Abstract
Lipid droplets (LDs) are organelles that contribute to various cellular functions that are vital for life. Aside from acting as a neutral lipid storage depot, they are also involved in building new membranes, synthesis of steroid hormones, and cell signaling. Many aspects of LD structure and function are not yet well-understood. Here we investigate the interaction of perilipin 3, a member of the perilipin family of LD binding proteins, and three N-terminal truncation mutants with lipid monolayers. The interaction is studied as a function of surface pressure for a series of systematically chosen lipids. We find that the C terminus of perilipin 3 has different insertion behavior from that of the longer truncation mutants and the full-length protein. Inclusion of N-terminal sequences with the C terminus decreases the ability of the protein construct to insert in lipid monolayers. Coupling of anionic lipids to negative spontaneous curvature facilitates protein interaction and insertion. The C terminus shows strong preference for lipids with more saturated fatty acids. This work sheds light on the LD binding properties and function of the different domains of perilipin 3.
Keywords: amphipathic alpha-helices, lipid droplets, protein-lipid interaction
Lipid droplets (LDs) are dynamic cell organelles that carry out a multitude of cellular functions vital for life, and protein-lipid interactions are crucial to the structure and function of LDs. While much work has focused on peripheral and integral membrane proteins, the mechanism by which LD binding proteins recognize and target to LDs is still poorly understood (1). This is especially true for dedicated LD binding proteins of the perilipin family, i.e., perilipin 1 through 5, that function in the biogenesis and metabolism (lipolysis) of LDs. No work has directly investigated the interaction of a perilipin family member with a phospholipid monolayer interface. In order to address how this family of proteins interacts with lipid interfaces, we investigated the interaction of perilipin 3 with phospholipid monolayers at the air-buffer interface. We chose perilipin 3 because it is found in the cytosol as well as on the LD surface, and because previous work has characterized the structure and LD association of the protein (1–3).
In addition to providing cellular energy, LDs take part in many other cellular functions, including signal transduction, formation of new cellular membranes, hormone synthesis, and lipid trafficking (4–8). Under certain physiological conditions, LDs have been found to act as storehouses for several different types of enzymes and proteins, including histones (9, 10), and they also facilitate virus replication (11–13). An understanding of how proteins associate with the LD surface is crucial for our understanding of LD biology.
Although the composition of the LD interior might vary from cell to cell, the basic structure consists of a neutral lipid core of triacylglycerols and cholesterol esters surrounded by a phospholipid-protein monolayer (14, 15). The phospholipid composition of the monolayer surrounding the LDs has not been clearly defined, but appears to consist mostly of phosphatidylcholine (PC) with a series of other phospholipids, diacylglycerol, and cholesterol (16, 17).
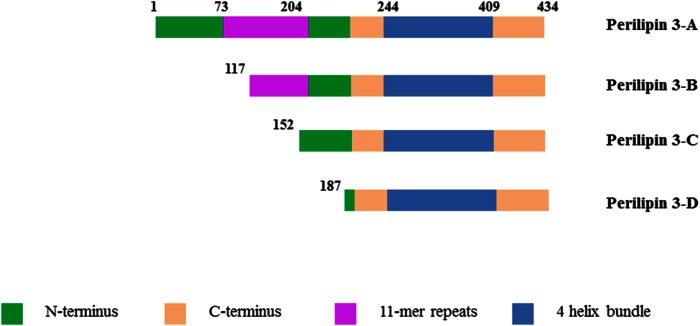
A major fraction of the proteins associated with the lipid monolayer belong to the perilipin family. The perilipins are synthesized on cytosolic ribosomes (18–20) and are thus sorted to the LD interface in a different manner than those proteins that originate from the endoplasmic reticulum, such as caveolin, for example (21). Sorting of caveolin is cooperatively mediated by a dual motif consisting of a hydrophobic sequence located close to a positively charged sequence (21). Targeting has been investigated for perilipins 1 through 5 in cellular studies and different regions of the proteins have been identified (1, 22–27). Based on currently available data, it appears likely that amphipathic sequences, especially the 11-mer repeat region of the proteins, target perilipins to LDs (1, 23, 28–31). The 11-mer repeat is a protein region with a repeating 11 amino acid pattern (32). These 11-mer repeats are found in the N terminus of all perilipins (see Fig. 1), with the largest such domain on perilipin 4 (33). The 11-mer repeats occur in other lipid binding proteins, such as α-synuclein and apoAI (34, 35). They form amphiphathic α-helices and have been ascribed a function in the reversible binding to lipid interfaces (34). However, the function of this domain in perilipin 3-lipid binding has not been explored in detail in model systems.
Fig. 1.
Domain model of the different protein constructs used in this study. Adapted from Hynson et al. (3) with the exception of the longer stretch of 11-mer repeats (see Discussion). Constructs are labeled for use in this paper. Perilipin 3D is the shortest construct, which is composed of a α/β domain and the amphipathic α-helix bundle. Perilipin 3C and 3B include parts of the 11-mer repeat region, while perilipin 3A refers to the full-length protein.
The mammalian perilipin family includes five distinct members that share sequence homology, especially in their N-terminal regions (20). The five mammalian proteins are perilipin 1, perilipin 2 (previously named adipophilin or ADRP), perilipin 3 (previously named TIP 47), perilipin 4 (previously named S3-12), and perilipin 5 (previously named OXPAT) (29, 36). Perilipins 1 and 2 are found exclusively on LDs and are classified as constitutively LD-associated proteins (18, 37–41), whereas perilipins 3, 4, and 5 circulate between LDs and the cytoplasm, and are therefore called exchangeable perilipin proteins (40, 42, 43). Some research suggests that perilipin 2 might also be found free in the cytosol (44, 45). The expression of perilipins 1 and 4 is confined to adipocytes and steroidogenic cells (18, 37–39, 41, 46), while perilipin 2 and perilipin 3 are ubiquitously expressed (19, 42, 43).
The structure of perilipin 3 has been investigated by two different groups. Hickenbottom et al. (2) resolved the crystal structure of the C-terminal region of murine perilipin 3 and revealed that it consists of an α/β domain and a bundle of four amphipathic α-helices. The helix bundle structure of the C terminus of perilipin 3 closely resembles the LDL receptor binding domain in the N-terminal region of apoE (2), and even shares sequence homology (47). Similar amphipathic α-helix bundle structures are found in other apolipoproteins, such as insect apolipophorin III (48). The function of this amphipathic α-helix bundle domain of perilipin 3 is unknown, and the detailed protein-lipid interactions have not been investigated (1).
Hynson et al. (3) recently studied the structure of full-length human perilipin 3 and showed that, as predicted by the crystal structure work, the protein has a highly helical C terminus in solution. This work additionally revealed a disordered N-terminal region and showed that the full-length protein assumes an extended conformation in solution, suggesting that the N and C termini might perform separate functions (3).
To shed light on the function of perilipin 3 and the mechanisms of lipid interaction, we studied the interaction of the C-terminal domain of perilipin 3 with model phospholipid monolayers at the air-buffer interface. Results are compared with those for two constructs that contain additional segments of the N-terminal portion of the protein, and those for the full-length protein (see Fig. 1). Model lipid monolayers formed at the air-buffer interface are especially useful model systems for the study of LD proteins. In vivo, LD proteins interact with a (phospho)lipid monolayer and the composition and physical properties of this monolayer can be made to closely resemble that surrounding the LD.
The Langmuir monolayer technique is a biophysical method frequently used to explore the lipid interaction of proteins and peptides, specifically protein insertion (49, 50).
