Abstract
apoA-I plays important structural and functional roles in reverse cholesterol transport. We have described the molecular structure of the N-terminal domain, Δ(185-243) by X-ray crystallography. To understand the role of the C-terminal domain, constructs with sequential elongation of Δ(185-243), by increments of 11-residue sequence repeats were studied and compared with Δ(185-243) and WT apoA-I. Constructs up to residue 230 showed progressively decreased percent α-helix with similar numbers of helical residues, similar detergent and lipid binding affinity, and exposed hydrophobic surface. These observations suggest that the C-terminal domain is unstructured with the exception of the last 11-residue repeat (H10B). Similar monomer-dimer equilibrium suggests that the H10B region is responsible for nonspecific aggregation. Cholesterol efflux progressively increased with elongation up to ∼60% of full-length apoA-I in the absence of the H10B. In summary, the sequential repeats in the C-terminal domain are probably unstructured with the exception of H10B. This segment appears to be responsible for initiation of lipid binding and aggregation, as well as cholesterol efflux, and thus plays a vital role during HDL formation. Based on these observations and the Δ(185-243) crystal structure, we propose a lipid-free apoA-I structural model in solution and update the mechanism of HDL biogenesis.
Keywords: apolipoprotein A-I, high density lipoprotein, cholesterol efflux
Heart disease remains the leading cause of death for both women and men in the United States according to the Centers for Disease Control and Prevention’s most recent 2010 vital statistics report (1). High blood pressure and elevated plasma cholesterol are two main risk factors for heart disease and lead to atherosclerosis (2). Plasma levels of HDLs are negatively correlated with the incidence of atherosclerosis due to its involvement in the pathways of reverse cholesterol transport (RCT) (3). However, it is now clear that it is not HDL cholesterol levels in plasma that are directly related to the anti-atherogenic role of HDL, but rather the cholesterol efflux ability of the HDL that determines its role in RCT (2).
apoA-I (243 amino acids, molecular mass 28 kDa) is the major protein component of HDL and plays an important role during RCT. apoA-I has four major functions during RCT: stabilization of the HDL structure, interaction with ABCA1 (4), activation of LCAT (5), and as a ligand for the hepatic HDL receptor, SR-BI (6). apoA-I can exist in three different states: lipid-free, lipid-poor, and lipid-bound, and, as a consequence, has a flexible and adaptable structure similar to the molten globular state (7). This flexible nature, in addition to the nonspecific aggregation properties, has hindered high-resolution structural studies of full-length apoA-I. Until now, the high-resolution structure of full-length lipid-free apoA-I has remained enigmatic. Knowledge of lipid-free apoA-I structure in full-length is crucial to understanding HDL formation and atherosclerosis development.
As shown in Fig. 1, mature apoA-I is encoded by two regions of the gene. Exon-3 encodes the first 43 residues, while exon-4 encodes the residues 44-243 (8). Sequence analysis has suggested that the first 43 residues form a G* helix, and the 44-243 sequence region contains 10 tandem 11/22-residue repeats (H1 to H10) thought to form lipid-binding class A amphipathic helices that represent the fundamental lipid-binding motif (9). In prior studies, we derived consensus sequences for the two types of 11-residue repeats (A and B) that divide the exon-4-encoded region into a series of putative helical segments with different homologies; H1(BB), H2(AA), H3(B), H4(AB), H5(AB), H6(AB), H7(AB), H8(BB), H9(B), and H10(AB) (10). Sequence analysis (9, 10), NMR assignments (11), hydrogen-deuterium exchange measurements (12, 13), and site-directed spin-label electron paramagnetic resonance spectroscopy (14–17) have provided different distributions, flexible regions, and positions for these putative helical tandem repeats in lipid-free or lipid-bound state. In addition, segment deletion and point mutation studies have further elucidated the possible conformation and function for each helical segment (18–27). MD simulations have provided important information about the flexibility of the apoA-I structure in the lipid-free and especially in the lipid-bound state that is hard to achieve through other methods (28–30).
Fig. 1.
Design of apoA-I elongation constructs extended by 11 residue repeating sequence based on consensus sequence analysis and secondary structure of crystal structure Δ(185-243). Green, exon-3 encoded region; ice-blue:,exon-4 encoded region; purple, CSP A homology sequence; cyan, CSP B homology sequence; yellow, proline.
apoA-I is believed to possess a two-domain structural organization with the N-terminal domain (residues 1-189) forming a helix bundle, while the C-terminal domain (190-243) forms a less organized structure (31). Initial binding to a lipid surface is thought to occur through the amphipathic C-terminal domain, followed by opening of the N-terminal helix bundle. This mechanism may represent a general feature for lipid interaction by exchangeable apolipoproteins because apoE behaves similarly. The two-domain structure of apoE has been verified by the full-length apoE3 NMR structure (32). Based on our previous mutation analysis (23–25, 33) and considering that residues 185 and 186 are both glycine, we proposed a slightly different two domain structure: N-terminal domain (1-184) and C-terminal domain (185-243) (34). This analysis led to the successful crystallization and crystallographic determination of the molecular structure of the N-terminal domain Δ(185-243) at 2.2 Å resolution (34).
Different laboratories have produced divergent results concerning the structure of the C-terminal domain. Borhani et al.’s (35) crystal structure that was thought to represent the lipid-bound state showed complete helical formation for the C-terminal domain except the first half of the H10 region. This helical conformation of the C-terminal domain is closely consistent with HX-derived secondary structure assignments for apoA-I in 10 nm discoidal and spherical HDL (36), which showed complete helical structure. The HX-derived secondary structure for lipid-free apoA-I suggested that the whole C-terminal domain lacks defined structure (12). In addition, lipid-free apoA-I models based on cross-linking experiments suggested that part of the H10 region formed helical structure (37, 38). Most recently, MD simulation of full-length apoA-I based on Borhani et al.’s (35) and our crystal structure suggested that parts of H8 and H10 form helical structure after equilibrium in solution (30). These divergent results imply that the C-terminal domain of apoA-I is a very dynamic region and may adopt different conformations and play important roles during the lipid-binding process.
In order to examine the C-terminal domain of apoA-I, successive elongation constructs of Δ(185-243), namely Δ(198-243), Δ(209-243), Δ(220-243), and Δ(231-243) with 11 residue increments of the sequence repeats were generated to probe the structure and function of the C-terminal domain of apoA-I in the lipid-free state and HDL formation as shown in Fig. 1.
MATERIALS AND METHODS
Materials
The n-octyl-β-D-glucopyranoside (BOG) was purchased from Dojindo Laboratories (Dojido Molecular Technologies, Inc., Gaithersburg, MD). Dimyristoylphosphatidylcholine (DMPC), (D+)-glucose monohydrate, and 8-anilino-1-naphthalenesulfonate (ANS) were purchased from Sigma. pDEST-hisMBPs were obtained from Addgene (http://www.addgene.org). Plasmids pDONOR 221, DH5α competent cells, and gateway recombinase were purchased from Invitrogen. BL21 (DE3) CodonPlus-RIL cells were purchased from Stratagene. Histrap and Superdex 75 columns were purchased from GE Healthcare. IPTG, antibiotics, and imidazole were purchased from American Bioanalytical. Complete EDTA-free protease-inhibitor cocktail was purchased from Roche. Amicon concentrators were purchased from Millipore. DMEM, RPMI 1640, MEM-HEPES, FBS, penicillin, and streptomycin were purchased from Invitrogen. BSA, methyl-β-cyclodextrin, 8-(4-chlorophenylthio)adenosine 3′,5′-cyclic monophosphate (cpt-cAMP), ACAT inhibitor (Sandoz 58-035), and HRP-conjugated antibodies to goat were purchased from Sigma. BODIPY-cholesterol (Topfluor) was purchased from Avanti. Genjet was purchased from SinaGen Laboratories. All chemicals used were of analytical grade purity. All solutions were made from doubly distilled water and were filtered by 0.2 μm Millipore filters.
