Abstract
Peroxisomes are subcellular organelles involved in various metabolic processes, including fatty acid and phospholipid homeostasis. The Zellweger spectrum disorders (ZSDs) represent a group of diseases caused by a defect in the biogenesis of peroxisomes. Accordingly, cells from ZSD patients are expected to have an altered composition of fatty acids and phospholipids. Using an LC/MS-based lipidomics approach, we show that the phospholipid composition is characteristically altered in cultured primary skin fibroblasts from ZSD patients when compared with healthy controls. We observed a marked overall increase of phospholipid species containing very long-chain fatty acids, and a decrease of phospholipid species with shorter fatty acid species in ZSD patient fibroblasts. In addition, we detected a distinct phosphatidylcholine profile in ZSD patients with a severe and mild phenotype when compared with control cells. Based on our data, we present a set of specific phospholipid ratios for fibroblasts that clearly discriminate between mild and severe ZSD patients, and those from healthy controls. Our findings will aid in the diagnosis and prognosis of ZSD patients, including an increasing number of mild patients in whom hardly any abnormalities are observed in biochemical parameters commonly used for diagnosis.
Keywords: peroxisomes, peroxisome biogenesis disorders, lipidomics, diagnosis
Peroxisomes are ubiquitous cell organelles, which, in humans, play an important role in lipid metabolism, including the β-oxidation of very long-chain fatty acids (VLCFAs) and bile acids, the α-oxidation of phytanic acid, and the synthesis of plasmalogens (1, 2). Defects in peroxisome function cause a variety of peroxisomal disorders. These include single enzyme deficiencies, affecting a specific peroxisome-dependent metabolic pathway, and the peroxisome biogenesis disorders, which affect multiple peroxisome-dependent metabolic pathways. The peroxisome biogenesis disorders are autosomal recessive disorders, and include the Zellweger spectrum disorders (ZSDs) and rhizomelic chondrodysplasia punctata (RCDP) type 1 (3, 4). The ZSDs constitute a spectrum of disease severity, which include three overlapping clinical phenotypes, formerly described as Zellweger syndrome, the most severe form, and neonatal adrenoleukodystrophy and infantile Refsum disease, two milder phenotypes (5). Cells of patients with the severe ZSD phenotype are characterized by the complete absence of functional peroxisomes, but cells of patients affected by one of the milder ZSD phenotypes often still have residual peroxisomal function (3).
The laboratory diagnosis of patients with a peroxisomal disorder usually starts with metabolite diagnostics in plasma, followed by enzymatic and biochemical studies in cultured fibroblasts (1). Among the biochemical parameters that may indicate a peroxisomal disorder, VLCFA, pristanic acid, phytanic acid, and the bile acid intermediates are often elevated in plasma and tissues of ZSD patients, whereas the levels of plasmalogens are usually decreased in tissues and erythrocytes (6–8).
In recent years, an increasing number of patients have been identified at a later age, who did not show clear clinical symptoms and/or the typical biochemical abnormalities indicating a peroxisomal disorder, which implies that some patients with a very mild phenotype may be difficult to diagnose (9–13). Hence, the identification of additional or more sensitive biomarkers for peroxisomal disorders would benefit the diagnosis of mildly affected ZSD patients and patients at an early stage of life. Furthermore, such biomarkers could potentially facilitate the monitoring of disease progression and response to possible treatments.
Because peroxisomes play a crucial role in lipid metabolism, we hypothesized that peroxisomal defects give rise to changes in phospholipid composition that may serve as diagnostic biomarkers for ZSD patients. Phospholipids are the major components of cell membranes, constituting the lipid bilayer matrix. In eukaryotes, phospholipids contain a glycerol backbone with a phosphate head group at the sn-3 position, and two esterified fatty acyl chains at the sn-1 and sn-2 positions (14). Phospholipids are classified by the structure of their head group, and each class consists of a variety of species that are specified by both the acyl chain length and the degree of saturations (15). In eukaryotic membranes, phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) constitute the major classes of phospholipids. In addition, ether phospholipids such as plasmalogens are important components of eukaryotic membranes. Plasmalogens contain a fatty alcohol at position sn-1 that is linked to a vinyl ether bond (16, 17).
Using a lipidomics approach, we show in this study that the phospholipid profiles are characteristically altered in cultured skin fibroblasts from ZSD patients when compared with cells from healthy controls, reflecting the role of peroxisomes in lipid metabolism. Furthermore, we identified specific sets of phospholipid ratios that may be useful for the diagnosis of ZSD patients.
