Abstract
Human T-cell leukemia virus type 1 (HTLV-1) is an oncogenic retrovirus that induces a fatal T-cell malignancy, adult T-cell leukemia (ATL). Among several regulatory/accessory genes in HTLV-1, HTLV-1 bZIP factor (HBZ) is the only viral gene constitutively expressed in infected cells. Our previous study showed that HBZ functions in two different molecular forms, HBZ protein and HBZ RNA. In this study, we show that HBZ protein targets retinoblastoma protein (Rb), which is a critical tumor suppressor in many types of cancers. HBZ protein interacts with the Rb/E2F-1 complex and activates the transcription of E2F-target genes associated with cell cycle progression and apoptosis. Mouse primary CD4+ T cells transduced with HBZ show accelerated G1/S transition and apoptosis, and importantly, T cells from HBZ transgenic (HBZ-Tg) mice also demonstrate enhanced cell proliferation and apoptosis. To evaluate the functions of HBZ protein alone in vivo, we generated a new transgenic mouse strain that expresses HBZ mRNA altered by silent mutations but encoding intact protein. In these mice, the numbers of effector/memory and Foxp3+ T cells were increased, and genes associated with proliferation and apoptosis were upregulated. This study shows that HBZ protein promotes cell proliferation and apoptosis in primary CD4+ T cells through activation of the Rb/E2F pathway, and that HBZ protein also confers onto CD4+ T-cell immunophenotype similar to those of ATL cells, suggesting that HBZ protein has important roles in dysregulation of CD4+ T cells infected with HTLV-1.
INTRODUCTION
Human T-cell leukemia virus type 1 (HTLV-1) is the etiological agent of a malignancy of CD4+CD25+ T cells, adult T-cell leukemia (ATL) and several inflammatory diseases such as HTLV-1-associated myelopathy/tropical spastic paraparesis and HTLV-1 uveitis.1,2 In HTLV-1-infected individuals, the provirus load, which corresponds to the number of infected cells in peripheral blood, is maintained at a constant level during the latent period, although viral replication is generally suppressed and viral particles cannot be detected in the serum.3 HTLV-1 propagates in vivo in two different ways: cell-to-cell transmission to uninfected cells (de novo infection) and clonal proliferation of infected cells (mitotic expansion).4,5 The fact that HTLV-1 causes infected cells to proliferate is probably related to the fact that it causes transformation of an infected clone, that is ATL, in a small fraction of carriers decades after the initial infection.
HTLV-1 regulatory/accessory genes are known to affect the expression and function of host factors.1 In particular, Tax and HTLV-1 bZIP factor (HBZ) expression in infected cells were shown to be important for leukemogenesis, because transgenic animal models expressing these viral genes developed malignant tumors.6 Tax is a potent activator of viral gene expression and of many oncogenic pathways such as nuclear factor-κB, PI3K/AKT and AP1, but its expression cannot be detected in 60% of ATL cases.1 HBZ is encoded by the anti-sense strand of the HTLV-1 provirus;7 it is the only viral gene that is genetically conserved and constitutively expressed in HTLV-1-infected cells and ATL cells, which suggests a role in pathogenesis.8,9 HBZ is unique in that it has functions associated with both its RNA and protein forms.8,10 We previously reported that HBZ RNA supports the proliferation of the IL-2-dependent human T-cell line, Kit225 and mouse primary CD4+ T cells.8,10 HBZ protein interacts with many host factors through several protein-binding motifs, such as LxxLL motifs and the bZIP domain to dysregulate cellular signaling pathways.11 We recently found that HBZ protein also promotes the proliferation of mouse primary CD4+ T cells, but it consequently induced apoptosis, unlike HBZ RNA.10 The significance and molecular mechanisms of the induction of apoptosis by HBZ protein have not been clearly defined.
Retinoblastoma (Rb) is a well-known tumor suppressor protein that has important roles in regulation of the cell cycle, DNA replication, differentiation and apoptosis.12 In cells in G0/G1 phase, hypophosphorylated Rb binds to E2F transcription factors and suppresses E2F-dependent gene expression. In response to growth-promoting signals, Rb is phosphorylated, and E2Fs are dissociated from the complex, resulting in the activation of E2F-mediated gene transcription. The E2F family induces expression of many genes associated with the G1/S transition, DNA replication and DNA repair. Overactive E2F-1 can also induce apoptosis,13 perhaps as part of a safety mechanism to prevent the malignant transformation of abnormal cells. Rb is frequently inactivated in many human cancers including virus-induced tumors, but the relationship between Rb and HBZ has not been evaluated prior to this study.
Here we show that HBZ protein interacts with Rb, dissociates histone deacetylase (HDAC) from the Rb/E2F-1 complex and induces transcription of E2F-target genes that are associated with the G1/S transition and apoptosis. In primary CD4+ T cells, HBZ protein strongly promotes cellular proliferation and induces apoptosis. These phenotypes are also observed in CD4+ T cells from HBZ transgenic (HBZ-Tg) mice, which develop diseases similar to those of HTLV-1 carriers.14 In contrast, HBZ RNA enhances cell growth but not apoptosis.10 These different modes of HBZ function are likely to be relevant to the oncogenic process in HTLV-1-infected cells.
RESULTS
HBZ protein binds to Rb protein
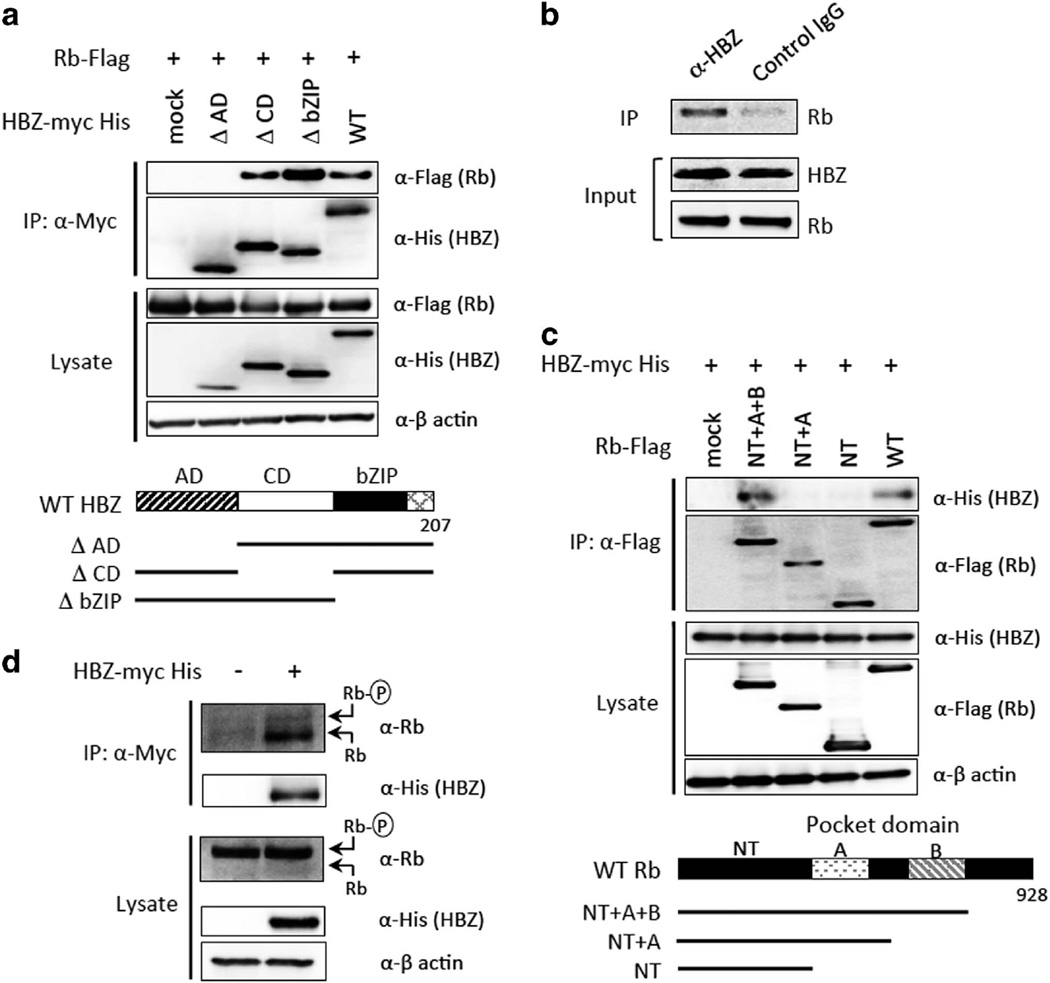
HBZ modulates several signaling pathways through interaction with host proteins; however, the important cellular target proteins of HBZ in cell cycle regulation remain unclear. We asked if Rb protein is a target of HBZ, because Rb is one of the key molecules in cell cycle regulation and many viral oncoproteins, including adenovirus E1A, human papilloma virus E7, SV40 T-antigen and also HTLV-1 Tax, are known to bind to Rb.15,16 A co-immunoprecipitation assay demonstrated that HBZ and Rb do interact with each other in 293FT cells (Figure 1a). By using a series of HBZ deletion mutants, we found that the activation domain of HBZ is required for its interaction with Rb (Figure 1a). We further performed an immunoprecipitation assay using MT4, an HTLV-1-infected human T-cell line, to confirm the interaction of endogenous HBZ with Rb. An anti-HBZ antibody co-precipitated endogenous Rb in MT4 cells, which indicated that the interaction occurs in HTLV-1 infected cells (Figure 1b).
