Abstract
Lipoproteins, namely high-density lipoproteins (HDL), transport a wide-variety of cargo in addition to cholesterol and lipids. In 2011, HDL and low-density lipoproteins (LDL) were reported to transport microRNAs (miRNA). Since the original discovery, there has been great excitement for this topic and a handful of follow-up publications. Here, we review the current landscape of lipoprotein transport of miRNAs. HDL-miRNAs have been demonstrated to be altered in cardiovascular disease (CVD), including hypercholesterolemia and atherosclerosis. As such, HDL and LDL-miRNAs may represent a novel class of disease biomarkers. Below, we review HDL-miR-92a and miR-486 levels in myocardial infarction and unstable angina, and HDL-miR-223 and miR-24 levels in coronary artery disease (CAD). Moreover, we address HDL’s contribution to the total pool of extracellular miRNAs in plasma and differential distribution of miRNAs across HDL sub-species. Finally, we address current and future challenges for this new field and the barriers to such work.
Introduction
microRNAs (miRNA) are small non-coding RNAs (approximately 22 nts in length) that suppress gene expression through post-transcriptional regulation of mRNA stability and translation1. Each cell-type contains 200–300 miRNAs out of the possible approximately 2000 reported miRNAs; however, not all mature miRNAs in a cell are likely to be transcribed or processed in the cell. Extracellular miRNAs likely serve as cellular messages and are transported between cells in an endocrine form of intercellular communication2, 3. The field of extracellular miRNAs has generated tremendous excitement, as they hold great potential as a novel class of disease biomarkers and possible drug targets. miRNAs have been found in all biological fluids from saliva to urine4–7. In plasma, they are protected from circulating ribonucleases through their association with lipid and protein carriers3. Little is known about how miRNAs are protected in other fluids, but lipid particles and vesicles are likely responsible. Much of the research on extracellular miRNAs has focused on exosomes, which are derived from multivesicular bodies in endosomes8. Exosomes contain both long RNAs (e.g. mRNAs) and small RNAs (e.g. miRNAs), and are composed of a phospholipid bilayer shell and hydrophilic core9–12. Similar in structure, but distinct in origin, are microvesicles13–15. These vesicles are generally larger than exosomes and are derived from shedding or blebbing of the plasma membrane. There have been reports of many other miRNA carriers, including apoptotic bodies from endothelial cells16. The isolation of each class can be tedious and most preparations are likely a heterogeneous mixture of extracellular vesicles (EV). Ribonucleoproteins have also been reported to transport and protect extracellular miRNAs. Most studies have investigated the role of extracellular Argonaute 2 (AGO2), which is the structure-function ribonucleoprotein in the RNA-Induced Silencing Complex (RISC)17, 18. Currently, it is unknown if circulating AGO is simply released from necrotic cells or exported in a regulated fashion. AGO-bound miRNAs have been reported to stand alone in plasma as well as be present in EVs17. Maybe the most critical carriers of miRNAs are the highly abundant lipoproteins, namely low-density lipoproteins (LDL) and high-density lipoproteins (HDL). Here, we review the current state-of-the-art in lipoprotein-miRNA research.
HDL-miRNAs
The idea that HDL could transport nucleic acids arose from a 2009 paper in which the authors demonstrated that small interfering RNAs (siRNA) could be effectively delivered to the liver using reconstituted apolipoprotein A-I (apoA-I), which is the main functional protein of HDL19. Following upon this study, we profiled approximately 700 miRNAs on pure human HDL using real-time PCR-based TaqMan Low Density Arrays (TLDA)20. After finding a consistent pattern in healthy subjects, we then sought to determine if HDL-miRNAs are significantly altered in hypercholesterolemia and we found significant HDL-miRNA changes in humans and mice20. Unlike EVs which have both mRNA and miRNAs, RNA circulating on HDL is predominantly small non-coding RNA (<40 nts); however, longer RNA species have been identified (approximately 60–90 nts in length), possibly full-length tRNAs. In addition to miRNAs, HDL likely transport other non-coding RNAs; however, this has not been published. Many of HDL’s top-ranked miRNAs likely originate from inflammatory cells. We have reported that macrophages (J774) export miR-223 to HDL in vitro20; however, it is highly likely that monocytes, neutrophils, and other inflammatory cells export miRNAs to HDL as well, as many of the most abundant miRNAs on HDL are highly abundant in these cell-types. The export of miR-223 to HDL was found to be inhibited by neutral sphingomyelinase 2 (nSMase2), as chemical inhibition of nSMase2 increased HDL-miR-223 export20. The roles of cholesterol and lipid transporters in miRNA export have largely not been studied and warrant investigation. Nevertheless, activation of liver-x-receptor to increase ATP-binding cassette transporter A1 (ABCA1) expression in macrophages failed to increase miR-223 export to HDL20. This does not preclude the possibility that ABCA1, ABCG1, and scavenger receptor BI (SR-BI) contribute to miRNA efflux to apoA-I or HDL in other cells or myeloid phenotypes. We did find that HDL delivers miRNAs to recipient Huh7 hepatoma cells and this process is dependent upon SR-BI20. Moreover, HDL-miRNA transfer capacity was greatly enhanced in baby hamster kidney (BHK) cells when SR-BI was over-expressed20. HDL-miRNA delivery to Huh7 cells was found to suppress target gene (mRNA) expression, and this likely occurs through canonical RNA silencing pathways, as HDL-miRNA transfer suppressed gene reporter (luciferase) activity in recipient BHK-SR-BI cells20. Nonetheless, it is likely that HDL can transfer miRNAs to recipient cells through SR-BI-independent mechanisms in other cell-types and possibly regulate gene expression in recipient cells through currently unknown non-canonical RNA silencing pathways.
