Abstract
The purpose of this study was to establish methods for isolation, culture, expansion, and characterization of rat hair follicle stem cells (rHFSCs). Hair follicles were harvested from 1-week-old Sprague–Dawley rats and digested with dispase and collagenase IV. The bulge of the hair follicle was dissected under a microscope and cultured in Dulbecco’s modified Eagle’s medium/F12 supplemented with KnockOut™ Serum Replacement serum substitute, penicillin–streptomycin, l-glutamine, non-essential amino acids, epidermal growth factor, basic fibroblast growth factor, polyhydric alcohol, and hydrocortisone. The rHFSCs were purified using adhesion to collagen IV. Cells were characterized by detecting marker genes with immunofluorescent staining and real-time polymerase chain reaction (PCR). The proliferation and vitality of rHFSCs at different passages were evaluated. The cultured rHFSCs showed typical cobblestone morphology with good adhesion and colony-forming ability. Expression of keratin 15, integrin α6, and integrin β1 were shown by immunocytochemistry staining. On day 1–2, the cells were in the latent phase. On day 5–6, the cells were in the logarithmic phase. Cell vitality gradually decreased from the 7th passage. Real-time PCR showed that the purified rHFSCs had good vitality and proliferative capacity and contained no keratinocytes. Highly purified rHFSCs can be obtained using tissue culture and adhesion to collagen IV. The cultured cells had good proliferative capacity and could therefore be a useful cell source for tissue-engineered hair follicles, vessels, and skin.
Keywords: Hair follicle stem cell, Q-PCR analysis, Immunocytochemistry, Wound healing, Regeneration
Introduction
Stem cells are undifferentiated cells with self-renewing, multi-lineage differentiation and good proliferative capacities. Hair follicle stem cells (HFSCs) are primarily found in the bulge of the outer root sheath of the hair follicle (Cotsarelis et al. 1990; Cotsarelis 2006). In vitro studies have shown that HFSCs have a high cloning capacity with good regenerative potential (Rochat et al. 1994). HFSCs can not only differentiate into hair follicles, but can also differentiate into nerve cells, smooth muscle cells, and epithelial cells, as well as sebaceous glands, sweat glands, and epidermis (Taylor et al. 2000; Yu et al. 2006; Liu et al. 2008). HFSCs are derived from the skin and hair follicles in abundance and autologous HFSCs can be easily isolated from patients. Furthermore, no serious complications have been reported during the harvest of HFSCs. HFSCs are therefore considered an ideal cell source for tissue-engineered skin (Liu et al. 2008).
Materials and animals
Animals
Six 1-week-old Sprague–Dawley rats, male and female, weighing 25 ± 2 g, were purchased from the SLACK company (Shanghai, China) (Certificate number: 2007000541281).
Reagents
Dulbecco’s modified Eagle’s medium (DMEM)/F-12 (1:1) medium (Cat: 11320-033), high glucose DMEM medium (Cat: 11965-092), KnockOut™ Serum Replacement (KSR, Cat: 10828010), collagenase IV (EC 3.4.24.24), dispase (EC 3.4.24.4), Coating Matrix (50×), TrypLE™ Express Enzyme (1×) (EC 3.4.21.4), l-glutamine, MEM non-essential amino acids (Cat: 11140050), and 2-mercaptoethanol (Cat: 21985-023) were obtained from (Gibco-Invitrogen, Carlsbad, CA, USA). Recombinant human epidermal growth factor (EGF) (Cat: PHG0315) and recombinant human basic fibroblast growth factor (bFGF) (Cat: 233-FB-025) were purchased from R&D (Minneapolis, MN, USA). Other reagents included penicillin–streptomycin mixture (100×; Solarbio, Beijing, China), solid hydrocortisone, trypan blue (Sangon Biotech, Shanghai, China), collagen IV (BD, Franklin Lakes, NJ, USA), Trizol RNA extraction reagent, reverse transcriptase (Invitrogen), SYBR Green PCR Master Mix (ABI, Foster City, CA, USA), antibodies against integrin β1 (Itgβ1), keratin 15 (Krt15) (Abcam, Cambridge, MA, USA), antibody against integrin α6 (Itgα6) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), secondary antibody (Jackson, Sacramento, CA, USA), and 4′,6-diamidino-2-phenylindole (DAPI; Roche, Basel, Switzerland). The primers used are listed in Table 1. This study was performed from August 2012 to December 2012 at the Huadong Stem Cell Center.
