Abstract
Snake venoms are mixtures of bioactive proteins and peptides that exhibit diverse biochemical activities. This wide array of pharmacologies associated with snake venoms has made them attractive sources for research into potentially novel therapeutics, and several venom-derived drugs are now in use. In the current study we performed a broad screen of a variety of venoms (61 taxa) from the major venomous snake families (Viperidae, Elapidae and “Colubridae”) in order to examine cytotoxic effects toward MCF-7 breast cancer cells and A-375 melanoma cells. MTT cell viability assays of cancer cells incubated with crude venoms revealed that most venoms showed significant cytotoxicity. We further investigated venom from the Red-bellied Blacksnake (Pseudechis porphyriacus); venom was fractionated by ion exchange fast protein liquid chromatography and several cytotoxic components were isolated. SDS-PAGE and MALDI-TOF mass spectrometry were used to identify the compounds in this venom responsible for the cytotoxic effects. In general, viper venoms were potently cytotoxic, with MCF-7 cells showing greater sensitivity, while elapid and colubrid venoms were much less toxic; notable exceptions included the elapid genera Micrurus, Naja and Pseudechis, which were quite cytotoxic to both cell lines. However, venoms with the most potent cytotoxicity were often not those with low mouse LD50s, including some dangerously venomous viperids and Australian elapids. This study confirmed that many venoms contain cytotoxic compounds, including catalytic PLA2s, and several venoms also showed significant differential toxicity toward the two cancer cell lines. Our results indicate that several previously uncharacterized venoms could contain promising lead compounds for drug development.
Keywords: Colubridae, Cytotoxicity, Drug development, Melanoma, Phospholipase A2, Three-finger toxin
Introduction
Snake venoms contain a broad diversity of organic and inorganic compounds, consisting primarily of toxins, enzymes, and other bioactive peptides (Mackessy 2010a; Calvete et al. 2006; Lomonte et al. 2014). Although a single venom may contain up to 100 different proteins (including isoforms), many of these compounds can be classified into a relatively limited number of protein families, including metalloproteases (Fox and Serrano 2005), phosphodiesterases (Mackessy 1998), phospholipases A2 (Kini 2003; Huang and Mackessy 2004; Doley et al. 2010), serine proteases (Mukherjee and Mackessy 2013), acetylcholinesterases (Anderson and Dufton 1998), disintegrins (Calvete et al. 2005; Saviola et al. 2013), and three-finger toxins (such as α-neurotoxins and cardiotoxins: Doley et al. 2008; Kini 2002; Kini and Doley 2010; Nirthanan and Gwee 2004). Millions of years of evolution have resulted in molecules with incredible selectivity for various physiological targets, leading to rapid immobilization and death of prey. However, the great diversity of molecules and biochemical activities found in snake venoms also makes these varied and complex mixtures attractive in the search for novel therapeutics.
The use of toxins as potential drugs has been a growing area of research in the last decade (Fox and Serrano 2007; Vonk et al. 2011), and numerous promising drugs have already been developed from snake venom proteins and peptides, with more currently under investigation (Earl et al. 2012; Fox and Serrano 2007; Koh and Kini 2012; Vink et al. 2012; Takacs and Nathan 2014). Classically, venom from Bothrops jararaca contributed to the development of the angiotensin-converting enzyme inhibitors, which are now widely used in the treatment of hypertension and kidney disease (Ferreira 1965; Ferreira et al. 1970; Koh and Kini 2012). Not surprisingly, several snake venom-derived drugs utilized for anticoagulation and hemostasis have also been developed. For example, the platelet receptor glycoprotein IIb/IIIa (integrin αIIbβ3) antagonist eptifibatide is derived from a disintegrin isolated from Sistrurus miliarius barbouri venom (Kereiakes et al. 1996), and several procoagulant drugs are currently under development (Earl et al. 2012). Additionally, venom disintegrins, which act by binding to and modulating the functions of integrins (Kamiguti et al. 1998), have demonstrated promising anti-angiogenic (Brown et al. 2008), anti-tumor (Swenson et al. 2005; Sánchez et al. 2009) and anti-metastatic effects (Lucena et al. 2011; McLane et al. 2008; Tian et al. 2007).
Although significant research has continued to examine novel sources for potential cancer treatments, many therapies are non-specific and often have severe side effects. Snake and other animal venoms comprise unique libraries of biological compounds offering vast arrays of pharmacological activities that recognize certain receptors with high specificity. In return, isolated venom compounds may be utilized in treatments for not only cancer, but disorders such as arthritis and thrombosis as well (Pal et al. 2002). In the current study, using a diverse collection of snake venoms representing the three major clades of venomous snakes (Elapidae, Viperidae and the rear-fanged “Colubridae”), we examined cytotoxic effects toward human breast (MCF-7) and melanoma (A-375) cancer cell lines. Pseudechis porphyriacus venom was chosen for further analysis, based on initial cytotoxicity assays, and isolation of two protein fractions is presented as an example of the cytotoxic effects of specific venom proteins. The methodologies we utilized in this study represent an effective approach for the initial screening of snake venoms and have the potential to uncover novel compounds (Chaim-Matyas and Ovadia 1987; El-Refael and Sarkar 2009; Oron et al. 1992), and many of the venoms analyzed here have not been previously investigated.
Materials and methods
Venom extraction and preparation
Venoms were manually extracted from snakes using established methods (Mackessy 1988; Hill and Mackessy 1997), with all procedures approved by the University of Northern Colorado Institutional Animal Care and Use Committee (protocols 0702 and 0901C). Venoms were then centrifuged, lyophilized and stored frozen at −20° C until use. Samples were reconstituted in ddH2O at 4 mg/mL, centrifuged at 10,000×g to pellet insoluble debris, and the supernatant was used in all experiments. Red-bellied Black Snake (P. porphyriacus) venom was collected using similar methods and was a gift of Venom Supplies Pty. Ltd. (Tanandu, AU). Venoms from most Bothrops species were a gift of Dr. C. Ownby. Specific materials were purchased from suppliers as noted below; all other biochemicals (analytical grade or better) were obtained from Sigma Chemical Co. (St. Louis, MO, USA).
Cell culture and viability assays
All cancer cells, media, cell viability assay kits, and fetal bovine serum were purchased from American Type Cell Culture (ATCC; Manassas, VA, USA). MCF-7 human breast adenocarcinoma cells (ATCC HTB-22) were maintained in Eagle’s minimum essential medium (EMEM) with 0.01 mg/mL bovine insulin and 10 % fetal bovine serum. A-375 human malignant melanoma cells (ATCC CRL-1619) were cultured in Dulbecco’s modified Eagle medium (DMEM) with 10 % fetal bovine serum. All cells were maintained in 75 mL flasks at <90 % confluence and incubated at 37 °C with 5 % CO2 in a humidified atmosphere. Cells were subcultivated according to instructions from ATCC, using trypsin–EDTA (0.05 % Trypsin and 0.02 % EDTA; ATCC PCS-999-003) and cryopreserved using 5 % dimethyl sulfoxide (DMSO) in the appropriate growth medium.
