Abstract
Lipoprotein (a) [Lp(a)] has attracted the interest of researchers and physicians due to its intriguing properties, including an intragenic multiallelic copy number variation in the LPA gene and the strong association with coronary heart disease (CHD). This review summarizes present knowledge of the structure, function, and genetics of Lp(a) with emphasis on the molecular and population genetics of the Lp(a)/LPA trait, as well as aspects of genetic epidemiology. It highlights the role of genetics in establishing Lp(a) as a risk factor for CHD, but also discusses uncertainties, controversies, and lack of knowledge on several aspects of the genetic Lp(a) trait, not least its function.
Keywords: copy number variation, Mendelian randomization, cardiovascular risk factor, lipoprotein metabolism
INTRODUCTION TO THE LIPOPROTEIN (a) TRAIT
Human lipoprotein (a) [Lp(a)] is a macromolecular complex in plasma that was first described in 1963 by the Norwegian physician Kåre Berg (1). Ever since its discovery, this enigmatic particle has intrigued basic researchers and clinicians due to its unknown physiological function and its association with atherosclerotic diseases, in particular coronary heart disease (CHD) [reviewed in (2)]. Lp(a) is composed of one molecule of a LDL-particle containing apoB-100 and one molecule of a large highly polymorphic glycoprotein named apo(a) (3–6). A characteristic feature of apo(a) is the presence of loop-like structures called kringles (7, 8). Kringle domains are triple loop structures stabilized by three internal disulfide bonds and are also present in other coagulation factors, such as plasminogen (PLG), prothrombin, urokinase, and tissue-type PLG activators (9–12). In contrast to PLG, the linker domain between kringles is glycosylated in apo(a). The apo(a) is synthesized by the liver (13). The two components of Lp(a) are covalently linked together by a disulfide bond between apoB-100 of the LDL moiety and one of the kringle domains in apo(a) (4, 5, 14–16). The assembly of Lp(a) is believed to occur at the hepatocyte cell membrane surface (17), but other scenarios have also been proposed [reviewed in (18, 19)].
Lp(a) was originally described as a dichotomous (Lp+, Lp−) genetic trait (20), but it soon became evident that it is quantitative rather than qualitative in nature (21–23). Lp(a) plasma concentrations are highly heritable (24–28). The major locus controlling the Lp(a) concentrations is the LPA gene (MIM 152200; ENSG00000198670) on the reverse strand of chromosome 6q27 (29–31), which encodes the apo(a) component of Lp(a) (25, 26, 32–34). Close LPA orthologs are found in all apes and in Old World monkeys.
INTRA- AND INTER-POPULATION DIFFERENCES IN Lp(a) CONCENTRATIONS
Plasma concentrations of Lp(a) show remarkable variation between individuals that exceeds those of other plasma lipoprotein components by far. Such variation in Lp(a) levels exists not only among individuals within a population, but also between different human populations (35–37) and has been observed in nonhuman primates too (38, 39). Human Lp(a) concentrations range from <0.1 mg/dl to more than 200 mg/dl, thus exhibiting up to three orders of magnitude difference among individuals. Between populations, up to a 3-fold difference in their mean values is observed. On average, Africans have 2- to 3-fold higher Lp(a) plasma concentrations than Europeans and most Asian populations. No explanation has yet been found as to why this trait shows such extensive variation, but some of the underlying genetic variation has been elucidated.
STRUCTURE AND EVOLUTION OF THE HUMAN LPA GENE
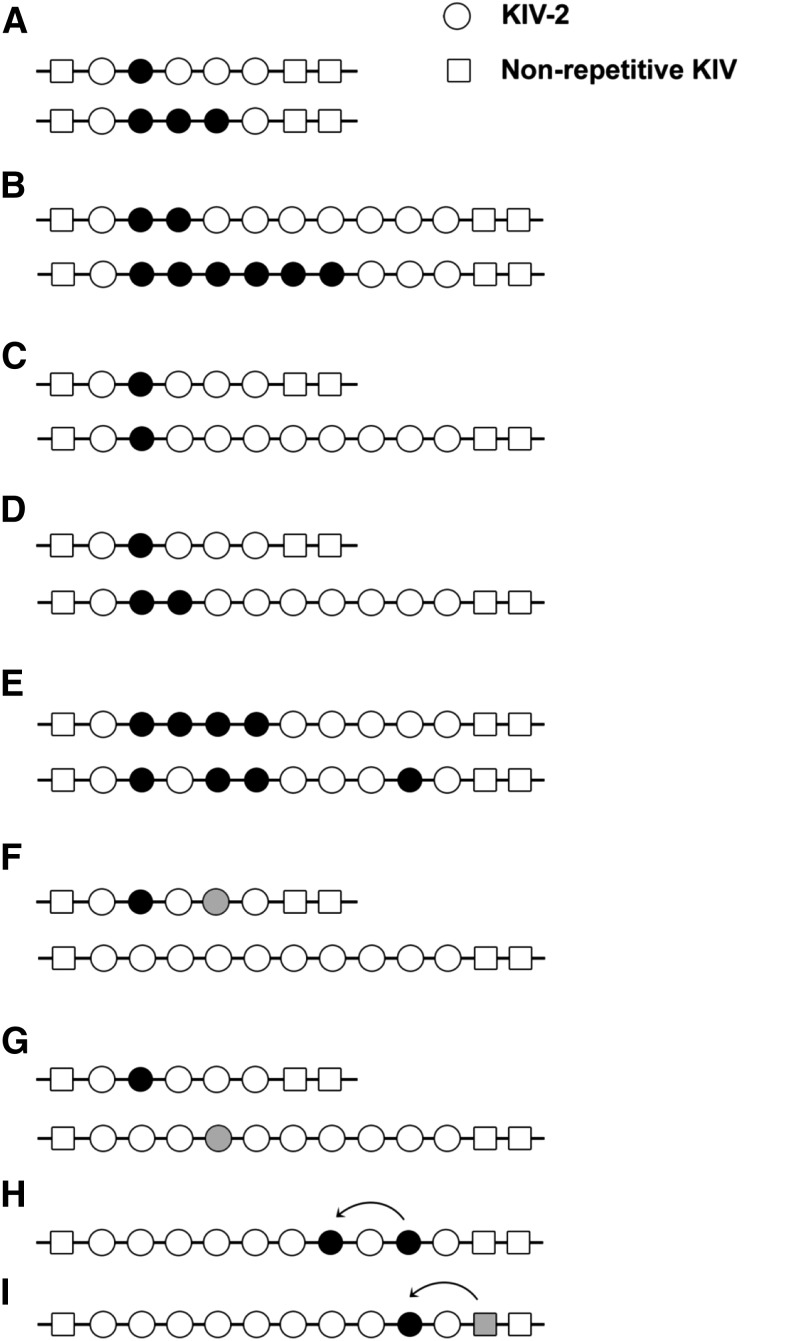
The LPA gene is closely related to PLG from which it has evolved by duplications, deletions, and gene conversions, as well as point mutations (40). PLG is characterized by five different paralogous kringle domains [kringles I–V (KI–KV)], each present as single copies. The two genes diverged during primate evolution about 40 million years ago (41). Human LPA shares a high sequence homology (78–100%) to human PLG in both the untranslated and coding regions (41). In the human lineage, an expansion and differentiation of the KIV domain in LPA resulted in 10 different types of KIV domains, all specific in their amino acid composition, while KI–KIII were lost by deletion. In macaques and baboons, KV was also lost (42, 43). Further expansion of one of the KIV domains [KIV type 2 (KIV-2)] resulted in the multiallelic (1 to >40 copies) intragenic copy number variation (CNV) known as the KIV-2 CNV (Figs. 1, 2A). The other KIV encoding domains (KIV-1 and KIV-3 to KIV-10) are present only as single copies (44). All kringle copies are transcribed and translated, hence the KIV-2 CNV leads to a size polymorphism of the encoded apo(a) (45). Isoform sizes ranging from 300 to 800 kDa have been determined by SDS-PAGE (6), but these values are not accurate because of, for example, the anomalous mobility of glycosylated proteins in SDS-PAGE. Each KIV unit consists of two exons that are separated by a long intron, which varies in length depending on the KIV type it is found on. The short introns, which separate the KIV copies between the second exon of one KIV copy and the first exon of the next KIV copy, are much more highly conserved in size. The KV and protease domains (PDs) are composed of two and six exons, respectively (Fig. 2).
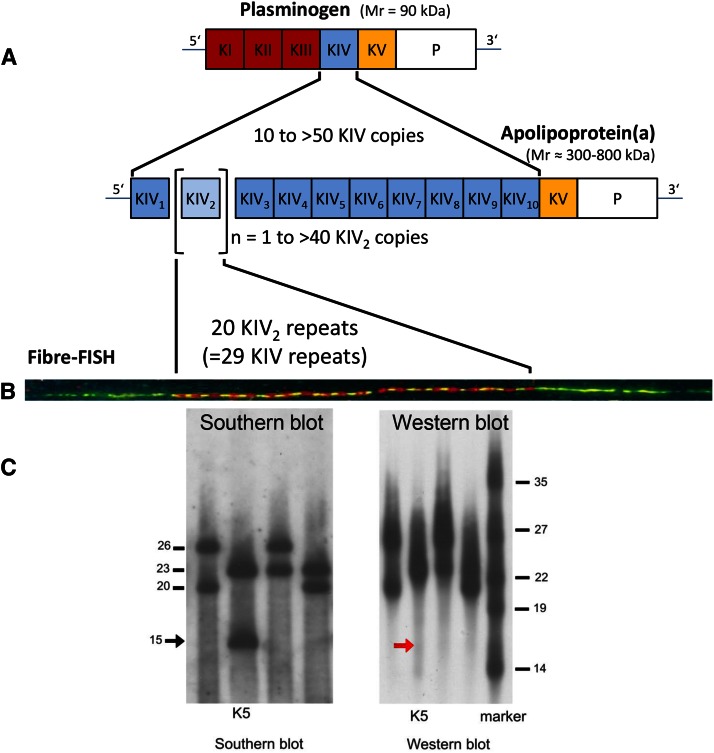
Fig. 1.
A: Schematic illustration of the structural homology between PLG and apo(a). PLG contains five different kringle structures (KI–KV) and a PD. The apo(a) is missing KI to KIII, but has a variable number of KIV copies. The minimum number is 10 (KIV-1 to KIV-10, including one copy of KIV-2). An individual can have more than 40 KIV-2 copies, which is the variable part of apo(a). Adapted and reprinted with permission of (2). B: Illustration of a two-color fiber-FISH image of the LPA KIV-2 domain from a 20-repeat allele using 4 kb (red) and 1.2 kb (green) intron probes, which enable the KIV-2 repeat number to be counted [for details see (139)]. C: Analysis of four samples from one family by PFGE/Southern blotting and Western blotting. This analysis demonstrates a null allele by simultaneous analysis of DNA by PFGE/Southern blotting (left) and plasma by Western blotting (right) from the same individuals. The arrow marks the absence of a signal corresponding to the allele with 15 KIV-2 repeats on the Western blot of the individual marked as K5. Figure adapted and reprinted with permission from (200).
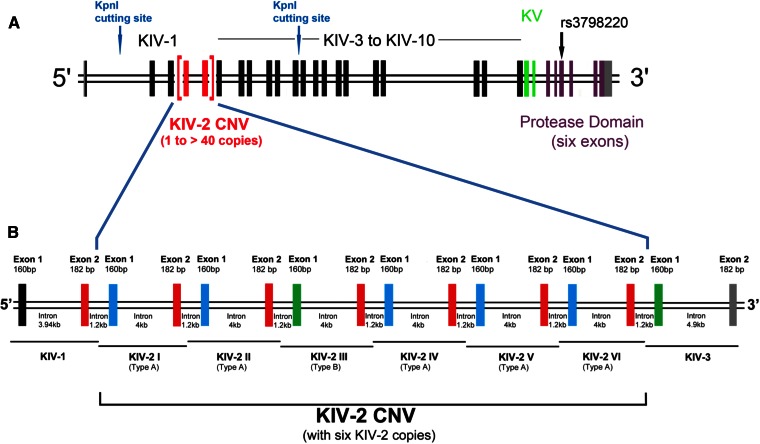
Fig. 2.
A: Exon-intron structure of different domains of the human LPA gene. Each KIV domain (KIV-2 in red, KIV-1 and KIV-3 to KIV-10 shown in black) and KV (green) consists of two exons, while the PD (purple) has six exons. The location of the KpnI cutting sites and the nonsynonymous SNP, rs3798220, is shown (137, 157). Modified from (201). B: Exon-intron structure of the KIV-2 domain and directly flanking KIV-1 and KIV-3 domains. The KIV-2 CNV is shown according to the LPA reference sequence (ENSG00000198670; GRCh38), which contains six KIV-2 copies. Each KIV-2 copy has a size of 5.5 kb and consists of two exons separated by a long intron of 4 kb. A short intron of 1.2 kb separates the two KIV-2 copies. Exon 2 (182 bp) of each KIV-2 copy is identical (red). Exon 1 (160 bp) is of three different types: type A (shown in blue), type B (shown in green), and type C (not depicted here). Types A, B, and C of exon 1 are differentiated by synonymous mutations. The third KIV-2 copy in the LPA reference sequence (ENSG00000198670; GRCh38) contains exon 1 of type B and is shown here accordingly. Note that the type B exon 1 of KIV-2 is 100% identical in its nucleotide sequence to KIV-3 exon 1 (both shown in green), and KIV-2 exon 2 has 100% identical nucleotide sequence as KIV-1 exon 2 (shown in red). This figure is not drawn to the scale.
The cluster of PLG and LPA on chromosome 6 also harbors the LPA-like gene, LPAL2 (also known as APOARGC; Ensembl:ENSG00000213071), which appears to be partially transcribed in the liver, but is subjected to nonsense-mediated decay (46).
STRUCTURE AND ASSEMBLY OF THE Lp(a) PARTICLE
apoB and apo(a) are present in Lp(a) in a molar ratio of 1:1, and apo(a) can be separated from the LDL-like moiety only by reductive cleavage (4, 5). Heterozygotes for two differently sized apo(a) isoforms have two distinct particles in plasma (47). The LDL moiety of Lp(a) is spherical and similar in lipid composition to LDL. The assembly of the Lp(a) particle occurs in two steps. The first is noncovalent docking of the KIV-5 to KIV-8 domains to the N terminus of apoB-100 [reviewed in (19)]. In the second step, the covalent binding of apo(a) to apoB occurs through the formation of a disulfide bond between the only unpaired cysteine in apo(a) in KIV-9 [Cys1568, in the old nomenclature Cys4057) (16, 48)2] with Cys4326 in apoB (49, 50).
Images from atomic force microscopy suggest that apo(a) is attached to LDL at two sites with its N- and C-terminal domains (51). This proposed structure is, however, difficult to reconcile with the well-established assembly process. Small angle X-ray scattering suggests that apo(a) is placed above the surface and wrapped around the LDL moiety. It underwent a conformational change from a compact to an extended form upon binding to a lysine analog. A study using hydrodynamic techniques and electron microscopy concluded, however, that the bulk of apo(a) is extended away from the lipoprotein surface (52). This model is attractive because it allows for ready interactions of the floating N-terminal “tail” of apo(a) with potential ligands.
FUNCTION OF Lp(a)
To date, the physiological function of Lp(a) remains mysterious, even more so given the huge inter-individual variation of Lp(a) levels and considering high heritability of the traits. A splice site variant, first described by Ogorelkova et al. (53) in Austrians and later in Finns, with a minor allele frequency of about 5%, was recently brought to prominence as a frequent loss-of-function (LoF) variant in a Finnish population study (54). Notably, the large number of identified homozygotes for the variant had no clinical signs or recognizable deficits in that Northern European population. Evaluation of clinical data from 227 homozygotes or compound heterozygotes for two splice variants in LPA resulted in no indication for an increased mortality or morbidity. This led the authors to conclude that Lp(a) is of no functional importance (54). However, Lp(a) may well have functional properties that are not (anymore) needed in the environment of Finland or Europe in general. On the other hand, several of the previously reported or hypothesized functions of apo(a)/Lp(a) seem unlikely considering this genetic evidence. Nevertheless, these functions will be summarized in this review.
Several functions have been proposed for apo(a)/Lp(a) in vitro, which might explain its pathogenic potential as well. The structural components of the Lp(a) particle have led to the suggestion that it may serve as a link between the cholesterol transport and the fibrinolytic system and may modulate blood clotting and fibrinolytic processes (55). Some studies have shown that Lp(a) and apo(a) indeed have an effect on many steps involved in coagulation and fibrinolysis cascades under in vitro conditions (56, 57). While apo(a) itself lacks fibrinolytic activity, it is reported to prevent the conversion of PLG to plasmin by inhibiting activators, such as streptokinase, urokinase, and tissue type PLG activator (t-PA) [reviewed in (19)]. However, genetic studies in several tens of thousands of individuals found that neither Lp(a) concentrations nor genetic variants associated with high Lp(a) concentrations were connected with the risk of venous thrombosis or venous thromboembolism (58, 59). The role of Lp(a) in coagulation is reviewed elsewhere in this Thematic Review series (57).
Another hypothesis is that Lp(a) is involved in wound healing and tissue repair (60). Lp(a), by interacting through apo(a), is recognized by different macromolecules and receptors present at the surface of macrophages, endothelial cells, fibroblasts, and platelets (61–63). Binding of Lp(a) to endothelial cells and smooth muscle cells is enhanced manifold by the protein, defensin (64), which is released by neutrophils. In vitro studies have shown an interaction between Lp(a) and components of the vascular wall and extra cellular matrix, including fibrin, fibronectin, glycosaminoglycans, proteoglycans, (65, 66), and developmental arteries and neural crest epidermal growth factor (EGF)-like protein (DANCE) [FIBULIN 5 (FBLN5)] (67). Lysine binding sites present in the kringle domains (KIV-6 to KIV-10) of apo(a) (19) partially mediate these interactions. Based on immunochemical studies (68), it is suggested that delivery of cholesterol to the sites of injury and wound healing may occur through binding of Lp(a) to fibrin (60). From structural homology of its domains, apo(a) might also possess growth factor-like properties as different growth factors, including the hepatocyte growth factor, which has evolved from an ancestral kringle containing serine protease (69).
