Abstract
APC mutation is the most common genetic changes in sporadic colorectal cancer (CRC). Despite deregulations of miRNAs have been frequently reported in this malignancy, APC regulated miRNAs have not been extensively documented. Here, by employing an APC inducible cell line and array analysis, we identified a total of 26 deregulated miRNAs. Among them members of miR-17-92 cluster were dramatically inhibited by APC and induced by enforced expression of β-catenin. Furthermore, we demonstrate that activated β-catenin resulted from APC loss binds to and activates the miR-17-92 promoter. Notably, enforced expression of miR-19a overrides APC tumour suppressor activity and knockdown of miR-19a in cancer cells with compromised APC function reduced their aggressive features in vitro. Finally, we observed that expression of miR-19a significantly correlates with β-catenin levels in colorectal cancer specimens, and it is associated to the aggressive stage of tumour progression. Thus our study reveals that miR-17-92 cluster is directly regulated by APC/β-catenin pathway and could be a potential therapeutic target in colon cancers with aberrant APC/β-catenin signaling.
Keywords: Adenomatous Polyposis Coli (APC), miRNA-17-92, β-catenin, CRC, transcription regulation
INTRODUCTION
Colorectal carcinoma (CRC) is one of the leading causes of cancer death in the developed world, with a disease-specific mortality rate estimated around 33%1. Over 85% of sporadic CRCs have loss-of-function mutations of the Adenomatous Polyposis Coli (APC) gene occurring at early stage of cell transformation2. APC is classified as tumour suppressor gene and plays a critical role in several cellular processes including cell division, adhesion and migration3,4, 5. At biochemical level, APC has been shown to integrate to canonical Wnt pathway whose stimulation triggers the translocation of the oncoprotein β-catenin from cell membrane to the cytoplasmic and nuclear compartments. Nuclear β-catenin acts as coactivator of T-cell and lymphoid enhancer (TCF/LEF) factors in the transcriptional activation of target genes 6, although a role of accumulated cytoplasmic β-catenin as stabilization factor of mRNA molecules has been recently suggested9. In epithelial cells, β-catenin associates at the cellular membrane with the adhesion molecule E-cadherin. Free cytoplasmic β-catenin is phosphorylated and targeted for ubiquitination-dependent degradation by a protein complex formed by APC, GSK-3, CKIα, and Axin7,8. Mutations of the APC gene in colorectal carcinomas results in unrestrained β-catenin signaling and contributes to a proinvasive gene expression profile along with cellular transformation10,11. It is worth notice that nearly half of colorectal tumours with intact APC genes were found to contain activating mutations of β-catenin6,12. Thus, mutation of APC or β-catenin represents the most common genetic change (>90%) in CRC, pointing to a driver activity of APC/β-catenin signaling in colon cancer development.
MiRNAs are short non-coding RNA molecules implicated in several cellular processes such as development, differentiation, proliferation, cell cycle progression, apoptosis, inflammation, and stress responses13,14. MiRNAs mechanism of action relies on inhibition of translation or induction of degradation of target mRNAs through direct binding to their 3′UTR15. Given their propensity to regulate numerous processes and target mRNAs, it is no surprising that aberrant expression of miRNAs has been linked to numerous pathological16-19. Several studies observed a frequent upregulation of miR-17-92 cluster, miR-31, miR-21 and miR-200 cluster in colorectal carcinoma, suggesting an oncogenic role of these miRNAs in this malignancy19,20. Despite a recent study reported miRNA profile in tumour from APC(Min/+) mouse21, APC regulated miRNAs remain largely uncharacterized in human CRC.