MATERIALS AND METHODS
Patient cell lines
We used primary skin fibroblast cell lines from seven anonymized healthy controls, seven anonymized ZSD patients with a severe phenotype (each homozygous for the severe c.2097insT mutation in PEX1), and seven anonymized ZSD patients with a mild phenotype (each homozygous for the hypomorphic c.2528G>A mutation in PEX1) (18). Fibroblasts were cultured in parallel in 162 cm2 flasks in Ham’s F-10 medium with L-glutamine, supplemented with 10% fetal calf serum (Invitrogen, Carlsbad, CA), 25 mM HEPES, 100 U/ml penicillin and 100 μg/ml streptomycin, and 250 μg/ml amphotericin in a humidified atmosphere of 5% CO2 at 37°C. After they reached confluence, the cells were harvested by trypsinization (0.5% trypsin-EDTA; Invitrogen) and washed once with phosphate-buffered saline and twice with 0.9% NaCl, followed by centrifugation at 4°C (16,100 g for 5 min) to obtain cell pellets. Pellets were stored at −80°C until analysis. To test the robustness of the method, we used cell pellets from other healthy controls and ZSD patients with a severe and mild phenotype, which were previously cultured and stored at −80°C for diagnostic purposes (five biological replicates per group).
Extraction of phospholipids
Fibroblast pellets were resuspended in water and sonicated on ice for 30 s at 8 W using a tip sonicator. Protein concentrations of the homogenates were determined using the bicinchoninic acid assay (19). Phospholipids were extracted using a single-phase extraction. We added a defined amount of internal standards [0.1 nmol of cardiolipin (CL)(14:0)4, 0.2 nmol of bis(monoacylglycero)phosphate (BMP)(14:0)2, 2.0 nmol of PC(14:0)2, 0.1 nmol of phosphatidylglycerol (PG)(14:0)2, 5.0 nmol of PS(14:0)2, 0.5 nmol of PE(14:0)2, 1.0 nmol of phosphatidic acid (PA)(14:0)2, 2.0 nmol of SM(d18:1/12:0), 0.02 nmol of lyso-PG(14:0), 0.1 nmol of lyso-PE (LPE)(14:0), 0.5 nmol of lyso-PC (LPC)(14:0), 0.1 nmol of LPA(14:0) (purchased from Avanti Polar Lipids, Alabaster, AL) dissolved in 120 μl of chloroform/methanol (1:1, v/v)] and 1.5 ml of chloroform/methanol (1:1, v/v) to 1 mg protein of the fibroblast homogenates. Subsequently, the mixture was sonicated in a water bath for 5 min, followed by centrifugation at 4°C (16,100 g for 5 min). The liquid phase was transferred to a glass vial and evaporated under a steam of nitrogen at 60°C. Subsequently, the residue was dissolved in 150 μl of chloroform/methanol (9:1, v/v), and 10 μl of the solution was injected into the HPLC-MS system. As no internal standard was available for phosphatidylinositol (PI), we normalized PI species on the PE internal standard. Because of this suboptimal quantification, we decided to exclude PI during further analysis. For completeness, annotated PI species are listed in supplemental Table S1.
HPLC-MS
Lipidomic analysis was performed as described previously with minor changes described in this section (20). The HPLC system consisted of an Ultimate 3000 binary HPLC pump, a vacuum degasser, a column temperature controller, and an autosampler (Thermo Scientific, Waltham, MA). The column temperature was maintained at 25°C. The lipid extract was injected onto a LiChrospher 2 × 250 mm silica-60 column, 5 μm particle diameter (Merck, Darmstadt, Germany). The phospholipids were separated from interfering compounds by a linear gradient between solution B (chloroform/methanol, 97:3, v/v) and solution A (methanol/water, 85:15, v/v). Solutions A and B contained 5 and 0.2 ml of 25% (v/v) aqueous ammonia per liter of eluent, respectively. The gradient (0.3 ml/min) was as follows: 0–1 min, 10% A; 1–4 min, 10–20% A, 4–12 min, 20–85% A; 12–12.1 min, 85–100% A; 12.1–14.0 min, 100% A, 14–14.1 min, 100–10% A; and 14.1–15 min, equilibration with 10% A. All gradient steps were linear, and the total analysis time, including the equilibration, was 15 min. A Q Exactive Plus (Thermo Scientific) was used in the negative and positive electrospray ionization mode. Nitrogen was used as the nebulizing gas. The spray voltage used was 2,500 V and the capillary temperature was 256°C (S-lens RF level, 50; auxiliary gas, 11; auxiliary gas temperature, 300°C; sheath gas, 48; sweep cone gas, 2). In both the negative and positive mode, mass spectra of phospholipid molecular species were obtained by continuous scanning from m/z 150 to m/z 2,000 with a resolution of 280,000.