Cytotoxicity of various crude venoms was examined using the colorimetric MTT [3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-teyrazolium bromide] assay, where cleavage of MTT by metabolically active cells produces a purple formazan product (Mosmann 1983). The amount of formazan formed is directly proportional to the number of viable cells, which can then be quantified at 570 nm. For all assays, one hundred μL aliquots of cells at a concentration of 106 cells/mL (MCF-7 cells) or 5 × 105 cells/mL (A-375 cells) were plated into 96-well plates and treated with venom (20 µg in 5 µL) or an identical volume of PBS as the control, in a single-blind fashion. Cells were incubated with venom or control treatments for 24 h and then 10 μL/well of MTT reagent (ATCC) was added. Cells were then returned to the incubator for 2 h before being treated with 100 μL of detergent reagent (ATCC) to rupture cells and dissolve formazan into a colored solution. Plates were stored overnight in the dark at room temperature. Finally, data were collected using a SpectraMax-190 96 well plate reader (Molecular Devices, Sunnyvale, CA, USA) at 570 nm. A standard curve was generated for each assay performed, which consisted of 106 cells/mL (A-375: 5 × 105) serially diluted 1:2 down to a concentration of 6.25 × 104 cells/mL. Linear regression analysis indicated a linear relationship between absorbance and cell density from 6.25 × 104 to 106 cells/mL with r2 values of >0.99. The equation for the line of best fit was used to calculate the concentration of viable cells after treatment with crude venom or fractionated venom components. For fast protein liquid chromatography (FPLC) fractionated P. porphyriacus venom, approximately 5 µg protein in 10 µL for each peak was assayed for cytotoxic effects toward MCF-7 cells, as mentioned above. All cell assays were performed with six replicates/sample and the percentage of viable cells was determined by the equation [(absorbance of treatment cells) − (absorbance of medium blank)/(absorbance of control cells) − (absorbance of medium blank)] × 100. All values are presented as mean ± standard error of the mean.
Venom fractionation by fast protein liquid chromatography (FPLC)
Venom samples from P. porphyriacus were fractionated by cation exchange liquid chromatography using an ÄKTA Purifier 900 (GE Biosciences, Inc. Piscataway, NJ, USA). Venom was solubilized in 20 mM MES HCl, pH 6.5, at a concentration of 12 mg/mL, vortexed, centrifuged at 10,000×g for 5 min, decanted and filtered through a 0.2 μm syringe filter. Five hundred µL (6 mg) of sample was injected into a Tricorn Mono S 5/50 GL column (GE Biosciences) at a flow rate of 1 mL/min, and the column was developed with a linear gradient of 20 mM MES HCl, pH 6.5 (buffer A) and 20 mM MES HCl with 2 M NaCl, pH 6.5, (buffer B): isocratic at 100 % buffer A for 10 min, then a linear gradient over 65 min to 20 % buffer B, followed by 20–100 % buffer B for 10 min, after which the mobile phase was returned to 100 % buffer A. Proteins were detected at 280 nm, collected with a Frac-920 fraction collector (GE Biosciences, Inc.) and frozen until further characterized.
Characterization of Pseudechis porphyriacus FPLC fractions
To estimate the number of proteins and their molecular masses, fractions obtained from the FPLC separation were characterized by reducing SDS polyacrylamide gel electrophoresis (SDS-PAGE) using 12 % NuPage Bis–Tris gels (Invitrogen Inc., San Diego, CA, USA). Venom fractions from the FPLC separation were dried with a Speed-Vac (Savant, Thermo Fisher Scientific, Waltham, MA, USA), re-solubilized in LDS buffer with 15 mM dithiothreitol (final concentration), heated at 70 °C for 15 min and approximately 10 µg protein per lane was applied to the gel. Standards for estimating molecular mass (Mark 12 unstained standards) were purchased from Invitrogen. To estimate protein families and obtain more accurate molecular masses, FPLC fractions of peaks 1 and 13 (fractions 3 and 148) were analyzed using matrix-assisted laser-desorption ionization time-of-flight mass spectroscopy (MALDI-TOF-MS), as these peaks demonstrated the highest cytotoxicity towards cells. Ten μL of selected FPLC fractions was desalted and concentrated with a ZipTip C4 micropipette tip. One μL of this sample was then spotted onto 1 μL sinapinic acid (10 mg/mL in 50 % acetonitrile with 0.1 % TFA) matrix. Data were collected using a Bruker (Billerica, MA, USA) Ultraflex mass spectrometer in linear mode. Samples were analyzed using a window of 5–16 kDa. Samples used in cytotoxicity assays were desalted and concentrated using spin columns (Amicon 3kD cutoff EMD Millipore Corp., Billerica, MA, USA), washed with 300 μL ddH2O × 2 spins and then lyophilized.