Several interleukin-6 (IL-6)-responsive elements have been identified in the promoter region of LPA (70), and some studies have suggested that apo(a)/Lp(a) could act as an acute phase protein (71, 72)). However, opposite effects have also been reported (73, 74). These inconsistent findings might be explained by opposing effects of different cytokines on apo(a) expression (75). The increased expression of apo(a) induced by IL-6 found in monkey hepatocyte cultures (75) was recently supported by studies demonstrating higher Lp(a) in individuals with increased IL-6 levels and the Lp(a)-lowering effect achieved by blockade of the IL-6 receptor through the monoclonal antibody, tocilizumab (76). Furthermore, utilizing reporter gene assays, this study confirmed a specific effect of IL-6 on apo(a) expression localized to the IL-6 responsive element at c.-46 to c.-40 in LPA.
Newer studies suggest that Lp(a) functions as a preferential carrier of oxidized phospholipids (OxPLs) and is a “sink” for OxPLs (77). It was suggested that the atherogenic potential of Lp(a) may be partially due to the observed correlation of Lp(a) levels with these pro-inflammatory OxPLs (78, 79). A lysine binding site present in the KIV-10 domain of apo(a) has been suggested to mediate the interaction between OxPLs and apo(a)/Lp(a) (43).
Whether the PD in LPA is completely inactive is still not resolved. The cleavage site generating active plasmin has mutated during the evolution that led to human LPA. Furthermore, the PD of LPA acquired a 27 amino acid long deletion. This predicts that it cannot be activated to plasmin. However, the amino acids forming the catalytic triad in plasmin are conserved in human apo(a), and some studies have reported that apo(a) has retained a proteolytic activity (80, 81).
Until recently, the absence or very low concentrations of Lp(a) in plasma had not been recorded to be associated with any deficiency syndrome. Recent studies, however, have reported the association between very low Lp(a) concentrations and an increased risk of type 2 diabetes mellitus (T2DM) (82–85).
METABOLISM OF Lp(a)
LPA is mainly transcribed in the liver (13). Significant amounts of apo(a) mRNA were consistently detected in hepatocytes from humans, baboons, and macaques (41, 42, 86, 87). Minor amounts of apo(a) mRNA have also been discovered in testes, brain, lung, and adrenal and pituitary glands from humans and monkeys (41, 42). Illegitimate expression of apo(a) mRNA in lymphocytes has been used in human genetic studies to circumvent the problem posed by the tissue expression pattern (53). However, it has been elegantly demonstrated that apo(a) on Lp(a) derives from the liver only, as apo(a) isoform sizes change to the donors’ genotype after liver transplantations (13).
The site of Lp(a) assembly from apo(a) and LDL(-like) particles remains controversial [reviewed in (18, 19)]. Different experimental approaches have resulted in different scenarios. No apo(a)/apoB complexes could be demonstrated in the endoplasmic reticulum or Golgi apparatus from HepG2 cells transfected with human LPA constructs, even when the exit of proteins from the Golgi was blocked, but such complexes were found in the cell media, suggesting an extracellular assembly (88) or assembly at the hepatocyte surface (17). In contrast, it has been concluded from kinetic turnover data that assembly occurs intracellularly (89). These contradictory findings have not yet been resolved.
The site and pathway of Lp(a) catabolism have not been identified and still remain a mystery. The role of the LDL receptor (LDLR) for the removal of Lp(a) from plasma remains unclear, although binding of Lp(a) to the LDLR (90, 91) and other members of the LDLR family (92) have been demonstrated. It has also been shown in vitro that Lp(a)-LDL complexes are recognized by the LDLR (91). Turnover studies have shown that the biological half-life of Lp(a) and LDL are similar (93), but one study concluded that the fractional catabolic rate of Lp(a) is 30% lower than that of LDL (90). Three independent studies demonstrated that differences in Lp(a) plasma concentrations are not due to differences in catabolism, but rather are due to different production rates (93–95). Fractional catabolic rates for Lp(a) with short and long isoforms were not significantly different (95). Conclusions on the role of the LDLR from in vivo turnover studies were controversial. Krempler et al. (90) concluded that LDL and Lp(a) are cleared by the same mechanism. However, turnover studies in homozygous familial hypercholesterolemia (FH) patients by Rader et al. (95) convincingly demonstrated that the LDLR is not physiologically important for Lp(a) catabolism. Studies in mice suggested that the clearance of Lp(a) is mediated by apo(a) and not the LDL moiety of Lp(a) (96). However, because mice do not have a gene for apo(a) and no Lp(a) in plasma and, hence, presumably no coevolved removal system, these studies have to be viewed with some caution. In view of the turnover data in FH patients, the elevated Lp(a) concentrations observed in patients with FH due to defective LDLR (97–102) and PCSK9 gain-of-function mutations (103) need an explanation. Though some studies (104, 105) did not confirm the earlier observations, these observations have been supported by more recent large studies, which also confirmed that Lp(a) is an independent risk factor for CVD in FH patients (106, 107). To explain the discrepant results from FH family studies, it has been suggested that the effect on Lp(a) levels depends on the type of mutation in the LDLR (108) or that other mechanisms are causing high Lp(a) concentrations in FH families (102, 104). A further conundrum is that PCSK9 inhibitors, which act through the LDLR pathway, can lower Lp(a) levels by 20–30% (109), whereas HMG-CoA inhibitors do not (110–112). Hence, which, if any, of the reported receptors acts as a binding site in vivo and removes Lp(a) from plasma is presently unclear.
The finding that Lp(a) levels are strongly influenced by chronic kidney disease (113, 114) and observations such as the presence of apo(a) fragments in urine (115, 116), as well as an arteriovenous difference in Lp(a) concentrations in the renal circulation (117), suggest that the kidneys may play a major role in Lp(a) catabolism.
OCCURRENCE OF LPA/Lp(a) IN OTHER SPECIES
Apart from humans, presence of a polymorphic protein having immunochemical properties and molecular mass similar to human apo(a) and/or a gene homologous to human LPA has been demonstrated in catarrhines (i.e., Old World monkeys and apes) (39–42, 118, 119). Today the corresponding genes have been partly sequenced and are accessible in online databases (Western common chimpanzee, ENSPTRG00000018770; Western lowland gorilla, ENSGGOG00000016065; Sumatran orangutan, ENSPPYG00000029841; olive baboon, ENSPANG00000014232; rhesus macaque, ENSMMUG00000016201). Apart from the above mentioned species, an apo(a)-like protein has also been detected in the European and African hedgehog (120, 121). Unlike primate apo(a), it is composed of multiple diversified tandem repeats of domains, which are homologous to PLG KIII, but lacks the PD and other kringles (121). Phylogenetic analysis and sequence comparisons of primate and hedgehog apo(a)/LPA indicate that both genes evolved independently, which is an example of convergent evolution at the molecular level (40). Reports on the occurrence of Lp(a) in a New World monkey species (122) and in guinea pigs (123) and derived hypotheses have not been confirmed and are considered to be based on immunochemical artifacts (39, 121).
GENETICS OF Lp(a)
Heritability of the Lp(a) trait
The heritability (h2) of the quantitative Lp(a) trait obtained from twin, family, and sib-pair studies is exceptionally high (70 to ≥90%) in all populations studied so far from Europe, Asia, and Africa (24–28, 32, 124–127). This genetic control is mainly exerted through the LPA locus. LPA alleles are expressed codominantly. Gene loci on chromosome 6q, other than LPA, explaining an additional small fraction of the variation in Lp(a) concentrations have been postulated (128), but have not been confirmed. Further loci identified to be associated with Lp(a) levels in some other linkage studies include regions on chromosomes 13q22-31, 11p14-15, and 1q23 (129, 130), but these have not been confirmed, either.
Genetic polymorphisms within LPA
KIV-2 CNV.
CNV is defined as changes of DNA segments typically more than 1 kb in size and present at a variable number in comparison with a reference genome (131). Copy number variants can involve deletions, insertions, duplications, and higher copy numbers, the latter termed as multi-allelic CNVs (131). By this definition, the KIV-2 CNV is a multi-allelic CNV harbored by the LPA locus. It was one of the first multiallelic CNVs to be described and extensively studied before the term was coined. The number of KIV-2 copies varies from 1 to >40 and the KIV-2 CNV exhibits >95% size heterozygosity in most populations (Fig. 1). Each KIV-2 copy has a size of ∼5.5 kb and consists of two exons (Fig. 2). The second exons of all the KIV-2 copies are 100% identical in their nucleotide sequence. The first exons are also identical in their amino acid sequence among each other, but can differ by three synonymous SNPs (41, 132) and have been classified accordingly as types A, B, and C. It is noteworthy that the KIV-2 exon 1 type B is completely identical to the first exon of KIV-3, and KIV-2 exons 2 share complete sequence identity with the second exon of KIV-1 (Fig. 2B), which probably has resulted in an erroneous allocation of SNPs in public databases (see below). The human reference sequence of LPA (ENSG00000198670; chromosome 6: 160,531,483-160,664,259 reverse strand. GRCh38:CM000668.2) includes six KIV-2 copies, one of which is of type B; all others are of type A. The apo(a) mRNA cloned by McLean et al. (41) in 1987, however, included 28 KIV-2 repeats, four among them of type B. Often the assignment of amino acids in apo(a) is still based on this longer molecule, and a nomenclature counting kringles from 1 to 37, with kringles 2 to 29 being the KIV-2 copies, is found in older publications on Lp(a)/apo(a), which has also resulted in some confusion.
The high internal homology in the KIV-2 CNV also extends to its introns (133). The underlying reason for this high degree of homology is still unclear. It has been suggested that this nearly perfect sequence identity of the KIV-2 units implies a recent evolution (41, 133, 134) or frequent expansions and contractions of the locus (41). Another tempting hypothesis is that purifying selection, in the sense that deleterious mutations are removed from the gene pool as a type of natural selection, is acting on the KIV-2 domains, but a high degree of sequence similarity can also result from frequent gene conversion (133), which can homogenize the sequences of the duplicated regions (135).
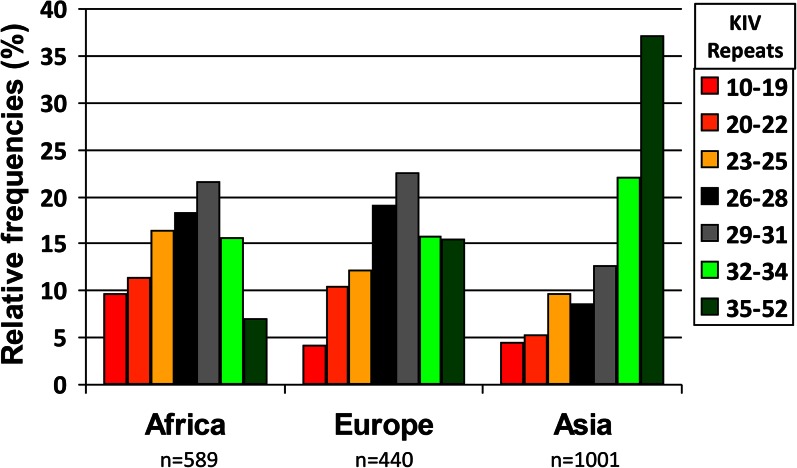
The frequency distributions of different sized LPA alleles are heterogeneous across different ethnic groups. In Asians, the frequency distribution of KIV-2 CNV sizes is shifted toward longer alleles as compared with Africans and Europeans, with the latter two showing rather similar size distributions (35–37) (Fig. 3). A further discussion of apo(a) in different ethnic groups can be found in this Thematic Review series (136).
Fig. 3.
Frequency distribution of KIV CNV alleles in three continental groups. Short repeats are more frequent in Africans and long repeats in Asians. Data are from (24, 32, 36) and unpublished observations.
Different techniques have been employed to determine the size of the KIV-2 CNV, which include pulsed-field gel electrophoresis (PFGE)/Southern blotting of genomic DNA (25, 137), quantitative PCR (qPCR) (138),Western blotting of plasma using apo(a)-specific antibodies, and fiber-fluorescence in situ hybridization (fiber-FISH) (139) (Fig. 1). While PFGE allows the determination of the number of KIV-2 copies in both alleles of an individual, qPCR can only give an estimate of the sum of KIV-2 copies on both alleles of the genomic sample. Fiber-FISH is the most precise technique available to determine the size of the KIV-2 CNV, whereby the number of KIV-2 copies on individual alleles can be counted under the fluorescence microscope. However, this method is not feasible for handling larger sample sizes, but served to define standards used in large population studies (138, 139). Both PFGE and fiber-FISH demand a high quantity of well-preserved nonfragmented DNA and, hence, are limited by conditions of sample collection and technical demand. Western blotting has been widely used to determine apo(a) isoform size and associated Lp(a) concentrations (see below for the latter) and, therefore, provides phenotypic information. Compared with PFGE, this method has advantages and drawbacks. Expressed alleles, i.e., alleles producing apo(a), are recognized and the amount of Lp(a) contributed by each allele can be determined in heterozygotes with two expressed alleles and exhibiting double band phenotypes. In these individuals, the CNV genotype can also be directly deduced from the phenotype. However, the frequencies of double band phenotypes reported in the literature vary considerably and range from 30 to 90% [see (136) in this Thematic Review series]. The reason for this is twofold. First, there are technical limitations. The number of double band phenotypes will depend on the sensitivity of the blotting system and the resolution of the gel system. Both will vary between studies. Second, it will depend on the genetic architecture of the Lp(a)/apo(a) trait in the population under study. As outlined below, populations differ in allele-associated Lp(a) concentrations. Hence, in populations where long alleles are associated with higher concentrations, e.g., Africans, more double band phenotypes are expected to be detected. Given an optimal detection system (that does not exist), the “true” number of double band phenotypes to be expected in a population will depend on the number of LoF alleles (null alleles) in that population, and that is presently unknown. Further, it is presently difficult to recognize truncated isoforms or splice variants at the protein level, if no prior knowledge from molecular analysis exists. PFGE on the other hand allows for determination of the CNV genotype, but provides no phenotypic information. The application of both methods simultaneously provides the most comprehensive information.
Phenotypic significance of the KIV-2 CNV
CNV is implicated as a major driving force during evolution, especially within the human and great ape lineage (140–143). Additional copies of genes provide the redundancy that allows some copies to acquire new or modified functions and diverse expression patterns, while the original function of the gene is maintained by other copies (144–146). However, CNV of a gene can also have pathological consequences, and changes in the copy number of genes have been related to autism, epilepsy, schizophrenia, Alzheimer’s and Parkinson’s disease, congenital anomalies, intellectual disability, and various other common and rare diseases (147).
While no function of the KIV-2 CNV has been established so far, kringle domains, in general, are known to be independent structural (148, 149) and functional domains (150–152) and their main function lies in the interaction with other proteins (153). Kringle domains of different proteins have the same triple loop structure, but are otherwise diversified and consequently bind different proteins and ligands (48, 63, 150, 151). So far, the identified ligands for the KIV-2 domain include the extracellular matrix protein, DANCE/FIBULIN 5 (67), and β2-glycoprotein I (154). However, the in vivo relevance and the functional implications of these interactions remain unclear.
Whether the marked differences in the KIV-2 CNV allele size distributions that are observed across populations (36, 37) are related to any functional effect of LPA is currently unclear. As the KIV-2 CNV is in the coding region, it might not be neutral to selection. Hence, it is tempting to speculate that natural selection has left its mark on the KIV-2 allele size distributions in populations. A strong argument for a significant function and possible selective advantage of the CNV is its presence, already, in nonhuman primates. However, where it has been studied, e.g., chimpanzees (39), baboons (38), or rhesus monkeys (155), the average size of apo(a) isoforms is shorter than in human populations. On the other hand, differences in allele frequencies might as well have resulted from neutral effects such as genetic drift alone. However, such questions cannot be addressed in the absence of data on the mutation rate and mode of evolution of this CNV. Currently, very little is known about the evolutionary dynamics of the KIV-2 CNV.
It has been observed in different studies that certain SNP haplotypes are associated with a narrow range of KIV-2 sizes (156, 157), possibly indicating that size changes in the KIV-2 CNV are mostly of comparatively restricted magnitude per mutation event. On the other hand, a size change of the KIV-2 CNV in one segregating allele described in a family study by Lackner, Cohen, and Hobbs (158) was rather large, with a loss of nine KIV-2 units, and was not accompanied with an exchange of markers up- and downstream of the CNV. These observations indicate that sister chromatid exchange and complex gene conversion events are involved in changing the number of KIV-2 repeats.
LPA/apo(a) size polymorphism and inverse correlation with Lp(a) levels
Though of unknown physiological or evolutionary function, the KIV-2 CNV of human LPA is of clinical interest due to the causal relationship between the number of KIV-2 repeats and Lp(a) levels, which in turn are a risk factor for CHD at elevated concentrations (159–162). Short KIV-2 CNV alleles have been shown to be associated with an increased risk for CHD in some populations (163–167).
An inverse correlation of CNV length with Lp(a) levels has been demonstrated in almost all analyzed populations [reviewed in (2)] (Fig. 4). Underlying the inverse correlation are differences in the processing of apo(a) isoforms during transit through the secretory pathway from the hepatocytes. Their retention time in the endoplasmic reticulum and the degree of presecretory degradation correlate with isoform size; hence, shorter apo(a) isoforms are secreted more efficiently than longer ones (88, 168, 169). It should be emphasized that these experiments have established that the association of Lp(a) plasma levels with KIV-2 CNV size reflects a general causal relationship. In line with these results from cell culture experiments, turnover studies in humans have demonstrated that inter-individual differences in Lp(a) levels reflect size-dependent synthesis rates of apo(a) isoforms rather than the rate of Lp(a) catabolism (93, 95).
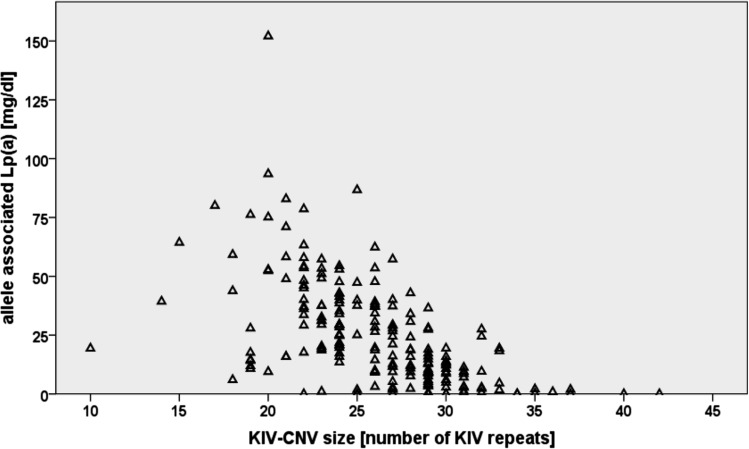
Fig. 4.
