Abstract
A reliable method for purifying envelope-stripped nuclei from immortalized murine embryonic fibroblasts (iMEFs) was established. Quantitative profiling of the glycerophospholipids (GPLs) in envelope-free iMEF nuclei yields several conclusions. First, we find the endonuclear glycerophospholipidome differs from that of bulk membranes, and phosphatidylcholine (PtdCho) and phosphatidylethanolamine species are the most abundant endonuclear GPLs by mass. By contrast, phosphatidylinositol (PtdIns) represents a minor species. We also find only a slight enrichment of saturated versus unsaturated GPL species in iMEF endonuclear fractions. Moreover, much lower values for GPL mass were measured in the iMEF nuclear matrix than those reported for envelope-stripped IMF-32 nuclei. The collective results indicate that the nuclear matrix in these cells is a GPL-poor environment where GPL occupies only approximately 0.1% of the total nuclear matrix volume. This value suggests GPL accommodation in this compartment can be satisfied by binding to resident proteins. Finally, we find no significant role for the PtdIns/PtdCho-transfer protein, PITPα, in shuttling PtdIns into the iMEF nuclear matrix.
Keywords: cell signaling, lipids, nuclear receptors/lipid ligands, phospholipids/metabolism, phospholipids/phosphatidylinositol
The mammalian nuclear matrix is now recognized as a site of lipid biosynthesis and signaling in both normal and pathological states (reviewed in 1–8). Radioisotope and stable isotope tracer studies consistently identify glycerophospholipids (GPLs), and the enzymatic activities that metabolize them, within purified nuclear fractions purportedly devoid of nuclear envelope and other cellular membrane contaminants (9–15). These findings suggest the existence of an independently-regulated GPL pool within the nuclear matrix, one distinct from the nuclear envelope and from bulk cellular membranes. In support of this interpretation, nuclear isoforms of a number of GPL metabolic enzymes have been described (10, 11, 16–22). The STAR-PAP RNA poly-A polymerase, the direct modification of a phosphoinositide headgroup presented to an inositide kinase by a nuclear receptor, and nuclear phosphoinositide control of the basal transcription machinery all provide compelling examples of GPL-regulated activities that discharge their functions within the nuclear matrix (23–27). At issue, however, is the scale of nuclear GPL metabolism and nuclear GPL load.
This difficult question can now be addressed using quantitative mass spectrometric methods. The single major study on this topic estimates that phosphatidylcholine (PtdCho) alone occupies 10–16% of the nuclear matrix of IRB-32 cells by volume (14). This is a startling conclusion considering that the genome itself is estimated to occupy some 39% of the nuclear volume in the IMF-32 cell line. Furthermore, the endonuclear GPL pool is reported to be unusual in that it is dominated by saturated PtdCho molecular species (14). The abundance of endonuclear GPLs, when coupled with their predominantly saturated nature, motivates speculation that phospholipids (PLs) profoundly influence the chemical properties of the nuclear matrix, i.e., by contributing to the formation of gel-like regions within the nuclear matrix (1, 14). This concept raises a fundamental question of how does the nuclear matrix accommodate such a large PL load? That is, how are lipids organized within the nuclear matrix?
Given the mounting evidence for endonuclear lipid signaling and the lingering questions regarding how lipids are organized in nuclear matrix, we reinvestigated the problem of endonuclear lipidomics. To this end, we developed a reliable and reproducible method for purification of envelope-free nuclei from immortalized murine embryonic fibroblasts (iMEFs). This method adheres to a stringent quality-control regime for assessing purity of isolated endonuclear compartments. Quantitative GPL profiling of these highly purified fractions describes an endonuclear GPL composition that is indeed distinct from that of bulk cellular membrane. Contrary to previous reports, however, the profile shows no particular enrichment of saturated GPL molecular species. Moreover, the mass measurements record drastically lower GPL contents in endonuclear compartments than those previously reported, at least for a neuroblastoma cell line. While the collective data confirm the nuclear matrix harbors a GPL pool of distinct composition from that of bulk membrane, the data also identify the iMEF nuclear matrix as a PL-poor environment that requires no unusual provisions for PL accommodation other than binding to resident proteins.
MATERIALS AND METHODS
Reagents and general notes
Chemicals and reagents were purchased from Fisher Scientific (Pittsburg, PA) or from Sigma-Aldrich (St. Louis, MO), unless otherwise stated. All lipid standards were purchased from Avanti Polar Lipids (Alabaster, AL). Organic solvents and supplies used to prepare samples for electron microscopy (EM) were obtained from Electron Microscopy Sciences (Hatfield, PA). The mass labels, myo-inositol-d6 and choline-d9, were obtained from C/D/N Isotopes (Pont-Claire, Quebec, Canada) and Sigma-Aldrich, respectively.
Media and antibodies
DMEM and antibiotics were obtained from Gibco/Invitrogen (Carlsbad, CA). FBS was obtained from Gemini Bio-products (Sacramento, CA). A mouse monoclonal antibody directed toward β-tubulin (product number T5293) and rabbit polyclonal antibody directed toward lamin A (product number L1293) were obtained from Sigma-Aldrich. A mouse monoclonal antibody toward α-tubulin was obtained from Neomarkers Inc. (Fremont, CA; product number MS-581-P). Other rabbit polyclonal antibodies utilized in this study include: an anti-calnexin antibody (Stressgen Assay Designs, Ann Arbor, MI; product number SPA-860), an anti-histone H3 antibody (generous gift of Brian Strahl, University of North Carolina-Chapel Hill), an anti-NURIM antibody (Santa Cruz Biotechnology, Santa Cruz, CA; product number sc-133260), and an anti-fibrillarin antibody (Abcam Inc., Cambridge, MA; product number ab5821). Goat-anti-mouse or goat-anti-rabbit HRP-conjugated secondary antibodies (Bio-Rad, Hercules, CA) were used for development in ECL assays. Donkey-anti-rabbit secondary antibody conjugated to IR Dye 800 was purchased from Rockland Immunochemicals Inc. (Gilbertsville, PA; product number 611-731-127) for use in Odyssey immunoblotting experiments.
Cell culture and transfection
iMEFs were derived from E14-E16 embryos, and immortalized iMEF lines were generated using the SV40 large T-antigen method (28). Unless otherwise specified, all primary and immortalized cell lines were cultured in complete DMEM containing 4.5 g/l glucose and supplemented with 10% FBS, 1 U/ml penicillin G, and 100 μg/ml streptomycin (complete DMEM). All cell culture was performed at 37°C in a 10% CO2 incubator.
Initial steps in purifying envelope-stripped iMEF nuclei
Envelope-stripped nuclei were prepared using the method of Martelli et al. (11) with essential modifications: iMEF cells were seeded to 150 mm tissue culture dishes and grown for 24–48 h. Approximately 107 iMEFs were pelleted (for a 1× preparation) and washed three times in Dulbecco’s PBS solution. After complete removal of PBS, the cell pellet was resuspended thoroughly in 500 μl of chilled buffer A [10 mM Tris-HCl (pH 7.4), 1% NP-40, 10 mM β-mercaptoethanol, 0.5 mM PMSF] supplemented with Complete Protease Inhibitor cocktail (Roche Biopharmaceuticals). Cells were incubated on ice for 8 min with occasional agitation. An equal volume of ice-cold ddH2O was added to swell the cells. Following a 3 min incubation, swollen cells were subsequently subjected to three passages through a 22 gauge needle. Removal of cellular debris was monitored by examination of several microliters of triturate by phase contrast microscopy. In sufficiently sheared samples, minimally 40 of 50 nuclei examined lacked significant cytosolic or membranous debris. After addition of 0.5 ml of chilled buffer B [10 mM Tris-HCl (pH 7.4), 2 mM MgCl2] supplemented with protease inhibitors, the nuclei were gently triturated and again examined by microscopy. Nuclei were returned to ice for 1 min in preparation for centrifugation, during which a portion of lysate (representing the whole cell fraction) was saved for immunoblot analysis of cellular markers and total protein quantification.
Ultimate steps in purifying envelope-stripped iMEF nuclei
Nuclei were sedimented at 86 g for 10 min at 4°C. The supernatant (cytosolic fraction) was either discarded or saved for quality control analysis as needed. The crude pellets were washed once with an excess of buffer B and sedimented again at 86 g for 10 min at 4°C. During an unscaled preparation, the purified nuclei were resuspended in buffer B and distributed as necessary for quality control protein analyses. When generating three times purified nuclear pellets for analysis of PLs by ESI LC-MS, the purified pellet was instead resuspended in 1 ml total of buffer B. Of this suspension, 85% of the final material was pelleted at 500 g for 2 min at 4°C. Following complete removal of the supernatant, the pellet was snap-frozen in liquid nitrogen and stored at −80°C in preparation for PL analysis by MS. Aliquots (150 μl) were collected for protein measurements and quality controls before the final pelleting step.
Immunoblot analyses of envelope-free nuclei
In preparation for immunoblot analysis, whole cell lysates, wash fractions, and nuclear pellets were homogenized in M-Per lysis buffer (Thermo Scientific) supplemented with Complete Protease Inhibitor Cocktail (Roche Biopharmaceuticals). Samples were triturated vigorously through a 25 gauge needle until complete sample disruption was achieved (as confirmed by phase contrast microscopy). Samples were clarified by centrifugation, and the supernatant was separated to a fresh tube for protein precipitation using the SDS-PAGE Clean-up kit (GE Life Sciences, Piscataway, NJ) according to the manufacturer’s directions. Precipitates were resuspended in CHAPS buffer (8 M urea, 2% CHAPS, and 50 mM DTT). These samples were solubilized in Laemmli sample buffer resolved by SDS-PAGE (10% gels), and nuclear and membrane markers were visualized by immunoblotting. Sample loading was normalized by “cell equivalents.” For Odyssey Westerns, the range of signal linearity was determined for each fraction with each antibody.
Resolved proteins were transferred to nitrocellulose membranes by standard methods. Membranes were blocked in the appropriate blocking reagent (as recommended by the manufacturer) for 1 h at room temperature and probed with primary antibody overnight at 4°C. Decorated membranes were washed three times for 10 min in TTBS and incubated for an additional 1–2 h with the corresponding HRP-conjugated secondary antibody diluted in 2% BSA in TTBS. Blots were again washed three times in TTBS and once in PBS before development using the ECL method (Amersham Biosciences). We define the threshold for acceptable purity as lack of detectable calnexin immunoreactivity in a nuclear preparation of 2.4 × 105 cell equivalents.
For detection of blotted proteins using the Odyssey platform, transferred membranes were blocked for 1 h at room temperature in Odyssey blocking buffer (LI-COR Biotechnology, Lincoln, NE). The appropriate primary and secondary antibodies were diluted in a 1:1 solution of Odyssey blocking buffer and PBS. Secondary incubations and terminal wash steps were performed in the dark. Decorated membranes were analyzed on the Odyssey® infrared imaging system using Odyssey® 2.0 software (LI-COR Biotechnology). Scan settings were high image quality, resolution was set to 169 μm, and the intensity of the scan was 5.0. Antibody signals were quantified as integrated intensities of the areas above and below the bands of interest.
