Abstract
B cell acute lymphoblastic leukaemia (B-ALL) cells express high levels of CXCR4 chemokine receptors for homing and retention within the marrow microenvironment. Bone marrow stromal cells (BMSC) secrete CXCL12, the ligand for CXCR4, and protect B-ALL cells from cytotoxic drugs. Therefore, the therapeutic use of CXCR4 antagonists has been proposed to disrupt cross talk between B-ALL cells and the protective stroma. Because CXCR4 antagonists can have activating agonistic function, we compared the genetic and pharmacological deletion of CXCR4 in B-ALL cells, using CRISPR-Cas9 gene editing and CXCR4 antagonists that are in clinical use (plerixafor, BKT140). Both genetic and pharmacological CXCR4 inhibition significantly reduced B-ALL cell migration to CXCL12 gradients and beneath BMSC, and restored drug sensitivity to dexamethasone, vincristine and cyclophosphamide. NOD/SCID/IL-2rγnull mice injected with CXCR4 gene-deleted B-ALL cells had significant delay in disease progression and superior survival when compared to control mice injected with CXCR4 wild-type B-ALL cells. These findings indicate that anti-leukaemia activity of CXCR4 antagonists is primarily due to CXCR4 inhibition, rather than agonistic activity, and corroborate that CXCR4 is an important target to overcome stroma-mediated drug resistance in B-ALL.
Keywords: B-ALL, CXCR4, CXCL12, CRISPR-Cas9, Bone marrow microenvironment
Introduction
B-cell acute lymphoblastic leukaemia (B-ALL) is the most common type of cancer in children and accounts for 20% of all adult acute leukaemia cases. Approximately 6000 new B-ALL patients are diagnosed annually in the United States, of whom 1400 succumb to their disease(Siegel, et al 2014), and B-ALL remains one of the leading causes of person-years of life lost in the United States (362,000 years in 2010)(Murphy, et al 2013). There have been major improvements in treatment outcome over the last decades with 5-year survival rates of 90% in patients below the age of 15 years although the rate is significantly lower (~40%) in adult B-ALL patients(Bhojwani and Pui 2013). Relapse has become the major challenge in the treatment of B-ALL; relapsed patients are commonly resistant to standard drugs and therefore the outcome generally is dismal(Fielding, et al 2007). Minimal residual disease (MRD) due to primary resistant sub-clones is considered the principal mechanism that paves the way to relapse, and the contribution of stroma-mediated drug resistance, also known as cell adhesion-mediated drug resistance (CAM-DR)(Damiano, et al 1999), has been established as a central mechanism responsible for MRD in B-ALL.
Stromal cell-mediated protection of B-ALL cells is a mechanism adapted from normal B cell development, in which contact between precursor B cells and bone marrow stromal cells (BMSC) is critical for the survival and expansion of selected B cell progenitors. Similarly, B-ALL cells undergo rapid spontaneous apoptosis in vitro in standard suspension culture conditions, unless they are co-cultured with BMSC, indicating that BMSC are critical for B-ALL survival(Manabe, et al 1992). Furthermore, the degree of BM infiltration and MRD disease are associated with relapses and inferior prognosis in B-ALL(Brüggemann, et al 2012), emphasizing that interactions between B-ALL cells and BMSC in the marrow microenvironment provide survival and drug resistance signals that should be targeted for better treatment outcome.
The chemokine CXCL12, previously called stromal cell-derived factor-1 (SDF-1), is constitutively secreted by BMSC and regulates the retention and migration of haematopoietic progenitor cells (HPC)(Peled, et al 1999), mature haematopoietic cells(Bleul, et al 1996) and various cancer cells(Burger and Kipps 2006), including B-ALL(Bradstock, et al 2000) and T-ALL(Pitt, et al 2015) cells. Besides being a potent chemokine, CXCL12 also has pro-survival and growth-promoting effects in normal and malignant B cells; in fact, CXCL12 originally was designated pre-B-cell growth-stimulating factor, before it was recognized as a chemokine family member(Nagasawa, et al 1996a). CXCL12 binds to the chemokine receptor CXCR4, a seven trans-membrane G protein coupled receptor, which is expressed at high levels on B-ALL cells, presumably to attract and confine B-ALL cells to BMSC. This function of CXCR4 in B cell precursors is further supported by CXCL12 and CXCR4 knockout (KO) mice, which have an identical phenotype with severe defects in early B lymphopoiesis, due to premature release of B cell progenitors from the marrow and their displacement into the blood(Ma, et al 1998, Nagasawa, et al 996b). Both normal B-cell precursors and B cell leukaemia cells share this mechanism for homing to CXCL12-secreting BMSC within the marrow. Clinically, high CXCR4 expression has been linked to an inferior outcome in B-ALL(Konoplev, et al 2011, van den Berk, et al 2014).
Small molecule inhibitors of CXCR4 have been tested as therapeutic agents in the pre-clinical setting(Burger and Peled 2009). For example, plerixafor (previously known as AMD3100) and BKT140 and its derivatives were shown to overcome stoma-mediated drug resistance, inhibit stroma-induced ALL cell growth/metabolism(Juarez, et al 2003) and inhibit disease progression in mouse models of B-ALL(Juarez, et al 2007). Besides inhibition of CXCR4 function, CXCR4 antagonists also can induce signalling after binding to its target, CXCR4. Plerixafor and ALX40-4C have been characterized as weak partial agonists, whereas the polyphemusin derivative peptide inhibitor BKT140 was characterized as an inverse CXCR4 agonist(Zhang, et al 2002). Signalling responses induced by stimulation of CXCR4 with high concentrations of plerixafor and ALX40-4C were less robust than those seen with its natural ligand, CXCL12, and hence plerixafor and ALX40-4C were characterized as weak partial CXCR4 agonists(Zhang, et al 2002). The agonistic activity of plerixafor and ALX40-4C raises concern that some of the activity seen with CXCR4 antagonists may be due to agonistic activity, rather than blockade of CXCR4 function. Along the same lines, preclinical work with BMS-936564/MDX-1338, a therapeutic anti-human CXCR4 monoclonal antibody, revealed that this CXCR4 antagonist also induced downstream signalling (Kuhne, et al 2013). The authors compared BMS-936564 with plerixafor in preclinical assays and noted marked differences; while BMS-936564 induced target cell apoptosis, plerixafor did not, suggesting that antibody binding to CXCR4 drives a signal to induce apoptosis that is independent from inhibition of CXCL12 binding(Kuhne, et al 2013). These differences in inhibitor-induced signalling raise the question of whether some of the anti-leukaemia activity seen in prior studies may be related to agonistic activity of the CXCR4 antagonist. Therefore, we compared pharmacological and genetic functional deletion of CXCR4 in preclinical models of B-ALL.