Table 1.
Sequences for primers
| Gene name | NCBI accession | Primer sequence | Product (bp) | |
|---|---|---|---|---|
| Krt10 | NM_001008804.1 | Forward | 5′-TTGGAAACCTGCAAATAACCC-3′ | 175 |
| Reverse | 5′-ATCATAGACGAAAGGACTCTACCC-3′ | |||
| Krt15 | NM_001004022.2 | Forward | 5′-AAAACCGTCGGGATGTAGAGG-3′ | 94 |
| Reverse | 5′-TTGCTGGTCTGGATCATTTCTGT-3′ | |||
| Krt19 | NM_199498.1 | Forward | 5′-CCAAGTTTGAGACAGAACAGGC-3′ | 156 |
| Reverse | 5′-CGTGGTTCTTCTTCAGGTAGGC-3′ | |||
| CD34 | NM_001107202.2 | Forward | 5′-CCTGCCGTCTGTCAATGTTTC-3′ | 146 |
| Reverse | 5′-GCACTCCTCGGATTCCTGAAC-3′ | |||
| Itgβ1 | NM_017022.2 | Forward | 5′-ATCATGCAGGTTGCAGTTTG-3′ | 72 |
| Reverse | 5′-CGTGGAAAACACCAGCAGT-3′ | |||
| Itgα6 | NM_053725.1 | Forward | 5′-CGTGGTTCTTCTTCAGGTAGGC-3′ | 188 |
| Reverse | 5′-CACATCTATGGACGCCCTCAC-3′ | |||
| ACTB | NM_031144.3 | Forward | 5′-GCTATGTTGCCCTAGACTTCGA-3′ | 173 |
| Reverse | 5′-GATGCCACAGGATTCCATACC-3′ |
Methods
Coating the dishes
Collagen IV (0.5 mL) and diluent (50 mL) were mixed and used to coat the 10 cm dishes (3.4 mL/100 mm) according to the manufacturer’s instruction. The dishes were placed at room temperature for 30–90 min, and the coating solution was discarded.
Isolation and culture of the primary HFSCs
One-week-old Sprague–Dawley rats were sacrificed by cervical dislocation and disinfected in 75 % ethanol. The skin near the beards was cut into 2 mm × 2 mm blocks using ophthalmic scissors, and rinsed with PBS three times. The tissue blocks were treated with 1 % dispase and 1 % collagenase IV at 37 °C for 90 min, and then washed with PBS three times. Under a stereomicroscope, the hair follicle was separated from the connective tissue sheath using a syringe needle. The two ends were cut off, leaving the bulge. Bulges were cultured in Matrigel Basement Membrane Matrix (BD, NY, USA) with 1 mL complete medium (869 μL/mL DMEM/F12, 100 μL/mL KSR, 10 μL/mL penicillin–streptomycin mixture, 10 μL/mL l-glutamine, 10 μL/mL MEM non-essential amino acids, 20 ng/mL EGF, 10 ng/mL bFGF, 1 μL/mL 2-mercaptoethanol, 10 ng/mL hydrocortisone) at 37 °C with 5 % CO2 for 2 days. After tissue adherence, 5 mL of complete medium was slowly added. Medium was changed every 3 days and cell migration, growth and proliferation were observed microscopically (Amoh et al. 2009; Nowak and Fuchs 2009).
Passage and expansion of HFSCs
HFSCs were digested with TrypLE™ Express Enzyme without phenol red at 37 °C for 8 min before being passaged. We seeded 2 × 106 primary cells in each 10 cm dish and passaged them when they reached 90 % of confluence. After purification step (see below), we seeded 2 × 106 rat follicle stem cells in 10 cm dish. The cells were cultured at 37 °C with 5 % CO2 and the medium was changed every 2–3 days.