Figure 1.
HBZ interacts with the B pocket of hypophosphorylated retinoblastoma (Rb) protein. (a) Interaction between Rb and wild-type HBZ and the HBZ deletion mutants. Schematic diagrams of the HBZ mutants are shown at the bottom. (b) Interaction between endogenous Rb and HBZ in MT4 cells. (c) Interaction between HBZ and wild-type Rb and Rb deletion mutants. Schematic diagrams of the Rb mutants are shown at the bottom. (d) HBZ preferentially binds to hypophosphorylated Rb. Expression vectors for the indicated proteins were transfected into 293FT cells, and protein interactions were analyzed by immunoprecipitation (IP). AD, activation domain; CD, central domain; NT, N terminus; WT, wild type. Representative data of three independent experiments are shown.
Rb is a member of the pocket protein family that conserves A and B-pocket domains. Viral and host proteins harboring the LxCxE-motif are known to interact with the B pocket of Rb and affect its activity.15 Our next co-immunoprecipitation experiment suggested that the B pocket of Rb is responsible for its binding to HBZ (Figure 1c). Although there is no sequence in HBZ that matches the LxCxE-motif, we found that 22 amino acids in the N terminus of HBZ were responsible for its interaction with Rb (Supplementary Figure S1). These observations suggest that HBZ can interfere with the association between the B-pocket domain of Rb and its cellular interactants.
In the immunoblot for Rb, two distinct bands represent its different phosphorylation states: the higher band is the hyperphosphorylated form and the lower is the hypophosphorylated one. When HBZ was overexpressed in 293FT cells, hypophosphorylated Rb was preferentially associated with HBZ, whereas mainly hyperphosphorylated Rb was present in the cell lysate (Figure 1d). We found no change in the total phosphorylation level or basal expression level of Rb in the presence of HBZ, suggesting that HBZ has no effect on the phosphorylation or stability of Rb.
HBZ inhibits the interaction between Rb and HDAC3
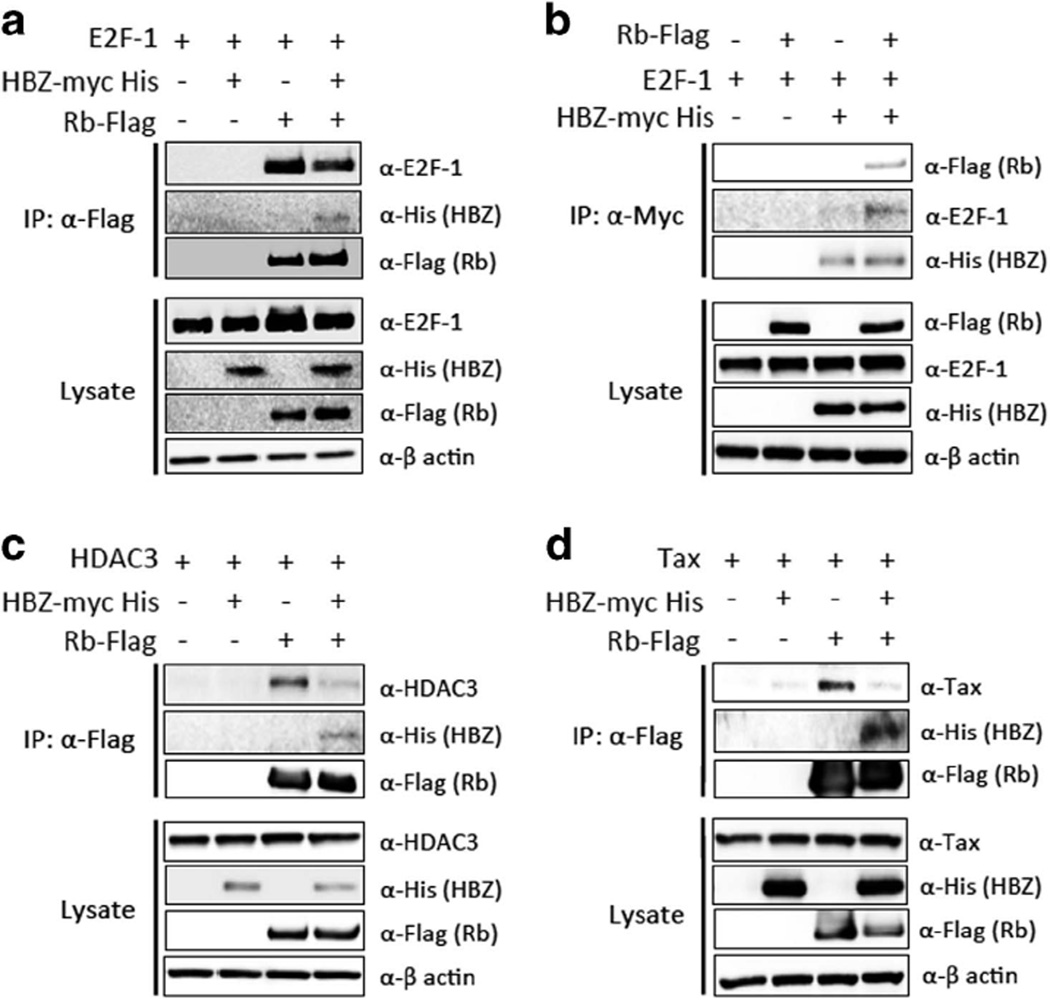
One of the major functions of Rb is inhibition of E2Fs, the critical transcription factors in cell cycle regulation. Rb suppresses E2F-mediated gene transcription through two different mechanisms.17 First, Rb binds to the transactivation domain of E2Fs, and inhibits their transcriptional activity. Second, the Rb/E2F complex recruits histone modifiers, such as HDACs, to the promoter region of E2F-target genes, and leads to transcriptional silencing by epigenetic means. Hypophosphorylation of Rb exposes its B-pocket region, allowing Rb to interact with E2Fs and HDACs.15 As HBZ appears to associate with the same domain of hypophosphorylated Rb (Figure 1b), we speculated that HBZ might affect the interaction between Rb and E2F and/or HDACs.