Inverse correlation of KIV-2 repeat length with Lp(a) plasma concentration. The apo(a) allele-associated Lp(a) concentrations were determined in individuals from an African population (Gabonese Bantu) by measuring total Lp(a) concentrations by ELISA and assigning the appropriate fraction to each of the 194 alleles by densitometric evaluation of immunoblots. Allele-associated levels are plotted against the number of KIV repeats. The figure demonstrates large differences in Lp(a) levels for KIV alleles of the same size and presence of short alleles with low Lp(a) concentrations. Data are from (24).
Population studies have shown that the inverse correlation is far from linear and varies in strength depending on the population. In general, it is weaker in populations of African descent than in Asians or Europeans (24, 35, 36, 170, 171), explaining 61–69% of the variance in Lp(a) levels in populations of European descent, but only 19–44% in African populations (24, 35, 36, 170, 172–174). Furthermore, differences in KIV-2 allele frequency distributions alone do not explain the vast differences observed in Lp(a) levels across populations. In general, a large variation in Lp(a) levels is observed for isoforms of the same size (94, 175). Hence, Lp(a) concentrations are not determined entirely by the KIV-2 CNV.
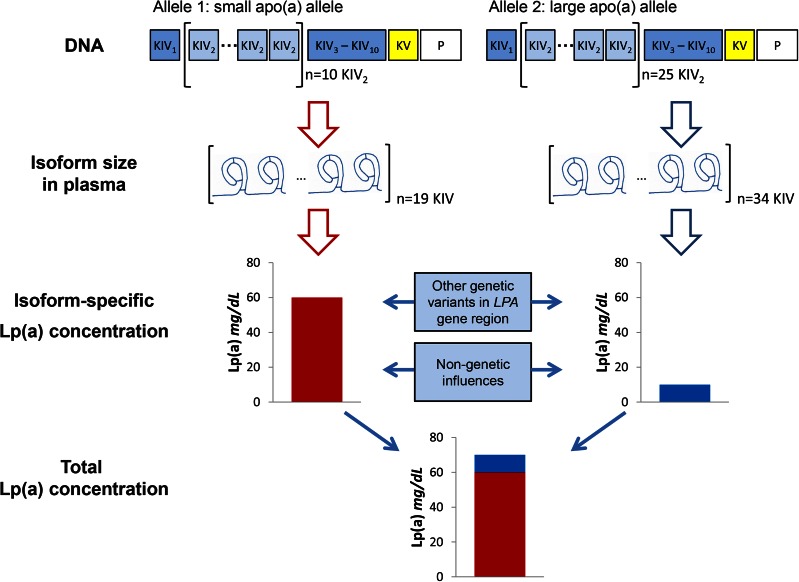
The Lp(a) concentrations associated with particular LPA alleles can be assessed experimentally when apo(a) isoform size is determined by SDS agarose gel electrophoresis followed by immunoblotting (Western blotting). Here, a (semi-)quantitative analysis of the blot by densitometry allows one to allocate the relative proportion of the total plasma concentration in an individual to a specific LPA allele, thus determining allele-specific Lp(a) concentrations (176) (Fig. 1). However, without prior knowledge of the actual KIV-2 CNV size of both alleles, neither can nonexpressed alleles be identified nor can samples with a single band in the Western blots be evaluated, as they could either be size homozygotes or carriers of one null allele. Consequently, a combination of genotyping of the KIV-2 CNV and phenotyping for apo(a) expression allows the most reliable assignment of allele-associated (or isoform-associated) Lp(a) concentrations.
Sequence variation in LPA affecting Lp(a) levels
As mentioned before, LPA is the major gene controlling the Lp(a) trait and explains 70–90% of the variance in Lp(a) levels (24, 26). However, the KIV-2 CNV alone explains only a considerably smaller fraction (19–77%) of the variation in Lp(a) concentrations depending on the population and methods used (35, 36, 173, 177). Many studies aimed at identifying sequence variation in the LPA gene, other than the KIV-2 size polymorphism, have been conducted in an effort to explain the fraction of variation in Lp(a) plasma concentrations not explained by the KIV-2 CNV. Sequence variations associated with Lp(a) levels have been identified in different regions of LPA, including the promoter. However, it should be kept in mind that association does not mean causality. Many reported variants only tag the underlying causal genetic variation (e.g., KIV-2 CNV) in LPA affecting the Lp(a) levels. A causal effect on Lp(a) levels has only been experimentally shown for a few variants, and for some other variants, this appears to be likely due to their type of mutation. In this review, we concentrate on variants for which a causal mechanism has either been described or appears plausible, and those which have been described extensively in the literature. Several of these SNPs explain, either alone or in combination, parts of the variation in Lp(a) levels between populations (53, 178, 179). As will be discussed, differences in the nature of the association of SNPs and KIV-2 CNV sizes between populations might contribute to this. Other SNPs might just discriminate the ethnic ancestry of LPA alleles and thus explain a large part of population differences in Lp(a) concentrations, but without suggesting or identifying the underlying causal variants (180).
The 5′pentanucleotide repeat polymorphism
A repeat polymorphism of the pentanucleotide sequence TTTTA [5′pentanucleotide repeat polymorphism (5′PNRP)] is found in the promoter region at −1373 upstream of the transcription start site of LPA (70). Alleles with four to twelve repeats have been reported. However, alleles with eight repeats are the most frequent in all populations (156, 175, 181). This polymorphism has been repeatedly shown to be associated with Lp(a) levels (175, 181–183) and explains, in a statistical sense, 3–14% of the variability in Lp(a) levels in Europeans (181). Higher Lp(a) levels have been reported for alleles harboring a lower number of pentanucleotide repeats in Europeans, but this association was not found in Africans (181). Also in Europeans, alleles with 10 or 11 repeats were found to be in significant linkage disequilibrium (LD) with short KIV-2 CNV alleles (175) and were associated with lower Lp(a) levels in Europeans (175, 181), which contrasted with the general inverse correlation between the KIV-2 CNV size and Lp(a) levels. However, in vitro analysis of the promoter activity for different 5′PNRP sizes has suggested that this polymorphism is not causally associated with Lp(a), but rather in LD with unknown causal sequence variation in LPA, as no differences in the promoter activities were observed for constructs carrying a different number of PNRP repeats (184, 185).
SNPs in the 5′ region of LPA
In addition to the 5′PNRP, the effect of SNPs in the 5′-flanking region of LPA on Lp(a) concentrations has been investigated. In particular, the two SNPs, rs1853021 (c.-49T>C, traditionally +93 C/T) and rs1800769 (c.-21G>A, traditionally +121 G/A), were identified (186, 187), which subsequently have been found to be associated with Lp(a) levels. For rs1853021, a C to T substitution introduces an alternative start codon (ATG). In vitro, this leads to a 60% lower translational activity for the rs1853021T allele, as compared with the wild-type rs1853021C and results in lower Lp(a) levels (183, 186–188). In sub-Saharan Africans, such an effect on Lp(a) levels has been reported, with variant carriers exhibiting 50–60% lower Lp(a) levels than expected (189). In contrast, no such association was found in Europeans, which might be explained by LD with long KIV-2/low Lp(a) alleles, thus masking any possible effect (189). For rs1800769, higher Lp(a) levels in vivo (156, 178, 183, 190) and a higher transcriptional activity in vitro (183) have been reported for the variant allele. In African Americans, this promoter variant was strongly associated with intermediate KIV-2 CNV sizes, while for European Americans it was more frequent on longer alleles, but in both populations an increase of CNV-size-adjusted Lp(a) levels was observed for the variant allele, which consequently could explain, in part, the higher Lp(a) concentrations observed in Africans (178). In Mexican Americans, however, the effect of the variant was masked by its LD with longer KIV-2 CNV alleles (191). Though the first study was conducted in dialysis patients, rs1800769 again illustrates the relevance of both ancestry and adjustment for KIV-2 CNV size in studies on Lp(a) concentrations.
In Europeans, variations in an enhancer region within a DNase I-hypersensitive site 20 kb upstream of LPA (192) have been shown to affect the activity of reporter-gene constructs either positively (rs9347440; 2.5-fold increase) or negatively (rs7758766 and rs7760010, 0.7- and 0.6-fold activity, respectively) (193). Again, LD with KIV-2 CNV size was observed, but effects remained detectable after correction for allele size, with rs9347440 being associated with 70% higher and rs7760010 with 40% lower Lp(a) concentrations than expected (193).
SNPs in other regions of LPA
Further studies have investigated SNPs in other coding and noncoding regions of the LPA locus. SNPs in the coding sequences of KIV-6 to KIV-10, with population-specific distributions in Africans and Europeans, have been studied by Ogorelkova et al. (179). Nonsynonymous substitutions in KIV-6 (rs200561706; p.Ser1193Phe) and in KIV-8 (p.Gly1393Arg, without rs number so far) were found to be associated with significantly lower Lp(a) levels in Africans (179). In contrast, a nonsynonymous substitution (p.Arg1508Trp) in KIV-9 (rs140720828), which has a frequency of 8% in Khoi San from South Africa, resulted in significantly increased Lp(a) levels. Residing in KIV-8, rs41272110 (p.Thr1399Pro; or Thr3888Pro) has been found to be associated with lower than expected Lp(a) levels in Europeans by several studies (178, 190), though with an inconsistent result to the study by Ogorelkova et al. (179). All these results were reported for Lp(a) levels corrected for KIV-2 CNV size.
The coding region of the KIV-10 harbors a nonsynonymous SNP (rs1801693), which results in a change of methionine to threonine (p.Met1679Thr) (194). No functional significance of this SNP has been reported (195), but it has been shown to be in LD with the KIV-2 CNV (195) as well as SNPs in the 5′-flanking region of LPA (156) and, because it was described early, it has been investigated extensively in studies on the genetic architecture of LPA.
Two other SNPs in LPA that have been extensively investigated include rs3798220, a nonsynonymous SNP (p.Ile1891Met, also often referred to as Ile4399Met) located in the protease-like domain of LPA, and rs10455872, which is located in the long intron of the KIV-7 domain. These two SNPs are associated with high Lp(a) concentrations and together explain about 36% of variation in Lp(a) levels in Europeans. Both are associated with short KIV-2 CNV alleles (157, 196, 197), which suggests that the association with high Lp(a) is due to LD with the CNV size. However, while the causal effect of the CNV size is not mediated by expression levels, for the rs10455872 variant allele higher mRNA levels have been reported (198).
Non-expressed apo(a) alleles (“null alleles”), i.e., where no isoform is detected by immunoblotting for the corresponding LPA allele (36) (Fig. 1), are found in all populations studied so far, but seem to be more frequent in Europeans than in other populations. Null alleles are distributed over the whole size range of the KIV-2 CNV (36). For some null alleles, the genetic basis has been revealed, and functional studies have been performed. This includes a +1 donor slice site mutation (rs41272114) in the intron of KIV-8, which reaches frequencies of up to 5% in individual populations from Europe and South Asia, and even up to 18% in South Americans (53) [data from the 1000 Genomes Project (1000G)] (199). The substitution, c.4289+1G>A, results in a truncated apo(a) isoform (53). This variant has recently been rediscovered in a large study from Finland in a search for low frequency LoF variants by exome sequencing (54). Other variants associated with null alleles were discovered in the KIV-2 CNV (132, 200) (see below).
Sequence variation within the KIV-2 CNV
Comparatively little is known about sequence variation within the KIV-2 CNV. Up to now, this region of the LPA gene has been left out in many studies aimed at screening the LPA domains for sequence variation, supposedly due to technical limitations posed by the high degree of intra-domain homology, which also still provides challenges for the large scale resequencing projects like the 1000G (199).
Considering the sheer length of the sequence composing the KIV-2 CNV, it might be expected that mutations accumulate in this region. From the ATG start codon to the 3′UTR and excluding the KIV-2 CNV, LPA encloses 99.7 kb. This correlates to the sequence length of a KIV-2 CNV with approximately 18 KIV-2 repeats. Hence, LPA alleles with at least 18 KIV-2 are almost 50% composed of KIV-2 CNV sequence. Likewise, all the nonrepetitive coding sequences in LPA sum up to 4,070 bp, which approximately equals the coding sequences of only 12 KIV-2 repeats. As most LPA alleles harbor more than 21 KIV repeats (35, 36), the KIV-2 CNV encodes the bulk of amino acids found in most apo(a) isoforms. It is plausible to expect that sequence variation within the KIV-2 repeats might explain more of the differences in the Lp(a) levels observed with the same sized apo(a) isoforms, and an example was given with the detection of a null-allele caused by a variant in the KIV-2 CNV (132). Also, variants within the KIV-2 copies may be functionally important, e.g., change the binding to ligands. It is noteworthy that in the chimpanzee reference sequence from Western common chimpanzees (Pan troglodytes verus), which comprises four KIV-2 orthologous copies (ENSPTRG00000018770; CHIMP2.1.4:CM000320.2), these copies considerably differ in their amino acid composition from each other, and only two are identical.
Variants in the CNV may be of different types and patterns as depicted in Fig. 5, which illustrates the complex nature of variation in the CNV. An example is a DraIII restriction site polymorphism that is found in the introns of some copies of KIV-2, but not in others (184). Depending on the distribution of the KIV-2 copies harboring the DraIII site, different patterns have been observed. Population-specific differences in the proportion of the KIV-2 copies containing this polymorphism exist. The frequency of the alleles with at least one KIV-2 copy having the DraIII site was shown to be 25% in Europeans, while the frequency of such KIV-2 copies in Chinese was as high as 50% (184). In Chinese, the patterns of the KIV-2 copies harboring this polymorphism show frequency distributions that are different from those found in Africans and Europeans. In Europeans, the distribution patterns of the KIV-2 copies with the DraIII site were found to be in LD with the TTTTA repeat polymorphism in the 5′-flanking region of LPA and variants located 3′ of the KIV-2 CNV, in the intron between KIV-6 and KIV-7. Moreover, the DraIII pattern was found to be in LD with the number of KIV-2 repeats and some patterns were restricted to a narrow range of CNV sizes. In Europeans, one particular pattern of this polymorphism was found to be associated with very low Lp(a) levels (184). The exact sites constituting the polymorphism have not yet been defined.
Fig. 5.
Possible distributions of sequence variations in the KIV-2 CNV. Different scenarios for the distributions of sequence variants (shown as filled circles) in KIV-2 copies of different sizes are illustrated. Low and high intra-allelic frequencies of a variant on short alleles (A) and longer alleles (B). C: The same number of KIV-2 copies harbors the variant on a short and a longer allele, i.e., the intra-allelic frequency of the variant is higher on the short allele. Thus detection of the variant is more likely if present on the short allele. D: Both alleles have the same intra-allelic frequency (20%), though the number of copies carrying the variant is different. Hence the probability of detection is the same. The order of variants carrying KIV-2 copies within the allele might vary, as shown in (E). These scenarios cannot be distinguished by present methods. Different variants can be allocated in cis (F) (shown for a genotype with one short and one longer allele) or in trans (G). While scenarios (F) and (G) cannot be distinguished in analyses based on diploid samples, this is possible if analysis is based on separated alleles. Panels H and I depict possible spreads of variants across the KIV-2 CNV (H) and between KIV-2 copies and the neighboring nonrepetitive KIV-3 (I) by gene conversion. Figure used and modified with permission from (200).
Two early studies have reported a near complete sequence conservation in the KIV-2 coding sequences (133, 134). However, these studies were restricted to samples of European descent only. On the other hand, a study by Parson et al. (132) identified a functionally important sequence variation in a European individual with an isoform much shorter than predicted from the KIV-2 CNV size determined by PFGE. This SNP was detected at the 60th nucleotide in the first exon of KIV-2, and introduces a stop codon resulting in a truncated protein that lacks the kringle domains needed to form an Lp(a) particle. Hence mutations that result in impaired complex formation represent another mechanism resulting in “null alleles” for Lp(a). This nonsense mutation has a frequency of 2% in Europeans (132).
Some SNPs within the KIV-2 domain have recently been reported by the 1000G and other large resequencing projects. However, for the samples covered in 1000G, the relevant phenotypic data about Lp(a) concentrations and the KIV-2 isoform sizes are missing and, consequently, it is impossible to analyze the effect of any such variation on Lp(a) levels and its association with the KIV-2 CNV size. Currently, the widely used next-generation sequencing technologies used in the 1000G project do not provide long enough sequence reads for precise alignment in case of the repetitive KIV-2 CNV. With short sequence reads, the assignment of any variation to the KIV-2 domain is not reliable due to a high degree of homology between the KIV-2 and the flanking nonrepetitive kringle domains (KIV-1 and KIV-3) in particular (Fig. 3). Moreover, the variants reported by 1000G are often allocated to specific KIV-2 copies. This assignment is questionable given the multi-allelic nature of the KIV-2 CNV and the fact that the human LPA reference sequence is composed of six KIV-2 copies only while the number of possible KIV-2 repeats ranges from 1 to >40 and, in most populations, the median is in the range of approximately 16–24 repeats (35, 36) (K. Schmidt et al., unpublished observations).
In a recent study, the coding exons and directly flanking intronic regions of the KIV-2 CNV were sequenced from single alleles of individuals from six populations, including Africans, Europeans, and Asians (200). Several synonymous and nonsynonymous variants, as well as two previously unreported splice site variants, were identified. Most of these variants were detected in African alleles. Among the variants found, an African-specific acceptor splice site variant (KIV-2 exon 2 −6T>G, with six dbSNP database entries, one for each KIV-2 copy: rs759106280, rs767154989, rs752434799, rs762062960, rs765664550, rs750828303) associated with short KIV-2 CNV size was present in all four analyzed African populations at high population frequencies (10–40%). This variant appears to be associated with lower Lp(a) levels than expected considering the KIV-2 CNV size of the alleles carrying it. In contrast to the frequent African-specific acceptor splice site variant, a rare donor splice site variant (KIV-2 exon 1 +1G>A, rs750988762, rs758511296, rs780283322, rs747169744, rs768983109, rs781627054) was found that was shared among Africans and Europeans and associated with nondetectable Lp(a). In Asians, only the synonymous variants that define the KIV-2 type B and type C were found at very high frequencies (70%) and observed with a broad range of different KIV-2 CNV sizes. These variants were rare in Europeans and Africans. A strong bottleneck suggested to have occurred during the migration of modern humans to East Asia might explain the enrichment of these variants and the lack of others in Asians. An intriguing observation was that the variants that were frequent in the population were shared among the KIV-2 copies of the alleles carrying them. The acceptor splice site variant as well as type B/C-defining variants were observed at high intra-allelic frequencies (relative proportion of the variant vs. wild-type carrying KIV-2 copies), i.e., they are shared among KIV-2 copies of an allele. Even more intriguing was the observation that similar intra-allelic frequencies were observed for the same variants carried on different sized KIV-2 alleles (200).