Purification of HFSCs
HFSCs were purified using a modified previously described method (Bickenbach and Chism 1998). In brief, collagen IV-coated 6-well dishes were placed at room temperature for 1 h. Primary cells were digested with TrypLE™ Express Enzyme and collected. The cells were pipetted into a single cell suspension and cultured for 15–20 min. The non-adherent cells were discarded with the culture medium and the adherent cells were further cultured in complete medium. Medium was changed every 3 days. Second passage cells were purified again using identical methods.
Real-time PCR
Third passage (P3) rHFSCs were used for real-time polymerase chain reaction (PCR) detection. The six target genes and single reference gene, beta-actin (ACTB), were amplified in different reaction tubes. Total RNA was extracted using the Trizol RNA extraction kit according to the manufacturer’s instructions. cDNA was obtained with reverse transcriptase. The primers were designed using Primier 5.0 software (Table 1). The PCR reaction system comprised 2× SYBR Green Mix (10 μL), primer Mix (1 μL), template (1 μL), and H2O (8 μL). The reaction system was loaded into Axygen PCR tubes, centrifuged briefly, and installed into the real-time PCR instrument. The SYBR Green method was used. The thermocycling program was as follows: 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and elongation at 72 °C for 20 s. Each cDNA sample was processed in triplicate. The copy number in each cDNA sample was calculated according to the calibration curve generated by the gene PCR products. The relative mRNA expression level of each gene was calculated using the −ΔΔCt method.
Immunocytochemistry
rHFSCs (P3) were cultured to logarithmic phase. Cells were then digested with TrypLE™ Express Enzyme trypsin for 8 min, centrifuged at 500g for 5 min, and then cultured on chamber slides at a density of 1 × 105 cells/well for 2 days. The culture medium was then discarded. The cells were washed with Phosphate Buffered Saline Tween (PBST) and fixed with 4 % PFA. Bovine serum albumin (5 %) (MP Biomedicals, Santa Ana, CA, USA) was then added at room temperature. Primary antibodies against integrin β1 (1:100), integrin α6 (1:50), and Krt15 (1:100) were then added separately to the samples. A negative control included PBST alone instead of the primary antibody. After washing with PBST, the secondary antibody was added in the dark for 30 min. DAPI (1:2,000) was added for 5 min to counterstain the nuclei, and the slides were air dried in the dark and mounted. Red fluorescence was observed under a fluorescence microscope.
Cell growth curve
rHFSCs (P3, P5, P7 and P9) in good condition were detached and seeded in 24-well plates at a density of 1 × 105 cells/well at 37 °C for 8 min. Cell counts were performed on days 1, 2, 3, 4, 5, 6, and 7 by a hemocytometer under the inverted microscope. Six wells were counted for each time point, and the mean cell number was calculated to plot the cell growth curve. A value of P < 0.05 was considered statistically significant.
Vitality of rHFSCs
rHFSCs (P1–P10) were used to calculate the vitality. Cells were digested using Express enzyme and pipetted into a single cell solution at a density of 1 × 105/mL. The cells were stained with trypan blue and counted using a hematocytometer. The mean of three repeated measurements were used. Cell vitality was calculated using the following formula: vital cell rate (%) = number of vital cells/(number of vital cells + number of dead cells) × 100 %.
Statistical analysis
Data are shown as the mean ± standard deviation (SD) and analyzed using SPSS software 18.0. For comparison between groups a Student’s t test was used. A value of P < 0.05 was considered statistically significant.
Results
Primary culture of HFSCs
On day 3 of hair follicle bulge culture, a few cells had migrated from the hair follicle bulge (Fig. 1). Approximately 60 % of the cells showed cobblestone morphology and were closely aligned, which is typical of epithelial cells. The remaining cells were round or spindle-shaped and firmly adherent to the 10 cm dish. On day 7 of culture, cell number was increased.
Fig. 1.
