JQ1 (n = 6 med. survival = 109.5 d) and combined SAHA + JQ1 (n = 6, median survival = 138.5 d) treatment groups. *P < 0.05; **P < 0.01; n.s., not significant by log-rank test for significance. (c) Quantification of tumor area per lung area. ***P < 0.001; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (d) Treatment schedule for administration of JQ1 and SAHA in a lung adenocarcinoma patient-derived xenograft. (e) Tumor volume quantification for lung adenocarcinoma xenografts in mice (n = 5 mice for each treatment group and vehicle control). ***P < 0.001 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m.
In vivo gene-editing identifies the role of p57
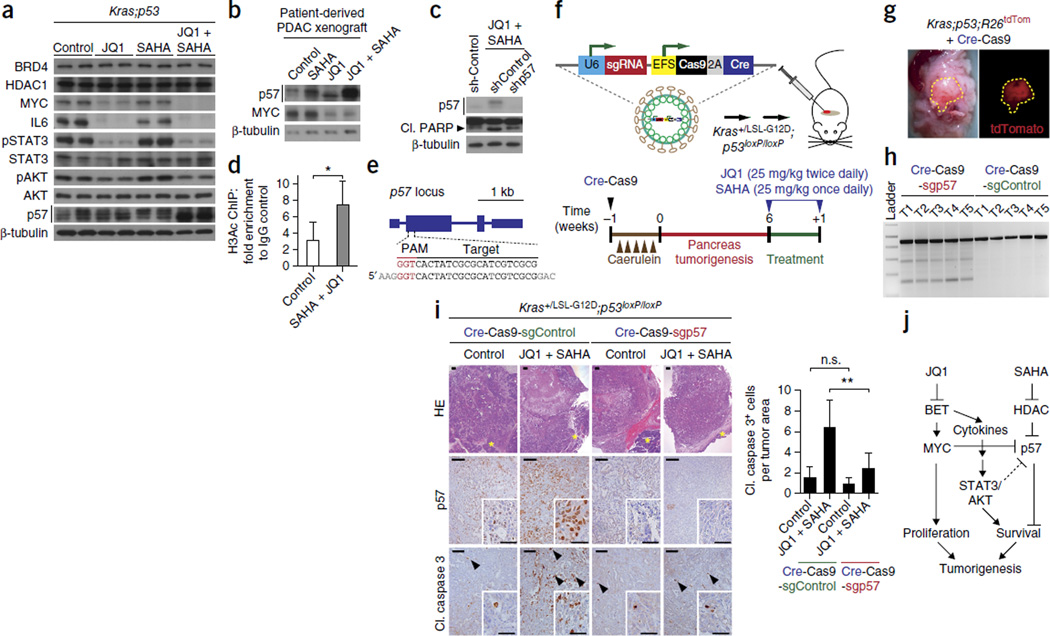
To investigate the molecular basis for the strong induction of apoptotic cell death in tumors treated with JQ1 and SAHA, we analyzed gene expression profiles of primary cells from Kras;p53 mutant mice and biopsy tissue from human PDX treated with JQ1 or SAHA alone or in combination. GSEA of microarray data showed decreased expression of HDAC targets in JQ1+SAHA-treated primary cells from Kras;p53 mutant mice and in human PDX, as well as a general decrease in transcription, as would be expected (Supplementary Fig. 14a). Signatures for increased apoptosis and decreased mitosis were also highly enriched in JQ1+SAHA-treated human and mouse cells (Supplementary Fig. 14a). p57 (officially known as Cdkn1c) was the most synergistically de-repressed gene seen in an analysis of microarray data from JQ1+SAHA-treated mouse PDAC cells (Supplementary Fig. 14b). Defining synergy as an effect in excess over an additive effect of the two drugs, p57 mRNA levels were increased on average 3.28-fold over additivity. Interestingly, p57 transcription is repressed by MYC45. p57 transcription is also often silenced in PDAC by epigenetic mechanisms, and its expression can be restored by HDAC inhibition46. This increase in p57 mRNA expression was validated at the protein level (Fig. 6a,b). Depletion of p57 using shRNA led to reduced levels of the apoptotic marker PARP in PDAC cells treated with JQ1+SAHA in culture (Fig. 6c), supporting a previously suggested role for p57 in promoting apoptosis47,48. We found increased acetylation of the p57 promoter by chromatin immunoprecipitation upon JQ1+SAHA treatment, correlating promoter acetylation with p57 expression (Fig. 6d).
Figure 6.
Identification of p57 as a key mediator of PDAC sensitivity to JQ1 and SAHA co-treatment using a gene editing platform in the pancreas of mice. (a) Immunoblot analysis of pancreatic tumor lysates dissected from Kras;p53 mutant mice in response to the indicated treatments and vehicle control (multiple independent mouse tumors were obtained and analyzed; two independent and representative samples are shown). (b) Immunoblot analysis of a patient-derived PDAC xenograft in response to the indicated treatments. (c) Immunoblot analysis of the human PDAC cell line CFPac1 following p57 knockdown (shp57) or scrambled shRNA (shControl) in response to JQ1 (250 nM) and SAHA (250 nM) treatment for 72 h. (d) Chromatin immunoprecipitation analysis of acetylated histone H3 (H3Ac) at the p57 promoter in JQ1 (250 nM) and SAHA (250 nM) or vehicle control–treated CFPac1 cells. *P < 0.05 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. of two independent experiments. (e) sgRNA design for targeting the mouse p57 locus. (f) Top, map of the pSECC lentivirus for the simultaneous expression of sgRNA, Cas9 and Cre, which allows genome editing and gene recombination when injected into the pancreatic parenchyma. Bottom, schematic of experimental caerulein- and lentiviral pSECC-induced tumorigenesis in Kras+/LSL-G12D;Trp53loxP/loxP mice with JQ1 and SAHA co-treatment. (g) Representative fluorescence (right, tdTomato) and bright-field (left) images of pancreatic tumors in Kras+/LSL-G12D;Trp53loxP/loxP;R26tdTomato mice following pSECC injection. (h) Surveyor assay for p57 on tumor biopsies from mice infected with pSECC viruses expressing sgControl or sgp57 guide RNAs. T, tumor, each number represents different mouse tumor biopsies. (i) Left, representative HE and IHC analysis (p57, cleaved caspase 3) from pancreatic tumors sections in Kras+/LSL-G12D;p53loxP/loxP mice infected with pSECC sgControl and shp57 lentiviruses and co-treated with JQ1 and SAHA or vehicle control (see also Supplementary Fig. 15d). Scale bars, 100 µm, insets 50 µm. Right, quantification of cleaved caspase 3–positive cells on tumor sections from control and treated mice (n = 4 for each experimental group). **P < 0.01; n.s., not significant (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. (j) A proposed model for synergistic JQ1 and SAHA co-treatment of PDAC. Inhibition of MYC and inflammatory signals by the inhibitor of BET family proteins JQ1 and inhibition of HDAC activity by SAHA affects several key signaling cascades in PDAC cells. JQ1 and SAHA co-treatment alters the expression of multiple gene programs, including that of p57, which results in a strong antitumoral response.