First, we investigated the interaction between HBZ, Rb and E2F-1. Contrary to our expectation, Rb could bind to E2F-1 even in the presence of HBZ, indicating that HBZ did not affect their binding (Figure 2a). Next, we evaluated the effect of Rb on the association between HBZ and E2F-1, and found that their binding looked stronger in the presence of Rb than in its absence (Figure 2b). These results suggest that Rb stabilizes the interaction between HBZ and E2F-1. However, this stabilization did not inactivate E2F-1 in the presence of HBZ, as shown in subsequent experiments. Next, we investigated whether HBZ affects the recruitment of HDAC to the Rb/E2F complex. In contrast to forming a complex with Rb and E2F-1, HBZ dissociated HDAC3 from Rb (Figure 2c).
Figure 2.
HBZ alters the interaction of Rb with HDAC3 and Tax. (a) Effect of HBZ on the interaction between Rb and E2F-1. (b) Effect of Rb on the interaction between HBZ and E2F-1. (c) Effect of HBZ on the interaction between Rb and HDAC3. (d) Effect of HBZ on the interaction between Rb and Tax. 293FT cells were transfected with indicated expression vectors, and protein interaction was analyzed by IP. Representative data of three independent experiments are shown.
It was reported that HTLV-1-encoded Tax protein also binds to Rb protein through the B pocket, and that this interaction results in the degradation of Rb.16 Interestingly, Tax/Rb interaction was suppressed by HBZ in cells transfected with both Tax and HBZ (Figure 2d). Indeed, Rb protein could be detected in HTLV-1-infected cell lines regardless of tax expression (Supplementary Figure S2), suggesting that HBZ modulates the effect of Tax on Rb levels.
HBZ activates E2F-1 and leads to G1/S transition in vitro
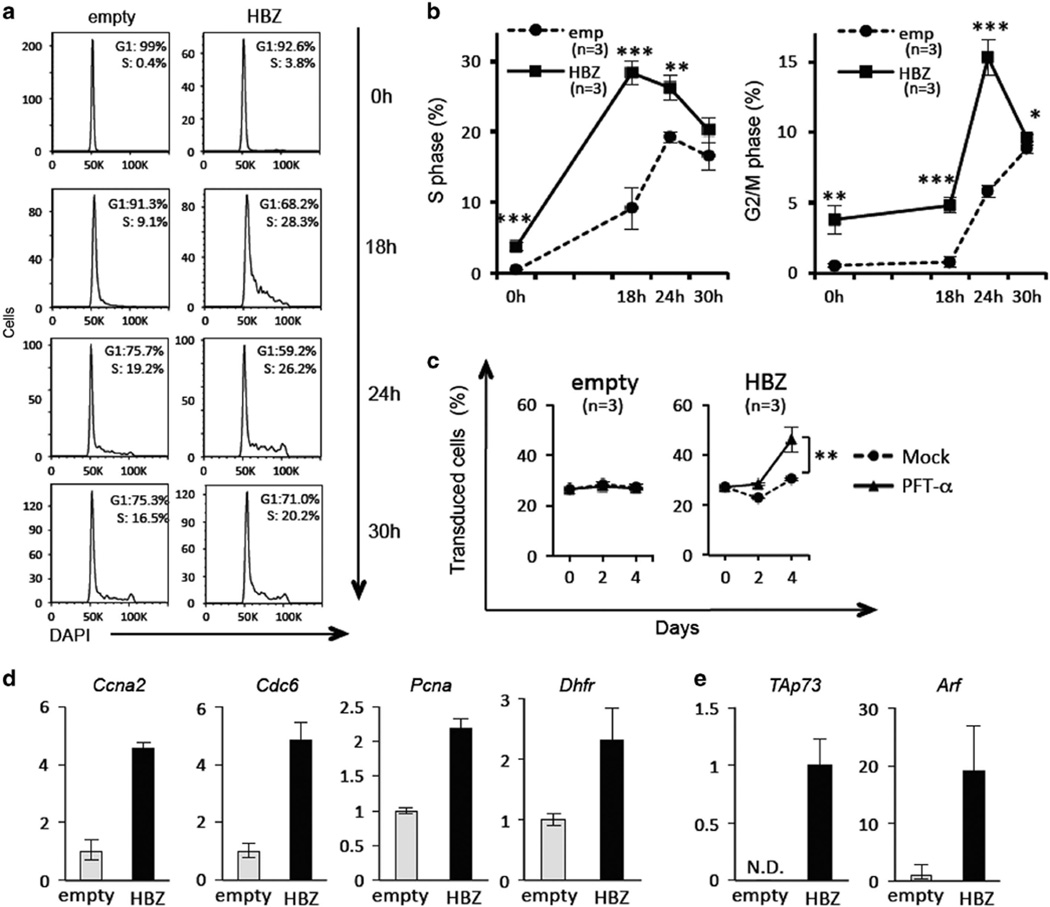
To study the effects of HBZ on the Rb/E2F pathway, we transduced mouse primary CD4+ T cells with a retroviral vector expressing HBZ and analyzed the impact of HBZ on cell cycle distribution. In unsynchronized culture, the HBZ-expressing population had a higher number of cells in S phase than empty vector-transduced cells (Supplementary Figure S3), suggesting that HBZ promotes G1/S transition. To observe G1/S transition more precisely, we synchronized empty vector- and HBZ-transduced cells in G1 phase by culturing them in medium containing a low concentration (10 U/ml) of rIL-2 (Figure 3a). After cells were released from G1 arrest by a higher concentration (40 U/ml) of rIL-2, HBZ-expressing cells entered S phase significantly faster than control cells (Figure 3a and b). At 18 h after release, the percentage of HBZ-expressing cells in S phase was 28%, and that of control cells was 9%. A consequent increase in G2/M cells was also observed (15% of HBZ-transduced vs 6% of empty vector-transduced) 24 h after release (Figure 3b).
Figure 3.
