Abstract
Background & Aims
A genome wide association study (GWAS) of 280 cases identified the hepatic cholesterol transporter ABCG8 as a locus associated with risk for gallstone disease, but findings have not been reported from any other GWAS of this phenotype. We performed a large-scale meta-analysis of GWASs of individuals of European ancestry with available prior genotype data, to identify additional genetic risk factors for gallstone disease.
Methods
We obtained per-allele odds ratio (OR) and standard error estimates using age- and sex-adjusted logistic regression models within each of the 10 discovery studies (8720 cases and 55,152 controls). We performed an inverse variance weighted, fixed-effects meta-analysis of study specific estimates to identify single nucleotide polymorphisms (SNPs) that were independently associated with gallstone disease. Associations were replicated in 6489 cases and 62,797 controls.
Results
We observed independent associations for 2 SNPs at the ABCG8 locus: rs11887534 (OR = 1.69; 95% confidence interval [CI], 1.54–1.86; P=2.44×10−60) and rs4245791 (OR=1.27; P=1.90×10−34). We also identified and/or replicated associations for rs9843304 in TM4SF4 (OR=1.12; 95% CI, 1.08–1.16; P=6.09×10−11), rs2547231 in SULT2A1 (encodes a sulfo-conjugation enzyme that acts on hydroxysteroids and cholesterol-derived sterol bile acids), (OR=1.17, 95% CI, 1.12– 1.21;P=2.24×10−10), rs1260326 in GCKR (encodes a glucokinase regulator) (OR=1.12; 95% CI, 1.07–1.17; P=2.55×10−10), and rs6471717 near CYP7A1 (encodes an enzyme that catalyzes conversion of cholesterol to primary bile acids) (OR=1.11; 95% CI, 1.08–1.15; P=8.84×10−9). Among individuals of African American and Hispanic American ancestry, rs11887534 and rs4245791 were positively associated with gallstone disease risk, while the association for the rs1260326 variant was inverse.
Conclusions
In this large-scale GWAS of gallstone disease, we identified 4 loci in genes that have putative functions in cholesterol metabolism and transport, and sulfonylation of bile acids or hydoxysteroids.
Keywords: genetics, risk factors, SNP, GWAS
Accounting for a substantial clinical burden in the United States, gallstone disease afflicts 6.3 million men and 14.2 million women between the ages of 20–74 years, leading annually to 700,000 cholecystectomies and an economic burden of 6.5 billion dollars.1 It was hypothesized as early as the 1960s that the composition of bile may play an important role in gallstone formation.2 Bile is formed by the transportation of cholesterol, bile acids and other organic molecules such as bilirubin from within the hepatocytes to the biliary canaliculi, and serves as a medium for excretion of lipid soluble products of metabolism. Precipitation of biliary constituents from their soluble state into their insoluble form, initiates the process of gallstone formation. Clinical conditions with chronic hemolytic states such as sickle cell disease have frequently been associated with pigmented gallstones,3 due to the increased delivery of unconjugated bilirubin into the bile via hepatocytes.4 However, the most common (80–90%) constituent of gallstones retrieved during cholecystectomy surgery or autopsy is biliary cholesterol. Studies that compared the constituents of lithogenic bile and normal bile observed that higher concentrations of cholesterol, or the alterations in relative proportions of other bile components such as bile salts and phospholipids can result in supersaturation of cholesterol.2,5 Redinger and Small further demonstrated a correlation between percentage saturation of biliary cholesterol in various ethnic groups and estimated gallstone prevalence rates in the same population in an ecological study. 6 Consequently, several lifestyle determinants such as female gender, greater parity, post-menopausal hormone therapy, Native American ancestry, high body mass index (BMI) and dyslipidemia are among the most important risk factors for gallstone disease, primarily due to their influence on cholesterol concentration in the bile.5,7
Based on familial clustering of gallstone disease, a 2–3 fold elevated risk among first-degree relatives8–10, and heritability estimates of 25–29% from twin studies,10,11 it has been suggested that genetic factors may play an important contributory role in cholelithiasis. More evidence to support this hypothesis was established using experimental crosses of inbred mice strains with varying prevalence of gallstones.12,13 Quantitative trait loci based approaches were utilized to generate a murine gallstone genetic map of several candidate lithogenic (lith) loci,12,14 with the idea that orthologous human LITH genes may be predicted due to homology between human and mouse genomes. These murine lith loci co-localized with about seven “likely”, and about twenty “plausible” candidate genes for gallstone disease, many of which are involved in cholesterol (e.g. ABCG5/ABCG8) and bile acid (e.g. ABCB11) synthesis, transport or metabolism.13
The identification of genetic risk factors of gallstone disease in humans was undertaken in 2007 in a discovery based genome wide association study (GWAS) of 280 cases and 360 controls.15 This study identified and replicated an approximately two-fold increased risk for carriers of the H-allele of D19H in the hepatic cholesterol transporter gene ABCG8 (rs11887534, risk allele frequency ~ 7%).15,16 Other studies that examined genetic associations with gallstone disease were based on biological insights of candidate loci or pathways. Buch et al. 17 investigated the association of known bilirubin loci18 with the incidence of gallstone disease, and observed a recessive mode of inheritance at the UGT1A1 SNP locus rs6742078, finding that carriers of the T/T genotype were predisposed to an increased risk of gallstone disease among men, but not among women.17 Moreover a recent study in women, examining associations of approximately 2000 gene centric loci in known lipid metabolism and obesity pathways,19 reported additional associations for the GCKR SNP rs1260326 and the TTC39B SNP rs686030 with gallstone disease; however these associations were not replicated.
Although there is strong evidence for genetic contribution towards the risk of gallstone disease, there are few replicated susceptibility loci identified from genome-wide, discovery based approaches, due to the limited size and scope of prior studies. In this study, we therefore conducted a large-scale GWAS meta-analysis in individuals with pre-existing genetic data on more than 2 million genetic variants, to discover additional loci associated with the risk of gallstone disease in individuals of European ancestry. We replicated the SNPs within each of the newly discovered loci in independent samples, and queried transcriptomic and metabolomic databases to derive clues about potential causal variants near the SNPs with highest evidence for association with gallstone disease.
MATERIALS AND METHODS
Study Participants
The study population for the discovery set consisted of individuals with extant genome-wide genotyping data available from previous studies, among whom we identified 8720 cases and 55,152 controls within the following 10 cohorts: the Study of Health in Pomerania (SHIP) and SHIP-TREND,20 the Nurses’ Health Study (NHS) I and II,21 the Health Professionals Follow-up Study (HPFS), Women’s Genome Health Study (WGHS),22 Atherosclerosis Risk in Communities Study (ARIC),23 the Framingham Heart Study (FHS) original and offspring cohorts,24 the Rotterdam study,25,26 community-based cases and controls from the Popgen biobank27,28 and a case-control cohort from the Vanderbilt DNA Biobank, BioVU.29 (Table 1) The validation set comprised of an additional 6,489 cases and 62,797 controls from the Copenhagen General Population Study and the Copenhagen City Heart Study, the Kiel Study (Germany) and from a subset of the samples from NHS1/NHSII and HPFS that did not overlap with the discovery set (Table 1). Details of study population, genotyping, quality control and imputation in each study are described in detail in the Supplementary Materials and Methods section and in Supplementary Figure 1. Definition and assessment of gallstone disease in each cohort is detailed in Supplementary Table 1. Briefly, gallstone disease cases were defined either by self-report in a questionnaire asking directly about gallstone disease or prior cholestectomy (WGHS, NHS, HPFS, FHS, ARIC, FHS, WHI) or ICD codes (Rotterdam study, BioVU, CCHS, CGPS), or abdominal ultrasonography (SHIP, SHIP-TREND, PopGen and Kiel)
Table 1.
