Abstract
Cilia and flagella play important roles in cell motility and cell signaling. These functions require that the cilium establishes and maintains a unique lipid and protein composition. Recent work indicates that a specialized region at the base of the cilium, the transition zone, serves as both a barrier to entry and a gate for passage of select components. For at least some cytosolic proteins, the barrier and gate functions are provided by a ciliary pore complex (CPC) that shares molecular and mechanistic properties with nuclear gating. Specifically, nucleoporins of the CPC limit the diffusional entry of cytosolic proteins in a size-dependent manner and enable the active transport of large molecules and complexes via targeting signals, importins, and the small G protein Ran. For membrane proteins, the septin protein SEPT2 is part of the barrier to entry whereas the gating function is carried out and/or regulated by proteins associated with ciliary diseases (ciliopathies) such as nephronophthisis, Meckel–Gruber syndrome and Joubert syndrome. Here, we discuss the evidence behind these models of ciliary gating as well as the similarities to and differences from nuclear gating.
Keywords: Cilia, Flagella, Primary cilium, Ciliary pore complex, Nuclear pore complex, Nucleoporin, Intraflagellar transport, Transition zone
Introduction
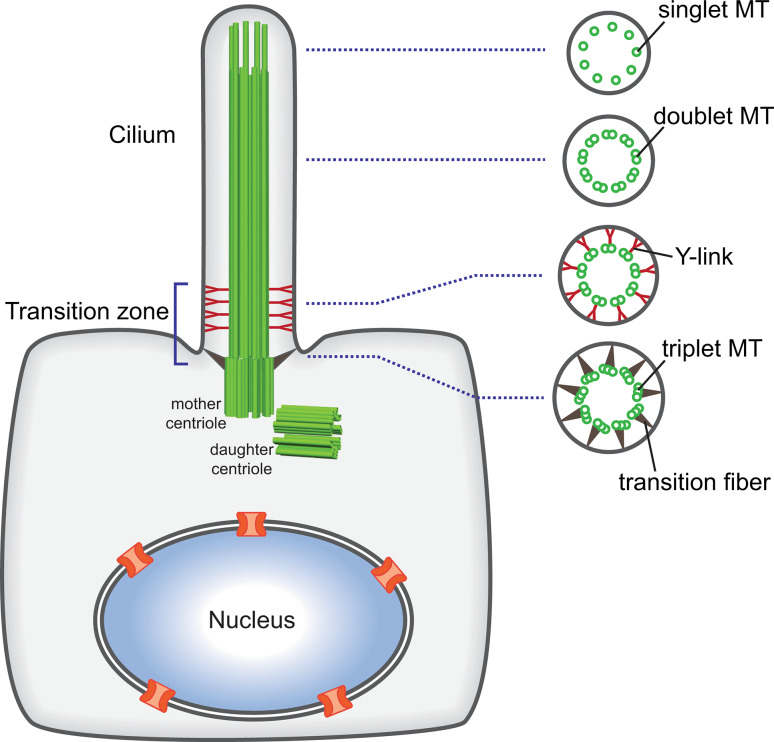
Eukaryotic cilia and flagella have many important functions. While motile cilia on epithelial cells generate external fluid flow and sperm flagella drive cell motility, non-motile cilia termed primary cilia are found on the surface of almost all mammalian cells and function as cellular antennae (reviewed in [1, 2]). The basic structure of eukaryotic cilia and flagella (hereafter both called cilia) is conserved across species and cell types. The microtubule-based skeletal structure, the axoneme, is comprised of an outer ring of 9 doublet microtubules (Fig. 1). Motile cilia usually also contain a central pair of singlet microtubules (the “9 + 2” pattern), while most non-motile cilia lack this central pair (the “9 + 0” pattern). Beyond its structural role, the axoneme serves as the track for an intraflagellar transport (IFT) system driven by kinesin (anterograde) and dynein (retrograde) motors (reviewed in [3, 4]). The 9 doublet microtubules arise from the 9 triplet microtubules of the mother centriole which is attached to the periciliary membrane via the largely uncharacterized transition fibers (reviewed in [5]).
Fig. 1.
General structure of the primary cilium. The mother centriole contains nine triplet microtubules (MT) and is anchored to the periciliary membrane by transition fibers. Nine doublet microtubules protrude from the triplet microtubules and form the axoneme. The proximal end of the cilium is termed the transition zone and is the site for gated entry into the ciliary compartment. Here, Y-shaped structures (the Y-links) connect the doublet microtubules to the ciliary membrane. At the distal end of the cilium in some cells and species, the doublet microtubules convert to singlet microtubules
The ciliary membrane is contiguous with the plasma membrane but has a unique protein and lipid composition; likewise, the composition of soluble proteins in the cilium is also unique (reviewed in [6–8]). Thus, gating mechanisms must exist to ensure that the proper proteins enter the ciliary compartment. Furthermore, the gating must be regulated as some ciliary proteins change their localization in response to extracellular cues; for example, in hedgehog signaling, entry and exit of the transmembrane proteins Patched, Smoothened and GPR161 is controlled by the hedgehog ligand (reviewed in [9]). Mislocalization of ciliary components, including both structural and signaling components, results in defects in ciliary motility and/or signaling pathways and consequentially is associated with a myriad of cilium-related diseases called ciliopathies (reviewed in [10, 11]).