In this study we profiled miRNA changes upon induction of APC expression in colorectal cancer cells. We found that mir-17-92 cluster, frequently upregulated in CRC, is significantly repressed by APC through induction of β-catenin degradation. We further unveiled that knockdown of miR-19a reduces aggressive features (cell growth, migration and invasion) in cancer cells with compromised APC function. Our study demonstrate that the reduction in miR-19a expression levels is a major mechanism by which APC exerts its tumour suppressor activity and suggest that miR-19a could be a potential therapeutic target in colon cancers with aberrant APC/β-catenin signaling.
RESULTS
MiRNA expression profile in APC-inducible expression colorectal cancer cells
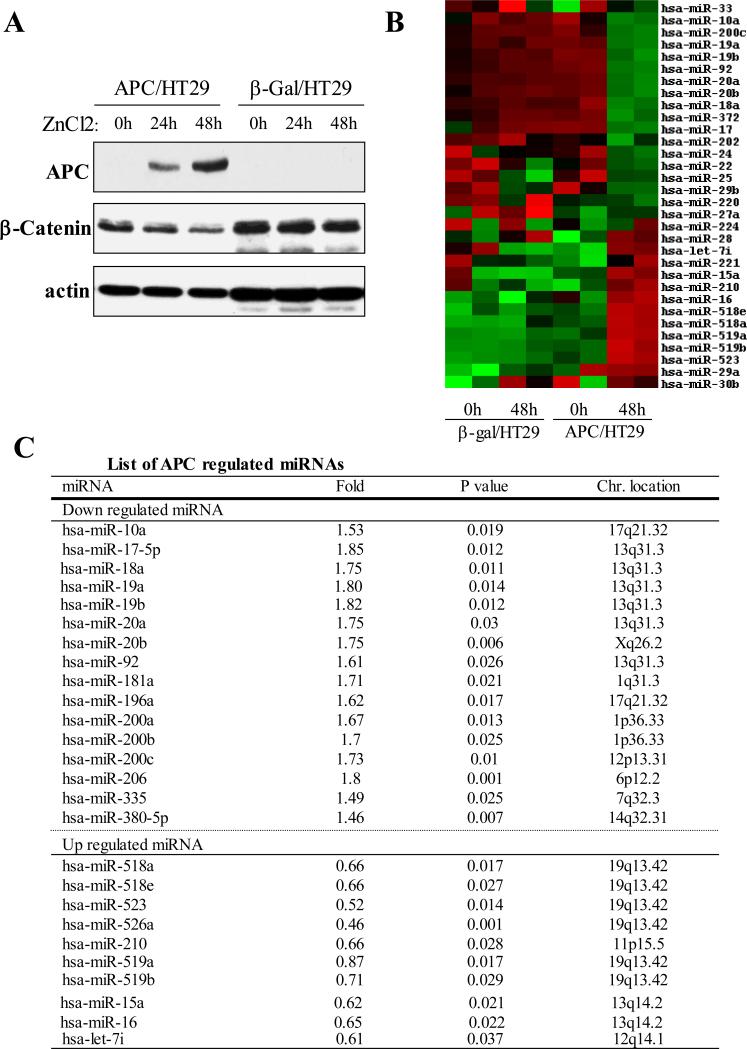
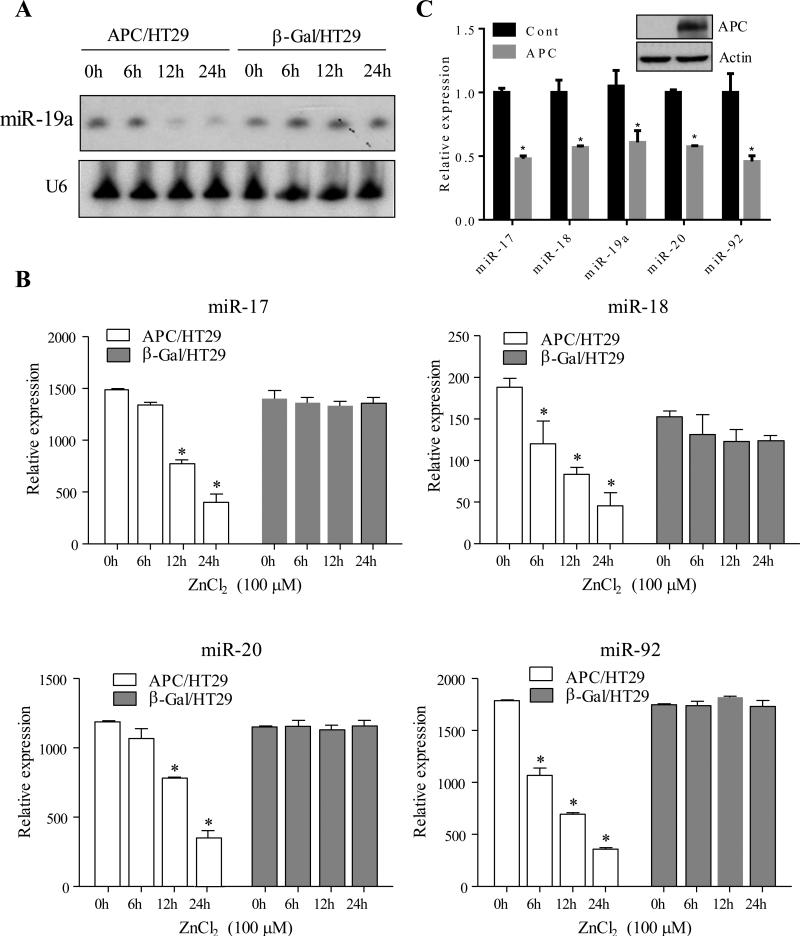
In HT29 cells the endogenous APC gene is mutated and not functional. In an attempt to identify the miRNAs regulated by APC in CRC, we analyzed miRNA profile in HT29 cells with ZnCl2-inducible APC expression (APC/HT29 cells). β-Gal/HT29 cells were used as controls. APC induction following ZnCl2 treatment for 0, 24 and 48 hours was validated by increased levels of full-length APC protein as well as by decreased β-catenin protein levels (Figure 1A). Following these conditions, RNA was isolated and hybridized to a custom miRNA array platform 22-24 that covers up to 650 miRNAs. More than a dozen of miRNAs were shown to be significantly differentially expressed between APC-induced and ctrl cells (Fig. 1B). Of these deregulated miRNAs (≥ 1.5 fold), 16 were reduced and 10 were induced by APC (Fig. 1C). Interestingly, APC repressed the expression levels of several members of miR-17-92 (e.g., miR-17, miR-18, miR-219, miR-20 and miR-92) and miR-200 (e.g., miR200a, miR200b and miR200c) families and increased the expression levels of miR-518 cluster (e.g., miR-518a/e, miR-519a/b, miR-523 and miR-526) and miR-15/16. These findings suggest that each cluster shares the same promoter, under the control of APC pathway. Previous studies have shown frequent upregulation of miR-17-92 family in human colorectal carcinoma25. The ability of APC to repress the expression of members of miR-17-92 cluster was confirmed by Northern blot and/or qRT-PCR analyses (Figure 2A and 2B). In addition, we expressed APC in another APC-mutant cell line HCT15 and found that miR-17-92 levels were significantly reduced in APC-transfected cells when compared to vector-treated cells (Figure 2C).
Figure 1. Profile of APC-regulated miRNAs.
(A) Western blot. APC/HT29 and β-Gal/HT29 cells were treated with ZnCl2 for indicated times and then subjected to immunoblot analysis with indicated antibodies (note: expression of APC leads to decrease in β-catenin level). (B and C) Heatmap (B) and table (C) show the miRNAs significantly regulated by expression of APC.
Figure 2. MiR-17-92 cluster is repressed by APC.
Northern bot (A) and real-time PCR (B) analysis revealed that expression of members of miR-17-92 cluster was decreased in APC/HT29 but not β-Gal/HT29 cells upon induction of APC expression. (C) HCT15 cells were transfected with APC and vector control. After incubation for 48 hours, the cells were subjected to immunoblot (insert panel) and real-time PCR analysis of levels of miR-17-92 cluster. Asterisk represents p<0.05.