These experiments in primary cells from Kras;p53 mutant mice and in human PDX led us to investigate whether de-repression of p57 was in part responsible for the induction of cell death observed following inhibition of BET proteins and HDACs. To this end, we developed an approach of lentiviral injections in the pancreas of adult mice based on reagents recently developed to manipulate gene expression using the CRISPR-Cas9 system in the lung49. We cloned a single-guide RNA (sgRNA) targeting the mouse p57 gene (sgp57) into the pSECC lentiviral backbone and examined the consequences of deleting p57 in mouse PDAC tumors treated with JQ1+SAHA (Fig. 6e,f). Cohorts of Kras;p53 mutant mice were infected with pSECC-sgp57 or pSECC-sgControl (with a scrambled sgRNA) lentiviral vectors by direct injection into the mouse pancreas (Supplementary Fig. 15a). Although adult mice are refractory to Kras-induced pancreas tumorigenesis50,51, this block was overcome by inducing moderate pancreatitis with caerulein29 in this model (Fig. 6f). Successful recombination of the alleles flanked by loxP sites by the Cre recombinase was confirmed by the activation of a reporter allele (Fig. 6g and Supplementary Fig. 15a). Induction of Cas9-mediated mutations in the p57 gene was verified using a Surveyor assay (Fig. 6h). Using this protocol, we observed that 3 of 8 mice developed tumors that lacked p57 expression, with 4 additional mice showing partial loss (at least 70%), upon JQ1+SAHA treatment. All treated sgRNA-control animals expressed p57 under the same conditions (Fig. 6i and Supplementary Fig. 15b,c). Notably, tumors with successful deletion of p57 showed a significant (P < 0.01) reduction in apoptosis when treated with JQ1+SAHA as compared to tumors treated with the combination therapy but infected with a control lentiviral vector (Fig. 6i), partially reversing the therapeutic effects of the combination therapy. Thus, p57 may be one of the key mediators of the anticancer action of JQ1 and SAHA in PDAC cells.
DISCUSSION
In this study we examined the possible use of small-molecule inhibitors of chromatin regulators in adenocarcinoma of the pancreas and the lung and found a potent synergistic tumor-suppressive effect between a specific inhibitor of the BET family proteins (JQ1, known as TEN-010 in clinical trials) and HDAC proteins (SAHA, also known as vorinostat, an FDA-approved drug). Although the mode of action of the two drugs is certainly multifaceted and certainly involves broad signaling and transcriptional changes in both tumor cells and the tumor stroma, we identified transcriptional induction of p57 as a key mediator of cell death upon JQ1+SAHA treatment in PDAC cells. We also noted faster migration for a pool of p57 protein upon treatment with JQ1+SAHA (see, for example, immunoblots in Fig. 6a,b), an effect which might be related to decreased phosphorylation of p57 by AKT, an event previously associated with higher p57 turnover rates52; this could provide an additional mechanism of p57 activation in this context. On the basis of these observations, we propose that MYC downregulation, reduced inflammation, and p57 induction are some of the processes affected by JQ1 and SAHA in tumors (Fig. 6j), and we suggest that p57 status may be used as a possible biomarker for how tumors respond to the combined treatment.
Recently, a combination of a HDAC inhibitor and a demethylase inhibitor was found to synergistically inhibit a subtype of glioma53. Similarly, loss of the chromatin remodeler EZH2 has been shown to amplify Ras-driven transcription in glioma and melanoma and enhance the sensitivity of tumor cells to BRD4-based strategies54. Thus, combinations of small-molecule inhibitors targeting chromatin regulators may have limited toxicity and strong inhibitory effects in many cancer types. Future studies will investigate whether the marked response to JQ1 and SAHA is specific to PDAC and LAC, two cancers with important stromal and inflammatory components. In particular, the inhibitory effects of JQ1 in a mouse model of Ras-driven lung adenocarcinoma are dependent on the tumor genotype, with loss of the Lkb1 tumor suppressor largely abolishing the anticancer effects of this drug55. Although we have performed our experiments with tumors bearing the most frequent genetic alterations in PDAC and LAC, it will be important in the future to investigate the effects of combined BET and HDAC inhibitors on subgroups of tumors with multiple genotypes. Nevertheless, our experiments identify a striking vulnerability to epigenetic-based targeting approaches in pancreatic and lung cancers, two tumor types that are largely resistant to current therapeutic approaches, and will hopefully facilitate the rapid clinical evaluation of this new strategy.
ONLINE METHODS
Ethics statement
Mice were maintained in accordance and compliance with policies approved by the Stanford University Administrative Panel on Laboratory Animal Care (Sage lab protocol #13565) and by the Institutional Animal Care and Use Committee at the Technical University of Munich.
Mouse strains
Ptf1a+/Cre, Kras+/LSL-G12D, Trp53loxP/loxP, p16/p19loxP/loxP, MycloxP/loxP, R26LSL-CAG-tdTomato and R26LSL-CAG-EYFP mice have been described before12,43,56–59. Mice were in a mixed C57BL/6;129/Sv background, and we systematically used littermates as controls in all the experiments (sex ratio per cohort balanced). All animals were numbered and experiments were conducted in a blinded fashion. After data collection, genotypes were revealed and animals assigned to groups for analysis. For treatment experiments mice were randomized. None of the mice with the appropriate genotype were excluded from this study. Histopathological analysis was conducted on de-identified slides by two pathologists (I.E. and B.S.). Animal experiments and care were in accordance with the guidelines of institutional authorities and approved by local authorities (see Ethics statement).