Results
Effects of crude venom MTT cell proliferation assay results
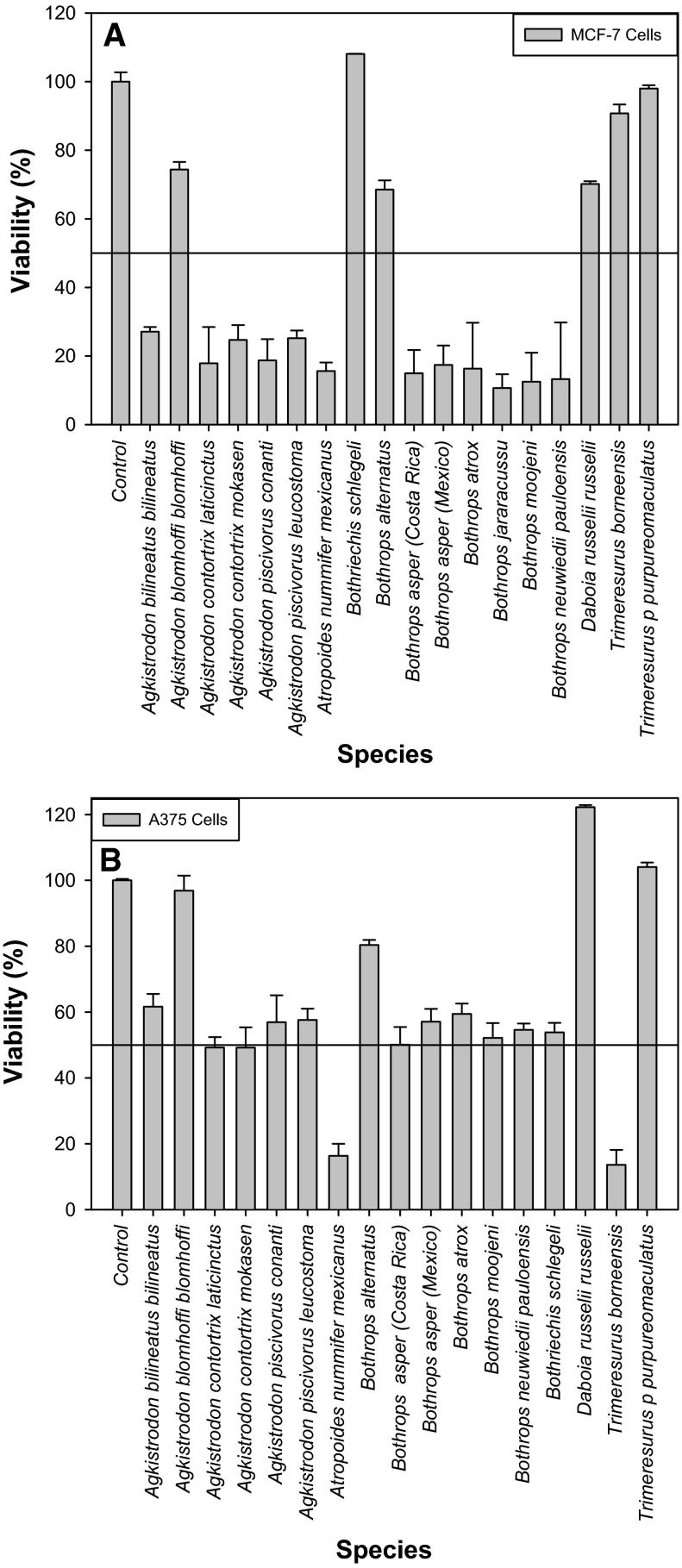
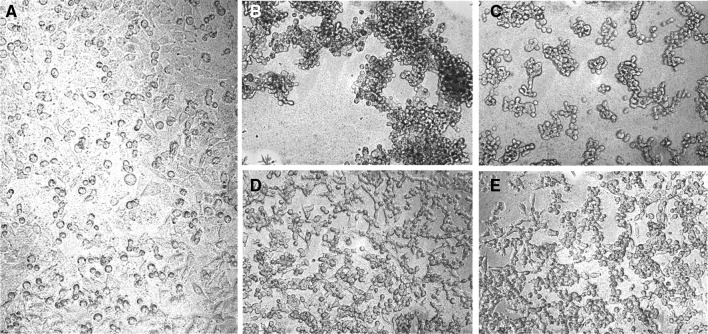
In general, rattlesnake venoms (Crotalus and Sistrurus) were more toxic to both breast cancer and melanoma cells (Fig. 1a, b) than were those from elapid and colubrid snakes (Fig. 3a, b). There were also, however, several highly cytotoxic elapid venoms and conversely, weakly cytotoxic rattlesnake venoms. Colubrid and elapid venoms demonstrated the highest degree of variation in cancer cell toxicity, though samples of some viperids (Agkistrodon, Atropoides, Bothriechis and Bothrops species) showed variable toxicities as well (Fig. 2a, b). Most crude venoms studied showed significant cytotoxicity, particularly toward MCF-7 cells (Fig. 4), although several, such as the hydrophiine elapids Hydrophis, Laticauda, Acanthophis and Notechis, did not (Figs. 3a, b, 4). Several venoms used to treat the A-375 melanoma cells caused a dramatic change in cell phenotype, developing a distinctly rounded morphology after treatment (Fig. 5b). In order to determine if cytotoxic effects were specific effects of the venoms or non-specific factors, dose–response curves were generated for four venoms of interest (Fig. 6a–d) and IC50 values were estimated. Crotalus oreganus cerberus and P. porphyriacus venoms demonstrated typical dose–response curves (Fig. 6a, b), in which increasing doses led to a decrease in viable cells, as indicated by decreased formazan production. However, venoms from both Bothrops alternatus and C. o. concolor, while exhibiting initial dose-dependent effects, appeared to reach a plateau at which greater doses of venom did not have significantly greater effects (Fig. 6c, d). One venom which showed high levels of cytotoxicity (P. porphyriacus venom) was used as an example to demonstrate cytotoxicity of specific venom components.
Fig. 1.
Effect of rattlesnake venoms (19 µg/100 µL medium) on human cancer cells. Cells (100 µL) were seeded into wells at 106 cells/mL (MCF-7) or 5 × 105 cells/mL (A-375), and all assays utilized six replicates. a MCF-7 breast cancer cells. b A-375 melanoma cells. Line represents 50 % viability
Fig. 3.
Effect of viperid venoms (19 µg/100 µL) on human cancer cells. Cells (100 µL) were seeded into wells at 106 cells/mL (MCF-7) or 5 × 105 cells/mL (A-375), and all assays utilized six replicates. a MCF-7 breast cancer cells. b A-375 melanoma cells. Line represents 50 % viability
Fig. 2.
Effect of rear-fanged snake (“colubrid”) and elapid venoms (19 µg/100 µL) on human cancer cells. Cells (100 µL) were seeded into wells at 106 cells/mL (MCF-7) or 5 × 105 cells/mL (A-375), and all assays utilized six replicates. a MCF-7 breast cancer cells. b A-375 melanoma cells. Line represents 50 % viability
Fig. 4.
Micrographs of control and venom-treated MCF-7 breast cancer cells after 24 h incubation with venoms (19 µg/100 µL). All assays utilized six replicates. a MCF-7 control—no venom; b P. porphyriacus venom—high cytotoxicity; c C. o. cerberus venom—high cytotoxicity and extreme cell damage (cell fragments); d B. alternatus venom—minimal cytotoxicity observed; e C. o. concolor venom—minimal cytotoxicity observed
Fig. 5.
Micrographs of control and venom-treated A-375 melanoma cells after 24 h incubation, showing differential effects of venoms. Treated cells (5 × 105 cells/mL) received 19 µg venom/100 µL medium, and all assays utilized six replicates. a A-375 control—no venom. b Effects of exposure to Bothrops alternatus venom—note the change in cell phenotype to a more rounded and clumped appearance. c Effects of exposure to Cryptelytrops (formerly Trimeresurus) purpureomaculatus purpureomaculatus venom—change in phenotype is similar to b. d Effects of exposure to Daboia russelii russelii venom—phenotypic changes are minimal. e Effects of exposure to Ahaetulla nasuta venom—again, phenotypic changes are minimal
Fig. 6.