APC repressed miR-17-92 expression through induction of β-catenin degradation
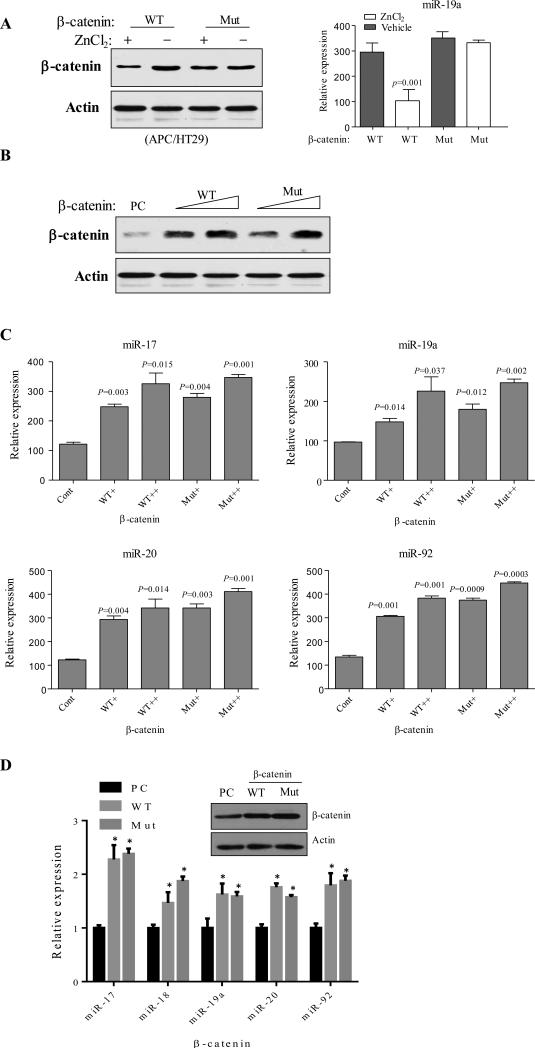
APC exerts tumour suppressor function by targeting oncoprotein β-catenin to degradation. This process requires β-catenin forming a destruction complex with APC, Axin and GSK-3β as well as phosphorylation by GSK-3β and CK1 at 4 serine/threonine sites within N-terminal armadillo domain of β-catenin26. Mutation of these serine/threonine residues occurs in CRC and leads to stabilization and cytoplasmic-nuclear translocation of β-catenin with subsequent activation of TCF/LEF transcription machinery. Therefore, we further investigated the involvement of β-catenin in APC-induced repression of miR-17-92. To this end, wild-type and mutant β-catenin were introduced into ZnCl2-inducible APC/HT29 cells. Following APC induction mutant β-catenin levels were unaffected, in contrast to wild-type β-catenin levels that were reduced (Figure 3A, left panel). Consequently, APC failed to repress miR-19a expression in cells transfected with mutant β-catenin (Figure 3A, right panel). These data suggest that miR-17-92 cluster down-regulation is the consequence of APC-induced β-catenin degradation. We further investigated whether ectopic expression of β-catenin induces miR-17-92 production in wild-type APC cells. Since most CRC cell lines have either APC or β-catenin mutation, we initially transfected HEK-293 cells, in which neither APC nor β-catenin is mutated, with different amounts of wild-type or constitutively active mutant β-catenin (Figure 3B). Real-time PCR analysis revealed that the members of miR-17-92 family were induced by enforced expression of wild-type and mutant β-catenin (Figure 3C). To confirm these results in colon cells, we repeated the experiments in NCI-H508 cells, in which both APC and β-catenin are wild-type, and found that the levels of miR-17-92 family members were significantly elevated upon expression of wild-type or constitutively active mutant β-catenin (Figure 3D). These data suggest that β-catenin induces miR-17-92 expression and that APC inhibition of miR-17-92 is mediated by β-catenin degradation.
Figure 3. β-catenin induces miR-17-92 cluster expression and mediates the APC action.
(A) APC/HT29 and β-Gal/HT29 cells were transfected with wild-type (WT) and constitutively active mutant (Mut) β-catenin and then treated with or without ZnCl2 to induce APC for 48 hours. β-catenin protein levels (left panel) and mirR-19a (right panel) were analyzed by Western blot (left) and qRT-PCR (right) analyses. (B) Western blot analysis of HEK293 cells that were transfected with increasing amount of wild-type and constitutively active mutant β-catenin. (C and D) Expression of miR-17-92 members in β-catenin-transfected HEK293 (C) and NCI-H508 (D) cells was analyzed by qRT-PCR. Asterisk represents p<0.05.
APC inhibits β-catenin binding to and transcription of miR-17-92 promoter
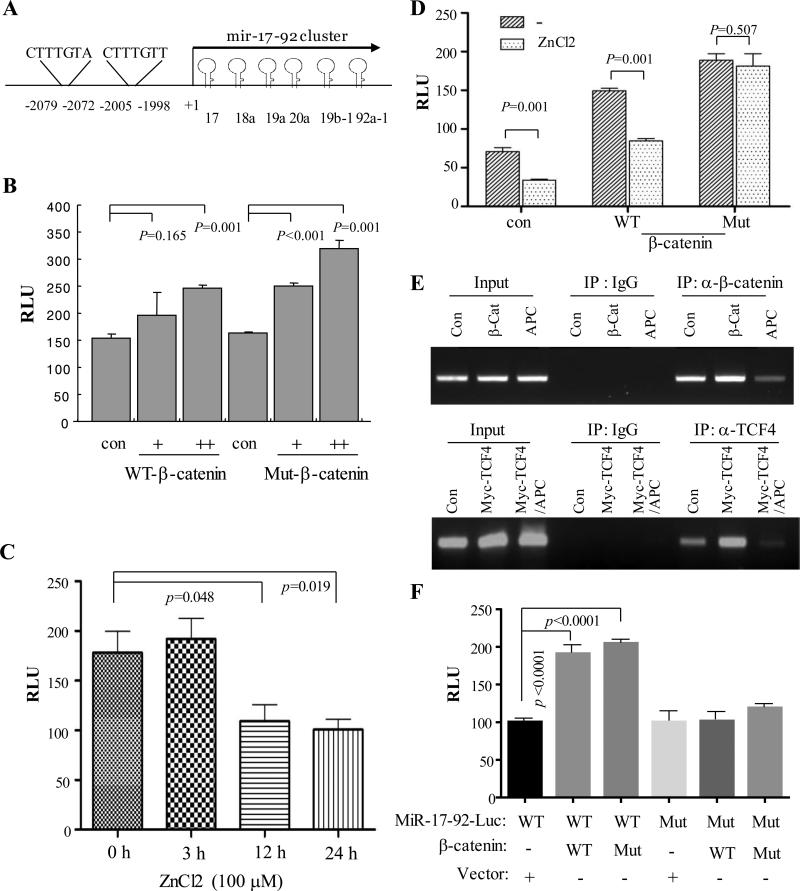
β-catenin is a co-activator of TCF/LEF transcription factors. In that regard, by bioinformatics analysis we identified two putative TCF/LEF binding consensus motifs within the human miR-17-92 promoter (Figure 4A). We next examined if β-catenin activates miR-17-92 promoter activity. For this purpose we transfected HEK293 cells with miR-17-92 promoter luciferase reporter construct along with different amount of wild-type or constitutively active β-catenin. Luciferase activity was assessed after 48 hours of incubation. Our data show that both wild-type and constitutively active β-catenin induce miR-17-92 promoter activity in a dose-dependent manner (Figure 4B). Consistent with these data, APC induction reduced miR-17-92 promoter activity (Figure 4C). Moreover, APC expression was unable to inhibit miR-17-92 promoter activity induced by mutant β-catenin (Figure 4D). We next investigated the effect of APC on affinity of β-catenin/TCF binding to miR-17-92 promoter. Since two β-catenin/TCF/LEF binding sites (−2079/−2072 and -2005/−1998) are close to each other (Figure 4A), we performed chromatin immunoprecipitation (ChIP) with a set of primers flanking both binding motifs. HEK293 cells were transfected with β-catenin or