Abstract
Nontypeable Haemophilus influenzae (NTHI) utilizes the Type IV pilus (Tfp) to adhere to respiratory tract epithelial cells thus colonizing its human host; however the host cell receptor to which this adhesive protein binds is unknown. From a panel of receptors engaged by Tfp expressed by other bacterial species, we showed that the majority subunit of NTHI Tfp, PilA, bound to intercellular adhesion molecule 1 (ICAM1) and that this interaction was both specific and of high affinity. Further, Tfp-expressing NTHI inoculated on to polarized respiratory tract epithelial cells that expressed ICAM1 were significantly more adherent compared to Tfp-deficient NTHI or NTHI inoculated on to epithelial cells to which ICAM1 gene expression was silenced. Moreover, pre-incubation of epithelial cells with recombinant soluble PilA (rsPilA) blocked adherence of NTHI, an outcome that was abrogated by admixing rsPilA with ICAM1 prior to application on to the target cells. Epithelial cells infected with adenovirus or respiratory syncytial virus showed increased expression of ICAM1, this outcome supported augmented adherence of Tfp-expressing NTHI. Collectively, these data revealed the cognate receptor for NTHI Tfp as ICAM1 and promote continued development of a Tfp-targeted vaccine for NTHI-induced diseases of the airway wherein upper respiratory tract viruses play a key predisposing role.
Introduction
Nontypeable Haemophilus influenzae (NTHI) is a common commensal of the human nasopharynx. NTHI also predominates as the causative agent of diseases of the upper and lower respiratory tracts of children and adults, including conjunctivitis, otitis media (OM), sinusitis, bronchitis and exacerbations of both cystic fibrosis and chronic obstructive pulmonary disease (Murphy, 2003, Sethi et al., 2008, Murphy et al., 2009b). Adherence to the mucus and epithelial cells that comprise the respiratory tract mucosa is critical for NTHI to colonize its human host; the necessary first step in the disease process. To accomplish this, the bacterium utilizes multiple adhesive proteins, including lipooligosaccharide and a number of surface-expressed proteins, including the Type IV pilus (Tfp). Expression of Tfp contributes significantly to the ability of NTHI to adhere to respiratory tract epithelial cells, exhibit motility via twitching and form biofilms, structures which further the recurrence and chronicity of NTHI-induced diseases (Bakaletz et al., 2005, Jurcisek et al., 2007, Carruthers et al., 2012, Novotny et al., 2015). Interestingly, dispersal of NTHI from a pre-formed biofilm in vitro upon exposure to antibodies directed against Tfp requires expression of both the majority subunit of this protein, PilA as well as LuxS, suggesting that both twitching motility and quorum signaling are integral to this dispersal (Novotny et al., 2015). Moreover, expression of Tfp is important for NTHI to exhibit competence, and the presence of each gene in the pil and com operons is required for uptake of exogenous DNA (Carruthers et al., 2012). In vivo, Tfp promote long-term colonization of the nasopharynx and robust biofilm formation within the middle ear in chinchilla models of experimental NTHI-induced OM (Jurcisek et al., 2007). Targeting Tfp by immunization with a recombinant protein designed to mimic PilA, the majority subunit of this protein, called ‘rsPilA’ for recombinant, soluble form of PilA (Novotny et al., 2009), admixed with the potent adjuvant LT(R192G/L211A) (Norton et al., 2011) induces the formation of antibodies that eradicate NTHI from middle ear fluids and mucosal biofilms when administered as a traditional preventative vaccine via a transcutaneous immunization strategy, yet also induces rapid resolution of disease with eradication of pre-established NTHI biofilms from the chinchilla middle ear when delivered in a therapeutic immunization regimen (Novotny et al., 2011).
As NTHI Tfp are linked to multiple key NTHI biological functions, pinpointing the cognate receptor for this adhesive protein is essential to gain a complete understanding of this important virulence factor and vaccine candidate. The receptors for several well-described adhesive factors expressed by NTHI are known and include host cellular proteoglycans which are utilized by the NTHI high molecular weight proteins (HMW) 1 and 2 (Noel et al., 1994), laminin and vitronectin are targeted by NTHI Protein E (Hallstrom et al., 2011) and laminin is bound by NTHI Protein F (Jalalvand et al., 2013), carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) is the receptor for NTHI outer membrane proteins P5 and P1 (Hill et al., 2001, Avadhanula et al., 2006b, Bookwalter et al., 2008, Tchoupa et al., 2015), intercellular adhesion molecule 1 (ICAM1) is the receptor for NTHI outer membrane protein P5 and platelet activating factor receptor (PAFr) is bound by NTHI lipooligosaccharide (Swords et al., 2000, Swords et al., 2001). However, the host cell receptor(s) for NTHI Tfp is not known. Therefore, this work was performed to identify the host cognate receptor for this critical NTHI virulence factor.
Herein, we demonstrated that ICAM1, a transmembrane glycoprotein expressed at the surface of many eukaryotic cell types, served as a primary cognate receptor for NTHI Tfp. These data also demonstrated a potential mechanism for the noted induction and/or exacerbation of NTHI-induced diseases of the airway following upper respiratory tract viral infection due to virus-induced increased expression of ICAM1 with concomitant increased adherence of NTHI.
Results
rsPilA bound to intercellular adhesion molecule 1 (ICAM1)
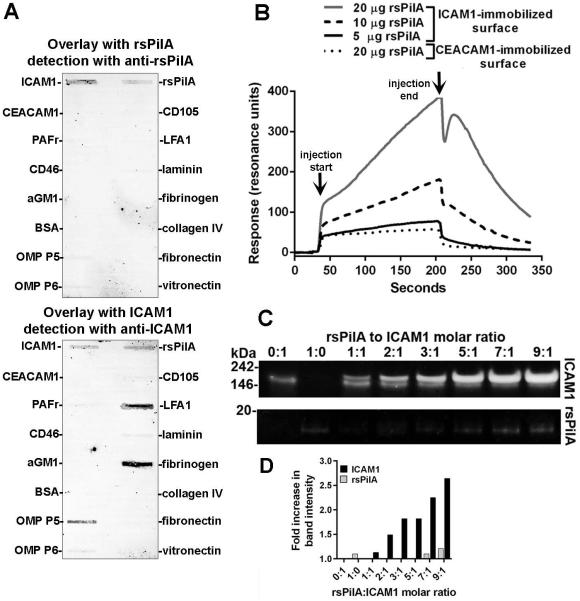
To identify the host cell cognate receptor for NTHI Tfp, recombinant proteins that represented candidate epithelial cell-surface receptors, in addition to several extracellular matrix (ECM) proteins were screened by far-Western slot blot. Potential receptors included those engaged by Tfp expressed by the respiratory tract pathogens Neisseria meningitidis (CD46) (Kallstrom et al., 1997) and Pseudomonas aeruginosa (asialo-GM1) (Craig et al., 2004); host receptors utilized by other known NTHI adhesins (ICAM1, CEACAM1, PAFr); and as a negative control, a protein expressed by endothelial cells (CD105) to which NTHI do not normally either have access to or adhere. Additionally, ECM proteins which NTHI could encounter at the epithelial cell surface and/or are bound by other NTHI outer membrane proteins were assayed including laminin, fibronectin, collagen IV, fibrinogen and vitronectin (Fink et al., 2002, Hallstrom et al., 2011, Jalalvand et al., 2013, Su et al., 2015). The purified recombinant protein rsPilA (for recombinant soluble PilA), which mimics PilA, the majority subunit of NTHI Tfp (Bakaletz et al.) , served as a surrogate for native pilin protein (Novotny et al., 2009). Incubation of rsPilA with host receptors and ECM proteins adsorbed on to PVDF membrane, then probed with rabbit anti-rsPilA yielded a band associated with ICAM1 [Fig. 1A, top membrane] and rsPilA previously adsorbed on to the PVDF membrane. An additional membrane was incubated with ICAM1 [Fig. 1A, bottom membrane] and showed a positive result for the converse interaction, that ICAM1 bound to adsorbed rsPilA. Moreover, as a positive control, ICAM1 interacted with LFA1 (Marlin et al., 1987), fibrinogen (Tsakadze et al., 2002) and NTHI OMP P5 (Avadhanula et al., 2006b), known binding partners for this molecule and thus validated this experimental procedure. Therefore of the panel of recombinant host receptors and ECM proteins tested, ICAM1 was identified as a primary receptor candidate for the NTHI Tfp; therefore we focused our efforts on further examination of this specific interaction.