The findings suggest that the LPA locus harbors more sequence variation in the KIV-2 region, explaining an additional fraction of variation in Lp(a) levels across ethnicities. They cast doubt on previous claims that most of the SNPs accounting for inter-ethnic differences in Lp(a) levels (i.e., African and European Americans) have been identified (178, 180).
In this context, it may be helpful to remember that the term “explains,” frequently used in a statistical sense, should not be confused with a molecular causal explanation. A rigorous proof of causality that SNPs in LPA affect plasma Lp(a) levels has only been provided in a few cases.
Population-specific differences exist in the association of SNPs with the KIV-2 CNV size, and the associated Lp(a) levels could be influenced by the differences in the LD patterns across populations. The SNP markers associated with Lp(a) levels or predicting the risk for CHD in one population may not be applicable to other populations. One example is rs3798220, where the variant allele is observed with vastly different KIV-2 CNV sizes in Europeans on the one hand and in nonEuropeans on the other hand (157, 201, 202). On the other hand, population-specific SNPs in LD with a narrow range of KIV-2 CNV sizes can be causally associated with Lp(a) levels. One example could be the frequent African-specific acceptor splice variant site detected in the screening of the KIV-2 region, which is associated with short KIV-2 alleles and comparatively low Lp(a) (K. Schmidt et al., unpublished observations). This variant might provide some explanation for the missing association of the short apo(a) size with CHD in individuals of African descent.
It remains to be determined whether CNV size changes can also explain the observation that the same variants are found on multiple KIV-2 copies on the same allele. Here, inter-locus gene conversion might also play an essential role (Fig. 5). The same applies for the observation that similar intra-allelic frequencies of specific KIV-2 variants are kept even between alleles of different KIV-2 CNV sizes. More extensive SNP haplotype data within the KIV-2 CNV may provide further insights into the mechanism of KIV-2 CNV size changes and their role in the spreading of variants across the KIV-2 copies. Recent technical breakthroughs, like single strand sequencing, will allow generation of data to answer these questions in the future.
Together, the genetic analysis of the LPA gene has shown that Lp(a) concentrations are determined by a complex interplay of the KIV-2 CNV with SNPs in all domains of the gene, including the CNV, and that other factors play only a minor part in the population at large (Fig. 6).
Fig. 6.
Scheme illustrating the determination of Lp(a) plasma levels by LPA gene variation and other factors. Major determinant is the KIV-2 CNV, which codes for isoform size in plasma. Because roughly 70–90% of all subjects express two apo(a) isoforms in plasma, a mixture of Lp(a) particles can be found in plasma. On average, small apo(a) alleles result in high Lp(a) concentrations and large apo(a) alleles result in low Lp(a) concentrations, as shown as isoform-specific Lp(a) concentration in this example. Therefore the total measured Lp(a) concentration in this individual is a mixture of particles consisting, to a major extent, of Lp(a) of small size. Besides the KIV-2 CNV, other genetic variants within the LPA gene region, as well as nongenetic factors, have an influence on Lp(a) concentrations.
GENETICS ESTABLISHES Lp(a) AS A RISK FACTOR FOR CVD: THE MENDELIAN RANDOMIZATION APPROACH
Numerous studies have repeatedly shown an association between elevated Lp(a) plasma levels and atherosclerotic CVDs, in particular CHD (159, 162, 164) and stroke (164). Retrospective case-control studies reported a significant association between high Lp(a) concentrations and CHD (203–206). However, such studies were disputed due to possible selection bias and also because they left the question unresolved whether high Lp(a) concentration was the cause or a result of the processes leading to CHD. The later scenario is also known as reverse causation. Prospective studies, on the other hand, initially challenged the view that Lp(a) is an independent risk factor. As we now know, the largest of these early studies (207) was biased (208). The method for Lp(a) quantification used in this study underestimates the concentration of Lp(a) for particles containing short apo(a) isoforms. These are exactly those contributing most to the elevated Lp(a) concentrations in patients. Even though further prospective studies and a large meta-analysis of prospective studies supported Lp(a) as a risk factor, final proof was lacking (162, 209). Only Mendelian randomization studies finally allowed the ability to rule out reversed causation as the reason for increased Lp(a) levels in CHD and established a causal role of Lp(a) levels in CHD (97, 138, 157, 163, 165, 210, 211). First studies of this kind had already been performed in parallel with prospective studies, but their impact was not fully recognized at the time. Meanwhile, this study type has been extended to include instrumental variable regression analysis (138, 212) and is widely used to support causality of biomarkers for various outcomes, which might allow the transition of a risk marker to a risk factor (213). On the other hand, this approach, if sufficiently powered studies are available, can be used to exclude causality, as was recently done for C-reactive protein and CHD (214, 215).
KIV-2 size and CHD risk
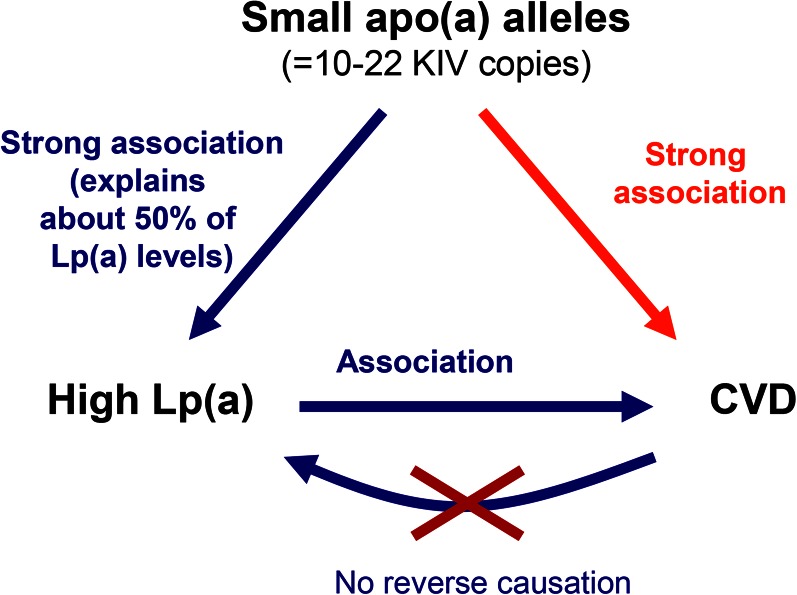
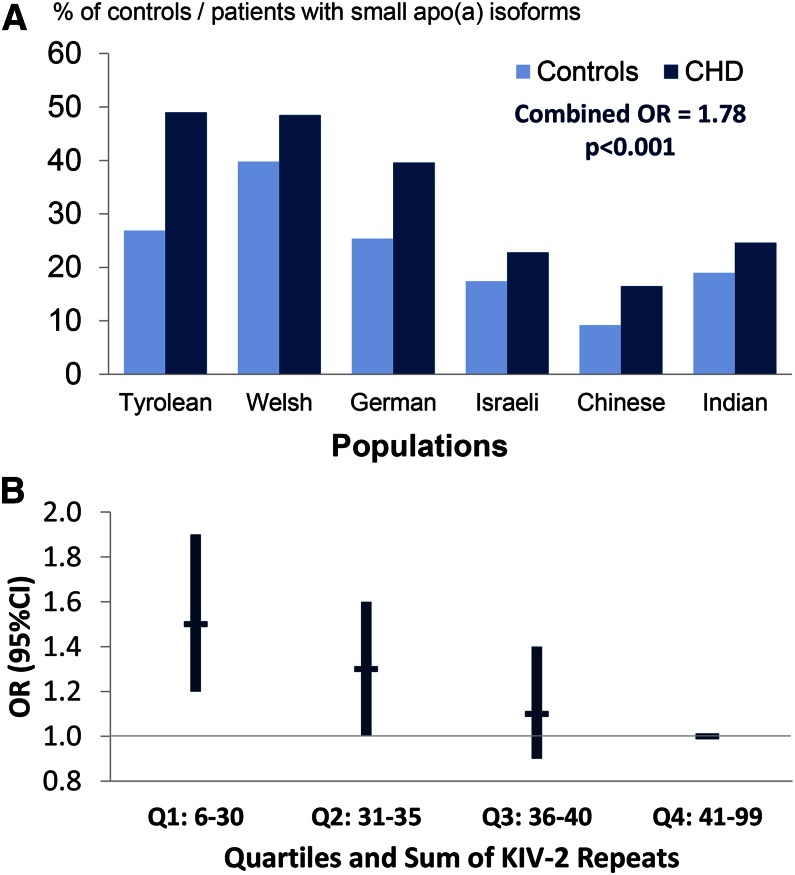
The idea of the type of study termed “Mendelian randomization” was first published by Katan (216), following discussions at a European Lipoprotein Club meeting, and was first put into practice to answer the question of whether or not Lp(a) is an independent risk factor for CHD/myocardial infarction (163, 210). The principal idea of the Mendelian randomization approach (216) applied to Lp(a) and its role in CHD is as follows. Given that high Lp(a) levels are associated with CHD in a causal manner, then any genetic variant in the LPA gene that affects Lp(a) levels must also be associated with CHD to the extent predicted by its influence on Lp(a) levels (Fig. 7). KIV-2 CNV size, which has a strong causal association with Lp(a) levels, has been used to test this hypothesis, and the association of the KIV-2 CNV with CHD was proven to not reflect reversed causation (97, 138, 157, 163, 165, 210, 211). A systematic review by Erquo et al. (164) of 40 studies involving 58,000 participants confirmed that short apo(a) size is indeed associated with CHD. This meta-analysis demonstrated that individuals with short isoforms are at an approximately 2-fold higher risk for CHD and stroke than individuals with longer apo(a) isoforms. Mendelian randomization studies using apo(a) isoforms were confirmed and complemented by studies using KIV-2 copy numbers determined by PFGE (165), and more recently by a large study in which copy numbers were determined by qPCR (138) (Fig. 8A, B).
Fig. 7.
Mendelian randomization approach to demonstrate a causal association between Lp(a) concentration and CHD. Because a low number of KIV copies (11–22 copies) is associated with high Lp(a) levels and high Lp(a) levels are associated with CHD, it follows that a low number of KIV copies will be associated with CHD if the association between Lp(a) and CHD is causal. As the latter is indeed the case, reverse causation [i.e., that CHD is secondarily causing an increase in Lp(a) levels] can be excluded. Figure reprinted with permission of (2).
Fig. 8.
KIV-2 CNV and risk for CHD. A: The apo(a) isoforms and risk for myocardial infarction. The percentage of short apo(a) isoforms (≤22 KIV-2 repeats) in 1,570 controls and 1,013 patients with myocardial infarction from six ethnic groups is shown. Note that short isoforms differ in frequency between populations, but are more frequent in patients in each ethnic group. Data are taken from (163). Figure adapted and reprinted with permission from (213). B: Risk of myocardial infarction by quartiles of genomic KIV-2 repeats in the Copenhagen City Heart Study. KIV-2 copies are determined by qPCR and therefore reflect the sum of KIV-2 repeats of both apo(a) alleles. The data are multivariable adjusted and show a stepwise increase in risk with decrease of KIV-2 copy number. Figure is created based on the data from (138).
SNPs as markers for CHD risk
Recent genome-wide association studies (GWASs) and candidate gene approaches using SNPs have both led to a further understanding of the association between genetic variation at the LPA locus with Lp(a) levels and CHD. Clarke et al. (157) used a custom genotyping chip containing 48,742 SNPs in 2,100 candidate genes to search for genetic associations with CHD in almost 8,000 CHD cases and 8,000 controls of European descent. The results revealed that several SNPs in the LPA region were associated with Lp(a) levels and CHD. The strongest association was observed for rs10455872 (an intronic SNP) and rs3798220 (a nonsynonymous SNP in the PD) (157). A subsequent meta-analysis of these findings reported that CHD risk in the carriers of the minor alleles of rs10455872 and rs3798220 was increased by 42 and 57%, respectively (217).
Another GWAS in Europeans (218) revealed that a haplotype composed of four SNPs, two in LPA (rs7767084 and rs10755578) and two in the neighboring genes, LPAL2 (rs3127599) and SLC22A3 (rs2048327), is strongly associated with CHD. The rare haplotype, CCTC, showed an 82% higher risk, while the more common haplotype, CTTG, increased the risk for CHD by 20% when compared with the most frequent TCTC haplotype. However, the association of these two haplotypes with CHD reflects the association of rs3798220 or rs10455872 with CHD. The haplotypes CCTC and CTTG showed an association with CHD only in carriers of the variant alleles of rs3798220 and rs10455872, respectively (219, 220). Moreover, the association of these two variants with the risk for CHD disappeared upon stratification for Lp(a) levels (157). Hence, all reported SNP associations so far can be explained by LD of the SNPs with short CNV alleles and the associated high Lp(a) levels. It is notable that the minor rs3798220 allele allows tagging of only a subgroup of high-risk short KIV CNV alleles in Europeans (202). In a reversed approach, it has recently been shown that the splice site variant, rs41272114, which results in low Lp(a), protects against CHD (54, 221).
Association of LPA variants with CHD and ethnicity
The association of KIV-2 CNV size and SNPs in LPA with Lp(a) levels is not identical in individuals of different ethnic backgrounds, which has to be considered in Mendelian randomization studies. A Mendelian randomization study in Asian Indians that failed to find an association (173) was taken as evidence for controversial outcomes of early Mendelian randomization studies (97, 138, 157, 163, 165, 210, 211). The reason for the different outcome of the study in Asian Indians is simple. One condition for a Mendelian randomization study was only insufficiently fulfilled. The association between short KIV-2 CNV alleles and high Lp(a) concentrations with CHD is strong in Europeans and white North Americans and very strong in East Asians (Chinese and Japanese) (164), but is very weak in Asian Indians (173). Hence any association, if present, is more difficult to detect in Asian Indians and requires much larger sample sizes. The same problem exists when studying African populations. While one study in African Americans reported an association between short apo(a) isoforms and high Lp(a) in men only (222), other studies found no association between Lp(a) or apo(a) and CHD in this ethnic group (223, 224). Genetic studies in African Americans can be complicated by the fact that they represent an admixed population, with substantial contribution of European and Asian ancestry to their gene pool [reviewed in (225)]. Furthermore, the African American population is composed of descendants from various genetically diverse African populations [reviewed in (226)]. Studies from African populations in Africa on the role of LPA in CHD are still lacking.
As outlined, GWASs and candidate gene approaches have identified SNPs within LPA associated with CHD (157, 196, 197, 217, 218). The question of whether the results of such association studies in one population (i.e., Europeans) are valid for other ethnic groups has recently been addressed for the nonsynonymous SNP, rs3798220 (201). This SNP has been promoted by commercial laboratories for CHD risk screening, as it tags high risk LPA alleles in Europeans, claiming that it was a cost-efficient way to assess the risk potential of LPA and provide guidance on the benefit of aspirin therapy for prevention of CVD (227). The variant allele of r33798220 is found at low frequencies in African Americans and Europeans, while at much higher frequencies in Asian populations and with highest frequencies in admixed Americans (Mexicans, Columbians, Puerto Ricans, and Peruvians), but it is absent in autochthonous Africans (199, 201). Almost all the studies that have investigated the association between rs3798220 and CHD were based on individuals of European descent. The only GWAS on genetic risk factors for CHD in East Asians (Japanese) that included rs3798220 did not find any association between the variant and coronary artery disease (CAD) (228). No association between the rs3798220 variant and high Lp(a) levels was reported in a small sample of Americans of East Asian and Hispanic descent (229) and Chinese CAD patients (230). In a large study conducted in various Asian populations, the variant allele neither associated with short KIV-2 CNV alleles nor with high Lp(a) concentrations in East and Southeast Asians (201). This finding suggests that rs379220 itself is not causal and its association with high Lp(a) and consequently increased CHD risk in Europeans could most likely be explained by the LD with short isoform size that comes with very high Lp(a) levels in Europeans. However, the LD pattern of rs3798220 with other SNPs in LPA appears to be shared between continental groups, thus highlighting the importance of also assessing KIV-2 CNV sizes and considering the genealogy of individuals when conducting SNP-based association studies at the LPA locus. Furthermore, rs3798220 is very rare in Asian Indians and it was shown that this SNP cannot explain Lp(a)-attributed risk for CAD in that population (173, 201).
IMPACT OF GENE LOCI INDEPENDENT FROM THE LPA LOCUS ON Lp(a) LEVELS
Lp(a) levels do not appear to be much influenced by nongenetic factors under physiological conditions in healthy individuals. However, some genetic conditions have strong effects on Lp(a) concentrations. Lp(a) is increased in FH (99) and familial defective apoB-100 (231). The increase in FH is dose-dependent, i.e., highest in FH homozygotes (98). In lipoprotein lipase deficiency (232) and abetalipoproteinemia (233), Lp(a) levels are decreased. A recent GWAS in African Americans identified the APOE gene locus to be associated with Lp(a) concentrations (234). This finding will require confirmation in different populations and functional elucidation. Other genes that have an influence on Lp(a) concentrations were not reported up to now because GWASs searching for Lp(a)-modulating genes have to be sufficiently powered to detect other “minor” genes besides the “giant” LPA locus (213).
NEW PHENOTYPIC ASSOCIATIONS OF Lp(a)
Lp(a) and the corresponding gene locus have been investigated in the context of various diseases with sometimes surprising results. Only recently and in the meanwhile confirmed by several studies including Mendelian randomization approaches, Lp(a) concentrations were found to be associated with the presence of aortic-valve calcification, incident aortic stenosis, aortic-valve replacement, and progression of aortic stenosis (235–238). Furthermore, very recently an association between Lp(a) concentrations and heart failure in more than 98,000 individuals from the general population has been described (239). In that study, when participants with a myocardial infarction or aortic valve stenosis were excluded in the analysis, the risk estimates were attenuated and mediation analysis revealed that 63% of heart failure risk was mediated via myocardial infarction or aortic valve stenosis (239).
