Abstract
Prokineticin 2 (PK2) has been indicated as an output signaling molecule for the suprachiasmatic nucleus (SCN) circadian clock. Most of these studies were performed with nocturnal animals, particularly mice and rats. In the current study, the PK2 and its receptor, PKR2, was cloned from a species of diurnal macaque monkey. The macaque monkey PK2 and PKR2 were found to be highly homologous to that of other mammalian species. The mRNA expression of PK2 and PKR2 in the macaque brain was examined by in situ hybridization. The expression patterns of PK2 and PKR2 in the macaque brain were found to be quite similar to that of the mouse brain. Particularly, PK2 mRNA was shown to oscillate in the SCN of the macaque brain in the same phase and with similar amplitude with that of nocturnal mouse brain. PKR2 expression was also detected in known primary SCN targets, including the midline thalamic and hypothalamic nuclei. However, the PKR2 expression was only detected in the dorsal SCN of the macaque brain, in contrast to the broad expression of PKR2 in the dorsal and ventral segments of the mouse SCN. The likely functional importance of this differential expression of PKR2 in the SCN segments in the diurnal monkey vs nocturnal mouse remains to be explored. In addition, we detected the expression of PKR2 mRNA in the dorsal raphe nucleus of both macaque and mouse brains. As a likely SCN to dorsal raphe projection has previously been indicated, the expression of PKR2 in the raphe nuclei of both macaque and mouse brain signifies a possible role of dorsal raphe nucleus as a previously unrecognized primary SCN projection target.
INTRODUCTION
In mammals, the endogenous pacemaker that drives circadian rhythms, such as activity and rest, resides in the suprachiasmatic nuclei (SCN). In the past two decades, the molecular mechanisms of the SCN circadian clock have been elucidated as interacting autoregulatory transcriptional/posttranslational negative feedback loops (Reppert & Weaver, 2002; Welsh et al., 2010). However, the mechanism how the information in the SCN is transmitted out of the SCN neurons to regulate the overt circadian rhythms is much less clear (Kalsbeek et al., 2006; Li et a., 2012; Welsh et al., 2010). Dye tracing have revealed that the primary efferent target areas of the SCN are quite limited and these projections predominantly end in the hypothalamus and the midline thalamus. Primary SCN target areas include lateral septum, bed nucleus of the stria terminalis, subparaventricular zone, paraventricular hypothalamic nucleus, dorsomedial hypothalamic nucleus as well as paraventricular thalamic nucleus (Buijs, 1996; Leak & Moore, 2001; Watts & Swanson, 1987; Watts et al., 1987). Lesion and transplant studies have indicated the diffusible nature of SCN output molecules (Lehman et al., 1987; Ralph et al., 1990; Silver et al., 1996).
To date, two diffusible molecules, vasopressin and prokineticin 2 (PK2), with both derived from two different clock-controlled genes, have been indicated as candidate SCN output molecules (Li et al., 2012; Reppert & Weaver, 2002; Welsh et al., 2010; Zhou & Cheng, 2005). Vasopressin was the first identified SCN output signal as vasopressin-deficient Brattleboro rats or vasopressin receptor-deficient mice display attenuated circadian rhythms (Jin et al., 1999; Kalsbeek et al., 2010; Li et al., 2009; Yamaguchi et al., 2013). Several lines of the evidence have emerged that supports the role of PK2 as an output signal for the SCN circadian clock: 1). PK2 expression in the SCN displays a high amplitude of circadian oscillation, and the PK2 oscillation in the SCN is dependent on the core SCN oscillators as it is abolished in mutant mice lacking the functional clockwork (Cheng et al., 2002; Cheng et al., 2005). 2). Receptor for PK2 (PKR2) is expressed in essentially all the primary SCN target sites, including the lateral septum, paraventricular thalamic nucleus, paraventricular hypothalamic nucleus and dorsomedial hypothalamic nucleus (Cheng et al., 2002; Cheng et al., 2006). 3). PK2 was shown to regulate the excitability of PKR2-positive neurons, such as the neurons of a primary SCN target, the paraventricular hypothalamic nucleus (Yuill et al., 2007) or the SCN neurons themselves (Burton et al., 2015; Ren et al., 2011). Coupling among SCN neurons is likely to be critical for the formation of coherent output signals from the SCN to drive overt circadian rhythms (Li et al., 2012; Loh et al., 2015; Mieda et al., 2015; Welsh et al., 2010). 4). Studies using the PK2-EGFP transgenic mice have revealed that PK2-expressing neurons in the SCN project to the known primary SCN targets (Zhang et al., 2009). 5). Intracranial delivery of PK2 into the lateral ventricle during subjective night, when endogenous PK2 is low, inhibited the nocturnal wheel running activity of rats (Cheng et al., 2002). 6). Genetic deletion of PK2 or its receptor PKR2 in mice leads to almost identical defects in circadian rhythms (Li et al., 2006; Prosser et al., 2007). In the absence of PK2 signaling, the amplitudes of circadian locomotor parameters were remarkably reduced, with rhythmicity amplitude of wheel-running activity of PK2-deficient or PKR2-deficient mice less than 20% of wild type mice in C57Bl6 inbred background (Li et al., 2006; Prosser et al., 2007). Rhythmicity of other circadian parameters, including sleep-wake cycle, body temperature, circulating glucocorticoid and glucose levels, as well as the expression of peripheral clock genes, was also significantly reduced (Li et al., 2006; Prosser et al., 2007). Additional studies have revealed the likely redundant signaling roles or interaction between vasopressin and PK2 in mediating SCN output. Detailed expression studies have indicated that PK2 expression in the SCN were scattered in both the dorsomedial and ventrolateral region of SCN and is partially co-expressed with vasopressin in the SCN neurons (Masumoto et al., 2006; Zhang et al., 2009). The amplitude of the circadian oscillation of PK2 mRNA in the SCN has been shown to be attenuated in the SCN of V1a-deficient mice and AVP neurons-specific Bmal deletion mice, indicating the likely interaction of vasopressin and PK2 in mediating SCN circadian clock output (Li et al., 2009; Mieda et al., 2015).