Characteristics of gallstone disease GWAS meta-analysis discovery and replication studies.
| Discovery studies | Study Design | Cases | Controls | %Female | Age, Mean ± SD | Genotyping platform |
Imputation platform |
||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||||
| WGHS | Nested case-control | 2853 | 20,436 | 100.0 | 100.0 | 55.6 ± 11.3 | 64.0 ± 7.1 | Illumina Duo | HapMap |
| NHS1/2/HPFS Affymetrix NHS1/2/HPFS Illumina |
Nested case-control | 1562 1019 |
6211 4400 |
72.2 85.5 |
53.2 75.7 |
60.5 ± 7.9 7.4 ±8.4 |
60.3 ± 8.1 56.3 ± 9.0 |
Affymetrix SNP 6.0, Illimina 550K, 660K |
1000G |
| SHIP | Nested case-control | 843 | 3134 | 65.6 | 47.1 | 60.3 ± 13.2 | 46.6 ± 15.8 | Affymetrix SNP 6.0 | 1000G |
| ARIC | Case-control (prevalent) | 832 | 8032 | 76.3 | 51.1 | 55.0±5.7 | 54.1±5.7 | Affymetrix 6.0 | HapMap |
| Rotterdam | Nested case-control | 705 | 5269 | 73.0 | 54.2 | 71.0 ± 8.8 | 68.7 ± 9.1 | Illimina 550K | HapMap |
| FHS | Nested case-control | 515 | 3783 | 71.3 | 53.2 | 67.2± 9.0 | 62.9± 9.6 | Affymetrix 550K | HapMap |
| BioVU | Hospital-based case- control |
202 | 2542 | 58.4 | 50.4 | 64.6 ±16.1 | 62.4 ±16.3 | Human660W–Quad BeadChip |
1000G |
| SPC (PopGen) | Nested case-control | 122 | 527 | 59.0 | 43.2 | 57.9 ± 12.7 | 62.5 ± 8.4 | Affymetrix 6.0 | 1000G |
| SHIP-TREND | Nested case-control | 67 | 818 | 64.2 | 53.6 | 56.6 ± 12.9 | 48.4 ± 13.4 | Illumina Omni 2.5 | 1000G |
| All discovery samples | 8720 | 55,152 | |||||||
| Replication studies | |||||||||
| CCHS and CGPS | Prospective cohort study | 3599 | 57,389 | 70.6 | 54.1 | 61.1±13.0 | 56.8±13.9 | Taqman/KASPar genotyping |
|
| Kiel University | Hospital-based case- control |
2104 | 2225 | 70.6 | 51.7 | 52.9 ± 11.2 | 39.7 ± 14.9 | TAQMAN genotyping | |
| NHS1/HPFS- Replication | Nested case-control | 786 | 3183 | 82.7 | 69.90 | 60.6 ± 7.4 | 59.5± 7.8 | Illumina OmniExpress | 1000G |
| All replication samples | 6489 | 62,797 | |||||||
| Combined Discovery + Replication | 15,209 | 117,949 | |||||||
| Replication in non-European ancestry individuals | |||||||||
| WHI (African American) | Nested case-control | 1384 | 6661 | 100.0 | 100.0 | 61.8 ± 6.9 | 61.5 ± 7.0 | Affymetrix 6.0 | 1000G |
| ARIC (African American) | Case-control (prevalent) | 115 | 2484 | Affymetrix 6.0 | HapMap | ||||
| WHI (Hispanic American) | Nested case-control | 1056 | 2403 | 100.0 | 100.0 | 60.9 ± 6.6 | 59.9 ± 6.7 | Affymetrix 6.0 | 1000G |
Statistical Analysis
Within each discovery study, we estimated the association between genotyped or imputed SNPs and the risk of gallstone disease by calculating beta coefficients and their standard errors using logistic regression models adjusted for age, sex and additional study specific covariates, assuming log-additive genetic effects. Prior to meta-analyses, we excluded imputed SNPs with imputation quality score and/or imputation R2 <0.3. We also employed a minor allele frequency (MAF) filter, excluding SNPs with a MAF of <0.01 for cohorts with more than 500 cases. For cohorts with <500 cases, we used a more stringent MAF threshold of 5 divided by the number of cases, thereby limiting analysis to SNPs expected to have 10 or more minor alleles within cases, to get robust estimates. Inverse variance weighted, fixed effects meta-analysis30 of study-specific estimates was performed to identify SNPs associated with gallstone disease, using METAL (http://genome.sph.umich.edu/wiki/METAL_Documentation). We selected the strongest independent markers at each locus, in order to attempt replication as well as to aid in functional/molecular interpretation, by performing conditional analyses in genomic regions (10 megabase windows using a less stringent nominal significance threshold for SNPs [discovery P < 5×10−06]), using the genome-wide complex trait analysis (GCTA) software31 (http://www.complextraitgenomics.com/software/gcta/). Conditional analysis is a mechanism to try to reduce the number of significant associations to the top most “independent” associations. We used 1753 healthy controls of European ancestry from the Type 2 Diabetes dataset within the NHS as reference population. Replication was performed for SNPs that were observed to be associated with gallstone disease risk at genome wide significance threshold of P < 5×10−8 following conditional analysis. We genotyped newly identified SNPs using the TAQMAN or KASPar assay in the replication datasets, except the NHS and HPFS studies, in which we had pre-existing genotype/imputation data. We reported fixed effects meta-analytic ORs and 95% CIs for combined associations from discovery and replication studies for all of the replicated SNPs. Heterogeneity of effect sizes between studies were determined using Cochran’s Q-test for heterogeneity32 as implemented in METAL30 and also by determining the I2 statistics33 that computes the proportion of overall variance that can be attributed due to differences in effect sizes between studies. For these SNPs, if discovery studies showed an evidence of heterogeneity (P<0.05), we reported association results using random effects meta-analysis in the combined discovery and replication studies.
In the replication studies, we additionally determined the strength of association for unit standard deviation increase in the weighted genetic risk score with gallstone disease risk. For the purpose of developing a genetic risk score, SNPs with missing information within the replication datasets were imputed by random sampling with replacement, from individuals with the SNP information available, and conditional on case-control status. We derived a genetic risk score for each study participant by assigning weights to each risk allele proportional to the logarithm of per allele relative risk estimate in the meta-analysis of discovery studies. The weighted genetic risk score (GRS) was standardized to have a zero mean and unit standard deviation.
We performed sensitivity analysis to exclude possible genetic associations mediated by BMI. Logistic regression models in each of the discovery studies were used to obtain beta coefficients and standard errors, after adjusting for BMI in additon to age and sex, followed by meta-analysis of study specific effect size estimates.
Post hoc analysis
We performed ancestry specific analyses to determine whether any of the variants with P<5×10−8 in the discovery and replication data sets show an association in African American or Hispanic American individuals, and whether they display differences in allelic frequencies across populations. Analysis was done in individuals of Afrcian American ancestry for 115 prevalent gallstone disease cases and 2,484 controls in the ARIC cohort and 1,384 incident and prevalent cases and 6,661 controls in the Women’s Health Initiative (WHI) cohort. Effect size estimates for Hispanic American ethnicity was done in 1,056 cases of incident or prevalent gallbladder disease and 2,403 controls within the WHI.