Primary culture of the rat hair follicle stem cells. a day 3, b day 7, c day 14. Scale bars a 200 μm, b-c 100 μm
Purification of HFSCs
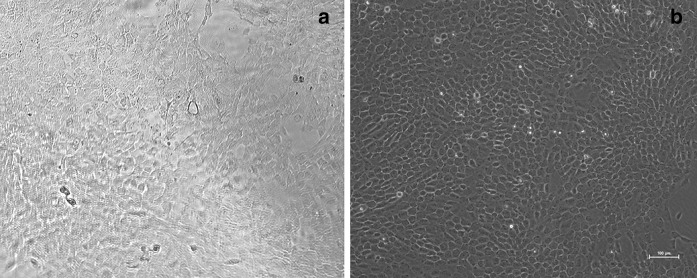
HFSCs (primary and then P2) were purified by culture on collagen IV (Fig. 2). Approximately 95 % of the cells showed typical morphology of stem cells, including a cobblestone and nest appearance, stereoscopic impression, high refractive index, high colony-forming ability, small cell size, and centralized, round and large nuclei. The cells readily formed colonies.
Fig. 2.
The second generation rHFSCs purified by rapid adhering on collagen IV. a Before purification, b After the second purification. Scale bars 100 μm
Real-time PCR
Expression levels of Krt19, Krt10, Krt15, CD34, Itgα6, and Itgβ1 in the purified HFSCs (P5) were detected using real-time PCR. Krt10 is a marker for keratinocytes, and Krt19, Krt15, Itgα6, and Itgβ1 are markers for HFSCs. The Krt19 gene was abundantly expressed, with medium expression for the Krt15 gene. Expression levels of Itgα6 and Itgβ1 were high, while CD34 and Krt10 expression were very low (Table 2). Significant differences were recorded between these genes (P < 0.05).
Table 2.
The Q-PCR quantitative results of the rat HFSCs (n = 3, )
| ACTB | Target gene | △Ct | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ct−1 | Ct−2 | Ct−3 | Av. | Dev. | Ct−1 | Ct−2 | Ct−3 | Av. | Dev. | ||
| Krt10 | 14.10 | 13.84 | 13.67 | 13.87 | 0.09 | 37.13 | 37.38 | 36.85 | 37.12 | 0.14 | 23.25 ± 0.26* |
| Krt15 | 14.10 | 13.84 | 13.67 | 13.87 | 0.09 | 23.75 | 23.92 | 23.67 | 23.78 | 0.03 | 9.91 ± 0.23* |
| Krt19 | 14.10 | 13.84 | 13.67 | 13.87 | 0.09 | 19.46 | 19.27 | 19.13 | 19.29 | 0.05 | 5.42 ± 0.05* |
| CD34 | 14.10 | 13.84 | 13.67 | 13.87 | 0.09 | 30.04 | 29.65 | 29.36 | 29.68 | 0.23 | 15.81 ± 0.13* |
| Itgβ1 | 14.10 | 13.84 | 13.67 | 13.87 | 0.09 | 20.70 | 20.82 | 21.00 | 20.84 | 0.05 | 6.97 ± 0.37* |
| Itgα6 | 14.10 | 13.84 | 13.67 | 13.87 | 0.09 | 20.98 | 20.89 | 21.16 | 21.01 | 0.04 | 7.14 ± 0.31* |
Krt keratin, Itg integrin, ACTB beta-actin
* Compared with ACTB, P < 0.05
Immunocytochemistry
Immunocytochemistry staining showed medium expression of Krt15, with positive expression of Itgα6 and Itgβ1 in the cytoplasm of HFSCs (P3, Fig. 3).
Fig. 3.