Mendelian randomization studies helped to shed some new light on the possible link of Lp(a) to the fibrinolytic system with blood clotting and fibrinolysis for which numerous, mostly in vitro, studies had been published in the past (58). Analyzing 41,000 individuals, neither Lp(a) tertiles nor the sum of K-IV repeats were found to be associated with the risk of venous thrombosis in general population studies (58). This finding has been confirmed by a further study that investigated the two LPA variants, rs10455872 and rs3798220, as surrogate markers for high Lp(a) levels (59). Both studies, however, found an association with atherosclerotic forms of CVD (58, 59). It is currently not clear whether the situation is different in children: a meta-analysis of eight studies including almost 600 children with venous thromboembolism and a suitable number of controls found elevated Lp(a) levels to be associated with a more than 4-fold increased risk for the first onset venous thromboembolism, but not recurrent cases (240). Large genetic studies in children are lacking.
Recent epidemiological studies (82–85) and a Mendelian randomization study surprisingly reported an association between very low Lp(a) concentrations and long apo(a) isoforms with an increased risk of T2DM (83) mostly in Europeans/Whites and Chinese subjects. Some studies used the SNP, rs10455872, to exclude a causal association with T2DM. This SNP is usually associated with high Lp(a) concentrations. These studies did not find an association between the noncarrier status of this SNP and T2DM (83, 84). However, as we discussed recently (241), the noncarrier status of rs10455872 includes about 86% of the European/White population with a very wide range in Lp(a) concentrations and with the inclusion of long and short apo(a) isoforms (83). It is therefore not a genetic marker of very low Lp(a) concentrations and therefore not an appropriate tool to support or exclude a causal role of Lp(a) in T2DM. Any functional mechanism underlying the association between low Lp(a) concentrations and T2DM is unclear, and no higher frequency of diabetes was reported for the Lp(a)-deficient individuals in the Finnish study by Lim et al. (54).
CONCLUSIONS
This review underscores the intriguing facets of the highly atherogenic Lp(a) and its genetic determination. The multi-allelic CNV in the LPA gene was the first example for the powerful Mendelian randomization studies, a method that became quite popular during the last decade. It provided very strong support for the causality of this lipoprotein for CHD. There are still large gaps in our understanding of the Lp(a)/LPA trait and open avenues to future research. Some new and surprising associations and sometimes contradictory results show that “there’s life in the old dog yet” (112).
Footnotes
Abbreviations:
- CAD
- coronary artery disease
- CHD
- coronary heart disease
- CNV
- copy number variation
- FH
- familial hypercholesterolemia
- fiber-FISH
- fiber-fluorescence in situ hybridization
- GWAS
- genome-wide association study
- IL-6
- interleukin-6
- KI–KV
- kringles I–V
- KIV-1–10
- kringle IV types 1–10
- LD
- linkage disequilibrium
- LDLR
- LDL receptor
- LoF
- loss-of-function
- OxPL
- oxidized phospholipid
- PD
- protease domain
- PFGE
- pulsed-field gel electrophoresis
- PLG
- plasminogen
- 5′PNRP
- 5′pentanucleotide repeat polymorphism
- qPCR
- quantitative PCR
- T2DM
- type 2 diabetes mellitus
- 1000G
- 1000 Genomes Project
REFERENCES
- 1.Berg K. 1963. A new serum type system in man–the Lp system. Acta Pathol. Microbiol. Scand. 59: 369–382. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg F., and Utermann G.. 2013. Lipoprotein(a): resurrected by genetics. J. Intern. Med. 273: 6–30. [DOI] [PubMed] [Google Scholar]
- 3.Ehnholm C., Garoff H., Renkonen O., and Simons K.. 1972. Protein and carbohydrate composition of Lp(a)lipoprotein from human plasma. Biochemistry. 11: 3229–3232. [DOI] [PubMed] [Google Scholar]
- 4.Gaubatz J. W., Heideman C., Gotto A. M. Jr., Morrisett J. D., and Dahlen G. H.. 1983. Human plasma lipoprotein(a): structural properties. J. Biol. Chem. 258: 4582–4589. [PubMed] [Google Scholar]
- 5.Utermann G., and Weber W.. 1983. Protein composition of Lp(a) lipoprotein from human plasma. FEBS Lett. 154: 357–361. [DOI] [PubMed] [Google Scholar]
- 6.Utermann G. 1989. The mysteries of lipoprotein(a). Science. 246: 904–910. [DOI] [PubMed] [Google Scholar]
- 7.Eaton D. L., Fless G. M., Kohr W. J., McLean J. W., Xu Q-T., Miller C. G., Lawn R. M., and Scanu A. M.. 1987. Partial amino acid sequence of apolipoprotein[a] shows that it is homologous to plasminogen. Proc. Natl. Acad. Sci. USA. 84: 3224–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kratzin H., Armstrong V. W., Niehaus M., Hilschmann N., and Seidel D.. 1987. Structural relationship of an apolipoprotein(a) phenotype (570 kDa) to plasminogen: homologous kringle domains are linked by carbohydrate-rich regions. Biol. Chem. Hoppe Seyler. 368: 1533–1544. [DOI] [PubMed] [Google Scholar]
- 9.Patthy L. 1985. Evolution of the proteases of blood coagulation and fibrinolysis by assembly from modules. Cell. 41: 657–663. [DOI] [PubMed] [Google Scholar]
- 10.Walz D. A., Hewett-Emmett D., and Seegers W. H.. 1977. Amino acid sequence of human prothrombin fragments 1 and 2. Proc. Natl. Acad. Sci. USA. 74: 1969–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Günzler W. A., Steffens G. J., Otting F., Kim S. M., Frankus E., and Flohé L.. 1982. The primary structure of high molecular mass urokinase from human urine. The complete amino acid sequence of the A chain. Hoppe Seylers Z. Physiol. Chem. 363: 1155–1165. [DOI] [PubMed] [Google Scholar]
- 12.Pennica D., Holmes W. E., Kohr W. J., Harkins R. N., Vehar G. A., Ward C. A., Bennett W. F., Yelverton E., Seeburg P. H., Heyneker H. L., et al. 1983. Cloning and expression of human tissue-type plasminogen activator cDNA in E. coli. Nature. 301: 214–221. [DOI] [PubMed] [Google Scholar]
- 13.Kraft H. G., Menzel H. J., Hoppichler F., Vogel W., and Utermann G.. 1989. Changes of genetic apolipoprotein phenotypes caused by liver transplantation. Implications for apolipoprotein synthesis. J. Clin. Invest. 83: 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer A., Gorges R., Kostner G. M., Paltauf F., and Hermetter A.. 1991. Sulfhydryl-selective fluorescence labeling of lipoprotein(a) reveals evidence for one single disulfide linkage between apoproteins(a) and B-100. Biochemistry. 30: 11245–11249. [DOI] [PubMed] [Google Scholar]
- 15.Guevara J. Jr., Spurlino J., Jan A. Y., Yang C., Tulinsky A., Venkataram Prasad B. V., Gaubatz J. W., and Morrisett J. D.. 1993. Proposed mechanisms for binding of apo[a] kringle type 9 to apo B-100 in human lipoprotein. [a] Biophys. J. 64: 686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunner C., Kraft H. G., Utermann G., and Müller H. J.. 1993. Cys(4057) of apolipoprotein(a) is essential for lipoprotein(a) assembly. Proc. Natl. Acad. Sci. USA. 90: 11643–11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White A. L., and Lanford R. E.. 1994. Cell surface assembly of lipoprotein(a) in primary cultures of baboon hepatocytes. J. Biol. Chem. 269: 28716–28723. [PubMed] [Google Scholar]
- 18.Dieplinger H., and Utermann G.. 1999. The seventh myth of lipoprotein(a): where and how is it assembled? Curr. Opin. Lipidol. 10: 275–283. [DOI] [PubMed] [Google Scholar]
- 19.Koschinsky M. L., and Marcovina S. M.. 2004. Structure-function relationships in apolipoprotein(a): insights into lipoprotein(a) assembly and pathogenicity. Curr. Opin. Lipidol. 15: 167–174. [DOI] [PubMed] [Google Scholar]
- 20.Berg K., and Mohr J.. 1963. Genetics of the LP system. Acta Genet. Stat. Med. 13: 349–360. [DOI] [PubMed] [Google Scholar]
- 21.Ehnholm C., Garoff H., Simons K., and Aro H.. 1971. Purification and quantitation of the human plasma lipoprotein carrying the Lp(a) antigen. Biochim. Biophys. Acta. 236: 431–439. [DOI] [PubMed] [Google Scholar]
- 22.Albers J. J., and Hazzard W. R.. 1974. Immunochemical quantification of human plasma Lp(a) lipoprotein. Lipids. 9: 15–26. [DOI] [PubMed] [Google Scholar]
- 23.Schultz J. S., Schreffler D. C., Sing C. F., and Harvie N. R.. 1974. The genetics of the Lp antigen. I. Its quantitation and distribution in a sample population. Ann. Hum. Genet. 38: 39–46. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt K., Kraft H. G., Parson W., and Utermann G.. 2006. Genetics of the Lp(a)/apo(a) system in an autochthonous Black African population from the Gabon. Eur. J. Hum. Genet. 14: 190–201. [DOI] [PubMed] [Google Scholar]
- 25.Kraft H. G., Köchl S., Menzel H. J., Sandholzer C., and Utermann G.. 1992. The apolipoprotein(a) gene: a transcribed hypervariable locus controlling plasma lipoprotein(a) concentration. Hum. Genet. 90: 220–230. [DOI] [PubMed] [Google Scholar]
- 26.Boerwinkle E., Leffert C. C., Lin J., Lackner C., Chiesa G., and Hobbs H. H.. 1992. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J. Clin. Invest. 90: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Austin M. A., Sandholzer C., Selby J. V., Newman B., Krauss R. M., and Utermann G.. 1992. Lipoprotein(a) in women twins: heritability and relationship to apolipoprotein(a) phenotypes. Am. J. Hum. Genet. 51: 829–840. [PMC free article] [PubMed] [Google Scholar]
- 28.Rao F., Schork A. J., Maihofer A. X., Nievergelt C. M., Marcovina S. M., Miller E. R., Witztum J. L., O’Connor D. T., and Tsimikas S.. 2015. Heritability of biomarkers of oxidized lipoproteins: twin pair study. Arterioscler. Thromb. Vasc. Biol. 35: 1704–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank S. L., Klisak I., Sparkes R. S., Mohandas T., Tomlinson J. E., McLean J. W., Lawn R. M., and Lusis A. J.. 1988. The apolipoprotein (a) gene resides on human chromosome 6q26-27 in close proximity to the homologous gene for plasminogen. Hum. Genet. 79: 352–356. [DOI] [PubMed] [Google Scholar]
- 30.Drayna D. T., Hegele R. A., Hass P. E., Emi M., Wu L. L., Eaton D. L., Lawn R. M., Williams R. R., White R. L., and Lalouel J. M.. 1988. Genetic linkage between lipoprotein(a) phenotype and a DNA polymorphism in the plasminogen gene. Genomics. 3: 230–236. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl G., Gersdorf E., Menzel H. J., Duba C., Cleve H., Humphries S., and Utermann G.. 1989. The gene for the Lp(a)-specific glycoprotein is closely linked to the gene for plasminogen on chromosome 6. Hum. Genet. 81: 149–152. [DOI] [PubMed] [Google Scholar]
- 32.Scholz M., Kraft H. G., Lingenhel A., Delport R., Vorster E. H., Bickeböller H., and Utermann G.. 1999. Genetic control of lipoprotein(a) concentrations is different in Africans and Caucasians. Eur. J. Hum. Genet. 7: 169–178. [DOI] [PubMed] [Google Scholar]
- 33.DeMeester C. A., Bu X., Gray R. J., Lusis A. J., and Rotter J. I.. 1995. Genetic variation in lipoprotein (a) levels in families enriched for coronary artery disease is determined almost entirely by the apolipoprotein (a) gene locus. Am. J. Hum. Genet. 56: 287–293. [PMC free article] [PubMed] [Google Scholar]
- 34.Mooser V., Scheer D., Marcovina S. M., Wang J. P., Guerra R., Cohen J., and Hobbs H. H.. 1997. The Apo(a) gene is the major determinant of variation in plasma Lp(a) levels in African Americans. Am. J. Hum. Genet. 61: 402–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandholzer C., Hallman D. M., Saha N., Sigurdsson G., Lackner C., Császár A., Boerwinkle E., and Utermann G.. 1991. Effects of the apolipoprotein(a) size polymorphism on the lipoprotein(a) concentration in 7 ethnic groups. Hum. Genet. 86: 607–614. [DOI] [PubMed] [Google Scholar]
- 36.Kraft H. G., Lingenhel A., Pang R. W. C., Delport R., Trommsdorff M., Vermaak H., Janus E. D., and Utermann G.. 1996. Frequency distributions of apolipoprotein(a) kringle IV repeat alleles and their effects on lipoprotein(a) levels in Caucasian, Asian, and African populations: The distribution of null alleles is non-random. Eur. J. Hum. Genet. 4: 74–87. [DOI] [PubMed] [Google Scholar]
- 37.Gaw A., Boerwinkle E., Cohen J. C., and Hobbs H. H.. 1994. Comparative analysis of the apo(a) gene, apo(a) glycoprotein, and plasma concentrations of Lp(a) in three ethnic groups: evidence for no common “null” allele at the apo(a) locus. J. Clin. Invest. 93: 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams-Blangero S., and Rainwater D. L.. 1991. Variation in Lp(a) levels and apo(a) isoform frequencies in five baboon subspecies. Hum. Biol. 63: 65–76. [PubMed] [Google Scholar]
- 39.Doucet C., Huby T., Chapman J., and Thillet J.. 1994. Lipoprotein[a] in the chimpanzee: relationship of apo[a] phenotype to elevated plasma Lp[a] levels. J. Lipid Res. 35: 263–270. [PubMed] [Google Scholar]
- 40.Lawn R. M., Schwartz K., and Patthy L.. 1997. Convergent evolution of apolipoprotein(a) in primates and hedgehog. Proc. Natl. Acad. Sci. USA. 94: 11992–11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLean J. W., Tomlinson J. E., Kuang W-J., Eaton D. L., Chen E. Y., Fless G. M., Scanu A. M., and Lawn R. M.. 1987. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 330: 132–137. [DOI] [PubMed] [Google Scholar]
- 42.Tomlinson J. E., McLean J. W., and Lawn R. M.. 1989. Rhesus monkey apolipoprotein(a). Sequence, evolution, and sites of synthesis. J. Biol. Chem. 264: 5957–5965. [PubMed] [Google Scholar]
- 43.Leibundgut G., Scipione C., Yin H., Schneider M., Boffa M. B., Green S., Yang X., Dennis E., Witztum J. L., Koschinsky M. L., et al. 2013. Determinants of binding of oxidized phospholipids on apolipoprotein (a) and lipoprotein (a). J. Lipid Res. 54: 2815–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haibach C., Kraft H. G., Köchl S., Abe A., and Utermann G.. 1998. The number of kringle IV repeats 3-10 is invariable in the human apo(a) gene. Gene. 208: 253–258. [DOI] [PubMed] [Google Scholar]
- 45.Utermann G., Menzel H. J., Kraft H. G., Duba H. C., Kemmler H. G., and Seitz C.. 1987. Lp(a) glycoprotein phenotypes: inheritance and relation to Lp(a)-lipoprotein concentrations in plasma. J. Clin. Invest. 80: 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byrne C. D., Schwartz K., Meer K., Cheng J. F., and Lawn R. M.. 1994. The human apolipoprotein(a)/plasminogen gene cluster contains a novel homologue transcribed in liver. Arterioscler. Thromb. 14: 534–541. [DOI] [PubMed] [Google Scholar]
- 47.Kraft H. G., Sandholzer C., Menzel H. J., and Utermann G.. 1992. Apolipoprotein(a) alleles determine lipoprotein(a) particle density and concentration in plasma. Arterioscler. Thromb. 12: 302–306. [DOI] [PubMed] [Google Scholar]
- 48.Koschinsky M. L., Côté G. P., Gabel B., and Van der Hoek Y. Y.. 1993. Identification of the cysteine residue in apolipoprotein(a) that mediates extracellular coupling with apolipoprotein B-100. J. Biol. Chem. 268: 19819–19825. [PubMed] [Google Scholar]
- 49.McCormick S. P. A., No J. K., Taylor S., Flynn L. M., Hammer R. E., and Young S. G.. 1995. Mutagenesis of the human apolipoprotein B gene in a yeast artificial chromosome reveals the site of attachment for apolipoprotein(a). Proc. Natl. Acad. Sci. USA. 92: 10147–10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callow M. J., and Rubin E. M.. 1995. Site-specific mutagenesis demonstrates that cysteine 4326 of apolipoprotein B is required for covalent linkage with apolipoprotein(a) in vivo. J. Biol. Chem. 270: 23914–23917. [DOI] [PubMed] [Google Scholar]
- 51.Xu S. 1998. Apolipoprotein(a) binds to low-density lipoprotein at two distant sites in lipoprotein(a). Biochemistry. 37: 9284–9294. [DOI] [PubMed] [Google Scholar]
- 52.Phillips M. L., Lembertas A. V., Schumaker V. N., Lawn R. M., Shire S. J., and Zioncheck T. F.. 1993. Physical properties of recombinant apolipoprotein(a) and its association with LDL to form an Lp(a)-like complex. Biochemistry. 32: 3722–3728. [DOI] [PubMed] [Google Scholar]
- 53.Ogorelkova M., Gruber A., and Utermann G.. 1999. Molecular basis of congenital Lp(a) deficiency: a frequent apo(a) ‘null’ mutation in Caucasians. Hum. Mol. Genet. 8: 2087–2096. [DOI] [PubMed] [Google Scholar]
- 54.Lim E. T., Wurtz P., Havulinna A. S., Palta P., Tukiainen T., Rehnstrom K., Esko T., Magi R., Inouye M., Lappalainen T., et al. 2014. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 10: e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miles L. A., and Plow E. F.. 1990. Lp(a): an interloper into the fibrinolytic system. Thromb. Haemost. 63: 331–335. [PubMed] [Google Scholar]