Figure 1.
rsPilA bound to ICAM1. (A) Far Western slot blot wherein incubation of rsPilA (top blot) or ICAM1 (bottom blot) with a panel of host cell receptors and extracellular matrix proteins adsorbed on to PVDF membranes revealed reactivity of rsPilA exclusively to ICAM1. Conversely, ICAM1 interacted with rsPilA, in addition to its known binding partners LFA1, NTHI OMP P5 and fibrinogen. (B) Sensorgram curves for surface plasmon resonance wherein ICAM1 immobilized to the surface of a sensor chip was exposed to rsPilA demonstrated a concentration-dependent increase in reactivity, a response not observed when rsPilA (at the greatest concentration) was assayed versus a CEACAM1-immobilized control surface. Arrows indicate start and stop of the injection cycle. (C) An increase in ICAM1 band intensity was shown by native PAGE upon incubation of ICAM1 with increasing molar ratios of rsPilA, a result quantitated in (D). Collectively, these data demonstrated that rsPilA bound to ICAM1.
We first used surface plasmon resonance to confirm the interaction between rsPilA and ICAM1 and to also determine the kinetics of the interaction between rsPilA in solution and recombinant ICAM1 immobilized to the sensor chip surface. Injection of increasing concentrations of rsPilA over the sensor chip showed a dose-dependent increase in association to ICAM1 [Fig. 1B]. From these data, the KD was calculated to be 70 nM, indicating an interaction of high affinity. This response was not duplicated by injection of rsPilA across a surface to which another putative receptor (CEACAM1) was immobilized, which confirmed the specificity of the interaction between rsPilA and ICAM1 and also complemented the results obtained by far-Western slot blot. Thus, rsPilA exhibited specific binding to, and a strong affinity for, ICAM1.
Next, the interaction between rsPilA and ICAM1 was examined under conditions where both proteins were admixed in solution so as to not restrict access to potential binding sites on either protein by immobilization on to a substrate. Samples consisting of an increasing molar ratio of rsPilA and a constant amount of ICAM1 were admixed and examined by native PAGE so as not to disrupt potential protein-protein binding. Similar to the results shown by surface plasmon resonance, rsPilA interacted with ICAM1, shown by an increase in band intensity and decrease in electrophoretic mobility compared to a sample containing only ICAM1 [Fig. 1C, top row], a result validated by quantitative densitometry [Fig. 1D, black bars]. Of note, two distinct protein bands were observed in samples that contained rsPilA at 1, 2, 3 or 5:1 molar ratios with ICAM1, which indicated the presence of both an ICAM1-rsPilA complex at ~205 kDa and free ICAM1 at ~190 kDa. This ~14 kDa increase in molecular mass correlated with the mass of rsPilA and therefore indicated that rsPilA bound to ICAM1. This premise was supported by the absence of an rsPilA protein band within the same samples [Fig. 1C, bottom row & Fig. 1D, absence of grey bars]. However, with further increase of rsPilA to 7:1 and 9:1 molar ratio with ICAM1, saturation of binding site(s) on ICAM1 was achieved, as a single band that represented the rsPilA-ICAM1 complex was visualized. Moreover, these data were synchronous with the appearance of an rsPilA protein band at 14 kDa which indicated an excess of rsPilA in the solution [Fig. 1C, bottom row & Fig. 1D, grey bars]. Thus, these data suggested that a molar excess of rsPilA was observed to interact with ICAM1 in solution. Collectively, this set of experiments revealed ICAM1 as a primary ligand for the NTHI Tfp surrogate, rsPilA, and by inference Tfp. This interaction was specific and of high affinity.
Demonstration that ICAM1 is expressed by polarized primary normal human bronchial epithelial cells
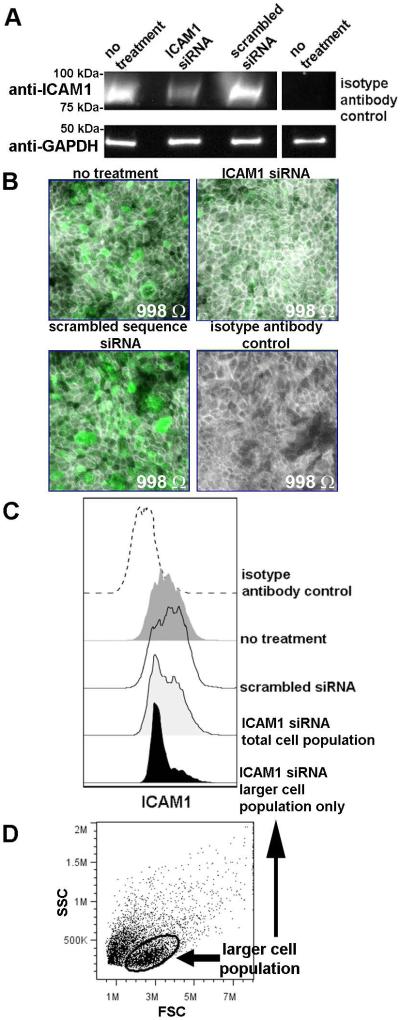
Whereas results with purified proteins identified ICAM1 as a receptor for rsPilA, we wanted to confirm these findings using ICAM1 expressed natively by respiratory tract epithelial cells. To do so, primary normal human bronchial epithelial cells (NHBEs) were established and cultured in vitro at an air-liquid interface to mimic the airway. Generation of polarized and differentiated cell cultures in this manner yields a pseudo-stratified tall respiratory epithelium, including basal cells, ciliated cells and mucus-producing goblet cells (Fulcher et al., 2005). We first demonstrated the presence of ICAM1 within NHBE cell lysates by SDS-PAGE and Western blot [Fig. 2A, top row, first column]. Therefore, ICAM1 was expressed by polarized NHBEs, as anticipated. In an effort to block expression of ICAM1, polarized cultures were transduced with ICAM1 siRNA. By Western blot, a 60% reduction in band intensity was detected, relative to cells transduced with a scrambled sequence siRNA or untreated NHBEs [Fig. 2A, top row, columns 1-3]. We also visualized native ICAM1 expression on intact cultures by fluorescent microscopy and observed robust labeling for this molecule [Fig. 2B, top left panel, green fluorescence]. Moreover, the ICAM1-specific fluorescent signal was reduced upon transduction of the epithelial cells with ICAM1 siRNA [Fig. 2B, top right panel]; a result not observed after incubation with scrambled sequence siRNA [Fig. 2B bottom left panel]. These data complemented our results shown by Western blot. As ICAM1 is important in cell-cell interactions, and disruption of these contacts could negatively influence the stability of the cultures and induce artifacts into the ensuing adherence assays, we stained cellular F-actin to examine the architecture of the cells in culture and additionally measured the transepithelial resistance to ascertain the integrity of the cellular junctions. Microscopy revealed cells that appeared to have tight interactions [Fig. 2B, F-actin labeling pseudocolored grey], an observation confirmed by comparable transepithelial resistance measurements among the untreated and siRNA-treated NHBEs [Fig. 2B, embedded values].
Figure 2.
Polarized NHBEs expressed ICAM1. (A) SDS-PAGE and western blot analysis for detection of ICAM1 in polarized NHBE cell lysates. Epithelial cells transfected with ICAM1 siRNA showed a reduction in band intensity, indicating a reduction in ICAM1 protein expression, a result not observed by treatment of cells with a scrambled sequence siRNA. Equivalent protein concentration per sample was indicated by probing for GAPDH. (B) Immunofluorescent labeling for native ICAM1 demonstrated robust expression of this molecule by polarized NHBEs in culture (green fluorescence) and NHBEs incubated with scrambled sequence siRNA, whereas epithelial cells transfected with ICAM1 siRNA had reduced ICAM1-specific fluorescence. Treatment with ICAM1 siRNA did not disrupt the polarized structure of the NHBE cultures, as shown by staining of F-actin (pseudocolored grey) and by the comparable transepithelial resistance measurements (values embedded within each image). (C) Flow cytometry histograms depicted expression of ICAM1 by intact NHBEs collected from polarized cultures. As ICAM1 expression was not completely silenced upon transfection with ICAM1 siRNA also shown by Western blotting and confocal microscopy, the population of cells that exhibited greater size by forward scatter (panel D, encircled population), and which likely represented the differentiated and apically positioned cells within the polarized cell culture, was further examined. These larger-sized cells showed a reduction in ICAM1 expression compared to untreated NHBEs or cells transfected with scrambled sequence siRNA. Therefore, ICAM1 was expressed by polarized NHBEs, the expression of which was reduced in the population of cells most likely exposed at the apical surface by treatment with ICAM1 siRNA.