Quantification of cellular protein
Prior to protein quantification, cellular fractions were reconstituted in 1% SDS and incubated at 95°C for 10 min. Cooled samples were homogenized by at least 30 passages through a 25 gauge needle. Satisfactory sample disruption was confirmed by phase microscopy and samples were subsequently clarified by centrifugation. In preparation for BCA analysis (Thermo Scientific, Rockland, IL), samples were diluted 1:10 or 1:20. Diluted fractions were analyzed in triplicate according to the protocol for BCA assay for microplate reader.
EM of envelope-free nuclei
Purified nuclei were pelleted and fixed in 2% glutaraldehyde, 1% tannic acid in 0.1 M sodium cacodylate, 2 mM CaCl2, and postfixed in 2% OsO4 in 0.1 M sodium cacodylate, 2 mM CaCl2. Samples were stained in 4% uranyl acetate in 50% ethanol, dehydrated in a 50–100% ethanol/water series, cleared with propylene oxide, and embedded in Embed-812 resin (Electron Microscopy Sciences). Ultrathin sections (50 μm) were prepared using a Leica UCT ultramicrotome, and images were acquired with a Tecnai 12 transmission electron microscope (FEI, Hillsboro, OR), equipped with a Gatan model 794 multiscan digital camera.
To examine nuclei at different stages of purification, samples were fixed during extraction, shearing, and “unveiling” stages (see Results) of separate 1× preparations. Nuclei undergoing the initial extraction in buffer A (500 μl total volume) were diluted to 10 ml with an excess of fixative [2% glutaraldehyde, 1% tannic acid, 2 mM CaCl2 in 0.1 M sodium cacodylate (pH 7.4)]. Nuclei were fixed at the 6 min time point. Sheared nuclei (1 ml) and “unveiled” nuclei (1.5 ml) were also diluted to the same final volume in fixative. One milliliter of each dilution was pelleted at 100 g for 10 min at 4°C. Fixed samples were washed two times for 10 min in 1 ml fixative. After washing thoroughly in buffer [2 mM CaCl2, 0.1 M sodium cacodylate(pH 7.4)], samples were processed for EM as described above.
Profiling of bulk iMEF PL
To generate iMEF pellets for bulk cellular GPL analysis, 1.2 × 106 cells were seeded onto 150 mm tissue culture dishes and grown for 24 h to 60–70% confluence. Some 107 cells were harvested by trypsinization, pelleted, and washed two times in HBSS without Ca2+ or Mg2+. Cells were then transferred to microfuge tubes in 1 ml of the same buffer and pelleted at 1,000 g for 5 min. Following complete removal of the supernatant, pellets were frozen in liquid nitrogen and stored at −80°C prior to lipid extraction and global PL quantification by MS.
GPL extraction and analyses
GPLs from whole cells or nuclear pellets were extracted using a modified Bligh and Dyer procedure (29). Approximately 1 × 107 iMEF cells or 3 × 107 nuclei per pellet in cold 1.5 ml microfuge tubes (Laboratory Product Sales, Rochester, NY) were vortexed with 800 μl of cold 0.1 N HCl:CH3OH (1:1) and 400 μl of cold CHCl3 was added. The extraction proceeded with vortexing (1 min) and centrifugation (5 min, 4°C, 18,000 g). Quantification of GPLs was achieved by the use of ESI LC-MS employing synthetic (non-naturally occurring) diacyl and lysophospholipid standards as communicated elsewhere (30). Typically, 200 ng of each odd-carbon standard was added to each sample. Identification of the individual PLs was accomplished by LC-MS/MS using an MDS SCIEX 4000 QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer and a Shimadzu HPLC system with a normal phase Luna Silica column (2 × 250 mm, 5 μm) using a gradient elution (30). Identification of the individual species was based on their chromatographic and mass spectral characteristics and comparison to these of chemically defined standards (30, 31). This analysis allows identification of both fatty acid moieties, but does not determine position on the glycerol backbone (sn-1 vs. sn-2).
Statistical analyses
Data are presented as means plus standard errors. Differences between percentages of the total GPL pool represented by different classes for whole cells versus nuclei were determined by Student’s t-test.
Molecular biology and site-directed mutagenesis
Rat PITPα cDNA was amplified by PCR and subcloned as a 0.85 kb HindIII-BamHI fragment into pEGFP-N1 (Clontech, Palo Alto, CA). A HA-tagged rat PITPα cDNA (C-terminal tag) was generated by amplification of the rat PITPα cDNA and insertion of the 0.85 kb product into the unique BamHI-NotI sites of pEF3HA, a derivative of pEF4 (Invitrogen, Carlsbad, CA). This construct contains a HA epitope that was incorporated into that construct as an XbaI-PmeI cassette (DNA sequence 5′-TATCCTTACGAC GTTCCAGACTATGCA-3′). Site-directed mutagenesis primers used in this study were from Fisher Scientific, and rat PITPα cDNAs were mutagenized according to QuickChange mutagenesis kit specifications (Stratagene, La Jolla, CA). All mutant constructs were confirmed by nucleotide sequence analysis.
Transient transfection of mammalian cell lines
Immortalized cell lines were transfected using the FuGene transfection reagent (Roche, Indianapolis, IN). Briefly, Cos7 cells or HeLa lines were seeded onto plastic dishes in complete DMEM 24 h prior to transfection. Once cells settled on the plastic surface, the medium was exchanged for antibiotic-free DMEM. One microgram aliquots of PITPα-EGFP constructs were incubated in 100 μl Opti-MEM (Invitrogen) premixed with 3 μl transfection reagent according to the manufacturer’s instructions. The complete transfection cocktail was incubated at room temperature for 1 h before distribution to the medium. At 12 h posttransfection, cells were split onto coverslips and cultured in complete DMEM.
Immunocytochemistry
HeLa or Cos7 cells, transiently transfected with appropriate PITPα-EGFP or PITPα-HA expression plasmids, were fixed 16–20 h posttransfection in 4% PFA in PBS. After permeabilization in 0.2% Triton X-100, cells expressing PITPα-EGFP constructs were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes) for 1 min. Cells transfected with PITPα-HA constructs were permeabilized similarly, then incubated for 1 h at room temperature in blocking buffer (2% BSA in PBS). Primary anti-HA antibodies (1:2,000 dilution in blocking buffer) were applied to blocked coverslips. After a 12–15 h incubation of coverslips with primary antibody at 4°C, cells were serially washed three times for 10 min in 1% BSA in PBS. Secondary antibodies (1:8,000 in blocking buffer) were then applied onto coverslips and fixed cells were incubated at 4°C for 5–8 h. After several washes (1% BSA in PBS), cells were counterstained with DAPI. Coverslips were mounted on glass slides, imaged on a Zeiss 510 META scanning laser confocal microscope, and images were processed using Adobe Photoshop 6.0.
Labeling iMEF cells in culture with deuterated GPL precursors
PITPα+/+ or pitpα0/0 iMEFs were seeded in complete DMEM and grown to a subconfluent (∼60%) density. Cells were washed in PBS and labeled for the desired time period in antibiotic-free DMEM containing 10% FBS, 80 μg/ml choline-d9, and 50 μg/ml myo-inositol-d6. For whole cell measurements, PITPα+/+ or pitpα0/0 iMEFs were collected in 800 μl HBSS for GPL extraction and analysis by LC-MS/MS. In experiments involving purified nuclei stripped of membranes, 3 × 107 cells were pelleted and washed in PBS prior to detergent-mediated removal of the nuclear envelope. Extracts pooled from three samples (9 × 107 nuclei) were isolated and analyzed by LC-MS.
RESULTS
Purification of envelope-free iMEF nuclei
Purification of quality envelope-free nuclei requires comprehensive removal of contaminating cellular membranes and nuclear envelope with minimal compromise of nuclear integrity or of the biochemical character of the nuclear matrix. Because of the experimental advantages offered by the genetically tractable murine embryonic fibroblast (MEF) system for addressing questions related to nuclear signaling, we developed a method for preparing highly purified envelope-free nuclei from these cells. The method generated purified endonuclear compartments suitable for quantitative lipidomic analyses and yielded reproducible data. The procedure employed serial stripping manipulations in the presence of detergent (1% NP-40), and hypotonic swelling and mechanical shearing in progressively more dilute detergent environments (1% → 0.5% → 0.33% → 0% NP-40) to arrive at envelope-free nuclear preparations.
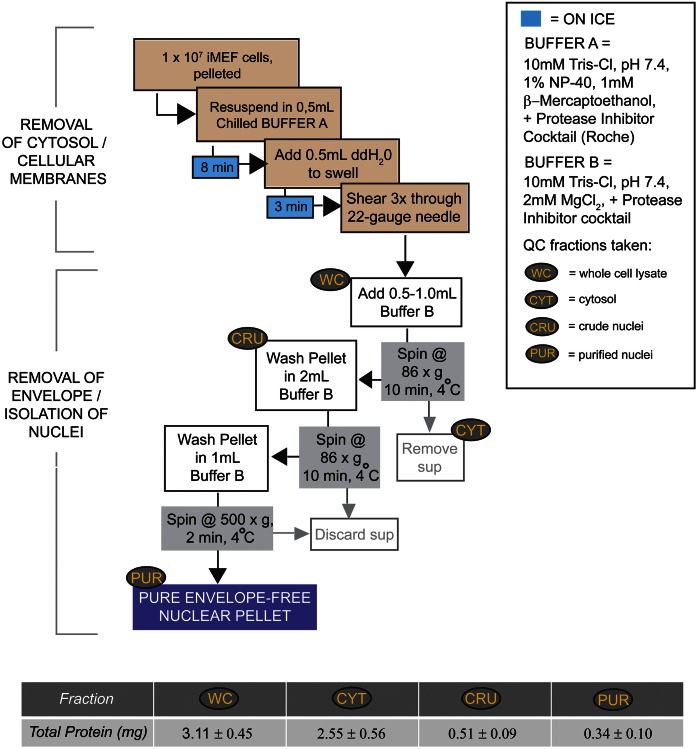
As a general comment, our protocol for purification of envelope-free nuclei worked optimally when 1 × 107 primary or iMEF cells were used as starting material (termed a 1× scale). The efficacy of the method is sensitive to scale and applications that require larger mass quantities of nuclei are best served by generating several smaller nuclear preparations in parallel and pooling the corresponding PL extracts. We recommend processing no more than 3 × 107 cells during a single purification. Buffer volumes used in preparing larger samples should be scaled accordingly. A methodological flowchart is depicted in Fig. 1. From nine independent 1× preparations of envelope-free nuclei, an average of 0.34 ± 0.10 mg of endonuclear protein was recovered. As the averaged 1× quantity of total cellular starting material was 3.11 ± 0.45 mg of total cellular protein, the final protein yield in the purified endonuclear fractions was approximately 10% of total starting material.