HDL-miRNA changes in mice
Upon finding significant HDL-miRNA changes in humans with familial hypercholesterolemia, an autosomal dominant disorder due to mutations in the LDL receptor (LDLR), we profiled HDL-miRNAs from Ldlr−/− mice compared to wild-type (WT) controls20. Mouse HDL was purified from plasma using anti-mouse apoA-I immunoprecipitation and HDL-miRNAs were quantified by real-time PCR-based TLDAs. We found many HDL-miRNA changes; however, very few miRNAs overlapped with what was differentially present on human HDL in hypercholesterolemia20. To further address mouse models of hypercholesterolemia and atherosclerosis, we injected Apoe−/− mice on either a high fat diet (HFD) or chow diet with human rHDL, which had very little to no miRNA before injections. After 6 h, we collected plasma and isolated human rHDL from mouse plasma by anti-human apoA-I immunoprecipitation. Using rodent TLDAs, we found significant changes in HDL-miRNAs in Apoe−/− mice fed a chow diet compared to WT mice on chow diet, thus suggesting that hypercholesterolemia alone may account for changes to HDL-miRNA signatures. We then found that Apoe−/− on a high-fat diet (HFD) had significant changes to HDL-miRNAs compared to Apoe−/− mice fed a chow diet, suggesting one possibility that the development or severity of atherosclerosis can have significant effects on circulating HDL-miRNAs20; however, this may also simply reflect changes to circulating cholesterol levels affecting HDL size, structure, and exchange potential. Again, we found very few miRNAs that were altered in both human hypercholesterolemia and a mouse model of hypercholesterolemia and atherosclerosis. We did find that HDL-miR-223 and miR-24 levels were increased and miR-135a* levels were decreased in the human study and the mouse study with Apoe−/− HFD compared to WT chow controls; however, some of these changes were not statistically significant (Table 1). The timing of 6 h suggests that HDL-miRNA levels are dynamic and detectable levels of exported miRNAs accumulate relatively quickly. This strategy to identify and quantify mouse miRNAs that human HDL collects during a short time-frame in vivo can be applied to any model of disease and has the benefit of sampling HDL-miRNA changes in temporal study designs. Most interestingly, we did find that 5 of the top-most abundant (top 10) HDL-miRNAs in healthy humans were also in the top 10 HDL-miRNAs in WT mice, suggesting that there is some level of conservation between humans and mice in health20. Yet, the general lack of overlap in humans and mice in hypercholesterolemia and atherosclerosis suggests that these animal models only partly reflect human disease.
Table 1.
Altered Lipoprotein miRNAs in Cardiovascular Disease
| Lipoprotein | miRNA | Direction | Disease | Ref. |
|---|---|---|---|---|
| HDL | miR-223 | Increased | Hypercholesterolemia | 20 |
| HDL | miR-24 | Increased | Hypercholesterolemia | 20 |
| HDL | miR-135* | Decreased | Hypercholesterolemia | 20 |
| HDL | miR-92a | Increased | Myocardial Infarction (MI) Unstable Angina (UA) Stable Angina (SA) |
25 |
| HDL | miR-486 | Increased | Myocardial Infarction (MI) Unstable Angina (UA) Stable Angina (SA) |
25 |
| LDL | miR-92a | Increased | Myocardial Infarction (MI) Stable Angina (SA) |
25 |
| LDL | miR-92a | Decreased | Unstable Angina (UA) | 25 |
| LDL | miR-146a | Increased | Stable Angina (SA) | 25 |
| LDL | miR-33 | Increased | Myocardial Infarction (MI) Unstable Angina (UA) |
25 |
HDL-miRNA biomarkers
The first independent validation that HDL transport miRNAs was reported in 201321. This study confirmed using quantitative PCR (qPCR) that miR-223, miR-92a, miR-126, miR-150, miR-146a, miR-30c, miR-378, miR-145 were present on human HDL. This study did find evidence that miR-155 was present on HDL which was missed in our earlier study20, 21. Using qPCR, the authors reported that HDL had >10,000 copies of miR-223 per µg of HDL total protein21. Furthermore, HDL-miR-223 levels were calculated as representing approximately 8% of all miR-223 in plasma, the highest of the miRNAs tested in this study21. Nonetheless, this study found a significant correlation between HDL-miRNA levels and total plasma miRNA levels. This calculation was based on 250 µg of HDL isolated from 250 µL of plasma which equates to approximately 1 mg HDL/ mL of plasma. This concentration is lower than the likely physiological concentrations for HDL circulating in plasma, which suggests that some HDL was possibly lost during the isolation and the reported HDL-miRNA percentages of total plasma miRNA levels may be underestimated. HDL proteins and total cholesterol account for approximately 55% and 15% of HDL mass, respectively22. Based on normal HDL-cholesterol ranges in humans (0.35 mg/mL–0.55 mg/mL), HDL total protein concentration in plasma ranges 1.3–2.0 mg/mL; however, this is difficult as HDL has to be isolated from plasma to quantify protein levels22–24. Nevertheless, a greater understanding of the total contribution of all HDL-miRNAs, and miRNAs from other lipoproteins, to the total plasma pool is needed, particularly in biomarker studies where plasma sampling without HDL isolation is preferred. In agreement with studies described above, a recent study confirmed that specific miRNAs are transported by lipoproteins in circulation25. Using real-time PCR, miR-486, miR-92a, miR-122, miR-125a, miR-146a, and miR-33 levels were quantified in HDL subspecies (HDL3 and HDL2)25. This was the first study to report miR-33 on HDL – miR-33 regulates HDL-cholesterol (HDL-C) levels through ABCA1 – as miR-33 was not previously detected on HDL using TaqMan TLDAs, which is not disconcerting for much of the variability between human HDL-miRNA profiles occurs in the lowly abundant miRNAs. This study reported that levels of miR-486 and miR-92a in large HDL2 are significantly increased in subjects with myocardial infarction (MI), unstable angina (UA), and stable angina (SA); the highest effect was HDL2-miR-486 changes associated with MI followed by UA and then SA subjects25. Conversely, HDL3-miR-92a levels were found to be significantly elevated in UA subjects compared to controls subjects25. To a lesser degree, miR-92a levels were also significantly elevated on HDL3 from MI and SA subjects25. In all CAD, miR-92a was the most abundant in HDL3 and miR-486 was the most abundant in HDL2, significantly higher than in HDL3, suggesting that HDL-miRNAs are differentially distributed across HDL subfractions and CAD likely influences their distribution and concentration. At this time, miR-486 and miR-92a have the greatest potential as markers of angina, both stable and unstable, as well as MI. Nevertheless, these studies are limited in that only a handful of candidate miRNAs were evaluated with CAD. Future studies will require HDL-miRNA profiling by PCR-based arrays or high-throughput small RNA sequencing, including more rigorous biomarker studies against standard indicators of CAD, UA, or MI.