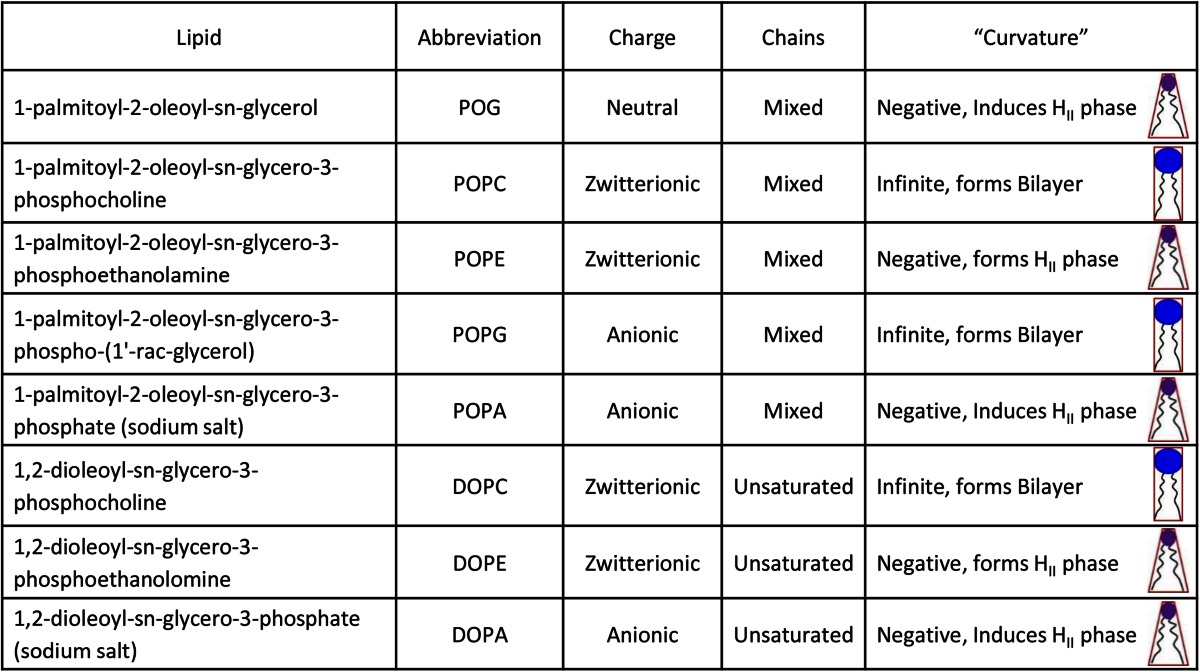
It must be stressed here that the lipids in this study are not necessarily those that occur naturally on the LD surface. Only limited data on the phospholipid composition of the LD monolayer is available, for a very limited set of cell types (16, 17). Instead, we chose those lipids with various properties that might, based on our previous work (50), influence the interaction of perilipin 3 with the phospholipid monolayer in vivo: spontaneous curvature (i.e., curvature stress of the monolayer, and access to hydrophobic interior of the LD) and charge.
We find that all constructs of perilipin 3 are surface active. The more of the protein N terminus that is included, the more surface active the construct is, as assessed from changes in the surface pressure of the self-assembled protein monolayer. The C-terminal α-helix bundle domain of perilipin 3 has markedly different insertion properties as compared with the full-length protein. Inclusion of more N-terminal sequences from the full-length protein reduces the maximum lipid monolayer pressure at which the protein construct remains able to insert into the lipid monolayer. At the same time, the affinity for the lipid monolayer is increased. The C terminus of perilipin 3 shows a high affinity for saturated fatty acids, as evidenced by a reduced insertion into a lipid monolayer containing unsaturated fatty acids. Taken together, our results suggest that the C terminus of perilipin 3 carries out distinct functions compared with the N terminus, which is dominated by the highly amphipathic 11-mer repeat region.
MATERIALS AND METHODS
Materials
The 1-palmitoyl-2-oleoyl-sn-glycerol (POG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (sodium salt) (POPA), and 1,2-dioleoyl-sn-glycero-3-phosphate (sodium salt) (DOPA) were purchased from Avanti Polar Lipids (Alabaster, AL). NaCl, EDTA, and Tris were purchased from Sigma-Aldrich. All of the chemicals were >99% pure. Chemicals for protein purification were purchased from Amresco. HPLC grade water was purchased from Fisher Scientific.
Protein expression and purification
In order to address our main question of what drives the interaction between perilipin 3 and a (phospho)lipid monolayer, we used four constructs (shown in Fig. 1) of the human protein (2, 3). Both Hynson et al. (3) and Hickenbottom et al. (2) showed that the C-terminal portion of perilipin 3 forms a well-defined structure in solution and can be readily expressed and purified. Additionally, the presence of the C terminus of the protein appears to be critical for high yield (∼15 mg pure protein from a 1 l culture) required for the biophysical work we carried out here. Production of large amounts (generally 10–15 mg/l cell culture) of proteins was carried out according to Hynson et al. (3). Briefly, pETHSUL vectors encoding full-length human perilipin 3 (AA1-434) and three N-terminal truncation mutants (AA117-434, 152-434, and 187-434) were expressed in Escherichia coli BL21 (DE3) Codon + RIL cells (Stratagene, La Jolla, CA) as fusion proteins carrying N-terminal His6-small ubiquitin-related modifier (SUMO) tags. Five milliliters of LB medium containing 100 μg/ml ampicillin was inoculated with a scraping from a glycerol stock and was grown at 37°C and 250 rpm for 8 h in an incubator (Innova 43 incubator shaker series; New Brunswick Scientific). One milliliter of this preculture was introduced into 1 l of auto-induction medium (51) containing 100 μg/ml ampicillin. The culture was allowed to grow for 16 h at 30°C, after which the cells were pelleted by centrifugation at 8,700 g for 7 min. The cell pellets were then resuspended in 20 ml of buffer A [50 mM HEPES (pH 7.5), 500 mM KCl, 10 mM imidazole, 10% glycerol, and 5 mM β-mercaptoethanol with an EDTA-free protease inhibitor tablet (Roche Life Sciences)]. The cells were then lysed by seven cycles of freezing and thawing followed by probe sonication (QSonica sonicator; 20 cycles of 15 s with 20 s on ice at 60% amplitude). The cell lysate was centrifuged at 29,000 g for 45 min at 4°C, and the supernatant was collected. The supernatant was first loaded onto a nickel-NTA column connected to a Bio-Rad Biologic LP system. The column was pre-equilibrated with buffer A and the bound proteins were eluted with 250 ml of a 10–500 mM gradient of imidazole. Five milliliter fractions were collected and tested for the presence of protein by SDS-PAGE. To cleave the His6-SUMO tag from the proteins, the pooled peak fractions from the first nickel column were added with 1 mM EDTA and 10 μg of an in-house made recombinant His6-tagged form of the catalytic domain of (dtUD1) SUMO hydrolase from Saccharomyces cerevisiae [see (52)]. The cleavage reaction was carried out for 4 h at 18°C and the mixture was then dialyzed overnight at 4°C in 2 l of buffer B [25 mM Tris-HCl (pH 8.0) and 5 mM β-mercaptoethanol]. After dialysis, the solution containing the cleavage mixture was purified on a second nickel column pre-equilibrated with buffer B. The flow through of this second column contained the cleaved perilipin 3 protein whereas His-tagged hydrolase enzyme, His6-SUMO tags, as well as any uncleaved proteins are retained by the Ni resin. Next, the resulting flow through was loaded onto a Q-Sepharose column pre-equilibrated with buffer B. The bound proteins were eluted with 250 ml of 0–100 mM gradient of KCl. The peak fractions were pooled and concentrated by filter centrifugation (Macrosep Advanced Centrifugal Devices, 10MWCO, PALL Corporation). The concentrated protein was then loaded onto a Superdex- 75 size-exclusion chromatography column (GE Healthcare) connected to a FPLC system and eluted in a buffer containing 50 mM Tris-HCl, 250 mM NaCl, and 2.5 mM Tris(2-carboxyethyl)-phosphine (pH 8.0).
The purified protein samples were sequenced and identified through mass spectroscopy at the proteomics core of the Cleveland Clinic (Learner Research Institute Proteomics Laboratory, Cleveland Clinic Foundation). After purification by four columns, three of our constructs were highly pure (≥95%) and one was ∼85% pure, and considered suitable for our biophysical characterization studies (see supplemental Fig. S1). The proteins could be stored (storage buffer: 50 mM Tris, 250 mM NaCl, 2.5 mM Tris(2-carboxyethyl)-phosphine, and 10% glycerol buffer at pH 8) at −80°C for about 6 months with no apparent degradation (the C-terminal domain of perilipin 3 was the most stable, data not shown).
Monolayer insertion experiments
The monolayer insertion experiments followed the procedure of Demel, van Doorn, and van der Hoorst (53). Briefly, the set-up consisted of a circular Teflon trough enclosed within a Plexiglas enclosure placed on a vibration isolation table. The trough had an inner diameter of 3 cm and a 5 cm outer diameter, and was 0.5 cm deep with space in the middle for a small magnetic stirrer bar (1 cm diameter × 0.5 cm deep). In the upper rim (1 cm) was a port through which protein was injected below the monolayer without disturbing the lipid monolayer. The trough was filled with 6.5 ml of Tris buffer (pH 7.2) (10 mM Tris, 150 mM NaCl, and 0.1 mM EDTA): the subphase for the experiment. Note that other common biological buffers (e.g., HEPES) and reducing agents (e.g., DTT and β mercaptoethanol) affect the surface pressure of lipid monolayers and should thus be avoided. A lipid monolayer was formed by dropwise addition of lipids (0.1 mM) dissolved in chloroform/methanol (2:1 vol ratio) until the desired surface pressure was reached. The solvent was allowed to evaporate for about 10 min until a stable pressure was reached, and protein was injected into the subphase through a port in the side of the trough. The monolayer experiments were carried out in a temperature-controlled room at 22.0 ± 1.0°C, and the subphase was continuously stirred with a magnetic bar to ensure proper mixing of the proteins in the subphase. All glassware, the Teflon trough, and magnetic stirrer were cleaned thoroughly before each monolayer experiment by a wash with KOH cleaning solution (164 g of ethanol, 24 g of water, and 25 g of KOH) and several rinses with deionized water before final rinsing with ultrapure water (HPLC grade water, catalog number W5-4; Fisher Scientific). The platinum Wilhelmy plate was rinsed with distilled water, KOH cleaning solution, and finally ultrapure water.