Materials & Methods
Reagents and Antibodies
Synthetic human CXCL12 (SDF1α) was purchased from R&D Systems (Minneapolis, MN, USA). The small molecule CXCR4 inhibitor, plerixafor, was purchased from SIGMA (St. Louis, MO, USA), and BKT140, a peptide CXCR4 inhibitor, was provided by Dr. Peled (Goldyne Savad Institute of Gene Therapy, Hadassah Hebrew University Hospital, Jerusalem, Israel). Dexamethasone and vincristine sulfate were purchased from SIGMA (St. Louis, MO, USA) and 4-Hydroperoxy cyclophosphamide was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-CXCR4-phycoerythrin (PE) antibodies (12G5 clone) were purchased from BD Pharmingen (San Jose, CA, USA) and anti-CXCR4-allophycocyanin (APC) antibodies (12G5 clone) were purchased from eBioscience (San Diego, CA, USA). Anti-human/mouse-CXCR7-PE antibodies (clone 8F11-M16) were purchased from Biolegend (San Diego, CA). All other antibodies and isotype controls (anti-CD44- FITC, anti-CD54-PE, anti-CXCR5-PE, anti-CXCR3-PE, anti-CD62L-FITC, anti-CD49D-PE and CCR7-PE) and respective isotype controls were purchased from BD Pharmingen.
Cell culture
The murine bone marrow stromal cell line 9–15C (from C3H/He mouse) and B-ALL cell lines were obtained from the RIKEN Cell Bank (Ibaraki, Japan). Cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY), 2.05 mM L-glutamine and 5% penicillin-streptomycin (Cellgro, Corning Inc, Corning, NY). Xenograft-expanded primary B-ALL cells were kindly provided by Dr. Müschen (University of California, San Francisco, CA) and were maintained in Alpha MEM medium supplemented with 20% FBS, GlutaMAX (Thermo Scientific, Grand Island, NY) and 5% penicillin streptomycin.
Chemotaxis assay
The migration of B-ALL cells in response to CXCL12 was evaluated using 24-well Corning chemotaxis chambers (Corning Life Sciences, Acton, MA) as described(Burger, et al 1999). In brief, Transwell chambers with 5-μm pore size polycarbonate inserts were used, and 5 × 106 B-ALL cells (in 100 μl RPMI 1640 with 0.5% bovine serum albumin [BSA]) were added to the top chamber, while CXCL12 (100 ng/ml) was added to the lower wells. The plates were incubated at 37°C after inserts were placed into the wells. Assay media (RPMI 1640 with 0.5% BSA) without CXCL12 was used as control. After 3 h, cells that had migrated to the lower chambers were collected and counted for 20 s by flow cytometry, using a FACSCalibur (BD Biosciences, San Jose, CA) at high flow. For comparison, a dilution of input cells (1:20) was counted under the same conditions.
Pseudoemperipolesis assay
Pseudoemperipolesis (PEP), the spontaneous migration of B-ALL cells beneath BMSCs, was tested and quantified as described(Burger, et al 1999). Briefly, on the day before the assay, BMSC (9–15C cells) were seeded in 24-well plates at a concentration of 9×104 cells per well. On the day of the experiment, the confluence of the BMSC layer was confirmed by phase-contrast microscopy, and B-ALL cells were added to each well to a final concentration of 2.5×106 ALL cells per well. ALL cells were pre-incubated with 10 μg/ml CXCR4 inhibitor (plerixafor, BKT140) for 1 h at 37°C. The plates were incubated for 4 h at 37°C and non-migrated cells in the supernatant were removed by thorough washing of each well 3 times with RPMI medium without disrupting the stromal cell layer. Stromal cell layers containing migrated B-ALL cells, which were considered to have undergone PEP, were trypsinized and the detached cells were suspended in 500 μl assay media (RPMI 1640 with 0.5% BSA) for counting by flow cytometry. To exclude stromal cells, a lymphocyte gate was set using lymphocyte forward and side scatter characteristics, and cell counting was performed as indicated above.
Measurement of Cell Viability
Cell viability was tested using 3,3′ dihexyloxacarbocyanine iodide (DiOC6) (Invitrogen, Carlsbad, CA) and propidium iodide (PI) staining, as described(Burger, et al 1999), followed by flow cytometry analysis (FACSCalibur, BD Biosciences, San Jose, CA). Apoptotic cells have low DiOC6 and PI fluorescence, whereas viable cells show high DiOC6 and low PI fluorescence; low DiOC6 and high PI fluorescence characterizes necrotic cells.
Cell Adhesion Mediated Drug Resistance (CAM-DR) Experiments
To quantify drug-induced cytotoxicity, co-cultures of ALL and BMSC were treated with dexamethasone (DEX), vincristine (VIN) or 4-Hydroperoxy cyclophosphamide (4HC), in the presence or absence of the CXCR4 antagonists plerixafor or BKT140. Briefly, the day before starting the assay, 2×104 cells/well 9–15C stromal cells were seeded in 24-well plates. After overnight incubation, the confluence of the stromal cell layer was determined by phase-contrast microscopy, and CXCR4 inhibitor (10 μg/ml) -treated and -untreated B-ALL cells were added into each well to a final concentration of 1×106 ALL cells per well. Cytotoxic drugs DEX or VIN were added at the indicated concentrations. For 4HC, cells were treated with indicated concentrations for 45 min and then washed with culture media before plating on stromal cells. ALL cells alone, in co-culture with 9–15C and with the cytotoxic drug were used as controls. Viability of cells in the supernatant was measured at 48 h. Where indicated, the viability of ALL cells that migrated under the 9–15C (through PEP) was also measured.
CRISPR/Cas9 based CXCR4 Knockout (KO)
We used the CRSIPR/cas9 technique for CXCR4 KO in the B-ALL lines. Briefly, the target sequence was designed using the website http://crispr.mit.edu/. A target sequence GCTTCTACCCCAATGACTTGTGG (TGG is the Protospacer adjacent motif (PAM)), located on the second extracellular domain of CXCR4, was chosen for targeted double strand break. The target site without the PAM sequence was cloned in the vecoe pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid #42230 kindly provided by Dr. Feng Zhang) which contains two expression cassettes, a human codon-optimized SpCas9 and the single guide RNA) following the protocol described at http://www.addgene.org/crispr/zhang/ (Cong, et al 2013). The plasmid (15 μg) was then electroporated into B-ALL cell-lines, using the Neon Transfection System (Life Technologies, NY) following single pulse with pulse voltage 1500V and pulse with 20 ms. After 72 h, CXCR4 expression was analysed by FACS. CXCR4 negative cell populations were enriched by MACS negative depletion, using anti-PE conjugated anti-CXCR4 and MACS anti-PE beads (Miltenyi Biotec, Auburn, CA) and then expanded. The Tanoue-CXCR4-KO cell line was sequenced to identify mutations introduced in the CXCR4 gene, sequence alignment with wild type CXCR4 is shown in Figure S1.