A key unanswered question in the circadian biology is the molecular mechanism underlying the diurnality or nocturnality. SCN, the master clock that restricts the activity of the mammals to day or night niche, clearly lies in the center for diurnality or nocturnality determination (Smale et al., 2003). As PK2 has been shown as a key output molecule for the SCN circadian clock in nocturnal animals, here we sought to clone and investigate the expression of PK2 and its receptor PKR2 in the brain of a diurnal monkey, Macaca mulatta, with particular attention paying to the SCN and its primary targets.
MATERIALS & METHODS
Monkey preparation
Six adult female monkeys (Macaca mulatta) were subjected to the standard 12 hr light: 12 hr dark (LD) cycle in the Oregon primate center facility. Lights were turned on at 7AM and turned off at 7PM. Monkeys were fed at libo, and were euthanized at 3AM, 7AM, 11AM, 3PM, 7PM, 11PM after being deeply anesthetized. Blood samples were drawn from the veins, and then monkeys were transcardially fixed with 4% paraformaldehyde. Brains were dissected, frozen, and separated into several blocks consisting of the cortex, the hypothalamus and thalamus, and the brainstem. Blocks were cut at twenty-five micrometer sections on a cryostat and sections were collected to slides and frozen at −70°C. Serum concentrations of melatonin were quantified with RIA. All animal procedures have been approved by IACUC.
Cloning of macaque PK2 and PKR2 and preparation of riboprobes for in situ hybridization
Full length cDNAs for both PK2 and PKR2 were obtained using PCR methods with reverse-transcribed RNA isolated from the monkey tissues. The in situ hybridization probe of PK2 is 1.2 kb in length and it contains the coding sequence and 3’UTR. The in situ hybridization probe of PKR2 is 620 bp in length and it contains mostly 3’UTR and a short stretch of coding sequence. Sense and antisense riboprobes were generated by T7 or SP6 RNA polymerases and were radioactively labeled with 35S-UTP, and the probes were purified using a G-50 Sephadex column (Roche, Indianapolis, IN). Concentrations of riboprobes were diluted to 2 × 107 cpm/ml in hybridization buffer solution (50% formaldehyde, 10% dextran sulfate, 0.02% Ficoll, 0.02% polyvinylpyrolidone, 0.02% BSA, 500 μg/ml tRNA, 10mM DTT, 0.3M NaCl, 10mM Tris, pH 8.0, 1mM EDTA, pH 8.0).
In situ hybridization
Tissue sections were fixed with 4% paraformaldehyde for 1 hour, followed by three washes of 0.1M phosphate buffer, air-dried, and stored at −20°C until use. Sections were processed for in situ hybridization as described (Cheng et al., 2002, Zhang et al., 2009). Briefly, sections were dried at room temperature for 2 hours, followed by pretreatment of proteinase K (1 μg/ml). Sections were air-dried and then hybridized with riboprobes by incubation at 60°C for 18 hours. After hybridization, tissue was treated to RNAase (20 μg/ml) (Sigma-Aldrich, St. Louis, MO), decreasing salinity washes and a 30 minute high stringency (68°C) wash. After dehydration and air-drying, tissue sections were exposed to Kodak Biomax film for 5–15 days. Specific hybridization signals were quantitatively analyzed using a videobased computer image analysis system (MCID, Imaging Research, Ontario, Canada). A calibration curve of optical density versus radioactivity (dpm/mg tissue wet weight) was constructed using 14C-standards. Figures were prepared by using Adobe Photoshop (Adobe Systems Inc.).
RESULTS
1. Macaque PK2 and PKR2 sequences
Macaque PK2 and PKR2 are highly conserved among mammalian species (Fig.1). There are only two conservative substitutions between the macaque and human PK2 sequences, and three conservative substitutions between the macaque and mouse PK2 sequences (Fig. 1A). Macaque PKR2 sequence is also more than 98% identical to the homology of either human or mouse (Fig. 1B).
Figure 1.

Alignment of PK2 and PKR2 sequences from human, macaque and mouse. A. Comparison of the amino acid sequences of PK2 from human, Macaca mulatta and mouse. The substitution residues are highlighted and the ten cysteines that are thought to form disulfide bonds are underlined. B. Comparison of the amino acid sequences of PKR2 from human, Macaca mulatta and mouse. The seven putative transmembrane segments are underlined.
2. Expression of PK2 and PKR2 in the macaque brain
We examined the expression of PK2 in the macaque brain by in situ hybridization. As with the mouse brain, PK2 expression was detected in the SCN of macaque brain (Fig. 2B). In addition to its expression in the SCN, PK2 mRNA was also detected in several other brain regions (Fig. 2A, C, D). Particularly, PK2 mRNA was observed in the nucleus accumbens shell (AcbSH) and medial preoptic area (MnPO), identical to the mouse brain (Cheng et al., 2002; Cheng et al., 2006). The PK2 mRNA was also detected in the following midline brain regions of macaque brain: paraventricular hypothalamic nucleus (PVN), paraventricular thalamic nucleus (PVT), the centromedial nucleus (CM) and the reunions nucleus (Re) of the thalamus. These midline structures are well known SCN primary targets that also express PKR2 the mouse brain (Cheng et al., 2002; Cheng et al., 2006) as well as in the macaque brain (see Fig. 3). The PK2 mRNA-positive signals in these regions are likely derived from the passing projection or terminals of PK2-expressing SCN projections.
Figure 2.