Although IFT is known to be responsible for transport of proteins within the ciliary compartment, it is largely unknown how ciliary proteins are selected for and gain entrance to the organelle. Ciliary entry takes place at a region at the base of the cilium termed the transition zone which can be defined structurally by the presence of electron-dense Y-shaped structures that span the space between the doublet microtubules and the ciliary membrane (Fig. 1, reviewed in [12–14]). The transition zone can also be defined molecularly, as many proteins whose mutations are associated with ciliopathies localize to this region of the cilium. A large body of work supports the idea that ciliopathy-associated gene products at the transition zone regulate the gated entry of ciliary proteins (reviewed in [15–17]). Gated entry of ciliary proteins has also been suggested to utilize, at least in part, the same molecules and mechanisms as gated entry into the nuclear compartment (reviewed in [18]). Specifically, it has been hypothesized that nucleoporins (NUPs), the building blocks of the nuclear pore complex (NPC), form a ciliary pore complex (CPC) at the transition zone that is responsible for the gated entry of cytosolic proteins. Here, we discuss recent work in uncovering the mechanisms of ciliary gating as well as the similarities to and differences from nuclear gating.
Diffusional entry of cytosolic proteins
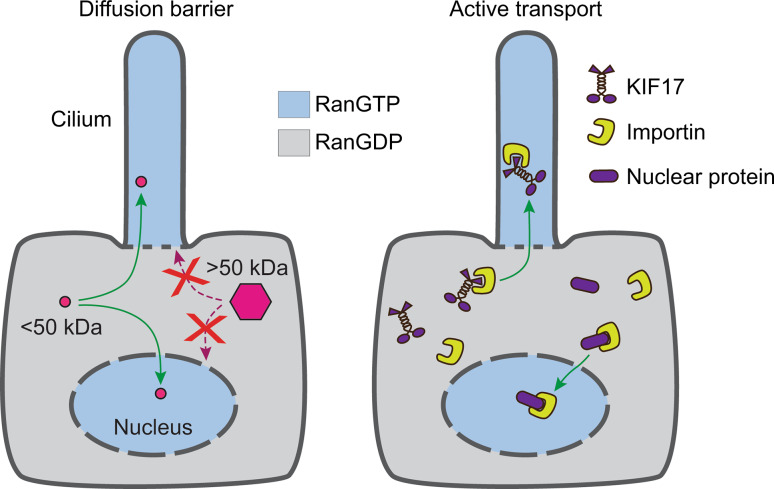
Unlike organelles such as mitochondria or the endoplasmic reticulum, the ciliary compartment is not isolated from the cytoplasm by a membrane barrier and yet the diffusional entry of soluble molecules is restricted at the base of the cilium (Fig. 2). In cultured mammalian cells, a size-dependent permeability barrier at the base of the primary cilium was shown to regulate the entry of soluble molecules [19–21]. Two studies found that restricted entry occurs for proteins in the 40–70 kDa range [19, 20], whereas a third study found that even larger proteins could enter, albeit with reduced kinetics [21]. Differences between these studies in the size cutoff of the barrier are likely due to different experimental methods as well as the fact that diffusional entry is likely governed by factors in addition to protein size, such as protein shape and surface charge distribution. Mathematical modeling of the kinetics of diffusional entry suggests that the ciliary barrier functions as a molecular sieve [21]. This is reminiscent of restricted entry into the nuclear compartment where NPCs act as sieve-like barriers, freely permeable to small molecules less than ~50 kDa and able to selectively import or export larger molecules in an energy-dependent manner (Fig. 2 and reviewed in [22]).
Fig. 2.
Gated entry of cytosolic molecules into the ciliary and nuclear compartments. Entry of cytosolic molecules is characterized by a size-exclusion barrier (left) and active transport across the barrier (right). The size-exclusion barrier limits the diffusional entry of soluble molecules larger than ~50 kDa. Large molecules gain entry via the active transport process in which importins bind to nuclear or ciliary proteins and mediate their transport across the barriers. The directionality of transport is specified by a RanGTP–RanGDP gradient across the ciliary-cytoplasmic and nuclear-cytoplasmic barriers
Diffusional entry into the ciliary compartment has also been investigated in photoreceptor cells. Again, a size-dependent distribution of proteins was found, with GFP multimers present in the outer segment (distal cilium) at lower abundance than single GFP molecules [23, 24]. GFP multimers are able to freely cross the diffusion barrier in mammalian cells [18] and their differential distribution in the outer segment can be explained by steric volume exclusion in which the membranous discs in the outer segment impose spatial restrictions on larger soluble molecules [24]. It may thus be that different types of cilia use different physical mechanisms to restrict localization of soluble proteins in the compartment.