As an additional assessment, we also examined ICAM1 expressed by NHBEs by flow cytometry [Fig. 2C, dark grey histogram]. However, in contrast to our microscopy results, by flow cytometry, no overall reduction in ICAM1-specific labeling was shown by the NHBEs treated with ICAM1 siRNA [Fig. 2C, light grey histogram] compared to untreated cells [Fig. 2C, dark grey histogram] or those incubated with scrambled sequence siRNA [Fig. 2C, clear histogram]. The histogram derived from ICAM1-specific siRNA treated cells did however demonstrate the presence of two differentially labeled populations of cells. Thereby, with the knowledge that ICAM1 is constitutively expressed by many cell types and plays an important role in cell-cell interactions, we hypothesized that application of ICAM1 siRNA on to the apical surface of the polarized NHBEs resulted in transduction of siRNA to only the surface-exposed cells within the multi-layered cell culture. To confirm this, scatter plots were examined which depicted two populations of cells based on forward scatter profile [Fig. 2D]. A gate was drawn to select the population that represented the larger and likely differentiated apically-situated cells [Fig. 2D, encircled], compared to the smaller basal cells. When only the population of cells within this gate were plotted, there was an 82% reduction in ICAM1-specific mean fluorescent intensity by the larger sized cells [Fig. 2C, black histogram], compared to the total population that included both small (basal) and large (differentiated) sized cells [Fig. 2C, light grey histogram]. This result was similar in trend to the 60% reduction in band intensity observed by Western blot of NHBE cell lysates treated with ICAM1 siRNA (Fig. 2A) and the reduction in ICAM1-specific staining visualized by fluorescent microscopy (Fig. 2B). Therefore, these data demonstrated that ICAM1 was expressed on the surface of polarized NHBEs and thereby would be accessible to binding by NTHI Tfp; moreover, the expression of ICAM1 could be silenced for those cells positioned at the apical surface and to which NTHI would most likely adhere.
NTHI adhered to NHBEs via the interaction between Tfp and ICAM1
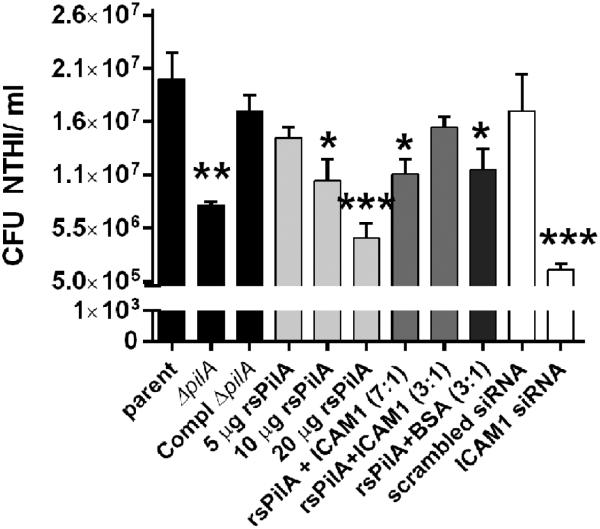
To examine the interaction between Tfp and ICAM1 in their native forms, Tfp-expressing NTHI were inoculated on to the apical surface of polarized NHBE cultures. As we’ve shown previously using submerged monolayer cultures of NHBEs (Jurcisek et al., 2007), compared to the parent strain, NTHI that could not express Tfp were approximately 58% less adherent to the polarized NHBEs assayed herein (P≤ 0.01) [Fig. 3, black bars]. Complementation of this mutation largely restored that ability of this strain to adhere to epithelial cells. Thus, expression of pilA, and by extension, Tfp was important for NTHI to adhere to polarized NHBEs grown at the air-liquid surface.
Figure 3.
NTHI Tfp mediated adherence to polarized epithelial cells via interaction with ICAM1. Compared to the parent strain, the pilA-deficient mutant was significantly less adherent to polarized primary respiratory tract epithelial cells, and adherence was largely restored upon complementation of pilA (black bars). Pre-incubation of NHBEs with rsPilA to block accessibility of NTHI Tfp to ICAM1 resulted in dose-dependent reduction in NTHI adherence (light grey bars). Admixing rsPilA with purified ICAM1 prior to application on to NHBEs mitigated the blockade effect when used at a 3:1 molar ratio (medium grey bars), a result not replicated by incubation of rsPilA with BSA at the same molar ratio (dark grey bars). Whereas transduction of polarized epithelial cells with scrambled sequence siRNA did not influence NTHI adherence, a significant reduction in NTHI adherence to ICAM1 siRNA transfected cells was observed. Results shown as CFU NTHI/ ml and significance relative to the parent strain indicated with asterisks: * P≤ 0.05, ** P≤ 0.01, *** P≤ 0.001.
To identify whether native ICAM1 served as a cognate receptor for NTHI Tfp, blockade of ICAM1 by rsPilA was attempted. Increasing concentrations of rsPilA at 5-, 10- and 20 μg per ml were applied to the apical surface of the NHBEs prior to inoculation with NTHI. A dose-dependent decrease in adherence was observed, and for samples incubated with 10- and 20 μg rsPilA per ml a significant reduction in adherence was shown compared to the parent strain (P≤ 0.05 and P≤ 0.001, respectively) [Fig. 3, light grey bars]. Thus, addition of rsPilA to the NHBE cell surface blocked a receptor utilized by NTHI, thereby limiting adherence. To determine whether the blockade was specific to an rsPilA-ICAM1 interaction, we now pre-incubated rsPilA with recombinant ICAM1 prior to application on to the NHBEs. Based on the observed binding between rsPilA and ICAM1 shown in Fig. 1, we presumed that ICAM1 would sequester rsPilA and thus limit rsPilA-specific blockade of native ICAM1. Admixing rsPilA and ICAM1 at a molar ratio of 7:1 resulted in a significant 46% reduction in adherence of NTHI, compared to adherence of NTHI to untreated NHBEs (P≤ 0.01). However, no significant difference was observed between NTHI inoculated on to NHBEs pre-incubated with a 3:1 molar ratio of rsPilA to ICAM1 compared to untreated cells [Fig. 3, medium grey bars]. These results are explained by the data shown by native PAGE in Fig. 1C, as an excess of rsPilA remained in solution when admixed with ICAM1 at a 7:1 molar ratio, whereas rsPilA was adsorbed out of solution when incubated at a 3:1 molar ratio. Therefore, admixing rsPilA and ICAM1 at a 7:1 ratio provided an excess of rsPilA available to bind to native ICAM1 on the NHBE cell surface thus block adherence of NTHI. Conversely, the solution consisting of a 3:1 molar ratio of rsPilA to ICAM1did not contain free rsPilA and as a result, native ICAM1 remained accessible for NTHI to bind via its Tfp. We confirmed the specificity of these observations by incubation of rsPilA with nonspecific BSA at a 3:1 molar ratio and observed a significant reduction in NTHI adherence (P≤ 0.05), as BSA did not interact and thus sequester rsPilA as observed by admixing rsPilA and ICAM1 at this molar ratio [Fig. 3 dark grey bar].
We next examined the ability of NTHI to adhere to NHBEs that did not express ICAM1 due to prior transfection with ICAM1 siRNA. Compared to untreated NHBEs, NTHI inoculated on to NHBEs transfected with a scrambled sequence siRNA were comparably adherent to NTHI inoculated on to untreated NHBEs, as anticipated [Fig. 3, white bar]. In contrast, 92% fewer NTHI adhered to polarized NHBEs in which expression of ICAM1 had been silenced (P≤ 0.0001). As it is known that NTHI also utilizes OMP P5 to bind ICAM1 (Avadhanula et al., 2006b), the significantly enhanced reduction in NTHI adherence to NHBEs transfected with ICAM1 siRNA is likely a result of the bacterium unable to engage ICAM1 with either of the critical adhesive proteins, Tfp or OMP P5. Collectively, data from these experiments demonstrated that expression of pilA, and by extension, expression of Tfp was required for robust bacterial adherence to polarized NHBEs grown in a Transwell system, thus extending our earlier observations with submerged monolayer cultures of NHBEs (Jurcisek et al., 2007). Moreover, adherence of Tfp-expressing NTHI was significantly reduced by blockade of natively ICAM1 with rsPilA and also by silencing expression of ICAM1. Taken together, these results revealed that NTHI Tfp engaged ICAM1 to adhere to the polarized primary human respiratory tract epithelial cells.