myc-TCF4 together with and without APC induction. ChIP assay was carried out with β-catenin and TCF4 antibody as well as IgG was used as control. Our data show that both β-catenin and TCF4 bind to miR-17-92 promoter and that enforced expression of APC significantly inhibits this binding (Figure 4E). Since β-catenin is a co-activator of TCF/LEF transcriptional factors, we further examined the requirement of TCF/LEF binding sites to activate miR-17-92 promoter by β-catenin. We mutated the TCF/LEF binding sites within the miR-17-92 promoter and showed that both wild type and constructively active β-catenin failed to induce the mutant miR-17-92 promoter activity (Figure 4F).
Figure 4. MiR-17-92 promoter is induced by β-catenin which is inhibited by APC.
(A) Diagram represents miR-17-92 promoter with the two TCF/LEF binding sites indicated. (B) β-catenin induces miR-17-92 luciferase promoter activity. HEK293 cells were transfected with pGL3-miR-17-92-Luc together with or without WT and Mut β-catenin. Luciferase assay was performed after 48 h incubation. (C) APC inhibits miR-17-92 promoter activity. pGL3-miR-17-92-Luc was introduced into APC/HT29 cells. Following induction of APC expression by ZnCl2, luciferase activity was measured. (D) APC represses wild-type but not constitutively active mutant β-catenin induced miR-17-92 promoter activity. Luciferase assay was carried out in APC/HT29 cells that were transfected with indicated plasmids and treated with/without ZnCl2. (E) β-catenin/TCF/LEF directly binds to miR-17-92 promoter, which is inhibited by APC. HEK293 cells were transfected with vector control, β-catenin or Myc-TCF4 together with and without APC, and then subjected to ChIP assay with β-catenin and TCF4 antibodies. IgG was used as control. (F) Mutation of TCF/LEF binding sites of the miR-17-92 promoter largely abrogates the miR-17-92 promoter activity induced by β-catenin. Following transfection of wild-type and mutant miR-17-92-Luc together with and without WT and constitutively active Mut β-catenin for 48 h, luciferase assay was performed.
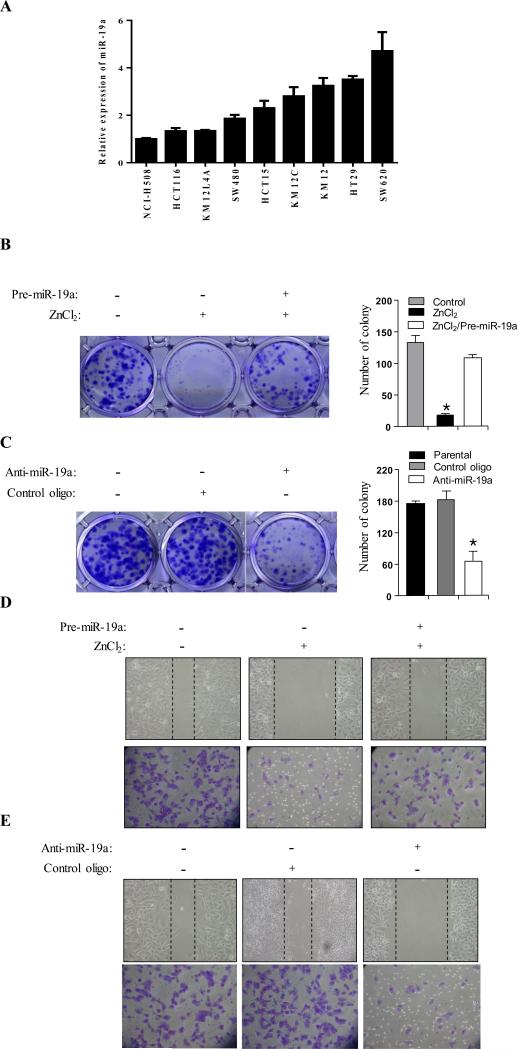
Enforced expression of miR-19a overrides APC tumour suppressor activity and knockdown of miR-19a in cancer cells with compromised APC function reduces aggressive features in vitro
MiR-19 has been shown to be a major oncogenic member of miR-17-92 cluster27. Furthermore, a recent study showed that expression of miR-19 in the context of the miR-17~92 cluster at medium levels promoted colon tumour progression28. By examination of a panel of colon cancer cell lines, we observed increase of miR-19a expression in either APC mutation or β-catenin mutation lines with variable levels when compared to wild type APC/β-catenin NCI-H508 cell line (Figure 5A). Since APC represses miR-17-92 at transcription level, we next asked if ectopic expression of miR-19a family member(s) is able to override APC tumour suppressor activity. Thus, we transfected APC/HT29 cells with pre-miR-19a. In agreement with previous reports29, induction of APC expression with ZnCl2 in APC/HT29 cells inhibited cell proliferation, cell migration and invasion. However, in pre-miR-19a treated cells, these APC-induced effects were strongly diminished (Figure 5B, 5D). As controls, we expressed miR-19a in β-Gal/HT29 cells and found no effect on colony growth and cell migration following treatment with ZnCl2 (Supplemental Figure 1). To further confirm these results, we introduced APC together with and without pre-miR-19a into HCT15 cells and observed that expression of APC alone repressed colony growth, cell migration and invasion. These phenotypes were largely overridden by co-expression of miR-19a (Supplemental Figure 2). On the other side, knockdown of miR-19a in parental HT29 cells reduced cell growth, migration and invasion (Figure 5C, 5E). We further verified these findings by knockdown of miR-19a in APC mutant HCT15 cells (Supplemental Figure 3) and ectopic expression of miR-19a in wild type APC/β-catenin NCI-H508 cells (Supplemental Figure 4). Collectively, these findings suggest that reduction in miR-19a expression levels is a major mechanism by which APC exerts its tumour suppressor activity.