GFP transfection
HEK293T cells were transfected with (5 μg) lentiviral plasmid and packaging plasmids pMD2.G (5 μg) and pPAX (5 μg) using Jet prime transfection reagents according to the manufacturer’s protocol (Polypus Transfection, Illkirch, France). The plasmids and HEK293T cells were kindly provided by Dr. Michael Andreeff (MD Anderson Cancer Center, Houston, TX). The medium containing the virus was collected after 48 h, centrifuged for 5 min at 1500 rpm and the supernatant was filtered through a 0.45-μm filter. NALM6 WT and NALM6-CXCR4-KO were re-suspended in 5 ml viral supernatant and incubated overnight at 37°C. Viral media was replaced with fresh media the next day and GFP expression was analysed after 48 h by florescent microscope. GFP positive cells were enriched and used for in vivo experiments.
In vivo experiments
All mouse experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center. To investigate the in vivo effects of genetic CXCR4 deletion in NALM6 cells, non-irradiated NOD/SCID/IL-2rγnull (NSG) mice (The Jackson Laboratory, Bar Harbor, ME) were injected via tail vein with 1×106 GFP positive NALM6 wild type or GFP-positive NALM6 CXCR4 KO cells. Bioluminescence imaging (BLI) was performed on days 6, 10, 14 and 17 to monitor tumour burden. Briefly, mice were anesthetized and imaged noninvasively with an in vivo imaging system (IVIS-200; Xenogen, Hopkinton, MA) after injection with luciferase substrate coelenterazine (Biotium, Heyward, CA). Total body bioluminescence was quantified in a region of interest drawn around each mouse. Three mice per group were sacrificed on day 17 to measure GFP positive leukaemic cells in different organs and femurs. The expression of surface CXCR4 protein was analysed by using a Gallios flow cytometer (Beckman Coulter, Brea, CA) and harvested cells were stained with antibodies against CXCR4-allophycocyanin (12G5). Overall survival and mean group survival times were estimated by the Kaplan-Meier method and compared with the log-rank test.
Data Analysis and Statistics
Results are depicted as mean ± standard error of the mean (SEM), of at least three experiments each. PEP experiments were conducted twice in duplicates. Statistical analysis was done with GraphPad Prism 6.0 for Macintosh (GraphPad Software, San Diego, CA, USA). Two-tailed student’s t-test was used for statistical comparison between groups and the P values were assigned as follows (*P < 0.05; **P < 0.01; ***P < 0.001). Flow Cytometry data were analysed using FLOWJO 9.4.11 software (TreeStar Inc., Ashland, OR).
Results
CXCR4 is highly expressed in ALL cells
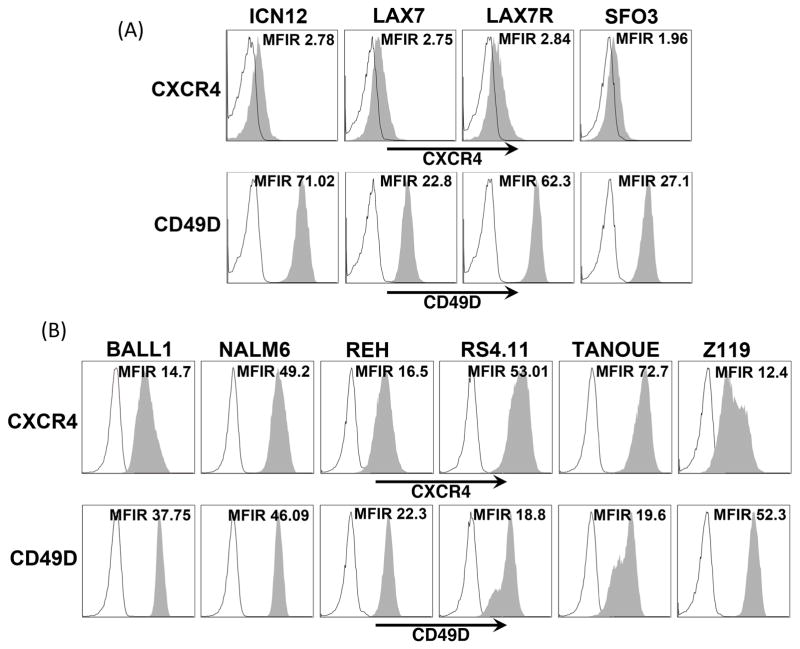
Surface expression of CXCR4 was lower on xenograft-expanded primary B-ALL cells as compared to B-ALL cell lines (Figure 1A, B and S1). All B-ALL cells expressed high CD49D (also termed ITGA4) levels. Other tested chemokine receptors were undetectable or had very low expression (Figure S2), suggesting that CXCR4 is the principal receptor for CXCL12 involved in migration of B-ALL cells to BMSC. Surface expression of adhesion molecules was variable across cell lines and xenografts, but there was consistent positivity for CD49D (VLA4, see Figure S2 and S3).
Figure 1. Expression of CXCR4 and CD49D in various ALL cell lines and xenograft cells.
Histograms show Mean Fluorescent Intensity Ratio (MFIR) of CXCR4 and CD49D (grey peaks) as compared to their respective isotype controls (black outlined peaks) in different B-ALL cell lines (A) and xenografts (B). Mean Fluorescent Intensity Ratio (MFIR) is shown individually for each histogram.
CXCR4 gene knockout inhibits B-ALL cell adhesion and migration
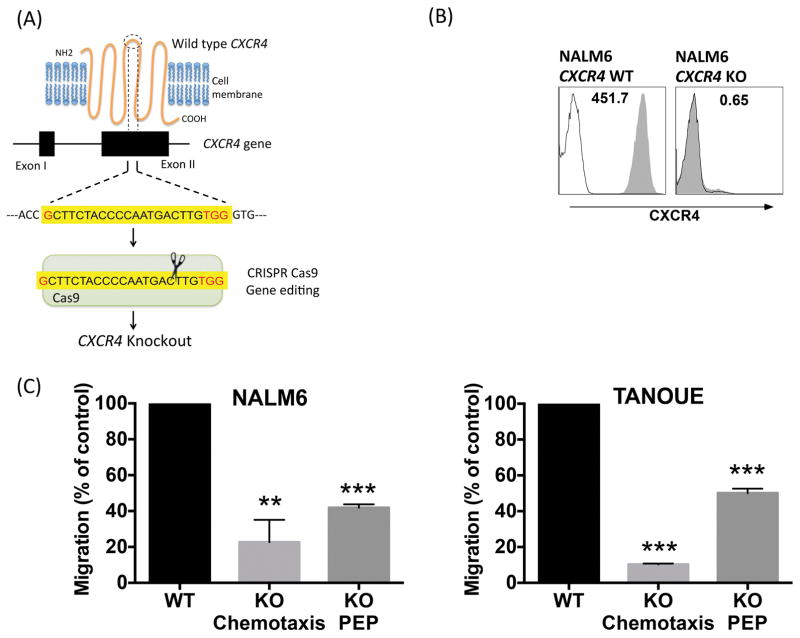
In order to study the function of CXCR4 in B-ALL, we selectively targeted it for KO in NALM6 and TANOUE cells (Figure 2A). Figure 2B shows representative histograms of CXCR4 surface expression in CXCR4 wild type and KO cells of the B-ALL line NALM6. CXCR4 KO NALM6 cells displayed significantly reduced chemotaxis and PEP, respectively reduced to 22.6 ± 12.5% or 41.8 ± 1.9% of wild-type controls (mean ± SEM, p<0.05). Similarly, chemotaxis and PEP of TANOUE CXCR4 KO cells also was significantly reduced to 10.3 ± 0.4% or 50.1 ± 2.5% of wild-type controls (mean ± SEM, p<0.05), respectively (Figure 2C).