LDL
Most of the work on lipoprotein miRNAs has been focused on HDL transport; however, each of the three studies also addressed LDL-miRNAs. For individual candidate miRNAs, e.g. miR-223, LDL levels were found to be considerably lower than HDL levels, with the exception of miR-155 which was found in greater abundance on LDL than HDL21. Wagner et al. reported that miR-223 was the most abundant of the miRNAs they tested on LDL (1500 copies/µg LDL); however, these levels were significantly lower than HDL-miR-223 (>10,000 copies/µg LDL)21. Moreover, they reported that the levels of miR-30c, miR-145, miR-150, miR-126, and miR-378 on LDL were minimally detected at <10 copies/µg LDL21. Niculescu et al. reported that miR-92a was the most abundant of the miRNAs they quantified on LDL25. While HDL-miRNAs were approximately 10-fold more abundant than LDL and IDL25. They did report that miR-92a levels were increased on LDL from SA and MI subjects and not detectable on LDL from UA subjects (Table 1)25. Furthermore, miR-146a was only detectable on LDL from SA subjects and miR-33 levels were increased on LDL from UA and MI subjects compared to SA and control subjects (Table 1)25. Wagner et al. found no differences in LDL-miRNA levels that they tested in healthy subjects compared to acute coronary syndrome (ACS) or CAD subjects21. In 2011, we found that HDL and LDL share many of the most abundant miRNAs; however, their profiles are statistically distinct from each other20. In agreement with Wagner et al., we found that miR-223 was the most abundant miRNA on LDL using real-time PCR-based profiling of all miRNAs20. The other top LDL-miRNAs include miR-150, miR-19b, miR-92a, miR-24, and miR-146a20. Collectively, HDL-miRNA transport appears to be more robust than LDL; however, future studies are needed to investigate the functional impact of miRNAs on apoB-containing particles (LDL and VLDL).
HDL-miRNA transfer to endothelial cells
MiRNAs have proven to be power regulators of endothelial gene expression and function, and endothelial miRNAs are critical factors in atherosclerosis26–29. Moreover, HDL is likely to have extensive contact with endothelial cells in circulation. As such, we sought to determine if HDL transfers miRNAs to endothelial cells and if delivered miRNAs regulate target gene expression. Using real-time PCR-based TLDAs, we found that miR-223 levels were significantly elevated in human coronary artery endothelial cells (HCAEC) after incubation with human HDL (1 mg) for 16–24 h30. Furthermore, we found that pre-treatment of HDL with RNaseA (to digest miRNAs) inhibited HDL’s ability to increase cellular miR-223 levels supporting HDL-miR-223 transfer30. Likewise, only native HDL (nHDL) treatments, not treatments with lipid-poor A-I, reconstituted HDL (rHDL), or small unilamellar vesicles (SUVs), resulted in increased miR-223 levels in recipient HCAECs, likely due to only native HDL containing miR-22330. In addition, we found that HDL-miR-223 levels were significantly reduced after incubation with HCAECs30. In validation studies, we found that cellular levels were significantly elevated approximately 2-fold within 1 h30. Most interestingly, miR-223 is not likely transcribed or processed in HCAECs as primary miR-223 transcripts and precursor miR-223 forms were minimally or not detected, and inhibition of transcription (Actinomycin D) failed to reduce mature miR-223 levels and failed to block the observed intracellular increase in mature miR-223 levels after HDL treatments30. Moreover, siRNA silencing of Dicer, a key miRNA processing enzyme, failed to reduce mature miR-223 levels in HCAECs suggesting that miR-223 is not processed by Dicer in endothelial cells. Wagner et al. reported that HDL complexed to exogenous C.elegans miRNA (cel-miR-39) was able to transfer cel-miR-39 to recipient human umbilical vein endothelial cells (HUVEC), albeit at low levels. Most interestingly, treatments with HDL complexed to single-stranded cel-miR-39 oligos, double-stranded cel-miR-39 mimetics, and precursor pre-cel-miR-39 were all able to result in detectable mature cel-miR-39 levels in recipient HUVECs21. This study also reported little to no increase in cellular miR-223 levels (HUVEC), or other miRNA candidates (miR-92a and miR-126), after HDL (1 mg/mL) treatments up to 24 h. To determine if endothelial cell-type (coronary artery vs. umbilical vein) affects HDL-miR-223 uptake and underlies the differences in the two studies, we treated both endothelial cell-types with native HDL (nHDL) 1 mg/mL for 16–24 h and found that nHDL treatments resulted in a >15-fold increase in HUVEC miR-223 levels; an effect even greater than HCAEC treatments (approximate 5-fold increase)30. Although there may be many factors that contributed to these discrepancies, differences in the concentration of miR-223 on the human HDL used for the treatments and possibly differences in cell culture conditions may factor in. Although these studies focused on HDL-miRNA delivery to endothelial cells, endothelial cells may also be a source (export) of HDL-miRNAs and contribute to HDL-miRNA signature in health and CVD. Further investigation of this hypothesis is warranted.