Generation of expression plasmids, expression and purification of apoA-I WT and mutant proteins
Elongation mutations were done following the QuickChange site-directed mutagenesis protocol. Truncated mutations were confirmed by DNA sequencing. Recombinant human WT apoA-I and elongation mutants were expressed in Escherichia coli and purified as described previously (34) with an additional glycine residue at the N terminus. Protein, at concentrations of <0.3 mg/ml, was denatured in 6 M GdnHCl and then dialyzed extensively against 5 mM phosphate buffer at pH 7.4 in the cold room to make sure it was properly folded.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were measured with an Aviv 62DS or Aviv 215 spectropolarimeter equipped with thermoelectric temperature control (Aviv Associates, Lakewood, NJ). Protein solutions at concentrations of ∼0.05–0.3 mg/ml in 5 mM phosphate buffer at pH 7.4 were placed in 1 mm quartz cuvettes for far-UV spectra measurements. Our previous studies (23) have demonstrated that at this protein concentration (e.g., <0.3 mg/ml), plasma apoA-I shows no sign of self-association. Far-UV (250–185 nm) spectra were recorded at 1 nm bandwidth and 1 nm step size with 5 s accumulation time for each data point at 25°C (25). Near-UV (325–250 nm) spectra were recorded at protein concentrations of ∼0.1–0.3 mg/ml in 10 mm quartz cuvettes at 1 nm bandwidth and 1 nm step size with 90 s accumulation time for each data point at 5°C (7). The spectra were normalized to protein concentration and expressed as mean residue ellipticity [Θ] using a mean residue weight. The α-helix content was estimated from the mean residue ellipticity at 222 nm [Θ222] according to a previous protocol (39). ORIGIN 7.5 (Microcal, Amherst, MA) was used for the CD data display and analysis.
Thermal and chemical unfolding.
Thermal unfolding was monitored by the protein ellipticity at 222 nm while heating the samples from 5 to 95°C with a temperature step size of 1°C with 90 s accumulation time for each data point. Van’t Hoff enthalpies (ΔHvs) of the transitions were determined by a conventional van’t Hoff analysis (40) with the assumption of a two-state unfolding model, with baselines that were obtained by linear extrapolation of the pre- and posttransitional regions, as previously described (25). The value of Tm was determined from the peak position of the first derivative function of d[Θ222]/dT 41.
Chemical unfolding was monitored by the protein ellipticity at 222 nm at 25°C, following incubation of protein samples with various concentrations of GdnHCl from 0 to 2 M to allow proteins to reach equilibrium. A linear extrapolation method (42) under the assumption of the two-state unfolding model was used to determine the apparent free energy difference between the native state and unfolded state of the protein in the presence of denaturant, ΔGD0, as previously described (25).
α-Helical induction by BOG
α-Helical content was monitored by protein ellipticity at 222 nm at 25°C, following incubation of protein samples with various concentrations of BOG, from 0 to 50 mM, after allowing the protein to reach equilibrium (27). For each protein, three samples were prepared independently and each sample was scanned three times with 5 s accumulation time for each data point. Three scans for each sample were averaged, the baseline subtracted, and α-helical content estimated.
Fluorescence spectroscopy
Fluorescence measurements were performed on a fluroMax-2 fluorescence spectrometer (Instruments S.A. Inc.) at 25°C using 5 nm excitation and 2.5 nm emission slit widths (33). The ANS fluorescence emission was recorded in the 5 mM phosphate buffer at pH 7.4 in the presence of WT apoA-I or apoA-I mutants with a protein concentration of 0.05 mg/ml and ANS concentration of 0.25 mM. ANS fluorescence was excited at a wavelength of 395 nm and the emission spectra were scanned at wavelengths from 400 to 600 nm. As a control, ANS fluorescence was also scanned in the same buffer in the absence of any protein or with 0.05 mg/ml BSA that is known to have hydrophobic binding pockets, as references.
DMPC turbidity clearance kinetic analysis
The kinetics of binding and remodeling of DMPC multilamellar liposomes by the WT and apoA-I elongation constructs was studied with a DMPC turbidity clearance method, as previously established (24). DMPC, 10 mg powder, was dissolved in a chloroform/methanol (2:1) solution and dried under nitrogen followed by resuspension in 2 ml phosphate buffer by vortexing to give 5 mg/ml DMPC stock. DMPC stock suspension (50 μl, 5 mg/ml, 250 μg total) was added into 1 ml, 0.1 mg/ml, WT or mutant apoA-I protein solution to achieve a lipid protein ratio of 2.5:1 (w/w). The DMPC clearance then was monitored by the decrease in absorbance at 325 nm at 24°C for 1 h.
Cholesterol efflux assay
Human embryonic kidney (HEK)293 cells were maintained in DMEM containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Mouse macrophage-derived J774 cells were maintained in RPMI containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. ABCA1-mediated efflux from HEK293 or J774 cells was determined as described with slight modifications (43, 44). HEK293 cells were plated in 24-well collagen-coated plates. Twenty-four hours later, cells were transiently transfected with cDNA encoding ABCA1 or the corresponding control empty vector using Genjet according to the manufacturer’s instructions. Cellular cholesterol pools were labeled 24 h posttransfection with 1 μM BODIPY-cholesterol in DMSO for 24 h. Cells were then washed and incubated with 10 μg/ml apoA-I or apoA-I mutant forms for 24 h. J774 cells were plated in 24-well plates. Twenty-four hours later, the cellular cholesterol pools were labeled with BODIPY-cholesterol complexed to methyl-β-cyclodextrin at a 1:30 ratio for 90 min in medium A (RPMI containing 0.2% BSA supplemented with 2 μg/ml ACAT inhibitor). ABCA1 was then unregulated by incubating the cells in medium A containing 0.3 mM cpt-cAMP for 20–24 h. Cells were washed and incubated with 10 μg/ml apoA-I forms in medium B (MEM-HEPES containing 0.01% BSA supplemented with 2 μg/ml ACAT inhibitor and 0.3 mM cpt-cAMP) for 24 h. Control cells were incubated in medium lacking cpt-cAMP. Efflux media from HEK293 and J774 cells were filtered to remove cell debris and cells were solubilized in 1% sodium cholate. Fluorescence in cells and media was measured using a plate reader (Tecan Infinite M1000). Percent efflux promoted by the apoA-I forms was calculated from the net fluorescence in medium after subtraction of fluorescence effluxed from cells incubated in medium not containing apoA-I as percent of total fluorescence in medium and cells in each well. Percent efflux from J774 or HEK cells upregulated to ABCA1 (where ABCA1 was not upregulated or transfected with control empty vector, respectively) was lower than the relative efflux calculated for cells not incubated with apoA-I. The HDL particles formed through the efflux in the media were separated by 4–15% native gel and immunoblotted with apoA-I antibodies.
Gel filtration assay
To determine the dimer and monomer distribution, purified protein constructs (0.5–1 mg/ml) were applied on the Superdex 75 10/300 gel filtration column equilibrated with 25 mM Tris and 150 mM NaCl at pH 8.0. The elution was monitored at UV 280 nm.