From the discovery GWAS meta-analyses summary statistics we determined the associations of (a) known non-alcoholic fatty liver disease variants, (b) previously reported variants associated with gallstone disease that did not reach genome wide significance in our data sets (UGT1A1 rs6742078 and TTC39B SNP rs686030) and (c) overlap with lith genes described from murine models.12–14
In post hoc analysis within the NHS and HPFS cohorts, for SNPs with P<5×10−08, we computed genotype specific associations with gallstone disease, and percentage population attributable risk for each genotype, as described previously.17 Additionally, we tested for associtions for these SNPs assuming different modes of inheritance (recessive and dominance effects), and for gene-gene interactions between these SNPs. For multiple independent associations at the same genetic locus (ABCG8 SNPs), we tested for associations of each haplotype combination with gallstone disease risk. We also evaluated for confounding effects of history of self reported hypercholesterolemia, use of cholesterol lowering drugs (ever/never) and post-menopausal hormone use (ever/never).
RNA sequencing of human gallbladder
We performed RNA sequencing from four human gallbladders (3 healthy controls and 1 patient with chronic gallstones) and 1 liver sample from the gallstone patient. RNA was obtained from gallbladder and liver of 1 female, age 71 with chronic cholecystitis and metastatic adenocarcinoma consistent with primary colon cancer (OriGene, CU0000000466). RNA was also obtained from 3 normal gallbladder samples, all female (ages 34, 46, 64) (BioChain, Lot Nos. A509245, A509248, A607331).
RNA Seq libraries were prepared using Ovation RNAseq v2 (NuGEN Technologies, Inc.) following guidelines for the Ovation SP Ultralow DR Multiplex System (NuGEN Technologies, Inc.). Library quality was verified for each sample using MiSeq (Illumina, Inc.) sequencing with 75bp paired-end reads. Samples were next sequenced using an Illumina HiSeq 2000 instrument (Illumina, Inc.) with 75bp paired-end reads. The raw reads in fastq format were mapped to human genome hg19 by Tophat (v2.0.9) with the parameter setting: -g 1 -N 2 -r 200. RefSeq transcripts reads count and RPKM were calculated by RSeQC (v2.3.6). The runs generated an average of 4,063,889 uniquely mapped reads per sample, with good mapping rates: cholecystitis gallbladder (89.5% uniquely mapped), cholecystitis liver (83.8%), and normal gallbladder samples (96.0%, 96.1%, and 84.9%, respectively). This data is available through GEO Accession number GSE66430, at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66430.
Expression QTL and ENCODE regulatory analyses
Proxy SNPs in linkage disequilibrium (R2>0.8) in populations of European ancestry were identified for gallstone index and replication SNPs using SNAP.34 Index SNPs and proxies were queried against a collected database of expression SNP (eSNP) results. The collected eSNP results met criteria for statistical thresholds for association with gene transcript levels as described in the original papers. A general overview of a subset of >50 eQTL studies has been published,35 with specific citations for >100 studies included in the current query following here. We assessed the concordance of the gallstone-identified eSNPs with the strongest eSNPs for each individual gene and dataset using linkage disequilibrium metrics (R2) and report results for either the index SNP or SNPs in LD with R2>0.8. The resulting eQTL SNPs with gene expression associations with P<5×10−06 were queried for overlap with ENCODE regulatory features using HaploReg v3 available at http://www.broadinstitute.org/mammals/haploreg/haploreg_v3.php.36 More details on eQTL and ENCODE regulatory analyses methods are available in the Supplementary Materials and Methods section.
Prior GWAS phenotype analysis
Gallstone index and replication SNPs and their proxies (as defined above) were queried against the NHLBI Genome-wide Repository of Associations between SNPs and Phenotypes (GRASP), version 2.0.0.0 available at http://apps.nhlbi.nih.gov/grasp/. Only results with p<5×10−8 were retained. The strongest SNP per GWAS phenotype per gallstone locus is reported.
RESULTS
Meta-analysis
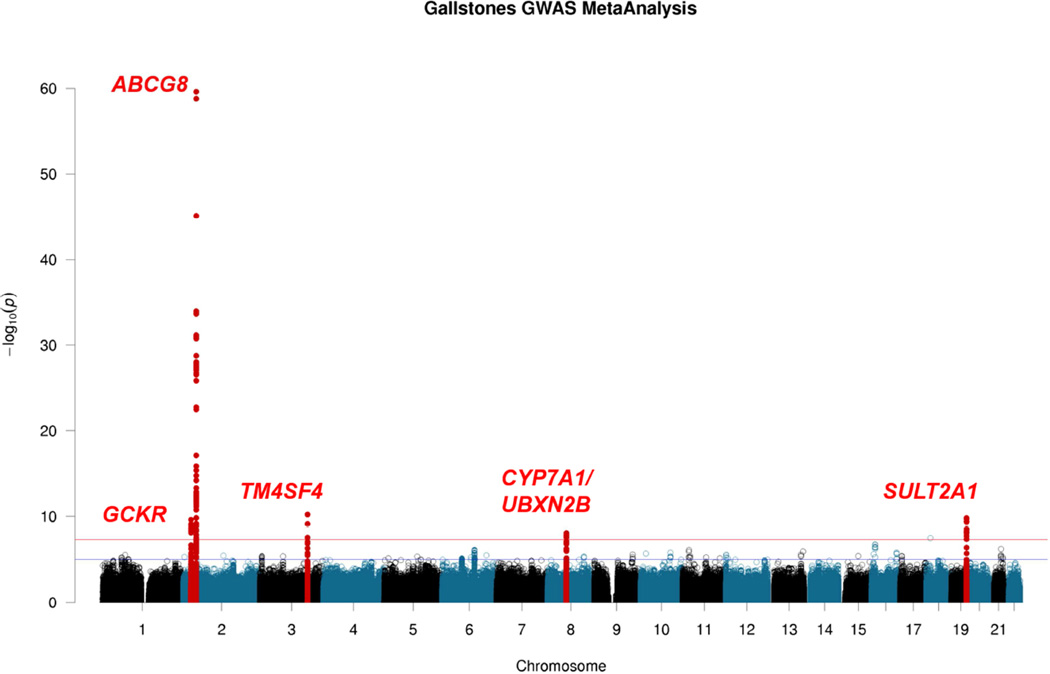
Fixed-effects meta-analysis,30 followed by conditional analyses within nominally significant regions31 (10Mb windows around SNPs with P<5×10−6), yielded seven SNPs from five genome-wide significant regions – ABCG5/8, TM4SF4, SULT2A1, UBXN2B/CYP7A1 and GCKR, independently associated with gallstone disease (P<5×10−8, Table 1, Figure 1 and Supplementary Table 2). There was no evidence of inflation of test statistics in the genome-wide meta-analysis (λ = 1.037, Q-Q plot in Supplementary Figure 2). The newly discovered SNPs had high imputation quality scores (> 0.80) in each of the discovery studies (Supplementary Table 3a). A sensitivity analysis adjusting for BMI prior to meta-analyses (to exclude genetic associations potentially mediated by BMI) yielded results that did not differ materially from those presented in Table 1 (Supplementary Table 3b). Regional association plots for the five independent loci are shown in Supplementary Figure 3. Except for the ABCG5 and ABCG8 loci, SNPs with P<1×10−4 in our discovery samples did not map to human orthologs of the candidate lith genes proposed in murine models. Although we did not observe a genome-wide significance for previously reported TTC39B SNP rs68603019, the A allele at the locus showed some evidence for an increased risk of gallstone disease (OR = 1.09, P = 0.000438).