Gated entry of cytosolic proteins
Ciliary formation and function require the import of a number of large multimeric cytosolic protein complexes including IFT particles and radial spoke proteins (RSPs) [25, 26]. Thus, mechanisms must exist to facilitate the transport of large soluble proteins across the permeability barrier in a selective manner. Recent work has uncovered a pathway that functions in the gated entry of at least several ciliary proteins [27, 28]. This pathway is again reminiscent of nuclear gating in which active transport of large proteins across the NPC utilizes nuclear localization sequences (NLS) that are recognized by transport receptors (importins, also called karyopherins) and shuttled across the NPC where the small G protein Ran in its GTP-bound form (RanGTP) promotes release of the transported proteins inside the nuclear compartment (Fig. 2). Thus, active maintenance of a Ran gradient across the nuclear envelope (RanGTP in the nucleoplasm and RanGDP in the cytoplasm) drives the directionality of transport (reviewed in [29, 30]).
For ciliary gating, a ciliary localization sequence (CLS) that is homologous to an NLS was identified for an IFT component, the kinesin-2 motor KIF17 [27], and a peripheral membrane protein, retinitis pigmentosa 2 (RP2) [28]. Both of these CLS motifs are recognized by importin-β2 (transportin-1) for transport across the ciliary barrier. Furthermore, the directionality of transport is driven by a RanGTP/GDP gradient as the ciliary compartment is enriched with RanGTP [27, 31] (Fig. 2). Disrupting the ciliary-cytoplasmic RanGTP/GDP gradient by increasing the cytosolic levels of RanGTP blocks ciliary import of KIF17 [27, 31]. These lines of evidence support the idea that active transport systems across the NPC and CPC utilize similar mechanisms.
It is not clear whether other cytosolic proteins contain a CLS-like sequence that confers ciliary localization and/or whether they gain entry by “piggy-backing” on KIF17 and associated IFT particles. It is important to note that not all sequences that look like a potential NLS (several lysine and/or arginine residues) function as NLS or CLS motifs. Indeed, KIF17 contains two highly similar basic patches but only one functions as a CLS [27]. In addition to further defining CLS and ciliary entry mechanisms, it will be interesting to determine whether ciliary export of cytosolic molecules functions in a manner similar to nuclear export. Two recent pieces of data suggest this may be the case. First, deletion of several predicted nuclear export sequences (NES) or treatment of cells with the exportin-1 (CRM1) inhibitor leptomycin B resulted in increased ciliary localization of the Hedgehog transcription factor Gli2 [32]. Second, phosphorylation of an NES in the N-terminus of huntington was shown to regulate ciliary-cytoplasmic and nuclear-cytoplasmic localization [33].
Thus, both nuclei and cilia utilize a barrier that restricts the diffusional entry of small cytosolic molecules and a gate that facilitates the entry of select larger molecules. It is still unclear how CLS and NLS signals are distinguished to target proteins to different organelles. One possibility is that cilium-specific targeting factors function in concert with the CLS. For example, truncated forms of KIF17 that contain the CLS but lack the kinesin motor domain localize to the nucleus [27]. Alternatively, similar yet distinct NLS and CLS signals may be present in the same molecule and regulate nuclear and ciliary localization, respectively, as shown for the Gli2 transcription factor [32].
Nucleoporins as components of the ciliary pore complex (CPC)
That the nuclear and ciliary barriers to cytosolic protein entry display similar physical properties (size-exclusion barrier and selective gate) suggests that there could be molecular similarities between the barriers. Indeed, a number of nucleoporins (NUPs), the proteins that form the barrier in the NPC, have been localized to the base of the cilium either by overexpression of fluorescently tagged proteins or immunostaining of endogenous proteins in mammalian cells [20]. That NUPs localize outside of the NPC and have functions beyond nuclear-cytoplasmic transport is not surprising as NUPs have been suggested to regulate gene expression and cell differentiation during interphase as well as centrosome integrity and spindle formation during mitosis (discussed further in [34–37]). However, another study failed to identify NUPs at the base of primary cilia [19], perhaps due to experimental differences. And although NUPs, importins, exportins and Ran are found in some ciliary proteomes, NUPs were not identified in the proteome of transition zone structures isolated from Chlamydomonas [38]. While negative results do not exclude the possibility that NUPs are present at the base of the cilium, these findings may reflect variations in the composition of the CPC or suggest the presence of NUP-independent gating mechanisms at the transition zone.