Cross-linking assay
Concentrations of purified protein were ∼1 mg/ml in 25 mM phosphate and 150 mM NaCl (pH 8.0) buffer for the cross-linking assay. Freshly solubilized BS3 was added to the protein solutions at a final concentration 0.25 mM. The mixed solution was cross-linked at room temperature for 30 min then 1 M Tris-HCl (pH 7.5) was added to quench the reaction for 15 min at room temperature and then loaded on a 4–15% SDS-PAGE.
RESULTS
α-Helical content and stability in the lipid-free state
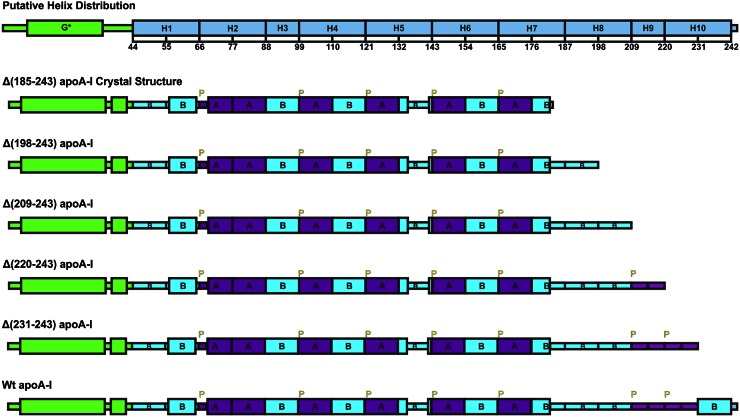
Normalized far-UV spectra of the WT and the apoA-I elongation variants were used to estimate the α-helical content in the secondary structure of the proteins. The number of residues in helical conformation was estimated from the α-helical content for each recombinant protein. As shown in Fig. 2A and Table 1, with the sequential elongation of the C-terminal domain up to residue 230, the helical content progressively decreased, while the number of residues in helical conformation remained approximately unchanged at ∼125 residues. These observations suggest that most of the residues in the C-terminal domain between residues 185 and 230 lack defined structure. The Δ(231-243) construct had similar helical content to WT full-length apoA-I, but the same number of helical residues (125) as the shorter constructs. Addition of the last 13 residues to give the full-length WT protein results in an increase in the number of helical residues to 132, suggesting that the last 11 residue repeat (H10B) contains helical structure.
Fig. 2.
Solution characteristics of apoA-I elongation constructs in lipid-free state by CD. A: Helical content and residues in helix probed by far-UV CD. B: Unfolding cooperativity probed by thermal unfolding normalized to WT apoA-I. C: Stability probed by GdnHCl denaturation normalized to WT apoA-I. D: Tertiary structures probed by near-UV CD.
TABLE 1.
α-Helical content and thermodynamic parameters of WT and elongation variants of lipid-free apoA-I determined by far-UV CD
| apoA-I | α-Helix (%)a | Number of Residues | Tm (°C)c | ΔHv (kcal/mol)c | ΔGD0 (kcal/mol)d | |
| In Protein | In Helixb | |||||
| Δ(185-243) | 68 ± 2e | 185 (−59) | 125 (−7) | 58 ± 1e | 40 ± 4e | 6.2 ± 0.5e |
| Δ(198-243) | 63 ± 1e | 198 (−46) | 125 (−7) | 63 ± 1e | 36 ± 2e | 6.1 ± 1.0 |
| Δ(209-243) | 60 ± 2e | 209 (−35) | 125 (−7) | 63 ± 1e | 35 ± 3e | 5.4 ± 1.0 |
| Δ(220-243) | 57 ± 1e | 220 (−24) | 125 (−7) | 62 ± 1e | 35 ± 3e | 5.1 ± 1.0 |
| Δ(231-243) | 54 ± 1 | 231 (−13) | 125 (−7) | 62 ± 1e | 30 ± 1e | 5.1 ± 0.3 |
| WT | 54 ± 1 | 244 | 132 | 64 ± 1 | 22 ± 2 | 4.6 ± 0.3 |
Values are the average ± SD from three to nine experiments for three independent preparations of each protein.
Estimated from the value [Θ222] at pH 7.4 and 25°C.
The number of residues in the helical conformation was estimated by multiplying the number of residues in the protein by its α-helical content. Values in parentheses show change in the number of residues compared with WT.
Parameters determined from the thermal unfolding curves. Tm is the maximum of the first derivative d(Θ222)/dT; the effective enthalpy, ΔHv, were determined from van’t Hoff analysis of the thermal unfolding curves.
The conformational stability, ΔGD0, was determined by the linear extrapolation method from the GdnHCl induced unfolding curves.
Significance of difference from the value for WT: P < 0.05.
As shown in Fig. 2B and Table 1, the successive C-terminal elongation constructs up to residue 230 exhibited similar unfolding cooperativity to Δ(185-243) that was greater than WT full-length apoA-I. These observations suggest that the final H10B sequence repeat in the C-terminal domain has an effect on the N-terminal helical bundle. Thus, the helical conformation in this region may result in interaction of H10B with the N-terminal helix bundle. Previous research has also suggested that the C-terminal helix is located near the N-terminal helix bundle and there is interaction between them (25, 26, 45). Due to the increase in the unfolding cooperativity, the stability of the elongation apoA-I variants is increased or remains similar compared with WT apoA-I (Fig. 2C, Table 1). Previous research also showed that Δ(191-220) exhibited lower α-helix content and thermal stability compared with WT apoA-I (45). Considering that the 191-220 segment may form a loop, this deletion might shift the H10B helix interaction with the N-terminal helix bundle causing the destabilization, while the elongation apoA-I variants remain similar or have increased stability.
Near-UV spectra
Near-UV spectra of the variant forms of apoA-I are shown in Fig. 2D. The major contribution to the near-UV spectra comes from the aromatic side chains (7). WT apoA-I and the C-terminal elongation constructs contain four Trp residues (W8, 50, 72, and 108), which are all located at the N-terminal domain helical bundle. All the elongation constructs and WT apoA-I have similar near-UV spectra after normalization to protein concentration and show a large negative peak at 292 nm (7) that corresponds to the Trp residues, suggesting that the WT and all the elongation constructs have similar tertiary structure. The C-terminal elongation constructs still possessed all four Trp, and showed similar near-UV spectra after normalization to protein concentration to WT apoA-I, but with a deeper peak at 292 nm. Thus, elongation of the C-terminal region decreased the Trp signal in the near-UV spectra, suggesting a more relaxed environment of the Trp (7). This observation suggests that elongation of the C terminus did not result in a major change in the N-terminal helical bundle organization, but made it somewhat less compact, consistent with the thermal unfolding data, with less unfolding cooperativity compared with Δ(185-243).