Two primary viral co-pathogens of NTHI-induced disease mediate increased expression of ICAM1 by NHBEs
As a preceding or concurrent upper respiratory tract viral infection predisposes to NTHI-induced diseases of the airway and is also known to dysregulate expression of multiple epithelial cell surface receptors to which NTHI adhere (Patel et al., 1992, Jiang et al., 1999, Bosch et al., 2013), we next examined the expression of ICAM1 by polarized NHBEs after infection with adenovirus or respiratory syncytial virus (RSV), two predominant viral co-pathogens of NTHI-induced diseases (Murphy et al., 2009a). Polarized NHBEs were inoculated apically with adenovirus serotype 1 or RSV strain A2 at multiplicity of infection (MOI) of 1 or 2 virus particles per epithelial cell and the viability of NHBEs within the cultures was determined after 72 h. NHBEs infected with adenovirus at MOI 1 or 2 remained 90% and 88% viable, respectively, based on exclusion of propidium iodide as assessed by flow cytometry (data not shown). Similarly, cells infected with RSV at MOI 1 or 2 were 89% and 88% viable, respectively. An MOI of 2 was therefore selected for further work with both viruses, as a significant decrease in viability of NHBEs was not observed compared to the lesser MOI and to also maximize the potential for virus-induced altered expression of ICAM1 by NHBEs.
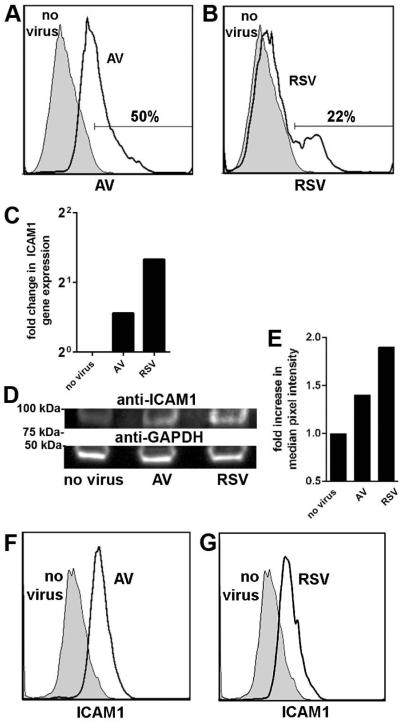
We confirmed viral infection of the polarized NHBEs for each virus and observed positive labeling for adenovirus in 50% of cells [Fig. 4A, clear histogram] and in 22% of cells inoculated with RSV [Fig. 4B, clear histogram], relative to uninfected cells [Figs. 4A & B, shaded histograms]. ICAM1 expression was next examined. By qRT-PCR, an increase in ICAM1-specific gene expression was detected in polarized cells inoculated with either adenovirus (1.5-fold) or RSV (2.0-fold), relative to uninfected cells [Fig. 4C]. This observed increase in ICAM1 expression was also observed at the protein level by SDS-PAGE and Western blot of NHBE cell lysates [Fig. 4D]. Infection of NHBEs with adenovirus showed a 1.4-fold increase in ICAM1-specific band intensity whereas a 1.9-fold increase was detected in lysates of RSV-infected cells compared to uninfected NHBEs, as determined by quantitative densitometry [Fig. 4E]. Examination of intact cells by flow cytometry showed a similar result, as compared to uninfected NHBEs, a 2-fold increase in ICAM1 expression was detected in adenovirus infected cells [Fig. 4F, clear histogram], and a 2.5-fold increase noted by RSV-infected cells [Fig. 4G, clear histogram]. Collectively, these data demonstrated increased expression of ICAM1 by polarized NHBEs after infection with either adenovirus or RSV.
Figure 4.
Inoculation of polarized epithelial cells with RSV or adenovirus resulted in increased expression of ICAM1. Incubation of NHBEs with (A) adenovirus or (B) RSV at MOI 2 for 72 h yielded positive labeling for respective viral antigen (clear histograms), compared to untreated cells (shaded histograms) as determined by flow cytometry. The proportion of cells that expressed viral antigen is indicated. (C) Fold change in ICAM1 gene expression normalized to GAPDH gene expression and relative to uninfected cells revealed a relative increase in ICAM1 transcript abundance by virus-infected NHBEs. (D) SDS-PAGE and western blot analysis of cell lysates probed for ICAM1 demonstrated an increase in band intensity after viral infection compared to uninfected cultures and (E) was quantitated by densitometry. Equivalent protein concentration per sample was indicated by probing for GAPDH (D, bottom row). ICAM1 expression increased on NHBEs infected with adenovirus (F, clear histogram) or RSV (G, clear histogram) compared to uninfected cells (F & G, shaded histograms).
Adherence of NTHI to virus-infected NHBEs
With the knowledge that NTHI that expressed PilA, and by extension Tfp, interacted with the epithelial cell surface receptor ICAM1 (see Fig. 3) and that infection with common respiratory viruses augmented expression of ICAM1 by NHBEs (see Fig. 4), we hypothesized that the greater availability of this host cell receptor would facilitate enhanced adherence of NTHI to virus-infected NHBEs compared to uninfected cells. We therefore inoculated NHBEs with adenovirus or RSV, followed 72 h later by NTHI. As postulated, significantly more NTHI adhered to NHBEs infected with adenovirus or RSV, compared to uninfected epithelial cells (P≤ 0.05) [Fig. 5, black bars]. In contrast, no variation in adherence was noted for the NTHI pilA mutant, regardless of viral infection [Fig. 5, clear bars]. Complementation of the pilA mutant largely restored the observed significantly greater adherence of NTHI to virus-infected NHBEs compared to uninfected cells (P≤ 0.05) [Fig. 5, grey bars]. Therefore, augmented availability of ICAM1 yielded a greater bacterial burden at the epithelial cell surface, particularly by NTHI that expressed the majority subunit of NTHI Tfp.
Figure 5.
Adherence of Tfp-expressing NTHI was greater to epithelial cells infected with adenovirus or RSV compared to uninfected cells. NTHI parent (black bars), pilA-deficient mutant (white bars) and complemented pilA mutant (grey bars) were inoculated on to uninfected NHBEs (no virus) or cells infected with adenovirus or RSV prior to bacterial challenge. Results show the CFU of each NTHI strain and significance relative to respective uninfected NHBEs indicated with asterisks: * P≤ 0.05.
Discussion
Adherence of NTHI within the human nasopharynx is critical for colonization and is the first step in induction of any disease caused by this heterogeneous microorganism. As such, NTHI express multiple surface-exposed proteins and lipooligosaccharide in order to establish a foothold within this anatomical niche. Tfp play many important roles in the biology and pathobiology of NTHI, including mediating adherence to respiratory tract epithelial cells and are the sole mechanism for NTHI to exhibit twitching motility (Bakaletz et al., 2005, Jurcisek et al., 2007, Novotny et al., 2015). Expression of pilA, which encodes the majority subunit of NTHI Tfp, is required for robust biofilm formation in vitro and in vivo (Jurcisek et al., 2007). Moreover, expression of Tfp is involved in dispersal of NTHI from established biofilms in vitro (Novotny et al., 2015); these communities of bacteria are known to contribute to the pathogenesis and chronic nature of diseases caused by this bacterium. Due to its established importance in these functions, NTHI Tfp, and specifically PilA, is under active assessment as a vaccine candidate (Novotny et al., 2009, Novotny et al., 2011, Murphy, 2015, Novotny et al., 2015).