HDL’s anti-inflammatory properties
HDL have long been considered anti-inflammatory, with a significant component being their ability to suppress endothelial cell activation and adhesion molecule expression31–33. In addition to profiling miRNAs in HCAEC after HDL treatments, gene (mRNA) arrays were used to quantify HDL-induced gene expression changes in HCAEC30. Significantly decreased genes (mRNA) after HDL treatments were filtered for putative miR-223 targets (mRNAs). Strikingly, we found multiple mRNA candidates to fit these criteria that have previously been associated with inflammation, including intercellular adhesion molecule-1 (Icam1) and colony-stimulating factor 2 (Csf2)30. Using this approach, we found that nHDL treatments were far superior to rHDL, lipidpoor apoA-I, or SUV treatments at reducing Icam1 and vascular cell adhesion molecule −1 (Vcam1) mRNA levels, thus suggesting that the key anti-inflammatory factor is on nHDL30. Based on these findings, we hypothesized that HDL-transfer of miR-223 to endothelial cells confers HDL’s ability to suppress ICAM-1 expression, and thus leukocyte adherence and inflammation. We first demonstrated that human ICAM-1 is a direct target of miR-223 using gene reporter (luciferase) assays containing the full-length 3’ untranslated region (3’ UTR) of ICAM1 and site-directed mutagenesis of the putative miR-223 target site30. This reporter was then tested in HCAECs to prove that HDL’s suppression of ICAM-1 is mediated, at least in part, through the suppression of the 3’ UTR, specifically at the predicted miR-223 target site. Mutation of the predicted miR-223 target site in ICAM1’s 3’UTR abolished HDL’s ability to suppress ICAM-1 3’UTR luciferase activity. Over-expression of miR-223 in HCAECs was also found to reduce ICAM1 mRNA and protein levels. Moreover, HDL from a WT mouse (containing miR-223) was able to reduce ICAM1 and CSF2 mRNA levels in HCAECs; however, HDL from Mir223−/− mice not only failed to reduce ICAM1 expression, but significantly increased ICAM1 or CSF2 mRNAs. These results suggest that HDL-miR-223 levels are indeed anti-inflammatory in HCAECs, but that HDL from a mouse deficient in miR-223 is pro-inflammatory. A critical component of HDL’s anti-inflammatory capacity is the ability to reduce tumor necrosis factor-α (TNF-α) activation of endothelial cells34. In our studies, we found that TNF-α-induced ICAM-1 levels were significantly reduced when miR-223 was over-expressed in stimulated HCAECs30. We then observed that pre-treating HCAECs with anti-sense miR-223 inhibitors blocked HDL’s ability to reduce ICAM-1 protein and TNF-α-stimulation of ICAM-1 protein levels30. To determine if HDL-miR-223 regulation of endothelial cell adhesion molecule expression had a functional impact, neutrophil adhesion assays were completed, and we found that both HDL treatments and miR-223 over-expression in HCAECs significantly inhibited TNF-α-induced neutrophil adhesion30. Collectively, these results suggest that HDL’s anti-inflammatory capacity in endothelial cells is mediated, in part, through its ability to transfer miR-223 to endothelial cells where it directly targets ICAM-1 at sites in the 3’UTR and suppresses gene expression and function, i.e. leukocyte adhesion. Moreover, to the best of our knowledge this may be the first or one of the earliest examples of an extracellular miRNA regulating genes in a cell where it is not transcribed or processed. In 1995, HDL and apoA-I were both demonstrated to dose-dependently suppress TNF-α-induced ICAM-1 protein expression in endothelial cells34. In this study, lipid-poor apoA-I would not be expected to contain miR-223 levels, as opposed to nHDL, however, both approaches inhibited ICAM-1 induction34. Although nHDL was far superior to lipid-poor apoA-I at inhibiting ICAM1 mRNA levels, apoA-I suppression of mRNA expression in non-activated cells may be a distinct mechanism from apoA-I attenuating TNF-α-induced ICAM-1 expression. Moreover, it’s also possible that apoA-I alone can post-translationally suppress ICAM-1 protein levels. In addition, HDL was not found to suppress TNFα-induced ICAM-1 expression in fibroblasts34, which supports cell specific regulation of ICAM-1 that could be linked to differential HDL-miR-223 delivery to different cell-types.