Forced expression of HBZ promotes G1/S transition and apoptosis in primary CD4+ T cells. (a, b) Mouse primary CD4+ T cells were transduced with HBZ and cultured in low IL-2 (10 U/ml) containing medium. Thirty-six hours after IL-2 withdrawal, transduced CD4+ T cells were purified using a FACSAria2 and cultured in fresh medium containing higher IL-2 (40 U/ml). Cell cycle distribution was monitored at the indicated times (0, 18, 24 and 30 h after release from G1 phase). Three independent experiments were performed in triplicate, and the representative data are shown. (c) GFP competition assay was carried out using bicistronic GFP expression vectors. After retroviral transduction of each construct, the proportion of transduced cells (GFP+ fraction) was measured by flow cytometry at the indicated date. Three independent experiments were performed in triplicate, and the representative data are shown. (d) Relative expression levels of cell cycle inducing genes and (e) proapoptotic genes were evaluated by qPCR. The expression level of genes was measured in triplicate. Statistical significance was determined by the two-tailed paired Student's t-test. N.D., not detectable. Asterisks indicate the statistical significance as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
HBZ enhances transcription of p53/p73 target genes and induces apoptosis in mouse CD4+ T cells
As over-activation of E2F-1 is known to induce p53/p73-dependent apoptosis,18 we evaluated apoptosis in HBZ-transduced cells and the involvement of the p53 and/or p73 pathways. We carried out a green fluorescent protein (GFP) competition assay with or without pifithrin-α (PFT-α; 2-(2-imino-4,5,6,7-tetrahydrobenzothiazol-3-yl)-1-p-tolylethanone), which inhibits apoptosis induced via p53 and p73. PFT-α was originally identified as an inhibitor of p53-induced transcription and apoptosis; thus is widely used as a p53 inhibitor.19 In addition, it was reported that PFT-α could prevent the p73 increase and apoptosis induced by a genotoxic agent.20 As shown in Figure 3c, the percentage of empty vector-transduced cells was unchanged 4 days after the treatment, and there was no effect of PFT-α on their proportion, indicating that empty vector-transduced cells, which are GFP-positive, did not have any growth advantage, nor did they undergo p53/p73-mediated apoptosis. Cells transduced with HBZ were almost equivalent at day 4 compared with day 0, and PFT-α significantly increased the percentage of HBZ-transduced cells. This result implies that HBZ has a growth-promoting activity on mouse CD4+ T cells, although the number of cells is simultaneously kept in check by apoptosis which likely is mediated by p53 and/or p73.
We next analyzed the effects of HBZ on the transcription of E2F-1 target genes by quantitative reverse transcriptase-PCR. Genes related to S phase progression, including Pcna, Ccna2, Dhfr and Cdc6, were upregulated in wild-type HBZ-transduced mouse CD4+ T cells (Figure 3d). In addition, the proapoptotic E2F-1 target genes, TAp73 and Arf, which act in p73- and p53-dependent apoptosis respectively, were markedly induced by HBZ (Figure 3e). These results suggest that HBZ enhances transcription of p53 and p73 target genes by activation of E2F-1, and subsequently induces apoptosis in primary CD4+ T cells.
The Rb/E2F-1 pathway is over-activated in HBZ-Tg mice
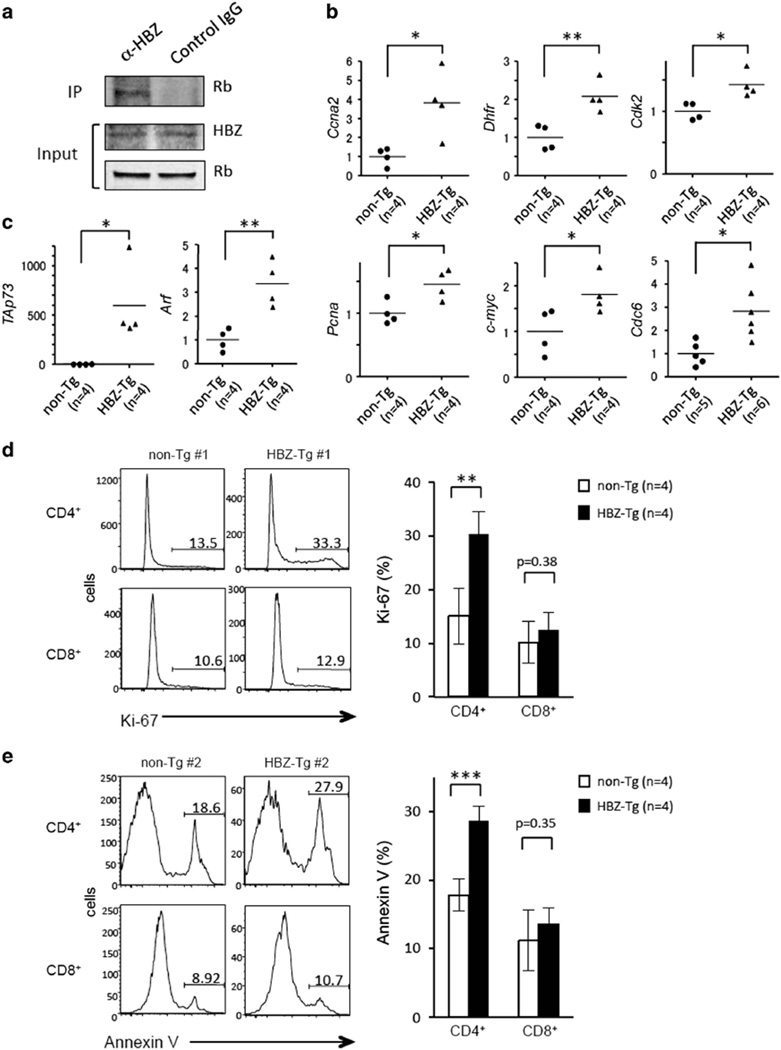
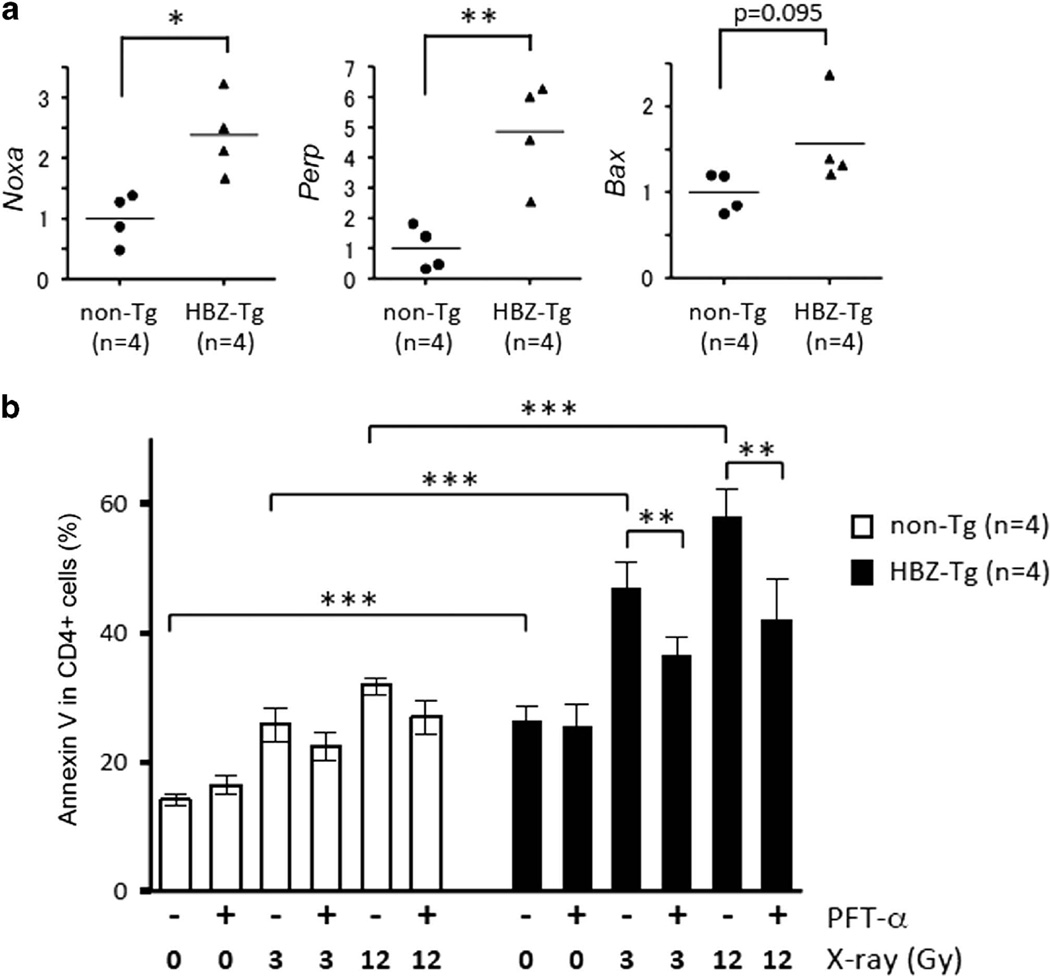
To analyze the effects of HBZ on the Rb/E2F-1 pathway in vivo, we used the HBZ-Tg mouse model that we previously established.8 We first isolated splenocytes from the HBZ-Tg mice and confirmed the interaction between HBZ and Rb by mmunoprecipitation (IP) (Figure 4a). Next we compared the expression levels of E2F-1 target genes in CD4+ T cells of HBZ-Tg with those of non-transgenic littermates. Like retrovirally transduced HBZ-expressing cells, HBZ-Tg CD4+ T cells had increased expression of genes associated with G1/S progression, such as Pcna, Ccna2, Dhfr, Cdk2, c-myc and Cdc6 (Figure 4b), and also of the proapoptotic genes TAp73 and Arf (Figure 4c). These observations suggest that HBZ enhances E2F-1 activity in vivo. We stained HBZ-Tg cells with anti-Ki-67, a marker for cell proliferation, and found that HBZ-Tg CD4+ T cells are more frequently in cell cycle progression compared with non-Tg cells (Figure 4d). In addition, when HBZ-Tg splenocytes were cultured ex vivo without any stimulation, increased apoptosis of CD4+ T cells was observed (Figure 4e), which is consistent with high levels of expression of TAp73 and Arf. We analyzed CD8+ T cells (which do not express HBZ) in those mice, and found that neither cell cycle progression nor apoptosis was increased in HBZ-Tg CD8+ cells (Figure 4d and e), suggesting that increased proliferation and apoptosis of CD4+ T cells was caused by HBZ, likely via activation of the Rb/E2F pathway and consequent activation of the p53/p73 pathways. Indeed, we found that expression of the proapoptotic genes regulated by p53 and p73, Noxa, Perp and Bax was upregulated in HBZ-Tg CD4+ T cells (Figure 5a).