Figure 5. MiR-19a mediates APC cellular function.
(A) Real-time qPCR analysis of miR-19a expression in a panel of colon cancer cell lines. (B and C) Colony formation was performed in APC/HT29 cells that were transfected with pre-miR-19a, anti-miR-19a or control oligo and then treated with (B) or without (C) ZnCl2. The number of colonies was quantified (right), (D and E) Cell migration and invasion assay. APC/HT29 cells were transfected and treated as above and subsequently subjected to cells migration (wound-healing) and invasion assays. Asterisk represents p<0.05.
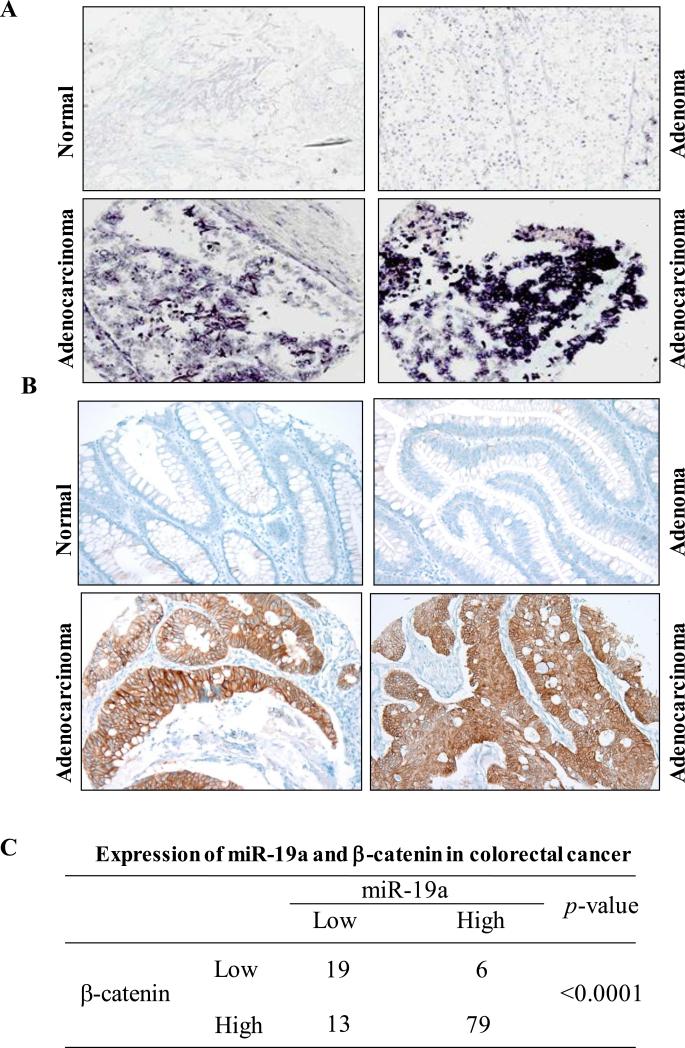
Coexpression of miR-19a and β-catenin in CRC
Mutations of APC/β-catenin pathway have been observed in more than 90% of sporadic CRCs. These mutations lead to stabilization and accumulation of β-catenin30. Having demonstrated that miR-17-92 is induced by APC mutations through β-catenin transcriptional activity, we further examined if this regulation occurs in vivo. We examined 117 colorectal tumours and 42 normal colon tissues for expression of miR-19a and β-catenin (Figure 6A-C). Among the 117 colorectal tumours, 85 had overexpression of miR-19a and 92 had overexpression of β-catenin. Among the 92 tumours with elevated β-catenin, 79 (86%) also had elevated miR-19a levels (p < 0.0001). These data suggest that there is a significant relationship of co-expression of β-catenin and miR-19a, which further support the findings of biochemical and functional links between APC/β-catenin and miR-17-92 cluster.
Figure 6. Co-expression of miR-19a and β-catenin in human colorectal cancer.
(A) LNA-miR-19a in situ hybridization analysis of colorectal cancer TMA. (B) Immunohistochemical staining of same colorectal cancer TMA with β-catenin antibody. Note: elevated β-catenin locates either cytoplasm or nucleus in tumours. (C) Overexpression of miR-19a is significantly correlated to elevated levels of β-catenin, p <0.0001.
DISCUSSION
Mutation of APC and elevated levels of miR-17-92 cluster have been frequently detected in CRC 25,31,32. In this report, we demonstrated a direct link between APC and miR-17-92. APC represses miR-17-92 through inhibition of β-catenin. Mutation of APC leads to stabilization of β-catenin, which in turn binds to and activates miR-17-92 promoter. Enforced expression of miR-19a largely overrides APC-inhibited cell growth, migration and invasion and knockdown of miR-19a in cancer cells with compromised APC function reduces aggressive features in vitro. Moreover, elevated level of β-catenin is significantly correlated with miR-19a overexpression in human colorectal carcinoma. These findings suggest that miR-17-92 cluster is one of key effects of APC/β-catenin/TCF4/LEF pathway.