RESULTS
Surface activity of perilipin 3
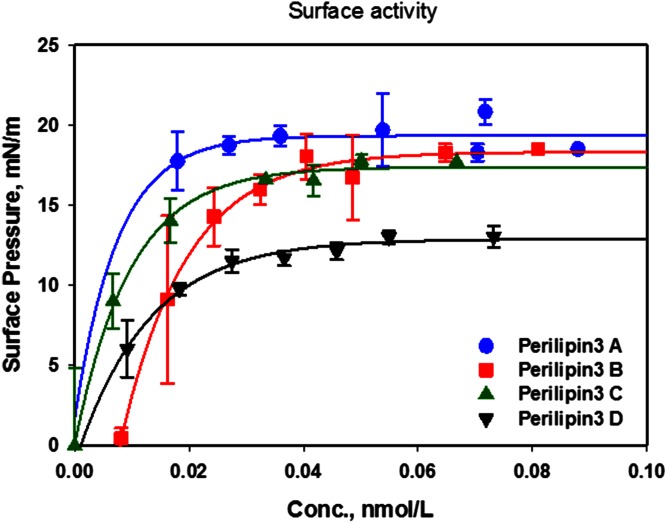
We first determined the surface activity of the amphipathic α-helix bundle of perilipin 3 (named perilipin 3D), followed by two additional N-terminal truncation mutants and finally the full-length recombinant protein (see Fig. 1). This surface-activity determination is an important control. If the change in lipid monolayer pressure, after insertion of the protein into the interface, is not larger than the surface pressure supported by the self-assembled protein alone, then it is possible that the protein will not insert into (i.e., in between) the lipid monolayer at all. It may, instead, simply form patches of protein at the interface, in equilibrium with a more-compressed lipid monolayer.
Figure 2 shows the increase in surface pressure of a bare buffer interface as a function of the bulk protein concentration. As expected, the amphipathic α-helix bundle of perilipin 3 (perilipin 3D) is highly surface active with a final surface pressure of ∼12 mN/m for the self-assembled [Gibbs (54)] monolayer. This is comparable to the maximum surface pressure reached by the self-assembled monolayer of the insect protein, apoLp-III, ∼14 mN/m, which consists entirely of an α-helix bundle that, in structure, resembles that of the C terminus of perilipin 3 (50). Addition of further domains from the N terminus (see Fig. 1) to the α-helix bundle of perilipin 3 increases the surface activity of the protein, such that the full-length protein (perilipin 3A) has the highest surface pressure of ∼19 mN/m. It is interesting to note here that addition of 35 amino acids to the sequence of the α-helix bundle essentially maximizes the surface activity. These data also show that a subphase concentration of 0.04 μM is required to maximize the surface pressure of the monolayer in our particular set-up. In order to assure that we used sufficient protein to maximize interaction with the lipid monolayers investigated, we used a subphase concentration of at least 0.07 μM in all subsequent experiments.
Fig. 2.
Surface activity of perilipin 3 and its truncation mutants. Increase in surface pressure of self-assembled monolayers of perilipin 3A–D as a function of subphase concentration (Conc.). Proteins were inserted beneath an air-buffer interface and surface pressure was allowed to equilibrate. Values shown are averages of three independent experiments and error bars represent the standard deviation. Buffer subphase: 10 mM Tris-HCl, 150 mM NaCl, and 0.2 mM EDTA at pH 7.2.
Perilipin 3D behaves similarly to apoLp-III, a model α-helix bundle protein
Next, we determined the interaction of the α-helix bundle of perilipin 3, perilipin 3D, with model lipid monolayers. The lipids we used in this study are listed in Table 1 together with the relevant biophysical properties. Specifically, these lipids were chosen such that they had the same acyl chain composition but differed in the headgroup (i.e., size, hydration, and charge), resulting in lipids that spanned a range of relevant physicochemical properties. The sn-1-palmitoyl and sn-2-oleoyl acyl chains were chosen, as they are representative of mammalian glycerophospholipid fatty acid composition (55).
TABLE 1.
Physical chemical properties of lipids used in this study
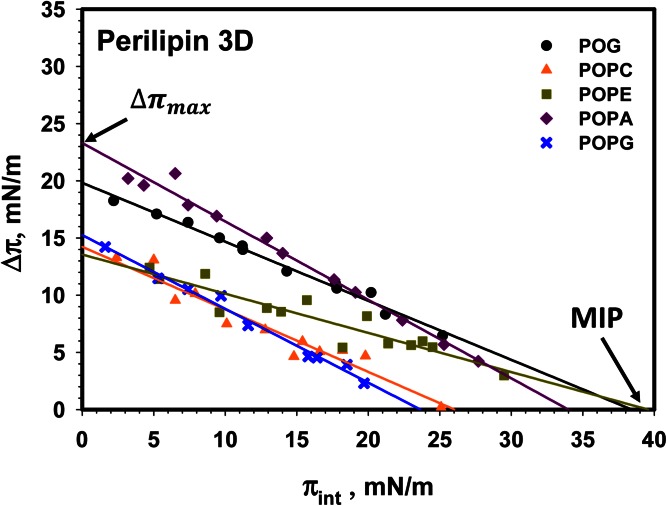
Figure 3 shows the cumulative data for the interaction of perilipin 3D with monolayers of POG, POPC, POPE, POPA, and POPG. Representative insertion kinetics are shown in supplemental Fig. S2. The resulting change in surface pressure of the lipid monolayer is plotted as a function of initial lipid monolayer pressure. Note that generally the Δπ values are larger than the surface pressure of the protein at the air-buffer interface alone (compare Figs. 2, 3). A possible exception is POPC, where the pressure of the resulting protein-lipid monolayer is comparable with that of the protein alone (see the discussion of acyl chain saturation below).
Fig. 3.
Insertion isotherms for perilipin 3D for a series of (phospho)lipids. Comparison of the insertion of perilipin 3-D into monolayers of POG (black circle), POPC (orange triangle), POPE (green square), POPA (purple diamond), and POPG (blue cross). MIP and Δπmax values are shown in Table 2. Buffer subphase: 10 mM Tris-HCl, 150 mM NaCl, and 0.2 mM EDTA at pH 7.2.
We extracted two key quantities from these insertion isotherms, namely the maximum insertion pressure (MIP) and the maximal change in monolayer pressure (Δπmax). The MIP is the lipid monolayer pressure at which the protein is no longer able to insert in between the lipids in the monolayer [see Fig. 3 and (49)]. This pressure has also been referred to as the exclusion pressure. The MIP value, therefore, is an indication of the propensity of insertion of a given protein into a given lipid monolayer. MIP values above 30–35 mN/m are used to indicate proteins with propensity to insert into a lipid bilayer (56, 57). The Δπmax is the maximal change in lipid monolayer pressure after protein insertion and is taken as the intercept of the change in lipid monolayer pressure axis at zero lipid monolayer pressure (see Fig. 3). This value is also used as an indication of affinity of the protein for the lipids in the monolayer.
The MIP and Δπmax values for perilipin 3D are shown in Table 2. The 95% confidence intervals for these values are calculated according to (49). We note that the insertion of the α-helix bundle of perilipin 3 is highest for POG and POPE, both of which have negative spontaneous curvature, and for POPA, which has both negative curvature and a negative charge (58–60). Note that the Δπmax for POPE is much reduced compared with POG and POPA, suggesting that the affinity of the helix bundle domain for POPE is limited. Interestingly, addition of a negative charge (POPA) significantly enhances the interaction with phospholipid monolayers, but negative charge alone is not sufficient (compare POPG, also negatively charged, to POPC and POPA). Only the MIPs for POG and POPE are sufficiently above the 30–35 mN/m typical of glycero(phospho)lipid bilayers (56) to suggest that this domain can interact with such hypothetical bilayers. Note that POG or POPE alone do not form fluid lipid bilayers under physiological conditions.
TABLE 2.
Maximum change in monolayer pressure Δπmax, and MIP data derived from data shown in Fig. 3 for the amphipathic α helix bundle of perilipin 3 (perilipin 3D)
| Perilipin 3D | Δπmax (mN/m) | MIP (mN/m) |
| POG | 19.8 ± 0.6 | 38.1 ± 1.4 |
| POPC | 14.2 ± 1.3 | 26.0 ± 0.5 |
| POPE | 13.6 ± 1.4 | 39.6 ± 0.4 |
| POPA | 23.3 ± 0.8 | 34.0 ± 1.2 |
| POPG | 15.3 ± 0.8 | 23.7 ± 1.2 |
TABLE 3.