Fig. 1.
Nuclear purification scheme. A schematic flow diagram for rapid purification of envelope-stripped iMEF nuclei is illustrated. Total protein in individual fractions generated during the nuclear purification process is shown (milligrams).
Visual landmarks for monitoring purification quality
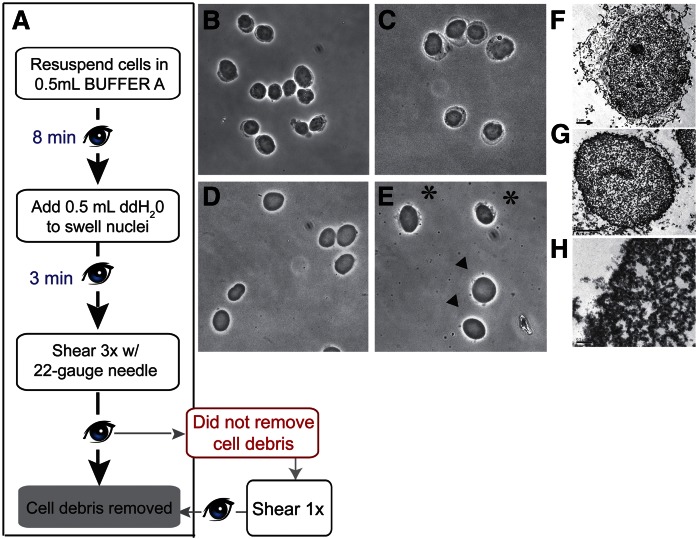
Several morphological transformations accompanied separation of the nuclear particle from contaminating cellular membranes and nuclear envelope. These transformations were readily observed during the purification and reliably diagnosed quality of the final preparation. As such, these morphological landmarks served as valuable real-time reporters of processing efficacy. The stages of processing at which these transformations were monitored are highlighted in Figs. 2, 3. Typical nuclear morphologies observed during the initial detergent extraction step (1% NP-40) are shown in Fig. 2B. The nuclear particles were readily distinguished from assorted cytosolic and organelle debris by light microscopic examination at 40× magnification. The efficacy of the hypotonic swelling step (NP-40 diluted to 0.5% at this step) was similarly interpretable as nuclei increased slightly in diameter (Fig. 2C).
Fig. 2.
Phase contrast monitoring of nuclear particles. A: Flow chart of the critical steps for removing extranuclear debris is indicated. All steps are executed on ice using chilled solutions and tubes. Eye symbols identify points where extraction efficacy is monitored by phase contrast microscopy. B–E: Phase contrast images of extracting nuclei at various steps during the purification. All images were taken using a 40× objective, and 3–5 μl of sample was typically viewed under a coverslip. B: Cells are resuspended in buffer A to initiate the extraction process. At this stage, nuclei are visible as the phase dense center of each cellular particle. Partially solubilized cytoplasm/plasma membrane contaminants are discerned as the less dense material surrounding the nuclear particle. C: Addition of an equal volume of chilled ddH2O swells the nuclei and further enhances contaminating material. D, E: Images of swollen nuclei sheared three times through a 22 gauge needle. D: Successfully sheared nuclei exhibit little to no visible debris attached. E: Nuclei marked with an asterisk retain attached debris. Nuclei marked with arrowheads are scored as at a suitable stage of purification. F–H: Electron micrographs of nuclear preparation stages. F: Electron micrograph of nucleus (1,100×) fixed after 6 min of extraction in buffer A (scale bar = 2 μm). G: Sheared nucleus (1,100×). Minor amounts of debris are still attached to the lamin boundary, which is clearly visible (scale bar = 2 μm). H: Higher magnification (15,000×) image of a nuclear border that has been successfully stripped of envelope (scale bar = 0.2 μm).
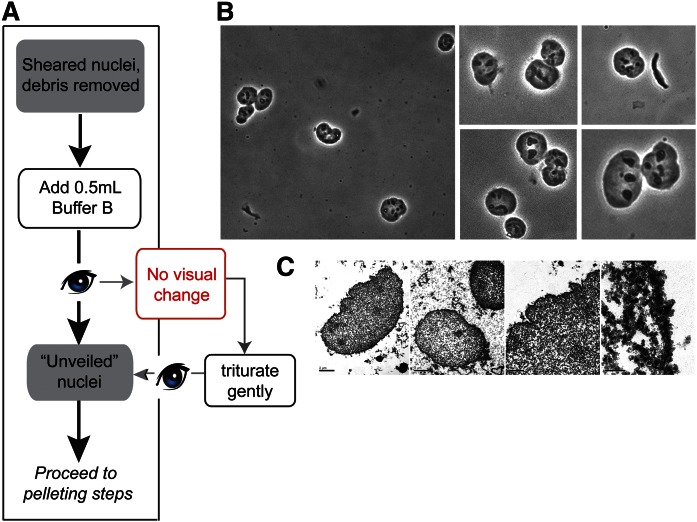
Fig. 3.
Unveiling of sheared nuclei. A: Diagram of prepelleting purification steps. B: Phase contrast images of nuclei following addition of buffer B. Images were taken using a 40× objective. Portions of images at this magnification were digitally enlarged and are shown to the right. At this stage, nuclear particles show increased contrast and nucleoli become visible. C: Electron micrographs of nuclei, fixed immediately after addition of buffer B. In the two leftmost panels, intact nuclei are observed with no attached debris at low magnification. High magnification images of nuclear borders (two rightmost panels) demonstrate removal of the nuclear envelope at this stage.
A subsequent and effective shearing step was an essential component of the purification. Rapid and vigorous trituration of crude nuclear fractions through a 22 gauge needle liberated nuclei of associated debris and left the sheared nuclei as oblate particles. Satisfactory outcomes at this stage were defined by lack of associated large debris in at least 40 of 50 nuclear particles examined. Examples of typical nuclear morphologies at this stage are shown in Fig. 2D, E. EM analyses demonstrated that cellular debris attached to the detergent-extracted nuclear particle (Fig. 2F) was mostly removed by the shearing step (Fig. 2G). Although a small amount of debris remained attached to nuclei at this stage, closer examination of nuclear borders revealed that shearing removed the majority of the nuclear envelope (Fig. 2H).
Upon further dilution of suitably sheared nuclei into Mg2+-containing buffer B (final NP-40 concentration at this stage is 0.33%), the nuclear envelope was effectively removed. This stripping event was monitored via phase contrast microscopy by what we term an unveiling process. Unveiling was marked by the nuclei becoming less opaque and the nucleoli assuming much sharper contrast relative to nucleoplasm (Fig. 3B). The loss of nuclear envelope was visible in electron micrographs of nuclei fixed immediately after addition of buffer B (Fig. 3C, left two panels). Borders of stripped nuclei appear fuzzy and without discernable traces of nuclear envelope (Fig. 3C, right two panels). Subsequent pelleting and washing steps in buffer B (no detergent present) completed the envelope stripping process (Fig. 4). Purified nuclei retained their morphologies and appearance throughout the purification.
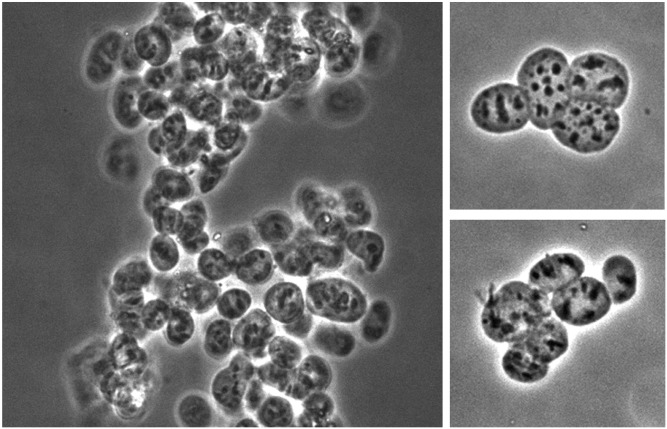
Fig. 4.
Morphology of purified nuclei. Purified envelope-free nuclear fractions were imaged by phase contrast using a 40× objective. Left panel: Nuclear particles are prone to aggregation after envelope removal. Right panels: Inspection of smaller groups of purified nuclear particles confirms that individual particles do not change shape or appearance during the final centrifugation steps of the purification process.
Imaging criteria for envelope-free nuclei
Visualization of envelope-stripped nuclei by EM directly interrogated the membrane content of nuclear preparations. When compared with in situ nuclear morphologies (Fig. 5A), low-magnification (<2,000×) electron micrographs of purified envelope-free nuclei (Fig. 5B) demonstrate efficient removal of extraneous material. Nuclear structure was nicely preserved, and contaminating organelles or other heterogeneous membrane debris were not apparent at this level of resolution. When inspected at higher magnification (>10,000×) by EM, the peripheries of individual nuclear particles presented a fuzzy border and the obvious membrane bilayer structures that constitute the nuclear envelope were no longer visible at these peripheries (compare Fig. 5C, E with Fig. 5D, F, respectively). However, the high resolution EM analyses detected two classes of impurities that commonly evade detection by lower magnification EM, i.e., the types of analyses typically presented as evidence for the envelope-free status of purified nuclear particles (14). First, small copurifying membrane-like strands, attached to the isolated nuclear particles, were occasionally observed (Fig. 5H). Second, these structures were also occasionally observed in the absence of an attached nucleus, and those were counted as well (Fig. 5G). Typically, two strands were recorded per 100 nuclei, and one small patch (average length ∼500 nm) was identified per 34 nuclei (Fig. 5H). We found this low level of potential contamination to be unavoidable. For purposes of reference, supplementary Table 1 compiles the contamination data over a series of independent nuclear preparations.