Dose-dependent cytotoxicity of elapid and viperid venoms toward MCF-7 breast cancer cells. Note that for a and b, toxicity curves follow a standard dose-dependent decrease approximating 0 % survivorship, while c and d appear to reach a threshold beyond which no effects of increased venom concentrations are observed. Crotalus o. cerberus is the least toxic (LD50) subspecies in the oreganus clade, while C. o. concolor is the most toxic
FPLC fractionation of Pseudechis porphyriacus venom and characterization of fractions
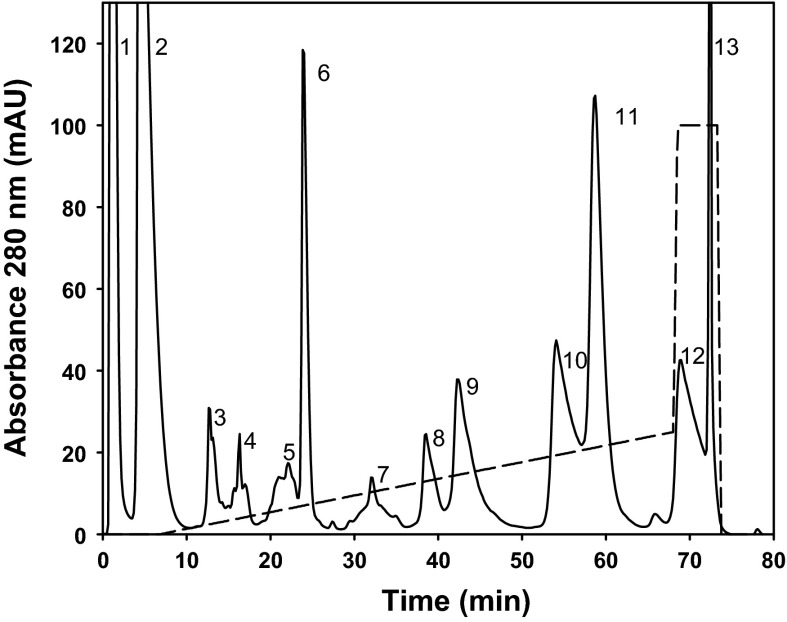
FPLC analysis of P. porphyriacus venom showed 13 well-defined peaks (Fig. 7a) that were further subjected to reducing SDS-PAGE in order to estimate purity and mass of the compounds. Gel bands were classified into protein families (Fig. 8a) based on previously identified venom proteins (see Mackessy 2010a). All 13 peaks were tested for cytotoxic effects towards MCF-7 cells, with peaks 1 and 13 exhibiting the highest cytotoxicity and peaks 2, 4–6 and 8–10 showing intermediate toxicity (Fig. 8b). The more potent cytotoxicity toward MCF-7 cells is likely due to the presence of a phospholipase A2 (PLA2) and a three-finger toxin (3FTx) in peak 1, and the PLA2 present in peak 13 also appears to be quite cytotoxic. Peak 1 also may contain an acetylcholinesterase as well as small amounts of a non-enzymatic cysteine-rich secretory protein (CRiSP); however, due to the known biological activities of these compounds it is not likely that they were responsible for the cytotoxic affects seen here. SDS-PAGE indicated the possibility of trace amounts of a CRiSP in peak 13, but upon further investigation using MALDI-TOF-MS, we suspect that this peak contains primarily a single PLA2. Further MALDI-TOF-MS analysis of peak 1 and peak 13 confirmed that peak 1 contained numerous different compounds (at least five), with several in the 6–7 kDa range, indicating the presence of 3FTx isoforms, and several in the 13–14.5 kDa range, expected masses of PLA2 isoforms (Fig. 8c). Peak 13 apparently contains a single PLA2 protein with a mass of 13,114.8 Da (Fig. 8 d). The presence of this protein species, and the absence of any proteins in the 21–31 kDa range (data not shown), indicates that this peak does not contain a CRiSP, as CRiSPs typically ionize well and have masses of ~25 kDa.
Fig. 7.
Fractionation of Pseudechis porphyriacus venom using cation exchange chromatography on a MonoS Tricorn column. Thirteen well-resolved peaks were observed
Fig. 8.

Characterization of fractionated P. porphyriacus venom. Numbers represents each of the 13 peaks obtained from cation exchange. a SDS-PAGE (reducing) analysis of each protein peak; protein family typically observed at given masses is indicated on the left. Note that phospholipases A2 and three-finger toxins (3FTxs) dominate the proteome of P. porphyriacus venom. b Cytotoxicity of each peak toward MCF-7 breast cancer cells; peaks 1 and 13 (asterisks) were most potent. c Mass spectrogram of peak 1 peptides using a 5–16 kDa window; 3FTxs (6–7 kDa) and PLA2s (13–14 kDa) dominate. d Mass spectrogram of peak 13 peptides using a 5–16 kDa window; only PLA2 (13,114.8 kDa) and trace amounts of a 3FTx are observed
Discussion
Research into venomous systems offers significant insights into the biological roles of venom compounds (Saviola et al. 2013) and provides useful information that can be utilized for effectively treating human snakebite (Gutiérrez et al. 2009). In addition, venom research provides potential avenues for novel drug discovery and design (Fox and Serrano 2007; Vonk et al. 2011). Venom composition varies between and among species depending on several factors, such as phylogenetic affinities (Mackessy 2010a), geographic localities (Alape-Girón et al. 2008; Núñez et al. 2009), snake age (Mackessy 1988, 1993; Mackessy et al. 2003; Calvete 2010) and diet (Gibbs and Mackessy 2009; Barlow et al. 2009). These often significant differences in venom composition, coupled with diverse and potent biological activities, make snake venoms attractive sources as pharmacological tools for understanding vertebrate physiological pathways and human diseases. As venoms are proving to be a excellent source of natural compounds exhibiting cytotoxic (Yalcın et al. 2014), apopototic (Samel et al. 2012) and anti-tumor (Lin et al. 2010) effects towards numerous cancerous cell lines, continued research may identify novel therapeutics for cancers or other diseases.
In the current study we examined the cytotoxic effects toward both MFC-7 breast cancer and A-375 melanoma cell lines of venoms representing three very different families of snakes, from the highly venomous front-fanged elapids and viperids to the relatively non-toxic, rear-fanged colubrids. Venoms from colubrids and elapids demonstrated the greatest variation in cytotoxicity and were generally less toxic to both MCF-7 and A-375 cells. For the rear-fanged colubrids, this finding was not unexpected, as various colubrid toxins and venoms are prey-specific, showing high toxicity against lizards or birds, but not against mammals (Mackessy 2002; Mackessy et al. 2006; Pawlak et al. 2006, 2009; Heyborne and Mackessy 2013). This varying toxin-receptor specificity, which is associated with different prey types, likely also influences the cytotoxicity documented in this study. Further, this family of snakes has been taxonomically challenging, and currently defined families and subfamilies (formerly all in the family Colubridae) are believed to be distantly related (e.g., Gauthier et al. 2012; Pyron et al. 2013; Wiens et al. 2012). It is therefore not unexpected that venom effects (and by extension composition) may vary significantly from the effects demonstrated by front-fanged snake venoms.
Viper venoms, on the other hand, tend to be quite toxic to a wide variety of prey (Gibbs and Mackessy 2009), suggesting that specificity of these venoms may be less pronounced and that viper venoms contain a diversity of toxins that act against numerous prey sources. It is well known that rattlesnakes and other vipers undergo an ontogenetic shift in prey preference; neonates tend to prey on small ectothermic prey, whereas adults feed primarily on larger, more metabolically advantageous endothermic prey (e.g. Mackessy 1988). This shift in prey preference is correlated with shifts in venom composition (Minton and Weinstein 1986; Mackessy 1988; Alape-Girón et al. 2008), providing vipers with an arsenal of venom compounds with differing receptor specificities and toxicities. Rattlesnake venoms are also classified into type I (high metalloproteinase activity and lower toxicity) or type II (low metalloproteinase activity, high toxicity) venoms, as toxicity and metalloproteinase activity are generally inversely correlated (Mackessy 2008).