The HDL-miRNA message
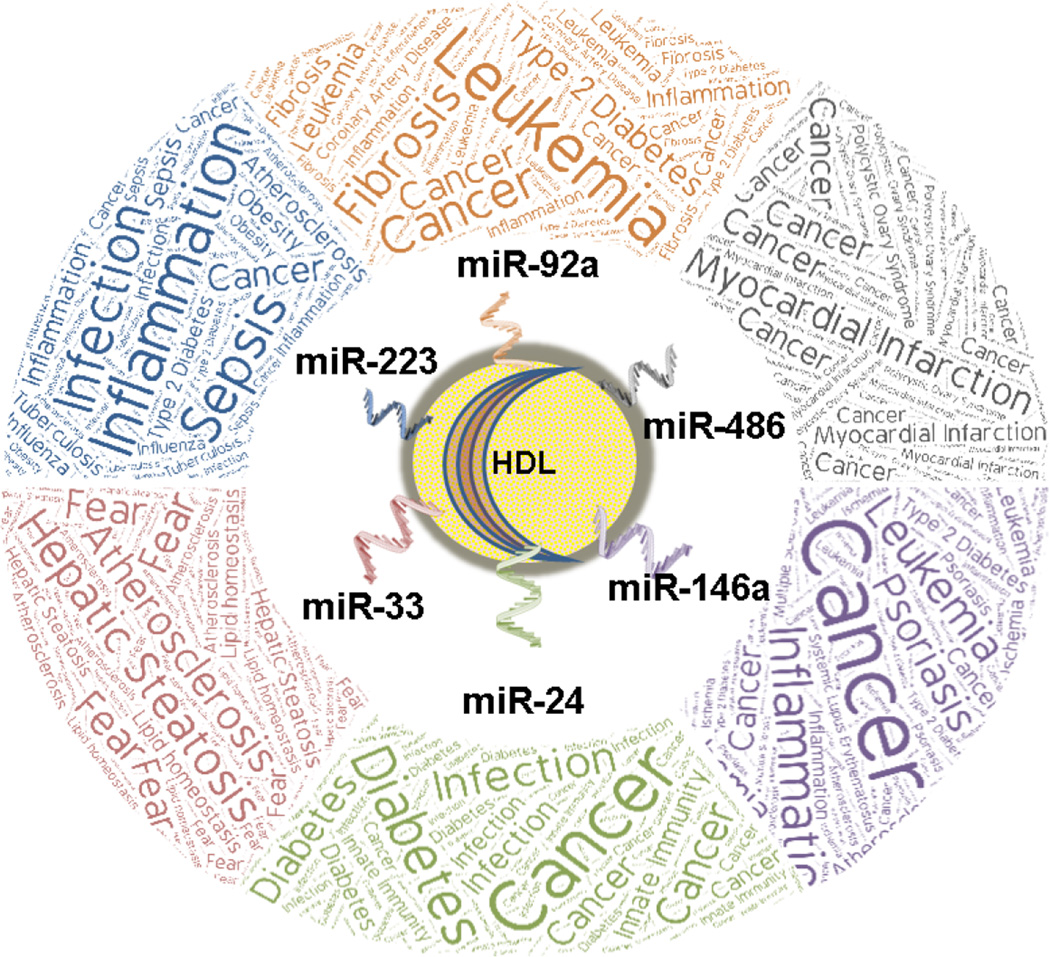
For lipoproteins, it has been convenient, albeit narrowed-minded, to classify LDL as the “bad” cholesterol (lipoprotein) and HDL as the “good” cholesterol, given the relationship between these particles and CVD risk. HDL have many beneficial properties, in addition to its role in reverse cholesterol transport, that antagonize atherosclerosis35. Recently, the concept of damaged or dysfunctional HDL has emerged as critical HDL functions have been reported to be lost or reduced in different metabolic diseases, including CVD36, 37. Currently, it is unclear whether changes in HDL-miRNAs are a consequence of or directly contribute to HDL dysfunction. Nonetheless, HDL-miRNA changes in disease (e.g. atherosclerosis) may hold biomarker potential and clues to the regulation of biological processes that promote or repress disease burden; however, speculating why specific miRNAs are changing and the biological impact of such changes is challenging due to the complexity of miRNA-mediated gene regulation (Figure 1). For example, predicting and then experimentally confirming the biological impact of miRNAs at the gene level (all gene expression changes) in a given tissue is difficult to fully grasp, as miRNAs can directly regulate many different mRNA targets in different tissues based on the stoichiometry of the miRNA and mRNA targets. Most interestingly, a majority of a miRNA’s impact on gene expression is likely mediated through indirect gene expression changes which pose a greater challenge to prediction. Furthermore, the biological impact of miRNAs at the pathway level is even more arduous to define as miRNAs can simultaneously suppress gene targets with opposing influences on a particular pathway. We have observed this with miR-27b regulation of triglyceride metabolism and lipid homeostasis38. As such, when cellular miRNA levels are altered in metabolism or in a disease context it is very difficult to predict changes at the pathway or systemic level as only a small fraction of gene expression changes associated with an altered miRNA would have been predicted. To these points, we are challenged to predict and then experimentally determine the biological impact of HDL-miRNA changes in disease, e.g. hypercholesterolemia. One hypothesis is that HDL-miRNA transport plays a key role in mediating the response to metabolic disease and helps to maintain systemic homeostasis through endocrine-like intercellular communication and distal or adjacent gene regulation, thus antagonizing the disease progress and promoting disease resolution. In other words, HDL-miRNAs may both reflect disrupted homeostasis in contributing cells/tissue and perpetuate the disease in recipient cells/tissues. An opposing hypothesis is that HDL is simply circulating with miRNA disease biomarkers and the biological activities of HDL-miRNA changes in the disease are minimal at the pathway or systemic level. Where HDL-miRNA changes fall in the spectrum between these hypotheses is likely unique for each miRNA and disease. Nevertheless, these barriers do not prevent the prediction of potential responses to HDL-miRNA changes in disease. For example, we reported that miR-223 levels are dramatically increased on HDL in hypercholesterolemia/atherosclerosis and that macrophages export miR-223 to HDL20. We have also found that HDL has the capacity to transfer miR-223 to recipient hepatoma cells in vitro20. In a separate study, we reported that miR-223 is a critical regulator of cholesterol homeostasis in hepatocytes and suppresses cholesterol biosynthesis in the liver, a key source for systemic cholesterol levels39. Connecting the data, one could hypothesize that macrophages may sense hypercholesterolemia or inflammatory cytokines associated with atherosclerosis and export miR-223 to HDL where it is transported to the liver to inhibit cholesterol synthesis and to the endothelium to reduce inflammation. As such, we would predict HDL-miR-223 to be anti-atherogenic and regulate counteractive biological processes to the disease, thus, aligning more towards the first hypothesis above. Nevertheless, this potential HDL-miR-223 axis has not been experimentally tested in vivo. As discussed above, miR-92a levels were reported to be increased on HDL from subjects with MI, UA, and CAD25. miR-92a has also been reported to be a pro-atherogenic miRNA in endothelial cells26. If HDL transfers the increased extracellular miR-92a levels to recipient endothelial cells, it would be expected to promote atherogenesis, not reduce the disease