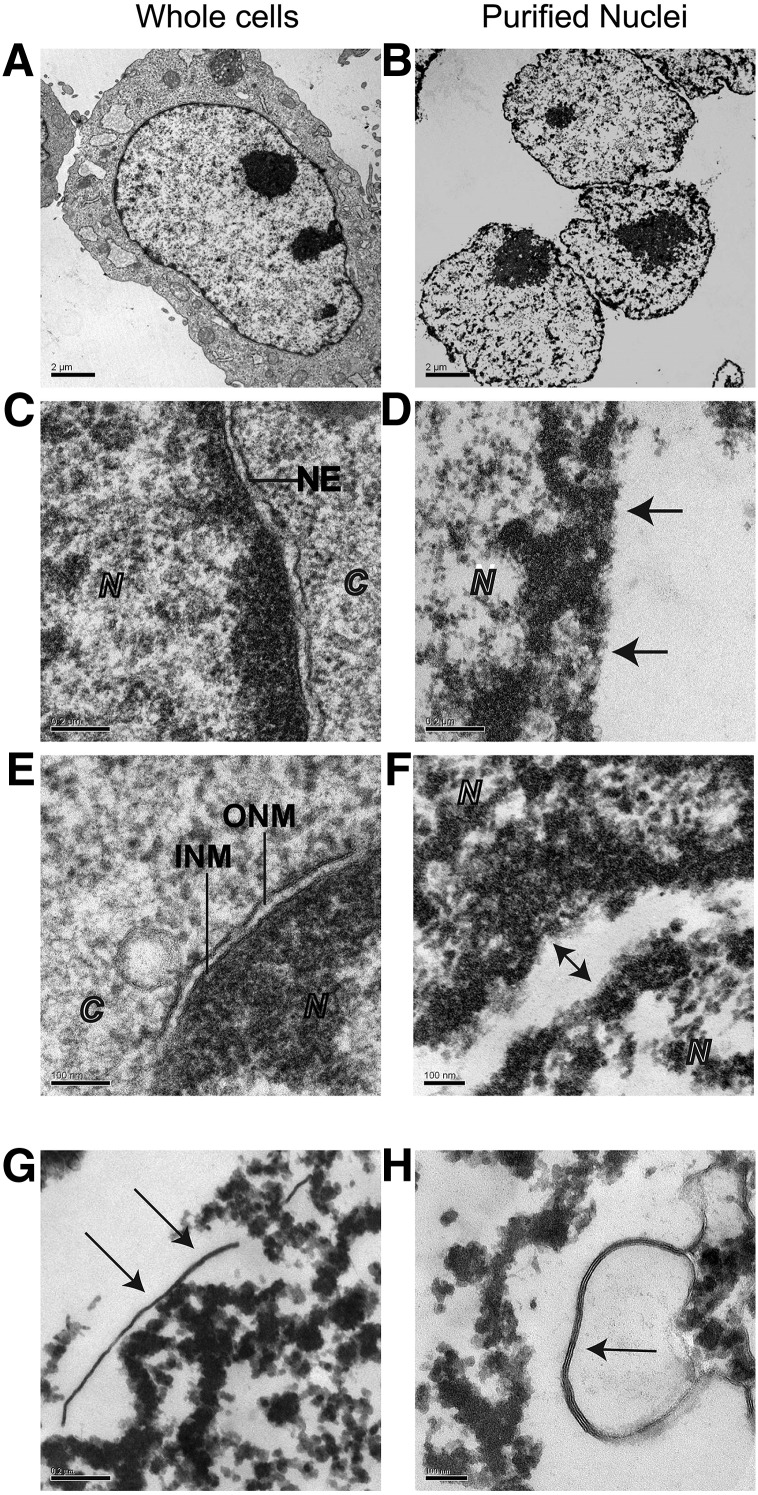
Fig. 5.
Electron microscopic analysis of envelope-free nuclear preparations. A: Whole iMEF cell. Nucleus is centered (1,100× magnification, scale bar = 2 μm). B: Three purified nuclear particles at 1,100× magnification. Nuclei are free of membrane and cellular debris, as evidenced by the clear background (scale bar = 2 μm). C: Nucleocytoplasmic boundary in whole iMEF cells. The nuclear envelope (NE) and its inner and outer leaflets are observed at 11,000× magnification, separating the nuclear contents (N) from the cytosolic portion of the cell (C) (scale bar = 0.2 μm). D: Nuclear boundary after envelope removal. At magnifications greater than 10,000×, envelope-stripped borders of nuclei appear dark and fuzzy (arrows). This is due to the osmophilic nature of the lamin boundary and condensed chromatin at the nuclear periphery, which is no longer attached to the nuclear envelope. E: High magnification images of nuclear boundary in an intact iMEF cell. At 30,000× magnification, the outer nuclear envelope (ONM) sits above the inner nuclear envelope (INM), which is bound tightly to the underlying lamin border (scale bar = 0.1 μm) F: High magnification image (30,000×) of two juxtaposed nuclei. Bidirectional arrow directs attention to the borders of the two purified nuclei, which lack discernable traces of bilayer (scale bar = 0.1 μm). G, H: Identifying membrane contaminants. Membrane strands are indicated by arrows. G: Scanning nuclear peripheries for membrane contaminants at magnifications higher than 10,000× enables detection of free membrane strands (15,000× magnification, scale bar = 0.2 μm). H: High magnification (42,000×) micrograph of two juxtaposed nuclei and a contaminating membrane patch (arrow) (scale bar = 50 nm). In quality preparations, no nuclei retain patches of membrane greater than 2 μm in length and contaminating membrane is most commonly a very small patch, on average 500 nm in length.
Biochemical criteria for purified envelope-free nuclei
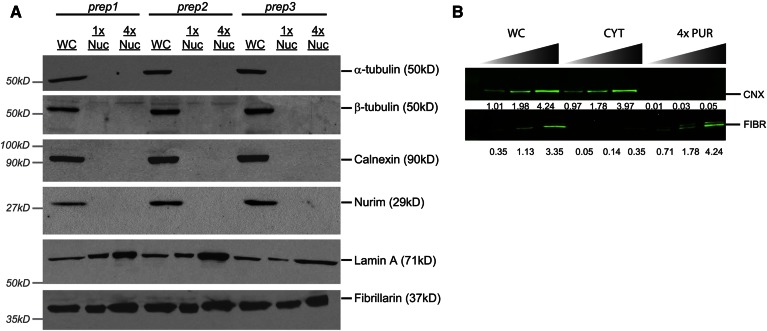
The accepted biochemical standard for envelope-free nuclei is the absence of the abundant cytosolic protein, β-tubulin, in the most purified fractions (14, 21, 32, 33). Unfortunately, this criterion does not adequately interrogate the preparation for contamination by the nuclear envelope, a membrane system physically contiguous with the highly abundant endoplasmic reticulum (ER). To control for these primary sources of contaminating membranes, we monitored membrane-free nuclear preparations for abundant integral membrane proteins of the ER and nuclear envelope. Endonuclear fractions purified by the protocol described herein exhibited undetectable levels of the abundant integral ER membrane protein, calnexin, and the inner nuclear envelope integral membrane protein, NURIM, in addition to undetectable levels of β-tubulin (Fig. 6A). We defined the threshold for acceptable nuclear matrix purity as lack of detectable calnexin immunoreactivity in a sample of 2.4 × 105 nuclei in ECL immunoblotting experiments. Odyssey immunoblotting experiments indicated that less than 0.3% of cellular calnexin remained in nuclear preparations (Fig. 6B), reporting a <300-fold enrichment of nuclear particles from contaminating ER membranes.
Fig. 6.
Immunoblot analyses of nuclear fractions. A: ECL immunoblot analysis of whole cell (WC) and purified nuclear fractions (NUC). Fractions collected from three separate nuclear preparations were prepared for gel electrophoresis as described in the Materials and Methods. Gels were loaded by cell equivalents based on the amount of whole cell lysate needed for blotting in the linear range. Comparison to a 4-fold excess of purified nuclei (4× PUR) highlights the absence/enrichment of cellular proteins in nuclear fractions. B: Odyssey immunoblot analysis of calnexin and fibrillarin content in whole cell, cytosolic, and 4× nuclear fractions from a single preparation. Each sample was loaded in triplicate. Gels were loaded with 15,000, 30,000, and 60,000 cell equivalents of both whole cell (WC) and cytosolic (CYT) protein fractions, whereas 60,000, 120,000, and 240,000 cell equivalents of purified nuclear protein were loaded for analysis. Relative intensity of each individual band was quantified using the Odyssey 2.0 software. These values are listed below the image underneath the corresponding bands. CNX, calnexin; FIBR, fibrillarin.
The endonuclear fractions were highly enriched for the nuclear matrix constituent, lamin A, however (Fig. 6A). Enrichment of the nucleolar protein, fibrillarin, was also monitored. Fibrillarin localizes to the nucleoplasm as well as to nucleoli (34), and thus served as a marker for loss of both nucleoplasm and nucleoli. Based on quantitative immunoblotting data, we again estimated at least a 250-fold purification of nuclear matrix components (i.e., fibrillarin) with respect to contaminating ER membrane protein (i.e., calnexin).
Total PL in cells and nuclei
The resident GPLs were quantified and their compositions profiled in purified endonuclear compartments. Based on a signal-to-noise ratio of >3 as limit of detection threshold, we found the nuclear matrix to be a GPL-poor compartment. PtdCho, phosphatidylethanolamine (PtdEtn), phosphatidylserine (PtdSer), phosphatidylinositol (PtdIns), phosphatidic acid (PtdOH), and phosphatidylglycerol (PtdGro) were all detected in the endonuclear compartment, in rank order of mass abundance. The total GPL mass measured per unit endonuclear compartment (0.077 ± 0.0075 nmol/106 cells) projected an estimated load of approximately 5.9 × 107 GPL molecules per nuclear particle. Based on this value, and our estimate that the average iMEF nucleus occupies a volume of approximately 600 μm3, we calculated that the GPL component occupies <0.1% of the iMEF nucleoplasm. As expected, endonuclear GPL load was small relative to whole cell amounts. The endonuclear GPL mass was almost two orders of magnitude less than that measured per unit iMEF cell (5.1 ± 0.43 nmol/106 cells). Thus, the iMEF endonuclear phospholipidome accounts for ∼1.4% of the total cellular GPL mass.
Lipidomic profiling of the major endonuclear PLs
With regard to composition, the bulk iMEF phospholipidome presented a diverse signature, with the leading GPL classes (by mass) represented by PtdCho, PtdEtn, and PtdSer molecular species, respectively (supplementrary Table 2). We note that 38:4 PtdIns was also represented within the 10 most abundant bulk GPL species. By contrast, the iMEF endonuclear phospholipidome was dominated (in mass terms) by PtdCho, PtdEtn, and PtdSer molecular species; whereas PtdIns, PtdOH, and PtdGro were poorly represented in endonuclear compartments. Interestingly, and as discussed in greater detail below, the differences in fractional representation of these minor PLs in bulk membranes versus the endonuclear compartment were highly significant (P < 0.001). By contrast, the PtdCho/PtdEtn ratios for these two systems were similar (approximately 1.85). We interpret the relative paucity of PtdIns, PtdOH, and PtdGro to argue against the measured endonuclear GPLs representing mere contamination from bulk ER (see Discussion).