Tfp expressed by Neisseria and Pseudomonas species are well-characterized and are comprised of a majority subunit protein (PilA and PilE, respectively) and a tip adhesin protein (PilC1 and PilC2 for Neisseria and PilY1 for Pseudomonas) (Giltner et al., 2012). NTHI Tfp are similarly comprised of a majority subunit protein, PilA, that demonstrates 87-100% amino acid sequence identity among NTHI strains examined (Bakaletz et al., 2005). Interrogation of the genome for NTHI, however, has not identified a gene with similarity to known Tfp tip adhesin proteins expressed by other bacterial pathogens. However, this observation is not unprecedented, as examination of the genome for Moraxella catarrhalis, which expresses a Tfp and resides within the same anatomical niche as NTHI, similarly has not yet identified a Tfp tip adhesin (Luke et al., 2004). We speculate that as a strictly human-adapted bacterium with a relatively small 1.9 M base pair genome (Harrison et al., 2005), NTHI has adapted to bind to receptors available within the human respiratory tract using PilA as the adhesive component. This type of interaction is an additional mechanism of adherence reported for Tfp expressed by Neisseria and Pseudomonas (Lee et al., 1994, Scheuerpflug et al., 1999).
We therefore utilized rsPilA as a purified surrogate protein for NTHI Tfp and screened a panel of potential cognate receptors and ECM proteins known to be utilized by either Tfp expressed by other respiratory tract pathogens or those engaged by several additional NTHI adhesins. Of the potential receptors tested, rsPilA interacted exclusively with ICAM1 and vice versa. Importantly, and by multiple assays, whether the recombinant proteins were immobilized to a substrate or mixed in solution, a specific interaction was demonstrated between ICAM1 and rsPilA.
We next wanted to assess the interaction between native proteins (e.g. ICAM1 expressed by epithelial cells and Tfp expressed by NTHI). We have previously shown that pilA-deficient NTHI are significantly less adherent to NHBEs cultured as a submerged monolayer (Jurcisek et al., 2007, Carruthers et al., 2012). To extend these findings, herein we inoculated NTHI on to differentiated NHBEs to more closely resemble an intact respiratory epithelium. To achieve this goal, we first confirmed that polarized NHBEs cultured at an air-liquid interface expressed ICAM1. Using these differentiated cells, we showed that an NTHI pilA mutant, that could not express Tfp, was significantly less adherent to polarized NHBEs, compared to the parent and pilA-complemented strains. These data demonstrated the importance of expression of PilA, and by inference Tfp, for NTHI to adhere to these host cells. Pre-incubation of NHBEs with rsPilA blocked adherence of NTHI, an effect that was abrogated if rsPilA was admixed with ICAM1 before adding to NHBE cultures. Thus, robust adherence of Tfp-expressing NTHI required access to ICAM1. Lastly, we silenced ICAM1 gene expression. As a result, NTHI were significantly less able to adhere to NHBEs. Therefore, expression of ICAM1 was necessary to facilitate Tfp-mediated adherence to differentiated NHBEs. Collectively, these data demonstrated that ICAM1 serves as a primary cognate receptor for NTHI Tfp.
ICAM1 (or CD54) is a transmembrane glycoprotein that is a member of the immunoglobulin superfamily and possesses an extracellular region that is arranged in five immunoglobulin-like domains (Bella et al., 1998). ICAM1 is expressed by multiple cells types, including epithelial cells, where it plays an important role in cell-to-cell interactions. ICAM1 is, however, also exploited by multiple pathogens as a receptor for adherence, such as rhinovirus (Staunton et al., 1989, Bella et al., 2000), RSV (Behera et al., 2001), Plasmodium falciparum (Chakravorty et al., 2005) and Porphyromonas gingivalis (Tamai et al., 2005, Kato et al., 2014). For NTHI, in addition to serving as a cognate receptor for Tfp, ICAM1 is one of two receptors engaged by outer membrane protein P5 (Avadhanula et al., 2006b). Redundancy in bacterial species, including NTHI as it possesses only a 1.9 M base pair genome (Harrison et al., 2005), is common to support critical functions such as adherence and acquisition of nutrients. Targeting a single host receptor for adherence via two critical adhesive proteins may be an adaptation by NTHI to provide a selective advantage, particularly given the increased expression of ICAM1 in response to respiratory tract virus infection (Roebuck et al., 1999, Gao et al., 2000, Arnold et al., 2005, Avadhanula et al., 2006a). Moreover, as the predicted amino acid sequences for PilA expressed by multiple clinical isolates of NTHI show ≥87% amino acid identity (Bakaletz et al., 2005), we anticipate that interaction between ICAM1 and Tfp expressed by other NTHI strains will be similar.
As this work demonstrated, ICAM1 expression increased after inoculation of NHBEs with two common respiratory tract viruses, adenovirus and RSV, both known co-pathogens in NTHI-induced respiratory tract disease (Ruohola et al., 2006, Murphy et al., 2009a). As a consequence, a greater number of Tfp-expressing NTHI bound to this cognate receptor. An increase in availability of ICAM1 in combination with immune suppressive effects associated with viral infection, may ultimately provide NTHI with the ideal microenvironment to multiply unrestricted and disseminate beyond the nasopharyngeal colonization site, resulting in disease. Thus, development of a means to block or abrogate NTHI colonization of the nasopharynx under normal conditions, and importantly, limit unrestricted growth and seeding of other niches within the upper and lower respiratory tracts under conditions of viral co-infection is essential. These data therefore provide significant support to the continued development of an NTHI Tfp-derived vaccine.
Experimental procedures
Bacterial strains
Nontypeable Haemophilus influenzae strain 86-028NP was isolated from the nasopharynx of a child undergoing tympanostomy and tube insertion due to chronic OM (Sirakova et al., 1994, Harrison et al., 2005). We deleted pilA to generate NTHI 86-028NP ΔpilA (Carruthers et al., 2012) and complementation of pilA on a plasmid resulted in strain NTHI 86-028NP Δpil/pPIL1 (Carruthers et al., 2012). The parent and ΔpilA strains were cultured on chocolate agar whereas the complemented ΔpilA variant was cultured on chocolate agar supplemented with 200 μg spectinomycin ml−1.
Culture of primary human respiratory tract epithelial cells
Normal human bronchial epithelial cells (Lonza) were cultured to 80% confluency in T-75 flasks (MIDSCI) in B-ALI growth medium (Lonza) prior to seeding onto collagen type IV (Sigma) coated 24-well Transwell membrane inserts, 0.4 μm pore size (Costar). After 2 days, the apical medium was removed and B-ALI differentiation medium (Lonza) was added exclusively to the basolateral chamber. Cells were cultured at the air-liquid interface for a minimum of 5 weeks prior to use and demonstrated the presence of functional cilia and production of mucus upon microscopic evaluation (data not shown).
Far Western slot blot
To identify the cognate ligand for NTHI Tfp, we used a recombinant N-terminally truncated soluble form of PilA, called rsPilA, as a surrogate for the native protein (Novotny et al., 2009). PVDF-LF membranes (BioRad) were prepared by immersion in methanol followed by incubation in cold Towbin buffer (25 mM Tris, 192 mM glycine, 20% methanol) then water for 5 min. Two micrograms of the following proteins were adsorbed on to PVDF by vacuum adsorption: recombinant human ICAM1 (R&D Systems), recombinant human CEACAM1 (R&D Systems), PAFr peptide (Biorbyt), human asialo-GM1 (EMD Millipore), human CD46 (Biorbyt), recombinant human CD105 (R&D Systems), laminin derived from human fibroblasts (Sigma-Aldrich), recombinant human vitronectin (Sigma-Aldrich), human placental collagen IV (Sigma-Aldrich), recombinant human vitronectin (Sigma-Aldrich) and human plasma fibrinogen (Sigma-Aldrich), NTHI OMP P6, NTHI OMP P6 and bovine serum albumin (BSA; Sigma-Aldrich). Membranes were then blocked with 5X casein blocking buffer (Sigma-Aldrich) plus 10% normal goat serum (Rockland Immunochemicals) then incubated with 50 μg of rsPilA or ICAM1 ml−1 Tris-buffered saline with 0.05% Tween 20 (TTBS). To detect binding of rsPilA, membranes were subsequently incubated with IgG-enriched rabbit anti-rsPilA (generated at Spring Valley labs); to detect ICAM1, membranes were incubated with mouse anti-ICAM1 (clone G5; Santa Cruz Biotechnology) then detected with goat anti-rabbit IgG-A488 or goat anti-mouse IgG-A488, respectively (Invitrogen) and visualized via FluorChem M imager (Protein Simple).