Figure 2. CXCR4 gene deletion significantly decreases chemotaxis and PEP of B-ALL cell lines.
(A) The mechanism of CXCR4 deletion through CRISPR-Cas9 gene editing system. The CRISPR Cas9 was directed towards the second extra cellular loop to introduce a frame-shift mutation resulting in lack of expression of the CXCR4 protein. (B) Histograms depict CXCR4 expression in NALM6 cells before and after CRISPR-Cas9 mediated CXCR4 knockout. Cells were stained with isotype control (black line) or CXCR4 antibody (shaded grey area). (C) NALM6 and TANOUE wild type (WT) and knockout (KO) cells (both untreated) were allowed to undergo chemotaxis towards 100 ng/ml CXCL12 or PEP beneath 9–15C Bone Marrow Stromal Cells and migrated cells were counted in flow cytometer for quantification. Bar diagrams representing mean chemotaxis/PEP (± SEM), with *p < 0.05, **p < 0.01 and ***p < 0.001.
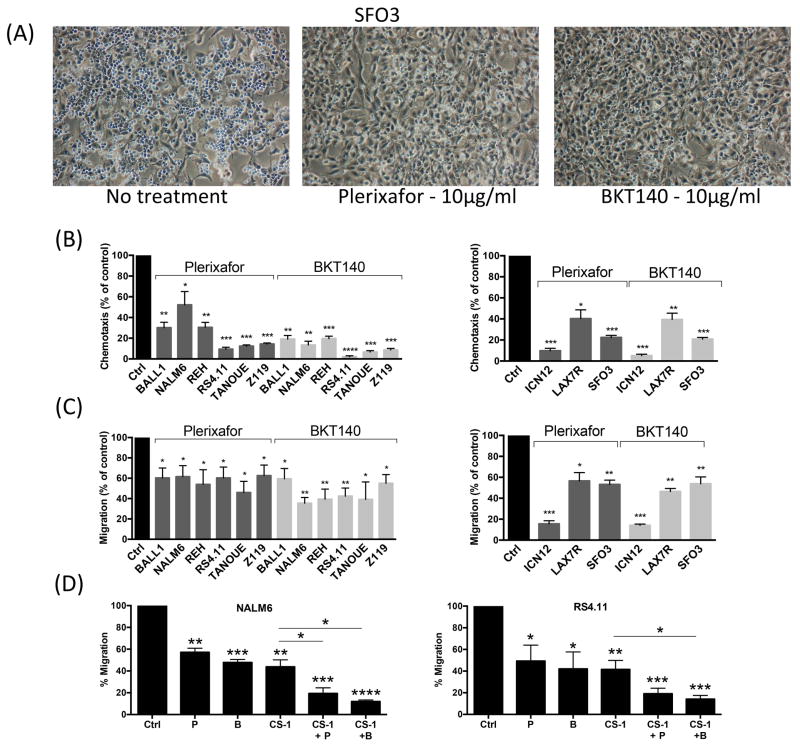
CXCR4 antagonists inhibit ALL cell migration and adhesion
Figure 3A depicts representative PEP images of SFO3 xenograft expanded primary B-ALL cells. Pre-incubation with CXCR4 antagonists (plerixafor or BKT140) effectively inhibited chemotaxis of B-ALL cell lines and primary B-ALL cells to CXCL12 (Figure 3B). For example, pre-treatment of NALM6 cells with plerixafor or BKT140 significantly reduced chemotaxis to 59.8 ± 5.6% or 13.4 ± 3.7% (mean ± SEM, p<0.05) of controls, respectively. After pre-incubation with plerixafor or BKT140, PEP was significantly inhibited (Figure 3C). For example, PEP of NALM6 cells was significantly reduced after plerixafor or BKT140 treatment to 61.4 ± 10.8% and 35.5 ± 5.5% of controls (mean ± SEM, p<0.05), respectively. To further understand the mechanism of PEP, we hypothesized that CD49D causes residual migration after CXCR4 inhibition. Interestingly, the combination of CXCR4 and CD49D inhibitors further significantly reduced PEP as compared to inhibition of CXCR4 or CD49D alone (Figure 3D).
Figure 3. CXCR4 inhibitors plerixafor (AMD3100) and BKT140 significantly reduce chemotaxis and Pseudoemperipolesis (PEP) of B-ALL cell lines and xenografts.
Representative phase contrast microscopy images (A) show SFO3 xenograft cells undergoing PEP in the presence or absence of CXCR4 inhibitor treatment (10 μg/ml). B-ALL cells were incubated in medium alone (control) or medium containing (10 μg/ml) plerixafor (AMD3100) or BKT140. The cells were allowed to (B) undergo chemotaxis towards 100 ng/ml CXCL12 or (A, C) undergo PEP beneath 9–15C Bone Marrow Stromal Cells and then counted in flow cytometer for quantification. The bar diagrams represents the mean chemotaxis/PEP (± SEM) of 6 B-ALL cell lines (left-hand graph) and 3 B-ALL xenografts (right-hand graph) in the presence or absence of CXCR4 inhibitors. Chemotaxis/PEP was significantly inhibited by both CXCR4 inhibitors (plerixafor/BKT140). (D) NALM6 (left panel) and RS4.11 (right panel) B-ALL cells undergoing PEP in the presence of plerixafor (P) or BKT140 (B) combined with CD49d antagonist CS-1, cells were incubated in medium alone (control), or medium supplemented with plerixafor (10 μg/ml), BKT140 (10 μg/ml), CS1 (10 μg/ml), or combinations of CS-1 with either CXCR4 antagonist, and then placed in BMSC co-cultures. Combination treatment with CXCR4 and CD49d antagonists inhibits B-ALL cell migration beneath BMSC (PEP) more effectively than single inhibitor treatment. The bar diagram represents the mean PEP (± SEM) (plerixafor/BKT140), with *p < 0.05, **p < 0.01 and ***p < 0.001.