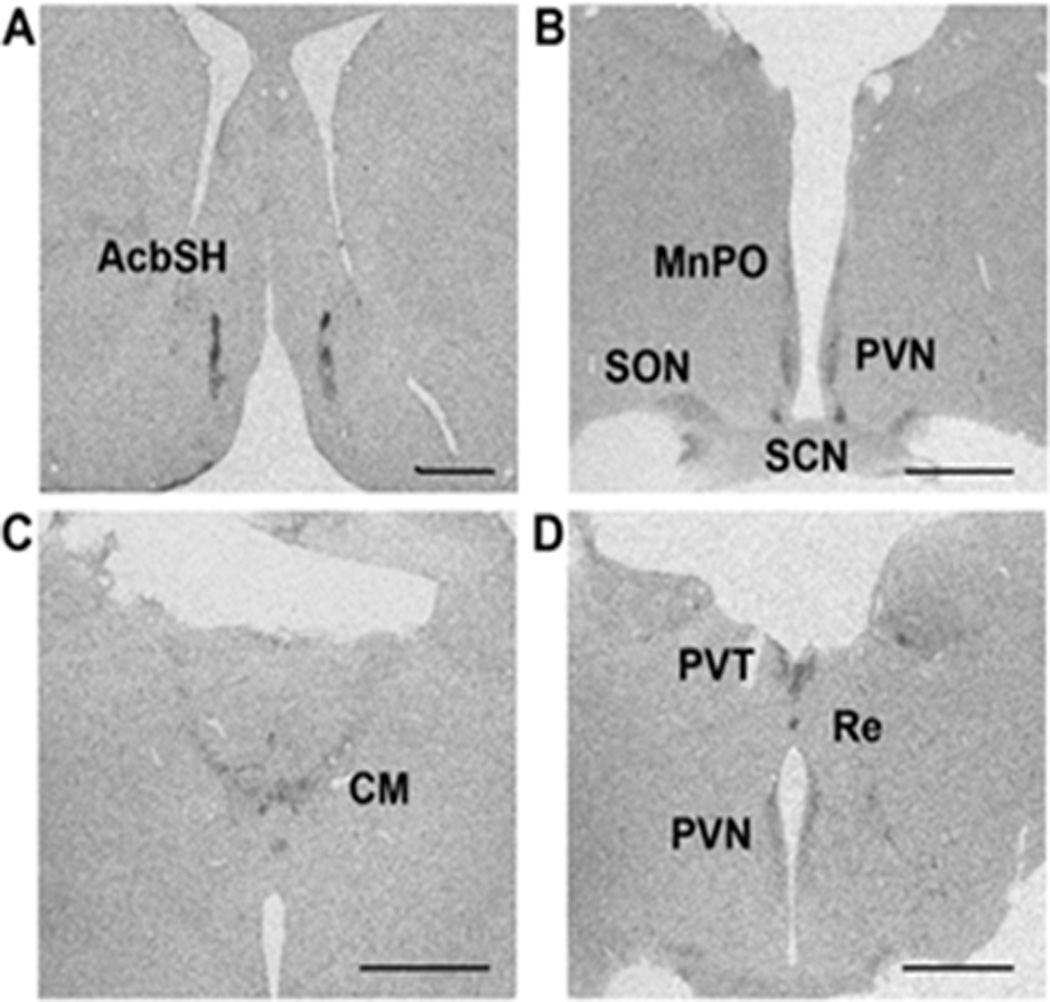
Expression of the PK2 mRNA in the macaque brain. Images A-D are shown from anterior to posterior in the hypothalamic regions of macaque brain. PK2 mRNA was detected in the nucleus accumbens shell (AcbSH), medial preoptic area (MnPO), paraventricular hypothalamic nucleus (PVN), supraoptic nucleus (SON), suprachiasmatic nucleus (SCN), centromedial nucleus of the thalamus (CM), paraventricular thalamic nucleus (PVT), and the reunions nucleus (Re). Scale bar, 2mm.
Figure 3.
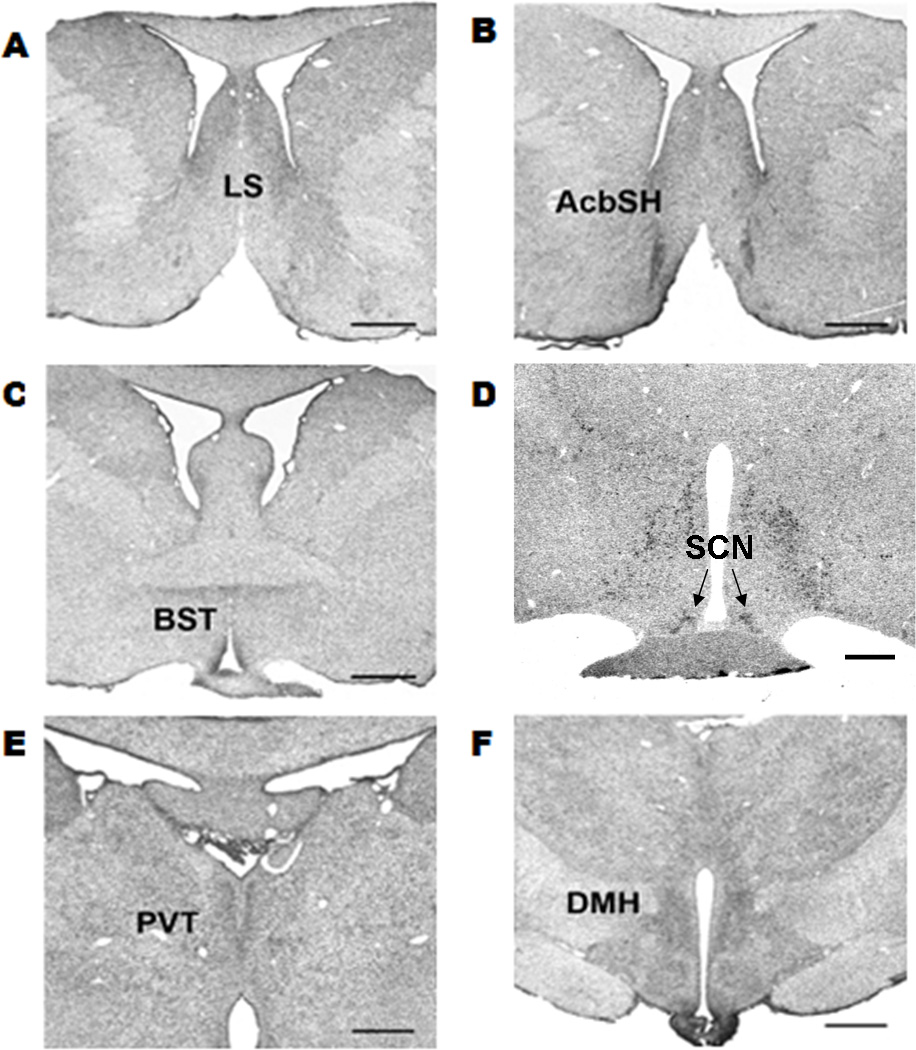
PKR2 mRNA expression in the macaque brain. PKR2 mRNA expression was detected in the (A) lateral septum (LS), (B) nucleus accumbens shell (AcbSH), (C) the bed nucleus of the stria terminalis (BST), (D) supraoptic nucleus (SON), suprachiasmatic nucleus (SCN), (E) paraventricular thalamic nucleus (PVT), and the (F) dorsomedial hypothalamic nucleus (DMH). Scale bar, 2mm.