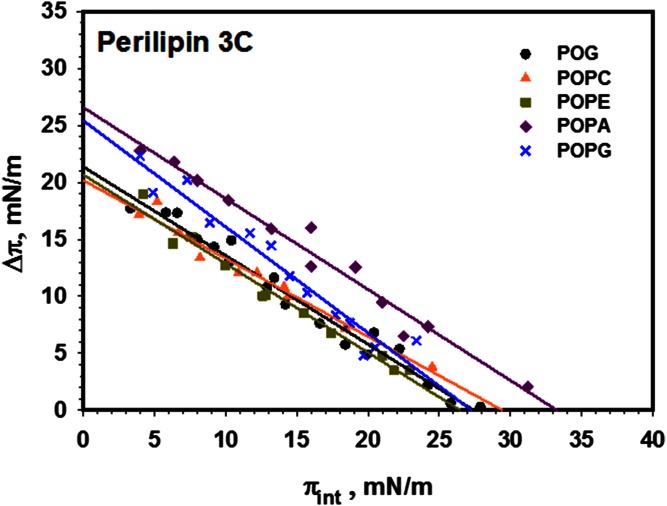
Maximum change in monolayer pressure Δπmax, and MIP data derived from data shown in Fig. 4 for construct perilipin 3C
| Perilipin 3C | Δπmax (mN/m) | MIP (mN/m) |
| POG | 21.4 ± 1.1 | 27.4 ± 0.6 |
| POPC | 20.2 ± 1.1 | 29.4 ± 0.7 |
| POPE | 20.7 ± 1.2 | 26.5 ± 1.0 |
| POPA | 26.6 ± 1.5 | 32.3 ± 0.7 |
| POPG | 25.4 ± 2.0 | 27.3 ± 0.8 |
Addition of further domains of the N terminus results in a reduced MIP and an increase in Δπmax
Next, we investigated the effect of addition of a small (35 amino acids, 152 through 187) portion of the N terminus to perilipin 3D (perilipin 3C). Figure 4 shows the resulting insertion isotherms and indicates a significant increase in the affinity of the protein for the lipid monolayer (higher Δπmax), consistent with the increase in surface activity of the protein (see Fig. 2). Interestingly, the addition of negative charge to the lipid monolayer significantly increases the affinity of the protein for the lipids (POPA and POPG), but the MIP is only increased (with respect to PC) for POPA, which has negative spontaneous curvature. Representative insertion kinetics are shown in supplemental Fig. S3.
Fig. 4.
Insertion isotherms for perilipin 3C for a series of (phospho) lipids. Comparison of the insertion of perilipin 3C into monolayers of POG (black circle), POPC (orange triangle), POPE (green square), POPA (purple diamond), and POPG (blue cross). MIP and Δπmax values are shown in Table 3. Buffer subphase: 10 mM Tris-HCl, 150 mM NaCl, and 0.2 mM EDTA at pH 7.2.
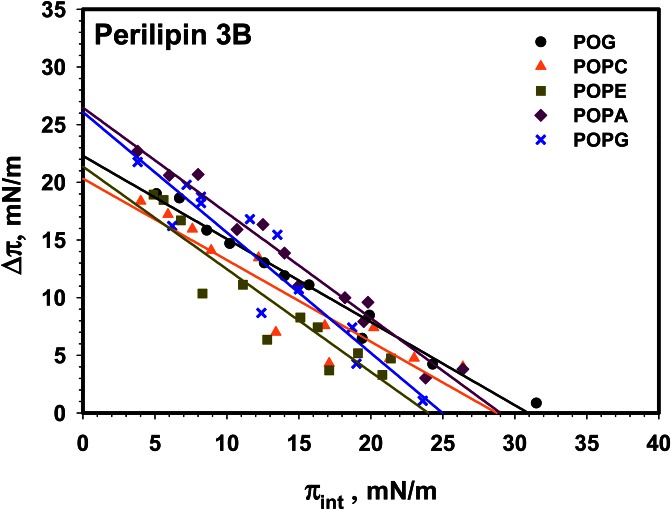
Addition of the 11-mer repeat region to the protein, to yield perilipin 3B, does not significantly alter its monolayer interaction except that it appears that POG has the highest MIP, compared with POPA, for perilipin 3C (see Fig. 5). It should be noted that the spread in the POG data at higher initial pressure is significant, resulting in a larger confidence interval. Representative insertion kinetics are shown in supplemental Fig. S4.
Fig. 5.
Insertion isotherms for perilipin 3B for a series of (phospho)lipids. Comparison of the insertion of perilipin 3B into monolayers of POG (black circle), POPC (orange triangle), POPE (green square), POPA (purple diamond), and POPG (blue cross). MIP and Δπmax values are shown in Table 4. Buffer subphase: 10 mM Tris-HCl, 150 mM NaCl, and 0.2 mM EDTA at pH 7.2.
TABLE 4.
Maximum change in monolayer pressure Δπmax, and MIP data derived from data shown in Fig. 5 for construct perilipin 3B
| Perilipin 3B | Δπmax (mN/m) | MIP (mN/m) |
| POG | 22.3 ± 1.2 | 31.0 ± 0.5 |
| POPC | 20 ± 3 | 29.4 ± 0.4 |
| POPE | 21 ± 3 | 24.0 ± 0.2 |
| POPA | 26.5 ± 1.7 | 29.0 ± 0.7 |
| POPG | 26 ± 3 | 25.0 ± 1.0 |
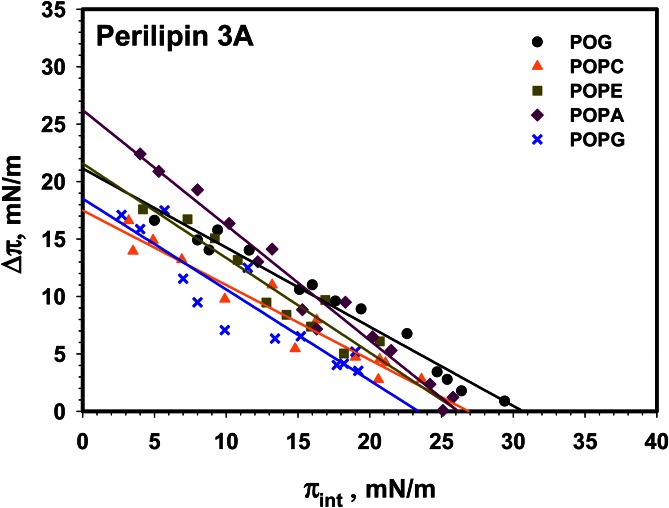
Like perilipin 3B, the full-length protein, perilipin 3A, does not show major differences in lipid insertion compared with perilipin 3C and 3B (see Fig. 6). Representative insertion kinetics are shown in supplemental Fig. S5.
Fig. 6.
Insertion isotherms for perilipin 3A (full-length protein) for a series of (phospho)lipids. Comparison of the insertion of full-length perilipin 3 into monolayers of POG (black circle), POPC (orange triangle), POPE (green square), POPA (purple diamond), and POPG (blue cross). MIP and Δπmax values are shown in Table 5. Buffer subphase: 10 mM Tris-HCl, 150 mM NaCl, and 0.2 mM EDTA at pH 7.2.
TABLE 5.
Maximum change in monolayer pressure Δπmax, and MIP data derived from data shown in Fig. 6 for construct perilipin 3A
| Perilipin 3A | Δπmax (mN/m) | MIP (mN/m) |
| POG | 21.1 ± 1.3 | 30.7 ± 0.5 |
| POPC | 17.5 ± 1.6 | 26.9 ± 0.4 |
| POPE | 22 ± 3 | 26.2 ± 1.2 |
| POPA | 26.2 ± 1.7 | 26.2 ± 0.5 |
| POPG | 19 ± 3 | 23.4 ± 0.2 |
Perilipin 3 prefers saturated acyl chains
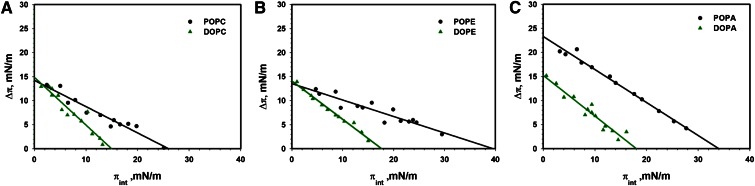
Our previous work on apoLp-III, a model amphipathic α-helix bundle protein, suggested that this protein prefers phospholipids with saturated acyl chains (50). We thus investigated the interaction of perilipin 3D with the unsaturated lipids, DOPC, DOPE, and DOPA. Figure 7A compares the results for DOPC and POPC (POPC data replicated from Fig. 3). Here it should be noted that it took a considerable time for perilipin 3D to start inserting into the DOPC monolayer (see supplemental Fig. S6 for insertion kinetics). This was previously observed for POPC (supplemental Fig. S2), but for DOPC this observation was especially noticeable. The data for DOPC is thus also taken at 40 min after protein insertion compared with 30 min for all other lipids. The MIP for perilipin 3D is significantly (>10 mN/m) lower for DOPC compared with POPC.
Fig. 7.