Bioinformatics and statistical analysis of lipidomics data
The raw LC-MS data were converted to mzXML format using MSConvert (21). The dataset was processed using an in-house developed metabolomics pipeline written in the R programming language (http://www.r-project.org). In brief, it consisted of the following five steps: 1) preprocessing using the R package “XCMS” (22); 2) identification of metabolites using an in-house database of (phospho)lipids, with known internal standards indicating the position of most of the PL clusters, matching m/z values within 3 ppm deviation; 3) isotope correction to obtain deconvoluted intensities for overlapping peak groups (23); 4) normalization on the intensity of the internal standard for PL classes for which an internal standard was available and scaling on measured protein content per sample; and 5) statistical analysis. Total phospholipid levels are defined as the summation of the relative abundance of all identified phospholipid species of the same class normalized to the corresponding internal standard, assuming identical response with respect to internal standard. Data in the figures are presented as mean ± SD. A Students t-test or one-way ANOVA with post hoc Bonferroni correction was used for statistical comparison between the groups. P = 0.01 or lower was considered as significant. Heat-maps of metabolites were created using the R programming language package “gplots.” Color in the heat-maps reflects the logarithm of the relative metabolite abundance with red being higher and blue lower than the mean abundance value per metabolite. Partial least squares regression discriminant analysis (PLS-DA) was performed using the R package “mixOMICS.”
RESULTS
Lipidomic profiling of ZSD patient fibroblasts
Because more than 60% of all ZSD patients harbor biallelic mutations in the PEX1 gene, we used primary skin fibroblasts from patients with the two most common PEX1 mutations (3, 18). These included fibroblasts from patients homozygous for the c.2097insT frame shift mutation in PEX1 (n = 7), which causes a severe phenotype (18), and fibroblasts from patients homozygous for c.2528G>A in PEX1 (n = 7), which causes a milder phenotype (9). Cells were cultured under standardized conditions and subsequently processed for lipidomic analysis. We identified 1,023 distinct phospholipid species (supplemental Table S1). We normalized PI species using the internal standard for PE, due to lack of an internal standard for PI. The thus annotated PI species can be found in the supplemental data (supplemental Table S1), but have not been used for further analysis. Because we used normal phase HPLC, phospholipid species with a higher number of carbon atoms in the fatty acyl chains eluted earlier from the column (lower retention time) and have a higher m/z than species with fewer carbon atoms in the fatty acyl chains (Fig. 1A; supplemental Fig. S1). Furthermore, phospholipid species with the same number of carbon atoms, but an increasing number of double bonds (unsaturations), had a lower retention time. We denoted identified phospholipids in our analysis as C(XX:Y), where XX denotes the total number of carbon atoms and Y the total number of double bonds in the fatty acyl chains, without the specification of which fatty acid is located at the sn-1 or sn-2 position. Without fragmentation data, it is not possible to distinguish between ether phospholipids with O-alkyl (plasmalogen) and O-alkenyl linkages. When the lipidomic profiles of cells from ZSD patients were compared with those of healthy controls, we observed an overall increase of phospholipid species containing VLCFAs with a total number of >40 carbon atoms in the fatty acyl chains, and a decrease of phospholipid species with shorter fatty acid species with ≤35 carbon atoms in the fatty acyl chains in ZSD patient fibroblasts.
Fig. 1.