We next examined the expression of PKR2 in macaque brain. Mild expression of PKR2 in the lateral septum (LS) was observed (Fig.3). Ventral to the anterior commissure, two circular patterns on either side of the third ventricle termed the bed nucleus stria terminalis (BST) showed PKR2 mRNA expression (Fig.3C). In the PVT of the thalamus, PKR2 mRNA is moderately expressed (Fig. 3E). PKR2-positive cells were also detected in the DMH and the ventrolateral ventromedial hypothalamus (VMH) (Fig. 3F). PKR2 mRNA expression was detected in the PVN. In all, the expression of PKR2 mRNA was detected in the known midline thalamus and hypothalamus primary SCN targets, similar to that was observed in the mouse brain (Cheng et al., 2002; Cheng et al., 2006). In the anterior hypothalamus, PKR2 was also detected in the SCN of macaque brain (Fig. 3D). However, in contrast to the broad expression of PKR2 in the dorsal and ventral segment of the mouse SCN, the PKR2 expression was only detected in the dorsal SCN of the macaque brain (Fig. 3D).
PKR2 expression was further examined in the brainstem of the macaque brain. As shown in Fig. 4, PKR2 mRNA was detected in the dorsal raphe nucleus (DR). As we have previously identified a likely SCN to DR projection in the PK2-EGFP transgenic mice (Zhang et al., 2009), the expression of PKR2 in the DR of the macaque brain prompted us to examine whether the PKR2 is also expressed in the mouse DR. As shown in Fig.5, robust PKR2 mRNA expression was indeed detected in the DR of the mouse brain. The evolutionary conservation of PKR2 expression in the DR and the previous detection of the SCN to DR projection in the PK2-EGFP transgenic mice (Zhang et al., 2009) may indicate a likely role of DR as a primary SCN target.
Figure 4.
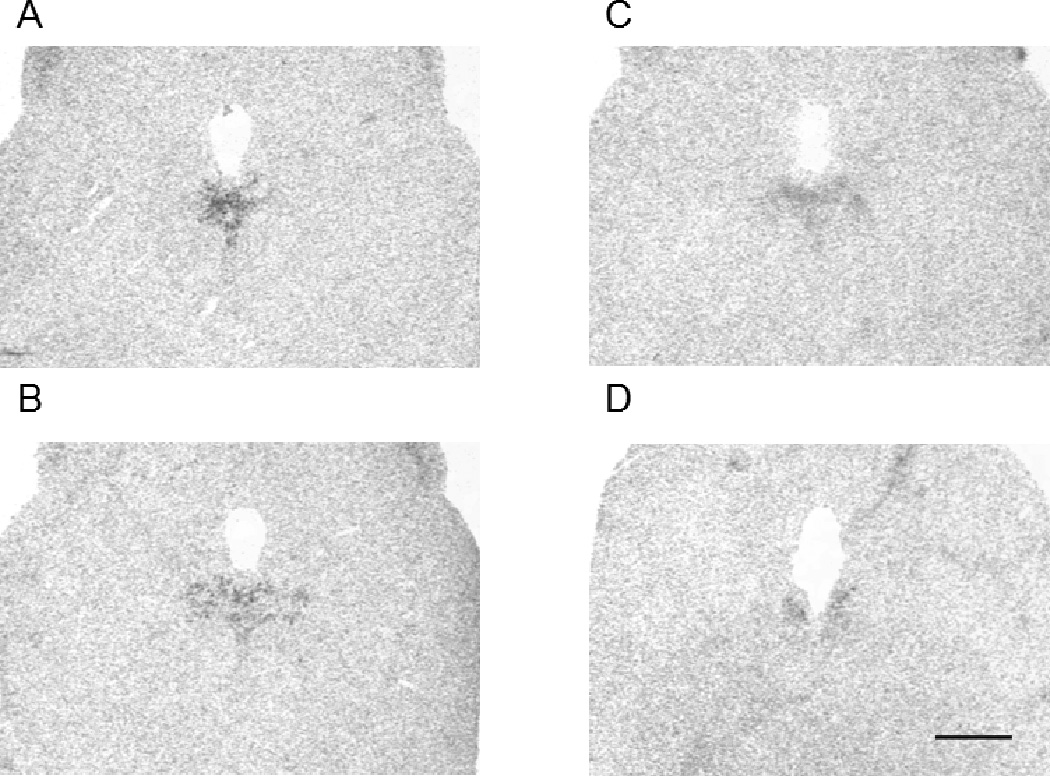
Expression of the PKR2 mRNA in the dorsal raphe nucleus of the macaque brain. PKR2 mRNA is detected in from anterior to posterior region of the dorsal raphe nucleus (DR) (A-D). Scale bar, 2mm.