- 56.Koschinsky M. L. 2006. Novel insights into Lp(a) physiology and pathogenicity: more questions than answers? Cardiovasc. Hematol. Disord. Drug Targets. 6: 267–278. [DOI] [PubMed] [Google Scholar]
- 57.Boffa M. B., and Koschinsky M. L.. 2015. Lipoprotein(a): truly a direct prothrombotic factor in cardiovascular disease? J. Lipid Res. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kamstrup P. R., Tybjaerg-Hansen A., and Nordestgaard B. G.. 2012. Genetic evidence that lipoprotein(a) associates with atherosclerotic stenosis rather than venous thrombosis. Arterioscler. Thromb. Vasc. Biol. 32: 1732–1741. [DOI] [PubMed] [Google Scholar]
- 59.Helgadottir A., Gretarsdottir S., Thorleifsson G., Holm H., Patel R. S., Gudnason T., Jones G. T., van Rij A. M., Eapen D. J., Baas A. F., et al. 2012. Apolipoprotein(a) genetic sequence variants associated with systemic atherosclerosis and coronary atherosclerotic burden but not with venous thromboembolism. J. Am. Coll. Cardiol. 60: 722–729. [DOI] [PubMed] [Google Scholar]
- 60.Brown M. S., and Goldstein J. L.. 1987. Plasma lipoproteins: teaching old dogmas new tricks. Nature. 330: 113–114. [DOI] [PubMed] [Google Scholar]
- 61.Pillarisetti S., Paka L., Obunike J. C., Berglund L., and Goldberg I. J.. 1997. Subendothelial retention of lipoprotein (a) - Evidence that reduced heparan sulfate promotes lipoprotein binding to subendothelial matrix. J. Clin. Invest. 100: 867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hughes S. D., Lou X. J., Ighani S., Verstuyft J., Grainger D. J., Lawn R. M., and Rubin E. M.. 1997. Lipoprotein(a) vascular accumulation in mice - in vivo analysis of the role of lysine binding sites using recombinant adenovirus. J. Clin. Invest. 100: 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keesler G. A., Gabel B. R., Devlin C. M., Koschinsky M. L., and Tabas I.. 1996. The binding activity of the macrophage lipoprotein(a) apolipoprotein(a) receptor is induced by cholesterol via a post-translational mechanism and recognizes distinct kringle domains on apolipoprotein(a). J. Biol. Chem. 271: 32096–32104. [DOI] [PubMed] [Google Scholar]
- 64.Higazi A. A. R., Lavi E., Bdeir K., Ulrich A. M., Jamieson D. G., Rader D. J., Usher D. C., Kane W., Ganz T., and Cines D. B.. 1997. Defensin stimulates the binding of lipoprotein (a) to human vascular endothelial and smooth muscle cells. Blood. 89: 4290–4298. [PubMed] [Google Scholar]
- 65.Kostner G. M., and Bihari-Varga M.. 1990. Is the atherogenicity of Lp(a) caused by its reactivity with proteoglycans? Eur. Heart J. 11: 184–189. [DOI] [PubMed] [Google Scholar]
- 66.van der Hoek Y. Y., Sangrar W., Côté G. P., Kastelein J. J. P., and Koschinsky M. L.. 1994. Binding of recombinant apolipoprotein(a) to extracellular matrix proteins. Arterioscler. Thromb. 14: 1792–1798. [DOI] [PubMed] [Google Scholar]
- 67.Kapetanopoulos A., Fresser F., Millonig G., Shaul Y., Baier G., and Utermann G.. 2002. Direct interaction of the extracellular matrix protein DANCE with apolipoprotein(a) mediated by the kringle IV-type 2 domain. Mol. Genet. Genomics. 267: 440–446. [DOI] [PubMed] [Google Scholar]
- 68.Walton K. W., Hitchens J., Magnani H. N., and Khan M.. 1974. A study of methods of identification and estimation of Lp(a) lipoprotein and of its significance in health, hyperlipidaemia and atherosclerosis. Atherosclerosis. 20: 323–346. [DOI] [PubMed] [Google Scholar]
- 69.Donate L. E., Gherardi E., Srinivasan N., Sowdhamini R., Aparicio S., and Blundell T. L.. 1994. Molecular evolution and domain structure of plasminogen-related growth factors (HGF/SF and HGF1/MSP). Protein Sci. 3: 2378–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wade D. P., Clarke J. G., Lindahl G. E., Liu A. C., Zysow B. R., Meer K., Schwartz K., and Lawn R. M.. 1993. 5′ control regions of the apolipoprotein(a) gene and members of the related plasminogen gene family. Proc. Natl. Acad. Sci. USA. 90: 1369–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noma A., Abe A., Maeda S., Seishima M., Makino K., Yano Y., and Shimokawa K.. 1994. Lp(a): an acute-phase reactant. Chem. Phys. Lipids. 67–68: 411–417. [DOI] [PubMed] [Google Scholar]
- 72.Lippi G., Braga V., Adami S., and Guidi G.. 1998. Modification of serum apolipoprotein A-I, apolipoprotein B and lipoprotein(a) levels after bisphosphonates-induced acute phase response. Clin. Chim. Acta. 271: 79–87. [DOI] [PubMed] [Google Scholar]
- 73.Chimienti G., Aquilino F., Rotelli M. T., Russo F., Lupo L., and Pepe G.. 2006. Lipoprotein(a), lipids and proinflammatory cytokines in patients undergoing major abdominal surgery. Br. J. Surg. 93: 347–353. [DOI] [PubMed] [Google Scholar]
- 74.Mooser V., Berger M. M., Tappy L., Cayeux C., Marcovina S. M., Darioli R., Nicod P., and Chiolero R.. 2000. Major reduction in plasma Lp(a) levels during sepsis and burns. Arterioscler. Thromb. Vasc. Biol. 20: 1137–1142. [DOI] [PubMed] [Google Scholar]
- 75.Ramharack R., Barkalow D., and Spahr M. A.. 1998. Dominant negative effect of TGF-b1 and TNF-a on basal and IL-6-induced lipoprotein(a) and apolipoprotein(a) mRNA expression in primary monkey hepatocyte cultures. Arterioscler. Thromb. Vasc. Biol. 18: 984–990. [DOI] [PubMed] [Google Scholar]
- 76.Müller N., Schulte D. M., Türk K., Freitag-Wolf S., Hampe J., Zeuner R., Schröder J. O., Gouni-Berthold I., Berthold H. K., Krone W., et al. 2015. IL-6 blockade by monoclonal antibodies inhibits apolipoprotein (a) expression and lipoprotein (a) synthesis in humans. J. Lipid Res. 56: 1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bergmark C., Dewan A., Orsoni A., Merki E., Miller E. R., Shin M. J., Binder C. J., Horkko S., Krauss R. M., Chapman M. J., et al. 2008. A novel function of lipoprotein [a] as a preferential carrier of oxidized phospholipids in human plasma. J. Lipid Res. 49: 2230–2239. [DOI] [PubMed] [Google Scholar]
- 78.Tsimikas S., Brilakis E. S., Miller E. R., McConnell J. P., Lennon R. J., Kornman K. S., Witztum J. L., and Berger P. B.. 2005. Oxidized phospholipids, Lp(a) lipoprotein, and coronary artery disease. N. Engl. J. Med. 353: 46–57. [DOI] [PubMed] [Google Scholar]
- 79.Kiechl S., Willeit J., Mayr M., Viehweider B., Oberhollenzer F., Kronenberg F., Wiedermann C. J., Oberthaler S., Xu Q., Witztum J. L., et al. 2007. Oxidized phospholipids, lipoprotein(a), lipoprotein-associated phospholipase A2 activity and 10-year cardiovascular outcomes: prospective results from the Bruneck study. Arterioscler. Thromb. Vasc. Biol. 27: 1788–1795. [DOI] [PubMed] [Google Scholar]
- 80.Salonen E. M., Jauhiainen M., Zardi L., Vaheri A., and Ehnholm C.. 1989. Lipoprotein(a) binds to fibronectin and has serine proteinase activity capable of cleaving it. EMBO J. 8: 4035–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jauhiainen M., Metso J., Koskinen P., and Ehnholm C.. 1991. Characterization of the enzyme activity of human plasma lipoprotein (a) using synthetic peptide substrates. Biochem. J. 274: 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mora S., Kamstrup P. R., Rifai N., Nordestgaard B. G., Buring J. E., and Ridker P. M.. 2010. Lipoprotein(a) and risk of type 2 diabetes. Clin. Chem. 56: 1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kamstrup P. R., and Nordestgaard B. G.. 2013. Lipoprotein(a) concentrations, isoform size, and risk of type 2 diabetes: a Mendelian randomisation study. Lancet Diabetes Endocrinol. 1: 220–227. [DOI] [PubMed] [Google Scholar]
- 84.Ye Z., Haycock P. C., Gurdasani D., Pomilla C., Boekholdt S. M., Tsimikas S., Khaw K. T., Wareham N. J., Sandhu M. S., and Forouhi N. G.. 2014. The association between circulating lipoprotein(a) and type 2 diabetes: is it causal? Diabetes. 63: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ding L., Song A., Dai M., Xu M., Sun W., Xu B., Sun J., Wang T., Xu Y., Lu J., et al. 2015. Serum lipoprotein (a) concentrations are inversely associated with T2D, prediabetes, and insulin resistance in a middle-aged and elderly Chinese population. J. Lipid Res. 56: 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Azrolan N., Gavish D., and Breslow J. L.. 1991. Plasma lipoprotein(a) concentration is controlled by apolipoprotein(a) (apo(a)) protein size and the abundance of hepatic apo(a) mRNA in a cynomolgus monkey model. J. Biol. Chem. 266: 13866–13872. [PubMed] [Google Scholar]
- 87.Koschinsky M. L., Beisiegel U., Henne-Bruns D., Eaton D. L., and Lawn R. M.. 1990. Apolipoprotein(a) size heterogeneity is related to variable number of repeat sequences in its mRNA. Biochemistry. 29: 640–644. [DOI] [PubMed] [Google Scholar]
- 88.Brunner C., Lobentanz E. M., Pethö-Schramm A., Ernst A., Kang C., Dieplinger H., Müller H. J., and Utermann G.. 1996. The number of identical kringle IV repeats in apolipoprotein(a) affects its processing and secretion by HepG2 cells. J. Biol. Chem. 271: 32403–32410. [DOI] [PubMed] [Google Scholar]
- 89.Su W., Campos H., Judge H., Walsh B. W., and Sacks F. M.. 1998. Metabolism of Apo(a) and ApoB100 of lipoprotein(a) in women: effect of postmenopausal estrogen replacement. J. Clin. Endocrinol. Metab. 83: 3267–3276. [DOI] [PubMed] [Google Scholar]
- 90.Krempler F., Kostner G. M., Roscher A., Haslauer F., Bolzano K., and Sandhofer F.. 1983. Studies on the role of specific cell surface receptors in the removal of lipoprotein(a) in man. J. Clin. Invest. 71: 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hofer G., Steyrer E., Kostner G. M., and Hermetter A.. 1997. LDL-mediated interaction of Lp[a] with HepG2 cells: a novel fluorescence microscopy approach. J. Lipid Res. 38: 2411–2421. [PubMed] [Google Scholar]
- 92.Niemeier A., Willnow T., Dieplinger H., Jacobsen C., Meyer N., Hilpert J., and Beisiegel U.. 1999. Identification of Megalin/gp330 as a receptor for lipoprotein(a) in vitro. Arterioscler. Thromb. Vasc. Biol. 19: 552–561. [DOI] [PubMed] [Google Scholar]
- 93.Krempler F., Kostner G. M., Bolzano K., and Sandhofer F.. 1980. Turnover of lipoprotein(a) in man. J. Clin. Invest. 65: 1483–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perombelon Y. F. N., Soutar A. K., and Knight B. L.. 1994. Variation in lipoprotein(a) concentration associated with different apolipoprotein(a) alleles. J. Clin. Invest. 93: 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rader D. J., Cain W., Ikewaki K., Talley G., Zech L. A., Usher D., and Brewer H. B.. 1994. The inverse association of plasma lipoprotein(a) concentrations with apolipoprotein(a) isoform size is not due to differences in Lp(a) catabolism but to differences in production rate. J. Clin. Invest. 93: 2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cain W. J., Millar J. S., Himebauch A. S., Tietge U. J., Maugeais C., Usher D., and Rader D. J.. 2005. Lipoprotein [a] is cleared from the plasma primarily by the liver in a process mediated by apolipoprotein [a]. J. Lipid Res. 46: 2681–2691. [DOI] [PubMed] [Google Scholar]
- 97.Seed M., Hoppichler F., Reaveley D., McCarthy S., Thompson G. R., Boerwinkle E., and Utermann G.. 1990. Relation of serum lipoprotein(a) concentration and apolipoprotein(a) phenotype to coronary heart disease in patients with familial hypercholesterolemia. N. Engl. J. Med. 322: 1494–1499. [DOI] [PubMed] [Google Scholar]
- 98.Kraft H. G., Lingenhel A., Raal F. J., Hohenegger M., and Utermann G.. 2000. Lipoprotein(a) in homozygous familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 20: 522–528. [DOI] [PubMed] [Google Scholar]
- 99.Utermann G., Hoppichler F., Dieplinger H., Seed M., Thompson G., and Boerwinkle E.. 1989. Defects in the low density lipoprotein receptor gene affect lipoprotein (a) levels: multiplicative interaction of two gene loci associated with premature atherosclerosis. Proc. Natl. Acad. Sci. USA. 86: 4171–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wiklund O., Angelin B., Olofsson S-O., Eriksson M., Fager G., Berglund L., and Bondjers G.. 1990. Apolipoprotein(a) and ischaemic heart disease in familial hypercholesterolaemia. Lancet. 335: 1360–1363. [DOI] [PubMed] [Google Scholar]
- 101.Mbewu A. D., Bhatnagar D., Durrington P. N., Hunt L., Ishola M., Arrol S., Mackness M., Lockley P., and Miller J. P.. 1991. Serum lipoprotein(a) in patients heterozygous familial hypercholesterolaemia, their relatives, and unrelated control populations. Arterioscler. Thromb. 11: 940–946. [DOI] [PubMed] [Google Scholar]
- 102.Friedlander Y., and Leitersdorf E.. 1995. Segregation analysis of plasma lipoprotein(a) levels in pedigrees with molecularly defined familial hypercholesterolemia. Genet. Epidemiol. 12: 129–143. [DOI] [PubMed] [Google Scholar]
- 103.Tada H., Kawashiri M. A., Yoshida T., Teramoto R., Nohara A., Konno T., Inazu A., Mabuchi H., Yamagishi M., and Hayashi K.. 2016. Lipoprotein(a) in familial hypercholesterolemia with proprotein convertase subtilisin/kexin type 9 (PCSK9) gain-of-function mutations. Circ. J. 80: 512–518. [DOI] [PubMed] [Google Scholar]
- 104.Soutar A. K., McCarthy S. N., Seed M., and Knight B. L.. 1991. Relationship between apolipoprotein (a) phenotype, lipoprotein (a) concentration in plasma, and low density lipoprotein (LDL) receptor function in a large kindred with familial hypercholesterolemia due to the Pro664-Leu mutation in the LDL-receptor gene. J. Clin. Invest. 88: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ghiselli G., Gaddi A., Barozzi G., Ciarrocchi A., and Descovich G.. 1992. Plasma lipoprotein(a) concentration in familial hypercholesterolemic patients without coronary artery disease. Metabolism. 41: 833–838. [DOI] [PubMed] [Google Scholar]
- 106.Jansen A. C., van Aalst-Cohen E. S., Tanck M. W., Trip M. D., Lansberg P. J., Liem A. H., van Lennep H. W., Sijbrands E. J., and Kastelein J. J.. 2004. The contribution of classical risk factors to cardiovascular disease in familial hypercholesterolaemia: data in 2400 patients. J. Intern. Med. 256: 482–490. [DOI] [PubMed] [Google Scholar]
- 107.Miller P. E., Martin S. S., Toth P. P., Santos R. D., Blaha M. J., Nasir K., Virani S. S., Post W. S., Blumenthal R. S., and Jones S. R.. 2015. Screening and advanced lipid phenotyping in familial hypercholesterolemia: the Very Large Database of Lipids study-17 (VLDL-17). J. Clin. Lipidol. 9: 676–683. [DOI] [PubMed] [Google Scholar]
- 108.Leitersdorf E., Friedlander Y., Bard J-M., Fruchart J-C., Eisenberg S., and Stein Y.. 1991. Diverse effect of ethnicity on plasma lipoprotein[a] levels in heterozygote patients with familial hypercholesterolemia. J. Lipid Res. 32: 1513–1519. [PubMed] [Google Scholar]
- 109.Raal F. J., Giugliano R. P., Sabatine M. S., Koren M. J., Langslet G., Bays H., Blom D., Eriksson M., Dent R., Wasserman S. M., et al. 2014. Reduction in lipoprotein(a) with PCSK9 monoclonal antibody evolocumab (AMG 145): a pooled analysis of more than 1,300 patients in 4 phase II trials. J. Am. Coll. Cardiol. 63: 1278–1288. [DOI] [PubMed] [Google Scholar]
- 110.Kostner G. M., Gavish D., Leopold B., Bolzano K., Weintraub M. S., and Breslow J. L.. 1989. HMG CoA reductase inhibitors lower LDL cholesterol without reducing Lp(a) levels. Circulation. 80: 1313–1319. [DOI] [PubMed] [Google Scholar]
- 111.Khera A. V., Everett B. M., Caulfield M. P., Hantash F. M., Wohlgemuth J., Ridker P. M., and Mora S.. 2014. Lipoprotein(a) concentrations, rosuvastatin therapy, and residual vascular risk: an analysis from the JUPITER trial (Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin). Circulation. 129: 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kronenberg F. 2014. Lipoprotein(a): there’s life in the old dog yet. Circulation. 129: 619–621. [DOI] [PubMed] [Google Scholar]
- 113.Kronenberg F., Utermann G., and Dieplinger H.. 1996. Lipoprotein(a) in renal disease. Am. J. Kidney Dis. 27: 1–25. [DOI] [PubMed] [Google Scholar]
- 114.Kronenberg F. 2014. Causes and consequences of lipoprotein(a) abnormalities in kidney disease. Clin. Exp. Nephrol. 18: 234–237. [DOI] [PubMed] [Google Scholar]
- 115.Kostner K. M., Maurer G., Huber K., Stefenelli T., Dieplinger H., Steyrer E., and Kostner G. M.. 1996. Urinary excretion of apo(a) fragments. Role in apo(a) catabolism. Arterioscler. Thromb. Vasc. Biol. 16: 905–911. [DOI] [PubMed] [Google Scholar]