CXCR4 gene deletion decreases leukaemia burden, bone marrow infiltration and increases survival in vivo
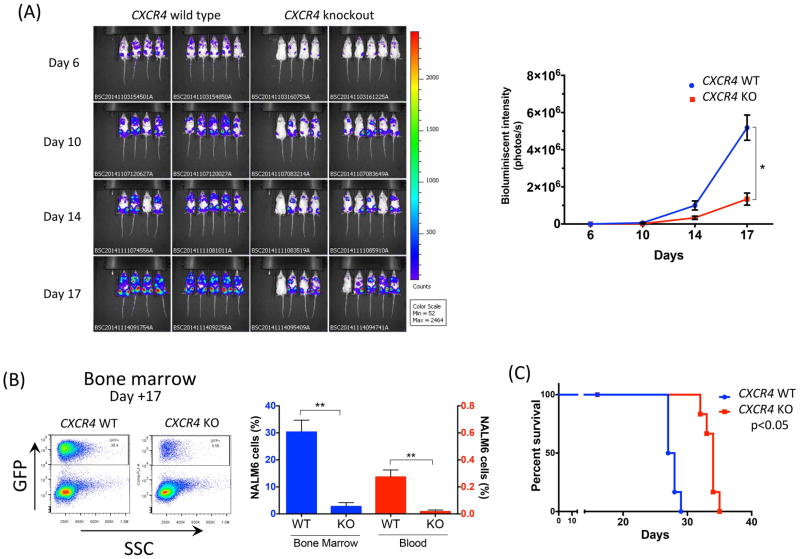
Mice injected with NALM6 CXCR4 KO cells had significantly less bioluminescent imaging (BLI) signal than with NALM6 WT cells throughout the course of the disease (p<0.001 on day 6, p<0.05 on day 10, p<0.05 on day 14 and p<0.001 on day 17) (Figure 4A). These differences became less significant at later time points (Figure S4). In three representative mice per group sacrificed on day 17, flow cytometry demonstrated that mice injected with NALM6 CXCR4 KO cells had less leukaemic infiltration in bone marrow than NALM6 WT (mean ± SEM, 2.8% ± 1.4% vs. 30.3% ± 4.4% respectively, p<0.001 (Figure 4B). Analysis of circulating NALM6 cells by FACS with CXCR4 12G5 antibody revealed the lack of CXCR4 expression in mice injected with NALM6 CXCR4 KO (mean fluorescent intensity/isotype, 2.1 ± 0.1) compared to NALM6 WT (120.1 ± 1.0, p<0.001) (data not shown). The proportion of circulating leukaemic cells was also measured in three representative mice from each group on day 17 and living mice (five mice per each group) on days 20 and 24. Mice injected with NALM6 CXCR4 KO cells had significantly lower numbers of circulating leukaemic cells than NALM6 WT (0.27% ± 0.05% vs. 0.02% ± 0.01%, p<0.05 on day 17; 0.78% ± 0.13% vs. 0.05%±0.004%, p<0.05 on day 20; 1.2% ± 0.34% vs. 0.1% ± 0.02%, p<0.05 on day 24)(Figure 4B, see Figure S5 for additional time points). Spleen engraftment by NALM6 CXCR4 KO and WT B-ALL cells was low in both groups and, in contrast to marrow and peripheral blood, the differences were not significant (Figure S6). These differences in leukaemia progression and disease burden translated into significant prolongation of survival in mice injected with NALM6 CXCR4 KO cells (median survival, 34 days vs. 27.5 days; p<0.05) (Figure 4C).
Figure 4. NALM6 CXCR4 Knockout cells display reduced leukaemia burden and increased survival.
(A) Nine mice per group were injected with GFP positive NALM6 CXCR4 wild type (WT) or NALM6 CXCR4 knockout (KO) cells and Bioluminescent Imaging (BLI) measured for all mice during the course of the experiment (on days 6, 10, 14 and 17). Right-hand graph depicts quantification of BLI (photons/second) for mice injected with NALM6 CXCR4 WT (blue line) vs. NALM6 CXCR4 KO (red line) (p<0.05). (B) Bone marrow and peripheral blood of three mice per group were analysed on day 17 for GFP positive leukaemia cells using flow cytometry. Representative dot plot shows leukaemia cells present in the bone marrow in mice injected with NALM6 CXCR4 WT or NALM6 CXCR4 KO cells. Bar graph shows quantification of number of GFP positive leukaemia cells in the bone marrow (red bars) or the blood (blue bars). *p < 0.05; **p < 0.01. (C) Kaplan-Meier survival curve shows the survival of mice injected with NALM6 CXCR4 WT (blue line) vs. NALM6 CXCR4 KO (red line) (p<0.05).
BMSCs confer drug resistance to B-ALL cells
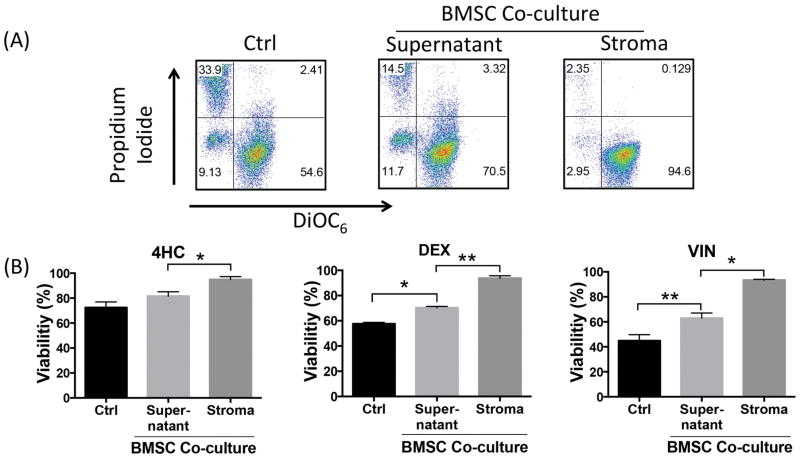
To test the effect of BMSC on drug-induced cytotoxicity, NALM6 cells were treated with DEX, or 4HC (the active metabolite of cyclophosphamide), or VIN in the presence or absence of 9–15C BMSC (Figure 5). Co-culture of NALM6 cells with BMSC significantly preserved B-ALL cell viability after DEX treatment at 70.3 ± 1% when compared to 57.6 ± 1.1% in suspension cultures without BMSC (mean ± SEM, p<0.05). Similarly, NALM6 viability remained higher in BMSC co-cultures after VIN treatment (63 ± 4.1%) when compared to 44.9 ± 4.8% in suspension cultures (mean ± SEM, p<0.05). Similar results were obtained with 4HC. Differences in drug resistance were also analysed based on localization of the B-ALL cells in relation to the BMSC. Interestingly, we noted that B-ALL cells that had migrated beneath and underneath the BMSC layer were highly resistant to DEX, 4HC and VIN, with viabilities 93.7 ± 1.9%, 94.9 ± 2.3% or 93.4 ± 0.5% (mean ± SEM, p<0.05, Figure 5B) respectively. Gating strategy for this experiment depicted in Figure S7.