Figure 5.
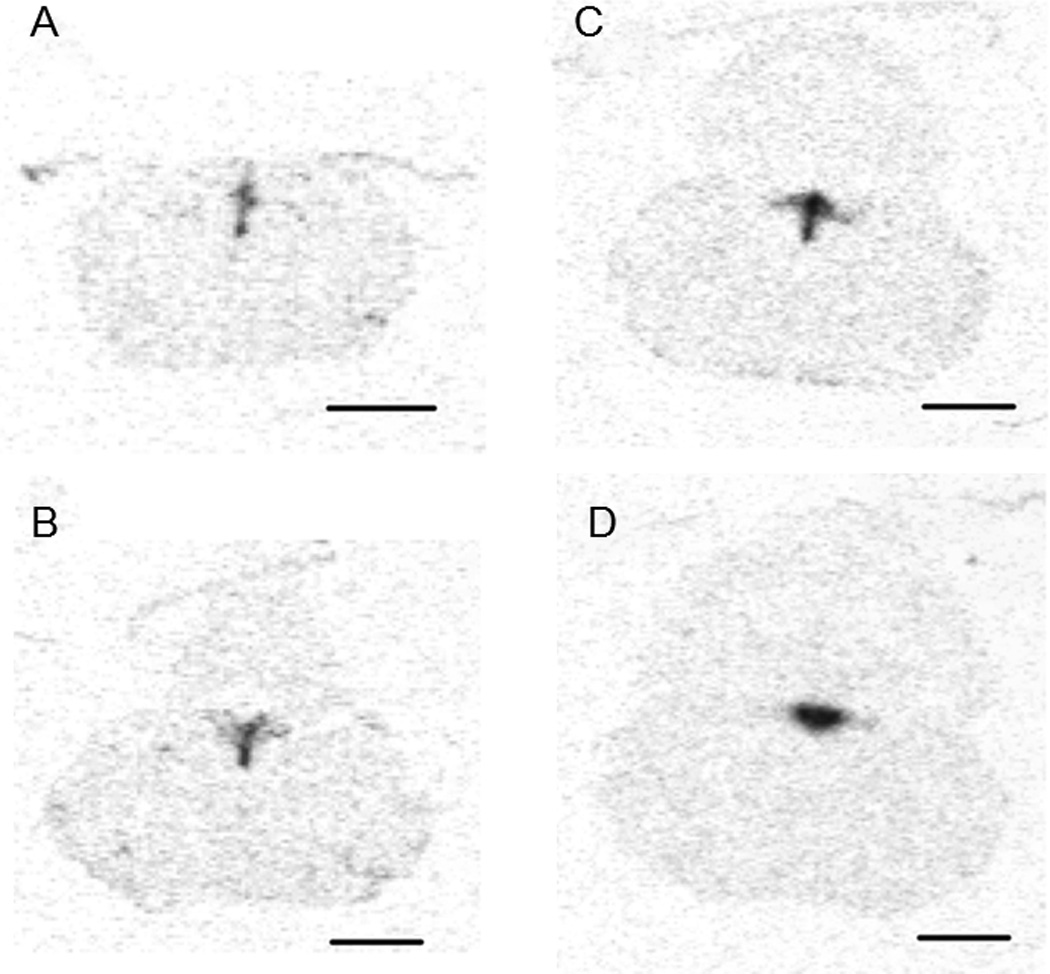
PKR2 mRNA expression in the dorsal raphe of the mouse brain. PKR2 mRNA expression is detected throughout the dorsal raphe (DR). Images (A-D) are shown from posterior to anterior mouse brainstem. Scale bar, 1mm.
3. Circadian oscillation of PK2 mRNA in the SCN of macaque brain
Oscillation of PK2 mRNA expression was next examined in the macaque brain over 6 time points in a 24 hour light-dark cycle. As shown in Fig. 6, PK2 mRNA levels in the SCN of the macaque brain oscillate during circadian time points, with the most robust expression at ZT4 (Fig. 6, 11AM time point). During the time period spanning the dark cycle, very low level of PK2 mRNA expression was observed in the SCN (Figs. 6, 7). The oscillation profile of PK2 mRNA showing a midday peak and a much lower level in the evening is synonymous to that of the mouse brain (Cheng et al., 2002). The amplitude of PK2 mRNA oscillation (peak/trough level ratio) is about ~50 fold, a figure also very similar to that of mouse as well (Cheng et al., 2002).
Figure 6.
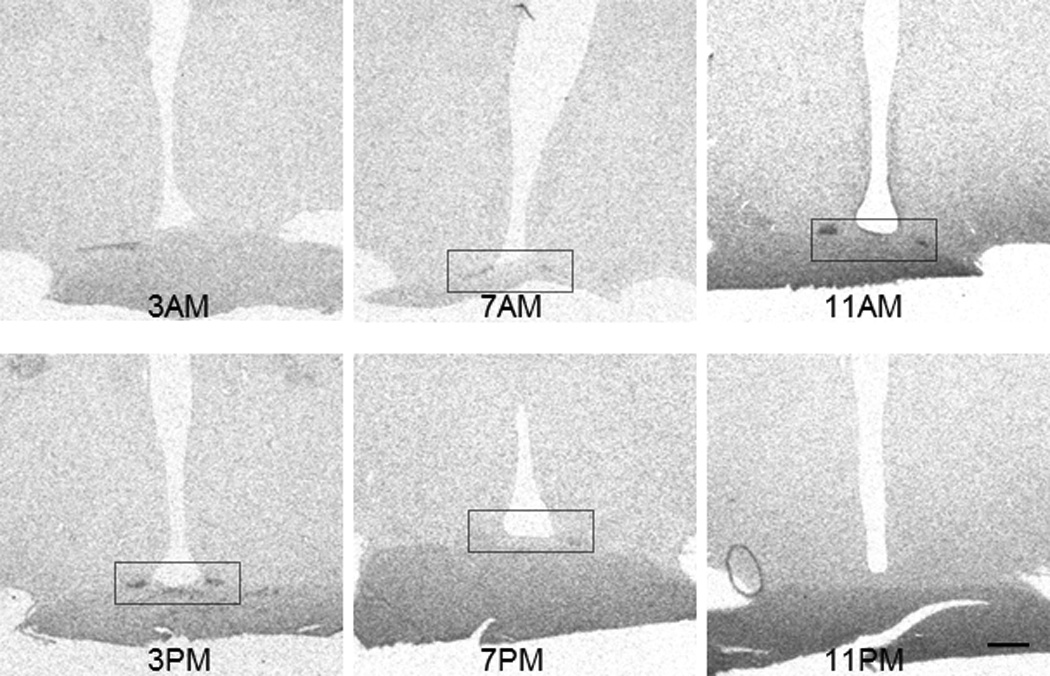
Circadian oscillation of the PK2 mRNA levels in the SCN of rhesus macaque monkeys. Shown are in situ hybridization images of PK2 from monkey brains that were sacrificed at 3AM, 7AM, 11AM, 3PM, 7PM, and 11PM. The circadian oscillation of PK2 mRNA level is apparent. Scale bar, 1mm.