burden. As such, the HDL-miR-92 axis would be expected to align closer to the second hypothesis proposed above. To further illustrate the challenge of predicting the impact of HDL-miRNA changes to disease processes, miR-92a has also been reported to suppress cytokine production and macrophage activation through the JNK pathway40 which would antagonize inflammation and atherosclerosis. Alternatively, the observed increase in HDL-miR-92 levels in CAD may simply represent endothelial dysfunction and atherosclerosis as a novel HDL-miRNA biomarker, in line with the second hypothesis. Similar to miR-92a, miR-486 levels were reported to be increased on HDL from subjects with CAD25. Recently, miR-486 was reported to promote cholesterol accumulation in macrophages through decreased ATP-binding cassette transporter A1 (ABCA1)-mediated cholesterol efflux41. Nonetheless, the level of miR-486 flux between HDL and macrophages and the impact on cholesterol accumulation in macrophages remains to be determined. miR-24 has been reported to be a critical factor in monocyte-macrophage and monocyte-dendritic cell differentiation, as miR-24 levels are inhibited in these cells during differentiation which is critical to cytokine secretion42. miR-24 levels were also reported to be increased on HDL in atherosclerosis in humans and mice20. If increased HDL-miR-24 levels result in increased miR-24 levels in monocytes during hypercholesterolemia/atherosclerosis, then we would predict that increased HDL-miR-24 levels may be beneficial and anti-atherogenic through suppression of macrophage and dendritic cell differentiation. HDL-miRNA changes offer unlimited potential for predictions and hypotheses (Figure 1), but as discussed, it is very difficult to accurately predict the impact or biological relevance of HDL-miRNA changes. As such, these hypotheses will require extensive testing, including loss-of-function studies for each miRNA in the setting of disease and the manipulation of HDL-miRNA intercellular communication in such settings. Nonetheless, HDL and other lipoprotein miRNAs are attractive biomarkers and potential drug targets in metabolic disease.
Figure 1.
Schematic of the potential impact for HDL-miRNA changes in pathophysiology.
Current and future challenges
The most important challenge to this new field is the demonstration of cell-to-cell communication in vivo. To accomplish this, new models and clever use of existing models will be needed. One simple approach would be to inject mouse or human HDL containing a specific miRNA into a miRNA knockout animal and trace HDL-miRNAs to different tissues/cells. This will likely be challenging as it is difficult to perfuse all blood and HDL from tissues and different cells likely have different uptake capacities for different HDL-miRNAs. One could overcome these issues by isolating primary cells (e.g. hepatocytes) after the injection to determine if the HDL-miRNAs were taken up by the hepatocytes in vivo. The limitation to this approach is that this only investigates HDL-miRNA delivery capacity in vivo and not intercellular communication. Strategies that take advantage of labeling specific miRNAs in specific cells will be useful in tracing miRNAs from donor cell-types to HDL and then to recipient cells. Bone-marrow or organ transplantation studies may also be viable approaches to address this problem. The next challenge is to understand if altered HDL-miRNA communication is causal or responsive to different cardiometabolic diseases. Likewise, it is important to understand if HDL-miRNA changes in disease contribute to disease progression (promotes) or resolution (antagonizes). This will be particularly difficult as directional changes in HDL-miRNA activity likely have dichotomous effects on different pathways in different tissues. Loss-of-function studies for specific HDL-miRNAs in models of disease will be required to determine the functional impact of HDL-miRNA communication in metabolic diseases. The ultimate goal of such research is to gain a level of understanding for HDL-miRNA changes in a given disease (e.g. atherosclerosis) that one could manipulate specific HDL-miRNA levels to prevent or treat the disease. Nonetheless, HDL-miRNAs may represent a novel class of disease biomarkers individually or in panels/cassettes that could be useful in multivariate analyses. Collectively, these large broad questions address the potential of HDL-miRNAs as biomarkers and bioactive molecules that may serve novel drug targets; however, much is left to understand about HDL-miRNA export, transport, and delivery.