ANS fluorescence analysis
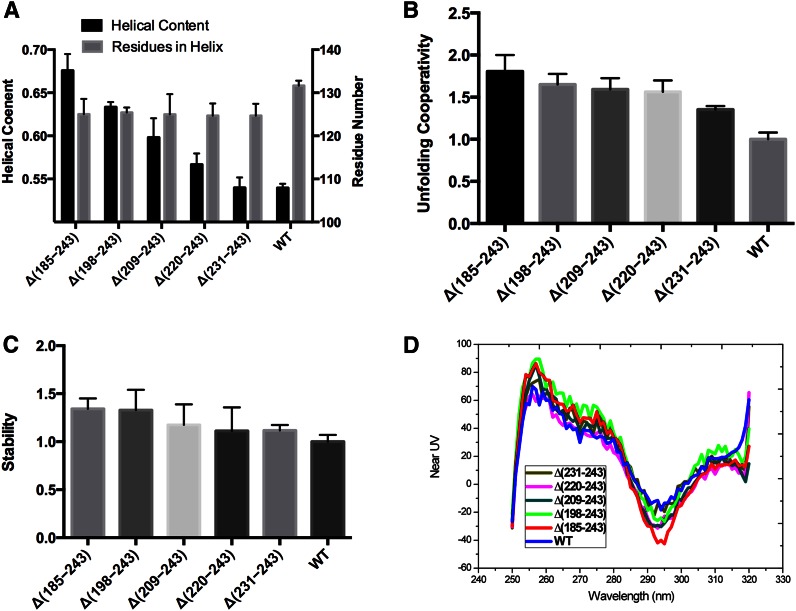
Fluorescence of ANS in the presence of the lipid-free WT and variant forms of apoA-I was measured to determine whether the C-terminal elongations affected the exposure of hydrophobic surfaces or cavities of apoA-I. The intrinsic fluorescence of ANS has been shown to be enhanced and blue shifted upon binding to hydrophobic surfaces or cavities, while the water-phase dye does not contribute to the emission (46). ANS fluorescence spectra recorded in the presence of WT and the apoA-I elongation variants, BSA, and in the buffer alone are shown in Fig. 3A. The wavelength of maximum fluorescence and relative intensity values determined from the spectra are listed in Table 2. In phosphate buffer, ANS fluorescence has a very low intensity and an emission maximum at 517 nm. In the presence of BSA, a fatty acid transporter that has multiple hydrophobic binding pockets, there is a significant (54 nm) blue shift and almost ∼7-fold enhancement in ANS fluorescence compared with the ANS in phosphate buffer alone. In the presence WT apoA-I, ANS emission shows a 40 nm blue shift and ∼4-fold increase in the intensity compared with the ANS in the buffer alone. In the presence of all the C-terminal elongation constructs up to residue 230, ANS emission shows only a 27–29 nm blue shift and similar fold (∼2-fold) increase in the intensity compared with the ANS in the buffer alone. These observations suggest that the C terminus of apoA-I has exposed hydrophobic surface due to the 231-241 residue (H10B) region. The C-terminal domain of apoA-I is generally considered to lack structure and be responsible for the aggregation and lipid binding (20). The combination of the CD data and the ANS fluorescence data suggests that formation of the helical structure in the 231-241 residue, H10B, region is required to obtain a large exposed hydrophobic surface or binding region because the preceding residues of the C-terminal domain have less effect. Previous research has also demonstrated that Δ(190-243) (31) and single (L230P) or triple (L230P/L233P/Y236P) proline insertion into the C-terminal helix decreases the ANS fluorescence (47), which is consistent with our data, suggesting that helical structure is required in the H10B region to obtain a large hydrophobic surface or binding region.
Fig. 3.
Exposed hydrophobic surface, detergent and lipid interaction of apoA-I elongation constructs. A: Exposed hydrophobic surface probed by ANS fluorescent intensity. B: Helical content induced by BOG. D1/2 is the BOG concentration required to induce half of the total induced helix. C: Helical residues before and after interaction with BOG. D: DMPC clearance assay monitored at 325 nm.
TABLE 2.
ANS fluorescence in the presence of WT and elongation variants of apoA-I
| Protein | WMF (ANS) (nm)a | IWTb | IPBc |
| Δ(185-243) | 488 (−29) | 0.54 ± 0.09 | 2.14 ± 0.45 |
| Δ(198-243) | 488 (−29) | 0.52 ± 0.08 | 2.06 ± 0.38 |
| Δ(209-243) | 490 (−27) | 0.50 ± 0.08 | 2.01 ± 0.39 |
| Δ(220-243) | 489 (−28) | 0.50 ± 0.08 | 1.99 ± 0.38 |
| Δ(231-243) | 488 (−29) | 0.53 ± 0.06 | 2.11 ± 0.31 |
| WT | 477 (−40) | 1.00 ± 0.06 | 3.98 ± 0.31 |
| BSA | 471 (−54) | 1.78 ± 0.05 | 7.08 ± 0.23 |
| ANS in PB alone | 517 | 0.25 ± 0.04 | 1.00 ± 0.18 |
WMF, wavelength of maximum fluorescence; PB, phosphate buffer.
ANS fluorescence in the presence of 0.05 mg/ml WT or the elongation variants of apoA-I or BSA or in phosphate buffer alone. Values in parentheses show change in WMF compared with ANS fluorescence in phosphate buffer alone.
ANS fluorescence intensity in relative units compared with the fluorescence in the presence of WT apoA-I.
ANS fluorescence intensity in relative units compared with the fluorescence in the buffer alone.
Interaction with BOG
BOG has been widely used as a mild lipid-mimicking detergent to induce and stabilize amphipathic α-helical structure in solution and during protein crystallization (48). The detergent has been used in numerous studies to investigate the effects of lipid environment in the helical conformation of the apolipoproteins (27). The critical micelle concentration (CMC) of BOG at 25°C is 20–25 mM. The α-helical content of the apoA-I variants as a function of BOG concentration is shown in Fig. 3B. The α-helical content of WT apoA-I increased from ∼54% to ∼64% as the BOG concentration increased from 0 mM to 50 mM and reached a maximum at the BOG CMC concentration, 25 mM. These results suggest that BOG can bind to WT apoA-I and induce α-helical structure by ∼10%. Induction of α-helical structure by BOG was also observed for the C-terminal elongation apoA-I variants, suggesting that BOG may bind to the C-terminal region and induce helical structure in this region, again reaching maximum effect after BOG micelle formation. Most interestingly, Δ(185-243) lost ∼5% helical content at the CMC, suggesting that BOG may partially disrupt the more compact structure in the N-terminal domain by binding with an exposed nonstructured region and affecting the structured region. All of the C-terminal elongation constructs form additional helical structure after binding to BOG. With the successive C-terminal elongation, more helical residues can be induced, suggesting that most of the C-terminal domain lacks helical structure, as shown in Fig. 3C. In addition, the BOG concentration to induce 50% of the total helical structure (D1/2) of the Δ(198-243), Δ(209-243), Δ(220-243), and Δ(231-243) forms are similar, ∼20 mM as shown in Fig. 3B. With the addition of the last H10b region, the D1/2 of the WT apoA-I significantly decreases to ∼15 mM BOG. This observation suggests that H10B with an exposed hydrophobic surface due to helical structure facilitates the binding with BOG and thus induces helical formation in the rest of the C-terminal domain of apoA-I.
DMPC turbidity clearance
Association of lipid-free apoA-I with DMPC multilamellar vesicles causes a decrease in the turbidity at 325 nm. This process reflects the kinetics of formation of DMPC-apoA-I complexes (49). Figure 3D shows the clearance curves. Compared with WT apoA-I, the C-terminal elongation constructs all showed decreased initial rate of clearance of DMPC liposome turbidity that almost removed the DMPC binding ability completely. This observation is consistent with data from other laboratories suggesting that the C-terminal domain of apoA-I is vital to initiate lipid binding (31). The elongation constructs demonstrate, for the first time, that the H10B region in the C-terminal domain is the vital region to initiate the DMPC binding and perhaps facilitate opening of the N-terminal folding domain for lipid solubilization. This result is consistent with the BOG binding and ANS fluorescence experiments.