Accumulating studies have shown that miR-17-92 cluster is one of the most elevated miRNA families in this malignancy33,34. Furthermore, this cluster has been suggested not only to be involved in progression of colorectal adenoma to adenocarcinoma but also to have diagnostic value in CRC35. For instance, analyzing of the exfoliated colonocytes isolated from feces of CRC patients and healthy volunteers, the expression of the miR-17-92 cluster was significantly higher in CRC patients than in healthy volunteers36. Additionally, up-regulation of miR-92 in serum was demonstrated to be a predictive value for CRC with a sensitivity of 89% and a specificity of 70%37. Increased miR-17-92 expression in CRC has also been shown to be associated to DNA copy number gain of miR-17-92 locus on 13q31 and c-MYC expression25. While MYC induces miR-17-92 promoter activity and expression38,39, the mechanism of elevated miR-17-92 in CRC remains elusive. We showed in this study that APC mutation induced miR-17-92 expression. Mechanistically APC mutation leads to stabilization and accumulation of β-catenin, which in turn binds to miR-17-92 promoter and promotes its expression. Indeed, we also document significant co-overexpression of miR-17-92 and β-catenin in CRC. Thus, we provided the evidence of miR-17-92 as a direct target of APC/β-catenin pathway in CRC.
It has been well documented that the genes regulated by β-catenin/TCF4/LEF such as CCND1, c-MYC, ID2 and MMP7, mediate APC cellular function40. Depletion of CCND1 or c-MYC significantly reduces phenotypes resulted from mutation of APC41. Previous studies identified miR-19 as the most important miRNA of the miR-17-92 cluster in c-Myc-induced lymphomagenesis27,42. A recent report showed that that quantitatively controlling expression of miR-17~92 determines colon tumour progression. Expression of miR-19 in the context of the miR-17~92 cluster at medium levels promoted tumour metastasis through induction of Wnt/β-catenin–mediated epithelial-mesenchymal transition by targeting to PTEN, whereas higher levels of miR-18a in the context of the miR-17~92 cluster inhibited tumour growth and metastasis by directly targeting β-catenin28. These findings not only further highlight the importance of miR-19 in promoting colon cancer progression but also suggest a fine-tone regulation between miR-17-92 and β-catenin to maintain certain levels of miR-19 and β-catenin in colon carcinogenesis. We demonstrated in this report that elevated levels of miR-19a were correlated with APC mutation and β-catenin expression in colon cancer cell lines and primary tumors, respectively (Figures 5A and 6). Enforced expression of miR-19a largely overrides the cellular phenotypes induced by wild-type APC. Further, we provide evidences that knockdown of miR-19a in APC-mutant cancer cells reduces cell growth, migration and invasion (Figure 5). Collectively, these data suggest that miR-19 is functionally important in APC pathway.
While alteration of APC is a very common event in CRC, no effective targeted therapy has been developed for APC mutation patients. Considerable efforts have been made to identify small molecule compounds that antagonize Wnt/β-catenin signaling 43,44. Previous studies reported that xanthothricin inhibits β-catenin/TCF4 transcriptional activity by disrupting β-catenin/TCF4 binding45. Furthermore, a series of acyl hydrazones destabilize β-catenin acting downstream of the β-catenin destruction complex46. However, these compounds have not clinically been tested in patients with APC mutation. Recent studies have shown that function of miRNAs can be efficiently and specifically inhibited by chemically modified anti-miR oligonucleotides, supporting their potential as targets for the development of novel therapies. In fact, antisense of miR-122, a key miRNA promoting HCV replication, is currently in phase II clinical trials for patients with chronic HCV infection 47. Our data show that miR-17-92 is a downstream target of APC and mediates APC cellular function. Thus, inhibition of miR-17-92, especially miR-19, using anti-miRs could be a potential therapeutic strategy for CRC patients with APC mutation.
MATERIAL AND METHODS
Cell Lines, Culture, Plasmid, Transfection and Antibodies
All the colon cancer cell lines were obtained from ATCC. ZnCl2 inducible and β-Gal/HT29 cell lines, kindly provided by Bert Vogelstein29, were maintained in McCoy's SA media (GIBCO/BRL) supplemented with 10% fetal bovine serum. NCI-H508 cell line was a gift from Dr. Thomas Ried (National Cancer Institute, USA). The rest colorectal cancer cell lines and HEK293 cells were obtained from the American Type Culture Collection and cultivated following the manufacture's instruction. The firefly luciferase reporter vectors pGL3 basic, pGL3 promoter, and Renilla luciferase vectors pRL-SV40 are from Promega (Madison, WI). The pGL3-miR-17-92 reporter plasmid was obtained from Dr. Tomas Stopka. Mutation of TCF/LEF binding sites of the miR-17-92 promoter was generated using QuikChange® Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA USA). Wild-type and constitutively active mutant β-catenin and Myc-TCF4 are from Addgene. Transfection was performed using Lipofectamine® reagents (Life Technologies). APC antibody was from Santa Cruz (C-20; Catalog #sc-896). Antibodies against β-catenin (Catalog #9562S) and TCF4 (Catalog #2565S) were purchased from Cell Signaling.
MicroRNA microarray analysis
Total RNA was isolated using Trizol reagent (Invitrogen). MiRNA profiling was performed as previously described48. Briefly, miRNA array was hybridized with [γ-32P]ATP–labeled small RNA probes. To ensure the accuracy of the hybridizations, each labeled RNA sample was hybridized with 3 separate arrays and quantified. In addition, 8 oligonucleotides with nonmatching any known miRNA were used as hybridization controls. Hybridization signals for each spot of the array and background values at 15 empty spots were measured. The signals that fail to exceed the average background value by more than 3 SDs were excluded. All statistical analyses on microarray data were performed using R software v2.5.0 http://www.r-project.org/ and the Bioconductor software package http://www.bioconductor.org/. The microarray data were initially background-corrected using a normal plus exponential convolution model, normalized a) within arrays using a method that normalizes the M-values for each single microarray using robustly fitted regression splines for each print-tip group and an empirical Bayesian approach in order to shrink the individual print-tip curves towards a common value, and subsequently b) between arrays using a method which ensures that the A-values (average intensities) have the same empirical distribution across arrays, leaving the M-values (log-ratios) unchanged49. After the normalization step the probes were pre-filtered on the basis of empty spots and negative control intensity distribution over all the arrays.