- 116.Mooser V., Marcovina S. M., White A. L., and Hobbs H. H.. 1996. Kringle-containing fragments of apolipoprotein(a) circulate in human plasma and are excreted into the urine. J. Clin. Invest. 98: 2414–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kronenberg F., Trenkwalder E., Lingenhel A., Friedrich G., Lhotta K., Schober M., Moes N., König P., Utermann G., and Dieplinger H.. 1997. Renovascular arteriovenous differences in Lp(a) plasma concentrations suggest removal of Lp(a) from the renal circulation. J. Lipid Res. 38: 1755–1763. [PubMed] [Google Scholar]
- 118.Hixson J. E., Britten M. L., Manis G. S., and Rainwater D. L.. 1989. Apolipoprotein(a) (Apo(a)) glycoprotein isoforms result from size differences in Apo(a) mRNA in baboons. J. Biol. Chem. 264: 6013–6016. [PubMed] [Google Scholar]
- 119.Rainwater D. L., and Manis G. S.. 1988. Immunochemical characterization and quantitation of lipoprotein(a) in baboons. Development of an assay depending on two antigenetically distinct proteins. Atherosclerosis. 73: 23–31. [DOI] [PubMed] [Google Scholar]
- 120.Laplaud P. M., Beaubatie L., Rall S. C. Jr., Luc G., and Saboureau M.. 1988. Lipoprotein[a] is the major apoB-containing lipoprotein in the plasma of a hibernator, the hedgehog (Erinaceus europaeus). J. Lipid Res. 29: 1157–1170. [PubMed] [Google Scholar]
- 121.Lawn R. M., Boonmark N. W., Schwartz K., Lindahl G. E., Wade D. P., Byrne C. D., Fong K. J., Meer K., and Patthy L.. 1995. The recurring evolution of lipoprotein(a). Insights from cloning of hedgehog apolipoprotein(a). J. Biol. Chem. 270: 24004–24009. [DOI] [PubMed] [Google Scholar]
- 122.Guo H. C., Michel J. B., Blouquit Y., and Chapman M. J.. 1991. Lipoprotein(a) and apolipoprotein(a) in a New World monkey, the common marmoset (Callithrix jacchus). Association of variable plasma lipoprotein(a) levels with a single apolipoprotein(a) isoform. Arterioscler. Thromb. 11: 1030–1041. [DOI] [PubMed] [Google Scholar]
- 123.Rath M., and Pauling L.. 1990. Immunological evidence for the accumulation of lipoprotein(a) in the atherosclerotic lesion of the hypoascorbemic guinea pig. Proc. Natl. Acad. Sci. USA. 87: 9388–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen T. J., Ji C. Y., and Hu Y. H.. 2009. Genetic and environmental influences on serum lipids and the effects of puberty: a Chinese twin study. Acta Paediatr. 98: 1029–1036. [DOI] [PubMed] [Google Scholar]
- 125.Ober C., Abney M., and McPeek M. S.. 2001. The genetic dissection of complex traits in a founder population. Am. J. Hum. Genet. 69: 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rao V. S., Khadrinarasimhaih N. B., Kanjilal S., Mukerjee M., and Kakkar V. V.. 2009. Genetic contribution to variation in atherothrombotic phenotypes in the Asian Indian population. The Indian Atherosclerosis Research Study. Thromb. Haemost. 102: 379–388. [DOI] [PubMed] [Google Scholar]
- 127.Hong Y., Dahlén G. H., Pedersen N., Heller D. A., McClearn G. E., and de Faire U.. 1995. Potential environmental effects on adult lipoprotein(a) levels: Results from Swedish twins. Atherosclerosis. 117: 295–304. [DOI] [PubMed] [Google Scholar]
- 128.Ober C., Nord A. S., Thompson E. E., Pan L., Tan Z., Cusanovich D., Sun Y., Nicolae R., Edelstein C., Schneider D. H., et al. 2009. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. J. Lipid Res. 50: 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Broeckel U., Hengstenberg C., Mayer B., Holmer S., Martin L. J., Comuzzie A. G., Blangero J., Nurnberg P., Reis A., Riegger G. A., et al. 2002. A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat. Genet. 30: 210–214. [DOI] [PubMed] [Google Scholar]
- 130.Barlera S., Specchia C., Farrall M., Chiodini B. D., Franzosi M. G., Rust S., Green F., Nicolis E. B., Peden J., Assmann G., et al. 2007. Multiple QTL influence the serum Lp(a) concentration: a genome-wide linkage screen in the PROCARDIS study. Eur. J. Hum. Genet. 15: 221–227. [DOI] [PubMed] [Google Scholar]
- 131.Feuk L., Carson A. R., and Scherer S. W.. 2006. Structural variation in the human genome. Nat. Rev. Genet. 7: 85–97. [DOI] [PubMed] [Google Scholar]
- 132.Parson W., Kraft H. G., Niederstatter H., Lingenhel A. W., Kochl S., Fresser F., and Utermann G.. 2004. A common nonsense mutation in the repetitive Kringle IV-2 domain of human apolipoprotein(a) results in a truncated protein and low plasma Lp(a). Hum. Mutat. 24: 474–480. [DOI] [PubMed] [Google Scholar]
- 133.Røsby O., Aleström P., and Berg K.. 2000. Sequence conservation in kringle IV-type 2 repeats of the LPA gene. Atherosclerosis. 148: 353–364. [DOI] [PubMed] [Google Scholar]
- 134.Rösby O., Aleström P., and Berg K.. 1997. High-degree sequence conservation in LPA kringle IV-type 2 exons and introns. Clin. Genet. 52: 293–302. [DOI] [PubMed] [Google Scholar]
- 135.Chen J. M., Cooper D. N., Chuzhanova N., Ferec C., and Patrinos G. P.. 2007. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8: 762–775. [DOI] [PubMed] [Google Scholar]
- 136.Enkhmaa B., Anuurad E., and Berglund L.. 2015. Lipoprotein(a): impact by ethnicity, environmental and medical conditions. J. Lipid Res. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lackner C., Boerwinkle E., Leffert C. C., Rahmig T., and Hobbs H. H.. 1991. Molecular basis of apolipoprotein (a) isoform size heterogeneity as revealed by pulsed-field gel electrophoresis. J. Clin. Invest. 87: 2153–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kamstrup P. R., Tybjaerg-Hansen A., Steffensen R., and Nordestgaard B. G.. 2009. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 301: 2331–2339. [DOI] [PubMed] [Google Scholar]
- 139.Erdel M., Hubalek M., Lingenhel A., Kofler K., Duba H. C., and Utermann G.. 1999. Counting the repetitive kringle-IV repeats in the gene encoding human apolipoprotein(a) by fibre-FISH. Nat. Genet. 21: 357–358. [DOI] [PubMed] [Google Scholar]
- 140.Stankiewicz P., Shaw C. J., Withers M., Inoue K., and Lupski J. R.. 2004. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 14: 2209–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nahon J. L. 2003. Birth of ‘human-specific’ genes during primate evolution. Genetica. 118: 193–208. [DOI] [PubMed] [Google Scholar]
- 142.Bailey J. A., and Eichler E. E.. 2006. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat. Rev. Genet. 7: 552–564. [DOI] [PubMed] [Google Scholar]
- 143.Dumas L., Kim Y. H., Karimpour-Fard A., Cox M., Hopkins J., Pollack J. R., and Sikela J. M.. 2007. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 17: 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Inoue K., Khajavi M., Ohyama T., Hirabayashi S., Wilson J., Reggin J. D., Mancias P., Butler I. J., Wilkinson M. F., Wegner M., et al. 2004. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat. Genet. 36: 361–369. [DOI] [PubMed] [Google Scholar]
- 145.Gonzalez E., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R. J., Freedman B. I., Quinones M. P., Bamshad M. J., et al. 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 307: 1434–1440. [DOI] [PubMed] [Google Scholar]
- 146.Perry G. H., Dominy N. J., Claw K. G., Lee A. S., Fiegler H., Redon R., Werner J., Villanea F. A., Mountain J. L., Misra R., et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39: 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Stankiewicz P., and Lupski J. R.. 2010. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61: 437–455. [DOI] [PubMed] [Google Scholar]
- 148.Castellino F. J., Ploplis V. A., Powell J. R., and Strickland D. K.. 1981. The existence of independent domain structures in human Lys77-plasminogen. J. Biol. Chem. 256: 4778–4782. [PubMed] [Google Scholar]
- 149.Ploplis V. A., Strickland D. K., and Castellino F. J.. 1981. Calorimetric evaluation of the existence of separate domains in bovine prothrombin. Biochemistry. 20: 15–21. [DOI] [PubMed] [Google Scholar]
- 150.Esmon C. T., and Jackson C. M.. 1974. The conversion of prothrombin to thrombin. IV. The function of the fragment 2 region during activation in the presence of factor V. J. Biol. Chem. 249: 7791–7797. [PubMed] [Google Scholar]
- 151.Lerch P. G., Rickli E. E., Lergier W., and Gillessen D.. 1980. Localization of individual lysine-binding regions in human plasminogen and investigations on their complex-forming properties. Eur. J. Biochem. 107: 7–13. [DOI] [PubMed] [Google Scholar]
- 152.Váradi A., and Patthy L.. 1981. Kringle 5 of human plasminogen carries a benzamidine-binding site. Biochem. Biophys. Res. Commun. 103: 97–102. [DOI] [PubMed] [Google Scholar]
- 153.Patthy L., Trexler M., Vali Z., Banyai L., and Varadi A.. 1984. Kringles: modules specialized for protein binding. Homology of the gelatin-binding region of fibronectin with the kringle structures of proteases. FEBS Lett. 171: 131–136. [DOI] [PubMed] [Google Scholar]
- 154.Köchl S., Fresser F., Lobentanz E., Baier G., and Utermann G.. 1997. Novel interaction of apolipoprotein(a) with beta-2 glycoprotein I mediated by the kringle IV domain. Blood. 90: 1482–1489. [PubMed] [Google Scholar]
- 155.Enkhmaa B., Abbuthalha A., Anuurad E., Zhang W., Tarantal A. F., and Berglund L.. 2015. Rhesus monkey (Macaca mulatta) lipoprotein(a) and apolipoprotein(a): high frequency of small size apolipoprotein(a) isoforms. J. Med. Primatol. 44: 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Puckey L. H., Lawn R. M., and Knight B. L.. 1997. Polymorphisms in the apolipoprotein(a) gene and their relationship to allele size and plasma lipoprotein(a) concentration. Hum. Mol. Genet. 6: 1099–1107. [DOI] [PubMed] [Google Scholar]
- 157.Clarke R., Peden J. F., Hopewell J. C., Kyriakou T., Goel A., Heath S. C., Parish S., Barlera S., Franzosi M. G., Rust S., et al. 2009. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361: 2518–2528. [DOI] [PubMed] [Google Scholar]
- 158.Lackner C., Cohen J. C., and Hobbs H. H.. 1993. Molecular definition of the extreme size polymorphism in apolipoprotein(a). Hum. Mol. Genet. 2: 933–940. [DOI] [PubMed] [Google Scholar]
- 159.Kamstrup P. R., Benn M., Tybaerg-Hansen A., and Nordestgaard B. G.. 2008. Extreme lipoprotein(a) levels and risk of myocardial infarction in the general population: the Copenhagen City Heart Study. Circulation. 117: 176–184. [DOI] [PubMed] [Google Scholar]
- 160.Berglund L., and Anuurad E.. 2008. Role of lipoprotein(a) in cardiovascular disease current and future perspectives. J. Am. Coll. Cardiol. 52: 132–134. [DOI] [PubMed] [Google Scholar]
- 161.Bennet A., Di A. E., Erqou S., Eiriksdottir G., Sigurdsson G., Woodward M., Rumley A., Lowe G. D., Danesh J., and Gudnason V.. 2008. Lipoprotein(a) levels and risk of future coronary heart disease: large-scale prospective data. Arch. Intern. Med. 168: 598–608. [DOI] [PubMed] [Google Scholar]
- 162.Danesh J., Collins R., and Peto R.. 2000. Lipoprotein(a) and coronary heart disease: meta-analysis of prospective studies. Circulation. 102: 1082–1085. [DOI] [PubMed] [Google Scholar]
- 163.Sandholzer C., Saha N., Kark J. D., Rees A., Jaross W., Dieplinger H., Hoppichler F., Boerwinkle E., and Utermann G.. 1992. Apo(a) isoforms predict risk for coronary heart disease: a study in six populations. Arterioscler. Thromb. 12: 1214–1226. [DOI] [PubMed] [Google Scholar]
- 164.Erqou S., Thompson A., Di A. E., Saleheen D., Kaptoge S., Marcovina S., and Danesh J.. 2010. Apolipoprotein(a) isoforms and the risk of vascular disease: systematic review of 40 studies involving 58,000 participants. J. Am. Coll. Cardiol. 55: 2160–2167. [DOI] [PubMed] [Google Scholar]
- 165.Kraft H. G., Lingenhel A., Köchl S., Hoppichler F., Kronenberg F., Abe A., Mühlberger V., Schönitzer D., and Utermann G.. 1996. Apolipoprotein(a) kringle IV repeat number predicts risk for coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 16: 713–719. [DOI] [PubMed] [Google Scholar]
- 166.Wild S. H., Fortmann S. P., and Marcovina S. M.. 1997. A prospective case-control study of lipoprotein(a) levels and apo(a) size and risk of coronary heart disease in Stanford Five-City Project participants. Arterioscler. Thromb. Vasc. Biol. 17: 239–245. [DOI] [PubMed] [Google Scholar]
- 167.Kronenberg F., Kronenberg M. F., Kiechl S., Trenkwalder E., Santer P., Oberhollenzer F., Egger G., Utermann G., and Willeit J.. 1999. Role of lipoprotein(a) and apolipoprotein(a) phenotype in atherogenesis: prospective results from the Bruneck study. Circulation. 100: 1154–1160. [DOI] [PubMed] [Google Scholar]
- 168.White A. L., Guerra B., and Lanford R. E.. 1997. Influence of allelic variation on apolipoprotein(a) folding in the endoplasmic reticulum. J. Biol. Chem. 272: 5048–5055. [DOI] [PubMed] [Google Scholar]
- 169.Lobentanz E-M., Krasznai K., Gruber A., Brunner C., Müller H-J., Sattler J., Kraft H-G., Utermann G., and Dieplinger H.. 1998. Intracellular metabolism of human apolipoprotein(a) in stably transfected Hep G2 cells. Biochemistry. 37: 5417–5425. [DOI] [PubMed] [Google Scholar]
- 170.Marcovina S. M., Albers J. J., Jacobs D. R. Jr., Perkins L. L., Lewis C. E., Howard B. V., and Savage P.. 1993. Lipoprotein[a] concentrations and apolipoprotein[a] phenotypes in Caucasians and African Americans: The CARDIA Study. Arterioscler. Thromb. 13: 1037–1045. [DOI] [PubMed] [Google Scholar]
- 171.Ali S., Bunker C. H., Aston C. E., Ukoli F. A., and Kamboh M. I.. 1998. Apolipoprotein A kringle 4 polymorphism and serum lipoprotein (a) concentrations in African blacks. Hum. Biol. 70: 477–490. [PubMed] [Google Scholar]
- 172.Helmhold M., Bigge J., Muche R., Mainoo J., Thiery J., Seidel D., and Armstrong V. W.. 1991. Contribution of the apo[a] phenotype to plasma Lp[a] concentrations shows considerable ethnic variation. J. Lipid Res. 32: 1919–1928. [PubMed] [Google Scholar]
- 173.Geethanjali F. S., Luthra K., Lingenhel A., Kanagasaba-Pathy A. S., Jacob J., Srivastava L. M., Vasisht S., Kraft H. G., and Utermann G.. 2003. Analysis of the apo(a) size polymorphism in Asian Indian populations: association with Lp(a) concentration and coronary heart disease. Atherosclerosis. 169: 121–130. [DOI] [PubMed] [Google Scholar]
- 174.Marcovina S. M., Albers J. J., Wijsman E., Zhang Z. H., Chapman N. H., and Kennedy H.. 1996. Differences in Lp[a] concentrations and apo[a] polymorphs between black and white Americans. J. Lipid Res. 37: 2569–2585. [PubMed] [Google Scholar]
- 175.Mooser V., Mancini F. P., Bopp S., Pethö-Schramm A., Guerra R., Boerwinkle E., Müller H-J., and Hobbs H. H.. 1995. Sequence polymorphisms in the apo(a) gene associated with specific levels of Lp(a) in plasma. Hum. Mol. Genet. 4: 173–181. [DOI] [PubMed] [Google Scholar]
- 176.Kronenberg F., Kuen E., Ritz E., Junker R., König P., Kraatz G., Lhotta K., Mann J. F. E., Müller G. A., Neyer U., et al. 2000. Lipoprotein(a) serum concentrations and apolipoprotein(a) phenotypes in mild and moderate renal failure. J. Am. Soc. Nephrol. 11: 105–115. [DOI] [PubMed] [Google Scholar]
- 177.Holmer S. R., Hengstenberg C., Kraft H. G., Mayer B., Poll M., Kurzinger S., Fischer M., Lowel H., Klein G., Riegger G. A., et al. 2003. Association of polymorphisms of the apolipoprotein(a) gene with lipoprotein(a) levels and myocardial infarction. Circulation. 107: 696–701. [DOI] [PubMed] [Google Scholar]
- 178.Chretien J. P., Coresh J., Berthier-Schaad Y., Kao W. H., Fink N. E., Klag M. J., Marcovina S. M., Giaculli F., and Smith M. W.. 2006. Three single-nucleotide polymorphisms in LPA account for most of the increase in lipoprotein(a) level elevation in African Americans compared with European Americans. J. Med. Genet. 43: 917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ogorelkova M., Kraft H. G., Ehnholm C., and Utermann G.. 2001. Single nucleotide polymorphisms in exons of the apo(a) kringles IV types 6 to 10 domain affect Lp(a) plasma concentrations and have different patterns in Africans and Caucasians. Hum. Mol. Genet. 10: 815–824. [DOI] [PubMed] [Google Scholar]
- 180.Deo R. C., Wilson J. G., Xing C., Lawson K., Kao W. H., Reich D., Tandon A., Akylbekova E., Patterson N., Mosley T. H. Jr., et al. 2011. Single-nucleotide polymorphisms in LPA explain most of the ancestry-specific variation in Lp(a) levels in African Americans. PLoS One. 6: e14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Trommsdorff M., Köchl S., Lingenhel A., Kronenberg F., Delport R., Vermaak H., Lemming L., Klausen I. C., Faergeman O., Utermann G., et al. 1995. A pentanucleotide repeat polymorphism in the 5′ control region of the apolipoprotein(a) gene is associated with lipoprotein(a) plasma concentrations in Caucasians. J. Clin. Invest. 96: 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Amemiya H., Arinami T., Kikuchi S., Yamakawa-Kobayashi K., Li L., Fujiwara H., Hiroe M., Marumo F., and Hamaguchi H.. 1996. Apolipoprotein(a) size and pentanucleotide repeat polymorphisms are associated with the degree of atherosclerosis in coronary heart disease. Atherosclerosis. 123: 181–191. [DOI] [PubMed] [Google Scholar]