Shift in the PC species in ZSD patient fibroblasts. Visualization of lipidomic analysis data for PC species. A: Scatter plot of m/z ratio and retention time (RT); PC species are represented as dots. Because we used normal phase HPLC, the summed carbon atom length in the phospholipid fatty acyl chains, ranging from C28:Y to C56:Y, increases with higher m/z. Similarly, the number of double-bonds in the phospholipid fatty acyl chains decreases with higher retention time. Green triangles pointing upwards indicate PC species that were significantly higher in ZSD patient fibroblasts compared with cells from healthy controls. Red triangles pointing downwards indicate PC species that were significantly decreased in ZSD patient fibroblasts when compared with cells from healthy controls. Triangles surrounded by a black line indicate significantly changed PC species in both mild and severe ZSD patient fibroblasts compared with healthy control cells, while filled triangles without surrounding line were significantly changed in severe ZSD patient fibroblasts when compared with healthy controls only. Open triangles indicate significantly changed PC species in mild ZSD patient fibroblasts compared with healthy control cells only. Circles indicate PC species that have been fragmented by MS/MS analysis (supplemental Table S5). B: Unsupervised hierarchical clustering plot of logarithm-transformed data. The upper panel indicates samples from severe ZSD patient fibroblasts, the middle panel samples from mild ZSD patient fibroblasts, and the lower panel samples from healthy control cells. From left to right, the summed carbon atom length in the phospholipid fatty acyl chains, ranging from C30:Y to C50:Y, increases. Color in the heat-maps reflects the logarithm of the relative metabolite abundance with red being higher and blue lower than the mean abundance value per metabolite. For visualization, PC species with the lowest peak abundance (≤5% of average peak abundance within PC class) are not displayed. See supplemental Fig. S3 for details. C: Cluster plot of computationally created peaks showing log-fold changes of both mild and severe ZSD patient fibroblasts relative to healthy control cells. Vertical bars indicate log-fold changes of severe ZSD patient fibroblasts relative to healthy control cells, while plus (+) symbols indicate log-fold changes of mild ZSD patient fibroblasts relative to healthy control cells.
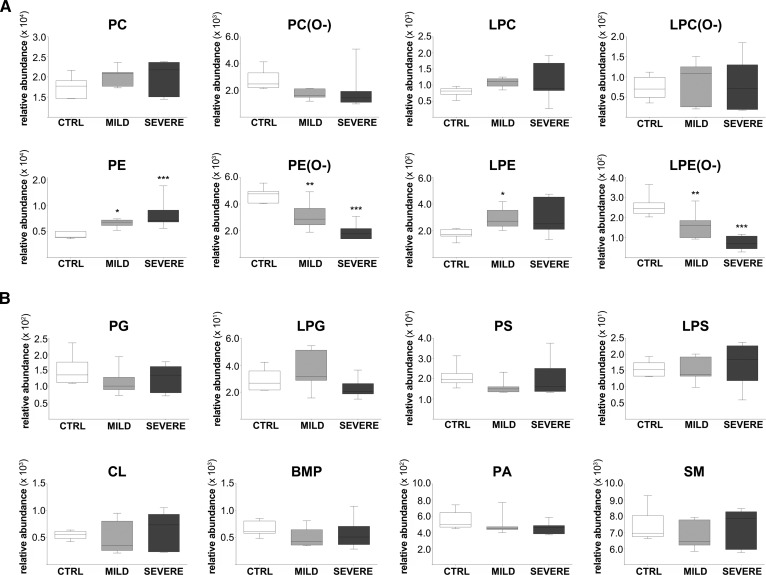
The most abundant PC species found in control fibroblasts had a total fatty acyl chain length ranging from C30 to C40 and were decreased in mild and severe ZSD patient fibroblasts (Fig. 1A, B). In contrast, PC species with fatty acyl chain lengths ranging from C42 to C58 were increased in ZSD patient fibroblasts when compared with control fibroblasts (Fig. 1B, C). The observation that the total PC levels were not changed in cells from mild or severe ZSD patients when compared with healthy controls (Fig. 2A) points to a shift in the PC species with long-chain fatty acids to VLCFAs in the ZSD patient fibroblasts. Analysis of LPC species, which are derivates of PC and only have one fatty acyl chain, also showed an increase of species with a fatty acyl chain length of ≥C24 in ZSD patient fibroblasts, while species with an acyl chain length of ≤C20 were decreased. Total LPC levels were not significantly changed (Fig. 2A).
Fig. 2.
Lipidomics profiles in mild and severe ZSD patient fibroblasts. Total phospholipid levels, sorted by phospholipid class. A: PC and PE, their derivates (LPC and LPE), and associated ether phospholipids (PC, LPC, PE, and LPE ether phospholipids). B: Other phospholipid classes, including PG, CL, BMP, PA, PS, SM, lyso-PG (LPG), and lyso-PS (LPS). Total phospholipid levels are defined as the summation of the relative abundance of all identified phospholipid species of the same class normalized to the corresponding internal standard, assuming identical response with respect to internal standard. Total phospholipid levels are shown as box-and-whisker plots, with white plots indicating the healthy control (CTRL) group, light gray plots the mild ZSD group, and dark gray plots the severe ZSD group; n = 7 per group. One-way ANOVA was performed to determine significant differences between the mild and severe ZSD group, respectively, when compared with the healthy control group (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
We detected similar changes in PE. The levels of PE species with a total fatty acyl chain length of ≥40 carbon atoms were significantly increased in the fibroblasts from mild and severe ZSD patients, whereas several PE species containing a total fatty acyl chain length of 30–33 carbon atoms were decreased (supplemental Fig. S1). In contrast to PC phospholipids, however, the total PE levels were significantly increased in fibroblasts of both mild and severe ZSD patients when compared with controls (Fig. 2A). The total LPE levels were increased only in severe ZSD patient fibroblasts when compared with healthy control cells.