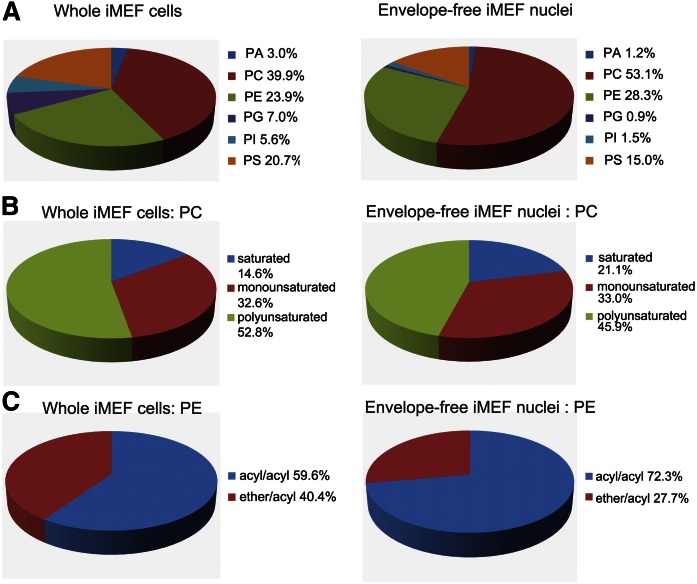
PtdCho was the most abundant endonuclear PL class, represented by 9 of the top 10 most abundant molecular species (supplementary Table S2). Greater than half of the endonuclear GPL mass was accounted for by PtdCho species (53.1 ± 2.8%). The proportion of the PtdCho pool represented by saturated molecular species was only modestly increased in nuclei relative to whole cells. In that regard, the 32:0 and 30:0 PtdCho were the only saturated PtdCho molecular species of sufficient abundance to justify quantification in the endonuclear fractions. Reciprocally, the polyunsaturated PtdCho species were only modestly reduced in abundance in the nuclear matrix relative to their representation in bulk GPL. The relative pool contributions of monounsaturated PtdCho species did not significantly differ in endonuclear compartments versus bulk cell material (Fig. 7B, middle row).
Fig. 7.
A: GPL composition of iMEF cells and purified envelope-free nuclei. Top row: GPL distribution by class for whole iMEFs and envelope-free iMEF nuclei (mean of n = 9 shown). The difference in composition between whole cells and nuclei for PtdOH (PA), PtdGro (PG), and PtdIns (PI) is highly significant (P < 0.001), while the PtdCho (PC)/PtdEtn (PE) ratio is ∼1.85 for both whole cells and nuclei, with no significant difference. PtdSer (PS). B: Percent contributions of saturated, monounsaturated, and polyunsaturated species to PtdCho pool. The proportions of polyunsaturated and saturated PtdCho differ (P < 0.01) between whole iMEFs and nuclei. C: The relative mass contribution of ether-linked PtdEtn species to the PtdEtn pool is substantially greater in whole iMEFs than in nuclei (P < 0.001).
These profiling data were inconsistent with strong enrichments of saturated PtdCho molecular species in envelope-free nuclear preparations. Indeed, unsaturated PtdCho molecular species compromised 79% of the total endonuclear PtdCho mass (Fig. 7). This pattern was observed across the other GPL classes as well. Inspection of the rank order of the 42 most abundant endonuclear GPLs revealed a general paucity in both diversity and mass abundance of saturated species (supplementary Table 2). For example, only 30% of the PtdSer mass was represented by saturated molecular species, and this fraction was much larger than for any other quantifiable endonuclear GPL class. PtdSer represented the third most abundant nuclear GPL class, comprising some 15% of the total endonuclear GPL mass. The preponderance of unsaturated GPLs was encouraging given the legitimate concern that detergent-mediated stripping of the nuclear envelope might generate artifactual detergent-insoluble lipid domains expected to be enriched in saturated GPL molecular species and sphingolipids. In that regard, SM was at the limit of detection in endonuclear fractions, indicating this sphingolipid class is a very minor constituent, at best, of the iMEF endonuclear PL pool (data not shown).
PtdEtn, the second most abundant GPL in the nuclear matrix, accounted for some 28% of total endonuclear GPL mass. We observed interesting distinctions between nonnuclear and nuclear PtdEtn species. Ether-linked PtdEtn was substantially reduced in endonuclear fractions when evaluated as a percentage of the overall PtdEtn pool. In comparison to a fractional representation of 40.4 ± 1.0% of bulk iMEF PtdEtn molecular species, only 27.7 ± 1.5% of the mass of endonuclear PtdEtn species was ether-linked (Fig. 7C, bottom row). The PtdCho/PtdEtn mass ratio, however, remained similar in endonuclear compartments when compared with that ratio in bulk iMEF GPL, further confirming ether-linked PtdEtn species were genuinely segregated from the nuclear matrix.
Lipidomic profiling of the minor endonuclear PLs
While PtdCho and PtdEtn were the most highly represented GPLs in the endonuclear PL profile, envelope-free nuclei were relatively deprived of other GPL species (supplementary Table 2). For example, PtdIns was depleted in the envelope-free nuclear fractions (1.5 ± 0.2% of nuclear GPL) as compared with whole iMEFs (5.6 ± 0.6% of bulk cellular GPL). The distribution by headgroup class (Fig. 7, top row) shows that the minor GPL classes (PtdOH, PtdIns, PtdGro) were all significantly reduced (in terms of fractional representation) in the most purified endonuclear fractions relative to PtdCho and PtdEtn. For purposes of comparison, endonuclear PtdCho, PtdEtn, and PtdSer species accounted for 1.63, 1.77, and 1.13% of the corresponding total cellular pools. By contrast, PtdGro was particularly sparse, only 0.02% of the total iMEF pool was recovered in the nuclear matrix.
Endonuclear PtdOH and PtdIns comprised only 0.53 and 0.42% of the corresponding total cellular pools. Interestingly, whereas the most abundant endonuclear PtdIns was the 38:4 molecular species, the only detectable PtdOH in this compartment was the 36:2 molecular species. In that regard, none of the three acyl chain configurations identified for endonuclear PtdIns were of the 36:2 variety. Those data were inconsistent with a simple nucleoplasmic phospholipase C-diacylglycerol kinase pathway. The action of such a metabolic sequence is expected to yield endonuclear 38:4 PtdOH (see Discussion).
Considerations for how PtdIns is imported into the nuclear matrix
The existence of endonuclear phosphoinositide biosynthetic and signaling pathways begs the question of how PtdIns is incorporated into the nuclear matrix. The nuclear matrix has no capacity to produce PtdIns in situ given that PtdIns synthase is an integral membrane protein of the ER. The PtdIns/PtdCho-transfer protein, PITPα, exhibits both cytoplasmic and endonuclear pools (33, 35, 36), raising the possibility that PITPα shuttles PtdIns into the nuclear matrix. The low steady-state mass quantities of PtdIns we measured in the nuclear matrix suggested that maintenance of a suitable endonuclear PtdIns supply could be achieved via the action of a shuttling lipid transfer protein, such as PITPα. Moreover, when coupled with the measured mass excess of endonuclear PtdCho to PtdIns in the nuclear matrix, a coupled endonuclear PtdIns-import/PtdCho-export cycle was also plausible. This shuttle hypothesis predicted that PITPα localization to the nucleus is facilitated by PtdIns binding, and that PITPα-deficient cells will be compromised for PtdIns import into the nucleus. We tested these two predictions in turn.
PtdIns-binding and PITPα localization in the nucleus
A combination of genetic and structural data identifies a set of residues critical for coordination of the PtdIns headgroup in the PITPα binding pocket (37–39). Four mutant versions with specific defects in PtdIns-binding were employed in these experiments, PitpαT59D, PitpαK61A, PitpαN90L, and PitpαE248K. The corresponding missense substitutions were incorporated into the PITPα-EGFP reporter for transient expression in HeLa cells and imaged. These constructs all produced stable proteins when expressed in cells, as confirmed by immunoblotting experiments (data not shown). Individual cells were scored as to whether the chimera localized predominantly to the nucleus or to the cytoplasm.
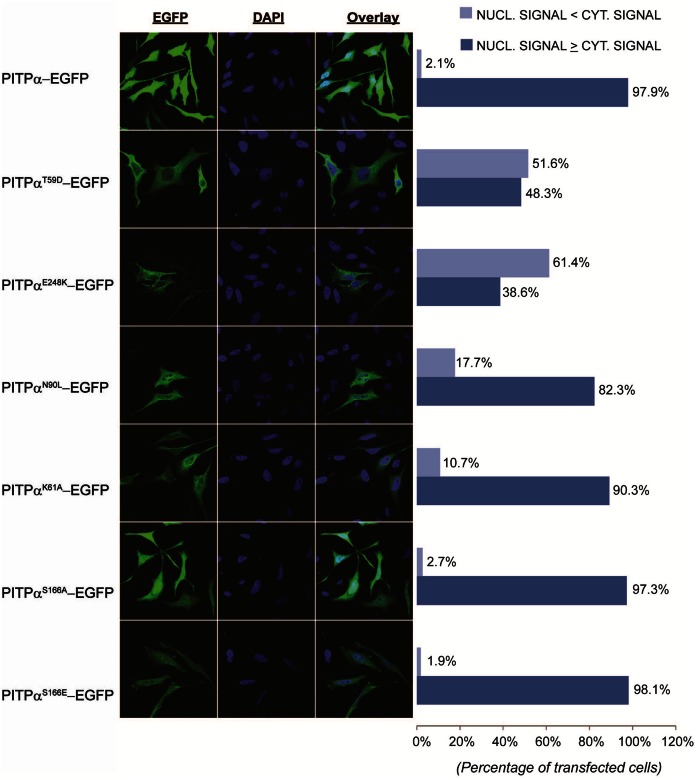
The PITPα-EGFP reporter exhibited a predominant nuclear localization in 97.9% of the cells imaged (Fig. 8A, B). By contrast, PitpαT59D-EGFP displayed a nuclear localization defect in 51.6% of the cells imaged (190/368). Similar localization defects were scored for cells expressing PitpαE248K-EGFP (61.4%, 189/308 cells). Milder, but nevertheless significant, nuclear localization defects were also recorded for the PitpαN90L-EGFP and PitpαK61A-EGFP chimeras (17.7%, 57/322 and 10.8%, 40/370 of cells imaged, respectively). Those data suggested that PtdIns-binding influences either PITPα import into or PITPα export from the nucleus. The relationship was not simple, however. PitpαS166A-EGFP and PitpαS166E-EGFP chimeras [i.e., proteins defective in both PtdIns and PtdCho-transfer activity (36, 39)] were fully competent for localization to the nucleus. Some 2.7% (11/404) and 1.9% (8/425) of the cells imaged scored as showing defects in nuclear EGFP fluorescence, respectively. While the collective data reported experiments performed in HeLa cells, the localization profiles of all of the chimeras were also evaluated in pitpα0/0 iMEFs and in Cos7 cells with similar results (data not shown).
Fig. 8.
Nuclear localization patterns of PITPα-EGFP and lipid binding mutants transiently overexpressed in HeLa cells. A: Confocal images of HeLa cells transiently expressing PITPα-EGFP and PITPα PL binding and transfer mutants. Cells were grown on coverslips, transfected, and fixed 18 h posttransfection. Cells are counterstained with DAPI to examine nuclear localization of mutant constructs. WT PITPα constructs produce nuclear fluorescence that is greater than or equivalent to cytoplasmic fluorescence in the vast majority of expressing cells. PtdIns-binding defective point mutants, PitpαT59D, PitpαK61A, PitpαN90L, PitpαE248K, and the PtdIns/PtdCho-binding defective point mutants, PitpαS166A and PitpαS166E, were scored for the relative strength of nuclear signal versus cytoplasmic fluorescence, as judged by coincidence with nuclear DAPI stain. B: Quantitation of nuclear localization defects in cells expressing PITPα-EGFP mutant constructs. Minimally 200 cells were scored for nuclear signal ≥ cytoplasmic signal or if nuclear signal < cytoplasmic signal.