The molecular similarities between NPC and CPC barriers include NUPs that contain up to 50 tandem sequences rich in phenylalanine and glycine residues (FG-repeats). FG-NUPs form a hydrogel-based barrier for nuclear-cytoplasmic transport and likely perform the same function in ciliary-cytoplasmic transport. A notable difference between the NPC and CPC barriers is the absence of transmembrane and nuclear basket NUPs at the base of the cilium in cultured cells [20]. It may thus be that the scaffold NUPs utilize unique interactions for anchoring at the base of the cilium (discussed further below). As the NUP composition of NPCs varies across species and even between tissues and during development in an animal, presumably endowing different NPCs with distinct functions (reviewed in [30, 39, 40]), it will not be surprising to find a unique NUP composition for the CPC.
A functional role for NUPs in regulating entry into the ciliary compartment was first suggested by the fact that global inhibition of NUP function blocked the import of new KIF17 kinesin-2 motors into the ciliary compartment [20]. To specifically test the role of a central FG-NUP in ciliary gating, an inducible homodimerization system was used to rapidly and specifically lock NUP62 in an unproductive conformation [41]. As expected, forced dimerization of NUP62 decreased the selective entry of an NLS-containing protein into nuclei and the kinesin-2 motor KIF17 into primary cilia. Further work showed that forced dimerization of NUP62 also decreased the ciliary entry of a variety of cytosolic proteins of varying sizes and functions, specifically the hedgehog transcription factor Gli2, the centrosomal component Tsga14 (CEP41), a potential transcription factor Gtl3, and the IFT-B component IFT88 [41]. Although this is reminiscent of the known role of central FG-NUPs in regulating the transport of select nuclear proteins across the NPC barrier [42–44], further work is needed to determine the mechanism of NUP-dependent ciliary import as homodimerization may cause a steric rather than mechanistic block to protein import.
Further work is also needed to determine whether and how the entry of axonemal proteins is gated. Blocking NUP62 function did not cause ciliary disassembly [41], suggesting that tubulins and other axonemal components may utilize a non-CPC mechanism to gain entry. However, homodimerization of NUP62 did not completely block the CPC [41] so the block to axonemal protein entry may not have been sufficient to cause axoneme instability. In addition, it is possible that tubulins can enter freely by diffusion as the Stokes radius of a tubulin heterodimer (4.3 nm [45]) is within the range of proteins that can access the ciliary compartment [19, 21]. How the entry of larger axonemal proteins (e.g., radial spoke proteins, dynein arms) is gated has not been explored.
Taken together, the evidence to date suggests that active transport of at least some cytosolic proteins into both the ciliary and nuclear compartments utilizes NLS/CLS motifs, importins, a RanGTP/GDP gradient, and FG-NUPs of a pore complex (Figs. 2, 3). Defects in CPC structure and function may cause ciliopathies as copy number variations in the scaffold nucleoporin NUP188 have been linked to heterotaxy [46].
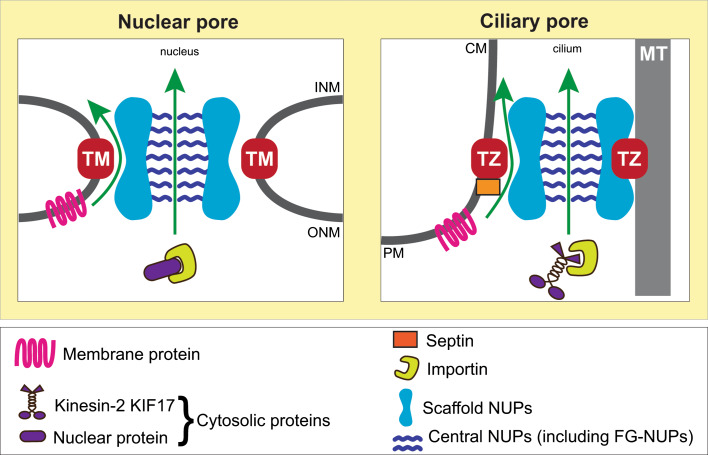
Fig. 3.
Models for nuclear and ciliary gating. Both nuclear and ciliary pores contain a central channel for the gated entry of cytosolic proteins with central pore NUPs (e.g., the FG-NUP NUP62) providing this sieve-like barrier. Gated entry of membrane proteins utilizes peripheral channels of the NPC for nuclear gating, while transition zone (TZ) proteins may form a peripheral channel for gated entry of ciliary membrane proteins. Both nuclear and ciliary pores contain scaffold NUPs (e.g., NUP93, NUP35). For the NPC, the scaffold is anchored by transmembrane (TM) NUPs whereas in the CPC, the scaffold may be anchored by transition zone proteins. TM transmembrane NUP, TZ transition zone protein, MT microtubule
Gated entry of membrane proteins
What mechanisms regulate the membrane protein composition of the ciliary compartment? Here, we find an important difference with how proteins access the nuclear and ciliary compartments. For inner nuclear membrane (INM) proteins, their entry involves lateral diffusion along the NPC membrane and selective retention by nucleoplasmic proteins (the diffusion–retention model, reviewed in [47–49]). Admission of INM proteins does not require central pore NUPs but is dependent on scaffold and transmembrane NUPs [50, 51] that presumably form a peripheral channel within the NPC [52, 53] (Fig. 3). Passage of INM proteins through this channel is sensitive to the size of their cytoplasmic domains [50, 54–58].