Gel filtration and cross-linking
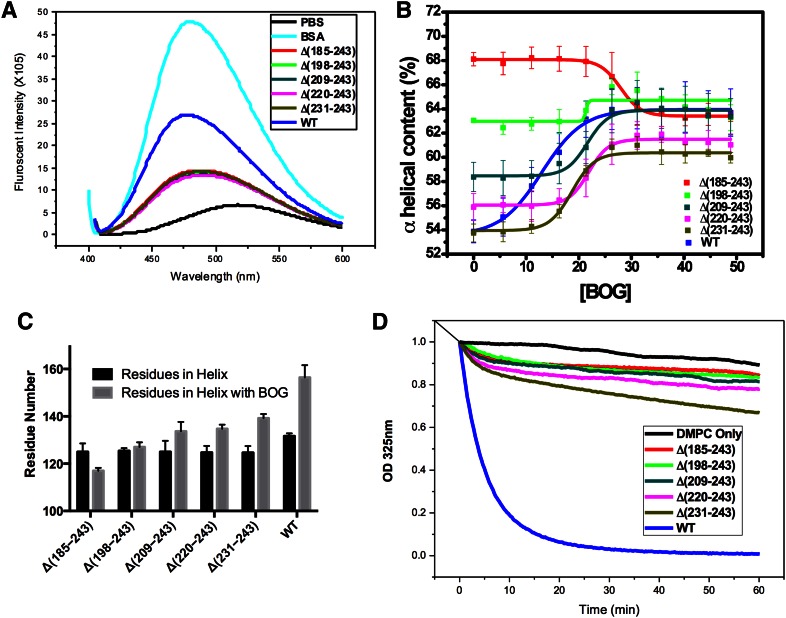
Previously, we have shown that after deletion of the C-terminal domain, Δ(185-243) undergoes a monomer-dimer equilibrium that facilitates the crystallization of Δ(185-243). As shown in Fig. 4A, the C-terminal elongation constructs show a similar monomer-dimer equilibrium to Δ(185-243) and progressively migrate toward the full-length apoA-I elution position due to the increment in the molecular weight. Full-length apoA-I exhibits a complex elution profile as a result of nonspecific self-association. In addition, cross-linking of the C-terminal elongation constructs with BS3 shows similar dimer/monomer forms on SDS-PAGE to Δ(185-243), while WT apoA-I can be cross-linked to higher oligomers, as shown in Fig. 4B. These data suggest that the H10B region not only forms helical structure but is also responsible for nonspecific aggregation of apoA-I.
Fig. 4.
Gel filtration (A) and cross-linking (B) of apoA-I elongation constructs.
Cholesterol efflux
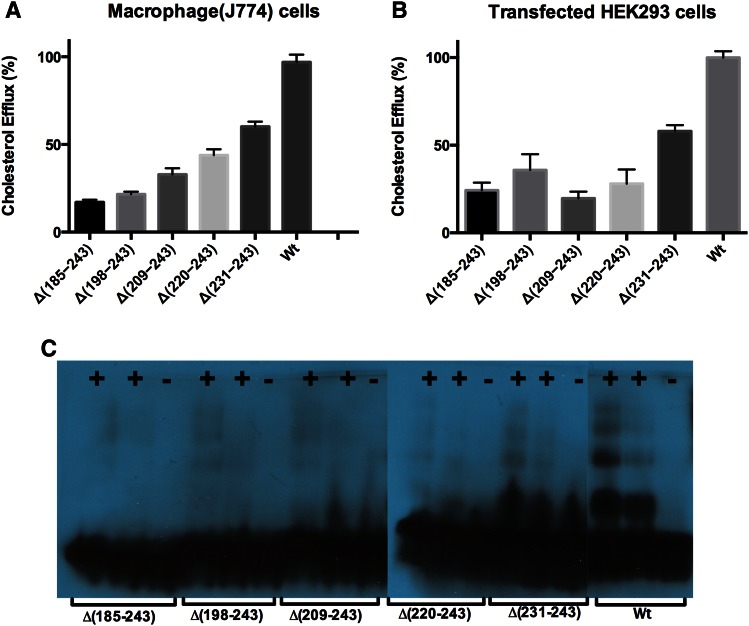
In order to test the role of the C-terminal domain of apoA-I in ABCA1-mediated cholesterol efflux, efflux assays were conducted using macrophages and transfected HEK cells. As shown in Fig. 5A, B, with the elongation of the C-terminal domain, the cholesterol efflux progressively increased. However, the cholesterol efflux ability of Δ(231-243) only achieved ∼60% compared with WT apoA-I. Again, these observations suggest that the last 11 residues, the H10B region, play a vital role during the interaction with ABCA1 to facilitate the cholesterol efflux. This result is consistent with previous research that showed that deletion [Δ(223-243)] or disruption of the C-terminal lipid binding domain (L230P or L230P233PY236P) of apoA-I drastically reduced cholesterol efflux, indicating that the C-terminal helix is essential during the cholesterol efflux process (50). Western blot analysis using anti-apoA-I antibodies shows that Δ(231-243) forms HDL-sized particles similar to WT apoA-I, whereas the shorter C-terminal constructs form less well-defined HDL particles due to decreased efflux, as shown in Fig. 5C.
Fig. 5.
Cholesterol efflux assay of apoA-I elongation constructs. A: Cholesterol efflux in macrophage J774 cells. B: Cholesterol efflux in transfected HEK293 cells. C: Western blots of HDL particles formed in the media of HEK293 cells using apoA-I antibodies. +, HEK293 cells transfected with ABCA-1 vectors; −, HEK293 cells transfected with empty vectors.
DISCUSSION
C-terminal domain structure in the lipid-free state
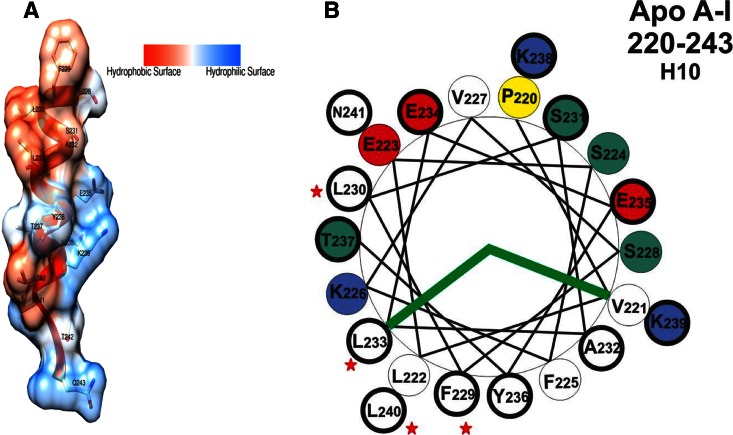
Based on the CD analysis, we concluded that most of the C-terminal domain of apoA-I, residues 185-243, adopts unstructured conformation with the exception of the final H10B repeat, residues 231-241, that forms helical structure. MMP-8-treated apoA-I digested at positions Glu191-Tyr192 and Val221-Leu222 also suggests that this region lacks structure in lipid-free apoA-I (51). Additionally, this analysis is consistent with the low-resolution crystal structure of Δ(1-43) from Borhani et al. (35). As shown in Fig. 6A, the H10B region is in helical conformation with a large exposed hydrophobic surface in this crystal structure. The helical-wheel representation of the H10 region shown in Fig. 6B demonstrates that the H10B region has a larger hydrophobic surface compared with other helical regions (52). Thus, not only hydrophobic residues, but also helical structure, are required to obtain a large exposed hydrophobic surface. A full-length molecular dynamics simulation of an apoA-I model by Segrest et al. (30) also suggested that the H10 region (residues 226-239) forms helical structure. Electron paramagnetic resonance studies of the C-terminal domain of apoA-I suggested that residues 221-236 form helical structure, while the rest lack definite structure in the lipid-free state (14). The entire C-terminal domain converts into helical structure upon lipid binding (14). Residues 191-220 play a role in enhancing the ability of apoA-I to bind to and solubilize lipids by forming helix upon lipid interaction (45). All of these studies suggest that the C-terminal domain has a dynamic structure and plays an important role during the lipid-binding process by shifting from disordered into helical structure.