Interestingly, our MTT assay results indicate that some venoms of high lethality to prey (including many of the elapid and viperid venoms evaluated) are not cytotoxic to cancer cells at doses assayed. For example, venom from C. o. concolor, which produces the most toxic venom (LD50 = 0.4 mg/kg) within the viridis/oreganus clade, and C. o. cerberus, which produces the least toxic venom (LD50 = 5.4 mg/kg) of this clade (Mackessy 2010b), showed a reverse relationship of cytotoxicity in our MCF-7 MTT assays (Fig. 6b, d). Crotalus durissus terrificus (LD50 = 0.13 mg/kg) and C. tigris (LD50 = 0.07 mg/kg) venoms, which are among the most toxic of viper venoms, showed very low cytotoxicity toward MCF-7 cells (Fig. 1a), while these same venoms showed variable cytotoxicity toward A-375 cells (Fig. 1b; C. tigris venom essentially non-cytotoxic). Similarly, among Australian elapids, Acanthophis antarcticus venom (LD50 = 0.34 mg/kg) and P. porphyriacus (LD50 = 2.53 mg/kg) venom have very different whole animal toxicities (Mirtschin et al. 1990), but for both cell lines tested, Acanthophis venom was essentially non-toxic, while Pseudechis venom was potently cytotoxic. These data again underscore the difficulty in attempting to model whole animal toxicity by an alternative in vitro assay.
While most venoms demonstrated similar toxicity against both MCF-7 and A-375 cells, there were significant differences in the cytotoxicities of several venoms toward the two cell lines. Venom from the colubrid Ahaetulla nasuta, in particular, was non-toxic toward A-375 cells, but it was quite cytotoxic toward MCF-7 cells. Differences in ontogeny and phenotype between these cell lines may explain the discrepancies observed in this study, and several differences have been documented. Mammary gland cells are derived from embryologic mesenchyme, while melanocytes originate from cells of the neural crest. MCF-7 breast cancer cells are known to produce insulin-like growth factors (Takahashi and Suzuki 1993) and are responsive to estradiol, as they express cytoplasmic estrogen receptors. Further, MCF-7 cells express the WNT7B oncogene (Huguet et al. 1994) and also contain the Tx-4 oncogene. A-375 cells express melanocyte-stimulating hormone receptors, which are not expressed on MCF-7 cells (Sharma et al. 1996). It is possible that differential expression of these (and other) receptors and oncogenes in the two cell types may be involved in the differential sensitivities observed.
Venoms that were highly cytotoxic, but which did not cause extensive cell lysis (as visualized by light microscopy) were of greatest interest for further characterization, because these venoms were suspected to contain fewer non-specific, highly toxic compounds. Venoms that demonstrated significantly greater toxicity than most other venoms within a family, and venoms that appeared to cause morphological changes in the cells, were also noted. Several viper venoms, from B. alternatus and Cryptelytrops (formerly Trimeresurus) purpureomaculatus purpureomaculatus, caused an intriguing change in A-375 cell morphology (see Fig. 5b–c), while D. r. russelii venom (Fig. 5d) did not. Recently, the serine protease russelobin and the phospholipase A2 RVAPLA2, both from D. r. russelii venom, were both demonstrated to lack cytotoxicity and did not induce morphological changes in MCF-7 or human colorectal adenocarcinoma (Colo-205) cells (Mukherjee and Mackessy 2013; Mukherjee 2014); however, an L-amino acid oxidase from the same venom showed potent cytotoxic effects toward MCF-7 cells (Mukherjee et al., in prep.). The most striking effects on cell morphology were observed after treatment with venom from B. alternatus, a venom that consists predominantly (43 %) of P-III snake venom metalloproteases (SVMPs) but includes other common viperid venom compounds such as serine proteases, PLA2s, L-amino acid oxidases, disintegrins, and C-type lectins (Öhler et al. 2010). Additional studies will be necessary to determine which specific venom compound(s) lead to the morphological changes.
Pseudechis porphyriacus venom was chosen for further investigation for several reasons. This elapid venom demonstrated a high degree of cytotoxicity without causing massive cellular destruction, suggesting the presence of anti-proliferative and cytotoxic compounds that may have a degree of selectivity. In addition, P. porphyriacus venom showed a clear dose–response curve (Fig. 6a), suggesting that its cytotoxicity was due to specific venom components, rather than nonspecific factors producing cell necrosis associated with venom compounds, such as some SVMPs.
MCF-7 cells treated with fractioned P. porphyriacus venom showed varying degrees of cytotoxicity, with peaks 1 and 13 demonstrating the highest cytotoxicity. Based on SDS-PAGE and MALDI-TOF analysis, both peaks 1 and 13 contained proteins with masses characteristic of PLA2s. Peak 1 (acidic peptides) contained numerous proteins, including a 56 kDa protein (possible acetylcholinesterase), an acidic PLA2 (mass = 14,048.3 Da) and several acidic three-finger toxins (3FTxs: masses of 6,648.2, 6,855.7 and 7,023.4 Da). Peak 13, containing a highly basic peptide that eluted during the high salt wash, consisted primarily of a basic PLA2 (mass = 13,114.8 Da). Snake venom PLA2’s have been shown to have a diversity of pharmacologic roles, including effects on cell proliferation and migration (Doley et al. 2009; Kini 2003). Although some PLA2 enzymes may not be cytotoxic (Mukherjee 2014), it is likely that the cytotoxicity of peaks 1 and 13 is due to the presence of a specific PLA2. Three-finger toxins also show a wide variety of pharmacologies (Kini and Doley 2010), including cytotoxicity, so the potential roles of these toxins in Peak 1 activity cannot be ruled out, but a similar sized (6,615 Da) three-finger toxin isolated from the same venom was demonstrated to show neurotoxic activity (Pierre et al. 2007). SVMPs also impact cultured cells, and some induce apoptosis (Fox and Serrano 2010; Masuda et al. 1998, 2000, 2001). However, Pseudechis venom has quite low SVMP activity which is localized in peaks 5 and 7 (data not shown), and which are only modestly cytotoxic and essentially non-toxic, respectively.
Continued investigation of unusual venoms and lead compounds identified through this work has promise to yield novel peptides for further evaluation as pharmaceuticals, and more effective treatments for cancers or other human related disorders is a major goal of natural compound research. While the field of rational drug design has exploded in recent years, natural products remain an excellent source of incredibly diverse bioactive molecules with potential for development into therapeutics. Snake venoms, and especially those of the understudied rear-fanged colubrids, represent an under-tapped resource of bioactive molecules with a wide variety of functions and potential applications (Saviola et al. 2014). Through more extensive screening of this “natural library” of venom compounds, it is likely that a number of novel therapeutics will emerge.