- 183.Suzuki K., Kuriyama M., Saito T., and Ichinose A.. 1997. Plasma lipoprotein(a) levels and expression of the apolipoprotein(a) gene are dependent on the nucleotide polymorphisms in its 5′-flanking region. J. Clin. Invest. 99: 1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mancini F. P., Mooser V., Guerra R., and Hobbs H. H.. 1995. Sequence microheterogeneity in apolipoprotein(a) gene repeats and the relationship to plasma Lp(a) levels. Hum. Mol. Genet. 4: 1535–1542. [DOI] [PubMed] [Google Scholar]
- 185.Bopp S., Kochl S., Acquati F., Magnaghi P., Petho-Schramm A., Kraft H. G., Utermann G., Muller H. J., and Taramelli R.. 1995. Ten allelic apolipoprotein[a] 5′ flanking fragments exhibit comparable promoter activities in HepG2 cells. J. Lipid Res. 36: 1721–1728. [PubMed] [Google Scholar]
- 186.Ichinose A. 1995. Characterization of the apolipoprotein(a) gene. Biochem. Biophys. Res. Commun. 209: 365–371. [DOI] [PubMed] [Google Scholar]
- 187.Ichinose A., and Kuriyama M.. 1995. Detection of polymorphisms in the 5′-flanking region of the gene for apolipoprotein(a). Biochem. Biophys. Res. Commun. 209: 372–378. [DOI] [PubMed] [Google Scholar]
- 188.Zysow B. R., Lindahl G. E., Wade D. P., Knight B. L., and Lawn R. M.. 1995. C/T polymorphism in the 5′ untranslated region of the apolipoprotein(a) gene introduces an upstream ATG and reduces in vitro translation. Arterioscler. Thromb. Vasc. Biol. 15: 58–64. [DOI] [PubMed] [Google Scholar]
- 189.Kraft H. G., Windegger M., Menzel H. J., and Utermann G.. 1998. Significant impact of the +93 C/T polymorphism in the apolipoprotein(a) gene on Lp(a) concentrations in Africans but not in Caucasians: confounding effect of linkage disequilibrium. Hum. Mol. Genet. 7: 257–264. [DOI] [PubMed] [Google Scholar]
- 190.Prins J., Leus F. R., Bouma B. N., and van Rijn H. J.. 1999. The identification of polymorphisms in the coding region of the apolipoprotein (a) gene–association with earlier identified polymorphic sites and influence on the lipoprotein (a) concentration. Thromb. Haemost. 82: 1709–1717. [PubMed] [Google Scholar]
- 191.Rainwater D. L., Kammerer C. M., VandeBerg J. L., and Hixson J. E.. 1997. Characterization of the genetic elements controlling lipoprotein(a) concentrations in Mexican Americans. Evidence for at least three controlling elements linked to LPA, the locus encoding apolipoprotein(a). Atherosclerosis. 128: 223–233. [DOI] [PubMed] [Google Scholar]
- 192.Wade D. P., Puckey L. H., Knight B. L., Acquati F., Mihalich A., and Taramelli R.. 1997. Characterization of multiple enhancer regions upstream of the apolipoprotein(a) gene. J. Biol. Chem. 272: 30387–30399. [DOI] [PubMed] [Google Scholar]
- 193.Puckey L. H., and Knight B. L.. 2003. Sequence and functional changes in a putative enhancer region upstream of the apolipoprotein(a) gene. Atherosclerosis. 166: 119–127. [DOI] [PubMed] [Google Scholar]
- 194.van der Hoek Y. Y., Wittekoek M. E., Beisiegel U., Kastelein J. J. P., and Koschinsky M. L.. 1993. The apolipoprotein(a) kringle IV repeats which differ from the major repeat kringle are present in variably-sized isoforms. Hum. Mol. Genet. 2: 361–366. [DOI] [PubMed] [Google Scholar]
- 195.Kraft H. G., Haibach C., Lingenhel A., Brunner C., Trommsdorff M., Kronenberg F., Müller H. J., and Utermann G.. 1995. Sequence polymorphism in kringle IV 37 in linkage disequilibrium with the apolipoprotein (a) size polymorphism. Hum. Genet. 95: 275–282. [DOI] [PubMed] [Google Scholar]
- 196.Luke M. M., Kane J. P., Liu D. M., Rowland C. M., Shiffman D., Cassano J., Catanese J. J., Pullinger C. R., Leong D. U., Arellano A. R., et al. 2007. A polymorphism in the protease-like domain of apolipoprotein(a) is associated with severe coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 27: 2030–2036. [DOI] [PubMed] [Google Scholar]
- 197.Lanktree M. B., Anand S. S., Yusuf S., and Hegele R. A.. 2010. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ Cardiovasc Genet. 3: 39–46. [DOI] [PubMed] [Google Scholar]
- 198.Lu W., Cheng Y. C., Chen K., Wang H., Gerhard G. S., Still C. D., Chu X., Yang R., Parihar A., O’Connell J. R., et al. 2015. Evidence for several independent genetic variants affecting lipoprotein (a) cholesterol levels. Hum. Mol. Genet. 24: 2390–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., Korbel J. O., Marchini J. L., McCarthy S., McVean G. A., and Abecasis G. R.. 2015. A global reference for human genetic variation. Nature. 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Noureen A., Fresser F., Utermann G., and Schmidt K.. 2015. Sequence variation within the KIV-2 copy number polymorphism of the human LPA gene in African, Asian, and European populations. PLoS One. 10: e0121582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Khalifa M., Noureen A., Ertelthalner K., Bandegi A. R., Delport R., Firdaus W. J., Geethanjali F. S., Luthra K., Makemaharn O., Pang R. W., et al. 2015. Lack of association of rs3798220 with small apolipoprotein(a) isoforms and high lipoprotein(a) levels in East and Southeast Asians. Atherosclerosis. 242: 521–528. [DOI] [PubMed] [Google Scholar]
- 202.Kronenberg F. 2014. Genetic determination of lipoprotein(a) and its association with cardiovascular disease. Convenient does not always mean better. J. Intern. Med. 276: 243–247. [DOI] [PubMed] [Google Scholar]
- 203.Dahlen G. H., Guyton J. R., Attar M., Farmer J. A., Kautz J. A., and Gotto A. M. Jr. 1986. Association of levels of lipoprotein Lp(a), plasma lipids, and other lipoproteins with coronary artery disease documented by angiography. Circulation. 74: 758–765. [DOI] [PubMed] [Google Scholar]
- 204.Kostner G. M., Avogaro P., Cazzolato G., Marth E., Bittolo-Bon G., and Qunici G. B.. 1981. Lipoprotein Lp(a) and the risk for myocardial infarction. Atherosclerosis. 38: 51–61. [DOI] [PubMed] [Google Scholar]
- 205.Orth-Gomér K., Mittleman M. A., Schenck-Gustafsson K., Wamala S. P., Eriksson M., Belkic K., Kirkeeide R., Svane B., and Rydén L.. 1997. Lipoprotein(a) as a determinant of coronary heart disease in young women. Circulation. 95: 329–334. [DOI] [PubMed] [Google Scholar]
- 206.Rhoads G. G., Dahlen G., Berg K., Morton N. E., and Dannenberg A. L.. 1986. Lp(a) lipoprotein as a risk factor for myocardial infarction. JAMA. 256: 2540–2544. [PubMed] [Google Scholar]
- 207.Ridker P. M., Hennekens C. H., and Stampfer M. J.. 1993. A prospective study of lipoprotein(a) and the risk of myocardial infarction. JAMA. 270: 2195–2199. [PubMed] [Google Scholar]
- 208.Rifai N., Ma J., Sacks F. M., Ridker P. M., Hernandez W. J., Stampfer M. J., and Marcovina S. M.. 2004. Apolipoprotein(a) size and lipoprotein(a) concentration and future risk of angina pectoris with evidence of severe coronary atherosclerosis in men: The Physicians’ Health Study. Clin. Chem. 50: 1364–1371. [DOI] [PubMed] [Google Scholar]
- 209.Erqou S., Kaptoge S., Perry P. L., Di A. E., Thompson A., White I. R., Marcovina S. M., Collins R., Thompson S. G., and Danesh J.. 2009. Lipoprotein(a) concentration and the risk of coronary heart disease, stroke, and nonvascular mortality. JAMA. 302: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sandholzer C., Boerwinkle E., Saha N., Tong M. C., and Utermann G.. 1992. Apolipoprotein(a) phenotypes, Lp(a) concentration and plasma lipid levels in relation to coronary heart disease in a Chinese population: Evidence for the role of the apo(a) gene in coronary heart disease. J. Clin. Invest. 89: 1040–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kronenberg F., Neyer U., Lhotta K., Trenkwalder E., Auinger M., Pribasnig A., Meisl T., König P., and Dieplinger H.. 1999. The low molecular weight apo(a) phenotype is an independent predictor for coronary artery disease in hemodialysis patients: a prospective follow-up. J. Am. Soc. Nephrol. 10: 1027–1036. [DOI] [PubMed] [Google Scholar]
- 212.Laschkolnig A., Kollerits B., Lamina C., Meisinger C., Rantner B., Stadler M., Peters A., Koenig W., Stöckl A., Dähnhardt D., et al. 2014. Lipoprotein(a) concentrations, apolipoprotein(a) phenotypes and peripheral arterial disease in three independent cohorts. Cardiovasc. Res. 103: 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kronenberg F. 2016. Human genetics and the causal role of lipoprotein(a) for various diseases. Cardiovasc. Drugs Ther. 30: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zacho J., Tybjaerg-Hansen A., Jensen J. S., Grande P., Sillesen H., and Nordestgaard B. G.. 2008. Genetically elevated C-reactive protein and ischemic vascular disease. N. Engl. J. Med. 359: 1897–1908. [DOI] [PubMed] [Google Scholar]
- 215.Wensley F., Gao P., Burgess S., Kaptoge S., Di A. E., Shah T., Engert J. C., Clarke R., Davey-Smith G., Nordestgaard B. G., et al. 2011. Association between C reactive protein and coronary heart disease: mendelian randomisation analysis based on individual participant data. BMJ. 342: d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Katan M. B. 1986. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1: 507–508. [DOI] [PubMed] [Google Scholar]
- 217.Li Y., Luke M. M., Shiffman D., and Devlin J. J.. 2011. Genetic variants in the apolipoprotein(a) gene and coronary heart disease. Circ Cardiovasc Genet. 4: 565–573. [DOI] [PubMed] [Google Scholar]
- 218.Trégouët D. A., Konig I. R., Erdmann J., Munteanu A., Braund P. S., Hall A. S., Grosshennig A., Linsel-Nitschke P., Perret C., DeSuremain M., et al. 2009. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat. Genet. 41: 283–285. [DOI] [PubMed] [Google Scholar]
- 219.Shiffman D., Louie J. Z., Rowland C. M., Malloy M. J., Kane J. P., and Devlin J. J.. 2010. Single variants can explain the association between coronary heart disease and haplotypes in the apolipoprotein(a) locus. Atherosclerosis. 212: 193–196. [DOI] [PubMed] [Google Scholar]
- 220.Koch W., Mueller J. C., Schrempf M., Wolferstetter H., Kirchhofer J., Schomig A., and Kastrati A.. 2013. Two rare variants explain association with acute myocardial infarction in an extended genomic region including the apolipoprotein(A) gene. Ann. Hum. Genet. 77: 47–55. [DOI] [PubMed] [Google Scholar]
- 221.Kyriakou T., Seedorf U., Goel A., Hopewell J. C., Clarke R., Watkins H., and Farrall M.. 2014. A common LPA null allele associates with lower lipoprotein(a) levels and coronary artery disease risk. Arterioscler. Thromb. Vasc. Biol. 34: 2095–2099. [DOI] [PubMed] [Google Scholar]
- 222.Paultre F., Pearson T. A., Weil H. F., Tuck C. H., Myerson M., Rubin J., Francis C. K., Marx H. F., Philbin E. F., Reed R. G., et al. 2000. High levels of Lp(a) with a small apo(a) isoform are associated with coronary artery disease in African American and White men. Arterioscler. Thromb. Vasc. Biol. 20: 2619–2624. [DOI] [PubMed] [Google Scholar]
- 223.Moliterno D. J., Jokinen E. V., Miserez A. R., Lange R. A., Willard J. E., Boerwinkle E., Hillis L. D., and Hobbs H. H.. 1995. No association between plasma lipoprotein(a) concentrations and the presence or absence of coronary atherosclerosis in African-Americans. Arterioscler. Thromb. Vasc. Biol. 15: 850–855. [DOI] [PubMed] [Google Scholar]
- 224.Guerra R., Yu Z., Marcovina S., Peshock R., Cohen J. C., and Hobbs H. H.. 2005. Lipoprotein(a) and apolipoprotein(a) isoforms. No association with coronary artery calcification in The Dallas Heart Study. Circulation. 111: 1471–1479. [DOI] [PubMed] [Google Scholar]
- 225.Mersha T. B., and Abebe T.. 2015. Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Hum. Genomics. 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Reed F. A., and Tishkoff S. A.. 2006. African human diversity, origins and migrations. Curr. Opin. Genet. Dev. 16: 597–605. [DOI] [PubMed] [Google Scholar]
- 227.Shiffman D., Slawsky K., Fusfeld L., Devlin J. J., and Goss T. F.. 2012. Cost-effectiveness model of use of genetic testing as an aid in assessing the likely benefit of aspirin therapy for primary prevention of cardiovascular disease. Clin. Ther. 34: 1387–1394. [DOI] [PubMed] [Google Scholar]
- 228.Takeuchi F., Yokota M., Yamamoto K., Nakashima E., Katsuya T., Asano H., Isono M., Nabika T., Sugiyama T., Fujioka A., et al. 2012. Genome-wide association study of coronary artery disease in the Japanese. Eur. J. Hum. Genet. 20: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Rowland C. M., Pullinger C. R., Luke M. M., Shiffman D., Green L., Movsesyan I., Devlin J. J., Malloy M. J., Kane J. P., and Undas A.. 2014. Lipoprotein (a), LPA Ile4399Met, and fibrin clot properties. Thromb. Res. 133: 863–867. [DOI] [PubMed] [Google Scholar]
- 230.Li Z. G., Li G., Zhou Y. L., Chen Z. J., Yang J. Q., Zhang Y., Sun S., and Zhong S. L.. 2013. Lack of association between lipoprotein(a) genetic variants and subsequent cardiovascular events in Chinese Han patients with coronary artery disease after percutaneous coronary intervention. Lipids Health Dis. 12: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.van der Hoek Y. Y., Lingenhel A., Kraft H. G., Defesche J. C., Kastelein J. J. P., and Utermann G.. 1997. Sib-pair analysis detects elevated Lp(a) levels and large variation of Lp(a) concentration in subjects with familiar defective ApoB. J. Clin. Invest. 99: 2269–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sandholzer C., Feussner G., Brunzell J., and Utermann G.. 1992. Distribution of apolipoprotein(a) in the plasma from patients with lipoprotein lipase deficiency with type III hyperlipoproteinemia: no evidence for a triglyceride-rich precursor of lipoprotein(a). J. Clin. Invest. 90: 1958–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Menzel H-J., Dieplinger H., Lackner C., Hoppichler F., Lloyd J. K., Muller D. R., Labeur C., Talmud P. J., and Utermann G.. 1990. Abetalipoproteinemia with an apoB-100-lipoprotein(a) glycoprotein complex in plasma: indication for an assembly defect. J. Biol. Chem. 265: 981–986. [PubMed] [Google Scholar]
- 234.Li J., Lange L. A., Sabourin J., Duan Q., Valdar W., Willis M. S., Li Y., Wilson J. G., and Lange E. M.. 2015. Genome- and exome-wide association study of serum lipoprotein (a) in the Jackson Heart Study. J. Hum. Genet. 60: 755–761. [DOI] [PubMed] [Google Scholar]
- 235.Thanassoulis G., Campbell C. Y., Owens D. S., Smith J. G., Smith A. V., Peloso G. M., Kerr K. F., Pechlivanis S., Budoff M. J., Harris T. B., et al. 2013. Genetic associations with valvular calcification and aortic stenosis. N. Engl. J. Med. 368: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kamstrup P. R., Tybjaerg-Hansen A., and Nordestgaard B. G.. 2014. Elevated lipoprotein(a) and risk of aortic valve stenosis in the general population. J. Am. Coll. Cardiol. 63: 470–477. [DOI] [PubMed] [Google Scholar]
- 237.Arsenault B. J., Boekholdt S. M., Dube M. P., Rheaume E., Wareham N. J., Khaw K. T., Sandhu M. S., and Tardif J. C.. 2014. Lipoprotein(a) levels, genotype, and incident aortic valve stenosis: a prospective mendelian randomization study and replication in a case-control cohort. Circ Cardiovasc Genet. 7: 304–310. [DOI] [PubMed] [Google Scholar]
- 238.Capoulade R., Chan K. L., Yeang C., Mathieu P., Bosse Y., Dumesnil J. G., Tam J. W., Teo K. K., Mahmut A., Yang X., et al. 2015. Oxidized phospholipids, lipoprotein(a), and progression of calcific aortic valve stenosis. J. Am. Coll. Cardiol. 66: 1236–1246. [DOI] [PubMed] [Google Scholar]
- 239.Kamstrup P. R., and Nordestgaard B. G.. 2016. Elevated lipoprotein(a) levels, LPA risk genotypes, and increased risk of heart failure in the general population. JACC Heart Fail. 4: 78–87. [DOI] [PubMed] [Google Scholar]
- 240.Young G., Albisetti M., Bonduel M., Brandao L., Chan A., Friedrichs F., Goldenberg N. A., Grabowski E., Heller C., Journeycake J., et al. 2008. Impact of inherited thrombophilia on venous thromboembolism in children: a systematic review and meta-analysis of observational studies. Circulation. 118: 1373–1382. [DOI] [PubMed] [Google Scholar]
- 241.Lamina C., and Kronenberg F.. 2013. The mysterious lipoprotein(a) is still good for a surprise. Lancet Diabetes Endocrinol. 1: 170–172. [DOI] [PubMed] [Google Scholar]