In addition to the major differences in the PC and PE phospholipids, we observed more specific differences in other phospholipid classes in mild and severe ZSD patient fibroblasts (Fig. 2A, B). For ether phospholipids, we detected species ranging from C30 to C48. Almost all of these were decreased in both mild and severe ZSD patient fibroblasts, which resulted in markedly decreased total levels of PC (P = 0.06), PE, and LPE ether phospholipids. The total LPC ether phospholipid levels were not changed. PS species with total chain length of ≥C40 were significantly increased in mild and severe ZSD patient fibroblasts, whereas the total PS levels were not changed. In SM, the most abundant species was SM(16:0), accounting for almost half of the total levels of this phospholipid class. SM species with a fatty acyl chain length of ≥C24 accumulated significantly in mild and severe ZSD patient fibroblasts, while several species with a fatty acyl chain length of <C24 were decreased. Total SM levels were not changed in any of the cells. The levels of several CL species were significantly decreased in ZSD patients, but the total CL levels were not altered. For BMP, several species with total chain length ≥C40 were significantly increased in ZSD patient fibroblasts. Finally, phospholipid species of PA, the precursor of all phospholipid classes, and PG were not significantly changed in ZSD patient fibroblasts compared with controls (Fig. 2B).
Discriminative phospholipid ratios for ZSD patients
To evaluate whether certain phospholipid species can be used as selective and/or sensitive biomarkers for ZSD patients, we applied PLS-DA to our lipidomics data. PLS-DA is a multivariate discrimination model that allows the exploration of high-dimensional data by identifying latent variables, the principal components (24). The PLS-DA model was created based on the fibroblast samples from severe ZSD patients and healthy controls, while the mild ZSD patient fibroblast samples were predicted using the model. Using the first two principal components, we visualized our lipidomics data in a two-dimensional score plot (supplemental Fig. S2). Subsequently, we developed a method to create phospholipid ratios (Fig. 3A). First, we ranked all phospholipid species by their contribution to the explained discrimination in the first principal component (PC1) (supplemental Table S2). PC and PE species with VLCFAs and PC and PE ether phospholipids, especially, were ranked among the metabolites explaining the highest variation between the groups. Next, we created ratios of the phospholipid species with the highest ranks in the PLS-DA (supplemental Fig. S2). We then selected the ratios based on their variance, favoring ratios with less variance (Fig. 3A). The final set of the most discriminative ratios clearly distinguished both the mild and severe ZSD patient samples from control fibroblast samples (Fig. 3B, supplemental Table S3). The phospholipid species constituting the majority of those selected ratios were ranked highest in the list of contribution to PC1 (supplemental Table S2).
Fig. 3.
The most discriminative ratios clearly distinguished both the mild and severe ZSD patient samples from control fibroblast samples. A: Flowchart for data processing and selection of the ratios. B: Examples of the final set of 218 most discriminative ratios of phospholipid species listed in supplemental Table S3 expressed as ratio values based on the relative abundance of the phospholipid species after normalization to the corresponding internal standards. Ratios are shown as box-and-whisker plots; n = 5 per group.
To test the robustness and reproducibility of our selected set of ratios, we repeated the lipidomic analysis using fibroblast cell pellets that were cultured and harvested at different time points and derived from different ZSD patients, but harboring the same mutations (n = 5 per group). We performed phospholipid profiling and the calculation of ratios as described before (Fig. 3A). Finally, we only selected ratios that were present in both data sets, thereby eliminating factors that may have been introduced by the synchronization of the cultured cells (Fig. 3A). The majority of the selected set of ratios was also found to discriminate between mild and severe ZSD patient and healthy control fibroblasts when they were not cultured in parallel under similar conditions. The most discriminative ratios we identified included ratios of PE and PC species, respectively, versus their corresponding PC and PE ether phospholipids (Fig. 3B; supplemental Tables S3, S4). Furthermore, we found several ratios of PC and SM species with a total chain length ≥C40 versus PC and SM species with shorter chain length that clearly distinguished both the mild and severe ZSD patient samples from control fibroblast samples.