Native PAGE
To demonstrate binding of rsPilA to ICAM1, native PAGE was performed. In a 25 μl volume, rsPilA was incubated with ICAM1 at molar ratios of 0, 1, 2, 3, 5, 7 and 9:1 (rsPilA:ICAM1) for 2.5 h at 37°C with shaking at 125 rpm. Samples were then mixed with native sample buffer (62.5 mM Tris-HCl at pH 6.8; 25% glycerol, 1% bromophenol blue) and applied to Mini-PROTEAN TGX gels (BioRad) in native running buffer (25 mM Tris, 192 mM glycine). Gels were fixed then stained with SYPRO Ruby Protein Gel stain (Molecular Probes) and visualized via FluorChem M imager. Band intensity and relative protein migration were determined using AlphaView SA software (Protein Simple).
Surface plasmon resonance
The interaction between rsPilA and ICAM1 was examined by surface plasmon resonance using a Biacore3000 instrument (GE Healthcare). All reagents were purchased from GE Healthcare. Flow cells on a CM5 reagent grade sensor chip were activated with a 1:1 mixture of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride and N-hydroxysuccinimide at a flow rate of 5 μl min−1 prior to immobilization of recombinant human ICAM1 suspended in sodium acetate, pH 4.0 to a response level of 5,000 resonance units (RU). HBS-N served as the running buffer. Unreacted sites were deactivated with ethanolamine-HCl, pH 8.5. As a negative control, CEACAM1 was immobilized to an additional flow cell. Multi-cycle kinetic analysis was performed wherein 15 μl of rsPilA at 5-, 10-, and 20 μg ml−1 HBS-N plus NSB Reducer was injected across each flow cell and binding calculated by comparing pre- and post-injection RU values. To determine binding rate kinetics, data were analyzed with BIAevaluation software.
Transfection of NHBEs with siRNA
Knockdown of the ICAM1 gene was mediated by transfection of polarized NHBEs with small interfering RNAs (siRNA). One day prior to transfection, the basolateral medium was replaced and the apical cell surface rinsed with B-ALI differentiation medium without gentamicin or amphotericin-B. To the apical surface, 100 pmol human ICAM1 siRNA or scrambled sequence siRNA (Santa Cruz Biotechnology, Inc.) diluted in transfection medium and transfection reagent was applied and the cells incubated for 24 h prior to use. To confirm integrity of polarized cultures after siRNA treatment, transepithelial resistance of each Transwell was measured in sterile Dulbecco’s phosphate buffered saline (DPBS) using an EVOM epithelial voltohmeter (World Precision Instruments).
Immunofluorescent microscopy
The expression of ICAM1 by NHBEs was revealed by immunofluorescent microscopy. Polarized NHBEs were fixed with 10% neutral buffered formalin then blocked with 10% normal goat serum in DPBS. ICAM1 was detected with mouse monoclonal antibody to ICAM1 (clone 15.2) conjugated to Alexa Fluor 488 (Santa Cruz Biotechnology) diluted in 1% normal goat serum in DPBS. Normal mouse IgG1-A488 served as a negative control. F-actin was stained with phalloidin-Alexa Fluor 594 (Invitrogen) diluted in DPBS plus 1% Tween 20. Transwell membranes were excised and placed specimen side down on to 10 μl ProLong Gold antifade reagent (Invitrogen) and viewed on a Zeiss 510 Meta-laser scanning confocal microscope (Carl Zeiss). Images were rendered with Zeiss Zen software.
SDS-PAGE and Western blotting
To examine relative expression of ICAM1 by polarized NHBEs, Western blotting was performed. The apical and basolateral surfaces of the polarized NHBEs were rinsed twice with DPBS prior to addition of TrypLE Select Enzyme 10X (Life Technologies) and incubation for 15 min at 37°C to release the cells from the transwell membrane. Cells were collected, washed by centrifugation and lysed by incubation in 300 mM NaCl, 50 mM Tris-HCl, pH 7.6 plus 0.5% Triton X-100 for 10 min at 37°C. After centrifugation at 20,000 x g for 10 min at 4°C, supernatants were collected, diluted in solubilizing buffer containing β-mercaptoethanol and applied to wells of Mini-PROTEAN TGX gels (BioRad). After electrophoretic separation in Tris/Glycine/SDS buffer (BioRad), proteins were transferred to nitrocellulose using iBlot dry transfer apparatus (Invitrogen) and blocked with TTBS plus 2% nonfat skim milk. ICAM1 was detected with a monoclonal antibody against ICAM1 (clone G5) or as a negative control, normal mouse IgG2a conjugated to HP (Santa Cruz Biotechnology, Inc.). To confirm that equivalent protein was applied to each well, mouse monoclonal antibody to GAPDH (Ambion) was utilized. Antibody was revealed with goat anti-mouse IgG-HP (Invitrogen) and ECL chemiluminescent substrate (GE Healthcare). Differences in relative ICAM1 protein quantity were determined using AlphaView SA software (Protein Simple).
Flow cytometry
To demonstrate expression of ICAM1 on the surface of polarized NHBEs, flow cytometry was performed. Cells were released from the transwell membrane as described using TrypLE Select Enzyme 10X, fixed in Cytofix buffer (BD Biosciences) and incubated with mouse monoclonal antibody to ICAM1 (clone 15.2) or normal mouse IgG1 conjugated to Alexa Fluor 488 (Santa Cruz Biotechnology, Inc.). A total of 20,000 events were collected using a BD Accuri C6 flow cytometer and data analyzed with FloJo software (FloJo, LLC). Assays were repeated a minimum of three times and representative histograms shown.
Adherence assays
To examine the relative adherence of NTHI to polarized NHBEs, one day prior to assessment, the basolateral medium was replaced and the apical cell surface rinsed with B-ALI differentiation medium without either gentamicin or amphotericin-B. NTHI strains were cultured on chocolate agar supplemented with antibiotic as appropriate for 18-22 h at 37°C, 5% CO2. To induce expression of NTHI Tfp, bacteria were suspended in brain heart infusion broth supplemented with 2 μg each β-NAD and heme ml−1 medium (sBHI), adjusted to an OD490nm of 0.6, then diluted 1:6 in sBHI and incubated static for 3 h at 37°C, 5% CO2 (Bakaletz et al., 2005). NTHI were inoculated at an MOI of 100 bacteria per epithelial cell in a 50 μl volume of B-ALI differentiation medium without antibiotics on to the apical surface of the NHBEs and incubated static for 1 h at 37°C, 5% CO2. The apical surface of NHBEs was then washed to remove non-adherent NTHI, the NHBEs released from the Transwell membrane as described and the NHBE-NTHI suspension serially diluted and plated on to chocolate agar to quantitate the relative number of adherence of NTHI.
To block the adherence of NTHI to NHBEs, prior to bacterial inoculation, rsPilA at 5-, 10- and 20 μg ml−1 in a 50 μl volume of B-ALI differentiation medium without antibiotics was applied to the apical surface of the NHBEs and incubated on a rotating rocker for 1 h. The cell surface was washed twice with DPBS prior to inoculation with NTHI. To mitigate the rsPilA-mediated blockade of NTHI adherence, 2 h prior to bacterial inoculation, rsPilA was incubated with recombinant human ICAM1 at molar ratios of 7:1 and 3:1 or to demonstrate specificity, admixed with the nonspecific protein BSA at a molar ratio of 3:1 for 1 h, and then applied to the surface of NHBEs as described. Each assay was repeated a minimum of three times and data are reported as CFU NTHI per ml.