Insertion isotherms for perilipin 3D in saturated and unsaturated phospholipids. Comparison of the insertion isotherms of perilipin 3D into a lipid monolayer of POPC and DOPC (A), POPE and DOPE (B), and POPA and DOPA (C). The change in lipid monolayer pressure was recorded after 30 min, except for DOPC where data was recorded after 40 min due to the slow insertion kinetics of the protein. Note the large difference in MIP for the more saturated (PO) and unsaturated (DO) lipids. Buffer subphase: 10 mM Tris-HCl, 150 mM NaCl, and 0.2 mM EDTA at pH 7.2
Because our previous work showed that negative spontaneous curvature affects protein insertion (see Figs. 3–6), we next compared the insertion of perilipin 3D into monolayers of POPE and DOPE (Fig. 7B; data for POPE taken from Fig. 3). In this case as well, the PO lipid shows significantly more insertion of the amphipathic α-helix bundle, with the MIP for POPE ∼22 mN/m higher than for DOPE. Figure 7C compares the interaction of perilipin 3D with monolayers of POPA and DOPA. Again, we observe significantly more (MIP >15 mN/m) insertion for monolayers of the more saturated lipid.
DISCUSSION
For the lipid monolayers, we utilized single lipids that systematically varied in physical chemical properties (charge, shape, hydration, acyl chain saturation, etc.), as shown in Table 1. The systematic variation of these properties allowed us to identify which physical and chemical properties of the lipid interface were important for perilipin 3 interaction. The use of our different constructs also allowed us to make conclusions on the role of the different perilipin 3 domains in lipid interaction.
The amphipathic α-helix bundle of perilipin 3 inserts more strongly into a phospholipid monolayer than the full-length protein
Our results show that the C-terminal domain (consisting of an α/β domain and 4-helix α-helix bundle) shows the largest insertion, i.e., highest MIP, into monolayers composed of lipids with negative spontaneous curvature, namely POG, POPE, and POPA (see Fig. 3). Lipids with negative spontaneous curvature are known to facilitate the interaction of peripheral membrane proteins, as the asymmetry in the headgroup area (small) and acyl chain area (larger; see Table 1) facilitates insertion of hydrophobic domains into the hydrophobic interior of the membrane bilayer (61, 62). A similar mechanism appears to be at play for the C terminus of perilipin 3 (perilipin 3D), where the negative curvature stress in the monolayer of POG, POPE, and POPA allows for further insertion of amino acids into the hydrophobic interior of the lipid monolayer. In the case of LDs, this may result in the interaction of the protein with the neutral lipids inside the droplet. Addition of further segments of the full-length protein to the C terminus (resulting in perilipin 3B, perilipin 3C, and finally the full-length protein) results in a dramatic decrease in MIP (>10 mN/m) for both POG and POPE, suggesting that the additional segments of the protein [i.e., regions of the N terminus, including the 11-mer repeats (see below)] results in less insertion of the protein into a phospholipid monolayer of neutral or zwitterionic lipids.
This is a puzzling finding because, energetically, the higher MIP value for the C terminus suggests that its interaction is more favorable than that of the full-length protein, even though the full-length protein still contains the C terminus. What could explain such perplexing behavior? One possibility is the following: The addition of N-terminal regions of the protein to the C terminus might sterically hinder the interaction of the C terminus with the phospholipid monolayer. The region between the α/β domain and 4-helix α-helix bundle is designated as the hydrophobic cleft (2). This region was suggested to be involved in lipid binding and it is possible that addition of N-terminal sequences close to this region affects the folding and rearrangement of the amphipathic α-helix bundle. Yet, in vivo, the hydrophobic cleft was shown to be dispensable for LD targeting (1). Another possibility is that, instead of steric hindrance, the addition of amino acids from the N terminus (35 amino acids are sufficient to essentially maximize the effect) increases the affinity of the C terminus for the aqueous phase. Clearly this finding needs to be further explored.
The exception to the decrease in MIP observed for POG and POPE is the lipid POPA, which, in addition to having negative spontaneous curvature, carries a negative charge in its headgroup. The charge of PA is usually considered to be −1 in membranes, but can easily reach −2 under favorable environmental conditions [pH, presence of hydrogen bond donors, e.g., the lipid PE (58, 63)]. The charge of PA will also be stabilized at −2 when it interacts with basic amino acid residues via the electrostatic hydrogen bond switch model (63). The preference for insertion of perilipin 3 into a PA monolayer does not diminish (surface pressure stays around 34 mN/m) with the addition of 35 amino acid residues to perilipin 3D (perilipin 3C, see Fig. 4). The MIP for perilipin 3B and the full-length protein (perilipin 3A) does reduce to 29 and 26 mN/m, respectively, for the POPA monolayer (see Figs. 5, 6). The effect of negative charge is also reflected in the Δπmax values, qualitative measures of lipid affinity that are highest for POPA for all four constructs investigated.
Negative charge influences the recruitment of perilipin 3 to the LD surface
The effect of charge is not novel in LD-associated proteins. For caveolins, it was shown that a cationic domain close to a hydrophobic region is required to sort the protein to LDs in the endoplasmic reticulum (21). Attaching these domains to non-LD binding proteins conferred to them LD localization (21).
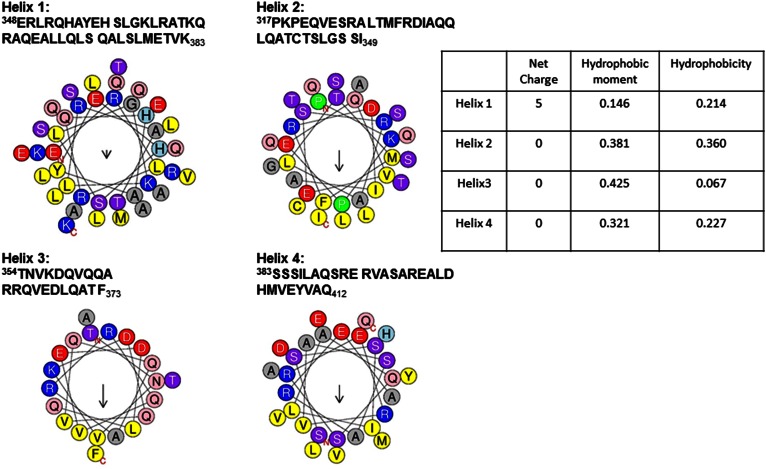
How can the effect of negative charge be explained for perilipin 3? Because all constructs show sensitivity to anionic lipids, we hypothesized that cationic residues in one or more of the amphipathic α-helices of the C-terminal domain of perilipin 3 facilitated the electrostatic interaction of the protein with the monolayer. Figure 8 shows the amino acid sequence and helical wheel representation (64) of all four helices [alignment taken from Hickenbottom et al. (2)] in the α-helix bundle of human perilipin 3, together with the calculated value for the hydrophobicity and hydrophobic moment of each of these helices (64). The first α-helix of the bundle is highly charged and is, in fact, not amphipathic (see Fig. 8). It contains nine cationic and four anionic amino acid residues, whereas the additional helices show an equal or lower number of cationic and anionic residues. In solution at neutral pH, helix 1 is likely to have a net charge of +3, as the two histidine residues are likely to be neutral (depending on local electrostatic environment in the protein structure and solution pH).
Fig. 8.
Helix wheel representation of the four α-helices in the amphipathic helix bundle (C terminus) of perilipin 3. To test for electrostatic and hydrophobic interactions, each helix is graphed separately. Note that only the first helix has a net (positive) charge, while the other three are neutral. In helices 2, 3, and 4, positively charged amino acids (R and K, R and R, and R and R, respectively) bound a hydrophobic lower face.
We propose that this substantial charge facilitates the initial electrostatic interaction with the lipid monolayer. This charge can potentially increase to +9 when we consider the following: An anionic lipid interface has a local surface pH that is considerably (potentially by 2 pH values or more) lower than that found in the bulk solution (65). At low pH the histidine residues in helix 1 will be positively charged and the anionic residues, aspartate and glutamate, might protonate and become neutral. The fact that histidine residues can act as a switch in local protein structure upon interaction of lipid binding domains to an anionic lipid interface is well-known (66, 67), but the likelihood of aspartate and glutamate protonation is often overlooked (B. de Kruijff, personal communication). While helix 1 of the amphipathic α-helix bundle carries the largest concentration of charge in the C terminus (again it is barely amphipathic), the additional helices are considerably more amphipathic, as evidenced by larger positive values of the hydrophobic moment (Fig. 8). However, protonation of aspartate and glutamate residues in the α-helices of the helix-bundle near anionic lipid interfaces can potentially contribute to the effect of negatively charged lipids on perilipin 3 binding to a lipid interface.
Negative charge and negative spontaneous curvature both contribute to lipid binding for the C terminus of perilipin 3
For the C-terminal domain, negative charge is not the only driving factor for monolayer insertion, as the MIP and Δπmax for POPG (anionic) are identical to those for POPC (zwitterionic), and both lipids have very small spontaneous curvatures (i.e., induce essentially no negative curvature stress in a lipid monolayer). Coupled with the results for POPA, POPE, and POG, this suggests that negative spontaneous curvature is important as well. This is also exemplified by the MIP and Δπmax values for the full-length protein (perilipin 3A, see Fig. 6), where the Δπmax is highest for the anionic POPA (which combines both negative charge and negative spontaneous curvature), but the MIP is significantly higher for neutral POG, although both induce negative curvature stress.