Figure 7.
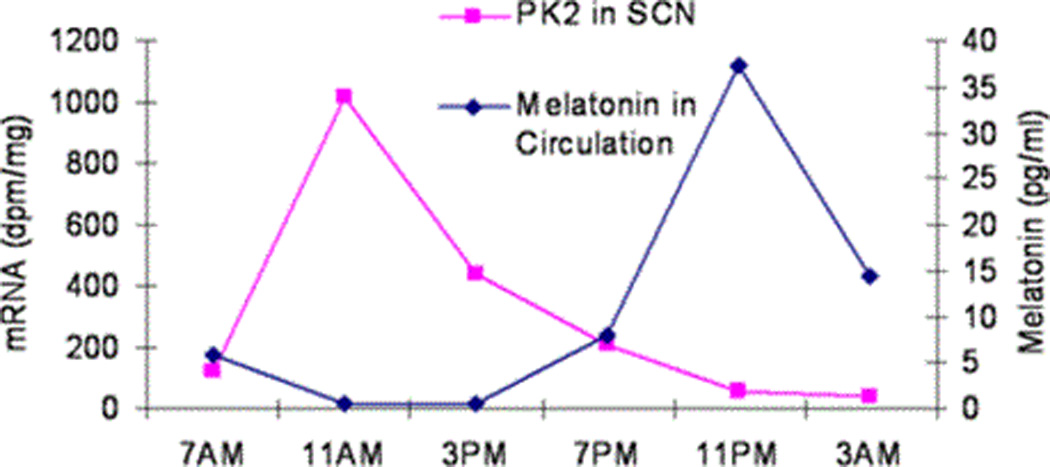
In all mammals, the duration of elevated melatonin secretion is equal to the extent of the dark phase. We simultaneously measured melatonin levels in the monkey blood circulation collected over the 24 hr circadian time period to correlate with the oscillating PK2 mRNA rhythm. As shown in Fig. 7, the concentrations of melatonin peaked during the dark hours. Taken together, these results indicate that PK2 mRNA oscillates in the diurnal macaque brain in the same phase and with similar amplitude as that of the nocturnal mouse.
DISCUSSIONS
Several lines of the evidence, obtained via morphological, pharmacological and genetic studies, have supported the role of PK2 as an output signaling for SCN circadian clock in nocturnal rodents (Li et al., 2012; Zhou & Cheng, 2005). In the current study, we cloned PK2 and PKR2 from a species of diurnal monkey, Macaca mulatta. The amino acid sequences of both PK2 and PKR2 are highly similar to that of human and mouse homologies (Li et al., 2001; Lin et al., 2002). We further examined the expression of PK2 and PKR2 in the macaque brain, with particular attention paying to the expression of PK2 or PKR2 in the circadian clock-related brain regions. In macaque monkey, PK2 mRNA expression is found in the SCN, AchSH, MnPO (Fig. 2), similar to that was observed in mouse brain (Cheng et al., 2002; Cheng et al., 2006). PKR2 is expressed in the SCN as well as several known primary SCN target nuclei such as LS, PVT, PVN, and DMH (Fig. 3). These results indicated that both PK2 and its receptor PKR2 have similar expression of PK2 and PKR2 in the mouse and macaque brains.
In the SCN of rhesus monkey, PK2 mRNA levels exhibit over 50-fold of diurnal oscillation, peaking during the middle day and being essentially absent during night hours (Figs. 6, 7). Thus the oscillation phase and amplitude of PK2 expression in the SCN of diurnal macaque is very similar to that of nocturnal mouse (Cheng et al., 2002) or hamster (Ji & Li, 2009). It should be cautioned that the quantification of PK2 expression in the SCN of the current study was from a single animal at each time point. Previously, Lambert et al studied the expression of PK2 in the SCN of a grass rat (Lambert et al., 2005), a largely diurnal rodent species. In grass rats, the oscillation of PK2 expression in the SCN was also observed in the same phase in nocturnal mouse. As earlier studies of other clockwork gene expression such as Per1, Per2 and Bmal1 also indicate that SCN operates quite in the same phase in nocturnal and diurnal species at molecular level (Dardente et al., 2002; Lincoln et al., 2002, Mrosovsky et al., 2001; Valenzuela et al., 2008;), it was not surprising to observe the similar PK2 oscillation in the macaque brain. As in addition molecular rhythm, the firing rate and the glucose utilization of SCN neurons are also in the same phase regardless of nocturnal and diurnal activity patterns (Smale et al., 2003), our studies are consistent with notion that the mechanism determining the active phase being day or night is likely located downstream from the signaling of SCN output molecules, such as PK2. However, we did notice the differential expression of PKR2 in the SCN between diurnal macaque brain and nocturnal moue brain. In contrast to the broad expression of PKR2 in the dorsal and ventral segments of the mouse SCN (Cheng et al., 2002; Cheng et al., 2005), the PKR2 expression was only detected in the dorsal SCN of the macaque brain (Fig. 3D). At least in nocturnal species, the likely functional difference between ventral and dorsal SCN has been implicated (Meijer et al., 2010). Obviously, the likely functional importance of the expression of PKR2 only in the dorsal SCN in the diurnal monkey and the broad expression of PKR2 in both ventral and dorsal SCN will need to be examined.