The effects of the various missense substitutions on PITPα localization profiles were not strictly dependent on the EGFP tag. HA-epitope-tagged versions of PITPα, PitpαT59D, PitpαK61A, PitpαN90L, and PitpαE248K were also individually expressed in HeLa cells. The cells were fixed, permeabilized, and HA-PITPα visualized by indirect immunofluorescence. The results largely recapitulated those obtained with EGFP-tagged chimeras (supplementary Fig. 2). The nuclear signal was dominant in cells expressing PITPα-HA, and this localization pattern was observed in 97.6% of transfected cells (980/1,004). Predominantly cytosolic distributions were observed for the PitpαT59D-HA (78.3%; 548/700), and PitpαE248K-HA expressing cells. For approximately 78% of cells expressing the PitpαT59D-HA fusion protein, the fluorescence signal was predominantly cytoplasmic, and a nuclear localization defect was observed in approximately 95.1% of cells transiently expressing the PitpαE248K-HA fusion proteins (448/471 cells; supplementary Fig. 2). Nuclear localization defects were less prominent in cells expressing PitpαK61A-HA, with only 15.6% of cells demonstrating a nuclear localization defect. We do note that the nuclear signal in cells expressing PitpαK61A-HA was most frequently of similar intensity to the cytoplasmic signal. It was rarely the dominant signal, as was observed for the PITPα-HA control. Nuclear localization defects were also scored in ∼25% of cells expressing PitpαS166E-HA (34/137 cells imaged; supplementary Fig. 2).
Quantitative lipidomics of pitpα0/0 iMEF nuclei
The capability to produce highly purified envelope-free MEF nuclei, when coupled with the availability of mutant MEF lines, made it possible to address previously intractable questions regarding mechanisms of endonuclear lipid signaling. To that end, we first compared endonuclear GPL composition and content in PITPα+/+ versus pitpα0/0 iMEFs to test whether the mutant GPL profile was dramatically altered relative to that of the WT case. By the criteria of marker protein enrichment (supplementary Fig. 3A) and examination by EM (supplementary Fig. 3B, C), nuclei from pitpα0/0 iMEFs were recovered to a comparably high purity as from WT cells. Satisfyingly, compositions of the whole cell and endonuclear phospholipidomes of both cell lines were also essentially indistinguishable (supplementary Fig. 4A, B). Moreover, the total PL content of the nuclear preparations was very similar for WT versus pitpα0/0 iMEFs (1,039.5 ± 226.7 pmol/mg vs. 1,094.2 ± 217.5 pmol/mg nuclear protein, respectively).
Dynamic lipidomics analysis of PtdIns import into nuclei
The general requirement for PtdIns binding for efficient localization of PITPα to the nucleus was consistent with a nuclear PtdIns-supply activity for this protein, perhaps as a mechanism for sustaining a nuclear phosphoinositide signaling cycle. Although qualitative inspection of steady-state nuclear PtdIns-4,5-P2 content by immunofluorescence staining did not suggest a major defect in nuclear pools of this phosphoinositide in PITPα-deficient cells, a direct test of the PtdIns shuttle model required kinetic analysis of the nuclear PtdIns-import process in the presence and absence of PITPα. To that end, standard pulse-chase strategies were modified to estimate endonuclear dynamics of PtdIns and PtdCho lipidomics in PITPα+/+ or pitpα0/0 MEFs. Deuterated PL precursors (Cho-d9 and Ins-d6) were used to label newly synthesized iMEF PtdCho and PtdIns in a pulse regimen, and precursor scan ESI-MS was used as readout to analyze the data. The ESI-MS analyses were conducted on whole cell lipid extracts as bulk control, and on lipids extracted from purified envelope-free nuclei as test case. Precursor scans of the Cho headgroup-derived m/z 184+ and m/z 193+ fragments over the mass range of 600–920 amu identified endogenous (i.e., preexisting) and newly synthesized PtdCho and SM species, respectively. Precursor scans of the Ins headgroup-derived m/z 241− and m/z 247− fragments over the mass range of 600–1,000 amu identified preexisting and newly synthesized PtdIns species, respectively. The light (pre-existing) species defined steady-state baselines for these measurements.
The fractional rates of Cho-d9 and Ins-d6 incorporation into whole cell lipid fractions were similar in PITPα+/+ and pitpα0/0 MEFs (supplementary Fig. 5A, B). Both newly synthesized Cho- and Ins-GPL pools were readily measured at all time-points analyzed, and those data reflect the capacity of MEFs to efficiently incorporate these soluble headgroup precursors into GPL. However, we reproducibly detected reduced fractional incorporation of Ins-d6 into newly synthesized PtdIns in pitpα0/0 MEFs. We do not presently understand either the underlying basis or the physiological significance of this observation. By contrast, fractional rates of incorporation of newly synthesized PtdIns in endonuclear compartments was much slower in both PITPα+/+ and pitpα0/0 MEFs relative to fractional rates of incorporation of newly synthesized PtdCho (supplementary Fig. 5C, D). Very little newly synthesized PtdIns was recovered from PITPα+/+ or pitpα0/0 endonuclear compartments during a 1 h pulse. Detectable incorporation required a pulse of at least 2 h. We interpret the data from these experiments to reflect the intrinsic ability of endonuclear compartments to produce PtdCho in situ, while PtdIns is only slowly imported from extranuclear sources. Those rates of fractional incorporation of newly synthesized PtdIns into pitpα0/0 MEF endonuclear compartments were only modestly reduced (at best) relative to those measured for the PITPα+/+ control (supplementary Fig. 5C), and we do not consider these modest differences to be significant.
DISCUSSION
Herein, we describe a facile and reliable method suitable for purifying quality envelope-free nuclei from iMEFs and from primary MEFs. The choice of cell model was driven by the wealth of MEF lines that can be derived from mutant strains of mice. The method incorporated an expanded repertoire of quality controls and defined criteria for purity. Particular attention was paid to the abundant integral membrane constituents of the membranes that constitute the greatest reservoirs for contamination (e.g., ER and nuclear envelope). This method reproducibly yielded purified nuclear particles that exhibited the dual properties of preserved ultrastructure and the essential absence of contaminating envelope. Quantitative phospholipidomics of the iMEF endonuclear compartment leads us to three basic conclusions: First, in agreement with previous reports (9, 13, 14), the composition of the endonuclear GPL pool is distinct from that of bulk cell GPL. Second, the endonuclear GPL pool is not substantially enriched in saturated molecular species relative to the saturated/unsaturated GPL-composition of the bulk cellular pool. Third, the nucleoplasm is a GPL-poor environment. Those latter two conclusions represent significant departures from previous ideas regarding the fundamental properties of the mammalian endonuclear phospholipidome.
A GPL-deprived nucleoplasm
Mass measurements of the 42 detectable endonuclear GPL species demonstrated the nuclear matrix to be a PL-poor environment. The endonuclear phospholipidome accounted for 1.4% of the total cellular GPL mass, and the GPL component occupied <0.1% of the volume of the iMEF nucleoplasm. Strict interpretations of those values are subject to several unavoidable caveats. First, removal of the nuclear envelope demanded manipulation of the organelle in progressively dilute detergent environments ranging from 1% NP-40 for the initial extraction, to 0.5 and 0.33% NP-40 in the intermediate stages, and no detergent in the final step. It is not clear how much of the baseline endonuclear GPL pool was lost during those obligate NP-40-containing steps. We view this as an unknowable factor because it is not possible to assess the ground state of endonuclear GPLs prior to purification of envelope-free nuclei for the comparison. But, the excellent reproducibility of the data through independent preparations suggests the perturbations imposed are consistent and manageable.
Second, high resolution EM analyses consistently detected trace amounts of membrane-like material in the most purified nuclear particle fractions. If those represented membrane remnants, such contaminants would have contributed to the GPL mass measured in the purified endonuclear fractions. The under-representation of PtdIns in the endonuclear GPL profiles relative to bulk cell membranes argues against contamination by bulk ER representing a major source of membrane contamination in these preparations, as ER is a PtdIns-rich membrane system. Rather, the inner nuclear envelope would be a more likely source of membrane contamination, and our analyses could not determine the extent to which the matrix leaflet of the inner nuclear envelope was removed. Because the lipids of this leaflet contact lamins and other abundant proteins of the nuclear periphery, this lipid pool is likely to be more resistant to stripping. What morphological state such remnants would assume during the preparation of envelope-free nuclei is unknown. These might represent the membrane remnants discussed above, or we might have failed to recognize such contamination altogether. Thus, the endonuclear GPL values reported herein, although low, may yet overestimate endonuclear GPL mass. However, we are encouraged with the reproducibility of the quantitative measurements.
The values we measured for endonuclear GPL load diverge from those reported for the IMR-32 neuroblastoma cell line. That is, the only other cell line of which we are aware for which a quantitative analysis of the nuclear matrix phospholipidome is described. A load of approximately 4 nmol PtdCho/106 nuclei was reported for those cells, along with the remarkable estimate that PtdCho alone occupies 12–16% of the IMR-32 nuclear volume (14). Why this large discrepancy between the two analyses? As both studies used 0.5–1.0% concentrations of nonionic detergent for solubilization of nuclear envelope, one formal possibility is that different cell lines genuinely exhibit manifestly different endonuclear GPL loads. While cell line-specific variations will almost certainly prove to be the case, we consider it unlikely that such variations fully account for the approximately two order of magnitude range that separates the two measurements. Rather, as ER/nuclear envelope remnants were not systematically monitored in the previous study (14), we suspect membrane contamination from these systems dominated the GPL mass. As a result, the reported mass and compositional measurements were misinterpreted as reporting strictly endonuclear GPL pools. Such contamination issues lead to large overestimates of endonuclear GPL mass if the nucleoplasm is a PL-poor environment. Indeed, as demonstrated herein, simple detergent solubilization steps do not efficiently remove ER and nuclear envelope contaminants from nuclear fractions.
Implications for organization of nuclear GPLs
The reported abundance of endonuclear GPL, particularly saturated PtdCho (14), in IMR-32 neuroblastoma cells posed the central question of how such a large PL load might be organized within the nuclear matrix. Standard solutions to the “PL-accommodation” problem, such as incorporation into membrane bilayers, are untenable. While invaginations of the nuclear envelope do extend into the nuclear interior, those structures are topologically distinct from the nuclear matrix itself (40, 41). Indeed, a hallmark feature of the nucleus is that it does not contain internal membrane-bound sub-compartments, despite the presence of morphologically and functionally distinct endonuclear domains (42). Alternatively, it is speculated that the large nuclear lipid-load provokes assembly of endonuclear PL into large aggregates or liquid crystalline phases (1, 3). In that regard, endonuclear PL “rafts” have been reported in hepatocytes (43–46), as have intranuclear lipid droplets, most strikingly under pathological conditions (47–49).