A major biochemical challenge is to better understand how HDL lipids and/or proteins interact with extracellular miRNAs. At this time, zwitterionic lipids (e.g. phosphatidylcholine) are the most likely binding factors. Furthermore, how specific miRNAs are selected for export and how they are exported to HDL are completely unknown. Compared to other carriers of miRNAs (e.g. extracellular vesicles), lipoproteins are substantially more concentrated in blood. Similar to HDL and LDL, it’s very likely that VLDL also contain miRNAs. The challenge now is to determine the stoichiometry of miRNAs to different types of lipoprotein particles and determine the biological relevance of miRNA cargo in each lipoprotein class to cardiometabolic disease. At the recipient cell, little is understood how HDL-miRNAs are transferred to recipient cells; however, HDL’s receptor SR-BI likely contributes to this process in hepatocytes20, and likely endothelial cells. Another critical question concerning HDL-miRNA function is determining if HDL-miRNAs silence predicted mRNA targets through canonical RISC using Argonaute family proteins or through novel ribonucleoprotein mechanisms for extracellular miRNAs. It is also likely that the route to the cytoplasm may influence lipoprotein miRNA activity in recipient cells, e.g. endocytosis and selective core uptake could result in two different miRNA activities in the cell.
Conclusions
Lipoprotein transport is a new and exciting field; however, there still remains a tremendous amount to discover. The biggest gap in this field is the need to prove that lipoprotein miRNAs are functionally important to disease resolution or progression. To do so, requires that we first demonstrate that lipoprotein transport of miRNAs is biologically relevant and has a functional impact through intercellular communication in vivo. The functional transfer of lipoprotein miRNAs to recipient cells (e.g. endothelial cells) supports a possible cell-to-cell communication pathway that confers HDL’s anti-inflammatory capacity. Nonetheless, miRNAs on HDL and LDL are altered in different cardiometabolic disease in humans and mice and likely hold value as new disease biomarkers. Collectively, lipoprotein transport of miRNAs represents a significant advancement for lipoproteins and extracellular miRNAs. Most importantly, lipoprotein miRNAs hold great potential as novel drug targets in new therapeutic strategies to prevent and treat cardiometabolic diseases.
Highlights.
HDL-miRNA changes in hypercholesterolemia and cardiovascular disease in humans.
HDL-miRNA changes in mouse models of hypercholesterolemia and atherosclerosis.
HDL-miRNA biomarkers of cardiovascular disease.
LDL-miRNAs in cardiovascular disease.
HDL-miRNA transfer to endothelial cells.
HDL’s anti-inflammatory properties mediated by miRNA transfer.
The HDL-miRNA message.
Current and future challenges to the lipoprotein miRNA field.
Acknowledgments
This work was supported by awards from the National Institutes of Health, National Heart, Lung and Blood Institute to K.C.V. HL128996, HL113039, and HL116263. This work was also supported by awards from the American Heart Association to K.C.V. CSA2066001 and D.L.M POST26630003.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol. 2013;33:186–192. doi: 10.1161/ATVBAHA.112.300139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23:91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutation research. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzimagiorgis G, Michailidou EZ, Kritis A, Markopoulos AK, Kouidou S. Recovering circulating extracellular or cell-free RNA from bodily fluids. Cancer epidemiology. 2011;35:580–589. doi: 10.1016/j.canep.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends in biochemical sciences. 2012;37:460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. Journal of proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr. 2007;1:156–158. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: Fit to deliver small RNA. Communicative & integrative biology. 2010;3:447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran S, Palanisamy V. Horizontal transfer of RNAs: exosomes as mediators of intercellular communication. Wiley interdisciplinary reviews RNA. 2012;3:286–293. doi: 10.1002/wrna.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aatonen M, Gronholm M, Siljander PR. Platelet-derived microvesicles: multitalented participants in intercellular communication. Seminars in thrombosis and hemostasis. 2012;38:102–113. doi: 10.1055/s-0031-1300956. [DOI] [PubMed] [Google Scholar]
- 14.Cantaluppi V, Gatti S, Medica D, Figliolini F, Bruno S, Deregibus MC, Sordi A, Biancone L, Tetta C, Camussi G. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney international. 2012;82:412–427. doi: 10.1038/ki.2012.105. [DOI] [PubMed] [Google Scholar]
- 15.Hulsmans M, Holvoet P. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc Res. 2013 doi: 10.1093/cvr/cvt161. [DOI] [PubMed] [Google Scholar]