Figure 4.
CD4+ T cells from HBZ-Tg mice show activation of the E2F pathway, progression of cell cycle and induction of apoptosis. (a) Interaction between endogenous Rb and HBZ in HBZ-Tg splenocytes. The result is representative of three independent experiments. The expression of E2F-1 target genes related to cell cycle (b) and apoptosis (c) in CD4+ T cells from non-Tg or HBZ-Tg mice was analyzed by qPCR. Each symbol represents an individual mouse. The horizontal lines indicate mean values. (d) Representative histograms of Ki-67 expression in CD4+ and CD8+ T cells from non-Tg (n=4) and HBZ-Tg (n=4) mice (left). The results are shown as the mean ± s.d. (right). (e) Apoptotic CD4+ T cells and CD8+ T cells from non-Tg (n=4) and HBZ-Tg (n=4) mice were analyzed by Annexin V staining. Representative histograms are shown (left), and the results are shown as the mean±s.d. (right). Statistical significance was determined by the two-tailed paired Student's t-test. Asterisks indicate the statistical significance as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 5.
p53 and p73-associated apoptotic signals are enhanced in CD4+ T cells of HBZ-Tg mice. (a) The expression of proapoptotic p53/p73 target genes in CD4+ T cells from non-Tg (n=4) and HBZ-Tg (n=4) mice was measured by qPCR. (b) Splenocytes from non-Tg (n=4) and HBZ-Tg (n=4) mice were treated with or without PFT-α for 30 min and then irradiated as indicated. The percentage of apoptotic cells in the CD4+ fraction was measured by Annexin V staining. Two independent experiments were performed and representative data are shown. Statistical significance was determined by the two-tailed paired Student's t-test. Asterisks indicate the statistical significance as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
In order to evaluate the effects of HBZ on apoptosis in more detail, we irradiated HBZ-Tg cells ex vivo and analyzed the number of apoptotic cells. As expected, HBZ-Tg cells were more sensitive to X-ray irradiation (Figure 5b). Furthermore, the inhibitory effect of PFT-α on irradiation-induced apoptosis was more obvious in CD4+ T cells from HBZ-Tg mice than those of non-Tg littermates (Figure 5b), suggesting that p53 and p73 are involved in the enhanced apoptosis of HBZ-Tg cells.
CD4+ T cells of HBZ SM-Tg mice showed similar phenotypes to those of HBZ-Tg mice
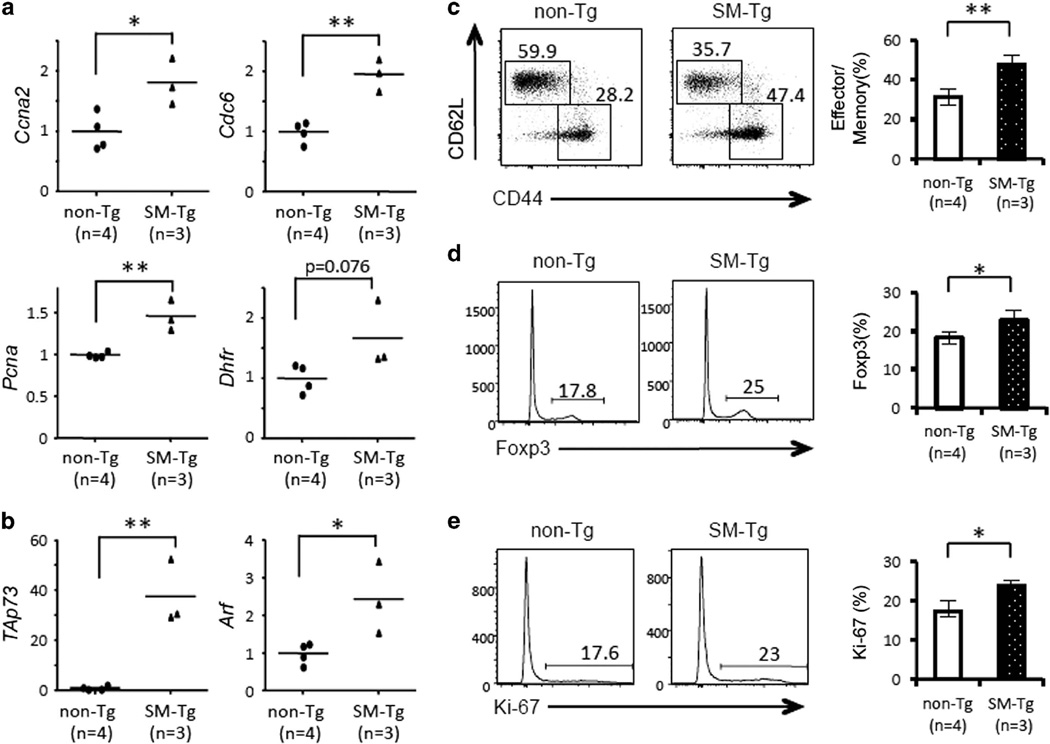
We have reported that HBZ RNA and HBZ protein have the distinct functions in primary CD4+ T cells; HBZ protein strongly induces apoptosis, whereas HBZ RNA inhibits it.10 In order to evaluate the functions of HBZ protein precisely in vivo, we generated a new transgenic strain expressing the HBZ SM mutant (SM-Tg), which contains silent mutations in the coding sequence (Supplementary Figure S4a). The SM mutant produces intact HBZ protein, but its RNA structure is completely different from that of wild type. Although the promoter of the HBZ SM is the same as the original HBZ-Tg mice, the expression level of the transgene in SM-Tg mice was about one-fifth of that in HBZ-Tg mice (Supplementary Figure S4b). We found that S phase-related genes, such as Ccna2, Cdc6, Pcna and Dhfr (Figure 6a), and the proapoptotic genes TAp73 and Arf (Figure 6b) were upregulated in CD4+ T cells from SM-Tg mice compared with non-Tg littermates. In SM-Tg mice, CD44+CD62L− effector/memory T cells and Foxp3+ cells were significantly increased among the CD4+ fraction (Figure 6c and d). Importantly, the number of Ki-67+ cells was also higher in SM-Tg mice than in non-Tg mice, suggesting that SM-Tg cells were more proliferative (Figure 6e). These features of CD4+ T cells in SM-Tg mice were quite similar to those of HBZ-Tg mice, indicating that HBZ protein, rather than RNA, is responsible for these immunophenotypic alterations observed in HBZ-Tg mice.