Immunofluorescence staining of passage 3 HFSCs. a Krt15, b integrin α6, c integrin β1. Scale bars 100 μm
Proliferation assay
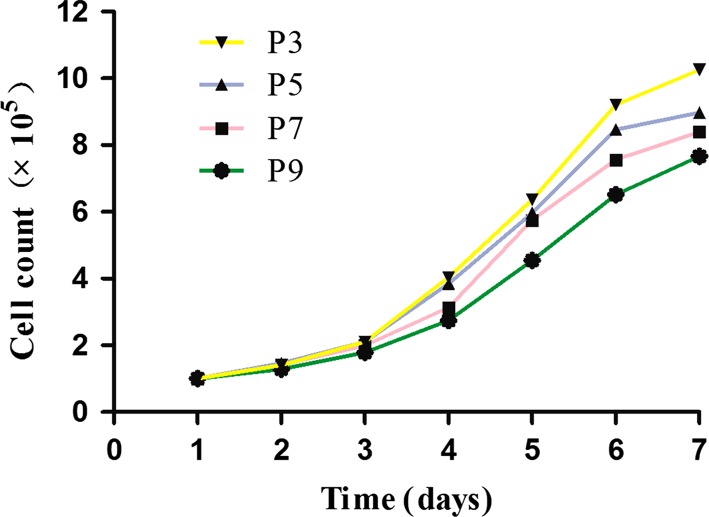
rHFSCs (P3, P5, P7 and P9) were seeded and on days 1–2, the cells were in the latent phase, proliferating slowly. Stem cell colonies appeared on day 3 and on days 5–6, cells were in the logarithmic phase, proliferating rapidly. Proliferation then slowed down gradually into the plateau phase. For HFSCs at P7, proliferation decreased slowly but remained quite rapid. The cells reached plateau phase or rest stage on day 7. The cell counts for rHFSCs at four different passages and at different culture days are shown in Fig. 4.
Fig. 4.
The growth curve of the different passage cells after purification
HFSCs vitality
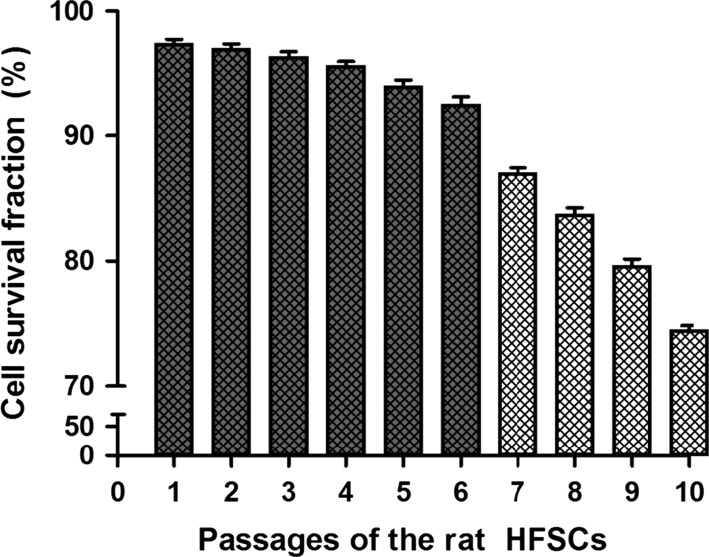
rHFSCs vitality at P1–P10 is shown in Fig. 5. Cell vitality at P1–P6 was very high, at >90 %. From P 7, cell vitality significantly decreased with increased passage. The vitality of P10 cells was <75 %. Therefore, cell vitality before P7 was highest.
Fig. 5.
The histogram of P1–P10 cell survival fraction
Discussion
Cell source is a major challenge for the development of tissue-engineered skin. The most commonly used cells are stem cells. HFSCs have a good proliferative capacity with differentiation potential into epidermis and related cells. In addition, HFSCs are a rich cell source and easy to obtain with minimal trauma. Autologous HFSCs are also free of immune rejection. Therefore, HFSCs may be the ideal cell source for tissue-engineered skin.
HFSCs are primarily found in the bulge of hair follicles. HFSCs are undifferentiated, multi-potent adult stem cells with good self-renewal ability and proliferative capacity (Cotsarelis et al. 1990; Cotsarelis 2006). Cotsarelis et al. (1990) labeled mouse skin with 3H-RdR and found that the hair matrix cells lost the labels, while over 95 % of the bulge cells remained labeled at week 4. Bickenbach and Chism (1998) injected rats subcutaneously with 5-bromodeoxyuridine and found that hair follicle epidermal stem cells and HFSCs were traced to the bulge of the outer root sheath of hair follicles. Furthermore, the bulge-derived HFSCs were found to be superior to epidermal stem cells in colony-formation. Lavker and Sun (2000) showed that HFSCs and epidermal stem cells were essentially the same group of stem cells, both giving rise to hair and skin. It is therefore well-established that HFSCs are located predominantly at the hair follicle bulge. This stem cell type is therefore the most promising source among epidermal stem cell types. Isolation and culture of hair follicle bulge cells was therefore the first step to obtain HFSCs for tissue-engineered skin. However, the current methods for their isolation and culture are unsatisfactory. The cell viability of HFSCs isolated by flow cytometry remains very poor and HFSC purity from tissue blocks is low. Furthermore, microdissection alone cannot isolate HFSCs with high purity.