- 16.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Science signaling. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 17.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA biology. 2012;9:1066–1075. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Kim SI, Shin D, Yoon Y, Choi TH, Cheon GJ, Kim M. Hepatic siRNA delivery using recombinant human apolipoprotein A-I in mice. Biochem Biophys Res Commun. 2009;378:192–196. doi: 10.1016/j.bbrc.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 20.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner J, Riwanto M, Besler C, Knau A, Fichtlscherer S, Roxe T, Zeiher AM, Landmesser U, Dimmeler S. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler Thromb Vasc Biol. 2013;33:1392–1400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 22.Skipski VP, Barclay M, Barclay RK, Fetzer VA, Good JJ, Archibald FM. Lipid composition of human serum lipoproteins. The Biochemical journal. 1967;104:340–352. doi: 10.1042/bj1040340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kontush A, Lindahl M, Lhomme M, Calabresi L, Chapman MJ, Davidson WS. Structure of HDL: particle subclasses and molecular components. Handbook of experimental pharmacology. 2015;224:3–51. doi: 10.1007/978-3-319-09665-0_1. [DOI] [PubMed] [Google Scholar]
- 24.Toth PP, Barter PJ, Rosenson RS, Boden WE, Chapman MJ, Cuchel M, D'Agostino RB, Sr, Davidson MH, Davidson WS, Heinecke JW, Karas RH, Kontush A, Krauss RM, Miller M, Rader DJ. High-density lipoproteins: a consensus statement from the National Lipid Association. Journal of clinical lipidology. 2013;7:484–525. doi: 10.1016/j.jacl.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Niculescu LS, Simionescu N, Sanda GM, Carnuta MG, Stancu CS, Popescu AC, Popescu MR, Vlad A, Dimulescu DR, Simionescu M, Sima AV. MiR-486 and miR-92a Identified in Circulating HDL Discriminate between Stable and Vulnerable Coronary Artery Disease Patients. PLoS One. 2015;10:e0140958. doi: 10.1371/journal.pone.0140958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loyer X, Potteaux S, Vion AC, Guerin CL, Boulkroun S, Rautou PE, Ramkhelawon B, Esposito B, Dalloz M, Paul JL, Julia P, Maccario J, Boulanger CM, Mallat Z, Tedgui A. Inhibition of microrna-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ Res. 2014;114:434–443. doi: 10.1161/CIRCRESAHA.114.302213. [DOI] [PubMed] [Google Scholar]
- 27.Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nature medicine. 2014;20:368–376. doi: 10.1038/nm.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Son DJ, Kumar S, Takabe W, Kim CW, Ni CW, Alberts-Grill N, Jang IH, Kim S, Kim W, Won Kang S, Baker AH, Woong Seo J, Ferrara KW, Jo H. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nature communications. 2013;4:3000. doi: 10.1038/ncomms4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 30.Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G, Catherinet C, Prado-Lourenco L, Levin MG, Thacker S, Sethupathy P, Barter PJ, Remaley AT, Rye KA. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nature communications. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Lenten BJ, Hama SY, de Beer FC, Stafforini DM, McIntyre TM, Prescott SM, La Du BN, Fogelman AM, Navab M. Anti-inflammatory HDL becomes pro-inflammatory during the acute phase response. Loss of protective effect of HDL against LDL oxidation in aortic wall cell cocultures. J Clin Invest. 1995;96:2758–2767. doi: 10.1172/JCI118345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Lenten BJ, Navab M, Shih D, Fogelman AM, Lusis AJ. The role of high-density lipoproteins in oxidation and inflammation. Trends in cardiovascular medicine. 2001;11:155–161. doi: 10.1016/s1050-1738(01)00095-0. [DOI] [PubMed] [Google Scholar]
- 33.Ansell BJ, Navab M, Watson KE, Fonarow GC, Fogelman AM. Anti-inflammatory properties of HDL. Rev Endocr Metab Disord. 2004;5:351–358. doi: 10.1023/B:REMD.0000045107.71895.b2. [DOI] [PubMed] [Google Scholar]
- 34.Cockerill GW, Rye KA, Gamble JR, Vadas MA, Barter PJ. High-density lipoproteins inhibit cytokine-induced expression of endothelial cell adhesion molecules. Arterioscler Thromb Vasc Biol. 1995;15:1987–1994. doi: 10.1161/01.atv.15.11.1987. [DOI] [PubMed] [Google Scholar]
- 35.Vickers KC, Remaley AT. HDL and Cholesterol: Life after the divorce? J Lipid Res. 2013 doi: 10.1194/jlr.R035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenson RS, Brewer HB, Jr, Ansell BJ, Barter P, Chapman MJ, Heinecke JW, Kontush A, Tall AR, Webb NR. Dysfunctional HDL atherosclerotic cardiovascular disease. Nature reviews Cardiology. 2016;13:48–60. doi: 10.1038/nrcardio.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaseda R, Jabs K, Hunley TE, Jones D, Bian A, Allen RM, Vickers KC, Yancey PG, Linton MF, Fazio S, Kon V. Dysfunctional high-density lipoproteins in children with chronic kidney disease. Metabolism: clinical and experimental. 2015;64:263–273. doi: 10.1016/j.metabol.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, Palmisano BT, Tabet F, Cui HL, Rye KA, Sethupathy P, Remaley AT. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci U S A. 2014;111:14518–14523. doi: 10.1073/pnas.1215767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai L, Song Y, Liu Y, Chen Q, Han Q, Chen W, Pan T, Zhang Y, Cao X, Wang Q. MicroRNA-92a negatively regulates Toll-like receptor (TLR)-triggered inflammatory response in macrophages by targeting MKK4 kinase. J Biol Chem. 2013;288:7956–7967. doi: 10.1074/jbc.M112.445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu D, Zhang M, Xie W, Lan G, Cheng HP, Gong D, Huang C, Lv YC, Yao F, Tan YL, Li L, Zheng XL, Tang CK. MiR-486 Regulates Cholesterol Efflux by Targeting HAT1. Biochem Biophys Res Commun. 2015 doi: 10.1016/j.bbrc.2015.11.128. [DOI] [PubMed] [Google Scholar]
- 42.Fordham JB, Naqvi AR, Nares S. Regulation of miR-24, miR-30b, and miR-142-3p during macrophage and dendritic cell differentiation potentiates innate immunity. J Leukoc Biol. 2015;98:195–207. doi: 10.1189/jlb.1A1014-519RR. [DOI] [PMC free article] [PubMed] [Google Scholar]