Figure 1. Manhattan plot of the results of genome-wide meta-analysis of gallstone disease in ten studies.
The plot shows −log10-transformed P values for all SNPs. The red horizontal line represents P = 5 × 10−8. The blue horizontal line represents P = 1 × 10−5.
Replication
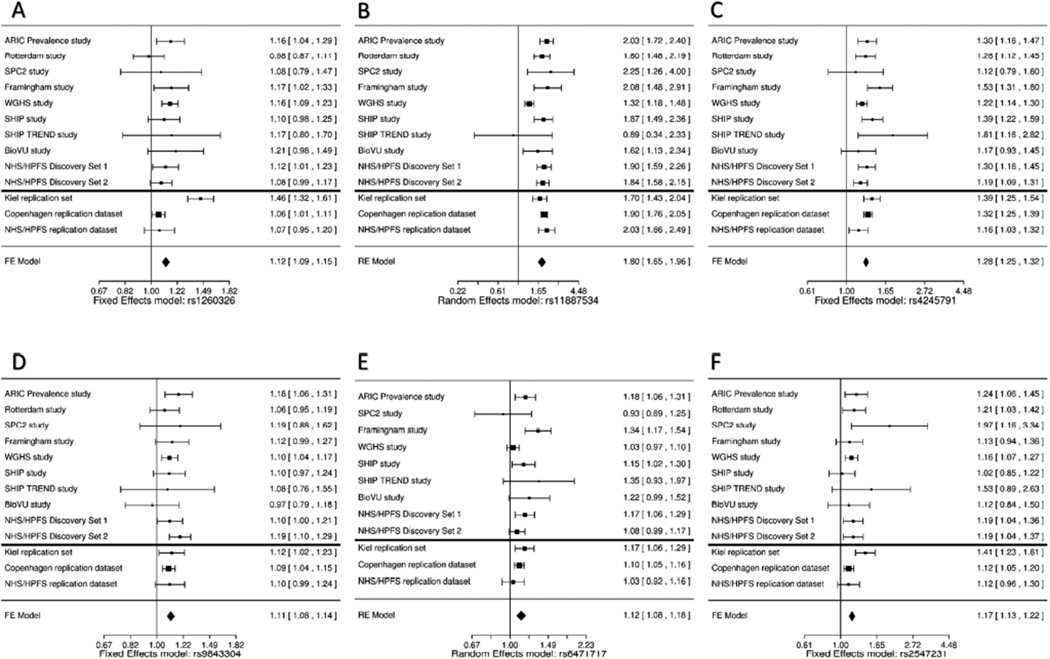
We selected six SNPs (rs11887534 and rs4245791 [ABCG8], rs6471717 [CYP7A1], rs9843304 [TM4SF4], rs2547231 [SULT2A1], and rs1260326 [GCKR]) for replication (Table 2) in an independent sample of 6,489 cases and 62,797 controls from three population-based studies and a case-control study (Table 1). The ABCG8 SNP rs4245791 (P-discovery = 1.90×10−34, R2 = 1.0 with rs4299376), and SULT2A1 SNP rs2547231 (P-discovery = 2.24×10−10, R2 = 0.90 with rs296391), have been previously shown to be strongly associated with hepatic ABCG837 and SULT2A138 expressions respectively, and therefore were selected for replication instead of the index SNPs. All of the selected SNPs were significantly associated with gallstone disease in meta-analysis from replication datasets. To account for heterogeneity of effect estimates for the ABCG8 locus SNP rs11887534 and for the UBXN2B/CYP7A1 SNP rs6471717 in the discovery meta-analysis (Table 1), we report their effect sizes using both fixed and random effects meta-analysis in the combined discovery and replication analyses (Table 2 and Figure 2). The fixed and random effects P-value for rs6471717 in combined discovery and replication analyses were 1.41×10−13 and 1.59×10−07 respectively. It is likely that evidence of heterogeneity reflects differences in magnitude of effect sizes of the susceptibility locus, possibly due to differences in study design or participant characteristics. However, the direction of effect was consistent for all replication SNPs across the studies (Figure 2F). Genetic risk scores (GRS) based on the six replicated SNPs and weighted on discovery stage beta-estimates were associated with an approximately 35% increased risk of gallstone disease for unit standard deviation increase in GRS, in all replication studies and provided modest improvement in area under the receiver operator characteristic curve (Supplementary Table 4 and Supplementary Figure 4).
Table 2.
Results of SNPs associated with gallstone disease in discovery and replication data sets.
| Replication stage | Combined – Discovery and Replication | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Hg38 / dbSNP 142 Location |
Gene variant |
Risk allele |
RAFa | ORb | Pvalue | HetcI2 | HetcP | RAFa | ORb | Pvalue | ORb(95% CI) |
| rs1260326 | chr2:27508073 |
GCKR, P446L |
C | 0.59 | 1.12 | 2.55×10−10 | <0.01 | 0.550 | 0.61 | 1.12 | 7.74×10−8 | 1.12 (1.09, 1.15) |
| rs1025447# | chr2:43795831 |
DYNC2LI1, intron |
T | 0.83 | 1.18 | 4.21×10−12 | <0.01 | 0.519 | 1.18 (1.13, 1.24) | |||
| rs11887534# | chr2:43839108 |
ABCG8, D19H |
C | 0.07 | 1.69 | 2.44×10−60 | 0.728 | 2.69×10−4 | 0.07 | 1.88 | 1.99×10−75 | 1.78 ( 1.70, 1.86) 1.80§ (1.65, 1.96) |
| rs4245791d# | chr2:43847292 |
ABCG8, intron |
T | 0.69 | 1.27 | 1.90×10−34 | 0.368 | 0.114 | 0.70 | 1.31 | 5.29×10−31 | 1.28 (1.25, 1.32) |
| rs9843304 | chr3:149493600 |
TM4SF4, intron |
C | 0.45 | 1.12 | 6.09×10−11 | <0.01 | 0.652 | 0.45 | 1.10 | 3.00×10−6 | 1.11 (1.08, 1.14) |
| rs6471717 | chr8:58464798 |
CYP7A1/, UBXN2B intergenic |
G | 0.35 | 1.11 | 8.84×10−9 | 0.573 | 0.016 | 0.34 | 1.10 | 3.16×10−6 | 1.11 (1.08, 1.14) 1.12§ (1.08, 1.18) |
| rs2547231e | chr19:47881800 |
SULT2A1, intron |
A | 0.84 | 1.17 | 2.24×10−10 | <0.01 | 0.537 | 0.84 | 1.17 | 1.09×10−7 | 1.17 (1.13, 1.22) |
RAF = risk allele frequency calculated using cases and controls.
OR = odds ratios. Odds ratio were obtained from fixed effect meta-analysis of study specific effect size estimates adjusted for age and gender in each discovery and replication study.
het = heterogeneity I2 and P-values from fixed effects meta-analysis
Proxy SNP for rs4299376 (P discovery stage = 1.18×10−34, R2 = 0.995, D’ = 0.999 among 1,753 Nurses’ Health Study participants )
Proxy SNP for rs296391 (P discovery stage = 1.59×10−10, R2 = 0.904, D’ = 0.969 among 1,753 Nurses’ Health Study participants)
Calculated using random effects meta-analysis (if discovery P-heterogeneity <0.05)
Conditioned on each other, discovery P-values for rs11887534, rs4245791 and rs1025447 were respectively 2.01×10−47, 3.39×10−21 and 6.14×10−10
Figure 2. Forest plots of meta-analyses of genome-wide significant SNPs in each of the discovery and replication data sets.