For the ciliary compartment, diffusion–retention appears unlikely to be the primary mechanism that regulates membrane protein composition for two reasons. First, imaging analysis in live cells shows that membrane proteins are highly mobile within the ciliary membrane [59–62]. Second, a septin-containing barrier restricts the diffusional passage of proteins between the plasma and ciliary membranes [59, 60]. Septins are known to form a diffusion barrier in budding yeast [63, 64] and presumably form a diffusion barrier around the proximal region of the flagellar membrane in mammalian sperm [65, 66]. Septin 2 (SEPT2) localizes to periciliary membrane in mammalian cells and a partial loss of SEPT2 function enabled the movement of membrane proteins between the plasma and ciliary membranes [59, 60, 67]. However, the role of septins at the ciliary base remains unclear [67] and it is possible that the effects of SEPT2 knockdown on protein mobility are due to indirect effects on the formation of barrier structures at the ciliary base.
The membrane protein composition of the ciliary compartment appears instead to be regulated by a gating mechanism at the transition zone. Gated entry of membrane proteins does not require components of the central channel of the CPC as forced dimerization of NUP62 did not affect the selective transport of membrane proteins into or out of the ciliary compartment, including the peripheral membrane protein retinitis pigmentosa 2 (RP2) and the transmembrane proteins Smoothened, Patched and GPR161 [41]. Rather, gated entry of membrane proteins requires a complex of ciliopathy-associated proteins that localize to the transition zone. Based on genetic and biochemical analysis, the transition zone proteins have been classified into two functional modules, the nephronophthisis (NPHP) and Meckel–Gruber syndrome/Joubert syndrome (MKS/JBTS) modules (reviewed in [12–14]). Components of the NPHP module contain lipid-binding and structural (e.g., coiled-coil) domains and have been implicated in regulating entry of both membrane and cytosolic proteins [68–71]. The MKS/JBTS module includes lipid-binding and transmembrane proteins and disruption of this module results in defects in ciliary membrane protein composition [59, 69–74], suggesting that this module functions to regulate the entry and exit of membrane proteins. Thus, the MKS/JBTS module may form a peripheral channel in the ciliary pore that regulates the entry of membrane proteins and is distinct from the NUP-containing central channel (Fig. 3).
Several other molecules and mechanisms have been suggested to regulate the membrane protein composition of cilia. First, gated entry of membrane proteins likely requires ciliary targeting sequences. Indeed, such signals have been identified in the cytoplasmic domains of several ciliary membrane proteins (reviewed in [7, 16, 75]) although it remains possible that these targeting sequences are primarily required for dynein-dependent transport of Golgi-derived vesicles to the ciliary base [69, 76, 77]. Second, retention in the plasma membrane can prevent the lateral diffusion of membrane proteins to the ciliary membrane [78]. Third, entry across the barrier is also regulated by SUMOylation [79, 80], phosphoinositide composition [81, 82], the Bardet–Biedl syndrome complex (BBSome), Tubby-like proteins (TULPs), and small GTPases of the Rab and Arf families (reviewed in [4, 16]). Indeed, defining the functions of septins, NPHP proteins, MKS/JBTS proteins, the BBSome, TULPS, and small GTPases in regulating the trafficking of proteins into the ciliary compartment remains an outstanding issue for the field.
Taken together, it appears that the passage of cytosolic and membrane proteins is gated in different ways for both the ciliary and nuclear compartments. Both organelles appear to utilize a NUP-containing pore as a barrier and gate for cytosolic protein entry (Fig. 3). Whereas nuclei utilize a diffusion–retention mechanism to regulate membrane protein composition, cilia appear to use septins as a barrier and transition zone proteins as a gate to regulate membrane protein passage (Fig. 3).
Structure of the ciliary gates
One of the most pressing questions in the field is the overall structural organization of the ciliary gate. The identification of NUP, septin, NPHP, and MKS/JBTS modules has provided important information on the molecules at the gate but the localization and organization of these molecules remain largely unclear. As scaffold and central pore NUPs are common between the NPC and CPC, it is tempting to think of these NUPs forming an NPC-like structure at the base of the cilium. However, the organization of these NUPs may differ between these locales. Variation in composition has already been noted across NPCs: central NUPs are shared across NPCs of human cell types, whereas there is significant variation in composition for peripheral NUPs [83]. Furthermore, the transmembrane NUPs are not strongly conserved across species [84–86]. There thus appear to be different ways in which scaffold NUPs can interact with their surrounding environment.