Nuclear lipid rafts and droplets are not common features of other cell types, however, so hepatocytes seem unusual in this regard. Even so, the purported nuclear rafts are seen only in heavily processed hepatocyte nuclei (43–46), and their identities as genuine rafts have not been directly confirmed by independent methods. If protein binding is a primary mechanism for nuclear lipid-organization, then the reported high nuclear abundance of GPL demands that a large fraction of nuclear protein be GPL-associated. All of these general ideas lead to speculations that PtdCho and other GPLs may play significant structural roles within the nuclear matrix (14, 50). Such complex issues surrounding mechanisms of PL-accommodation largely evaporate if the nuclear matrix proves to be a PL-poor compartment, as our data indicate it to be. With the stoichiometries we measure, GPL-binding to resident proteins alone could satisfy the PL-accommodation problem without the need for occupying a major fraction of nuclear protein with bound GPL. A GPL-deprived nuclear matrix also circumvents issues associated with a global gel-like physical environment formed by an abundance of saturated GPL. Our conclusions are consistent with the results of multiple studies that describe the nucleoplasm as a highly dynamic environment where naive molecules, such as GFP, exhibit comparable diffusion coefficients in the cytoplasm versus the nuclear matrix [e.g., (34, 51)]. While involvements of GPLs in structural or organizational capacities within the nuclear matrix remain tenable, our data indicate any such involvements are most likely highly constrained and of a small scale.
Endonuclear PL molecular species
In our hands, iMEF endonuclear GPLs are predominantly of the unsaturated variety with few saturated molecular species. Indeed, of the major endonuclear GPL classes (PtdCho, PtdEtn, PtdSer), >70% of the total PL mass is represented by unsaturated molecular species and most of these are of the polyunsaturated variety. While the relative contributions of unsaturated molecular species to the total pool are modestly reduced for iMEF nucleoplasmic GPLs (as compared with bulk GPLs), the collective data do not support previous conclusions that saturated PtdCho dominates the endonuclear phospholipidome (14).
Implications for nuclear PL signaling
There is an abundance of evidence demonstrating that the nucleus is an active compartment of lipid signaling, however. The nucleus contains the necessary components of a complete phosphoinositide cycle (10, 11, 22): PtdOH and diacylglycerol metabolism (15, 20, 52), phosphoinositide-dependent enzymatic activities that require the intact GPL as a cofactor (21, 53, 54), and SM-dependent sphingolipid signaling pathways (9, 11, 12, 55–60). The barren landscape of the endonuclear phospholipidome, at least under steady-state conditions, holds interesting implications for nucleoplasmic signaling in iMEFs. First, the data suggest nuclear signaling in these cells may involve only small numbers of GPL molecules. Execution of such “pico-scale” signaling implies either an intimate channeling between signaling PLs (or their products) and effector, or that PL action is mediated by direct association with a catalytic activity that efficiently amplifies signaling. One example of the latter scenario is the STAR-PAP poly-A RNA polymerase that uses PtdIns-4,5-P2 as essential cofactor (23, 24). Other examples include direct modification of a phosphoinositide bound to the nuclear receptor by inositide multikinase kinase (25, 26) and control of the basal transcription machinery by nuclear phosphoinositides (27).
Second, if the signaling scheme consumes many GPL molecules, then the bulk of the endonuclear GPL-driven signaling events likely occur on the nucleoplasmic surface of the inner nuclear envelope. It remains formally possible, however, that in situ GPL synthesis also fuels endonuclear lipid signaling pathways. In that regard, the nucleus houses isoforms of the PtdCho-biosynthetic enzymes of the CDP-choline pathway (14, 18). Moreover, robust pathways for GPL import into the nucleoplasm might also exist. PtdIns-driven signaling pathways are outstanding candidates for this last type of supply mechanism, as it is generally accepted that PtdIns synthase activity is excluded from the nuclear compartment (14). Similarly, the paucity of SM in iMEF nucleoplasm implies a requirement for active import pathways if these cells genuinely execute endonuclear SM-signaling pathways.
No matter the scale, consumption of signaling lipids is essential for proper regulation of both the gain and the persistence of any lipid signaling response. Perhaps downregulation is a primary rationale for why lipid metabolic enzymes are found in the nucleoplasm, i.e., to consume signaling lipids by channeling these into production of more inert molecular species. Our identification of 38:4 PtdIns as, by far, the most abundant endonuclear PtdIns molecular species, when only 36:2 PtdOH is detected in this same compartment in an essentially 1:1 stoichiometry, is interesting from this perspective. A simple nucleoplasmic phospholipase C-diacylglycerol kinase pathway will yield endonuclear 38:4 PtdOH. Perhaps this PtdOH is generated, but is rapidly and quantitatively remodeled to 36:2 PtdOH in the nucleoplasm. Such a remodeling could occur via an endonuclear Lands cycle employing the sequential actions of phospholipase A2 and lyso-PtdOH acyltransferase (57). Alternatively, 38:4 PtdOH may be channeled into 38:4 PtdCho or 38:4 PtdEtn synthesis by nuclear isoforms of enzymes of the CDP-choline and CDP-ethanolamine pathways (14, 18). In either case, the physical context for how such enzymes register their substrates in the GPL-depleted nucleoplasm remains an open and fundamental question. How (or even whether) the corresponding biosynthetic products are exported from the nucleoplasm also remains to be established.
Endonuclear lipid dynamics and metabolism
The open questions regarding the scale and nature of GPL signaling and metabolism in the nucleus, and how (or whether) GPLs shuttle into and out of the nucleoplasm, are difficult ones to experimentally address. As suitably noninvasive methods with which to investigate these issues do not exist, the most tractable experimental approaches demand reliable methods to rapidly and effectively purify envelope-stripped nuclear particles. These methods must yield endonuclear fractions with consistent properties, and the analytical platforms must be chaperoned by rigorously defined sets of quality controls. Herein, we describe one such method. Whether this particular protocol translates to other cell types, or whether it requires modification as a function of the specific application, requires further investigation. The method does, however, provide a useful blueprint, and a set of quality control parameters, for development of nuclear matrix purification regimes suited for cell lines of interest to individual researchers.
Supplementary Material
Acknowledgments
E.K.T. and V.A.B. thank Alan Hunt and Anthony Postle (University of Southampton, UK) for introducing them to this interesting biological problem, and for their interest and encouragement in the early stages of this work. David Myers and Stephen Milne (Vanderbilt) are also acknowledged for technical assistance with MS and data processing.
Footnotes
Abbreviations:
- DAPI
- 4′,6-diamidino-2-phenylindole
- EM
- electron microscopy
- ER
- endoplasmic reticulum
- GPL
- glycerophospholipid
- iMEF
- immortalized murine embryonic fibroblast
- MEF
- murine embryonic fibroblast
- PL
- phospholipid
- PtdCho
- phosphatidylcholine
- PtdEtn
- phosphatidylethanolamine
- PtdGro
- phosphatidylglycerol
- PtdIns
- phosphatidylinositol
- PtdOH
- phosphatidic acid
- PtdSer
- phosphatidylserine
This research was supported by Foundation for the National Institutes of Health Grants R01-NS37723 and R01-GM112591, and Welch Foundation Grant BE-0017 awarded to V.A.B. P.T.I. and H.A.B. were supported by National Institutes of Health Large Scale Collaborative Initiative LIPID MAPS Grant U54 GM069338 and the James S. McDonnell Foundation. I.F. and L.C. were supported by Italian MIUR-FIRB Grant RBAP10447J. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no financial conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Irvine R. F. 2006. Nuclear inositide signalling–expansion, structures and clarification. Biochim. Biophys. Acta. 1761: 505–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke J. H., Letcher A. J., D’Santos C. S., Halstead J. R., Irvine R. F., and Divecha N.. 2001. Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukaemia cells. Biochem. J. 357: 905–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunt A. N. 2006. Dynamic lipidomics of the nucleus. Biochim. Biophys. Acta. 1761: 577–587. [DOI] [PubMed] [Google Scholar]
- 4.Monserrate J. P., and York J. D.. 2010. Inositol phosphate synthesis and the nuclear processes they affect. Curr. Opin. Cell Biol. 22: 365–373. [DOI] [PubMed] [Google Scholar]
- 5.Barlow C. A., Laishram R. S., and Anderson R. A.. 2010. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 20: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albi E., and Viola-Magni M. P.. 2004. The role of intranuclear lipids. Biol. Cell. 96: 657–667. [DOI] [PubMed] [Google Scholar]
- 7.Fiume R., Stijf-Bultsma Y., Shah Z. H., Keune W. J., Jones D. R., Jude J. G., and Divecha N.. 2015. PIP4K and the role of nuclear phosphoinositides in tumour suppression. Biochim. Biophys. Acta. 1851: 898–910. [DOI] [PubMed] [Google Scholar]
- 8.Divecha N. 2016. Phosphoinositides in the nucleus and myogenic differentiation: how a nuclear turtle with a PHD builds muscle. Biochem. Soc. Trans. 44: 299–306. [DOI] [PubMed] [Google Scholar]
- 9.Viola-Magni M. P., Gahan P. B., and Pacy J.. 1985. Phospholipids in plant and animal chromatin. Cell Biochem. Funct. 3: 71–78. [DOI] [PubMed] [Google Scholar]
- 10.Cocco L., Gilmour R. S., Ognibene A., Letcher A. J., Manzoli F. A., and Irvine R. F.. 1987. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem. J. 248: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martelli A. M., Gilmour R. S., Bertagnolo V., Neri L. M., Manzoli L., and Cocco L.. 1992. Nuclear localization and signalling activity of phosphoinositidase C beta in Swiss 3T3 cells. Nature. 358: 242–245. [DOI] [PubMed] [Google Scholar]
- 12.Albi E., and Magni M. V.. 1999. Sphingomyelin synthase in rat liver nuclear membrane and chromatin. FEBS Lett. 460: 369–372. [DOI] [PubMed] [Google Scholar]