Pancreatic cancer mouse models
Pancreatitis-induced tumorigenesis
Acute pancreatitis was induced at 6 to 8 weeks of age in Ptf1a+/Cre;Kras+/LSL-G12D (Kras) mice by administration of 8 hourly intraperitoneal (IP) injections of caerulein (100 µg/kg body weight) (Sigma-Aldrich) over 2 consecutive days as described previously25,60. Mice were treated as indicated with JQ1 (25 mg/kg twice daily, IP) and/or SAHA (25 mg/kg, once daily, Selleckchem) or vehicle 10% (2-hydroxypropyl)-β-cyclodextrin (Sigma-Aldrich). The twice-daily injection regiment for JQ1 is based on in vivo drug stability data61. IL6 injections were performed intravenously (i.v.) (mouse recombinant IL6, 1 µg per mouse). Pancreatic lesions were analyzed 7 d after the last injection. In parallel animals were analyzed at day 0 to confirm the caerulein-induced damage. None of the treatments affected the initial pancreatic injury (data not shown).
Spontaneous model of pancreatic intraepithelial neoplasia (PanIN) development
PanIN progression was analyzed in Ptf1a+/Cre;Kras+/LSL-G12D (Kras) and Ptf1a+/Cre;Kras+/LSL-G12D; MycloxP/loxP (Kras;Myc) mice aged 6 months. Quantification of low- (PanIN1a and 1b) and high-grade (PanIN2 and 3) lesions was performed by a pathologist (B.S.). Histopathological analysis was conducted on de-identified slides based on the classification consensus62. Five images (×100) were taken in standardized positions (so as to cover the whole section) for each slide. PanINs were counted from 8 independent animals for each group. Error bars represent s.e.m.
To calculate the relative normal acini area, Kras and Kras;Myc tumor sections were stained for amylase. Positive regions on six random, non-overlapping, ×100 images were collected from 3 mice per genotype. For each image positive amylase area was normalized to total pancreas tissue area using ImageJ software. Error bars represent s.e.m.
Model of aggressive PDAC—long-term treatment
For survival studies, we used Ptf1a+/Cre;Kras+/LSL-G12D;Trp53loxP/loxP (Kras;p53) mice, which develop aggressive, quickly fatal disease. Mice were followed for signs of disease progression. At the end of the experiment, tumors were processed for biochemical, histological and immunohistochemical analysis.
Mice were treated as indicated with JQ1 (25 mg/kg twice daily, IP) and/or SAHA (25 mg/kg, once daily, Sigma-Aldrich) or vehicle 10% (2-hydroxypropyl)-β-cyclodextrin (Sigma-Aldrich). Gemcitabine (Sigma) was dissolved in saline and dosed at 100 mg/kg of gemcitabine on a Q3Dx4 schedule (every third day for 4 cycles) as previously described30,63.
Model of aggressive PDAC—acute treatment
To study the short-term effects of JQ1 treatment on Kras;p53 mutant tumors, 35d old animals with fully developed PDAC were treated with JQ1 (25 mg/kg twice daily, IP) over 5 d.
CRISPR-Cas9–mediated direct genome editing in the mouse pancreas
Plasmid and lentivirus preparation
The U6-sgRNA-EFS-Cas9-2A–Cre (pSECC) lentiviral vector was described previously49. For sgRNA cloning, the pSECC vector was digested with BsmBI and ligated with annealed oligos with complementary BsmBI fragments. Specific sgRNAs were designed using CRISPR Design64,65 to avoid potential off-target sites. A Cdkn1c specific guide RNA (PAM sequence underlined) pre-validated in vitro was used: GCGC TGCTACGCGCTATCACTGG. Lentivirus was produced and titered as described previously66. Briefly, the pSECC vector was co-transfected with packaging vectors into 293T cells using polyethylenimine (PEI) transfection. The supernatant was collected at 48, 72 and 96 h. Concentrated virus was recovered by ultracentrifugation at 25,000 r.p.m. for 120 min and re-suspended in 25 µl PBS with 1% (vol/vol) Matrigel (BD Matrigel Growth Factor Reduced, BD Biosciences).
Lentiviral injections to the pancreas
pSECC allows expression of target sgRNA and the Cas9 nuclease with the Cre recombinase. This results in simultaneous genome editing and Cre-mediated activation of KrasG12D and ablation of Trp53 in infected mouse pancreata. This model enables rapid, flexible, and scalable investigation of gene function in vivo. Cohorts of Kras+/LSL-G12D;Trp53loxP/loxP mice were infected with pSECC-sgp57 or pSECC-sgControl lentiviral vectors by direct injection into the pancreas. The recipient mice were anesthetized with isoflurane. Anesthesia depth was assessed based on withdrawal reflex immediately before making an incision and monitored continuously during the surgery. The hair on the ventral surface of the mouse was shaved and the incision site was disinfected with iodophors (Betadine scrub). A disposable surgical drape (precut and sterilized) was placed, and mice were subjected to laparotomy. The incision size was ~1 cm long depending on the size of the individual animal. The splenic part of the pancreas was exposed and the viral solution was slowly injected into the pancreas parenchyma using a 30-gauge needle over 30 s to avoid backward flow. The addition of Matrigel to the injected virus solutions resulted in immediate increased viscosity of the injected fluid and mitigated leakage to prevent transduction outside of the pancreatic tissue (see Supplementary Fig. 15). This injection procedure mimics the previously described protocols for xenograft cell implantation67. The successful introduction of lentiviral particles into the pancreas parenchyma does not lead to significant side effects, such as pancreatitis, as previously described for intraductal viral administration68. Risk of vascular damage can be minimized by passing the needle parallel to any visible vessels, along the length of the pancreas. Finally, the abdominal muscle layer was closed with interrupted suture and the overlying skin was closed using Monocryl sutures (Ethicon).
Pancreatitis induction and treatment
Adult mice normally become refractory to KrasG12D-induced pancreas tumorigenesis, but if these mice are challenged with a mild form of chronic pancreatitis, they develop the full spectrum of PanINs and invasive PDAC50. Thus, mice were “primed” with 250 µg/kg caerulein twice daily for 5 consecutive days29 after lentiviral injection. All successfully transduced animals developed pancreatic tumors. We did not observe sarcoma development in the pancreas, however, in one case we noticed a mesenteric tumor and in few cases neoplastic growth in the peritoneal cavity around the area where the injection was performed.
Analysis of p57 positive areas
Tumor sections were immunostained for p57. For each tumor, the p57-positive area was normalized to the total tumor area using ImageJ software. The distribution of p57 staining status in all infected animals was divided in three groups: negative (<30% of the tumor area stained positive for p57), mixed (30–70% of tumor area stained positive for p57) and positive tumors (70–100% of the tumor area stained positive for p57).