(A) Random effects meta-analysis: rs11887534. (B) Fixed effects meta-analysis: rs4245791. (C) Fixed effects meta-analysis: rs2547231. (D) Fixed effects meta-analysis: rs9843304. (E) Fixed effects meta-analysis: rs1260326. (F) Random effects meta-analysis: rs6471717.
SNP Associations in African American and Hispanic American populations
We observed that three SNPs from two loci – rs1260326, rs11887534 and rs4245791 were significantly associated (P<0.05) with gallstone disease among African American and Hispanic American individuals (Table 4). However, the direction of association was opposite to what we observed in the European population for rs1260326. We did not observe an association in these ethnicities for rs9843304, rs6471717 and rs2547231 SNPs. Moreover, we also observed marked differences in allele frequencies – for e.g. the T allele at rs1260326 is the major allele in individuals of European ancestry (frequency = 0.59), but minor allele in African American individuals (frequency = 0.14) and individuals of Hispanic American ancestry (frequency = 0.22). Similarly, the C allele at rs9843304 has a frequency of 0.45 in individuals of European ancestry, but about 0.8 in African Americans and 0.42 in Hispanic Americans.
Table 4.
Results of SNPs associated with gallstone disease in African American and Hispanic American ethnicities.
| SNP | Risk/ Other Allele |
European Ancestry Meta-analysis |
African American Ancestry (ARIC) Cases: 115 Controls: 2484 |
African American Ancestry (WHI) Cases: 1384 Controls: 6661 |
Hispanic American Ancestry (WHI) Cases: 1056 Controls: 2403 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAFa | ORb (95%CI) |
RAFa | OR (95%CI) | P | RAFa | ORb (95%CI) |
P | RAFa | ORb (95%CI) |
P | ||
| rs1260326 | T/C | 0.59 | 1.12 (1.09, 1.15) |
0.16 | 0.90 (0.61,1.34) |
0.616 | 0.15 | 0.86 (0.76,0.97) |
0.018 | 0.35 | 0.85 (0.76,0.95) |
0.004 |
| rs11887534 | C/G | 0.07 | 1.78 (1.70, 1.86) |
0.06 | 0.58 (0.26,1.32) |
0.196 | 0.09 | 1.22 (1.08,1.38) |
0.002 | 0.20 | 1.13 (1.02,1.24) |
0.017 |
| rs4245791 | T/C | 0.69 | 1.28 (1.25, 1.32) |
0.86 | 1.03 (0.69,1.54) |
0.877 | 0.86 | 1.30 (1.15,1.47) |
4.52×10−5 | 0.78 | 1.35 (1.19,1.54) |
6.82×10−6 |
| rs9843304 | C/T | 0.45 | 1.11 (1.08, 1.14) |
0.85 | 0.93 (0.61,1.42) |
0.737 | 0.78 | 1.08 (0.98,1.18) |
0.104 | 0.42 | 1.06 (0.96,1.18) |
0.253 |
| rs6471717 | G/A | 0.35 | 1.11 (1.08, 1.14) |
0.21 | 1.05 (0.74,1.47) |
0.801 | 0.22 | 0.93 (0.84,1.04) |
0.196 | 0.23 | 1.04 (0.92,1.18) |
0.513 |
| rs2547231 | A/C | 0.84 | 1.17 (1.13, 1.22) |
0.90 | 0.77 (0.47, 1.26) |
0.300 | 0.90 | 0.92 (0.81,1.06) |
0.239 | 0.90 | 1.12 (0.94, 1.33) |
0.205 |
RAF = risk allele frequency calculated using cases and controls.
OR = odds ratios. Odds ratio adjusted for age and gender.
Post hoc analyses
Supplementary Table 5 shows the associations for dominant and recessive models and population attributable risks for each genotype of the 6 GWAS-significant variants within the NHS and HPFS cohort samples. We did not observe substantially stronger dominance/recessive effects for any of the SNPs compared to the log-additive models that we used for our discovery analyses. We conducted haplotype analysis for the two independent associations in the ABCG8 locus. In Supplementary Table 6, we show the associations of 6 different haplotype combinations at rs11887534 (C/G) and rs4245791 (T/C). We observed that the presence of at least one C-T haplotype at this locus, i.e. the C allele at rs11887534 and T allele at rs4247591 was associated with a substantial increase in the risk of gallstone disease in both males and females, compared to individuals without the CT haplotype. We confirmed using the haplotype analysis that rs11887534 is likely to be the main driver of the ABCG8 association with gallstone disease risk. We did not observe any evidence for gene-gene interactions (Supplementary Table 7), after correcting for multiple comparisons. There was no evidence of confounding of genetic associations after adjusting for self-reported hypercholesterolemia, intake of cholesterol lowering drugs (ever/never) in the NHS and HPFS cohorts or for post-menopausal hormone therapy in the NHS cohort (Supplementary Table 8).
The UGT1A1 SNP rs6742078 did not show an overall association with gallstone disease in log-additive models of our discovery data set (P<0.114). However, we replicated in the NHS and the HPFS cohorts, the previously reported recessive mode of effect for rs6742078 TT genotype carriers with stronger evidence for association among size among males (OR = 1.45, 95% CI = 1.14, 1.85, P = 0.00284), compared to females (OR = 1.16, 95% CI = 1.00–1.34, P=0.0498). 17,39(Supplementary Table 9)
After multiple comparisons correction, genetic variants associated with nonalcoholic fatty liver disease were not observed to be associated with overall gallstone disease in our GWAS meta-analysis (data not shown).
Expression QTL and ENCODE regulatory analyses of discovered loci
Queries of gallstone index and proxy (R2>0.8 and P<5×10−6) SNPs revealed that several are strong eQTLs (Supplementary Table 10) with some of these located within ENCODE regulatory elements (Supplementary Table 11). Few gene expression studies, and no eQTL studies, have been conducted in gallbladder tissues. Gallstone index SNPs or proxies were the strongest eQTL for TM4SF4 (in liver), ABCG8 (in adipose), SULT2A1 (in liver, brain, and lung), C2orf16 (in liver), and LITAF (in liver, brain, and adipose) (Supplementary Table 12). Studies that have examined associations between SNPs and metabolite levels or ratios in blood, show that rs2547231 and rs1260326 are highly significantly associated with ratios of metabolites in the cholesterol metabolism pathway (Supplementary Table 13).40 Results of RNA sequencing from four human gallbladders (3 healthy controls and 1 patient with chronic gallstones) and 1 liver sample from the gallstone patient are reported in Table 3. The top GWAS loci ABCG5/8, SULT2A1, GCKR and CYP7A1 had higher expression in liver, compared to the gallbladder, suggesting they may influence the composition of bile. In contrast, TM4SF4 showed higher expression in gallbladder than the liver, with expression nearly twice as high in the chronic gallstones gallbladder as in the 3 normal samples(Table 3, Supplementary Figure 5), suggesting a local mechanism of action for this gene in gallbladder.
Table 3.