Fig. 6.
Exposed hydrophobic surface of the H10B region. A: Surface hydrophobicity of residues 228-243 from Borhani et al.’s (35) crystal structure (Protein Data Bank accession number: 1AV1). B: Helical wheel of the H10 region of apoA-I. Bold circle, H10B residues 231-241; white, hydrophobic residue; yellow, proline; red, acidic residue; blue, basic residue; green, neutral residue; red star, hydrophobic residues of H10B in or near the hydrophobic surface of the helical wheel.
The H10B region is responsible for lipid binding and aggregation
All of the elongation constructs lacking the H10B region show decreased hydrophobic exposure to ANS and slow DMPC clearance similar to Δ(185-243). These results are consistent with previous research demonstrating that the C-terminal domain is responsible for lipid binding (20). Here we pinpoint this function to the H10B region. The helical structure in this region in solution forms a large exposed hydrophobic surface that is responsible for lipid binding.
In addition, gel filtration and cross-linking demonstrate that all the constructs lacking the H10B region exhibit monomer and dimer equilibrium in contrast to the nonspecific aggregation of WT apoA-I. Thus, for the first time, we identify that the region of apoA-I responsible for aggregation corresponds to that responsible for initiation of lipid binding. This result is consistent with previous studies that show that a peptide representing the apoA-I α-helix composed of amino acids 220-241 has the highest affinity for lipids (19) compared with other sequence repeats, and an apoA-I mutation lacking the C-terminal residues (residues 187-243) has reduced ability to bind with lipids to form particles (22).
The H10B region is vital for the formation of HDL through cholesterol efflux
The large hydrophobic surface area of the H10B region plays a vital role during cholesterol efflux. The elongation constructs up to residue 230 restore some cholesterol efflux ability compared with Δ(185-243), but only achieve approximately 60% of full-length apoA-I efflux ability. This observation suggests that the H10B region with exposed hydrophobic surface facilitates the interaction with lipid membrane. WT apoA-I has significantly lower BOG concentration required to induce the secondary structure of apoA-I. This observation suggests that the H10B region forming hydrophobic helix facilitates the integration between apoA-I and the cell membrane, thus facilitating cholesterol efflux. Recently, a novel apoA-I truncation (apoA-I Mytilene), due to a heterozygous nonsense mutation (Gln216), has been reported to be associated with markedly decreased levels of HDL cholesterol and decreased total cholesterol efflux consistent with the effect of the truncation mutations (53). Furthermore, treatment of apoA-I and HDL with MMP-8 resulted in a significant reduction (up to 84%) in their ability to facilitate cholesterol efflux from cholesterol-loaded macrophages (51). In addition, apoA-I Nichinan (ΔE235) is associated with low plasma HDL levels due to the disruption of the ability of the C-terminal region to form helical structure, resulting in reduced efficiency in lipid binding and cholesterol efflux ability (54).
Lipid-free apoA-I in solution and HDL formation mechanism
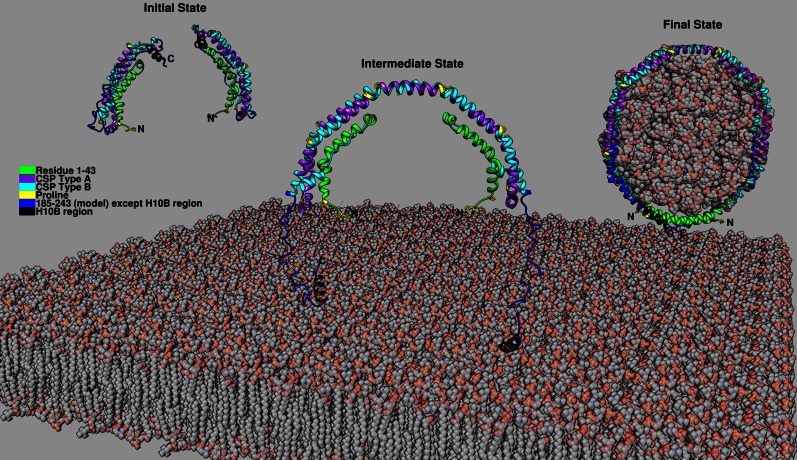
Previously, we have proposed a model for the monomer conformation of Δ(185-243) based on the crystal dimer of Δ(185-243). In this “domain swap” model, the helix 5 region converts from helical structure into a loop leading to the H6 and H7 regions folding back to form the helical bundle with the N-terminal region. With the knowledge of C-terminal domain being unstructured with the exception of the H10B region, we propose a model for full-length apoA-I monomer in solution. In this model, the C-terminal domain can extend out or fold back onto the helix bundle. Considering that all of the C-terminal elongation constructs are not degraded during expression, the C-terminal folded-back conformation is more likely. This concept is consistent with Segrest et al.’s (30) simulation model in which the H10 region forms helical structure. In hydrogen-deuterium exchange experiments, the last helix could not be detected due to rapid hydrogen-deuterium exchange because of unstable helical structure in the measurement time frame (12). Based on this full-length apoA-I model and the two-step lipid binding model (31), we have updated our previous proposed mechanism for HDL formation (55) as shown in Fig. 7. In the initial state, monomeric apoA-I adopts the helix bundle conformation with the H6 and H7 regions folded back with the C-terminal H10B region helical forming exposed hydrophobic surface. The interaction between the N- and C-terminal domains results in a molten globular structure of apoA-I in solution. This interaction makes the N-terminal helical bundle less stable and more readily accessible for interaction with lipids. In the intermediate state, when the monomeric apoA-I is close to the cell membrane, the C-terminal H10B region may bind to the pleating region of the membrane formed through ABCA1 action (56) either spontaneously (57) or facilitated through the interaction with ABCA1. Further binding of an additional apoA-I, also perhaps through ABCA1 interaction, can position the two monomeric apoA-Is close enough to facilitate the domain swap interaction and result in dimerization, while the N-terminal domain remains in the aqueous phase. Additional lipid interaction with the H5 region might facilitate this process. In the final state, lipids are solubilized by the double belt dimer with additional helical conformation forming in the unstructured region of the C-terminal domain, further stabilizing the interaction and resulting in the release of discoidal HDL from the cell membrane.
Fig. 7.
Proposed full-length apoA-I in the lipid-free state and a possible three-state mechanism of HDL formation. Full-length apoA-I in lipid-free state: residues 1-184 from Δ(185-243) crystal structure (Protein Data Bank accession number: 3R2P), residues 185-243 from Δ(1-43) crystal structure (Protein Data Bank accession number: 1AV1) with relaxation from helix to random coil except the H10B region. Initial state: Monomeric apoA-I adopts the helix bundle conformation with the C-terminal helical H10B region forming exposed hydrophobic surface. Intermediate state: Two monomeric apoA-I close enough to facilitate the domain swap interaction and result in dimerization due to binding to lipids spontaneously or facilitated through ABCA1. Final state: Lipids are solubilized by the double belt dimer with additional helical conformation forming in the unstructured region of the C-terminal domain further stabilizing the interaction and resulting in the release of discoidal HDL from the cell membrane.