Conclusion
We have shown that venoms from snakes of three distinct families have varying and sometimes exceptional levels of cytotoxicity towards MCF-7 and A-375 cell lines. Although a number of venoms with low cytotoxicity against both of these cell lines could be excluded from further investigation, specific compounds demonstrated significant anti-metastatic activity without undue cytotoxicity and should be investigated further. However, the many venoms that did show high cytotoxicity can be examined further for the compound(s) responsible for such effects and also warrant ongoing investigation. Our data suggest that compounds such as PLA2s and perhaps 3FTxs may be central to cytotoxic activities seen in P. porphyriacus venom, and the highly stable molecular scaffolds of these proteins may be of significant use for future therapeutic drug design (e.g. Fruchart-Gaillard et al. 2012). To the best of our knowledge, this study represents the first cytotoxicity screening of many of the snake venoms tested and further illustrates the utility of snake venoms in biomedical research.
Acknowledgments
Funding for this project was provided by a grant (to SPM) from the Colorado Office of Economic Development and Trade (COEDIT), Bioscience Discovery Evaluation Grant Program. Additional funding was provided by the UNC Office of Sponsored Programs. We thank Peter J. Mirtschin of Venom Supplies Pty. Ltd. for providing P. porphyriacus venom, Dr. Charlotte Ownby of Oklahoma State University for numerous elapid and viperid venoms, and A. Ah-Young, B. Heyborne, J. LeRoy Waite, and A. Wastell for assistance with venom extractions (rattlesnakes and colubrids).
Conflict of interest
The authors state that there are no conflicts of interest.
Ethics Standard
All vertebrate animal manipulations (venom extractions of snakes) were in accordance with protocols approved by the UNC IACUC.
References
- Alape-Girón A, Sanz L, Escolano J, Flores-Díaz M, Madrigal M, Sasa M, Calvete JJ. Snake venomics of the lancehead pitviper Bothrops asper: geographic, individual, and ontogenetic variations. J Proteome Res. 2008;7:3556–3571. doi: 10.1021/pr800332p. [DOI] [PubMed] [Google Scholar]
- Anderson LA, Dufton MJ. Acetylcholinesterases. In: Bailey GS, editor. Enzymes from snake venoms. Ft. Collins: Alaken; 1998. pp. 545–578. [Google Scholar]
- Barlow A, Pook CE, Harrison RA, Wüster W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc Biol Sci. 2009;276:2443–2449. doi: 10.1098/rspb.2009.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Staniszewska I, Valle LD, Tuszynski GP, Marcinkiewicz C. Angiostatic activity of obtustatin as alpha1beta1 integrin inhibitor in experimental melanoma growth. Int J Cancer. 2008;123:2195–2203. doi: 10.1002/ijc.23777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete JJ. Snake venomics, antivenomics, and venom phenotyping: the ménage à trois of proteomic tools aimed at understanding the biodiversity of venoms. In: Kini RM, Clemetson KJ, Markland FS, McLane MA, Morita T, editors. Toxins and hemostasis: from bench to bedside. Dordrecht: Springer; 2010. pp. 45–72. [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Monleon D, Esteve V, Celda B, Juarez P, Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Sanz L. Snake venomics of Bitis gabonica gabonica. Protein family composition, subunit organization of venom toxins, and characterization of dimeric disintegrins bitisgabonin-1 and bitisgabonin-2. J Proteome Res. 2006;6:326–336. doi: 10.1021/pr060494k. [DOI] [PubMed] [Google Scholar]
- Chaim-Matyas A, Ovadia M. Cytotoxic activity of various snake venoms on melanoma, B16F10 and chondrosarcoma. Life Sci. 1987;40:1601–1607. doi: 10.1016/0024-3205(87)90126-3. [DOI] [PubMed] [Google Scholar]
- Doley R, Pahari S, Mackessy SP, Kini RM. Accelerated exchange of exon segments in viperid three-finger toxin genes (Sistrurus catenatus edwardsii; Desert Massasauga) BMC Evol Biol. 2008;8:196. doi: 10.1186/1471-2148-8-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doley R, Zhou X, Kini RM. Snake venom phospholipase A2 enzymes. In: Mackessy SP, editor. Handbook of venoms and toxins of reptiles. Boca Raton: Taylor and Francis/CRC Press; 2010. pp. 173–206. [Google Scholar]
- Earl STH, Masci PP, Jersey JD, Lavin MF, Dixon J. Drug development from Australian elapid snake venoms and the Venomics pipeline of candidates for haemostasis: Textilinin-1 (Q8008), Haempatch™ (Q8009) and CoVase™ (V0801) Toxicon. 2012;59:456–463. doi: 10.1016/j.toxicon.2010.12.010. [DOI] [PubMed] [Google Scholar]
- El-Refael MF, Sarkar NH. Snake venom inhibits the growth of mouse mammary tumor cells in vitro and in vivo. Toxicon. 2009;54:33–41. doi: 10.1016/j.toxicon.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Ferreira SH. A bradykinin-potentiating factor (BPF) present in the venom of Bothrops jararaca. Br J Pharmacol Chemother. 1965;24:163–169. doi: 10.1111/j.1476-5381.1965.tb02091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira SH, Bartelt DC, Greene LJ. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry. 1970;9:2583–2593. doi: 10.1021/bi00815a005. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SM. Approaching the golden age of natural product pharmaceuticals from venom libraries: an overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr Pharm Des. 2007;13:2927–2934. doi: 10.2174/138161207782023739. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SMT. Snake venom metalloproteinases. In: Mackessy SP, editor. Handbook of venoms and toxins of reptiles. Boca Raton: Taylor and Francis/CRC Press; 2010. pp. 115–138. [Google Scholar]
- Fruchart-Gaillard C, Mourier G, Blanchet G, Vera L, Gilles N, Ménez R, Marcon E, Stura EA, Servent D. Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors. PLoS One. 2012;7:39166. doi: 10.1371/journal.pone.0039166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier JA, Kearney M, Maisano JA, Rieppel O, Behlke ADB. Assembling the squamate tree of life: perspectives from the phenotype and the fossil record. Bull Peabody Mus Nat Hist. 2012;53:3–308. doi: 10.3374/014.053.0101. [DOI] [Google Scholar]