MiRNA qRT-PCR Detection and Quantification
Expression of miR-17-92 family members and U6 was detected using TaqMan microRNA Reverse Transcription kit (Applied Biosystem, Foster City, CA, USA). Briefly, 200 ng of total RNA from each cell line and tumour RNA were used for primer specific reverse transcriptase (RT) in both miR-17-92 and U6, and then 2 μl of the RT product was used for subsequent qPCR. The qPCR was performed on ABI HT9600 (ABI, Foster City, CA, USA) and data were collected and analyzed using ABI SDS version 2.3. To calculate relative concentration, miR-17-92 and U6 CT values for all samples were obtained. A normalized expression for each sample was obtained by subtracting CT of has-miR by the same sample's U6 CT and designated as ΔCT. This value is then transformed by performing 2^-(ΔCT).
Cell Proliferation, Migration and Invasion Assay
Cell growth was determined using colony formation assay. Cell migration was evaluated by wound-healing assay. Cell invasion assay was performed as we previously described14. Briefly, β-Gal/HT29 cells were transfected with anti-miR19a whereas APC/HT29 cells were treated with pre-miR-19a. As controls, β-Gal/HT29 and APC/HT29 cells were transfected with control oligonucleotides against GFP. Following culture with and without additional ZnCl2 for 24 h, cells were seeded in upper chamber of Boyden Chambers coated with Matrigel. All chambers contained normal culture medium containing ZnCl2. After incubation for 48 h, invasion and migration were examined under a Nikon inverted light microscope.
Luciferase and Chromatin Immunoprecipitation (ChIP) Assay
Luciferase assay was performed using the Luciferase Assay System (Promega), and activities were normalized to β-galactosidase activity. ChIP assay was performed essentially as previously described14. Solubilized chromatin was prepared from a total of 2 × 107 asynchronously growing cells. The chromatin solution was diluted 10-fold with ChIP dilution buffer, and precleared with protein-A beads blocked with 2 μg of sheared salmon sperm DNA and preimmune serum. The precleared chromatin solution was divided and utilized in immunoprecipitation assays with anti-β-catenin, -TCF4 antibodies or IgG. Following wash, the antibody-protein-DNA complex was eluted from the beads by resuspending the pellets in 1% SDS, 0.1 M NaHCO3 at room temperature for 20 min. After cross-linking, protein and RNA were removed by digestion. Purified DNA was subjected to PCR with primers specific for 2 putative TCF/LEF-binding sites within the human miR-17-92 promoter region (forward miR-17-92 chip-F 5-AGCGCCTCCAGAACAAAGCGGC-3, miR-17-92 chip-R 5-TCCCGCGCCACACTCCCAGCAA-3). Amplified PCR products (343 bp) were resolved by 1.5% of agarose gel electrophoresis and visualized by ethidium bromide staining.
Colorectal Tumour Tissue Microarray (TMA), MiRNA Locked Nucleic Acid in Situ Hybridization (ISH) and Immunohistochemical Staining
Primary colon cancer, adenoma and normal tissue specimens were obtained from patients who underwent surgery at H. Lee Moffitt Cancer Center and approved by the Institutional Review Board and in accord with an assurance filed with and approved by the US Department of Health and Human Services. Informed consent was obtained from all subjects. MiRNA ISH was performed in colorectal cancer TMAs, which contains 117 colon cancers and 20 adenomas, and analyzed as described previously23,50. The probe sequences used were as follows: LNA-miR-19a; 5'-digoxigenin-tCAgTTtTGcatAGatttgcaca-3'. Immunohistochemistry analysis and immunofluorescence staining were performed following our routine procedures22. Immunohistochemistry analysis of colorectal cancer TMAs with β-catenin antibody was carried out as we previously14.
Statistical Analysis
Statistical comparisons were based upon unpaired Student's t test. Correlation of expression of miR-19a and β-catenin was analyzed with Chi-square test. P ≤ 0.05 was considered to be statistically significant.
Supplementary Material
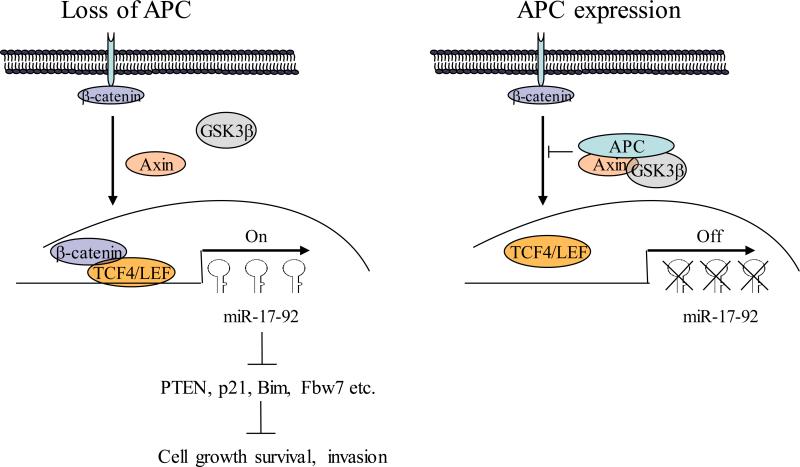
Figure 7.
Diagram represents the proposed model of APC regulation of miR-17-92 cluster.