DISCUSSION
Peroxisomes play an important role in fatty acid oxidation and ether phospholipid biosynthesis. In ZSD patients, peroxisomes are nonfunctional in the severe phenotype, but have residual activity in the mild phenotype. Among other biochemical parameters, impaired peroxisomal functioning results in the accumulation of VLCFAs and a marked decrease in plasmalogens (7). In accordance with this, our lipidomic data showed a global accumulation of phospholipid species containing VLCFA chains (>C40) in cultured ZSD patient fibroblasts. Indeed, we found that the most abundant PC species with long-chain fatty acids in healthy control fibroblasts were decreased in fibroblasts from ZSD patients, while PC and PE species with VLCFAs accumulated in ZSD patient cells. Abe et al. (25) recently also reported elevated levels of PC species with VLCFA chains in cells from severe ZSD patients with mutations in PEX13 and PEX16. In contrast to our findings, they did not report a decrease in PC species containing medium- and long-chain fatty acids (25). In our study, the total levels of PE, the second most abundant phospholipid in eukaryotic membranes, was significantly increased in cells of mild and severe ZSD patients, while the PE ether phospholipid levels were decreased. The upregulation of PE in combination with a decrease in PE ether phospholipid levels has also been reported in fibroblasts derived from RCDP and severe ZSD patients (25, 26). Finally, we found an accumulation of SM species with C26 and C28 in severe ZSD patients, which was also observed in an earlier study with fibroblasts from severe ZSD patients (27).
Using specific phospholipid species identified in our analysis, we were able to define a set of ratios that significantly discriminate between cells from ZSD patients and healthy controls. The use and power of metabolite ratios rather than individual levels as biochemical markers have been described for other inborn errors of metabolism, such as the ratio of monolysocardiolipin to CL for the diagnosis of Barth syndrome (28) and the ratio of hexacosanoic acid (C26:0) to docosanoic acid (C22:0) for patients with adrenoleukodystropy and ZSD (29). Furthermore, Pettus et al. (30) proposed the ratio of C26:1/0-ceramide/C22:0-ceramide as a potential marker for the diagnosis of peroxisome dysfunction.
Hubbard et al. (31) reported that LPC(26:0) can serve as a diagnostic marker for the diagnosis of adrenoleukodystrophy and peroxisomal disorders of β-oxidation in dried blood spots, and developed a LC-MS/MS method suitable for newborn screening. Moreover, they proposed that LPC(26:0) can also be used as a diagnostic marker for ZSD patients. Our data show that this is true for severe ZSD patients, but this may not be suitable for ZSD patients with a very mild phenotype. Here, we present a set of phospholipid ratios that can be used for diagnostics of ZSD patients. The most discriminative ratios we identified mainly included ratios of PE and PE ether phospholipids, and PC and PC ether phospholipid species, which were also found to be highly ranked in our PLS-DA analysis. These ratios reflect biochemical parameters that are often abberant in ZSDs, including elevated levels of VLCFAs and decreased levels of plasmalogens. Furthermore, we found several highly discriminative ratios of phospholipid species with VLCFAs versus phospholipid species with shorter chain length, which indicates a shift in phospholipid species with long-chain fatty acids to species with VLCFAs in ZSD patient fibroblasts. The selected set of ratios we present in this study is robust, has a high reproducibility, and is not dependent on fibroblast cells that were cultured under synchronized conditions, but is also applicable for cell pellets that have been cultured at different time points. Although it has to be taken into consideration that the phospholipid profiles presented in this study were determined in fibroblasts derived from patients with a mutation in the PEX1 gene only, the set of ratios discriminates clearly between cells from mild and severe ZSD patients and those from healthy controls. Because the clinical and biochemical presentation of ZSD patients with a defect in any of the other PEX genes is indistinguishable from patients with a mutation in the PEX1 gene (3), these ratios will likely serve as a valuable and robust tool for diagnostic purposes of ZSD in general.
In conclusion, we identified characteristically altered lipidomic profiles in cultured fibroblasts from ZSD patients when compared with healthy control cells, which reflect the important role of peroxisomes in lipid metabolism. Based on these findings, we identified a discriminative set of phospholipid ratios that may be a useful tool for diagnostic purposes of ZSD patients.
Supplementary Material
Acknowledgments
The authors thank Henk van Lenthe for helpful discussions and advice.