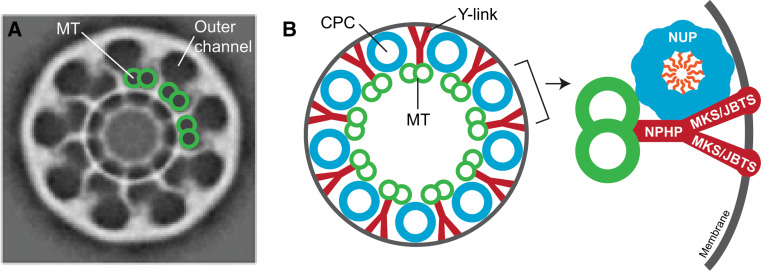
In the NPC, the scaffold NUPs are anchored in the nuclear envelope via protein–protein interactions with transmembrane NUPs (e.g., [87–89] and references therein). For the CPC, the NUPs may be anchored at the base of the cilium via interactions with transition zone proteins (Figs. 3, 4). One attractive possibility is that the NPHP module proteins localize to the Y-links and anchor the NUPs of the central channel. Several lines of evidence support this possibility. First, the CEP290 gene product in Chlamydomonas (homolog of NPHP6) localizes to the Y-links by immunoelectron microscopy and loss of CEP290 function results in defects in microtubule–membrane connections at the transition zone [70]. Second, pull-down of the transition zone proteins NPHP4 and NPHP5 identified several scaffold NUPs present in the protein complexes by mass spectrometry [90]. Third, recent cryo-electron tomography of the terminal plate region at the base of Tetrahymena cilia revealed the presence of 9 pores at the base of the cilium, with each pore located peripheral to a doublet microtubule and between the stems of the Y-links (Fig. 4a, [91]). These pores have a diameter comparable to that of the NPC (~50 nm [92]) and were suggested to be the conduits for passage of cytosolic IFT particles (~38 nm [91]). Thus, the Tetrahymena pores could be the CPCs, although no direct evidence has been reported. Furthermore, pores have not been observed at the ciliary base in other species.
Fig. 4.
Possible structural relationship between the ciliary pore and the transition zone. a Electron cryotomography reveals nine pores (outer channels) in isolated basal bodies from Tetrahymena. Each outer channel is located adjacent to a doublet microtubule (MT) and may be a pore for the entry of ciliary components. Reprinted from Ounjai et al. [91], with permission from Elsevier. b Schematic drawings of the putative arrangement of the CPC and Y-link structures. The CPCs are comprised of scaffold and central NUPs and form a barrier for entry of cytosolic proteins, whereas the Y-links are comprised of NPHP, MKS, and JBTS proteins and form a barrier for entry of membrane proteins. NPHP nephronophthisis, MKS Meckel–Gruber syndrome, JBTS Joubert syndrome, MT microtubule
Conclusions and future directions
Gating mechanisms that determine the protein complement of the cilium are fundamental for ciliary and thus cellular function. Evidence to date suggests a model whereby a NUP-containing central channel (the CPC) functions, like the NPC, as a sieve-like barrier and gate for the passage of cytosolic molecules and an NPHP- and MKS/JBTS-containing peripheral channel functions to regulate the passage of membrane proteins to and from the adjacent plasma membrane (Fig. 4). Further studies are required to verify that these are distinct gates/pathways and to delineate the mechanisms of each pathway. For example, do cytosolic proteins other than KIF17 contain a CLS or do they piggy-back on kinesins and IFT particles for entry through the NUP-containing CPC? Is the entry of core axonemal components, such as tubulin, axonemal dynein and radial spoke proteins, gated and if so, how? How is the entry and exit of membrane proteins regulated, particularly in response to ligand such as Hedgehog?
Although many proteins that localize to the base of the primary cilium have been identified, their specific roles in the structure and/or function of the ciliary gate are largely unknown. For example, do FG-containing NUPs of the CPC function as they do in the NPC? Although forced dimerization of NUP62 blocked active transport of cytosolic proteins into the ciliary compartment [41], the mechanism of inhibition is unclear. The structural organization of the ciliary gate is a particularly pressing problem. Where exactly are the gates located and is there actually a NUP-containing pore like that of the NPC? What are the protein components of the Y-links and are these structural hubs for organizing both the membrane gate at the transition zone and the cytosolic gate at the CPC? When during ciliary assembly are the NUP, NPHP, MKS/JBTS components assembled and when is gating function established? High-resolution imaging methods including electron microscopy and super-resolution microscopy will likely be critical for answering these questions.
Acknowledgments
We thank members of the Verhey lab for helpful discussions. DT is supported by a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science. Work in KJV’s lab is supported by NIH RO1GM070862.