It has been indicated that HFSCs are essentially epidermal stem cells. Epidermal stem cells have good adhesion ability compared to other cell types, with a faster adhesion speed to collagen IV, laminin (the main component of basement membrane), and extracellular matrix (ECM). However, most epidermal stem cells adhere to the ECM, such as collagen, within 10 min. In comparison, most transiently proliferating cells adhere in 20–60 min (Jones and Watt 1993). This simple and effective differential adhesion method has been successfully used to isolate epidermal stem cells.
In this study, this method, together with microdissection, dispase and collagenase IV digestion, and Matrix coating, was successfully used to obtain HFSCs with good proliferative capacity and purity. The cells isolated using this method showed typical cobblestone and nest morphology, with high colony-forming ability.
Specific markers of HFSCs are currently available. HFSCs are commonly characterized with two or more markers. CD200, CD34, P63, Krt10, Krt15, Krt19, integrin β1, and integrin α6 are considered as potential specific HFSCs markers (Kloepper et al. 2008; Ohyama et al. 2006; Amoh et al. 2005). It has been reported that as cells proliferate, the reduction in Krt15 expression appears earlier than the reduction in Krt19 expression, and therefore Krt15− Krt19+ cells may proliferate transiently in the early phase. Therefore, it is proposed that Krt15 may be more specific than Krt19 in characterizing HFSCs (Lavker and Sun 1982). In this study, we used immunocytochemistry and real-time PCR to detect the specific gene markers for the characterization of HFSCs. In real-time PCR analysis, a ΔΔCt < 7 usually denotes abundant expression of a gene, while a ΔΔCt > 7 denotes medium or low expression of a gene (Schefe et al. 2006). This study found that expression of Krt19 was highest, with medium expression of Krt15. Expression levels of integrin β1 and integrin α6 were high, while CD34 expression was low. Immunofluorescence staining of the purified rHFSCs showed positive expression of Krt15, integrin β1 and integrin α6. Therefore, this method can be used to successfully isolate and culture rHFSCs from the hair follicle bulge. The obtained rHFSCs were high in purity with good proliferative ability.
We found that the proliferation ability and vitality of rHFSCs decreased with increased passage. HFSCs went through the latent phase, proliferation phase, and then the plateau phase. The cells showed good proliferation and colony-forming ability. From P7, the cells showed a gradual decrease in proliferation ability, but continued to proliferate rapidly. The rHFSC vitality rate was significantly higher up to P6 than that of later passages. Vitality decreased significantly from P7 with increased passage. The vitality rate was <75 % in P10 cells.
HFSCs are promising stem cells with good proliferative and multi-lineage differentiation capacities. Furthermore, HFSCs can be transfected with various genes to obtain additional biological features (Da-hai et al. 2012; Takamiya et al. 2002). These genes include vascular endothelial growth factor, EGF, angiopoietin, and angiotensin. Genetically modified HFSCs may therefore be used in tissue-engineered skin, which could be highly beneficial for the treatment of larger skin defects caused by burns, trauma and cutting.
Conclusions
Highly purified rHFSCs can be obtained using tissue culture and adhesion to collagen IV. The cultured cells had good proliferative capacity and could therefore be a useful cell source for tissue-engineered hair follicles, vessels, and skin.
Acknowledgments
This study was supported by the Social Welfare and Technology Development program, Department of Science and Technology of Zhejiang Province (2010C33133).
Contributor Information
Renfu Quan, Phone: +8657183812008, Email: quanrenfu@126.com, Email: quanrenf@163.com.