Figure 5. Bone Marrow Stromal Cell (BMSC) co-culture overcomes drug-induced cytotoxicity of B-ALL cell line and CXCR4 deletion reverses this effect.
(A) Dot plots show representative experiment depicting viability of dexamethasone (DEX) treated NALM6 cells in the presence and absence of BMSC co-culture. In the NALM6-BMSC co-culture sample, viability of NALM6 cells in supernatant and those that migrated beneath the stroma was measured separately. (B) Bar graphs show mean of three separate experiments illustrated in dot plots. In addition to DEX treatment, the experiment was also performed with 2.5 μM 4-HydroperoxyCyclophosphamide (4HC) and 2 nM vincristine (VIN) in the presence and absence (control) of BMSC co-culture. Bar diagrams represent mean drug induced cytotoxicity (± SEM) of three separate experiments. Viability was measured using PI/DiOC6 staining at 48 h; asterisks indicate significant differences in cytotoxicity (*p < 0.05; **p < 0.01).
Pharmacological and genetic CXCR4 inhibition partially overcomes BMSC-mediated drug resistance
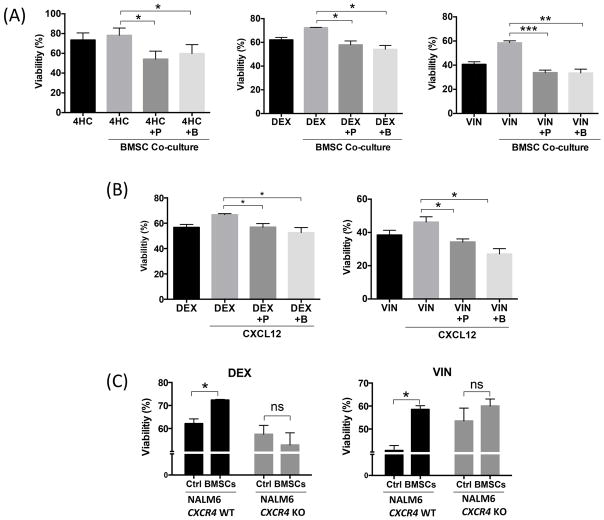
In BMSC co-culture, the addition of plerixafor or BKT140 significantly increased DEX-induced apoptosis of NALM6 cells, from 72.38 ± 0.25% to 57.9 ± 3.2% or 54.2 ± 3.2% viable cells respectively (mean ± SEM, p<0.05) (Figure 6A). Similarly, plerixafor or BKT140 addition to the NALM6-BMSC co-culture sensitized NALM6 cells to VIN treatment, increasing VIN-induced cytotoxicity to 33.8 ± 2.08% and 33.5 ± 3.1% viable cells respectively as compared to NALM6-BMSC alone (58.5 ± 1.6%, mean ± SEM, p<0.05). Lastly, plerixafor or BKT140 addition to NALM6-BMSC co-cultures increased 4HC-induced cytotoxicity to 66.03 ± 5.1% and 67.1 ± 1.2% viable cells respectively as compared to NALM6-BMSC alone (82.9 ± 1%, mean ± SEM, p<0.05) (Figure 6A). This reversal by CXCR4 inhibitors of BMSC-mediated resistance of NALM6 outlines the critical role that CXCR4 plays in making cells resistant to chemotherapy. Similar results were obtained for DEX-treated ICN12 xenograft cells, indicating that this is not strictly a cell line phenomenon, and for REH and Z119 cell lines (Figure S8).
Figure 6. CXCR4 inhibitors sensitize NALM6 cells to chemotherapy.
(A) Bar graphs depict viability of NALM6 cells in supernatant, 48 h after treatment with 100 nM Dexamethasone (DEX), 2 nM Vincristine (VIN) and 2.5 μM 4-Hydroperoxy Cyclophosphamide (4HC) in the presence and absence (control) of bone marrow stromal cell (BMSC) co-culture. Additionally, before plating on stromal cells, NALM6 cells were incubated in medium alone or medium containing (10 μg/ml) plerixafor (AMD3100, P) or BKT140 (B). (B) Viability of NALM6 cells was analysed 48 h after treatment with 100 nM DEX or 2 nM VIN in the presence and absence (control) of CXCR4 ligand, CXCL12. Additionally, before the addition of CXCL12, NALM6 cells were incubated in medium alone or medium containing (10μg/ml) plerixafor (AMD3100) or BKT140. (C) NALM6 CXCR4 Wild type (WT) (black bars) and NALM6 CXCR4 Knockout (KO) (grey bars) were treated with 100 nM DEX (left hand side graph) or 2 nM VIN (right hand side graph) in the presence and absence (control) of BMSC co-culture and viability measured for cells in the supernatant. Bar diagrams represent mean drug induced cytotoxicity (± SEM) of three separate experiments. Viability was measured using PI/DiOC6 staining at 48 h; asterisks indicate significant differences in cytotoxicity (*p < 0.05; **p < 0.01; ***p < 0.001).
To further dissect the mechanism by which BMSCs provide leukaemia cells with resistance, we tested whether recombinant CXCL12 could provide the same degree of resistance as BMSC (Figure 6B). Synthetic CXCL12 partially rescued NALM6 cells from DEX-induced cytotoxicity (66.7 ± 0.8% viability) as compared to monoculture alone (56.7 ± 2.3% viability, mean ± SEM, p<0.05). This was a specific effect, because the addition of 10 μg/ml plerixafor or BKT140 to CXCL12-supplemented monocultures abrogated this rescue from DEX-induced cytotoxicity (56.9 ± 2.7 and 52.6 ± 3.9% viability, respectively, mean ± SEM, p<0.05), to a point similar to DEX treatment alone. Similar results were observed with vincristine, corroborating the findings of the previous experiments and indicating CXCR4 as one of the critical mechanisms used by B-ALL cells for chemo-resistance.
To further corroborate the findings of pharmacological inhibition of CXCR4 in the co-culture setting, NALM6 CXCR4 KO cells were treated with DEX and VIN in the presence and absence of BMSCs (Figure 6C). Unlike wild type NALM6 cells, NALM6 CXCR4 KO cells showed no significant effect on cytotoxicity of the absence or presence of BMSCs (57.5 ± 3.8% vs. 52.8 ± 5.2% for DEX and 53.5 ± 5.5% vs. 60.06 ± 2.9% for VIN, respectively, mean ± SEM, p=not significant). The failure of BMSCs to rescue NALM6 CXCR4 KO cells from DEX- or VIN-induced cytotoxicity further indicates the importance of CXCR4 in BMSC-mediated chemo-resistance.