Viral infection of NHBEs
Adenovirus, serotype 1 is a pediatric clinical isolate (Suzuki et al., 1994) and respiratory syncytial virus strain A2 (RSV) was purchased from American Type Culture Collection. NHBEs were infected apically with RSV or adenovirus at MOI of 1 or 2 viral particles per epithelial cell (based on number of cells exposed at the apical surface) in a 50 μl volume of B-ALI basal medium and incubated for 2 h at 37°C, 5% CO2 on a rotating rocker to allow for virus adsorption. The cells were then washed with medium and incubated static for an additional 72 h. To determine cell viability, NHBEs were released from the Transwell membranes by incubation in TrypLE Select Enzyme 10X, washed in DPBS and assessed for exclusion of propidium iodide by flow cytometry. As cell viability was noted to be ≥88% for each virus regardless of MOI, subsequent experiments were performed with adenovirus and RSV at MOI 2 viral particles per epithelial cell. To confirm viral infectivity, NHBEs were released from the Transwell membranes as described, fixed with Cytofix buffer and incubated with goat anti-adenovirus or goat anti-RSV conjugated to FITC (ViroStat) diluted in Perm/Wash buffer (BD Biosciences) overnight at 4°C. Uninfected cells served as a negative control. Additionally, to demonstrate an increase in ICAM1 expression by virally-infected NHBEs, flow cytometry and Western blotting were performed as described.
Relative quantification of ICAM1 mRNA transcript abundance after viral infection of NHBEs
To determine ICAM1-specific transcript abundance in virus-infected NHBEs relative to uninfected cells, polarized NHBEs in triplicate were lysed with Trizol and RNA isolated using Qiagen RNeasy kit and mini columns. Quantitative RT-PCR was performed using SuperScript III Platinum SYBR Green one-step qRT-PCR kit (Invitrogen) and the following primers for ICAM1 as the gene of interest and GAPDH as a reference gene: ICAM1F 5’-CTGCAGACAGTGACCATC-3’, ICAM1R 5’-GTCCAGTTTCCCGGACAA-3’, GAPDHF 5’-CCTCTGACTTCAACAGCGACA-3’, GAPDHR 5’-TTACTCCTTGGAGGCCATGTG-3’ (Integrated DNA Technologies) (Cheng et al., 2014, Gulraiz et al., 2015). mRNA expression of ICAM1 was normalized to GAPDH levels and fold change in ICAM1 levels between uninfected and virally-infected cells calculated using 2−ΔΔCt method (Livak et al., 2001).
Adherence of NTHI to virus-infected NHBEs
NHBEs were seeded into 96-well plates as described (Jurcisek et al., 2007). After washing the cell surface with B-ALI basal medium, NHBEs were infected apically with RSV or adenovirus at MOI of 2 viral particles per epithelial cell in a 50 μl volume of B-ALI basal medium and incubated for 2 h at 37°C, 5% CO2 on a rotating rocker to allow for virus adsorption. The cells were then washed to remove unbound virus, antibiotic free medium applied and cells were incubated static for an additional 72 h. Two h prior to inoculation with NTHI, B-ALI basal medium containing 1% heat-inactivated fetal bovine serum (Lonza) was applied to wells to limit non-specific interactions. NTHI were prepared to promote Tfp expression as described above and inoculated on to the NHBEs at an MOI of 100 bacteria per epithelial cell. One hour later, cells were released with TrypLE Select Enzyme 10X and the NHBE-NTHI suspension serially diluted and plated on to chocolate agar to quantitate the relative number of adherence of NTHI. Triplicate wells were assayed and each experiment was repeated a minimum of three times. Data are reported as CFU NTHI per ml.
Statistical analyses
To demonstrate differences in adherence of NTHI to polarized epithelial cells, a one way analysis of variance (ANOVA) was performed and comparisons among experimental groups determined with Dunnett’s multiple comparison test using GraphPad Prism software. P-values ≤0.05 were considered statistically significant.
Acknowledgements
This work was funded by NIDCD/NIH R01 003915. We thank Kenneth L. Brockman and Michael O. Ward, Jr. for technical assistance and Ms. Jennifer Neelans for assistance with manuscript preparation and submission.
Footnotes
The authors have no conflicts of interest to declare.
References
- Arnold R, Konig W. Respiratory syncytial virus infection of human lung endothelial cells enhances selectively intercellular adhesion molecule-1 expression. Journal of immunology. 2005;174:7359–7367. doi: 10.4049/jimmunol.174.11.7359. [DOI] [PubMed] [Google Scholar]
- Avadhanula V, Rodriguez CA, Devincenzo JP, Wang Y, Webby RJ, Ulett GC, Adderson EE. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. Journal of virology. 2006a;80:1629–1636. doi: 10.1128/JVI.80.4.1629-1636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avadhanula V, Rodriguez CA, Ulett GC, Bakaletz LO, Adderson EE. Nontypeable Haemophilus influenzae adheres to intercellular adhesion molecule 1 (ICAM-1) on respiratory epithelial cells and upregulates ICAM-1 expression. Infect Immun. 2006b;74:830–838. doi: 10.1128/IAI.74.2.830-838.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakaletz LO, Baker BD, Jurcisek JA, Harrison A, Novotny LA, Bookwalter JE, et al. Demonstration of Type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect Immun. 2005;73:1635–1643. doi: 10.1128/IAI.73.3.1635-1643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera AK, Matsuse H, Kumar M, Kong X, Lockey RF, Mohapatra SS. Blocking intercellular adhesion molecule-1 on human epithelial cells decreases respiratory syncytial virus infection. Biochemical and biophysical research communications. 2001;280:188–195. doi: 10.1006/bbrc.2000.4093. [DOI] [PubMed] [Google Scholar]
- Bella J, Kolatkar PR, Marlor CW, Greve JM, Rossmann MG. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J, Rossmann MG. ICAM-1 receptors and cold viruses. Pharmaceutica acta Helvetiae. 2000;74:291–297. doi: 10.1016/S0031-6865(99)00056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookwalter JE, Jurcisek JA, Gray-Owen SD, Fernandez S, McGillivary G, Bakaletz LO. A carcinoembryonic antigen-related cell adhesion molecule 1 homologue plays a pivotal role in nontypeable Haemophilus influenzae colonization of the chinchilla nasopharynx via the outer membrane protein P5-homologous adhesin. Infect Immun. 2008;76:48–55. doi: 10.1128/IAI.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch AA, Biesbroek G, Trzcinski K, Sanders EA, Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS pathogens. 2013;9:e1003057. doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers MD, Tracy EN, Dickson AC, Ganser KB, Munson RS, Jr., Bakaletz LO. Biological roles of nontypeable Haemophilus influenzae type IV pilus proteins encoded by the pil and com operons. J Bacteriol. 2012;194:1927–1933. doi: 10.1128/JB.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty SJ, Craig A. The role of ICAM-1 in Plasmodium falciparum cytoadherence. European journal of cell biology. 2005;84:15–27. doi: 10.1016/j.ejcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Cheng SE, Lee IT, Lin CC, Hsiao LD, Yang CM. Thrombin induces ICAM-1 expression in human lung epithelial cells via c-Src/PDGFR/PI3K/Akt-dependent NF-kappaB/p300 activation. Clin Sci (Lond) 2014;127:171–183. doi: 10.1042/CS20130676. [DOI] [PubMed] [Google Scholar]
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- Fink DL, Green BA, St Geme JW., 3rd The Haemophilus influenzae Hap autotransporter binds to fibronectin, laminin, and collagen IV. Infect Immun. 2002;70:4902–4907. doi: 10.1128/IAI.70.9.4902-4907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods in molecular medicine. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- Gao J, Choudhary S, Banerjee AK, De BP. Human parainfluenza virus type 3 upregulates ICAM-1 (CD54) expression in a cytokine-independent manner. Gene expression. 2000;9:115–121. doi: 10.3727/000000001783992641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiology and molecular biology reviews : MMBR. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulraiz F, Bellinghausen C, Bruggeman CA, Stassen FR. Haemophilus influenzae increases the susceptibility and inflammatory response of airway epithelial cells to viral infections. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2015;29:849–858. doi: 10.1096/fj.14-254359. [DOI] [PubMed] [Google Scholar]
- Hallstrom T, Singh B, Resman F, Blom AM, Morgelin M, Riesbeck K. Haemophilus influenzae protein E binds to the extracellular matrix by concurrently interacting with laminin and vitronectin. The Journal of infectious diseases. 2011;204:1065–1074. doi: 10.1093/infdis/jir459. [DOI] [PubMed] [Google Scholar]