In addition to the observed PKR2 expression in the consensus middle thalamic and hypothalamic SCN target nuclei, PKR2 expression is observed in the dorsal raphe nuclei of the macaque brain (Fig. 4), which is also true for the mouse brain (Fig. 5). As a long SCN to raphe projection was previously observed in PK2-EGFP transgenic mice (Zhang, et al., 2009), these observations may indicate a possible SCN projection pathway with raphe nuclei as a target. Previous studies have revealed that the primary targets receiving SCN projection are very limited, and are primarily located in the midline thalamus and hypothalamus region (Buijs, 1996; Leak & Moore, 2001; Watts & Swanson, 1987; Watts et al., 1987). Our studies suggest that dorsal raphe nucleus may likely be an unrecognized primary SCN target that lie outside these midline thalamus and hypothalamus regions. As afferent projections to the SCN from the raphe nuclei in rodents are well established (Dudley et al., 1999; Glass et al., 2000; Glass et al., 2003; Hay-Schmidt et al., 2003; Moore & Speh, 2004; Welsh et al., 2010), the connections between SCN and DR are likely to be mutual and could involve in both the input and output pathways for the SCN circadian clock.
Acknowledgments
The research is supported in part by grants from the NIH (MH67753), AG029612, AG036670, and OD011092.
REFERENCES
- Buijs RM. The anatomical basis for the expression of circadian rhythms: the efferent projections of the suprachiasmatic nucleus. Prog Brain Res. 1996;111:229–240. doi: 10.1016/s0079-6123(08)60411-2. [DOI] [PubMed] [Google Scholar]
- Burton KJ, Li XH, Li JD, Hu WP, Zhou QY. Rhythmic Trafficking of TRPV2 in the Suprachiasmatic Nucleus is Regulated by Prokineticin 2 Signaling. Journal of Circadian Rhythms. 2015;13:2. doi: 10.5334/jcr.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Bittman EL, Hattar S, Zhou QY. Regulation of prokineticin 2 expression by light and the circadian clock. BMC Neurosci. 2005;6:17. doi: 10.1186/1471-2202-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MY, Leslie FM, Zhou QY. Expression of prokineticins and their receptors in the adult mouse brain. J Comp Neurol. 2006;498:796–809. doi: 10.1002/cne.21087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardente H, Klosen P, Caldelas I, Pévet P, Masson-Pévet M. Phenotype of Per1- and Per2-expressing neurons in the suprachiasmatic nucleus of a diurnal rodent (Arvicanthis ansorgei): comparison with a nocturnal species, the rat. Cell Tissue Res. 2002;310:85–92. doi: 10.1007/s00441-002-0609-9. [DOI] [PubMed] [Google Scholar]
- Dudley TE, Dinardo LA, Glass JD. In vivo assessment of the midbrain raphe nuclear regulation of serotonin release in the hamster suprachiasmatic nucleus. J Neurophysiol. 1999;81:1469–1477. doi: 10.1152/jn.1999.81.4.1469. [DOI] [PubMed] [Google Scholar]
- Glass JD, DiNardo LA, Ehlen JC. Dorsal raphe nuclear stimulation of SCN serotonin release and circadian phase-resetting. Brain Res. 2000;859:224–232. doi: 10.1016/s0006-8993(00)01963-6. [DOI] [PubMed] [Google Scholar]
- Glass JD, Grossman GH, Farnbauch L, DiNardo L. Midbrain raphe modulation of nonphotic circadian clock resetting and 5-HT release in the mammalian suprachiasmatic nucleus. J Neurosci. 2003;23:7451–7460. doi: 10.1523/JNEUROSCI.23-20-07451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay-Schmidt A, Vrang N, Larsen PJ, Mikkelsen JD. Projections from the raphe nuclei to the suprachiasmatic nucleus of the rat. J Chem Neuroanat. 2003;25:293–310. doi: 10.1016/s0891-0618(03)00042-5. [DOI] [PubMed] [Google Scholar]
- Ji Y, Li X. Cloning and developmental expression analysis of prokineticin 2 and its receptor PKR2 in the Syrian hamster surpachiasmatic nucleus. Brain Res. 2009;1271:18–26. doi: 10.1016/j.brainres.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Palm IF, La Fleur SE, Scheer FA, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Fliers E, Hofman MA, Swaab DF, Buijs RM. Vasopressin and the output of the hypothalamic biological clock. J. Neuroendocrinol. 2010;22:362–372. doi: 10.1111/j.1365-2826.2010.01956.x. [DOI] [PubMed] [Google Scholar]
- Lambert CM, Machida KK, Smale L, Nunez AA, Weaver DR. Analysis of the prokineticin 2 system in a diurnal rodent, the unstriped Nile grass rat (Arvicanthis niloticus) J Biol Rhythms. 2005;20:206–218. doi: 10.1177/0748730405275135. [DOI] [PubMed] [Google Scholar]
- Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol. 2001;433:312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- Lehman MN, Silver R, Gladstone WR, Kahn RM, Gibson M, Bittman EL. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci. 1987;7:1626–1638. doi: 10.1523/JNEUROSCI.07-06-01626.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JD, Hu WP, Boehmer L, Cheng MY, Lee AG, Jilek A, Siegel JM, Zhou QY. Attenuated circadian rhythms in mice lacking the prokineticin 2 gene. J Neurosci. 2006;26:11615–11623. doi: 10.1523/JNEUROSCI.3679-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JD, Burton KJ, Zhang C, Hu SB, Zhou QY. Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am J Physiol Regul Integr Comp Physiol. 2009;296:R824–R830. doi: 10.1152/ajpregu.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JD, Hu WP, Zhou QY. The circadian output signals from the suprachiasmatic nuclei. Prog Brain Res. 2012;199:119–127. doi: 10.1016/B978-0-444-59427-3.00028-9. [DOI] [PubMed] [Google Scholar]