RNA sequencing RPKM (Reads Per Kilobase of transcript per Million mapped reads) values observed for genes near regions of discovered SNPs.
| Locus/gene | Normal Gallbladder (n=3)* |
Cholelithiasis Gallbladder (n=1) |
Cholelithiasis Liver (n=1) |
|---|---|---|---|
| ABCG5/8 | <10 | <10 | 47.3 (ABCG5) |
| TM4SF4 | 348.07 | 634 | 107.7 |
| GCKR | <10 | <10 | 143 |
| SULT2A1 | <10 | <10 | 217 |
| CYP7A1 | <10 | <10 | 20.6 |
For normal gallbladder samples the values reflect the mean RPKM across samples.
DISCUSSION
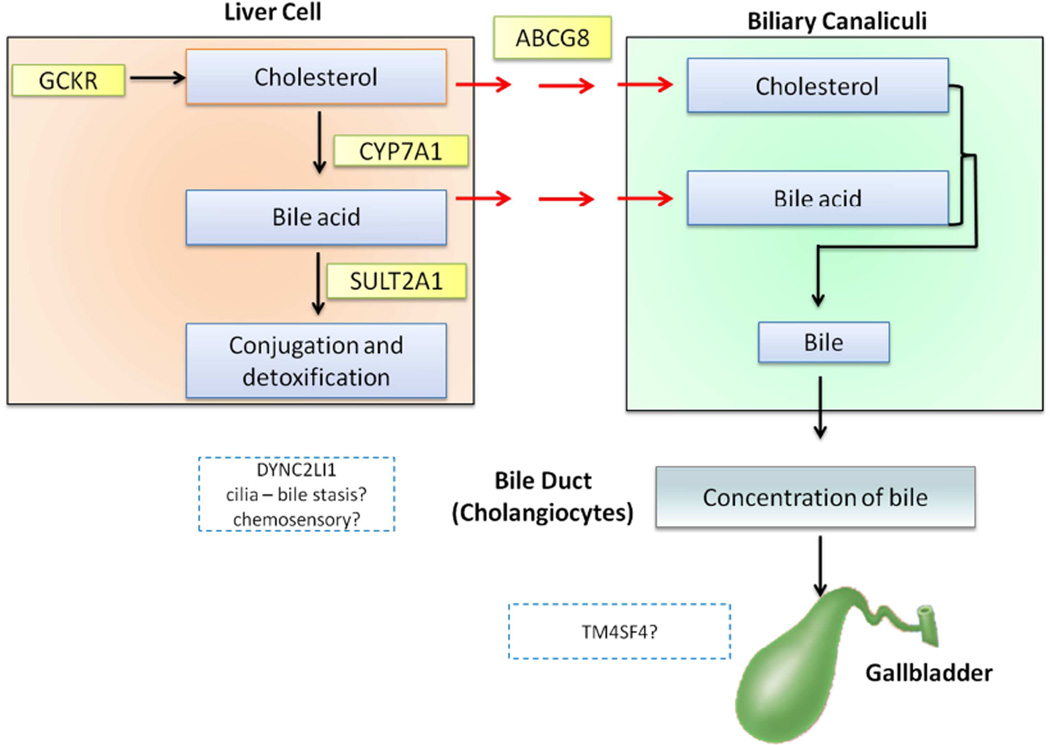
In this large-scale genome-wide association meta-analysis, we discovered 4 novel susceptibility loci (SULT2A1, TM4SF4, GCKR, and CYP7A1) and confirmed one known locus (ABCG8). The only previous GWAS of gallstone disease, comprising 280 cases and 360 controls in the discovery cohort, identified rs11887534 in ABCG8 as associated with gallstone disease.15 In addition to confirming this association, we observed an independent association of rs4245791, an intronic variant in ABCG8, consistent with results from previous fine-mapping efforts.41 Thus, there are at least two independent gallstone risk variants at the ABCG8 locus. The biological role of ABCG5/8 is to facilitate efflux of cholesterol from enterocytes and hepatocytes into the intestine and bile, respectively.42 Therefore, genetic variants in ABCG5/8 that increase the risk of gallstone disease would be expected to confer a gain-of-function since high bile cholesterol concentration promotes the formation of cholesterol gallstones7. Indeed, the gallstone-associated H-allele of D19H has been shown to increase cholesterol efflux ~3 fold in vitro, and the gallstone-associated allele of rs4245791 has been associated with increased mRNA levels (i.e., a gain-of-function effect).37,43 A third independent association within 5 Mb of rs11887534, mapped to DYNC2LI1, was identified, but was not carried forward to replication due to limited capacity. DYNC2LI1 is a component of cilia structure, and potentially relevant since primary cilia of cholangiocytes regulate osmolarity, and flow of bile.44
Several of the newly discovered loci are in or near genes known to play a role in cholesterol or bile acid metabolism (Supplementary Table 8 and Supplementary Figure 6). Association of the discovered SNPs with the genes was made on the basis of (a) missense mutations as a result of the variant such as D19H in ABCG8 and P446L in GCKR, or (b) due to mapping of the SNP in the intron of the gene, coupled with strong evidence of association from eQTL (TM4SF4, and SULT2A1) and mQTL data (GCKR and SULT2A1), or (c) genomic proximity to genes with strong evidence of relevance in cholesterol/bile acid metabolism pathways (e.g. CYP7A1). The glucokinase regulatory protein (GCKR) regulates the conversion of glucose to glucose-6-phosphate in the liver. The GCKR P446L variant associated with gallstone disease, even after adjustment for BMI, has been associated with other phenotypes/traits, including lipid levels, glycemic traits, and type 2 diabetes. We postulate that P446L may influence risk of gallstone disease by increasing the availability of cholesterol to the liver (via high endogenous synthesis), thereby increasing cholesterol concentration in the bile.45–47 We also identified rs6471717 near CYP7A1, associated with gallstone disease. Inside the liver, the rate-limiting step in the conversion of cholesterol to primary bile acids is catalyzed by the enzyme CYP7A1.48 Thus, genetic variation influencing CYP7A1 activity may influence gallstone disease both via increased cholesterol and decreased bile acid levels. In support of this, individuals homozygous for deleterious mutations in CYP7A1 suffer from premature gallstone disease.49 SULT2A1 catalyzes the conjugation of sulfates to a wide range of steroids and bile acids before biliary excretion.50 Bile acids help to solubilize biliary cholesterol, and thus prevent gallstone formation. Altered hepatic sulfation of bile acids due to genetic variation in SULT2A1 may influence bile acid metabolism and, in turn, biliary levels of bile acids, and ultimately the risk of gallstone formation. The rs2547231 variant near SULT2A1 has been associated with SULT2A1 expression 38, and with the ratio of two products of SULT2A1 (X-11440 and androsten-3beta,17beta-diol disulfate 2). 40 Finally, we found that an intronic variant in TM4SF4 was significantly associated with gallstone disease. TM4SF4 encodes transmembrane 4 L six family member 4, which has been implicated in liver regeneration as well as pancreas development.51 The role of TM4SF4 in gallstone disease is yet to be examined. TM4SF4 was identified as expressed in liver via eQTL results, with evidence for binding of liver-regulatory elements in ENCODE project data. Furthermore, our RNA sequencing data demonstrates that TM4SF4 is highly expressed in gallbladder tissue, particularly in the chronic gallstone disease sample. Queries of the Protein Atlas also confirm the TM4SF4 RNA and protein is most highly expressed in glandular cells of the gallbladder, duodenum and small intestine as well as liver bile duct and hepatocytes.52
The major strength of this study is the large discovery and replication datasets compared to the only prior gallstone GWAS. However, several limitations are noteworthy. First, we did not have information on gallstone composition (cholesterol/pigment/mixed), and could not discern between stone types. Second, gallstone case definitions varied across cohort settings. However, this concern is minimized by the observation that ABCG8 D19H, a known susceptibility locus, displayed similar risk associations in most sub-cohorts. Third, the majority of studies defined gallstones as a history of gallstones or prior cholecystectomy. We expect this led to under-representation of asymptomatic gallstones (~80% of all gallstones are asymptomatic) and would bias toward the null hypothesis. However, since symptomatic gallstone cases require medical interventions, their overrepresentation may lead to discovery of markers that have more clinical relevance. Fourth, in ethnicity specific analyses, we observed opposite direction of association among European versus African/Hispanic ancestry individuals for rs1260326, which suggests that this variant may not be truly causal, but may be tagging the true causal SNPs – and due to differences in linkage disequilibrium patterns or haplotype structures across populations, this correlation may be direct in one population and inverse in the other. Nevertheless, the replication of these loci in diverse populations reinforces the importance of these loci in gallstone disease due to marginal consistent associations across ethnicities. Fifth, another limitation of this study is the relatively small sample size of available RNA sequencing data, which limits our ability to determine whether cis genes are expressed in our tissues of interest. However, to our knowledge, there is no database that reports eQTL results for gallbladder tissue and with this small sample, we could not derive a conclusive evidence of comparative expression levels in gallbladder versus liver. Sixth, in the absence of functional studies, the hypothesized associations between SNPs and the genes based on bioinformatics/eQTL data may be speculative, and the true mechanisms by which these SNPs may impact gallstone disease may have been missed. Seventh, we used log-additive models to assess associations with gallstone disease. This may have reduced our ability to detect genetic associations that follow other modes of inheritance. Finally, we may not have been able to detect rare causal alleles in LD with the most significant GWAS SNPs, because conditional analysis using GCTA requires a large reference sample to estimate linkage disequilibrium.