References
- 1.Basten SG, Giles RH. Functional aspects of primary cilia in signaling, cell cycle and tumorigenesis. Cilia. 2013;2:6. doi: 10.1186/2046-2530-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverman MA, Leroux MR. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 2009;19:306–316. doi: 10.1016/j.tcb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Sung CH, Leroux MR. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat Cell Biol. 2013;15:1387–1397. doi: 10.1038/ncb2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Q, Ling K, Hu J. The essential roles of transition fibers in the context of cilia. Curr Opin Cell Biol. 2015;35:98–105. doi: 10.1016/j.ceb.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emmer BT, Maric D, Engman DM. Molecular mechanisms of protein and lipid targeting to ciliary membranes. J Cell Sci. 2010;123:529–536. doi: 10.1242/jcs.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madhivanan K, Aguilar RC. Ciliopathies: the trafficking connection. Traffic. 2014;15:1031–1056. doi: 10.1111/tra.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nozawa YI, Lin C, Chuang PT. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr Opin Genet Dev. 2013;23:429–437. doi: 10.1016/j.gde.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novarino G, Akizu N, Gleeson JG. Modeling human disease in humans: the ciliopathies. Cell. 2011;147:70–79. doi: 10.1016/j.cell.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czarnecki PG, Shah JV. The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 2012;22:201–210. doi: 10.1016/j.tcb.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J Cell Biol. 2012;197:697–709. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szymanska K, Johnson CA. The transition zone: an essential functional compartment of cilia. Cilia. 2012;1:10. doi: 10.1186/2046-2530-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim YS, Tang BL. Getting into the cilia: nature of the barrier(s) Mol Membr Biol. 2013;30:350–354. doi: 10.3109/09687688.2013.842003. [DOI] [PubMed] [Google Scholar]
- 16.Malicki J, Avidor-Reiss T. From the cytoplasm into the cilium: bon voyage. Organogenesis. 2014;10:138–157. doi: 10.4161/org.29055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiter JF, Blacque OE, Leroux MR. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 2012;13:608–618. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kee HL, Verhey KJ. Molecular connections between nuclear and ciliary import processes. Cilia. 2013;2:11. doi: 10.1186/2046-2530-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YC, et al. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat Chem Biol. 2013;9:437–443. doi: 10.1038/nchembio.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Calvert PD, Schiesser WE, Pugh EN., Jr Diffusion of a soluble protein, photoactivatable GFP, through a sensory cilium. J Gen Physiol. 2010;135:173–196. doi: 10.1085/jgp.200910322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Najafi M, Maza NA, Calvert PD. Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc Natl Acad Sci USA. 2012;109:203–208. doi: 10.1073/pnas.1115109109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown JM, Cochran DA, Craige B, Kubo T, Witman GB. Assembly of IFT trains at the ciliary base depends on IFT74. Curr Biol. 2015;25:1583–1593. doi: 10.1016/j.cub.2015.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dishinger JF, et al. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurd TW, Fan S, Margolis BL. Localization of retinitis pigmentosa 2 to cilia is regulated by Importin beta2. J Cell Sci. 2011;124:718–726. doi: 10.1242/jcs.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113(Pt 10):1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- 30.Raices M, D’Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 31.Fan S, et al. Induction of Ran GTP drives ciliogenesis. Mol Biol Cell. 2011;22:4539–4548. doi: 10.1091/mbc.E11-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos N, Reiter JF. A central region of Gli2 regulates its localization to the primary cilium and transcriptional activity. J Cell Sci. 2014;127:1500–1510. doi: 10.1242/jcs.139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiuri T, Woloshansky T, Xia J, Truant R. The huntingtin N17 domain is a multifunctional CRM1 and Ran-dependent nuclear and cilial export signal. Hum Mol Genet. 2013;22:1383–1394. doi: 10.1093/hmg/dds554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatel G, Fahrenkrog B. Dynamics and diverse functions of nuclear pore complex proteins. Nucleus. 2012;3:162–171. doi: 10.4161/nucl.19674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashizume C, Moyori A, Kobayashi A, Yamakoshi N, Endo A, Wong RW. Nucleoporin Nup62 maintains centrosome homeostasis. Cell Cycle. 2013;12:3804–3816. doi: 10.4161/cc.26671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascual-Garcia P, Jeong J, Capelson M. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep. 2014;9:433–442. doi: 10.1016/j.celrep.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Diener DR, Lupetti P, Rosenbaum JL. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr Biol. 2015;25:379–384. doi: 10.1016/j.cub.2014.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Field MC, Koreny L, Rout MP. Enriching the pore: splendid complexity from humble origins. Traffic. 2014;15:141–156. doi: 10.1111/tra.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- 41.Takao D, Dishinger JF, Kee HL, Pinskey JM, Allen BL, Verhey KJ. An assay for clogging the ciliary pore complex distinguishes mechanisms of cytosolic and membrane protein entry. Curr Biol. 2014;24:2288–2294. doi: 10.1016/j.cub.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, Kufe D. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 44.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Bassam J, van Breugel M, Harrison SC, Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left–right patterning. Proc Natl Acad Sci USA. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katta SS, Smoyer CJ, Jaspersen SL. Destination: inner nuclear membrane. Trends Cell Biol. 2014;24:221–229. doi: 10.1016/j.tcb.