Discussion
B-ALL cells express high levels of CXCR4 for migration and adhesion to BMSC, which in turn cause adhesion-mediated resistance to drugs commonly used in the treatment of B-ALL patients. Therefore, CXCR4 has become a therapeutic target in B-ALL, and CXCR4 antagonists have been used in the preclinical setting as an adjuvant to sensitize B-ALL cells to chemotherapeutic agents(Juarez, et al 2003, Sison, et al 2013, Sison, et al 2014). Given the differences between CXCR4 antagonists, for example in terms of partial agonist activity, we compared pharmacological and genetic CXCR4 inhibition in B-ALL. We found that both pharmacological and genetic CXCR4 disruption resulted in impaired migration and adhesion of B lymphoblasts. Consequently, B-ALL cells treated with CXCR4 antagonists were more susceptible to the cytotoxicity induced by dexamethasone, vincristine and cyclophosphamide. Also, CXCR4 wild type, but not CXCR4 KO cells were rescued from drug-induced cytotoxicity by co-culture with BMSC (Figure 6C). Therefore, agonistic activity of CXCR4 antagonists, as demonstrated for plerixafor(Trent, et al 2003, Zhang, et al 2002), does not seem to impact the anti-tumour activity in any major fashion. Accordingly, in vivo experiments confirmed that CXCR4-KO in B-ALL cells significantly reduced leukaemia burden and delayed disease progression, resulting in a modest survival benefit (Figure 4C). These findings corroborate the importance of CXCR4 in B-ALL cell engraftment and disease progression in vivo and confirm the results generated with CXCR4 antagonists in prior mouse models of B-ALL(Juarez, et al 2003; Juarez, et al 2007).
We noted that xenograft-expanded primary B-ALL cells showed markedly lower CXCR4 expression than B-ALL cell lines (Figure S2), with preserved CXCR4 function. This low CXCR4 surface expression presumably is caused by chronic CXCR4 down-regulation due to CXCR4 endocytosis in BMSC co-cultures, in which primary B-ALL cells are routinely maintained, in response to BMSC-secreted CXCL12. Similar observations were made by van den Berk et al (2014), who showed significantly lower CXCR4 surface expression on leukaemia cells isolated from the bone marrow when compared to peripheral blood leukaemia cells. Conversely, co-culture of B-ALL cell lines with BMSC resulted in CXCR4 down-regulation (Figure S9). An additional chemokine receptor of interest was the second CXCL12 receptor, CXCR7 (also termed ACKR3), which previously has been characterized as a decoy receptor for CXCL12(Naumann, et al 2010). Our expression analysis using anti-CXCR7 monoclonal antibodies indicate that B-ALL cells are negative for CXCR7 protein expression, arguing against a role of CXCR7 in B-ALL. Melo Rde et al (2014) recently reported high ACKR3/CXCR7 expression in ALL, based on reverse transcription polymerase chain reaction screening, but substantial CXCR7 protein levels were only found in T-ALL cells.
Leukaemia-BMSC co-culture experiments have previously suggested that non-tumoural cells in the microenvironment, especially BMSC, protect leukaemia cells from drug-induced cytotoxicity. We expanded on this by analysing separately B-ALL cells that had undergone PEP to reside under the BMSC layer, and comparing them to B-ALL cells that remained on top of the BMSC. In contrast to B-ALL cells residing on top of BMSC, the viability of B-ALL cells underneath the BMSC was not affected by drug treatment, revealing that chemo-protection was highest for those B-ALL cells that had crawled underneath the stromal layer.
CXCR4 antagonists have previously been explored as adjuvants to sensitize leukaemia cells to conventional chemotherapy in CLL(Burger, et al 2005) and AML(Nervi, et al 2009) in the preclinical and clinical setting, without increased toxicity to the normal haematopoiesis. Our data indicate that CXCR4 antagonists, with or without intrinsic agonistic activity, sensitize B-ALL cells to chemotherapy due to inhibition of CXCR4, and hence could be tested in a similar fashion.
In summary, our study corroborates the importance of CXCR4 in the pathogenesis of B-ALL by demonstrating its effect on migration, adhesion and survival of leukaemia cells, in vitro and in vivo. Our data suggest that targeting of CXCR4 in B-ALL may help to reduce MRD and support the clinical testing of CXCR4 antagonists in B-ALL to sensitize leukaemia cells that are normally protected in the stroma microenvironment to the cytotoxicity of standard chemotherapy.
Supplementary Material
Acknowledgments
The study was supported in part by a Cancer Prevention and Research Institute of Texas (CPRIT) grant (to JAB), a Leukemia & Lymphoma Society Scholar Award in Clinical Research (to JAB), the National Institute of Health 5R01-CA155056-03 (to MK) and the MD Anderson Cancer Center Support Grant CA016672.
Footnotes
Authorship contribution
S.R. performed the in vitro experiments, analysed the data, designed the figures and wrote the paper; B.S.K conducted and M.K supervised mice experiments and reviewed the paper; D.G. and R.E.D provided CRISPR Cas9 supervision and reagents; M.S. and S.K. discussed in vitro experimental data and reviewed the paper; MM developed and provided B-ALL cells samples and reviewed the manuscript, A.P. kindly provided BKT140 CXCR4 antagonist and J.A.B. designed the research, supervised the study, analysed the data and wrote the paper.
Conflict-of-interest disclosure
Amnon Peled is CEO/CSO and founder of Biokine Therapeutics Ltd. All other authors declare no competing financial interests.