In summary, this GWAS meta-analysis of previously genotyped cohorts discovered novel SNPs associated with gallstone disease in European ancestry individuals from four distinct and biologically plausible loci. These genetic variants were replicated in independent samples, bringing the total number of GWAS-identified lithogenic loci to five. Further studies addressing the functionality of these novel candidate genes are warranted to establish their causal role in gallstone development.
Supplementary Material
Figure 3.
Schematic figure showing possible role of novel susceptibility loci in gallstone formation.
Acknowledgments
The authors wish to thank all study participants and researchers, clinicians, technicians and administrative staff who contributed to this study. The meta-analysis was supported by NIH grants K24DK098311 (ATC). RNA sequencing, FHS and eQTL analyses were supported with NHLBI Intramural funds. Acknowledgments for discovery GWAS studies and replication datasets are available in the online version.
Abbreviations used in this paper
- ARIC
Atherosclerosis Risk in Communities Study
- BioVU
Vanderbilt DNA Biobank
- BMI
body mass index
- CI
confidence intervals
- eSNP
expression single nucleotide polymorphism
- eQTL
expression quantitative trait loci
- FHS
Framingham Heart Study
- GCTA
genome-wide complex trait analysis
- GWAS
genome-wide association studies
- HPFS
Health Professionals Follow-up Study
- MAF
minor allele frequency
- NHS
Nurses’ Health Study
- OR
odds ratio
- RPKM
reads per kilobase per million
- SHIP
Study of Health in Pomerania
- SNP
single nucleotide polymorphism
- WGHS
Women’s Genome Health Study
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Andrew D. Johnson (ADJ), Charlotte Andersson (CA), Amit D. Joshi (AJ) and Andrew T. Chan (ATC) conceived, designed and coordinated the study and performed the statistical analysis. Manuscript preparation and drafting: ADJ, CA, AJ, ATC, SS. Manuscript editing: All co-authors. Genotype and imputation data: DIC (WGHS); PK, CC, ATC, HC, GC, IDV, CF, FH, LRP, ER, RT, DJH, JLW, JHK, MG (NHS, NHSII and HPFS); SDM, BHS, AH, HLAJ, AU (Rotterdam); ATH (CCHS and CGPS); UV and AT (SHIP and SHIP TREND); WT, LCW(ARIC); JCD, DMR (BioVU); PLA, JH, CK, APR (WHI). Phenotype ascertainment: LR, DIC (WGHS); AJ, CC, ATC (NHS, NHS II and HPFS); SDM, BHS, AH, HLAJ, AU (Rotterdam); BGN (CCHS and CGPS); HV (SHIP and SHIP TREND); ARF, PLL (ARIC); PLA, APR (WHI)
GWAS data analyses: LR, DIC (WGHS); AJ, CC, PK (NHS, NHS II and HPFS); AT (SHIP, SHIP-TREND); WT, LCW (ARIC); RN, BHS (Rotterdam Study); ADJ (FHS); PEW, JCD (BioVU); SB (SPC); AT (SHIP and SHIP TREND); SS (CCHS and CGPS); PLA, JH, CK, APR (WHI). ADJ conceived the gallbladder and liver RNA sequencing experiments, and created the eQTL database. YW and JZ performed RNA sequencing. JDE and ADJ conducted RNA sequencing read mapping and analysis and eQTL analysis.
AJ, CA, SB, SS, RN, LCW and PEW equally contributed to this manuscript as first co-authors. DMR, BHS, WT, AT, JH, ATH, DIC, ATC and ADJ equally contributed to this manuscript as senior co-authors. All authors critically reviewed the manuscript and approved the final version.
Accession number for publicly accessible data repository: RNA sequencing data is available through GEO Accession number GSE66430, at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66430.
POTENTIAL CONFLICTS OF INTERESTS
The authors declare no potential conflicts of interests.