ACKNOWLEDGEMENT
We are grateful for the Molecular Biology and Analytic Microscopy Core and Tissue Core facilities at H. Lee Moffitt Cancer Center. We also thank Dr. Thomas Ried for providing NCI H508 cell line. This work was supported in part by CA137041, CA160455 (JQC) and the Moffitt Cancer Center Foundation. The H. Lee Moffitt Cancer Center & Research Institute is supported in part by NCI Cancer Center Support Grant #P30 CA076292. ML was supported by “Fondazione del Monte di Bologna e di Ravenna” (Bologna, Italy).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 2.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mimori-Kiyosue Y, Matsui C, Sasaki H, Tsukita S. Adenomatous polyposis coli (APC) protein regulates epithelial cell migration and morphogenesis via PDZ domain-based interactions with plasma membranes. Genes Cells. 2007;12:219–233. doi: 10.1111/j.1365-2443.2007.01045.x. [DOI] [PubMed] [Google Scholar]
- 4.Phelps RA, Broadbent TJ, Stafforini DM, Jones DA. New perspectives on APC control of cell fate and proliferation in colorectal cancer. Cell Cycle. 2009;8:2549–2556. doi: 10.4161/cc.8.16.9278. [DOI] [PubMed] [Google Scholar]
- 5.Barth AI, Caro-Gonzalez HY, Nelson WJ. Role of adenomatous polyposis coli (APC) and microtubules in directional cell migration and neuronal polarization. Semin Cell Dev Biol. 2008;19:245–251. doi: 10.1016/j.semcdb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 7.Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI. Mechanism of phosphorylation- dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 9.D'Uva G, Bertoni S, Lauriola M, De Carolis S, Pacilli A, D'Anello L, et al. Beta-catenin/HuR post-transcriptional machinery governs cancer stem cell features in response to hypoxia. PLoS One. 2013;8:e80742. doi: 10.1371/journal.pone.0080742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudo Y, Kitajima S, Ogawa I, Hiraoka M, Sargolzaei S, Keikhaee MR, et al. Invasion and metastasis of oral cancer cells require methylation of E-cadherin and/or degradation of membranous beta-catenin. Clin Cancer Res. 2004;10:5455–5463. doi: 10.1158/1078-0432.CCR-04-0372. [DOI] [PubMed] [Google Scholar]
- 11.Pendas-Franco N, García JM, Peña C, Valle N, Pálmer HG, Heinäniemi M, et al. DICKKOPF-4 is induced by TCF/beta-catenin and upregulated in human colon cancer, promotes tumour cell invasion and angiogenesis and is repressed by 1alpha,25- dihydroxyvitamin D3. Oncogene. 2008;27:4467–4477. doi: 10.1038/onc.2008.88. [DOI] [PubMed] [Google Scholar]
- 12.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2009;2:pe1. doi: 10.1126/scisignal.252pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tysnes BB. Tumour-initiating and -propagating cells: cells that we would like to identify and control. Neoplasia. 2010;12:506–515. doi: 10.1593/neo.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou JJ, Zheng S, Sun LF, Zheng L. MicroRNA regulation network in colorectal cancer metastasis. World J Biol Chem. 2014;5:301–307. doi: 10.4331/wjbc.v5.i3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Necela BM, Carr JM, Asmann YW, Thompson EA. Differential expression of microRNAs in tumours from chronically inflamed or genetic (APC(Min/+)) models of colon cancer. PLoS One. 2011;6:e18501. doi: 10.1371/journal.pone.0018501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 Negatively Regulates Estrogen Receptor{alpha} and Is Associated with Tamoxifen Resistance in Breast Cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 24.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, Meijerink WJ, et al. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707–714. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumour suppressor, beta-catenin and GSK3 beta. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 27.van Haaften G, Agami R. Tumourigenicity of the miR-17-92 cluster distilled. Genes Dev. 2010;24:1–4. doi: 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang H, Wang P, Wang Q, Wang B, Mu J, Zhuang X, et al. Quantitatively controlling expression of miR-17~92 determines colon tumor progression in a mouse tumor model. Am J Pathol. 2014;184:1355–1368. doi: 10.1016/j.ajpath.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morin PJ, Vogelstein B, Kinzler KW. Apoptosis and APC in colorectal tumourigenesis. Proc Natl Acad Sci USA. 1996;93:7950–7954. doi: 10.1073/pnas.93.15.7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyazawa K, Iwaya K, Kuroda M, Harada M, Serizawa H, Koyanagi Y, et al. Nuclear accumulation of beta-catenin in intestinal-type gastric carcinoma: correlation with early tumour invasion. Virchows Arch. 2000;437:508–513. doi: 10.1007/s004280000283. [DOI] [PubMed] [Google Scholar]
- 31.Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11:501–506. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanza G, Ferracin M, Gafà R, Veronese A, Spizzo R, Pichiorri F, et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, et al. The miR-17-92 cluster of microRNAs confers tumourigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–8557. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu G, Tang JQ, Tian ML, Li H, Wang X, Wu T, et al. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol. 2012;106:232–237. doi: 10.1002/jso.22138. [DOI] [PubMed] [Google Scholar]
- 35.Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20:1272–1286. doi: 10.1158/1055-9965.EPI-11-0035. [DOI] [PubMed] [Google Scholar]
- 36.Koga Y, Yasunaga M, Takahashi A, Kuroda J, Moriya Y, Akasu T, et al. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila) 2010;3:1435–1442. doi: 10.1158/1940-6207.CAPR-10-0036. [DOI] [PubMed] [Google Scholar]
- 37.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 38.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 39.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 41.Hulit J, Wang C, Li Z, Albanese C, Rao M, Di Vizio D, et al. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumour number in ApcMin mice. Mol Cell Biol. 2004;24:7598–7611. doi: 10.1128/MCB.24.17.7598-7611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi-Yanaga F, Sasaguri T. Drug development targeting the glycogen synthase kinase-3beta (GSK-3beta)-mediated signal transduction pathway: inhibitors of the Wnt/beta-catenin signaling pathway as novel anticancer drugs. J Pharmacol Sci. 2009;109:179–183. doi: 10.1254/jphs.08r28fm. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 45.Lepourcelet M, Chen YN, France DS, Wang H, Crews P, Petersen F, et al. Small- molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell. 2004;5:91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 46.Song S, Christova T, Perusini S, Alizadeh S, Bao RY, Miller BW, et al. Wnt inhibitor screen reveals iron dependence of beta-catenin signaling in cancers. Cancer Res. 2011;71:7628–7639. doi: 10.1158/0008-5472.CAN-11-2745. [DOI] [PubMed] [Google Scholar]
- 47.Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol. 2012;199:407–412. doi: 10.1083/jcb.201208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solmi R, Lauriola M, Francesconi M, Martini D, Voltattorni M, Ceccarelli C, et al. Displayed correlation between gene expression profiles and submicroscopic alterations in response to cetuximab, gefitinib and EGF in human colon cancer cell lines. BMC Cancer. 2008;8:227. doi: 10.1186/1471-2407-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 50.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.