Confirmation of Cas9 targeted loci mutation by T7E1 assay (Surveyor assay)
Mutation detection using the T7 (Surveyor) nuclease was performed as previously described69. Briefly, PCR products spanning the potential repair/damage sites were amplified and a mixture of 9 µl of PCR products and 1 µl of NEB buffer 2 was melted and re-annealed in a thermal cycler under the following conditions: 95 °C for 10 min, 95 °C to 85 °C at 2 °C/s, 85 °C for 1 min, 85 °C to 25 °C at 0.1 °C/s, 25 °C for 1 min and 4 °C for an indefinite period. Exactly 6 µl of the re-annealed mixture was treated with 0.5 µl of T7 endonuclease I (5 units) by addition of 0.4 µl of NEB buffer 2 and 3.1 µl of ddH2O at 37 °C for 20 min. Analysis by 2% agarose-gel electrophoresis followed. The gel was stained with 1 µg/ml ethidium bromide in Tris-acetate-EDTA buffer for 5 min and then imaged with a gel-imaging system (Bio-Rad).
In vivo imaging and tumor volume measurements
As a model of locally advanced PDAC for imaging-guided tumor response evaluation, we used Kras;p53 mutant mice. MR imaging experiments were initiated at an age of approximately 30 d and were performed weekly. Before imaging, mice were anesthetized by continuous gaseous infusion of 2% isoflurane (Abbott) for at least 10 min using a veterinary anesthesia System (Vetland Medical). During imaging, the dose was kept at 2% isoflurane, animal temperature was maintained and continuously monitored and eyes were protected with an eye ointment. Tumor growth kinetics changes were followed with T2 weighted imaging protocol using microscopy surface coil inside a Philips 1.5T or 3.0T clinical scanner. An axial multi-slice T2-weighted (T2w) TSE sequence (resolution 0.3 × 0.3 × 0.7 mm3, minimum 30 slices, TE = 90 ms, TR>3 s) was applied for tumor detection. Solid tumor volumes were calculated using in house optimized ImageJ based software that differentiates between solid and cystic parts of the tumor (Supplementary Fig. 3a). By analyzing multiple mice at sequential time points, we found PDAC development occurring at 4–6 weeks in Kras;p53 mutant mice with 100% penetrance. On average, tumors of animals at age 41 d met inclusion criteria of 200–400 mm3 of tumor mass (Supplementary Fig. 3b).
Preparation of pancreatic epithelial explants culture
Pancreatic epithelial explants from 4- to 6-week old wild-type mice were established by modification of previously published protocols60. In brief, the whole pancreas was harvested and treated twice with 1.2 mg/ml collagenase-VIII (Sigma-Aldrich). Following multiple wash steps with McCoy’s medium containing soybean trypsin inhibitor (SBTI, 0.2 mg/ml), digested samples were filtered through a 100 µm filter, resuspended in culture medium (Waymouth’s MB 752/1 supplemented with 0.1% BSA, 0.2 mg/ml SBTI; 50 µg/ml bovine pituitary extract, 10 µg/ml Insulin, 5 µg/ml transferrin, 6.7 ng/ml selenium in 30% FCS) and allowed to recover for 1h at 37 °C. Thereafter, cells were pelleted and resuspended in culture medium supplemented with penicillin G (1000 U/ml), streptomycin (100 µg/ml), amphotericin B, 0.1% FCS, and an equal volume of rat tail collagen and immediately plated on plates pre-coated with 2.5 mg/ml of rat tail collagen type I. In stimulation experiments, recombinant human EGF (rhEGF, Invitrogen) was added at final concentration of 25 ng/ml. Cells were treated with JQ1 at a concentration of 250 nM. For quantification, acinar explants were seeded in triplicates. Cell clusters were counted from at least 3 optical fields/well and reported as a percentage of acinar clusters and duct-like spheres. The quantification was performed in two independent experiments; the number of mice is reported in the main text.
Lung adenocarcinoma (LAC) mouse model
Kras+/LSL-G12D;Trp53loxP/loxP (Kras;p53) mice were treated with 5 × 106 pfu of adenovirus expressing Cre (University of Iowa adenovirus core) by intratracheal infection as previously described70.
Patient fresh PDAC biopsies and patient-derived xenograft (PDX) tumors
Tissue samples were collected from patients following pancreatic resection for PDAC at the Technische Universität München and Ruhr-University Bochum. All patients were informed and written consent was obtained. The tissue collection was performed according to a protocol approved by the Ethics Committee of the Ruhr-University Bochum (permission no. 3534-09 and 3841-10) and the Ethics Committee of the Technische Universität München (Project number: 5510/12). The lung adenocarcinoma (LAC) patient-derived xenograft (Fig. 5) was obtained from the Stanford University medical center following IRB approval. The LAC sample bears a KrasG12V mutation and is wild-type for EGFR and ALK. PDAC samples have KrasG12V mutations and did not express CDKN2A transcripts.
The fresh tumor samples were initially grown directly in the mouse as a tissue fragment. Then tumors were minced using a razor blade and digested in collagenase/dispase digestion buffer (Roche) for 1 h at 37 °C. Cells were passed through 10 µm and 40 µm cell strainers and centrifuged for 1,200 rpm for 8 min. Cells were incubated in RBC lysis buffer for 2 min and then resuspended in 6 ml of media and spun through 0.5 ml of serum layered on the bottom of the tube to remove cellular debris. Cells were depleted of lineage-positive cells using biotin conjugated anti-mouse CD45, CD31 and Ter119 (eBiosciences) and depleted on a MACS LS column (Miltenyi Biotec) using anti-biotin microbeads per the manufacturer’s directions. After this procedure, single cells (500,000) were mixed with Matrigel (BD Biosciences) and injected into 6- to 8-week-old female NOD/scid mouse flank. Tumor volume was measured at the times indicated and calculated using the ellipsoid formula (length × width2).
Xenograft tumors (Supplementary Fig. 11e–i) from early passage human PDACs were grown in female NMRI-nu/nu mice treated with vehicle or JQ1 and SAHA (JQ1 50 mg/kg, SAHA 25 mg/kg once daily) for 21 d.
Serum analysis and cytokine ELISA
Cytokine concentrations in serum were measured using the Mouse Inflammatory Cytokines & Chemokines Multi-Analyte ELISArray Kit (Qiagen) according to the manufacturer’s specifications. Data are presented as relative to the WT and treatment controls. Sera were collected from the blood samples of individual mice at the end of the experiment under terminal anesthesia following a protocol for cardiac puncture. Serum samples were separated from blood within 1 h following blood collection by centrifugation at 500g for 10 min. Separated samples were aliquotted and stored at −80 °C for subsequent testing. Media concentration of IL-6 was measured using human-specific ELISA kits (Pierce) according to the manufacturer’s protocols.