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Laba JK, Steen A, Veenhoff LM. Traffic to the inner membrane of the nuclear envelope. Curr Opin Cell Biol. 2014;28:36–45. doi: 10.1016/j.ceb.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Lusk CP, Blobel G, King MC. Highway to the inner nuclear membrane: rules for the road. Nat Rev Mol Cell Biol. 2007;8:414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- 50.Ungricht R, Klann M, Horvath P, Kutay U. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J Cell Biol. 2015;209:687–703. doi: 10.1083/jcb.201409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boni A, Politi AZ, Strnad P, Xiang W, Hossain MJ, Ellenberg J. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J Cell Biol. 2015;209:705–720. doi: 10.1083/jcb.201409133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 53.Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 54.Ohba T, Schirmer EC, Nishimoto T, Gerace L. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soullam B, Worman HJ. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29:2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuleger N, Kelly DA, Richardson AC, Kerr AR, Goldberg MW, Goryachev AB, Schirmer EC. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J Cell Biol. 2011;193:109–123. doi: 10.1083/jcb.201009068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2012;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- 60.Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–439. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell. 2011;22:4694–4703. doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye F, Breslow DK, Koslover EF, Spakowitz AJ, Nelson WJ, Nachury MV. Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. Elife. 2013;2:e00654. doi: 10.7554/eLife.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. doi: 10.1016/S1097-2765(00)80324-X. [DOI] [PubMed] [Google Scholar]
- 64.Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 65.Ihara M, et al. Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev Cell. 2005;8:343–352. doi: 10.1016/j.devcel.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 66.Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 67.Ghossoub R, et al. Septins 2, 7 and 9 and MAP4 colocalize along the axoneme in the primary cilium and control ciliary length. J Cell Sci. 2013;126:2583–2594. doi: 10.1242/jcs.111377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Awata J, Takada S, Standley C, Lechtreck KF, Bellve KD, Pazour GJ, Fogarty KE, Witman GB. Nephrocystin-4 controls ciliary trafficking of membrane and large soluble proteins at the transition zone. J Cell Sci. 2014;127:4714–4727. doi: 10.1242/jcs.155275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cevik S, et al. Active transport and diffusion barriers restrict Joubert Syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet. 2013;9:e1003977. doi: 10.1371/journal.pgen.1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams CL, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dowdle WE, et al. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet. 2011;89:94–110. doi: 10.1016/j.ajhg.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roberson EC, et al. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J Cell Biol. 2015;209:129–142. doi: 10.1083/jcb.201411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pazour GJ, Bloodgood RA. Targeting proteins to the ciliary membrane. Curr Top Dev Biol. 2008;85:115–149. doi: 10.1016/S0070-2153(08)00805-3. [DOI] [PubMed] [Google Scholar]
- 76.Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/S0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 77.Mazelova J, et al. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28:183–192. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol. 2011;193:219–233. doi: 10.1083/jcb.201009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y, Zhang Q, Wei Q, Zhang Y, Ling K, Hu J. SUMOylation of the small GTPase ARL-13 promotes ciliary targeting of sensory receptors. J Cell Biol. 2012;199:589–598. doi: 10.1083/jcb.201203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McIntyre JC, Joiner AM, Zhang L, Iniguez-Lluhi J, Martens JR. SUMOylation regulates ciliary localization of olfactory signaling proteins. J Cell Sci. 2015;128:1934–1945. doi: 10.1242/jcs.164673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garcia-Gonzalo FR, Phua SC, Roberson EC, Garcia G, 3rd, Abedin M, Schurmans S, Inoue T, Reiter JF. Phosphoinositides regulate ciliary protein trafficking to modulate hedgehog signaling. Dev Cell. 2015;34:400–409. doi: 10.1016/j.devcel.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chavez M, Ena S, Van Sande J, de Kerchove d’Exaerde A, Schurmans S, Schiffmann SN. Modulation of ciliary phosphoinositide content regulates trafficking and sonic hedgehog signaling output. Dev Cell. 2015;34:338–350. doi: 10.1016/j.devcel.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 83.Ori A, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 85.Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Onischenko E, Weis K. Nuclear pore complex-a coat specifically tailored for the nuclear envelope. Curr Opin Cell Biol. 2011;23:293–301. doi: 10.1016/j.ceb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eisenhardt N, Redolfi J, Antonin W. Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J Cell Sci. 2014;127:908–921. doi: 10.1242/jcs.141739. [DOI] [PubMed] [Google Scholar]
- 88.Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA. 2014;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sang L, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ounjai P, Kim KD, Liu H, Dong M, Tauscher AN, Witkowska HE, Downing KH. Architectural insights into a ciliary partition. Curr Biol. 2013;23:339–344. doi: 10.1016/j.cub.2013.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elad N, Maimon T, Frenkiel-Krispin D, Lim RY, Medalia O. Structural analysis of the nuclear pore complex by integrated approaches. Curr Opin Struct Biol. 2009;19:226–232. doi: 10.1016/j.sbi.2009.02.009. [DOI] [PubMed] [Google Scholar]