Yueming Ni, Email: niyueming1@126.com.
References
- Amoh Y, Li L, Katsuoka K, Penman S, Hoffman RM (2005) Multipotent nestin-positive, keratin-negative hair-follicle bulge stem cells can form neurons. Proc Natl Acad Sci USA 102:5530–5534 [DOI] [PMC free article] [PubMed]
- Amoh Y, Kanoh M, Niiyama S Hamada Y, Kawahara K, Sato Y, Hoffman RM, Katsuoka K (2009) Human hair follicle pluripotent stem (hfPS) cells promote regeneration of peripheral-nerve injury: an advantageous alternative to ES and iPS cells. J Cell Biochem 107:1016–1020 [DOI] [PubMed]
- Bickenbach JR, Chism E. Selection and extended growth of murine epidermal stem cells in culture. Exp Cell Res. 1998;244:184–195. doi: 10.1006/excr.1998.4163. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G. Epithelial stem cells: a folliculocentric view. J Invest Dermatol. 2006;126:1459–1468. doi: 10.1038/sj.jid.5700376. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-C. [DOI] [PubMed] [Google Scholar]
- Hu DH, Zhang ZF, Zhang YG, Zhang WF, Wang HT, Cai WX, Bai XZ, Zhu HY, Shi JH, Tang CW (2012) A potential skin substitute constructed with hEGF gene modified HaCaT cells for treatment of burn wounds in a rat model. Burns 38:702–712 [DOI] [PubMed]
- Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-K. [DOI] [PubMed] [Google Scholar]
- Kloepper JE, Tiede S, Brinckmann J, Reinhardt DP, Meyer W, Faessler R, Paus R (2008) Immunophenotyping of the human bulge region: the quest to define useful in situ markers for human epithelial hair follicle stem cells and their niche. Exp Dermatol 17:592–609 [DOI] [PubMed]
- Lavker RM, Sun TT. Heterogeneity in epidermal basal keratinocytes: morphological and functional correlations. Science. 1982;215:1239–1241. doi: 10.1126/science.7058342. [DOI] [PubMed] [Google Scholar]
- Lavker RM, Sun TT. Epidermal stem cells: properties, markers and location. Proc Natl Acad Sci USA. 2000;97:12373–12375. doi: 10.1073/pnas.250380097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Peng HF, Andreadis ST. Contractile smooth muscle cells derived from hair-follicle stem cells. Cardiovasc Res. 2008;79:24–33. doi: 10.1093/cvr/cvn059. [DOI] [PubMed] [Google Scholar]
- Nowak JA, Fuchs E. Isolation and culture of epithelial stem cells. Methods Mol Biol. 2009;482:215–232. doi: 10.1007/978-1-59745-060-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama M, Terunuma A, Tock CL, Radonovich MF, Pise-Masison CA, Hopping SB, Brady JN, Udey MC, Vogel JC (2006) Characterization and isolation of stem cell-enriched human hair follicle bulge cells. J Clin Invest 116:249–260 [DOI] [PMC free article] [PubMed]
- Rochat A, Kobayashi K, Barrandon Y. Location of stem cells of human hair follicles by clonal analysis. Cell. 1994;76:1063–1073. doi: 10.1016/0092-8674(94)90383-2. [DOI] [PubMed] [Google Scholar]
- Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H (2006) Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. J Mol Med (Berl) 84:901–910 [DOI] [PubMed]
- Takamiya M, Saigusa K, Aoki Y. Immunohistochemical study of basic fibroblast growth factor and vascular endothelial growth factor expression for age determination of cutaneous wounds. Am J Forensic Med Pathol. 2002;23:264–267. doi: 10.1097/00000433-200209000-00012. [DOI] [PubMed] [Google Scholar]
- Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM (2000) Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell 102:451–461 [DOI] [PubMed]
- Yu H, Fang D, Kumar SM, Li L, Nguyen TK, Acs G, Herlyn M, Xu X (2006) Isolation of a novel population of multipotent adult stem cells from human hair follicles. Am J Pathol 168:1879–1888 [DOI] [PMC free article] [PubMed]