Figure 6.
CD4+ T cells of SM-Tg mice show activation of the E2f pathway. The expression of E2F-1 target genes related to the cell cycle (a) and apoptosis (b) in CD4+ T cells from non-Tg (n=4) or SM-Tg (n=3) mice was analyzed by qPCR. Each symbol represents an individual mouse. The horizontal lines indicate mean values. (c) Representative dot plots of CD44 and CD62L expression in CD4+ T cells from non-Tg (n=4) and SM-Tg (n=3) mice (left) and mean percent of CD4+ T cells that are effector/memory T cells (CD44+CD62L−) (right). (d) Representative histograms of Foxp3 expression in CD4+ T cells from non-Tg (n=4) and SM-Tg (n=3) mice (left) and mean results (right). (e) Representative histograms of Ki-67 expression in CD4+ T cells from non-Tg (n=4) and SM-Tg (n=3) mice (left) and mean results (right). Statistical significance was determined by the two-tailed paired Student's t-test. Asterisks indicate the statistical significance as follows: *P < 0.05; **P < 0.01.
DISCUSSION
Many oncoviruses are known to target Rb, resulting in activation of Rb/E2F-1-mediated gene transcription. Oncoviral suppression of Rb is generally due to destabilization of Rb itself or of the Rb/E2F complex. Human papilloma virus E7 protein,21 hepatitis C virus NS5B22 and Epstein–Barr virus EBNA3C23 subject Rb to ubiquitin-mediated proteasomal degradation. Polyoma virus-encoded large T antigens (LTs), such as SV40 LT and Merkel cell polyomavirus LT,24 interact with Rb and upregulate E2F-target genes by destroying the Rb/E2F complex. In addition, Tax was reported to induce ubiquitin-independent degradation of Rb by tethering it to proteasome subunit α2.16 In this study, we found that HBZ protein associates with the Rb/E2F complex without dissociating it, but removes HDAC from the complex instead, whereas HBZ has no effect on the stability of Rb protein. These findings indicate that HTLV-1 utilizes novel ways to suppress the function of Rb.
It is known that Tax and HBZ tend to target the same cellular proteins/pathways and regulate them in opposite ways, suggesting that these two viral oncoproteins can fine-tune many signaling pathways by their counter-action in HTLV-1-infected cells.2 Here, we found that HBZ and Tax competitively interact with Rb through its B-pocket motif, but both of them suppress the function of Rb. Tax expression is driven by the 5′ long terminal repeat (LTR), and HBZ is transcribed from the 3′LTR. Recent studies using LTR-reporter plasmids showed that each LTR is differentially regulated in T cells. The activity of the 5′LTR is ordinarily low but is strongly enhanced by stimuli such as TCR stimulation and Tax.25,26 In contrast, the 3′LTR is consistently activated through Sp1-responsive motifs.26,27 These findings suggest that Tax expression is transiently induced, whereas HBZ is continuously expressed in HTLV-1-infected T cells. We therefore hypothesize that HBZ and Tax collaborate to consistently activate the Rb/E2F pathway regardless of the activation status of HTLV-1-infected cells.
E2F-1 is known to be an inducer of apoptosis as well as a cell cycle accelerator.18 In response to various stresses, especially DNA damage, E2F-1 is stabilized by ATM, CHK1 and CHK2,18 and over-activation of E2F-1 leads cells to apoptosis through p53- and p73-dependent pathways.28,29 It has been also reported that Rb loss induces the accumulation of replication-dependent DNA double-strand breaks.30 Thus, E2F-1-induced apoptosis is thought to contribute to the elimination of pre-malignant cells that have an accumulation of DNA damage. In this study, we demonstrate that HBZ protein triggers both proliferation and apoptosis of primary CD4+ T cells (Figures 3c, 4d and e), suggesting that interaction between HBZ protein and Rb over-activates the E2F-1 pathway. This concurrence of cell proliferation and apoptosis resembles the oncogenic stress responses triggered by activation of proto-oncoproteins such as Ras and Myc.31 In cancer cells, this failsafe mechanism is overridden in a variety of ways. Interestingly, it was reported recently that HBZ downregulates a cellular gene, OBFC2A, which is involved in detection and repair of DNA damage.32 Moreover, we recently found that HBZ RNA possesses antiapoptotic activity through induction of survivin in mouse primary CD4+ T cells; this activity opposes the proapoptotic activity of the HBZ protein.10 These effects of HBZ might cause genetic instability and enable cells to evade apoptosis triggered by HBZ protein and/or DNA damage.
CD4+ T cells from wild-type HBZ-Tg mice demonstrated similar phenotypes to HBZ-transduced cells: enhanced proliferation, increased apoptosis and transcriptional activation of p53/p73 target genes. These findings suggest that the effects of HBZ protein, rather than HBZ RNA, are dominant in HBZ-Tg mice. Indeed, the immunophenotypes of CD4+ T cells from transgenic mice expressing the HBZ SM mutant gene were similar to CD4+ T cells from transgenic mice expressing wild-type HBZ: the effector/memory T-cell and Foxp3+ T-cell populations in both types of transgenic mice were increased compared with those in non-Tg mice. HBZ protein is known to interact with the transcriptional coactivator p300 and enhance the expression Foxp3, resulting in the increase in Foxp3+ T cells in HBZ-Tg mice.14,33 As the increase in Foxp3+ T cells and the concurrent induction of IFN-γ-producing Th1-like CD4+ T cells have been implicated in inflammation in HBZ-Tg mice,34 HBZ protein might have pathogenic properties by itself. To understand the precise roles of HBZ protein in HBZ-mediated inflammation and oncogenesis, further analysis of HBZ SM-Tg mice will be required.