- 13.Divecha N., Letcher A. J., Banfic H. H., Rhee S. G., and Irvine R. F.. 1995. Changes in the components of a nuclear inositide cycle during differentiation in murine erythroleukaemia cells. Biochem. J. 312: 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt A. N., Clarke G. T., Attard G. S., and Postle A. D.. 2001. Highly saturated endonuclear phosphatidylcholine is synthesized in situ and colocated with CDP-choline pathway enzymes. J. Biol. Chem. 276: 8492–8499. [DOI] [PubMed] [Google Scholar]
- 15.Jones D. R., D’Santos C. S., Merida I., and Divecha N.. 2002. T lymphocyte nuclear diacylglycerol is derived from both de novo synthesis and phosphoinositide hydrolysis. Int. J. Biochem. Cell Biol. 34: 158–168. [DOI] [PubMed] [Google Scholar]
- 16.Smith C. D., and Wells W. W.. 1983. Phosphorylation of rat liver nuclear envelopes. I. Characterization of in vitro lipid phosphorylation. J. Biol. Chem. 258: 9360–9367. [PubMed] [Google Scholar]
- 17.Smith C. D., and Wells W. W.. 1983. Phosphorylation of rat liver nuclear envelopes. II. Characterization of in vitro lipid phosphorylation. J. Biol. Chem. 258: 9368–9373. [PubMed] [Google Scholar]
- 18.Wang Y., Sweitzer T. D., Weinhold P. A., and Kent C.. 1993. Nuclear localization of soluble CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 268: 5899–5904. [PubMed] [Google Scholar]
- 19.Freyberg Z., Sweeney D., Siddhanta A., Bourgoin S., Frohman M., and Shields D.. 2001. Intracellular localization of phospholipase D1 in mammalian cells. Mol. Biol. Cell. 12: 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boronenkov I. V., Loijens J. C., Umeda M., and Anderson R. A.. 1998. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell. 9: 3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topham M. K., Bunting M., Zimmerman G. A., McIntyre T. M., Blackshear P. J., and Prescott S. M.. 1998. Protein kinase C regulates the nuclear localization of diacylglycerol kinase-zeta. Nature. 394: 697–700. [DOI] [PubMed] [Google Scholar]
- 22.Divecha N., Rhee S. G., Lechter A. J., and Irvine R. F.. 1993. Phosphoinositide signalling enzymes in rat liver nuclei: phosphoinositidase C isoform beta 1 is specifically, but not predominantly, located in the nucleus. Biochem. J. 289: 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellman D. L., Gonzales M. L., Song C., Barlow C. A., Wang P., Kendziorski C., and Anderson R. A.. 2008. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 451: 1013–1017. [DOI] [PubMed] [Google Scholar]
- 24.Gonzales M. L., Mellman D. L., and Anderson R. A.. 2008. CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J. Biol. Chem. 283: 12665–12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blind R. D., Miyuki Suzawa M., and Ingraham H. A.. 2012. Direct modification and regulation of a nuclear receptor-phosphoinositide complex by the inositol-lipid kinase IPMK and PTEN. Sci. Signal. 5: ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sablin E. P., Blind R. D., Uthayaruban R., Chiu H. J., Deacon A. M., Das D., Ingraham H. A., and Fletterick R. J.. 2015. Structure of liver receptor homolog-1 (NR5A2) with PIP3 hormone bound in the ligand binding pocket. J. Struct. Biol. 192: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stijf-Bultsma Y., Sommer L., Tauber M., Baalbaki M., Giardoglou P., Jones D. R., Gelato K. A., van Pelt J., Shah Z., Rahnamoun H., et al. . 2015. The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol. Cell. 58: 453–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shay J. W., and Wright W. E.. 1989. Quantitation of the frequency of immortalization of normal human diploid fibroblasts by SV40 large T-antigen. Exp. Cell Res. 184: 109–118. [DOI] [PubMed] [Google Scholar]
- 29.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 30.Ivanova P. T., Milne S. B., Byrne M. O., Xiang Y., and Brown H. A.. 2007. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 432: 21–57. [DOI] [PubMed] [Google Scholar]
- 31.Milne S., Ivanova P., Forrester J., and Brown H. A.. 2006. Lipidomics: an analysis of cellular lipids by ESI-MS. Methods. 39: 92–103. [DOI] [PubMed] [Google Scholar]
- 32.Manzoli L., Billi A. M., Gilmour R. S., Martelli A. M., Matteucci A., Rubbini S., Weber G., and Cocco L.. 1995. Phosphoinositide signaling in nuclei of Friend cells: tiazofurin down-regulates phospholipase C beta 1. Cancer Res. 55: 2978–2980. [PubMed] [Google Scholar]
- 33.Rubbini S., Cocco L., Manzoli L., Lutterman J., Billi A. M., Matteucci A., and Wirtz K. W. A.. 1997. Phosphoinositide signalling in nuclei of Friend cells: DMSO-induced differentiation reduces the association of phosphatidylinositol-transfer protein with the nucleus. Biochem. Biophys. Res. Commun. 230: 302–305. [DOI] [PubMed] [Google Scholar]
- 34.Phair R. D., and Misteli T.. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404: 604–609. [DOI] [PubMed] [Google Scholar]
- 35.De Vries K. J., Westerman J., Bastiaens P. I., Jovin T. M., Wirtz K. W., and Snoek G. T.. 1996. Fluorescently labeled phosphatidylinositol transfer protein isoforms (alpha and beta), microinjected into fetal bovine heart endothelial cells, are targeted to distinct intracellular sites. Exp. Cell Res. 227: 33–39. [DOI] [PubMed] [Google Scholar]
- 36.Phillips S. E., Ile K., Boukhelifa M., Huijbregts R. P. H., and Bankaitis V. A.. 2006. Specific and nonspecific membrane binding determinants cooperate in targeting phosphatidylinositol transfer protein β-isoform to the murine trans-Golgi network. Mol. Biol. Cell. 17: 2498–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alb J. G. Jr., Gedvilaite A., Cartee R. T., Skinner H. B., and Bankaitis V. A.. 1995. Mutant rat phosphatidylinositol/phosphatidylcholine transfer proteins specifically defective in phosphatidylinositol transfer: implications for the regulation of phosphatidylinositol transfer activity. Proc. Natl. Acad. Sci. USA. 92: 8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoder M. D., Thomas L. M., Tremblay J. M., Oliver R. L., Yarbrough L. R., and Helmkamp G. M. Jr. 2001. Structure of a multifunctional protein. Mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J. Biol. Chem. 276: 9246–9252. [DOI] [PubMed] [Google Scholar]
- 39.Tilley S. J., Skippen A., Murray-Rust J., Swigart P. M., Stewart A., Morgan C. P., Cockcroft S., and McDonald N. Q.. 2004. Structure-function analysis of phosphatidylinositol transfer protein alpha bound to human phosphatidylinositol. Structure. 12: 317–326. [DOI] [PubMed] [Google Scholar]
- 40.Fricker M., Hollinshead M., White N., and Vaux D.. 1997. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J. Cell Biol. 136: 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Echevarría W., Leite M. F., Guerra M. T., Zipfel W. R., and Nathanson M. H.. 2003. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat. Cell Biol. 5: 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misteli T. 2005. Concepts in nuclear architecture. BioEssays. 27: 477–487. [DOI] [PubMed] [Google Scholar]
- 43.Rossi G., Magni M. V., and Albi E.. 2007. Sphingomyelin-cholesterol and double stranded RNA relationship in the intranuclear complex. Arch. Biochem. Biophys. 459: 27–32. [DOI] [PubMed] [Google Scholar]
- 44.Rossi G., Viola-Magni M., and Albi E.. 2007. Signal transducer and activator of transcription 3 and sphingomyelin metabolism in intranuclear complex during cell proliferation. Arch. Biochem. Biophys. 464: 138–143. [DOI] [PubMed] [Google Scholar]
- 45.Albi E., Lazzarini R., and Viola Magni M.. 2008. Phosphatidylcholine/sphingomyelin metabolism crosstalk inside the nucleus. Biochem. J. 410: 381–389. [DOI] [PubMed] [Google Scholar]
- 46.Cascianelli G., Villani M., Tosti M., Marini F., Bartoccini E., Viola Magni M., and Albi E.. 2008. Lipid microdomains in cell nucleus. Mol. Biol. Cell. 19: 5289–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alb J. G. Jr., Cortese J. D., Phillips S. E., Albin R. L., Nagy T. R., Hamilton B. A., and Bankaitis V. A.. 2003. Mice lacking phosphatidylinositol transfer protein-alpha exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J. Biol. Chem. 278: 33501–33518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Layerenza J. P., Gonzales P., Garcia de Bravo M. M., Polo M. P., Sisti M. S., and Ves-Losada A.. 2013. Nuclear lipid droplets: a novel nuclear domain. Biochim. Biophys. Acta. 1831: 327–340. [DOI] [PubMed] [Google Scholar]
- 49.Uzbekov R., and Roingeard P.. 2013. Nuclear lipid droplets identified by electron microscopy of serial sections. BMC Res. Notes. 6: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorski S. A., Dundr M., and Misteli T.. 2006. The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol. 18: 284–290. [DOI] [PubMed] [Google Scholar]
- 51.Yagisawa H. 2006. Nucleocytoplasmic shuttling of phospholipase C-delta1: a link to Ca2+. J. Cell. Biochem. 97: 233–243. [DOI] [PubMed] [Google Scholar]
- 52.D’Santos C. S., Clarke J. H., Irvine R. F., and Divecha N.. 1999. Nuclei contain two differentially regulated pools of diacylglycerol. Curr. Biol. 9: 437–440. [DOI] [PubMed] [Google Scholar]
- 53.Gozani O., Karuman P., Jones D. R., Ivanov D., Cha J., Lugovskoy A. A., Baird C. L., Zhu H., Field S. J., Lessnick S. L., et al. . 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 114: 99–111. [DOI] [PubMed] [Google Scholar]
- 54.Mortier E., Wuytens G., Leenaerts I., Hannes F., Heung M. Y., Degeest G., David G., and Zimmermann P.. 2005. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J. 24: 2556–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamiya-Koizumi K., Umekawa H., Yoshida S., and Kojima K.. 1989. Existence of Mg2+-dependent, neutral sphingomyelinase in nuclei of rat ascites hepatoma cells. J. Biochem. 106: 593–598. [DOI] [PubMed] [Google Scholar]
- 56.Alessenko A., and Chatterjee S.. 1995. Neutral sphingomyelinase: localization in rat liver nuclei and involvement in regeneration/proliferation. Mol. Cell. Biochem. 143: 169–174. [DOI] [PubMed] [Google Scholar]
- 57.Neitcheva T., and Peeva D.. 1995. Phospholipid composition, phospholipase A2 and sphingomyelinase activities in rat liver nuclear membrane and matrix. Int. J. Biochem. Cell Biol. 27: 995–1001. [DOI] [PubMed] [Google Scholar]
- 58.Tsugane K., Tamiya-Koizumi K., Nagino M., Nimura Y., and Yoshida S.. 1999. A possible role of nuclear ceramide and sphingosine in hepatocyte apoptosis in rat liver. J. Hepatol. 31: 8–17. [DOI] [PubMed] [Google Scholar]
- 59.Mizutani Y., Tamiya-Koizumi K., Nakamura N., Kobayashi M., Hirabayashi Y., and Yoshida S.. 2001. Nuclear localization of neutral sphingomyelinase 1: biochemical and immunocytochemical analyses. J. Cell Sci. 114: 3727–3736. [DOI] [PubMed] [Google Scholar]
- 60.Albi E., Pieroni S., Viola Magni M. P., and Sartori C.. 2003. Chromatin sphingomyelin changes in cell proliferation and/or apoptosis induced by ciprofibrate. J. Cell. Physiol. 196: 354–361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.