- Harrison A, Dyer DW, Gillaspy A, Ray WC, Mungur R, Carson MB, et al. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J Bacteriol. 2005;187:4627–4636. doi: 10.1128/JB.187.13.4627-4636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DJ, Toleman MA, Evans DJ, Villullas S, Van Alphen L, Virji M. The variable P5 proteins of typeable and non-typeable Haemophilus influenzae target human CEACAM1. Mol Microbiol. 2001;39:850–862. doi: 10.1046/j.1365-2958.2001.02233.x. [DOI] [PubMed] [Google Scholar]
- Jalalvand F, Su YC, Morgelin M, Brant M, Hallgren O, Westergren-Thorsson G, et al. Haemophilus influenzae protein F mediates binding to laminin and human pulmonary epithelial cells. The Journal of infectious diseases. 2013;207:803–813. doi: 10.1093/infdis/jis754. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Nagata N, Molina E, Bakaletz LO, Hawkins H, Patel JA. Fimbria-mediated enhanced attachment of nontypeable Haemophilus influenzae to respiratory syncytial virus-infected respiratory epithelial cells. Infect Immun. 1999;67:187–192. doi: 10.1128/iai.67.1.187-192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bookwalter JE, Baker BD, Fernandez S, Novotny LA, Munson RS, Jr., Bakaletz LO. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol Microbiol. 2007;65:1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- Kallstrom H, Liszewski MK, Atkinson JP, Jonsson AB. Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol Microbiol. 1997;25:639–647. doi: 10.1046/j.1365-2958.1997.4841857.x. [DOI] [PubMed] [Google Scholar]
- Kato Y, Hagiwara M, Ishihara Y, Isoda R, Sugiura S, Komatsu T, et al. TNF-alpha augmented Porphyromonas gingivalis invasion in human gingival epithelial cells through Rab5 and ICAM-1. BMC microbiology. 2014;14:229. doi: 10.1186/s12866-014-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Sheth HB, Wong WY, Sherburne R, Paranchych W, Hodges RS, et al. The binding of Pseudomonas aeruginosa pili to glycosphingolipids is a tip-associated event involving the C-terminal region of the structural pilin subunit. Mol Microbiol. 1994;11:705–713. doi: 10.1111/j.1365-2958.1994.tb00348.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luke NR, Howlett AJ, Shao J, Campagnari AA. Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation. Infect Immun. 2004;72:6262–6270. doi: 10.1128/IAI.72.11.6262-6270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin SD, Springer TA. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1) Cell. 1987;51:813–819. doi: 10.1016/0092-8674(87)90104-8. [DOI] [PubMed] [Google Scholar]
- Murphy TF. Respiratory infections caused by non-typeable Haemophilus influenzae. Current opinion in infectious diseases. 2003;16:129–134. doi: 10.1097/00001432-200304000-00009. [DOI] [PubMed] [Google Scholar]
- Murphy TF. Vaccines for Nontypeable Haemophilus influenzae: the Future Is Now. Clinical and vaccine immunology : CVI. 2015;22:459–466. doi: 10.1128/CVI.00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TF, Bakaletz LO, Smeesters PR. Microbial interactions in the respiratory tract. The Pediatric infectious disease journal. 2009a;28:S121–126. doi: 10.1097/INF.0b013e3181b6d7ec. [DOI] [PubMed] [Google Scholar]
- Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, et al. Nontypeable Haemophilus influenzae as a pathogen in children. The Pediatric infectious disease journal. 2009b;28:43–48. doi: 10.1097/INF.0b013e318184dba2. [DOI] [PubMed] [Google Scholar]
- Noel GJ, Love DC, Mosser DM. High-molecular-weight proteins of nontypeable Haemophilus influenzae mediate bacterial adhesion to cellular proteoglycans. Infect Immun. 1994;62:4028–4033. doi: 10.1128/iai.62.9.4028-4033.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton EB, Lawson LB, Freytag LC, Clements JD. Characterization of a mutant Escherichia coli heat-labile toxin, LT(R192G/L211A), as a safe and effective oral adjuvant. Clinical and vaccine immunology : CVI. 2011;18:546–551. doi: 10.1128/CVI.00538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, Adams LD, Kang DR, Wiet GJ, Cai X, Sethi S, et al. Epitope mapping immunodominant regions of the PilA protein of nontypeable Haemophilus influenzae (NTHI) to facilitate the design of two novel chimeric vaccine candidates. Vaccine. 2009;28:279–289. doi: 10.1016/j.vaccine.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, Clements JD, Bakaletz LO. Transcutaneous immunization as preventative and therapeutic regimens to protect against experimental otitis media due to nontypeable Haemophilus influenzae. Mucosal immunology. 2011;4:456–467. doi: 10.1038/mi.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, Jurcisek JA, Ward MO, Jr., Jordan ZB, Goodman SD, Bakaletz LO. Antibodies against the majority subunit of Type IV pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS-dependent manner and confer therapeutic resolution of experimental otitis media. Mol Microbiol. 2015 doi: 10.1111/mmi.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J, Faden H, Sharma S, Ogra PL. Effect of respiratory syncytial virus on adherence, colonization and immunity of non-typable Haemophilus influenzae: implications for otitis media. International journal of pediatric otorhinolaryngology. 1992;23:15–23. doi: 10.1016/0165-5876(92)90075-z. [DOI] [PubMed] [Google Scholar]
- Roebuck KA, Finnegan A. Regulation of intercellular adhesion molecule-1 (CD54) gene expression. Journal of leukocyte biology. 1999;66:876–888. doi: 10.1002/jlb.66.6.876. [DOI] [PubMed] [Google Scholar]
- Ruohola A, Meurman O, Nikkari S, Skottman T, Salmi A, Waris M, et al. Microbiology of acute otitis media in children with tympanostomy tubes: prevalences of bacteria and viruses. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2006;43:1417–1422. doi: 10.1086/509332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerpflug I, Rudel T, Ryll R, Pandit J, Meyer TF. Roles of PilC and PilE proteins in pilus-mediated adherence of Neisseria gonorrhoeae and Neisseria meningitidis to human erythrocytes and endothelial and epithelial cells. Infect Immun. 1999;67:834–843. doi: 10.1128/iai.67.2.834-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. The New England journal of medicine. 2008;359:2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- Sirakova T, Kolattukudy PE, Murwin D, Billy J, Leake E, Lim D, et al. Role of fimbriae expressed by nontypeable Haemophilus influenzae in pathogenesis of and protection against otitis media and relatedness of the fimbrin subunit to outer membrane protein A. Infect Immun. 1994;62:2002–2020. doi: 10.1128/iai.62.5.2002-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- Su YC, Mukherjee O, Singh B, Hallgren O, Westergren-Thorsson G, Hood D, Riesbeck K. Haemophilus influenzae P4 Interacts With Extracellular Matrix Proteins Promoting Adhesion and Serum Resistance. The Journal of infectious diseases. 2015 doi: 10.1093/infdis/jiv374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Bakaletz LO. Synergistic effect of adenovirus type 1 and nontypeable Haemophilus influenzae in a chinchilla model of experimental otitis media. Infect Immun. 1994;62:1710–1718. doi: 10.1128/iai.62.5.1710-1718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords WE, Buscher BA, Ver Steeg Ii K, Preston A, Nichols WA, Weiser JN, et al. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol Microbiol. 2000;37:13–27. doi: 10.1046/j.1365-2958.2000.01952.x. [DOI] [PubMed] [Google Scholar]
- Swords WE, Ketterer MR, Shao J, Campbell CA, Weiser JN, Apicella MA. Binding of the non-typeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signalling. Cellular microbiology. 2001;3:525–536. doi: 10.1046/j.1462-5822.2001.00132.x. [DOI] [PubMed] [Google Scholar]
- Tamai R, Asai Y, Ogawa T. Requirement for intercellular adhesion molecule 1 and caveolae in invasion of human oral epithelial cells by Porphyromonas gingivalis. Infect Immun. 2005;73:6290–6298. doi: 10.1128/IAI.73.10.6290-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchoupa AK, Lichtenegger S, Reidl J, Hauck CR. Outer membrane protein P1 is the CEACAM-binding adhesin of Haemophilus influenzae. Mol Microbiol. 2015 doi: 10.1111/mmi.13134. [DOI] [PubMed] [Google Scholar]
- Tsakadze NL, Zhao Z, D'Souza SE. Interactions of intercellular adhesion molecule-1 with fibrinogen. Trends in cardiovascular medicine. 2002;12:101–108. doi: 10.1016/s1050-1738(01)00157-8. [DOI] [PubMed] [Google Scholar]