Acknowledgments
The authors thank Dr. Olga Gursky and Dr. Shobini Jayaraman for advice for the CD experiment and helpful discussion.
Footnotes
Abbreviations:
- ANS
- 8-anilino-1-naphthalenesulfonate
- BOG
- n-octyl-β-D-glucopyranoside
- CD
- circular dichroism
- CMC
- critical micelle concentration
- DMPC
- dimyristoylphosphatidylcholine
- HEK
- human embryonic kidney
- RCT
- reverse cholesterol transport
- [Θ]
- mean residue ellipticity
This work was supported by National Heart, Lung, and Blood Institute Grant HL116518-02. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Murphy S. L., Xu J., and Kochanek K. D.. 2013. Deaths: Final Data for 2010. National vital statistics reports; vol 61 no 4. National Center for Health Statistics, Hyattsville, MD. [PubMed] [Google Scholar]
- 2.Rosamond W., Flegal K., Friday G., Furie K., Go A., Greenlund K., Haase N., Ho M., Howard V., Kissela B., et al. 2007. Heart disease and stroke statistics–2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 115: e69–e171. [Erratum. 2010. Circulation 122: e9.] [DOI] [PubMed] [Google Scholar]
- 3.Fielding C. J., and Fielding P. E.. 1995. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 36: 211–228. [PubMed] [Google Scholar]
- 4.Lee J-Y., and Parks J. S.. 2005. ATP-binding cassette transporter AI and its role in HDL formation. Curr. Opin. Lipidol. 16: 19–25. [DOI] [PubMed] [Google Scholar]
- 5.Fielding C. J., Shore V. G., and Fielding P. E.. 1972. A protein cofactor of lecithin:cholesterol acyltransferase. Biochem. Biophys. Res. Commun. 46: 1493–1498. [DOI] [PubMed] [Google Scholar]
- 6.Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., and Krieger M.. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 271: 518–520. [DOI] [PubMed] [Google Scholar]
- 7.Gursky O., and Atkinson D.. 1996. Thermal unfolding of human high-density apolipoprotein A-1: implications for a lipid-free molten globular state. Proc. Natl. Acad. Sci. USA. 93: 2991–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcel Y. L., and Kiss R. S.. 2003. Structure-function relationships of apolipoprotein A-I: a flexible protein with dynamic lipid associations. Curr. Opin. Lipidol. 14: 151–157. [DOI] [PubMed] [Google Scholar]
- 9.Segrest J. P., Jackson R. L., Morrisett J. D., and Gotto A. M. Jr. 1974. A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 38: 247–258. [DOI] [PubMed] [Google Scholar]
- 10.Nolte R. T., and Atkinson D.. 1992. Conformational analysis of apolipoprotein A-I and E-3 based on primary sequence and circular dichroism. Biophys. J. 63: 1221–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okon M., Frank P. G., Marcel Y. L., and Cushley R. J.. 2002. Heteronuclear NMR studies of human serum apolipoprotein A-I. Part I. Secondary structure in lipid-mimetic solution. FEBS Lett. 517: 139–143. [DOI] [PubMed] [Google Scholar]
- 12.Chetty P. S., Mayne L., Lund-Katz S., Stranz D., Englander S. W., and Phillips M. C.. 2009. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 19005–19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sevugan Chetty P., Mayne L., Kan Z. Y., Lund-Katz S., Englander S. W., and Phillips M. C.. 2012. Apolipoprotein A-I helical structure and stability in discoidal high-density lipoprotein (HDL) particles by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 109: 11687–11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oda M. N., Forte T. M., Ryan R. O., and Voss J. C.. 2003. The C-terminal domain of apolipoprotein A-I contains a lipid-sensitive conformational trigger. Nat. Struct. Biol. 10: 455–460. [DOI] [PubMed] [Google Scholar]
- 15.Lagerstedt J. O., Cavigiolio G., Budamagunta M. S., Pagani I., Voss J. C., and Oda M. N.. 2011. Structure of apolipoprotein A-I N terminus on nascent high density lipoproteins. J. Biol. Chem. 286: 2966–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagerstedt J. O., Budamagunta M. S., Liu G. S., DeValle N. C., Voss J. C., and Oda M. N.. 2012. The “beta-clasp” model of apolipoprotein A-I–a lipid-free solution structure determined by electron paramagnetic resonance spectroscopy. Biochim. Biophys. Acta. 1821: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda M. N., Budamagunta M. S., Borja M. S., Petrlova J., Voss J. C., and Lagerstedt J. O.. 2013. The secondary structure of apolipoprotein A-I on 9.6-nm reconstituted high-density lipoprotein determined by EPR spectroscopy. FEBS J. 280: 3416–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorci-Thomas M., Kearns M. W., and Lee J. P.. 1993. Apolipoprotein A-I domains involved in lecithin-cholesterol acyltransferase activation. Structure:function relationships. J. Biol. Chem. 268: 21403–21409. [PubMed] [Google Scholar]
- 19.Palgunachari M. N., Mishra V. K., Lund-Katz S., Phillips M. C., Adeyeye S. O., Alluri S., Anantharamaiah G. M., and Segrest J. P.. 1996. Only the two end helixes of eight tandem amphipathic helical domains of human apo A-I have significant lipid affinity. Implications for HDL assembly. Arterioscler. Thromb. Vasc. Biol. 16: 328–338. [DOI] [PubMed] [Google Scholar]
- 20.Laccotripe M., Makrides S. C., Jonas A., and Zannis V. I.. 1997. The carboxyl-terminal hydrophobic residues of apolipoprotein A-I affect its rate of phospholipid binding and its association with high density lipoprotein. J. Biol. Chem. 272: 17511–17522. [DOI] [PubMed] [Google Scholar]
- 21.Rogers D. P., Roberts L. M., Lebowitz J., Datta G., Anantharamaiah G. M., Engler J. A., and Brouillette C. G.. 1998. The lipid-free structure of apolipoprotein A-I: effects of amino-terminal deletions. Biochemistry. 37: 11714–11725. [DOI] [PubMed] [Google Scholar]
- 22.Burgess J. W., Frank P. G., Franklin V., Liang P., McManus D. C., Desforges M., Rassart E., and Marcel Y. L.. 1999. Deletion of the C-terminal domain of apolipoprotein A-I impairs cell surface binding and lipid efflux in macrophage. Biochemistry. 38: 14524–14533. [DOI] [PubMed] [Google Scholar]
- 23.Gorshkova I. N., Liadaki K., Gursky O., Atkinson D., and Zannis V. I.. 2000. Probing the lipid-free structure and stability of apolipoprotein A-I by mutation. Biochemistry. 39: 15910–15919. [DOI] [PubMed] [Google Scholar]
- 24.Gorshkova I. N., Liu T., Zannis V. I., and Atkinson D.. 2002. Lipid-free structure and stability of apolipoprotein A-I: probing the central region by mutation. Biochemistry. 41: 10529–10539. [DOI] [PubMed] [Google Scholar]
- 25.Fang Y., Gursky O., and Atkinson D.. 2003. Structural studies of N- and C-terminally truncated human apolipoprotein A-I. Biochemistry. 42: 6881–6890. [DOI] [PubMed] [Google Scholar]
- 26.Fang Y., Gursky O., and Atkinson D.. 2003. Lipid-binding studies of human apolipoprotein A-I and its terminally truncated mutants. Biochemistry. 42: 13260–13268. [DOI] [PubMed] [Google Scholar]
- 27.Zhu H. L., and Atkinson D.. 2004. Conformation and lipid binding of the N-terminal (1-44) domain of human apolipoprotein A-I. Biochemistry. 43: 13156–13164. [DOI] [PubMed] [Google Scholar]