In conclusion, we show here that HBZ protein interacts with the Rb/E2F-1 complex and induces transcription of E2F-target genes. Activation of the Rb/E2F pathway by HBZ protein accelerates G1/S transition and apoptosis of primary CD4+ T cells. Both HBZ and Tax can bind to Rb and suppress its inhibitory effect on E2F-mediated gene transcription, suggesting that E2F is continuously activated in HTLV-1-infected cells. HBZ RNA also induces cellular proliferation, and potentially could suppress apoptosis induced by HBZ protein. These properties of HBZ might be manifestations of important strategies of HTLV-1 to promote clonal proliferation of infected cells, and as a side effect, induce the leukemogenesis of ATL.
MATERIALS AND METHODS
Cell lines
Human T-cell lines infected with HTLV-1 (MT1, MT2, MT4, HPB-ATL-T, HPB-ATL-2, TL-Om1 and ED)35,36 and uninfected T-cell lines (Jurkat and SupT1) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics at 37 °C under 5% CO2 atmosphere. Kit225 cells were maintained in RPMI 1640 medium supplemented with 10% FBS, antibiotics and 85 U/ml recombinant IL-2 (WAKO, Osaka, Japan).8 The human embryonic kidney cell line 293FT (Thermo Fisher Scientific, Waltham, MA, USA) was cultured in Dulbecco’s modified Eagle medium supplemented with 10% FBS and 500 µg/ml G418.
Plasmids
An HBZ-expressing plasmid, pcDNA3.1mycHis-HBZ was generated as previously described.37 The coding sequences of Rb, E2F-1 and HDAC3 were amplified by reverse transcriptase-PCR and subcloned into pCAG-Flag,38 pcDNA3 and pcDNA3.1, respectively. Retroviral expression vector for HBZ was described previously.14 The HBZ mutant that functions only in protein form (silent mutations: SM) was also used in the previous study.8 pCG-Tax was kindly provided by Dr Fujisawa, Kansai Medical University, Osaka, Japan.
Mice
Transgenic (Tg) mice expressing wild-type HBZ under the murine CD4 promoter have been previously described.8 Transgenic mice expressing the HBZ SM mutant, which functions only in protein form, were generated using the same promoter. In brief, the sequence of the HBZ SM mutant was cloned into the H/M/T-CD4 vector,39 and the expression cassette was microinjected into C57BL/6 J embryos. An F0 founder mouse was crossed with wild-type C57BL/6 J mice, and transgenic progeny were obtained. Tg mice at 7–14 weeks of age were randomly analyzed, and compared with the age-matched controls. All data obtained from mice were subjected to the statistical analysis without exclusion. Three to six non-Tg and Tg mice were analyzed in each experiment, and the exact numbers of mice (n) are shown in the figures and figure legends. No blinded test was done in this study. Animal experiments were approved by the Institutional Animal Research Committee of Kyoto University (Permit numbers D13-02, D14-02 and D15-02).
IP and immunoblotting
Lysates of 293FT cells transfected with the expression vectors were incubated with anti-c-myc (clone 9E10, Sigma-Aldrich, St Louis, MO, USA) or anti-Flag M2 antibodies (Sigma-Aldrich) for 1 h at 4 °C, and immune complexes were incubated with protein G-sepharose (GE Healthcare, Little Chalfont, UK) for 1 h at 4 °C. For immunoprecipitation of HBZ-binding proteins, rabbit polyclonal anti-HBZ antibody40 was incubated with the lysates of MT4 or splenocytes from HBZ-Tg mice. The following antibodies were used for immunoblotting: anti-Flag (M2, Sigma-Aldrich), anti-His (PM002, MBL, Nagoya, Japan), anti-Actin (AC-40, Sigma-Aldrich); anti-Rb (clone 4H1 from Cell Signaling Technology (Danvers, MA, USA) and clone EPR17512 from Santa Cruz Biotechnologies, Dallas, TX, USA), anti-E2F-1 (clone c-20, Santa Cruz Biotechnologies), anti-HDAC3 (polycolonal, Abcam, Cambridge, UK) and anti-Tax (clone MI73).35 Peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG were purchased from GE Healthcare. All IP and immunoblotting in this study were repeated independently at least three times, and the representative results are shown.
Flow cytometric analysis
Single-cell suspensions of mouse splenocytes were prepared in phosphate buffered saline containing 1% FBS and stained with monoclonal antibodies for CD4 (clone RM4–5, BD, San Jose, CA, USA), CD8 (clone 53.6.7, BioLegend, San Diego, CA, USA), CD44 (clone IM7, BD), CD62L (clone MEL-14, eBioscience, San Diego, CA, USA) and Ki-67 (clone B56, BD). For intracellular staining, we used the mouse Foxp3 staining kit (eBioscience) and monoclonal antibodies to Foxp3 (clone FJK-16 s, eBioscience). Flow cytometric analysis and cell sorting were carried out using a FACSVerse and a FACSAriaII (BD), respectively, and the data was analyzed using FlowJo software (Treestar, Ashland, OR, USA). For detection of apoptosis, mouse spleen cells were incubated in RPMI 1640 medium supplemented with 10% FBS, antibiotics and 50 µm 2-ME for 5 h. The annexin V-APC staining kit (eBioscience) was used according to the manufacturer’s instructions. Pifithrin-α (Santa Cruz Biotechnology) was added to culture medium for 30 min before irradiation. For cell cycle analysis, retrovirally transduced CD4+ T cells were sorted by FACSAriaII and stained with 4′6-diamidino-2-phenylinodole. Cell cycle distributions were analyzed by FACSVerse immediately after the sorting or after additional culture in fresh medium containing IL-2 (40 U/ml). The LIVE/DEAD Fixable Dead Cell Stain Kit (Thermo Fisher Scientific) was used to distinguish viable from dead cells. Fixation and permeabilizaion of cells were performed with Fixation/Permeabilization working solution (eBioscience). The GFP competition assay was also performed to determine the viability of transduced cells as reported previously.10 The numbers of the mice (n) analyzed with each flow cytometric analysis are shown in the figures and figure legends.
Retroviral transduction
A packaging cell line, Plat-E, was transfected with pMXs-Ig vectors as described previously.14 CD4+ cells were prepared with the CD4 Enrichment Kit (BD) and activated by 0.5 mg/ml anti-CD28 antibody (clone 37.51, BioLegend), plate-bounded anti-CD3e antibody (clone 145-2C11, R&D systems, Minneapolis, MN, USA) and 40 U/ml rIL-2. After 2 days, activated T cells were transduced with viral supernatant and 4 µg/ml polybrene, and centrifuged at 3000 rpm for 60 min. GFP-positive cells were analyzed by FACSVerse with FlowJo software. All experiments using retrovirally transduced cells were independently repeated three times to confirm the reproducibility, and the representative results are shown.
Synthesis of complementary DNA and qPCR
RNA of mouse CD4+ splenocytes was extracted with Trizol Reagent (Thermo Fisher Scientific). Complementary DNA was synthesized using the ReverTra Ace kit (TOYOBO, Osaka, Japan) after DNaseI treatment according to the manufacturer’s instructions. Quantitative PCR (qPCR) was carried out using SybrGREEN (Thermo Fisher Scientific) and the Applied Biosystems StepOnePlus real-time PCR system (Thermo Fisher Scientific) according to the manufacturer’s instructions. The sequences of gene-specific primers used in this study are described in the supporting information (Supplementary Table S1). The expression level of genes in each sample was measured in triplicate, and the mean value is shown in the figures. The numbers of the samples (n) analyzed with qPCR are shown in the figures and figure legends.