Footnotes
Abbreviations:
- LPC
- lyso-phosphatidylcholine
- LPE
- lyso-phosphatidylethanolamine
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PLS-DA
- partial least squares regression discriminant analysis
- PS
- phosphatidylserine
- VLCFA
- very long-chain fatty acid
- ZSD
- Zellweger spectrum disorder
This work was supported by the FP-7-PEOPLE-2012-Marie Curie-ITN316723 PERFUME (K.H. and H.R.W.). The funding source had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Wanders R. J., and Waterham H. R.. 2006. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75: 295–332. [DOI] [PubMed] [Google Scholar]
- 2.Van Veldhoven P. P. 2010. Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J. Lipid Res. 51: 2863–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waterham H. R., and Ebberink M. S.. 2012. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim. Biophys. Acta. 1822: 1430–1441. [DOI] [PubMed] [Google Scholar]
- 4.Steinberg S. J., Dodt G., Raymond G. V., Braverman N. E., Moser A. B., and Moser H. W.. 2006. Peroxisome biogenesis disorders. Biochim. Biophys. Acta. 1763: 1733–1748. [DOI] [PubMed] [Google Scholar]
- 5.Aubourg P., and Wanders R.. 2013. Peroxisonal disorders. Handb. Clin. Neurol. 113: 1593–1609. [DOI] [PubMed] [Google Scholar]
- 6.Wanders R. J., Purvis Y. R., Heymans H. S., Bakkeren J. A., Parmentier G. G., van Eldere J., Eyssen H., van den Bosch H., Tager J. M., and Schutgens R. B.. 1986. Age-related differences in plasmalogen content of erythrocytes from patients with the cerebro-hepato-renal (Zellweger) syndrome: implications for postnatal detection of the disease. J. Inherit. Metab. Dis. 9: 335–342. [DOI] [PubMed] [Google Scholar]
- 7.Steinberg S., Jones R., Tiffany C., and Moser A.. 2008. Investigational methods for peroxisomal disorders. Curr. Protoc. Hum. Genet. Chapter 17: 17.6.. [DOI] [PubMed]
- 8.Wanders R. J., Ferdinandusse S., Brites P., and Kemp S.. 2010. Peroxisomes, lipid metabolism and lipotoxicity. Biochim. Biophys. Acta. 1801: 272–280. [DOI] [PubMed] [Google Scholar]
- 9.Weller S., Gould S. J., and Valle D.. 2003. Peroxisome biogenesis disorders. Annu. Rev. Genomics Hum. Genet. 4: 165–211. [DOI] [PubMed] [Google Scholar]
- 10.Steinberg S. J., Snowden A., Braverman N. E., Chen L., Watkins P. A., Clayton P. T., Setchell K. D., Heubi J. E., Raymond G. V., Moser A. B., et al. 2009. PEX10 defect in a patient with no detectable defect in peroxisome assembly or metabolism in cultured fibroblasts. J. Inherit. Metab. Dis. 32: 109–119. [DOI] [PubMed] [Google Scholar]
- 11.Régal L., Ebberink M. S., Goemans N., Wanders R. J., De Meirleir L., Jaeken J., Schrooten M., Van Coster R., and Waterham H. R.. 2010. Mutations in PEX10 are a cause of autosomal recessive ataxia. Ann. Neurol. 68: 259–263. [DOI] [PubMed] [Google Scholar]
- 12.Ebberink M. S., Koster J., Visser G., Spronsen F. v., Stolte-Dijstra I., Smit G. P., Fock J. M., Kemp S., Wanders R. J., and Waterham H. R.. 2012. A novel defect of peroxisome division due to a homozygous non-sense mutation in the PEX11β gene. J. Med. Genet. 49: 307–313. [DOI] [PubMed] [Google Scholar]
- 13.Mignarri A., Vinciguerra C., Giorgio A., Ferdinandusse S., Waterham H., Wanders R., Bertini E., Dotti M. T., and Federico A.. 2012. Zellweger spectrum disorder with mild phenotype caused by PEX2 gene mutations. JIMD Rep. 6: 43–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vance D. E., and Vance J. E.. 2008. Phospholipid biosynthesis in eukaryotes. In Biochemistry of Lipids, Lipoproteins and Membranes. 5th edition. D. E. Vance and J. E. Vance, editors. Elsevier Science, Amsterdam, The Netherlands. 213–244. [Google Scholar]
- 15.Holthuis J. C., and Menon A. K.. 2014. Lipid landscapes and pipelines in membrane homeostasis. Nature. 510: 48–57. [DOI] [PubMed] [Google Scholar]
- 16.Brites P., Waterham H. R., and Wanders R. J.. 2004. Functions and biosynthesis of plasmalogens in health and disease. Biochim. Biophys. Acta. 1636: 219–231. [DOI] [PubMed] [Google Scholar]