- Gibbs HL, Mackessy SP. Functional basis of a molecular adaptation: prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon. 2009;53:672–679. doi: 10.1016/j.toxicon.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Gutiérrez JM, Lomonte B, León G, Alape-Girón A, Flores-Díaz M, Sanz L, Angulo Y, Calvete JJ. Snake venomics and antivenomics: proteomic tools in the design and control of antivenoms for the treatment of snakebite envenoming. J Proteomics. 2009;72:165–182. doi: 10.1016/j.jprot.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Heyborne WH, Mackessy SP. Isolation and characterization of a taxon-specific three-finger toxin from the venom of the Green Vinesnake (Oxybelis fulgidus; family Colubridae) Biochimie. 2013;95:1923–1932. doi: 10.1016/j.biochi.2013.06.025. [DOI] [PubMed] [Google Scholar]
- Hill RE, Mackessy SP. Venom yields from several species of colubrid snakes and differential effects of ketamine. Toxicon. 1997;35:671–678. doi: 10.1016/S0041-0101(96)00174-2. [DOI] [PubMed] [Google Scholar]
- Huang P, Mackessy SP. Biochemical characterization of phospholipase A2 (trimorphin) from the venom of the Sonoran Lyre Snake Trimorphodon biscutatus lambda (family Colubridae) Toxicon. 2004;44:27–36. doi: 10.1016/j.toxicon.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Huguet EL, McMahon JA, McMahon AP, Bicknell R, Harris AL. Differential expression of human Wnt genes 2, 3, 4, and 7B in human breast cell lines and normal and disease states of human breast tissue. Cancer Res. 1994;54:2615–2621. [PubMed] [Google Scholar]
- Kamiguti A, Zuzel M, Theakston R. Snake venom metalloproteinases and disintegrins: interactions with cells. Braz J Med Biol Res. 1998;31:853–862. doi: 10.1590/S0100-879X1998000700001. [DOI] [PubMed] [Google Scholar]
- Kereiakes DJ, Kleiman NS, Ambrose J, Cohen M, Rodriguez S, Palabrica T, Herrmann TC, Sutton JM, Weaver WD, McKee DB, Fitzpatrick V, Sax FL, Higby N, Ratner D, Slatylak S, DeAngelo, D, Trainor K, Rose D, Johnson S, Miele R, Cowfer J, Martin J (1996) Randomized, double-blind, placebo-controlled dose-ranging study of tirofiban (MK-383) platelet IIb/IIIa blockade in high risk patients undergoing coronary angioplasty. J Am Coll Cardiol 27:536–542 [DOI] [PubMed]
- Kini RM. Molecular molds with multiple missions: functional sites in three-finger toxins. Clin Exp Pharmacol Physiol. 2002;29:815–822. doi: 10.1046/j.1440-1681.2002.03725.x. [DOI] [PubMed] [Google Scholar]
- Kini RM. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Kini RM, Doley R. Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon. 2010;56(2010):855–867. doi: 10.1016/j.toxicon.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Koh CY, Kini RM. From snake venom toxins to therapeutics–cardiovascular examples. Toxicon. 2012;59:497–506. doi: 10.1016/j.toxicon.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Lin E, Wang Q, Swenson S, Jadvar H, Groshen S, Ye W, Markland FS, Pinski J. The disintegrin contortrostatin in combination with docetaxel is a potent inhibitor of prostate cancer in vitro and in vivo. Prostate. 2010;70:1359–1370. doi: 10.1002/pros.21173. [DOI] [PubMed] [Google Scholar]
- Lomonte B, Tsai WC, Ureña-Diaz JM, Sanz L, Mora-Obando D, Sánchez EE, Fry BG, Gutiérrez JM, Gibbs HL, Sovic MG, Calvete JJ. Venomics of New World pit vipers: genus-wide comparisons of venom proteomes across Agkistrodon. J Proteomics. 2014;96:103–116. doi: 10.1016/j.jprot.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena S, Sanchez EE, Perez JC. Anti-metastatic activity of the recombinant disintegrin, r-mojastin 1, from the Mohave rattlesnake. Toxicon. 2011;57:794–802. doi: 10.1016/j.toxicon.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackessy SP. Venom ontogeny in the Pacific rattlesnakes Crotalus viridis helleri and C. v. oreganus. Copeia. 1988;1988:92–101. doi: 10.2307/1445927. [DOI] [Google Scholar]
- Mackessy SP. Fibrinogenolytic proteases from the venoms of juvenile and adult northern Pacific rattlesnake (Crotalus viridis oreganus) Comp Biochem Physiol. 1993;106B:181–189. doi: 10.1016/0305-0491(93)90025-z. [DOI] [PubMed] [Google Scholar]
- Mackessy SP. Phosphodiesterases, ribonucleases and deoxyribonucleases. In: Bailey GS, editor. Enzymes from snake venoms. Ft. Collins: Alaken; 1998. pp. 361–404. [Google Scholar]
- Mackessy SP. Biochemistry and pharmacology of colubrid snake venoms. J Toxicol Toxin Rev. 2002;21:43–83. doi: 10.1081/TXR-120004741. [DOI] [Google Scholar]
- Mackessy SP. Venom composition in rattlesnakes: trends and biological significance. In: Hayes WK, Cardwell MD, Beaman KR, Bush SP, editors. The biology of rattlesnakes. Loma Linda: Loma Linda University Press; 2008. pp. 495–510. [Google Scholar]
- Mackessy SP. The field of reptile toxinology. Snakes, lizards, and their venoms. In: Mackessy SP, editor. Handbook of venoms and toxins of reptiles. Boca Raton: Taylor and Francis/CRC Press; 2010. pp. 3–24. [Google Scholar]
- Mackessy SP. The evolution of venom composition in the Western Rattlesnakes (Crotalus viridis sensu lato): toxicity versus tenderizers. Toxicon. 2010;55:1463–1474. doi: 10.1016/j.toxicon.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Mackessy SP, Williams K, Ashton K. Characterization of the venom of the midget faded rattlesnake (Crotalus viridis concolor): a case of venom paedomorphosis? Copeia. 2003;2003:769–782. doi: 10.1643/HA03-037.1. [DOI] [Google Scholar]
- Mackessy SP, Sixberry NM, Heyborne WH, Fritts T. Venom of the Brown Treesnake, Boiga irregularis: ontogenetic shifts and taxa-specific toxicity. Toxicon. 2006;47:537–548. doi: 10.1016/j.toxicon.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Masuda S, Hayashi H, Araki S. Two vascular apoptosis-inducing proteins from snake venom are members of the metalloprotease/disintegrin family. Eur J Biochem. 1998;253:36–41. doi: 10.1046/j.1432-1327.1998.2530036.x. [DOI] [PubMed] [Google Scholar]
- Masuda S, Ohta T, Kaji K, Fox JW, Hayashi H, Araki S. cDNA cloning and characterization of vascular apoptosis-inducing protein 1. Biochem Biophys Res Commun. 2000;278:197–204. doi: 10.1006/bbrc.2000.3770. [DOI] [PubMed] [Google Scholar]
- Masuda S, Hayashi H, Atoda H, Morita T, Araki S. Purification, cDNA cloning and characterization of the vascular apoptosis-inducing protein, HV1, from Trimeresurus flavoviridis. Eur J Biochem. 2001;268:3339–3345. doi: 10.1046/j.1432-1327.2001.02246.x. [DOI] [PubMed] [Google Scholar]
- McLane MA, Joerger T, Mahmoud A. Disintegrins in health and disease. Front Biosci. 2008;13:6617–6637. doi: 10.2741/3177. [DOI] [PubMed] [Google Scholar]