Statistical analysis
Statistical significance was determined by the two-tailed paired Student’s t-test in all experiments performed in this study. The data are presented as the means of samples ± s.d. P-values <0.05 are considered statistically significant. Asterisks indicate the statistical significance as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Linda Kingsbury and Kathleen Hayes-Ozello for editorial comments and proofreading. This study was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan to MM (22114003) and JY (26460554) and a grant from the Takeda Science Foundation (JY) and a grant from the National Institutes of Health (CA100730) to PLG. This study was performed as a research program of the Platform for Dynamic Approaches to Living System from the Ministry of Education, Culture, Sports, Science and Technology, Japan (AK), and also supported in part by the JSPS Core-to-Core Program A, Advanced Research Networks.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
REFERENCES
- 1.Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 2.Matsuoka M, Yasunaga JI. Human T-cell leukemia virus type 1: replication, proliferation and propagation by Tax and HTLV-1 bZIP factor. Curr Opin Virol. 2013;3:684–691. doi: 10.1016/j.coviro.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Gillet NA, Malani N, Melamed A, Gormley N, Carter R, Bentley D, et al. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117:3113–3122. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melamed A, Laydon DJ, Gillet NA, Tanaka Y, Taylor GP, Bangham CR. Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog. 2013;9:e1003271. doi: 10.1371/journal.ppat.1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philip S, Zahoor MA, Zhi H, Ho YK, Giam CZ. Regulation of human T-lymphotropic virus type I latency and reactivation by HBZ and Rex. PLoS Pathog. 2014;10:e1004040. doi: 10.1371/journal.ppat.1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasunaga J, Matsuoka M. Molecular mechanisms of HTLV-1 infection and pathogenesis. Int J Hematol. 2011;94:435–442. doi: 10.1007/s12185-011-0937-1. [DOI] [PubMed] [Google Scholar]
- 7.Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J Virol. 2002;76:12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci USA. 2006;103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan J, Ma G, Nosaka K, Tanabe J, Satou Y, Koito A, et al. APOBEC3G generates nonsense mutations in human T-cell leukemia virus type 1 proviral genomes in vivo. J Virol. 2010;84:7278–7287. doi: 10.1128/JVI.02239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitobe Y, Yasunaga J, Furuta R, Matsuoka M. HTLV-1 bZIP Factor RNA and Protein Impart Distinct Functions on T-cell Proliferation and Survival. Cancer Res. 2015;75:4143–4152. doi: 10.1158/0008-5472.CAN-15-0942. [DOI] [PubMed] [Google Scholar]
- 11.Satou Y, Matsuoka M. Molecular and cellular mechanism of leukemogenesis of ATL: emergent evidence of a significant role for HBZ in HTLV-1-induced pathogenesis. Leuk Res Treat. 2012;2012:213653. doi: 10.1155/2012/213653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 13.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 14.Satou Y, Yasunaga J, Zhao T, Yoshida M, Miyazato P, Takai K, et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011;7:e1001274. doi: 10.1371/journal.ppat.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehn K, Fuente Cde L, Strouss K, Berro R, Jiang H, Brady J, et al. The HTLV-I Tax oncoprotein targets the retinoblastoma protein for proteasomal degradation. Oncogene. 2005;24:525–540. doi: 10.1038/sj.onc.1208105. [DOI] [PubMed] [Google Scholar]
- 17.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 18.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- 19.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 20.Davidson W, Ren Q, Kari G, Kashi O, Dicker AP, Rodeck U. Inhibition of p73 function by Pifithrin-alpha as revealed by studies in zebrafish embryos. Cell Cycle. 2008;7:1224–1230. doi: 10.4161/cc.7.9.5786. [DOI] [PubMed] [Google Scholar]
- 21.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 22.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, et al. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–1347. doi: 10.1371/journal.ppat.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight JS, Sharma N, Robertson ES. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci USA. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borchert S, Czech-Sioli M, Neumann F, Schmidt C, Wimmer P, Dobner T, et al. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J Virol. 2014;88:3144–3160. doi: 10.1128/JVI.02916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaims AY, Khani F, Zhang Y, Roberts AI, Devadas S, Shi Y, et al. Immune activation induces immortalization of HTLV-1 LTR-Tax transgenic CD4+ T cells. Blood. 2010;116:2994–3003. doi: 10.1182/blood-2009-07-231050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arpin-Andre C, Laverdure S, Barbeau B, Gross A, Mesnard JM. Construction of a reporter vector for analysis of bidirectional transcriptional activity of retrovirus LTR. Plasmid. 2014;74:45–51. doi: 10.1016/j.plasmid.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida M, Satou Y, Yasunaga J, Fujisawa J, Matsuoka M. Transcriptional control of spliced and unspliced human T-cell leukemia virus type 1 bZIP factor (HBZ) gene. J Virol. 2008;82:9359–9368. doi: 10.1128/JVI.00242-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 30.Bosco EE, Mayhew CN, Hennigan RF, Sage J, Jacks T, Knudsen ES. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 2004;32:25–34. doi: 10.1093/nar/gkg919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 32.Vernin C, Thenoz M, Pinatel C, Gessain A, Gout O, Delfau-Larue MH, et al. HTLV-1 bZIP factor HBZ promotes cell proliferation and genetic instability by activating OncomiRs. Cancer Res. 2014;74:6082–6093. doi: 10.1158/0008-5472.CAN-13-3564. [DOI] [PubMed] [Google Scholar]
- 33.Zhao T, Satou Y, Sugata K, Miyazato P, Green PL, Imamura T, et al. HTLV-1 bZIP factor enhances TGF-beta signaling through p300 coactivator. Blood. 2011;118:1865–1876. doi: 10.1182/blood-2010-12-326199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto-Taguchi N, Satou Y, Miyazato P, Ohshima K, Nakagawa M, Katagiri K, et al. HTLV-1 bZIP factor induces inflammation through labile Foxp3 expression. PLoS Pathog. 2013;9:e1003630. doi: 10.1371/journal.ppat.1003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeda S, Maeda M, Morikawa S, Taniguchi Y, Yasunaga J, Nosaka K, et al. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int J Cancer. 2004;109:559–567. doi: 10.1002/ijc.20007. [DOI] [PubMed] [Google Scholar]
- 36.Katoh T, Harada T, Morikawa S, Wakutani T. IL-2- and IL-2-R-independent proliferation of T-cell lines from adult T-cell leukemia/lymphoma patients. Int J Cancer. 1986;38:265–274. doi: 10.1002/ijc.2910380218. [DOI] [PubMed] [Google Scholar]
- 37.Zhao T, Yasunaga J, Satou Y, Nakao M, Takahashi M, Fujii M, et al. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood. 2009;113:2755–2764. doi: 10.1182/blood-2008-06-161729. [DOI] [PubMed] [Google Scholar]
- 38.Peloponese JM, Jr, Yasunaga J, Kinjo T, Watashi K, Jeang KT. Peptidylproline cis-trans-isomerase Pin1 interacts with human T-cell leukemia virus type 1 tax and modulates its activation of NF-kappaB. J Virol. 2009;83:3238–3248. doi: 10.1128/JVI.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawada S, Gowrishankar K, Kitamura R, Suzuki M, Suzuki G, Tahara S, et al. Disturbed CD4+ T cell homeostasis and in vitro HIV-1 susceptibility in transgenic mice expressing T cell line-tropic HIV-1 receptors. J Exp Med. 1998;187:1439–1449. doi: 10.1084/jem.187.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold J, Zimmerman B, Li M, Lairmore MD, Green PL. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood. 2008;112:3788–3797. doi: 10.1182/blood-2008-04-154286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.