- 17.Braverman N. E., and Moser A. B.. 2012. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta. 1822: 1442–1452. [DOI] [PubMed] [Google Scholar]
- 18.Ebberink M. S., Mooijer P. A., Gootjes J., Koster J., Wanders R. J., and Waterham H. R.. 2011. Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Hum. Mutat. 32: 59–69. [DOI] [PubMed] [Google Scholar]
- 19.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., and Klenk D. C.. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150: 76–85. [DOI] [PubMed] [Google Scholar]
- 20.Houtkooper R. H., Rodenburg R. J., Thiels C., van Lenthe H., Stet F., Poll-The B. T., Stone J. E., Steward C. G., Wanders R. J., Smeitink J., et al. 2009. Cardiolipin and monolysocardiolipin analysis in fibroblasts, lymphocytes, and tissues using high-performance liquid chromatography-mass spectrometry as a diagnostic test for Barth syndrome. Anal. Biochem. 387: 230–237. [DOI] [PubMed] [Google Scholar]
- 21.Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., et al. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30: 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith C. A., Want E. J., O’Maille G., Abagyan R., and Siuzdak G.. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78: 779–787. [DOI] [PubMed] [Google Scholar]
- 23.Liebisch G., Vizcaíno J. A., Köfeler H., Trötzmüller M., Griffiths W. J., Schmitz G., Spener F., and Wakelam M. J.. 2013. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 54: 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaz F. M., Pras-Raves M., Bootsma A. H., and van Kampen A. H.. 2015. Principles and practice of lipidomics. J. Inherit. Metab. Dis. 38: 41–52. [DOI] [PubMed] [Google Scholar]
- 25.Abe Y., Honsho M., Nakanishi H., Taguchi R., and Fujiki Y.. 2014. Very-long-chain polyunsaturated fatty acids accumulate in phosphatidylcholine of fibroblasts from patients with Zellweger syndrome and acyl-CoA oxidase1 deficiency. Biochim. Biophys. Acta. 1841: 610–619. [DOI] [PubMed] [Google Scholar]
- 26.Dorninger F., Brodde A., Braverman N. E., Moser A. B., Just W. W., Forss-Petter S., Brügger B., and Berger J.. 2015. Homeostasis of phospholipids - the level of phosphatidylethanolamine tightly adapts to changes in ethanolamine plasmalogens. Biochim. Biophys. Acta. 1851: 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hama K., Nagai T., Nishizawa C., Ikeda K., Morita M., Satoh N., Nakanishi H., Imanaka T., Shimozawa N., Taguchi R., et al. 2013. Molecular species of phospholipids with very long chain fatty acids in skin fibroblasts of Zellweger syndrome. Lipids. 48: 1253–1267. [DOI] [PubMed] [Google Scholar]
- 28.Kulik W., van Lenthe H., Stet F. S., Houtkooper R. H., Kemp H., Stone J. E., Steward C. G., Wanders R. J., and Vaz F. M.. 2008. Bloodspot assay using HPLC-tandem mass spectrometry for detection of Barth syndrome. Clin. Chem. 54: 371–378. [DOI] [PubMed] [Google Scholar]
- 29.Moser A. E., Singh I., Brown F. R. 3rd, Solish G. I., Kelley R. I., Benke P. J., and Moser H. W.. 1984. The cerebrohepatorenal (Zellweger) syndrome. Increased levels and impaired degradation of very-long-chain fatty acids and their use in prenatal diagnosis. N. Engl. J. Med. 310: 1141–1146. [DOI] [PubMed] [Google Scholar]
- 30.Pettus B. J., Baes M., Busman M., Hannun Y. A., and Van Veldhoven P. P.. 2004. Mass spectrometric analysis of ceramide perturbations in brain and fibroblasts of mice and human patients with peroxisomal disorders. Rapid Commun. Mass Spectrom. 18: 1569–1574. [DOI] [PubMed] [Google Scholar]
- 31.Hubbard W. C., Moser A. B., Liu A. C., Jones R. O., Steinberg S. J., Lorey F., Panny S. R., Vogt R. F. Jr., Macaya D., Turgeon C. T., et al. 2009. Newborn screening for X-linked adrenoleukodystrophy (X-ALD): validation of a combined liquid chromatography-tandem mass spectrometric (LC-MS/MS) method. Mol. Genet. Metab. 97: 212–220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.