Aspartate transaminase (AST), alanine transaminase (ALT) and blood urea nitrogen (BUN) levels were measured in serum prepared as indicated above at the Animal Diagnostic Laboratory at Stanford Veterinary Service Center.
Histology and immunohistochemistry
Tissue specimens were fixed in 4% buffered formalin for 24 h and stored in 70% ethanol until paraffin embedding. 3-µm sections were stained with hematoxylin and eosin (HE), Sirius Red or used for immunohistochemical studies.
Immunohistochemistry was performed on formalin-fixed, paraffin embedded mouse and human tissue sections using a biotin-avidin method as described before71. The following antibodies were used (at the indicated dilutions): Albumin (#PA5–27707, Thermo Scientific, 1:1,000), αSMA (#5694, Abcam, 1:500), β-catenin (#610154, BD Bioscience, 1:1 000), BRD4 (#13440, Cell Signaling, 1:200), CK19 (#TromaIII, Developmental Studies Hybridoma Bank, 1:500), cleaved caspase 3 (#9664, Cell Signaling, 1:200), CD45 (#ab10558, Abcam, 1:100), CD31 (#550274, BD Pharmingen, 1:100), F4/80 (#48000, Invitrogen, 1:500), IAP (#ab137879, Abcam, 1:250), Ki67 (#550609, BD Bioscience, 1:1,000), MUC5AC (#145P1, NeoMarkers, 1:500), MUC2 (#sc-15335, Santa Cruz Biotechnology, 1:100), MYC (#39688, Abcam, 1:250), p57/Kip2 (#ab75974, Abcam, 1:200), pAKT (#2965, Cell Signaling, 1:100), pERK1/2 (#4370, Cell Signaling, 1:500), pSTAT3 (#9145, Cell Signaling, 1:500), PDX1 (a generous gift of Dr. C. V. Wright, 1:10,000), SOX9 (#AB5535, EMD Millipore, 1:250). Sections were developed with DAB and counterstained with hematoxylin. Pictures were taken using a Zeiss microscope equipped with the Axiovision software. Analysis of the tumor area and IHC analysis was done using ImageJ software by measuring pixel units.
Cell culture, reagents and transfections
Authenticated cancer cells (of various genotypes): CFPac1 (KrasG12V;p53C242R), Panc1 (KrasG12D;p53R273H), CaPan1 (KrasG12V;p53A159V;Smad4S343*), CaPan2 (KrasG12D;p53T125C), HPFAII (KrasG12D;p53T151S;Cdkn2aR29_A34del), Colo357 (KrasG12V;Smad4mut), Panc89 (p53mut;Cdkn2amut), PaTu8902 (KrasG12V;p53C176S), SW1990 (KrasG12D;p53P191del), AsPC1 (KrasG12D;p53C135fs*35:Smad4R100T), MiaPaCa2 (KrasG12C;p53R248W), YPAC (KrasG12V;p53H179R;Smad4R515fs*22), BxPC1 (p53Y220C), 293T were obtained from the American Type Culture Collection and DanG (KrasG12V;p53993+16del.38;Smad4mut) cells were obtained from CLS Cell Lines Service GmbH and cultured in RPMI Medium supplemented with 10% FCS (Life Technologies), 100 units/ml penicillin/streptomycin and glutamine (Life Technologies). Primary mouse pancreatic cancer lines were obtained from Kras;p53 mouse PDAC. Cells were grown in Dulbecco’s modified Eagle’s medium (Life Technologies) supplemented with 10% FCS (Life Technologies), 100 units/ml penicillin/streptomycin and glutamine (Life Technologies). All cells were cultured at 37 °C in a humidified incubator with 5% CO2. All cell lines were routinely evaluated for mycoplasma contamination.
Lentiviral shRNA constructs
shRNA sequences targeting MYC, BRD4 and p57, published previously72, were cloned in a pLKO vector carrying a puromycin resistance gene. Lentiviruses were generated in 293T cells using second-generation packaging vectors. Target cells were infected and selected using puromycin selection (2 µg/ml puromycin for 1 week, Invitrogen).
Cell assays
Cell viability
Cells were seeded in 96-well plates at 2,000 cells per well (optimal density for growth) in a total volume of 100 µl media containing 2% FBS. Serially diluted compounds in 100 µl media were added to the cells 12 h later. After 72 h incubation, cell viability was assessed by an MTT assay (Roche) according to the manufacturer’s instructions.
Invasion assays
Twenty-four hours after 250 nM JQ1 treatment, 100,000 of the indicated cells were seeded in a Transwell chamber pre-coated with Matrigel (BD Biosciences). Medium containing 10% FBS in the lower chamber served as chemoattractant. JQ1 was added to both upper and lower chambers. After 48 h, the non-invading cells and extracellular matrix were gently removed with a cotton swab and invasive cells located on the lower side of the chamber were stained with crystal violet and photographed. For colorimetric assays, the stained wells were treated with 500 µl 10% acetic acid and the absorbance was measured at 560 nm using a spectrophotometer.
Colony-formation assay
For long-term colony-formation assay, 10,000–50,000 cells per well were seeded in 6-well plates and treated with JQ1 or DMSO. After 12 d, cells were fixed with methanol, stained with crystal violet, photographed and quantified.
Senescence associated-X-gal staining
Staining was carried out as described previously73. Briefly, cells were grown subconfluently on cell culture chamber slides, treated with indicated concentration of JQ1 for 48 h and then incubated with fixative (glutaraldehyde) followed by the chromogenic SA-β-galactosidase substrate X-gal in a buffer at pH 6.0. After a blue color developed (12 h), the cells were washed with phosphate-buffered saline (PBS) and viewed by bright field or phase contrast microscopy.
Annexin V apoptosis staining
Staining was performed according to the manufacturer’s protocol (BD, APC annexin V). Cells were grown in 10 cm plates and treated with varying concentrations of JQ1. Annexin V was quantified in various cells selectively for an early apoptotic cells (annexin V+, DAPI−), excluding accumulated dead cells (annexin V+, DAPI+). All analyses were performed using Flowing Software 2.1 and independently repeated.
Synergy screening and calculations
Cells were seeded in 96-well plates at 2,000 cells per well (optimal density for growth) in a total volume of 100 µl media containing 2% FBS. The following day, cells were treated in duplicate with single agents and their fixed-ratio combination for 72 h over a 5-point, 32-fold concentration range, which was centered on the single-agent concentrations that inhibited viability by 50% (IC50). Cell viability was measured by MTT assay (Roche) according to the manufacturer’s instructions.