References
- Bhojwani D, Pui CH. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14:e205–217. doi: 10.1016/S1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
- Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) [see comments] J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradstock KF, Makrynikola V, Bianchi A, Shen W, Hewson J, Gottlieb DJ. Effects of the chemokine stromal cell-derived factor-1 on the migration and localization of precursor-B acute lymphoblastic leukemia cells within bone marrow stromal layers [In Process Citation] Leukemia. 2000;14:882–888. doi: 10.1038/sj.leu.2401729. [DOI] [PubMed] [Google Scholar]
- Brüggemann M, Raff T, Kneba M. Has MRD monitoring superseded other prognostic factors in adult ALL? Blood. 2012;120:4470–4481. doi: 10.1182/blood-2012-06-379040. [DOI] [PubMed] [Google Scholar]
- Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23:43–52. doi: 10.1038/leu.2008.299. [DOI] [PubMed] [Google Scholar]
- Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94:3658–3667. [PubMed] [Google Scholar]
- Burger M, Hartmann T, Krome M, Rawluk J, Tamamura H, Fujii N, Kipps TJ, Burger JA. Small peptide inhibitors of the CXCR4 chemokine receptor (CD184) antagonize the activation, migration, and antiapoptotic responses of CXCL12 in chronic lymphocytic leukemia B cells. Blood. 2005;106:1824–1830. doi: 10.1182/blood-2004-12-4918. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano JS, Cress AE, Hazlehurst LA, Shtil AA, Dalton WS. Cell adhesion mediated drug resistance (CAM-DR): role of integrins and resistance to apoptosis in human myeloma cell lines. Blood. 1999;93:1658–1667. [PMC free article] [PubMed] [Google Scholar]
- Fielding AK, Richards SM, Chopra R, Lazarus HM, Litzow MR, Buck G, Durrant IJ, Luger SM, Marks DI, Franklin IM, McMillan AK, Tallman MS, Rowe JM, Goldstone AH. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood. 2007;109:944–50. doi: 10.1182/blood-2006-05-018192. [DOI] [PubMed] [Google Scholar]
- Juarez J, Bradstock KF, Gottlieb DJ, Bendall LJ. Effects of inhibitors of the chemokine receptor CXCR4 on acute lymphoblastic leukemia cells in vitro. Leukemia. 2003;17:1294–1300. doi: 10.1038/sj.leu.2402998. [DOI] [PubMed] [Google Scholar]
- Juarez J, Dela Pena A, Baraz R, Hewson J, Khoo M, Cisterne A, Fricker S, Fujii N, Bradstock KF, Bendall LJ. CXCR4 antagonists mobilize childhood acute lymphoblastic leukemia cells into the peripheral blood and inhibit engraftment. Leukemia. 2007;21:1249–1257. doi: 10.1038/sj.leu.2404684. [DOI] [PubMed] [Google Scholar]
- Konoplev S, Jorgensen JL, Thomas DA, Lin E, Burger J, Kantarjian HM, Andreeff M, Medeiros LJ, Konopleva M. Phosphorylated CXCR4 is associated with poor survival in adults with B-acute lymphoblastic leukemia. Cancer. 2011;117:4689–4695. doi: 10.1002/cncr.26113. [DOI] [PubMed] [Google Scholar]
- Kuhne MR, Mulvey T, Belanger B, Chen S, Pan C, Chong C, Cao F, Niekro W, Kempe T, Henning KA, Cohen LJ, Korman AJ, Cardarelli PM. BMS-936564/MDX-1338: a fully human anti-CXCR4 antibody induces apoptosis in vitro and shows antitumor activity in vivo in hematologic malignancies. Clin Cancer Res. 2013;19:357–366. doi: 10.1158/1078-0432.CCR-12-2333. [DOI] [PubMed] [Google Scholar]
- Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe A, Coustan-Smith E, Behm FG, Raimondi SC, Campana D. Bone marrow-derived stromal cells prevent apoptotic cell death in B-lineage acute lymphoblastic leukemia. Blood. 1992;79:2370–2377. [PubMed] [Google Scholar]
- de Melo RC, Longhini AL, Bigarella CL, Baratti MO, Traina F, Favaro P, de Melo Campos P, Saad ST. CXCR7 is highly expressed in acute lymphoblastic leukemia and potentiates CXCR4 response to CXCL12. PLoS One. 2014;9:e85926. doi: 10.1371/journal.pone.0085926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SL, Xu J, Kochanek KD. Deaths: final data for 2010. Natl Vital Stat Rep. 2013;61:1–117. [PubMed] [Google Scholar]
- Nagasawa T, Nakajima T, Tachibana K, Iizasa H, Bleul CC, Yoshie O, Matsushima K, Yoshida N, Springer TA, Kishimoto T. Molecular cloning and characterization of a murine pre-B-cell growth-stimulating factor/stromal cell-derived factor 1 receptor, a murine homolog of the human immunodeficiency virus 1 entry coreceptor fusin. Proc Natl Acad Sci USA. 1996a;93:14726–14729. doi: 10.1073/pnas.93.25.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S-i, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. doi: 10.1038/382635a0. 996b. [DOI] [PubMed] [Google Scholar]
- Naumann U, Cameroni E, Pruenster M, Mahabaleshwar H, Raz E, Zerwes HG, Rot A, Thelen M. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5:e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, Prior JL, Piwnica-Worms D, Bridger G, Ley TJ, DiPersio JF. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113:6206–6214. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T, Nagler A, Ben-Hur H, Many A, Shultz L, Lider O, Alon R, Zipori D, Lapidot T. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- Pitt LA, Tikhonova AN, Hu H, Trimarchi T, King B, Gong Y, Sanchez-Martin M, Tsirigos A, Littman DR, Ferrando AA, Morrison SJ, Fooksman DR, Aifantis I, Schwab SR. CXCL12-Producing Vascular Endothelial Niches Control Acute T Cell Leukemia Maintenance. Cancer Cell. 2015;27:755–768. doi: 10.1016/j.ccell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- Sison EA, Rau RE, McIntyre E, Li L, Small D, Brown P. MLL-rearranged acute lymphoblastic leukaemia stem cell interactions with bone marrow stroma promote survival and therapeutic resistance that can be overcome with CXCR4 antagonism. Br J Haematol. 2013;160:785–797. doi: 10.1111/bjh.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sison EAR, Magoon D, Li L, Annesley CE, Rau RE, Small D, Brown P. Plerixafor as a chemosensitizing agent in pediatric acute lymphoblastic leukemia: efficacy and potential mechanisms of resistance to CXCR4 inhibition. Oncotarget. 2014;5:8947. doi: 10.18632/oncotarget.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent JO, Wang ZX, Murray JL, Shao W, Tamamura H, Fujii N, Peiper SC. Lipid bilayer simulations of CXCR4 with inverse agonists and weak partial agonists. J Biol Chem. 2003;278:47136–47144. doi: 10.1074/jbc.M307850200. [DOI] [PubMed] [Google Scholar]
- van den Berk LC, van der Veer A, Willemse ME, Theeuwes MJ, Luijendijk MW, Tong WH, van der Sluis IM, Pieters R, den Boer ML. Disturbed CXCR4/CXCL12 axis in paediatric precursor B-cell acute lymphoblastic leukaemia. Br J Haematol. 2014;166:240–249. doi: 10.1111/bjh.12883. [DOI] [PubMed] [Google Scholar]
- Zhang WB, Navenot JM, Haribabu B, Tamamura H, Hiramatu K, Omagari A, Pei G, Manfredi JP, Fujii N, Broach JR, Peiper SC. A point mutation that confers constitutive activity to CXCR4 reveals that T140 is an inverse agonist and that AMD3100 and ALX40-4C are weak partial agonists. J Biol Chem. 2002;277:24515–24521. doi: 10.1074/jbc.M200889200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.