- Minton SA, Weinstein SA. Geographic and ontogenic variation in venom of the western diamondback rattlesnake Crotalus atrox. Toxicon. 1986;24:71–80. doi: 10.1016/0041-0101(86)90167-4. [DOI] [PubMed] [Google Scholar]
- Mirtschin PJ, Crowe GR, Davis R. Dangerous snakes of Australia. In: Gopalakrishnakone P, Chou LM, editors. Snakes of medical importance (Asia-Pacific Region) Singapore: Venom and Toxin Research Group, NUS; 1990. pp. 1–174. [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Mukherjee AK. A major phospholipase A2 from Daboia russelii russelii venom shows potent anticoagulant action via thrombin inhibition and binding with plasma phospholipids. Biochimie. 2014;99:153–161. doi: 10.1016/j.biochi.2013.11.026. [DOI] [PubMed] [Google Scholar]
- Mukherjee AK, Mackessy SP. Biochemical and pharmacological properties of a new thrombin-like serine protease (Russelobin) from the venom of Russell’s Viper Daboia russelii russelii and assessment of its therapeutic potential. BBA Gen Subj. 2013;1830:3476–3488. doi: 10.1016/j.bbagen.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Nirthanan S, Gwee MCE. Three-finger neurotoxins and the nicotinic acetylcholine receptor, forty years on. J Pharmacol Sci. 2004;94:1–17. doi: 10.1254/jphs.94.1. [DOI] [PubMed] [Google Scholar]
- Núñez V, Cid P, Sanz L, De La Torre P, Angulo Y, Lomonte B, Gutiérrez JM, Calvete JJ. Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J Proteomics. 2009;73:57–78. doi: 10.1016/j.jprot.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Öhler M, Georgieva D, Seifert J, von Bergen M, Arni RK, Genov N, Betzel C. The venomics of Bothropsalternatus is a pool of acidic proteins with predominant hemorrhagic and coagulopathic activities. J Proteome Res. 2010;9:2422–2437. doi: 10.1021/pr901128x. [DOI] [PubMed] [Google Scholar]
- Oron U, Chaim-Matyas A, Ovadia M. Histopathological changes in WEHI-3B leukemia cells following intoxication by cytotoxin P4 from Naja nigricollis nigricollis venom. Toxicon. 1992;30:1122–1126. doi: 10.1016/0041-0101(92)90058-D. [DOI] [PubMed] [Google Scholar]
- Pal SK, Gomes A, Dasgupta SC, Gomes A. Snake venom as therapeutic agents: from toxin to drug development. Indian J Exp Biol. 2002;40:1353–1358. [PubMed] [Google Scholar]
- Pawlak J, Mackessy SP, Fry BG, Bhatia M, Mourier G, Fruchart-Gaillard C, Servent D, Ménez R, Stura E, Ménez A, Kini RM. Denmotoxin: a three-finger toxin from colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity. J Biol Chem. 2006;281:29030–29041. doi: 10.1074/jbc.M605850200. [DOI] [PubMed] [Google Scholar]
- Pawlak J, Mackessy SP, Sixberry NM, Stura EA, Le Du MH, Ménez R, Foo CS, Ménez A, Nirthanan S, Kini RM. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 2009;23:534–545. doi: 10.1096/fj.08-113555. [DOI] [PubMed] [Google Scholar]
- Pyron RA, Burbrink FT, Wiens JJ. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 2013;13:93. doi: 10.1186/1471-2148-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samel M, Trummal K, Siigur E, Siigur J. Effect of HUVEC apoptosis inducing proteinase from Vipera lebetina venom (VLAIP) on viability of cancer cells and on platelet aggregation. Toxicon. 2012;60:648–655. doi: 10.1016/j.toxicon.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Sánchez EE, Rodríguez-Acosta A, Palomar R, Lucena SE, Bashir S, Soto JG, Pérez JC. Colombistatin: a disintegrin isolated from the venom of the South American snake (Bothrops colombiensis) that effectively inhibits platelet aggregation and SK-Mel-28 cell adhesion. Arch Toxicol. 2009;83:271–279. doi: 10.1007/s00204-008-0358-y. [DOI] [PubMed] [Google Scholar]
- Saviola AJ, Chiszar D, Busch C, Mackessy SP. Molecular basis for prey relocation in viperid snakes. BMC Biol. 2013;11:20. doi: 10.1186/1741-7007-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviola AJ, Peichoto ME, Mackessy SP (2014) Rear-fanged snake venoms: an untapped source of novel compounds and potential drug leads. Toxin Rev. doi: 10.3109/15569543.2014.942040. (in press)
- Sharma SD, Jiang J, Hadley ME, Bentley DL, Hruby VJ. Melanotropic peptide-conjugated beads for microscopic visualization and characterization of melanoma melanotropinreceptors. Proc Natl Acad Sci. 1996;93:13715–13720. doi: 10.1073/pnas.93.24.13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Pierre L, Fischer H, Adams DJ, Schenning M, Lavidis N, de Jersey J, Masci PP, Lavin MF. Distinct activities of novel neurotoxins from Australian venomous snakes for nicotinic acetylcholine receptors. Cell Mol Life Sci. 2007;64:2829–2840. doi: 10.1007/s00018-007-7352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson S, Costa F, Ernst W, Fujii G, Markland F. Contortrastatin, a snake venom disintegrin with anti-angiogenic and anti-tumor activity. Pathophysiol Haemost Thromb. 2005;34:169–176. doi: 10.1159/000092418. [DOI] [PubMed] [Google Scholar]
- Takacs Z, Nathan S. Animal venoms in medicine. In: Wexler P, editor. Encyclopedia of toxicology. Third. Amsterdam: Elsevier; 2014. pp. 252–529. [Google Scholar]
- Takahashi K, Suzuki K. Association of insulin-like growth-factor-I-induced DNA synthesis with phosphorylation and nuclear exclusion of p53 in human breast cancer MCF-7 cells. Int J Cancer. 1993;55:453–458. doi: 10.1002/ijc.2910550322. [DOI] [PubMed] [Google Scholar]
- Tian J, Paquette-Straub C, Sage EH, Funk SE, Patel V, Galileo D, McLane MA. Inhibition of melanoma cell motility by the snake venom disintegrin eristostatin. Toxicon. 2007;49:899–908. doi: 10.1016/j.toxicon.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink S, Jin AH, Poth KJ, Head GA, Alewood PF. Natriuretic peptide drug leads from snake venom. Toxicon. 2012;59:434–445. doi: 10.1016/j.toxicon.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Vonk FJ, Jackson K, Doley R, Madaras F, Mirtschin PJ, Vidal N. Snake venom: from fieldwork to the clinic. BioEssays. 2011;33:269–279. doi: 10.1002/bies.201000117. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Hutter CR, Mulcahy DG, Noonan BP, Townsend TM, Sites JW, Jr, Reeder TW. Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species. Biol Lett. 2012;8:1043–1046. doi: 10.1098/rsbl.2012.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcın HT, Ozen MO, Gocmen B, Nalbantsoy A. Effect of Ottoman viper (Montivipera xanthina (Gray, 1849)) venom on various cancer cells and on microorganisms. Cytotechnology. 2014;66:87–94. doi: 10.1007/s10616-013-9540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]