Combination index (CI) scores were calculated as previously described74 using CalcuSyn software (Biosoft)75. This software uses the Chou-Talalay combination index method, which is based on the median-effect equation, itself a derivation from the mass-action law. For this analysis, Drug1 was combined with Drug2 at a constant ratio determined by IC50Drug1/IC50Drug2. We entered the resulting proliferation data, along with the data obtained from single drug treatments, into CalcuSyn to determine a combination index value (CI) for each combination point, which quantitatively defines additivity (1 < CI > 1.5), synergy (CI < 1), and antagonism (CI > 1.5). The resulting values were used to construct a plot of CI values over a range of fractions affected. The drug synergy screen was performed in CFPac1 cells using 9 targeted inhibitors (range of concentrations tested): BRDi (JQ1, 125–4,000 nM), HDACi (SAHA, 125–4,000 nM), MEKi (CI1040, 31.25–1,000 nM), PI3Ki (GDC0941, 31.25–1,000 nM), IGF1Ri (OSI–906, 31.25–1,000 nM), EGFRi (erlotinib, 12.5–400 nM), STAT3i (Stattic, 250–8,000 nM), SRCi (fasatinib 12.5–400 nM) and CDK4i (PD0332991, 250–8,000 nM).
Cell extracts and immunoblot analysis
For total cell extracts, cells were lysed in RIPA buffer (10 mM Tris-HCl pH 8, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton, 0.1% SDS, 1 mM PMSF, protease inhibitors (Roche) and a phosphatase inhibitor cocktail (Sigma-Aldrich)) for 15 min. Protein concentration was determined by the BCA assay (Pierce).
Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane and analyzed by immunoblot. Antibodies used were as follows (with the indicated dilutions): pAKT (#2965, 1:1,000), AKT (#4685, 1:1,000), pERK1/2 (#4370, 1:3,000), ERK1/2 (#4695, 1:3,000), pSTAT3 (#9145, 1:1,000), STAT3 (#12640, 1:1,000), BRD4 (#13440, 1:1,000), BRD2 (#5848, 1:1,000), BCL-XL (#2764, 1:1,000), cleaved caspase 3 (#9664, 1:1,000), cleaved PARP (#9546, 1:1,000, human specific; 9548, 1:1,000, mouse specific), IL6 (#12912, 1:1,000) MYC (#9402, 1:1,000) and HDAC1 (#2062, 1:1,000) were purchased from Cell Signaling Technology; cyclinD1 (#16663, 1:1,000) and p57Kip2 (#ab75974, 1:500) from Abcam; BRD3 (#a302–367, 1:500) from Bethyl, beta-tubulin (#05–661, 1:10,000) from Millipore, MCL1 (#sc-819, 1:1,000) from Santa Cruz Biotechnology, and beta-actin (#A5316, 1:5,000) and GAPDH (#G8795, 1:5,000) from Sigma-Aldrich.
qRT-PCR
RNA was isolated using the Qiagen RNeasy Isolation Kit followed by cDNA synthesis (SuperScript II, Invitrogen). Real-Time PCR was performed with 800 nM primers diluted in a final volume of 20 µl in SYBR Green Reaction Mix (Applied Biosystems). RT-PCRs were performed as follows: 95 °C for 10 min, 35 cycles of 95 °C for 15 s and 60 °C for 1 min. qRT-PCR data are representative of 4 independent mouse tumor biopsy isolations per treatment. All samples were analyzed in triplicate. Gapdh expression was used for normalization. The following primers were used: hIL6 For 5′ GGTACATCCTCGACGGCATCT; hIL6 Rev 5′ GTGCCTCTTTGCTGCTTTCAC; hIL1A For 5′ TGTATGTGACTGCC CAAGATGAAG; hIL1A Rev 5′ AGAGGAGGTTGGTCTCACTACC; hGAPDH For 5′ CAACGGATTTGGTCGTATTGG; hGAPDH; Rev 5′ TGATGGCAACA ATATCCACTTTACC; mIl6 For 5′ CTCTGGGAAATCGTGGAAAT; mIl6 Rev 5′ CCAGTTTGGTAGCATCCATC; mMyc For 5′AGCTGTTTGAAGGCTGG ATT; mMyc Rev 5′ AATAGGGCTGTACGGAGTCG; mGapdh For 5′ AAGG TCATCCCAGAGCTGAA mGapdh Rev 5′ CTGCTTCACCACCTTCTTGA.
Quantitative chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described76. Briefly, to crosslink DNA with chromatin, CFPac1 (1 × 107 cells) were incubated for 15 min in 1% formaldehyde at room temperature. Fixation was quenched by the addition of glycine to a final concentration of 0.125 M for 5 min at room temperature. Fixed cells were washed in ice-cold PBS, pelleted, and frozen at −80 °C or resuspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0, with protease inhibitor), and the DNA with chromatin was broken into 200- to 1,000-bp fragments by sonication using a Virsonic probe sonicator at setting 2 at 20% output power for 8 cycles of 15 s. The chromatin was precleared with protein A Sepharose beads (GE Healthcare) before being diluted and bound by 4 µg of the primary antibody overnight at 4 °C. Following antibodies were used for immunoprecipitation: BRD4 clone E2A7X (#13440, Cell Signaling), H3Ac (#06–599, Millipore) or IgG as background control. After precipitation, the DNA was purified by spin columns (Qiagen). Quantitative PCR was performed using a real-time thermal cycler (Bio-Rad). Reactions were set up in triplicate for each sample with 800 nM primers diluted in a final volume of 20 µl in SYBR Green Reaction Mix (Applied Biosystems). qPCRs were performed as follows: 95 °C for 10 min, 35 cycles of 95 °C for 15 s and 60 °C for 1 min. For BRD4 ChIP, six sets of primers located from −1141 to +118 bp relative to the IL-6 transcriptional start site (+1) were used: Region A: −1141 to −1061; IL6-A For 5′ GGGCTTCTGAACCAGCTTGA, IL6-A Rev 5′ CAGGCACGGCTCTAGGCTC. Region B: −981 to −900; IL6-B For 5′ AAGATGCCACAAGGTCCTCCT; IL6-B Rev 5′ CCACTTGGTTCAGGGC AGA. Region C: −720 to −621; IL6-C For 5′ CAGCAGCCAACCTCCTCTAAGT, IL6-C Rev 5′ CAAGGCGTCTCCAGGTGG. Region D: −600 to −500; IL6-D For 5′ AGGATGGCCAGGCAGTTCTA, IL6-D Rev 5′ AAGCCTGGGATTATGA AGAAGGTA. Region E: −327 to −225; IL6-E For 5′ TGCATGACTT CAGCTTTACTC, IL6-E Rev 5′ GCAGAACCACTCTTCCTTTAC. Region F: +18 to +118; IL6-F For 5′ ACCGGGAACGAAAGAGAAGC, IL6-F Rev 5′ CTGGCAGTTCCAGGGCTAAG.