While the effect of positive charge (highest Δπmax value) in the protein is largest for POPA (constructs perilipin 3B and 3C also show higher Δπmax for POPG), hydrophobic interactions, as exemplified by the role of negative spontaneous curvature, are thus important as well. While LDs are mainly covered by a PC monolayer, it is known that PC interfaces have a slight negative charge [PC vesicles show a negative zeta potential (68)]. Furthermore, the structure of the lipid monolayer surrounding LDs is not known and it is likely that patches of neutral lipids become exposed, at least part of the time, to the aqueous environment (69). Oil (hydrophobic) interfaces in water are known to be negatively charged due to the accumulation of hydroxyl ions from solution (70). We demonstrated this previously in a study on oil droplet fusion. Pure triolein droplets in water do not want to fuse [supplemental data to (71)] and screening by salt dramatically reduced the docking time (time before fusion takes place) of two triolein droplets. The role of these observations for the interaction of perilipin 3 with LDs is unknown.
Recruitment and targeting of perilipin 3
Multiple studies have addressed the recruitment and targeting of perilipins to the LD surface. Cells expressing perilipin 1, 2, and 3 show perilipin 3 localized on small droplets at the cell periphery. Increasingly larger LDs become coated with perilipin 2, and eventually the largest LDs at the center of these cells contain predominantly perilipin 1 (40). The reason for this distribution of perilipins is unknown, but may be due to maturation of LDs and movement to the cell center. LDs are dynamic organelles and much is still unclear concerning their lipid composition and how this changes during the lifecycle of a single droplet.
There is no universal agreement on the exact region (amino acid sequence) on perilipins required for recruitment and targeting to the LD surface. However, an overview of the literature suggests that amphipathic α-helices in the N terminus of the protein are necessary and sufficient (1, 25, 28). This region overlaps with the so-called 11-mer repeat region. The 11-mer repeats found in the N terminus of perilipin 3 are imperfect, in that the sequence homology within the repeating sequences is not as conserved as in the repeats found in perilipin 4 (33).
Different authors have indicated different stretches of amino acids as belonging to these so-called 11-mer repeats (2, 28). Careful examination of the perilipin 3 sequence suggests that the imperfect 11-mer repeats span the entire region from amino acid 73 to amino acid 204. Supplemental Fig. S7 shows the helical wheel representations (assuming that this part of the protein indeed folds into α-helices) of this region, represented as a perfect α-helix as well as a 3/11 α-helix (3 full turns per 11 amino acids). We show both of these conformations because the amphipathic α-helices of the 11-mer repeats in apolipoproteins and α-synuclein were proposed to adopt this 3/11 conformation (34). Comparison of the hydrophobic moment of these helices with those of the α-helix bundle shows that they are comparable. What is striking is the presence of four proline residues in the first part of the 11-mer repeat-like region. The role of these residues is unknown, but is likely to affect the secondary structure of this segment of the protein. If folded in an amphipathic helix or helices, most of the N terminus could potentially interact with a (phospho)lipid monolayer via hydrophobic interactions. Indeed, we now show that the Δπmax (a qualitative measure for the affinity of the protein for the lipid monolayer) increases upon addition of N-terminal sequences (perilipin 3C, perilipin 3B, and the full-length protein). This agrees with the cell-based studies that show that the N terminus is necessary and sufficient for LD targeting and binding (1, 25, 28). For example, compare the insertion isotherms (Δπmax) for perilipin 3D with those of perilipin 3C and 3B. Interestingly, inclusion of the entire N terminus, i.e., the so-called PAT domain (perilipin 3B and 3C compared with the full-length protein) decreases Δπmax again, at least for most of the lipids investigated (the only exception to this decrease in Δπmax is the lipid POPA, which combines negative charge with negative spontaneous curvature, i.e., access to the hydrophobic interior of the droplet.) The PAT domain was named after the founding members of the perilipin family, perilipin 1 through 3, and is based on the old names for these proteins. The name PAT-domain has since been used for the N terminus of these proteins and constitutes a region of ∼100 amino acids that is conserved among the perilipin family members, except for perilipin 4 (40, 72). In human perilipin 3, the PAT domain is located between amino acid 21 and amino acid 120, and thus includes a significant part of the 11-mer repeat region (see Fig. 1 and discussion above) (72, 73).
The function of the C- and N-terminal domain of perilipin 3 is mostly unknown. Our data clearly suggest that each of these domains contributes separately and independently to lipid binding. The N terminus with net positive charge (see above and Fig. 8) is likely responsible for initial recruitment to negatively charged lipid interfaces. Once the initial interaction commences, it is the N terminus that takes over lipid binding and firmly anchors the protein to the phospholipid-oil interface of the LD. Because the MIP of the full-length protein is so much lower than the MIP of perilipin 3D for POG, POPA, and POPE, one possible scenario is that as soon as the N terminus of the protein interacts, the C-terminal α-helix bundle detaches from the LD surface. Another possibility is that the α-helix bundle is easily displaced from this surface by additional perilipin 3 molecules that bind. Our data also suggests that the full-length protein, while electrostatically attracted to anionic lipid interfaces, does not bind to other membranes inside the cell, as the MIP is lower than that expected for a lipid bilayer (56, 57).
Role of phospholipid saturation on perilipin 3 LD interaction
Previously, we showed that the insertion of the amphipathic α-helix bundle protein, apoLp-III from Locusta migratoria, is sensitive to the saturation of lipid acyl chains. Replacing the palmitic acid from the sn-1 position with an oleic acid decreased the insertion of apoLp-III for each of the lipids investigated (50). We now show that the helix bundle domain of perilipin 3 (perilipin 3D) is also acutely sensitive to the saturation of the phospholipid acyl chain. The di-oleoyl lipids, DOPC, DOPE, and DOPA, showed a dramatic decrease of MIP by more than 10 mN/m compared with the PO species. This decrease is much more significant than what we observed for apoLp-III.
What are the biological implications of these findings? Very little is known about the phospholipid composition of the lipid monolayer surrounding LDs (16, 17). What has been published shows great differences in lipid composition. Tauchi-Sato et al. (17) show that LDs contain a significant fraction of lyso-lipids, something not observed for biomembranes (74). On the other hand, Storey et al. (16) show that LDs containing mainly perilipin 1 have phospholipids with more saturated fatty acids, while LDs containing mainly perilipin 2/perilipin 3 have phospholipids with more unsaturated fatty acids. The differences between these two studies are perhaps not surprising, as the phospholipid composition may depend sensitively on cell type and maturation state (i.e., function) of the LD. Thus, the phospholipid composition is likely to vary spatially and temporally in the cell. Previous work by the Atshaves group seems to suggest that LDs coated with perilipin 3 contain a lipid monolayer with more unsaturated chains than the pool of LDs that are primarily coated with perilipin 1 (16). If this is indeed the case, then it is unlikely that the amphipathic α-helix bundle of perilipin 3 is bound to the phospholipid monolayer. Future work will investigate the role of lipid acyl chain saturation for the full-length protein.
CONCLUSIONS
The results discussed above raise a number of substantial questions that need to be addressed before the interaction of perilipin 3 with LDs can be fully understood. Our data appear to be at odds with those from cell biological studies. For example, we find that the C terminus of perilipin 3 has a higher MIP than any of the other constructs tested, including the full-length protein. On the other hand, cell biological work shows that the C terminus is not required for targeting and binding to LDs (1, 28). Additionally, the C terminus interacts less favorably with unsaturated lipids but Atshaves and colleagues appear to show that perilipin 3 is enriched on LDs with a more unsaturated phospholipid monolayer (16).
Can these findings be reconciled? One possibility is that, indeed, the C terminus does not bind to the phospholipid monolayer surrounding the LD (28). However, in light of our data, we find this possibility unlikely. Bulankina et al. (1) show that both the C terminus and the full-length protein are able to transform lipid vesicles (of dimyristoylphosphatidylcholine and dimyristoylphosphatidylglycerol at or very near their main phase transition temperature) into lipid discs. This clearly indicates that the full-length protein, as well as the C terminus, interacts with lipid interfaces. Also, the amphipathic α-helix bundle domain is the only domain present in the insect protein, apoL-III, whose function in lipophorin stability and, thus, lipid interaction is extensively studied and well-established (48, 75–78). When perilipin 3 first interacts with the surface of a LD, it is likely that the amphipathic α-helix bundle domain opens up so that the hydrophobic side of the four helices is able to insert into the phospholipid interface, identically to what has been described for apoLp-III.