References
- 1.Shaffer EA. Gallstone disease: Epidemiology of gallbladder stone disease. Best Pract Res Clin Gastroenterol. 2006;20:981–996. doi: 10.1016/j.bpg.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Admirand WH, Small DM. The physicochemical basis of cholesterol gallstone formation in man. Journal of Clinical Investigation. 1968;47:1043–1052. doi: 10.1172/JCI105794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarnaik S, Slovis TL, Corbett DP, et al. Incidence of cholelithiasis in sickle cell anemia using the ultrasonic gray-scale technique. J Pediatr. 1980;96:1005–1008. doi: 10.1016/s0022-3476(80)80626-3. [DOI] [PubMed] [Google Scholar]
- 4.Cahalane MJ, Neubrand MW, Carey MC. Physical-chemical pathogenesis of pigment gallstones. Semin Liver Dis. 1988;8:317–328. doi: 10.1055/s-2008-1040553. [DOI] [PubMed] [Google Scholar]
- 5.Wang DQ, Carey MC. Characterization of crystallization pathways during cholesterol precipitation from human gallbladder biles: identical pathways to corresponding model biles with three predominating sequences. J Lipid Res. 1996;37:2539–2549. [PubMed] [Google Scholar]
- 6.Redinger RN, Small DM. BIle composition, bile salt metabolism and gallstones. Archives of Internal Medicine. 1972;130:618–630. [PubMed] [Google Scholar]
- 7.Portincasa P, Moschetta A, Palasciano G. Cholesterol gallstone disease. Lancet. 2006;368:230–239. doi: 10.1016/S0140-6736(06)69044-2. [DOI] [PubMed] [Google Scholar]
- 8.Sarin SK, Negi VS, Dewan R, et al. High familial prevalence of gallstones in the first-degree relatives of gallstone patients. Hepatology. 1995;22:138–141. [PubMed] [Google Scholar]
- 9.Hsing AW, Bai Y, Andreotti G, et al. Family history of gallstones and the risk of biliary tract cancer and gallstones: a population-based study in Shanghai, China. Int J Cancer. 2007;121:832–838. doi: 10.1002/ijc.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakeeb A, Comuzzie AG, Martin L, et al. Gallstones: genetics versus environment. Ann Surg. 2002;235:842–849. doi: 10.1097/00000658-200206000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsika D, Grjibovski A, Einarsson C, et al. Genetic and environmental influences on symptomatic gallstone disease: a Swedish study of 43,141 twin pairs. Hepatology. 2005;41:1138–1143. doi: 10.1002/hep.20654. [DOI] [PubMed] [Google Scholar]
- 12.Lammert F, Carey MC, Paigen B. Chromosomal organization of candidate genes involved in cholesterol gallstone formation: a murine gallstone map. Gastroenterology. 2001;120:221–238. doi: 10.1053/gast.2001.20878. [DOI] [PubMed] [Google Scholar]
- 13.Lyons MA, Wittenburg H. Cholesterol gallstone susceptibility loci: a mouse map, candidate gene evaluation, and guide to human LITH genes. Gastroenterology. 2006;131:1943–1970. doi: 10.1053/j.gastro.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 14.Paigen B, Schork NJ, Svenson KL, et al. Quantitative trait loci mapping for cholesterol gallstones in AKR/J and C57L/J strains of mice. Physiol Genomics. 2000;4:59–65. doi: 10.1152/physiolgenomics.2000.4.1.59. [DOI] [PubMed] [Google Scholar]
- 15.Buch S, Schafmayer C, Volzke H, et al. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet. 2007;39:995–999. doi: 10.1038/ng2101. [DOI] [PubMed] [Google Scholar]
- 16.Goodloe R, Brown-Gentry K, Gillani NB, et al. Lipid trait-associated genetic variation is associated with gallstone disease in the diverse Third National Health and Nutrition Examination Survey (NHANES III) BMC Med Genet. 2013;14:120. doi: 10.1186/1471-2350-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buch S, Schafmayer C, Völzke H, et al. Loci From a Genome-Wide Analysis of Bilirubin Levels Are Associated With Gallstone Risk and Composition. Gastroenterology. 2010;139:1942–1951. e2. doi: 10.1053/j.gastro.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Johnson AD, Kavousi M, Smith AV, et al. Genome-wide association meta-analysis for total serum bilirubin levels. Human Molecular Genetics. 2009;18:2700–2710. doi: 10.1093/hmg/ddp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez S, Gaunt TR, Guo Y, et al. Lipids, obesity and gallbladder disease in women: insights from genetic studies using the cardiovascular gene-centric 50K SNP array. Eur J Hum Genet. 2016;24:106–112. doi: 10.1038/ejhg.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volzke H, Alte D, Schmidt CO, et al. Cohort profile: the study of health in Pomerania. Int J Epidemiol. 2011;40:294–307. doi: 10.1093/ije/dyp394. [DOI] [PubMed] [Google Scholar]
- 21.Colditz GA, Manson JE, Hankinson SE. The Nurses’ Health Study: 20-year contribution to the understanding of health among women. J Womens Health. 1997;6:49–62. doi: 10.1089/jwh.1997.6.49. [DOI] [PubMed] [Google Scholar]
- 22.Ridker PM, Chasman DI, Zee RY, et al. Rationale, design, and methodology of the Women’s Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem. 2008;54:249–255. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 23.The Atherosclerosis Risk in Communities (ARIC) Study: design objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 24.Kannel WB, Feinleib M, McNamara PM, et al. An investigation of coronary heart disease in families The Framingham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 25.Hofman A, Grobbee DE, de Jong PT, et al. Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol. 1991;7:403–422. doi: 10.1007/BF00145007. [DOI] [PubMed] [Google Scholar]
- 26.Hofman A, Brusselle GG, Darwish Murad S, et al. The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol. 2015;30:661–708. doi: 10.1007/s10654-015-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nothlings U, Krawczak M. [PopGen. A population-based biobank with prospective follow-up of a control group] Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55:831–835. doi: 10.1007/s00103-012-1487-2. [DOI] [PubMed] [Google Scholar]
- 28.Krawczak M, Nikolaus S, von Eberstein H, et al. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 29.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype- phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Lee SH, Goddard ME, et al. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 33.Higgins JPT, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ : British Medical Journal. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson AD, Handsaker RE, Pulit SL, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Gierman HJ, Levy D, et al. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC genomics. 2014;15:532. doi: 10.1186/1471-2164-15-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teupser D, Baber R, Ceglarek U, et al. Genetic regulation of serum phytosterol levels, risk of coronary artery disease Circulation. Cardiovascular genetics. 2010;3:331–339. doi: 10.1161/CIRCGENETICS.109.907873. [DOI] [PubMed] [Google Scholar]
- 38.Zhai G, Teumer A, Stolk L, et al. Eight common genetic variants associated with serum DHEAS levels suggest a key role in ageing mechanisms. PLoS genetics. 2011;7:e1002025. doi: 10.1371/journal.pgen.1002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stender S, Frikke-Schmidt R, Nordestgaard BG, et al. Extreme bilirubin levels as a causal risk factor for symptomatic gallstone disease. JAMA Intern Med. 2013;173:1222–1228. doi: 10.1001/jamainternmed.2013.6465. [DOI] [PubMed] [Google Scholar]
- 40.Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stender S, Frikke-Schmidt R, Nordestgaard BG, et al. The ABCG5/8 cholesterol transporter and myocardial infarction versus gallstone disease. J Am Coll Cardiol. 2014;63:2121–2128. doi: 10.1016/j.jacc.2013.12.055. [DOI] [PubMed] [Google Scholar]
- 42.Graf GA, Yu L, Li WP, et al. ABCG5 and ABCG8 are obligate heterodimers for protein trafficking and biliary cholesterol excretion. J Biol Chem. 2003;278:48275–48282. doi: 10.1074/jbc.M310223200. [DOI] [PubMed] [Google Scholar]
- 43.von Kampen O, Buch S, Nothnagel M, et al. Genetic and functional identification of the likely causative variant for cholesterol gallstone disease at the ABCG5/8 lithogenic locus. Hepatology. 2013;57:2407–2417. doi: 10.1002/hep.26009. [DOI] [PubMed] [Google Scholar]
- 44.Larusso NF, Masyuk TV. The role of cilia in the regulation of bile flow. Dig Dis. 2011;29:6–12. doi: 10.1159/000324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nature genetics. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kottgen A, Albrecht E, Teumer A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nature genetics. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paumgartner G, Sauerbruch T. Gallstones: pathogenesis. Lancet. 1991;338:1117–1121. doi: 10.1016/0140-6736(91)91972-w. [DOI] [PubMed] [Google Scholar]
- 49.Pullinger CR, Eng C, Salen G, et al. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J Clin Invest. 2002;110:109–117. doi: 10.1172/JCI15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinshilboum RM, Otterness DM, Aksoy IA, et al. Sulfation and sulfotransferases 1: Sulfotransferase molecular biology: cDNAs and genes. FASEB J. 1997;11:3–14. [PubMed] [Google Scholar]
- 51.Anderson KR, Singer RA, Balderes DA, et al. The L6 domain tetraspanin Tm4sf4 regulates endocrine pancreas differentiation and directed cell migration. Development. 2011;138:3213–3224. doi: 10.1242/dev.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.