H3Ac27 ChIP was performed across the CDKN1C transcription start site from −72 to +89: p57 For 5′ GTATAAAGGGGGCGCAGGCGGGCT, p57 Rev 5’TGGTGGACTCTTCTGCGTCGGGTTC.
Calculation of average cycle threshold (Ct) and s.d. for triplicate reactions was performed, and each DNA fraction was normalized to the input to account for chromatin sample preparation differences: ΔCtnorm ChIP = CtChIP – Ctinput – log2(dilution), where “dilution” is input dilution factor = 100. Normalized background was then subtracted using the following equation: ΔΔCt = ΔCtnorm ChIP – ΔCtnorm IgG. The s.d. was calculated using propagation of error:
Calculation of relative quantity of amplified sequence (fold enrichment) was carried out according to the following equation:
Q = 2−ΔΔCt and calculation of error for relative quantity: Qerror ± = Q ± 2−(ΔΔCt ± SDΔΔCt).
Meta-analysis of public PDAC data sets
We downloaded raw data for 7 publicly available PDAC gene expression studies from the NCBI GEO and EBI ArrayExpress. After re-annotating the probes, each data set was separately normalized using GCRMA. We applied two meta-analysis approaches to the normalized data77. Briefly, the first approach combines effect sizes from each data set into a meta-effect size to estimate the amount of change in expression across all data sets. For each gene in each data set, an effect size was computed using Hedges’ adjusted g. If multiple probes mapped to a gene, the effect size for each gene was summarized using the fixed effect inverse-variance model. We combined study-specific effect sizes to obtain the pooled effect size and its standard error using the random effects inverse-variance technique. We computed z-statistics as a ratio of the pooled effect size to its standard error for each gene, and compared the result to a standard normal distribution to obtain a nominal P-value. P-values were corrected for multiple hypotheses testing using FDR78. We used a second non-parametric meta-analysis that combines P-values from individual experiments to identify genes with a large effect size in all datasets. Briefly, we calculated a t-statistic for each gene in each study. After computing one-tail P-values for each gene, they were corrected for multiple hypotheses using FDR. Next, we used Fisher’s sum of logs method, which sums the logarithm of corrected P-values across all data sets for each gene, and compares the sum against a chi-square distribution with 2k degrees of freedom, where k is the number of data sets used in the analysis.
Gene Chip analysis and GSEA
For expression profiling, primary mouse PDAC cells treated with vehicle, JQ1 (1 µM), SAHA (2 µM) or JQ1+SAHA (1 µM/2 µM) for 6 h and JQ1+SAHA-treated and control tissue samples of the patient-derived xenografts (Supplementary Fig. 11e–i) were used.
Total RNA was isolated using the Qiagen RNeasy Isolation Kit. RNA quality was assessed on an Agilent 2100 Bioanalyser, RNA 6000 Pico Series II Chips (Agilent). A total of 6 µg of labeled RNA was hybridized to mouse (Mouse Genome 430 p.m. 16-Array Plate; Affymetrix) or human (Agilent Human Genome Oligo Microarrays) expression gene chip arrays according to the manufacturer’s protocols. The microarray protocols and data reported in this article have been deposited in NCBI’s Gene Expression Omnibus (GEO) database, accession numbers GSE 64836 and 64837. GSEA software was provided by the Broad Institute of the Massachusetts Institute of Technology and Harvard University (http://www.broad.mit.edu/gsea/)79. For both gene sets, we used the default parameters of the GSEA software package; the genes set permutation was used. In brief, the normalized enrichment score (NES) provides “the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes.” The false discovery rate q-value (FDR) is “the estimated probability that a gene set with a given NES represents a false positive finding”. In general, given the lack of coherence in most expression data sets and the relatively small number of gene sets being analyzed, an FDR cutoff of 25% was considered appropriate.
Gene expression correlation
Heatmaps represent the Pearson correlation between expression of the noted genes in GSE15471 human pancreatic cancer expression data set. Heatmaps were created in R using the “pheatmap” package.
Statistics
Kaplan-Meier survival curves were calculated using the survival time for each mouse from all littermate groups. The log-rank test was used to test for significant differences between the groups. For image quantification and gene expression analysis, statistical significance was assayed by Student’s t-test with Prism GraphPad software (two-tailed unpaired and paired t-test depending on the experiment—variance was first systematically examined using an F-test). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns: not significant. Data are represented as mean ± s.e.m.
Supplementary Material
Acknowledgments
P.K.M. and A.H. significantly contributed to the work and are listed as co-first authors based on a previous agreement. We thank C. Vakoc (Cold Spring Harbor Laboratory) for sharing shRNA plasmids, M. Winslow (Stanford University) for the R26CAG-tdTomato reporter mice and I. Moreno de Alboran (Spanish National Biotechnology Centre) for MycloxP mice. This work was supported by the German Research Foundation (SFB824/C4 to J.T.S.), the European Union’s Seventh Framework Program for research, technological development and demonstration (FP7/CAM-PaC) under grant agreement 602783, the German Cancer Consortium (DKTK) (to R.M.S. and J.T.S.), and the Lustgarten Foundation (J.S.). P.K.M. was supported by the Tobacco-Related Disease Research Program, a Dean’s Fellowship from Stanford University, and the Child Health Research Institute and Lucile Packard Foundation for Children’s Health at Stanford. J.S. is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases. T.K. was supported by a Boehringer Ingelheim Fonds MD Fellowship.