- 28.Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., and Segrest J. P.. 2010. Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285: 4652–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segrest J. P., Jones M. K., Catte A., and Thirumuruganandham S. P.. 2012. Validation of previous computer models and MD simulations of discoidal HDL by a recent crystal structure of apoA-I. J. Lipid Res. 53: 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segrest J. P., Jones M. K., Shao B., and Heinecke J. W.. 2014. An experimentally robust model of monomeric apolipoprotein A-I created from a chimera of two X-ray structures and molecular dynamics simulations. Biochemistry. 53: 7625–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito H., Dhanasekaran P., Nguyen D., Holvoet P., Lund-Katz S., and Phillips M. C.. 2003. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 278: 23227–23232. [DOI] [PubMed] [Google Scholar]
- 32.Chen J., Li Q., and Wang J.. 2011. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. USA. 108: 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorshkova I. N., Liu T., Kan H. Y., Chroni A., Zannis V. I., and Atkinson D.. 2006. Structure and stability of apolipoprotein a-I in solution and in discoidal high-density lipoprotein probed by double charge ablation and deletion mutation. Biochemistry. 45: 1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mei X., and Atkinson D.. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286: 38570–38582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borhani D. W., Rogers D. P., Engler J. A., and Brouillette C. G.. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94: 12291–12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chetty P. S., Nguyen D., Nickel M., Lund-Katz S., Mayne L., Englander S. W., and Phillips M. C.. 2013. Comparison of apoA-I helical structure and stability in discoidal and spherical HDL particles by HX and mass spectrometry. J. Lipid Res. 54: 1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard R. D., Fulp B., Samuel M. P., Sorci-Thomas M. G., and Thomas M. J.. 2013. The conformation of lipid-free human apolipoprotein A-I in solution. Biochemistry. 52: 9470–9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva R. A., Hilliard G. M., Fang J., Macha S., and Davidson W. S.. 2005. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 44: 2759–2769. [DOI] [PubMed] [Google Scholar]
- 39.Mao D., and Wallace B. A.. 1984. Differential light scattering and absorption flattening optical effects are minimal in the circular dichroism spectra of small unilamellar vesicles. Biochemistry. 23: 2667–2673. [DOI] [PubMed] [Google Scholar]
- 40.Pace C. N., Shirley B. A., and Thomas J. A.. 1989. Measuring the conformational stability of a protein. In Protein Structure: A Practical Approach. T. E. Creighton, editor. IRL Press, Oxford, UK. 311–329. [Google Scholar]
- 41.John D. M., and Weeks K. M.. 2000. van’t Hoff enthalpies without baselines. Protein Sci. 9: 1416–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pace C. N., and Vanderburg K. E.. 1979. Determining globular protein stability: guanidine hydrochloride denaturation of myoglobin. Biochemistry. 18: 288–292. [DOI] [PubMed] [Google Scholar]
- 43.Wang N., Silver D. L., Costet P., and Tall A. R.. 2000. Specific binding of ApoA-I, enhanced cholesterol efflux, and altered plasma membrane morphology in cells expressing ABC1. J. Biol. Chem. 275: 33053–33058. [DOI] [PubMed] [Google Scholar]
- 44.Sankaranarayanan S., Kellner-Weibel G., de la Llera-Moya M., Phillips M. C., Asztalos B. F., Bittman R., and Rothblat G. H.. 2011. A sensitive assay for ABCA1-mediated cholesterol efflux using BODIPY-cholesterol. J. Lipid Res. 52: 2332–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagao K., Hata M., Tanaka K., Takechi Y., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., and Saito H.. 2014. The roles of C-terminal helices of human apolipoprotein A-I in formation of high-density lipoprotein particles. Biochim. Biophys. Acta. 1841: 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lacowicz J. R. 1999. Principles of Fluorescence Spectroscopy. 2nd edition. Plenum Press, New York. [Google Scholar]
- 47.Tanaka M., Dhanasekaran P., Nguyen D., Ohta S., Lund-Katz S., Phillips M. C., and Saito H.. 2006. Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry. 45: 10351–10358. [DOI] [PubMed] [Google Scholar]
- 48.Newby Z. E., O’Connell J. D. 3rd, Gruswitz F., Hays F. A., Harries W. E., Harwood I. M., Ho J. D., Lee J. K., Savage D. F., Miercke L. J., et al. 2009. A general protocol for the crystallization of membrane proteins for X-ray structural investigation. Nat. Protoc. 4: 619–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pownall H. J., Massey J. B., Kusserow S. K., and Gotto A. M. Jr. 1978. Kinetics of lipid-protein interactions: interaction of apolipoprotein A-I from human plasma high density lipoproteins with phosphatidylcholines. Biochemistry. 17: 1183–1188. [DOI] [PubMed] [Google Scholar]
- 50.Vedhachalam C., Liu L., Nickel M., Dhanasekaran P., Anantharamaiah G. M., Lund-Katz S., Rothblat G. H., and Phillips M. C.. 2004. Influence of ApoA-I structure on the ABCA1-mediated efflux of cellular lipids. J. Biol. Chem. 279: 49931–49939. [DOI] [PubMed] [Google Scholar]
- 51.Salminen A., Astrom P., Metso J., Soliymani R., Salo T., Jauhiainen M., Pussinen P. J., and Sorsa T.. 2015. Matrix metalloproteinase 8 degrades apolipoprotein A-I and reduces its cholesterol efflux capacity. FASEB J. 29: 1435–1445. [DOI] [PubMed] [Google Scholar]
- 52.Segrest J. P., Jones M. A., Mishra V. K., and Anantharamaiah G. M.. 2002. Experimental and computational studies of the interactions of amphipathic peptides with lipid surfaces. In Peptide-Lipid Interactions. S. A. Simon and T. J. McIntosh, editors. Academic Press, San Diego, CA. 397–435. [Google Scholar]
- 53.Anthanont P., Polisecki E., Asztalos B. F., Diffenderfer M. R., Barrett P. H., Millar J. S., Billheimer J., Cuchel M., Rader D. J., and Schaefer E. J.. 2014. A novel ApoA-I truncation (ApoA-IMytilene) associated with decreased ApoA-I production. Atherosclerosis. 235: 470–476. [DOI] [PubMed] [Google Scholar]
- 54.Kono M., Tanaka T., Tanaka M., Vedhachalam C., Chetty P. S., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., and Saito H.. 2010. Disruption of the C-terminal helix by single amino acid deletion is directly responsible for impaired cholesterol efflux ability of apolipoprotein A-I Nichinan. J. Lipid Res. 51: 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mei X., and Atkinson D.. 2015. Lipid-free Apolipoprotein A-I Structure: Insights into HDL Formation and Atherosclerosis Development. Arch. Med. Res. 46: 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segrest J. P., Jones M. K., Catte A., Manchekar M., Datta G., Zhang L., Zhang R., Li L., Patterson J. C., Palgunachari M. N., et al. 2015. Surface density-induced pleating of a lipid monolayer drives nascent high-density lipoprotein assembly. Structure. 23: 1214–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kono M., Okumura Y., Tanaka M., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., and Saito H.. 2008. Conformational flexibility of the N-terminal domain of apolipoprotein a-I bound to spherical lipid particles. Biochemistry. 47: 11340–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]