What is not clear is the exact role this binding and opening up of the helix bundle has for the function of the full-length protein. One possibility is that the amphipathic α-helix bundle domain further stabilizes the interaction of the protein with the phospholipid-neutral lipid (i.e., triacylglycerol and cholesterol ester) interface and further mediates the exchangeability of the protein, as has been proposed by Narayanaswami and Ryan (79). The ability to exchange between the cytosol and LD surface might be mediated by the saturation of LD (phospho)lipids and could thus be related to maturation of the LD. We show that the interaction of the amphipathic α-helix bundle domain is less favorable for unsaturated lipids than it is for saturated lipids. In fact, the amphipathic α-helix bundle is not likely to insert at all into a fully formed PC monolayer with unsaturated acyl chains, as the MIP for this interaction is well below 30 mN/m.
It is also possible that the interaction partner for the amphipathic α-helix bundle domain is not the phospholipid monolayer at all, but the neutral lipids underlying this monolayer. Further work on the full-length protein and more sophisticated model systems (a buffer-phospholipid-oil interface) are needed to address these questions and are currently underway.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr. Robert Hynson for providing helpful insights regarding protein expression and purification. The authors would also like to thank Dr. Patrick J. Loll for providing vectors for SUMO hydrolase expression.
Footnotes
Abbreviations:
- DOPA
- 1,2-dioleoyl-sn-glycero-3-phosphate (sodium salt)
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- LD
- lipid droplet
- MIP
- maximum insertion pressure
- PC
- phosphatidylcholine
- POG
- 1-palmitoyl-2-oleoyl-sn-glycerol
- POPA
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (sodium salt)
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- SUMO
- small ubiquitin-related modifier
- Δπmax
- maximal change in monolayer pressure
This work was supported by the Ohio Board of Regents and funding from the National Science Foundation for the NSF-REU Program (CHE-1263087) (T.A.). E.E.K. is supported by a Farris Family Fellowship and Kent State University.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bulankina A. V., Deggerich A., Wenzel D., Mutenda K., Wittmann J. G., Rudolph M. G., Burger K. N., and Honing S.. 2009. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185: 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hickenbottom S. J., Kimmel A. R., Londos C., and Hurley J. H.. 2004. Structure of a lipid droplet protein; the PAT family member TIP47. Structure. 12: 1199–1207. [DOI] [PubMed] [Google Scholar]
- 3.Hynson R. M., Jeffries C. M., Trewhella J., and Cocklin S.. 2012. Solution structure studies of monomeric human TIP47/perilipin-3 reveal a highly extended conformation. Proteins. 80: 2046–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farese R. V. Jr., and Walther T. C.. 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 139: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimoto T., and Parton R. G.. 2011. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3: a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy D. J. 2012. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma. 249: 541–585. [DOI] [PubMed] [Google Scholar]
- 7.Olofsson S. O., Bostrom P., Andersson L., Rutberg M., Perman J., and Boren J.. 2009. Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim. Biophys. Acta. 1791: 448–458. [DOI] [PubMed] [Google Scholar]
- 8.Zehmer J. K., Huang Y., Peng G., Pu J., Anderson R. G., and Liu P.. 2009. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 9: 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cermelli S., Guo Y., Gross S. P., and Welte M. A.. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16: 1783–1795. [DOI] [PubMed] [Google Scholar]
- 10.Welte M. A. 2007. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 17: 363–369. [DOI] [PubMed] [Google Scholar]
- 11.Herker E., and Ott M.. 2011. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol. Metab. 22: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., and Shimotohno K.. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9: 1089–1097. [DOI] [PubMed] [Google Scholar]
- 13.Samsa M. M., Mondotte J. A., Iglesias N. G., Assuncao-Miranda I., Barbosa-Lima G., Da Poian A. T., Bozza P. T., and Gamarnik A. V.. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5: e1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown D. A. 2001. Lipid droplets: proteins floating on a pool of fat. Curr. Biol. 11: R446–R449. [DOI] [PubMed] [Google Scholar]
- 15.Murphy D. J., and Vance J.. 1999. Mechanisms of lipid-body formation. Trends Biochem. Sci. 24: 109–115. [DOI] [PubMed] [Google Scholar]
- 16.Storey S. M., McIntosh A. L., Senthivinayagam S., Moon K. C., and Atshaves B. P.. 2011. The phospholipid monolayer associated with perilipin-enriched lipid droplets is a highly organized rigid membrane structure. Am. J. Physiol. Endocrinol. Metab. 301: E991–E1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., and Fujimoto T.. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277: 44507–44512. [DOI] [PubMed] [Google Scholar]
- 18.Brasaemle D. L., Barber T., Kimmel A. R., and Londos C.. 1997. Post-translational regulation of perilipin expression. Stabilization by stored intracellular neutral lipids. J. Biol. Chem. 272: 9378–9387. [DOI] [PubMed] [Google Scholar]
- 19.Brasaemle D. L., Barber T., Wolins N. E., Serrero G., Blanchette-Mackie E. J., and Londos C.. 1997. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 38: 2249–2263. [PubMed] [Google Scholar]
- 20.Londos C., Brasaemle D. L., Schultz C. J., Segrest J. P., and Kimmel A. R.. 1999. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 10: 51–58. [DOI] [PubMed] [Google Scholar]
- 21.Ingelmo-Torres M., Gonzalez-Moreno E., Kassan A., Hanzal-Bayer M., Tebar F., Herms A., Grewal T., Hancock J. F., Enrich C., Bosch M., et al. 2009. Hydrophobic and basic domains target proteins to lipid droplets. Traffic. 10: 1785–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia A., Sekowski A., Subramanian V., and Brasaemle D. L.. 2003. The central domain is required to target and anchor perilipin A to lipid droplets. J. Biol. Chem. 278: 625–635. [DOI] [PubMed] [Google Scholar]
- 23.McManaman J. L., Zabaronick W., Schaack J., and Orlicky D. J.. 2003. Lipid droplet targeting domains of adipophilin. J. Lipid Res. 44: 668–673. [DOI] [PubMed] [Google Scholar]
- 24.Ohsaki Y., Maeda T., Maeda M., Tauchi-Sato K., and Fujimoto T.. 2006. Recruitment of TIP47 to lipid droplets is controlled by the putative hydrophobic cleft. Biochem. Biophys. Res. Commun. 347: 279–287. [DOI] [PubMed] [Google Scholar]
- 25.Orlicky D. J., Degala G., Greenwood C., Bales E. S., Russell T. D., and McManaman J. L.. 2008. Multiple functions encoded by the N-terminal PAT domain of adipophilin. J. Cell Sci. 121: 2921–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Targett-Adams P., Chambers D., Gledhill S., Hope R. G., Coy J. F., Girod A., and McLauchlan J.. 2003. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 278: 15998–16007. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian V., Garcia A., Sekowski A., and Brasaemle D. L.. 2004. Hydrophobic sequences target and anchor perilipin A to lipid droplets. J. Lipid Res. 45: 1983–1991. [DOI] [PubMed] [Google Scholar]
- 28.Rowe E. R., Mimmack M. L., Barbosa A. D., Haider A., Isaac I., Ouberai M. M., Thiam A. R., Patel S., Saudek V., Siniossoglou S. , et al. 2016. Conserved amphipathic helices mediate lipid droplet targeting of perilipins 1-3. J. Biol. Chem. 291: 6664–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brasaemle D. L. 2007. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48: 2547–2559. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura N., and Fujimoto T.. 2003. Adipose differentiation-related protein has two independent domains for targeting to lipid droplets. Biochem. Biophys. Res. Commun. 306: 333–338. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi T., Matsushita S., Motojima K., Hirose F., and Osumi T.. 2006. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 281: 14232–14240. [DOI] [PubMed] [Google Scholar]
- 32.Dure L., 3rd 1993. A repeating 11-mer amino acid motif and plant desiccation. Plant J. 3: 363–369. [DOI] [PubMed] [Google Scholar]
- 33.Scherer P. E., Bickel P. E., Kotler M., and Lodish H. F.. 1998. Cloning of cell-specific secreted and surface proteins by subtractive antibody screening. Nat. Biotechnol. 16: 581–586. [DOI] [PubMed] [Google Scholar]
- 34.Bussell R. Jr., and Eliezer D.. 2003. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 329: 763–778. [DOI] [PubMed] [Google Scholar]
- 35.Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., and Harvey S. C.. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274: 31755–31758. [DOI] [PubMed] [Google Scholar]