Footnotes
Accession codes. Protein Data Bank/NCBI: Coordinates have been deposited with accession code GSE64836 and GSE64837.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
P.K.M. conceived the study; designed and performed the in vivo studies of pancreatitis-induced PDAC and the Myc knockout studies, and established and performed the five-arm patient-derived PDAC xenograft assays; performed the drug screen, the tissue organoids experiments, the cell culture studies, the ChIP analysis, and the target validation assays in cell lines and in vivo; designed and performed survival treatment of mouse models of PDAC and LAC; designed and performed in vivo CRISPR-Cas9 studies; and coordinated the project, interpreted data, and drafted the manuscript. A.H. performed survival treatment of the Kras;p53 mouse model of PDAC, contributed to MRI animal imaging and tumor volume analysis, contributed to analysis of PDAC xenografts and Myc knockout studies, generated mouse expression arrays, and contributed to cell culture studies and interpretation of the data. S.S.M. and L.D.A. contributed to design and testing of intrapancreatic viral injections. M.W. and G.S. contributed to cell culture studies and interpretation of data. S.H., M.E. and J.K. obtained and prepared surgical tissue samples of PDAC for xenograft and tissue studies. M.T.-A., A.G., R.B., I.H. and P.B.N. performed MRI animal imaging and interpretation and tumor volume analysis. S.M.L. and P.K. performed bioinformatics analyses. S.A.H. and D.V. established and performed treatment of the two-arm PDAC xenograft study and generated human expression arrays. F.J.S.-R. and T.J. designed and constructed pSECC lentivirus. B.S. and I.E., pathologists, performed histological and IHC tissue evaluation and lesion progression quantification. M.H. contributed to IHC analysis. E.H. and V.E. contributed to data interpretation. T.K. contributed to plasmid and lentivirus preparation. L.C.S. and E.A.S.-C. obtained surgical tissue samples of LAC and developed PDX models. J.E.B. contributed to JQ1 inhibitor study protocols and interpretation of data. R.M.S. performed data interpretation. J.S. and J.T.S were equally responsible for supervision of research, data interpretation and manuscript preparation.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 3.Ghaneh P, Costello E, Neoptolemos JP. Biology and management of pancreatic cancer. Gut. 2007;56:1134–1152. doi: 10.1136/gut.2006.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maitra A, Hruban RH. Pancreatic cancer. Annu. Rev. Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 6.Collins MA, Pasca di Magliano M. Kras as a key oncogene and therapeutic target in pancreatic cancer. Frontiers Physiol. 2013;4:407. doi: 10.3389/fphys.2013.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Infante JR, et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur. J. Cancer. 2014;50:2072–2081. doi: 10.1016/j.ejca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat. Rev. Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 9.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bardeesy N, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl. Acad. Sci. USA. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 13.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morton JP, et al. Mutant p53 drives metastasis and overcomes growth arrest/ senescence in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook N, Jodrell DI, Tuveson DA. Predictive in vivo animal models and translation to clinical trials. Drug Discov. Today. 2012;17:253–260. doi: 10.1016/j.drudis.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Witkiewicz AK, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCleary-Wheeler AL, et al. Insights into the epigenetic mechanisms controlling pancreatic carcinogenesis. Cancer Lett. 2013;328:212–221. doi: 10.1016/j.canlet.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Means AL, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132:3767–3776. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 25.Morris JP, Cano DA, Selkine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahai V, et al. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol. Cancer Ther. 2014;13:1907–1917. doi: 10.1158/1535-7163.MCT-13-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman MH, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ardito CM, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh M, et al. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat. Biotechnol. 2010;28:585–593. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 32.Garcia PL, et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene. 2015 doi: 10.1038/onc.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy N, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015;29:658–671. doi: 10.1101/gad.256628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schleger C, Verbeke C, Hildenbrand R, Zentgraf H, Bleyl U. c-MYC activation in primary and metastatic ductal adenocarcinoma of the pancreas: Incidence, mechanisms, and clinical significance. Mod. Pathol. 2002;15:462–469. doi: 10.1038/modpathol.3880547. [DOI] [PubMed] [Google Scholar]
- 35.Lesina M, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Fukuda A, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAllister F, et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell. 2014;25:621–637. doi: 10.1016/j.ccr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21:836–847. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodir NM, et al. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25:907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rielland M, et al. Senescence-associated SIN3B promotes inflammation and pancreatic cancer progression. J. Clin. Invest. 2014;124:2125–2135. doi: 10.1172/JCI72619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corcoran RB, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71:5020–5029. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obata J, et al. p48 subunit of mouse PTF1 binds to RBP-Jkappa/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells. 2001;6:345–360. doi: 10.1046/j.1365-2443.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- 43.Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldser DM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468:572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adhikary S, et al. Miz1 is required for early embryonic development during gastrulation. Mol. Cell. Biol. 2003;23:7648–7657. doi: 10.1128/MCB.23.21.7648-7657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato N, Matsubayashi H, Abe T, Fukushima N, Goggins M. Epigenetic down-regulation of CDKN1C/p57KIP2 in pancreatic ductal neoplasms identified by gene expression profiling. Clin. Cancer Res. 2005;11:4681–4688. doi: 10.1158/1078-0432.CCR-04-2471. [DOI] [PubMed] [Google Scholar]
- 47.Yan Y, Frisen J, Lee MH, Massague J, Barbacid M. Ablation of the CDK inhibitor p57(Kip2) results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 48.Vlachos P, Nyman U, Hajji N, Joseph B. The cell cycle inhibitor p57(Kip2) promotes cell death via the mitochondrial apoptotic pathway. Cell Death Differ. 2007;14:1497–1507. doi: 10.1038/sj.cdd.4402158. [DOI] [PubMed] [Google Scholar]
- 49.Sánchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 51.Guerra C, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao RY, et al. CDK inhibitor p57(Kip2) is downregulated by Akt during HER2-mediated tumorigenicity. Cell Cycle. 2013;12:935–943. doi: 10.4161/cc.23883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grasso CS, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med. 2015;21:555–559. doi: 10.1038/nm.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Raedt T, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–251. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- 55.Shimamura T, et al. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin. Cancer Res. 2013;19:6183–6192. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 56.Kawaguchi Y, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 57.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 58.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 59.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazur PK, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283–287. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matzuk MM, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hruban RH, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 63.Jameson KL, et al. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat. Med. 2013;19:626–630. doi: 10.1038/nm.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 67.Kim MP, et al. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat. Protoc. 2009;4:1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.José A, et al. Intraductal delivery of adenoviruses targets pancreatic tumors in transgenic Ela-myc mice and orthotopic xenografts. Oncotarget. 2013;4:94–105. doi: 10.18632/oncotarget.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu P, et al. Mutation detection using Surveyor™ nuclease. Biotechniques. 2004;36:702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 70.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazur PK, et al. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. USA. 2010;107:13438–13443. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 74.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 75.Chou TC. Drug combination sand their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 76.Carey MF, Peterson CL, Smale ST. Chromatin immunoprecipitation (ChIP) Cold Spring Harbor Protoc. 2009 doi: 10.1101/pdb.prot5279. [DOI] [PubMed] [Google Scholar]
- 77.Khatri P, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J. Exp. Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Storey JD. A direct approach to false discovery rates. J. R. Stat. Soc. B. 2002;64:479–498. [Google Scholar]
- 79.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.