- Li M, Bullock CM, Knauer DJ, Ehlert FJ, Zhou QY. Identification of two prokineticin cDNAs: recombinant proteins potently contract gastrointestinal smooth muscle. Mol Pharmacol. 2001;59:692–698. doi: 10.1124/mol.59.4.692. [DOI] [PubMed] [Google Scholar]
- Lin DC, Bullock CM, Ehlert FJ, Chen JL, Tian H, Zhou QY. Identification and molecular characterization of two closely related G protein-coupled receptors activated by prokineticins/endocrine gland vascular endothelial growth factor. J Biol Chem. 2002;277:19276–19280. doi: 10.1074/jbc.M202139200. [DOI] [PubMed] [Google Scholar]
- Lincoln G, Messager S, Andersson H, Hazlerigg D. Temporal expression of seven clock genes in the suprachiasmatic nucleus and the pars tuberalis of the sheep: evidence for an internal coincidence timer. Proc Natl Acad Sci U S A. 2002;99:13890–13895. doi: 10.1073/pnas.212517599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh DH, Kudo T, Colwell CS. Short circuiting the circadian system with a new generation of precision tools. Neuron. 2015;85:895–898. doi: 10.1016/j.neuron.2015.02.037. [DOI] [PubMed] [Google Scholar]
- Masumoto KH, Nagano M, Takashima N, Hayasaka N, Hiyama H, Matsumoto S, Inouye ST, Shigeyoshi Y. Distinct localization of prokineticin 2 and prokineticin receptor 2 mRNAs in the rat suprachiasmatic nucleus. Eur J Neurosci. 2006;23:2959–2970. doi: 10.1111/j.1460-9568.2006.04834.x. [DOI] [PubMed] [Google Scholar]
- Meijer JH, Michel S, Vanderleest HT, Rohling JH. Daily and seasonal adaptation of the circadian clock requires plasticity of the SCN neuronal network. Eur J Neurosci. 2010;32:2143–2151. doi: 10.1111/j.1460-9568.2010.07522.x. [DOI] [PubMed] [Google Scholar]
- Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, Sakurai T. Cellular Clocks in AVP Neurons of the SCN Are Critical for Interneuronal Coupling Regulating Circadian Behavior Rhythm. Neuron. 2015;85:1103–1116. doi: 10.1016/j.neuron.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Moore RY, Speh JC. Serotonin innervation of the primate suprachiasmatic nucleus. Brain Res. 2004;1010:169–173. doi: 10.1016/j.brainres.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Mrosovsky N, Edelstein K, Hastings MH, Maywood ES. Cycle of period gene expression in a diurnal mammal (Spermophilus tridecemlineatus): implications for nonphotic phase shifting. J Biol Rhythms. 2001;16:471–478. doi: 10.1177/074873001129002141. [DOI] [PubMed] [Google Scholar]
- Prosser HM, Bradley A, Chesham JE, Ebling FJ, Hastings MH, Maywood ES. Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc Natl Acad Sci U S A. 2007;104:648–653. doi: 10.1073/pnas.0606884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Ren P, Zhang H, Qiu F, Liu Y-Q, Gu H, et al. Prokineticin 2 Regulates the Electrical Activity of Rat Suprachiasmatic Nuclei Neurons. PLoS ONE. 2011;6:e20263. doi: 10.1371/journal.pone.0020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- Smale L, Lee T, Nunez AA. Mammalian diurnality: some facts and gaps. J Biol Rhythms. 2003;18:356–366. doi: 10.1177/0748730403256651. [DOI] [PubMed] [Google Scholar]
- Valenzuela FJ, Torres-Farfan C, Richter HG, Mendez N, Campino C, Torrealba F, Valenzuela GJ, Serón-Ferré M. Clock gene expression in adult primate suprachiasmatic nuclei and adrenal: is the adrenal a peripheral clock responsive to melatonin? Endocrinology. 2008;149:1454–1461. doi: 10.1210/en.2007-1518. [DOI] [PubMed] [Google Scholar]
- Watts AG, Swanson LW, Sanchez-Watts G. Efferent projections of the suprachiasmatic nucleus: I. Studies using anterograde transport of Phaseolus vulgaris leucoagglutinin in the rat. J Comp Neurol. 1987;258:204–229. doi: 10.1002/cne.902580204. [DOI] [PubMed] [Google Scholar]
- Watts AG, Swanson LW. Efferent projections of the suprachiasmatic nucleus: II. Studies using retrograde transport of fluorescent dyes and simultaneous peptide immunohistochemistry in the rat. J Comp Neurol. 1987;258:230–252. doi: 10.1002/cne.902580205. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, Dong X, Tsujimoto G, Okuno Y, Doi M, Okamura H. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science. 2013;342:85–90. doi: 10.1126/science.1238599. [DOI] [PubMed] [Google Scholar]
- Yuill EA, Hoyda TD, Ferri CC, Zhou QY, Ferguson AV. Prokineticin 2 depolarizes paraventricular nucleus magnocellular and parvocellular neurons. Eur J Neurosci. 2007;25:425–434. doi: 10.1111/j.1460-9568.2006.05293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Truong KK, Zhou QY. Efferent projections of prokineticin 2 expressing neurons in the mouse suprachiasmatic nucleus. PLoS One. 2009;4:e7151. doi: 10.1371/journal.pone.0007151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Cheng MY. Prokineticin 2 and circadian clock output. FEBS J. 2005;272:5703–5709. doi: 10.1111/j.1742-4658.2005.04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