- 36.Kimmel A. R., Brasaemle D. L., McAndrews-Hill M., Sztalryd C., and Londos C.. 2010. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 51: 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., and Londos C.. 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266: 11341–11346. [PubMed] [Google Scholar]
- 38.Greenberg A. S., Egan J. J., Wek S. A., Moos M. C. Jr., Londos C., and Kimmel A. R.. 1993. Isolation of cDNAs for perilipins A and B: sequence and expression of lipid droplet-associated proteins of adipocytes. Proc. Natl. Acad. Sci. USA. 90: 12035–12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Servetnick D. A., Brasaemle D. L., Gruia-Gray J., Kimmel A. R., Wolff J., and Londos C.. 1995. Perilipins are associated with cholesteryl ester droplets in steroidogenic adrenal cortical and Leydig cells. J. Biol. Chem. 270: 16970–16973. [DOI] [PubMed] [Google Scholar]
- 40.Wolins N. E., Brasaemle D. L., and Bickel P. E.. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580: 5484–5491. [DOI] [PubMed] [Google Scholar]
- 41.Wolins N. E., Skinner J. R., Schoenfish M. J., Tzekov A., Bensch K. G., and Bickel P. E.. 2003. Adipocyte protein S3–12 coats nascent lipid droplets. J. Biol. Chem. 278: 37713–37721. [DOI] [PubMed] [Google Scholar]
- 42.Miura S., Gan J. W., Brzostowski J., Parisi M. J., Schultz C. J., Londos C., Oliver B., and Kimmel A. R.. 2002. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277: 32253–32257. [DOI] [PubMed] [Google Scholar]
- 43.Wolins N. E., Rubin B., and Brasaemle D. L.. 2001. TIP47 associates with lipid droplets. J. Biol. Chem. 276: 5101–5108. [DOI] [PubMed] [Google Scholar]
- 44.Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., and Brown D. A.. 2007. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J. Lipid Res. 48: 2751–2761. [DOI] [PubMed] [Google Scholar]
- 45.Sletten A., Seline A., Rudd A., Logsdon M., and Listenberger L. L.. 2014. Surface features of the lipid droplet mediate perilipin 2 localization. Biochem. Biophys. Res. Commun. 452: 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., and Londos C.. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 36: 1211–1226. [PubMed] [Google Scholar]
- 47.Faustino A. F., Carvalho F. A., Martins I. C., Castanho M. A., Mohana-Borges R., Almeida F. C., Da Poian A. T., and Santos N. C.. 2014. Dengue virus capsid protein interacts specifically with very low-density lipoproteins. Nanomedicine. 10: 247–255. [DOI] [PubMed] [Google Scholar]
- 48.Weers P. M., and Ryan R. O.. 2006. Apolipophorin III: role model apolipoprotein. Insect Biochem. Mol. Biol. 36: 231–240. [DOI] [PubMed] [Google Scholar]
- 49.Calvez P., Bussieres S., Eric D., and Salesse C.. 2009. Parameters modulating the maximum insertion pressure of proteins and peptides in lipid monolayers. Biochimie. 91: 718–733. [DOI] [PubMed] [Google Scholar]
- 50.Rathnayake S. S., Mirheydari M., Schulte A., Gillahan J. E., Gentit T., Phillips A. N., Okonkwo R. K., Burger K. N., Mann E. K., Vaknin D., et al. 2014. Insertion of apoLp-III into a lipid monolayer is more favorable for saturated, more ordered, acyl-chains. Biochim. Biophys. Acta. 1838: 482–492. [DOI] [PubMed] [Google Scholar]
- 51.Studier F. W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41: 207–234. [DOI] [PubMed] [Google Scholar]
- 52.Weeks S. D., Drinker M., and Loll P. J.. 2007. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr. Purif. 53: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demel R. A., van Doorn J. M., and Van Der Horst D. J.. 1992. Insect apolipophorin III: interaction of locust apolipophorin III with diacylglycerol. Biochim. Biophys. Acta. 1124: 151–158. [DOI] [PubMed] [Google Scholar]
- 54.Adamson A. W., and Gast A. P.. 1997. Physical Chemistry of Surfaces. 6th edition. Wiley, New York. [Google Scholar]
- 55.Luckey M. Membrane Structural Biology: With Biochemical and Biophysical Foundations. 2nd edition. Cambridge University Press, New York. [Google Scholar]
- 56.Marsh D. 1996. Lateral pressure in membranes. Biochim. Biophys. Acta. 1286: 183–223. [DOI] [PubMed] [Google Scholar]
- 57.Brockman H. 1999. Lipid monolayers: why use half a membrane to characterize protein-membrane interactions? Curr. Opin. Struct. Biol. 9: 438–443. [DOI] [PubMed] [Google Scholar]
- 58.Kooijman E. E., Carter K. M., van Laar E. G., Chupin V., Burger K. N., and de Kruijff B.. 2005. What makes the bioactive lipids phosphatidic acid and lysophosphatidic acid so special? Biochemistry. 44: 17007–17015. [DOI] [PubMed] [Google Scholar]
- 59.Kooijman E. E., Chupin V., de Kruijff B., and Burger K. N.. 2003. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 4: 162–174. [DOI] [PubMed] [Google Scholar]
- 60.Kooijman E. E., Chupin V., Fuller N. L., Kozlov M. M., de Kruijff B., Burger K. N., and Rand P. R.. 2005. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 44: 2097–2102. [DOI] [PubMed] [Google Scholar]
- 61.de Kruijff B. 1997. Lipid polymorphism and biomembrane function. Curr. Opin. Chem. Biol. 1: 564–569. [DOI] [PubMed] [Google Scholar]
- 62.van den Brink-van der Laan E., Killian J. A., and de Kruijff B.. 2004. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta. 1666: 275–288. [DOI] [PubMed] [Google Scholar]
- 63.Kooijman E. E., Tieleman D. P., Testerink C., Munnik T., Rijkers D. T., Burger K. N., and de Kruijff B.. 2007. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 282: 11356–11364. [DOI] [PubMed] [Google Scholar]
- 64.Gautier R., Douguet D., Antonny B., and Drin G.. 2008. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 24: 2101–2102. [DOI] [PubMed] [Google Scholar]
- 65.Cevc G. 1990. Membrane electrostatics. Biochim. Biophys. Acta. 1031: 311–382. [DOI] [PubMed] [Google Scholar]
- 66.He J., Vora M., Haney R. M., Filonov G. S., Musselman C. A., Burd C. G., Kutateladze A. G., Verkhusha V. V., Stahelin R. V., and Kutateladze T. G.. 2009. Membrane insertion of the FYVE domain is modulated by pH. Proteins. 76: 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee S. A., Eyeson R., Cheever M. L., Geng J., Verkhusha V. V., Burd C., Overduin M., and Kutateladze T. G.. 2005. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA. 102: 13052–13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freire J. M., Domingues M. M., Matos J., Melo M. N., Veiga A. S., Santos N. C., and Castanho M. A.. 2011. Using zeta-potential measurements to quantify peptide partition to lipid membranes. Eur. Biophys. J. 40: 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thiam A. R., Farese R. V. Jr., and Walther T. C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14: 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marinova K. G., Alargova R. G., Denkov N. D., Velev O. D., Petsev D. N., Ivanov I. B., and Borwankar R. P.. 1996. Charging of oil-water interfaces due to spontaneous adsorption of hydroxyl ions. Langmuir. 12: 2045–2051. [Google Scholar]
- 71.Ghimire C., Koirala D., Mathis M. B., Kooijman E. E., and Mao H.. 2014. Controlled particle collision leads to direct observation of docking and fusion of lipid droplets in an optical trap. Langmuir. 30: 1370–1375. [DOI] [PubMed] [Google Scholar]
- 72.Lu X., Gruia-Gray J., Copeland N. G., Gilbert D. J., Jenkins N. A., Londos C., and Kimmel A. R.. 2001. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm. Genome. 12: 741–749. [DOI] [PubMed] [Google Scholar]
- 73.Nielsen R. L., Andersen M. H., Mabhout P., Berglund L., Petersen T. E., and Rasmussen J. T.. 1999. Isolation of adipophilin and butyrophilin from bovine milk and characterization of a cDNA encoding adipophilin. J. Dairy Sci. 82: 2543–2549. [DOI] [PubMed] [Google Scholar]
- 74.Gennis R. B. 1989. Biomembrane, Molecular Structure and Function. Spirnger-Verlag, New York. [Google Scholar]
- 75.Soulages J. L., and Arrese E. L.. 2000. Dynamics and hydration of the alpha-helices of apolipophorin III. J. Biol. Chem. 275: 17501–17509. [DOI] [PubMed] [Google Scholar]
- 76.Soulages J. L., Salamon Z., Wells M. A., and Tollin G.. 1995. Low concentrations of diacylglycerol promote the binding of apolipophorin III to a phospholipid bilayer: a surface plasmon resonance spectroscopy study. Proc. Natl. Acad. Sci. USA. 92: 5650–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weers P. M., Narayanaswami V., Kay C. M., and Ryan R. O.. 1999. Interaction of an exchangeable apolipoprotein with phospholipid vesicles and lipoprotein particles. Role of leucines 32, 34, and 95 in Locusta migratoria apolipophorin III. J. Biol. Chem. 274: 21804–21810. [DOI] [PubMed] [Google Scholar]
- 78.Weers P. M., and Ryan R. O.. 2003. Apolipophorin III: a lipid-triggered molecular switch. Insect Biochem. Mol. Biol. 33: 1249–1260. [DOI] [PubMed] [Google Scholar]
- 79.Narayanaswami V., and Ryan R. O.. 2000. Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta. 1483: 15–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.