Abstract
Both environmental agents and spontaneous cellular events cause serious DNA damage, threatening the integrity of the genome. In response to replication stress or genotoxic agents triggered DNA damage, degradation of p12 subunit of DNA polymerase delta (Pol δ) results in an inter-conversion between heterotetramer (Pol δ4) and heterotrimer (Pol δ3) forms and plays a significant role in DNA damage response in eukaryotic cells. In this work, we used mass spectrometry-based proteomic approach to identify those cellular stress response protein changes corresponding to the degradation of p12 in DNA-damaged HeLa cells by the treatment with hydroxyurea (HU). A total of 736 ± 13 proteins in non-treated control group and 741 ± 19 protein spots in HU-treated cells were detected, of which 34 proteins (17 up-regulated and 17 down-regulated) exhibited significantly altered protein expression levels. Their physiological roles are mainly associated with cellular components, molecular functions, and biological processes by gene ontology analysis, among which 21 proteins were mapped to KEGG pathways. They are involved in 5 primary pathways with the subsets involving 16 secondary pathways by further KEGG analysis. More interestingly, the up-regulation of translationally controlled tumor protein was further identified to be associated with p12 degradation by Western blot analysis. Our works may enlarge and broaden our view for deeply understanding how global cellular stress responds to DNA damage, which could contribute to the etiology of human cancer or other diseases that can result from loss of genomic stability.
Keywords: Hydroxyurea treatment, DNA damage response, Two-dimensional gel electrophoresis, Mass spectrometry, Down-regulation of p12 subunit, Up-regulation of TCTP
Introduction
Cell and tissue culture has become a core technology in the modern life sciences in recent years, offering a possibility for genetic diagnosis and therapy as well as tissue engineering. In addition to the study of the cellular homeostasis, cell culture has also provided the basis for investigating the regulation of these processes from genetic level to individual protein molecules. The culture conditions are crucial for favoring cell adhesion, proliferation, and differentiation. Apart from the nutrient limitation, a primary cause of cell death during the stationary and death phase of the growth (Mercille and Massie 1994), a particularly important aspect is the accumulation of toxic waste products both from endogenous and from exogenous sources over time, which leads to the induction of DNA damage, formation of DNA lesions, and then premature apoptotic cell death (Al-Rubeai and Singh 1998; Kaina 2003; Roos and Kaina 2006).
There are estimated to be more than 10,000 of DNA lesions every day from endogenous sources alone (Derks et al. 2014). However, in most cases, DNA damage arises from exogenous sources, such as ultraviolet (UV) light from the sun, ionizing radiation, and numerous environmental chemicals (Roos and Kaina 2006). If these lesions cannot be repaired in time or damaged DNAs are incorrectly repaired, it could lead to serious consequences. Accumulation of unrepaired DNA damage or incorrect repair significantly contributes to the etiology of human cancer or other diseases that can result from the loss of genomic stability (Hoeijmakers 2009).
In response to adverse effects of DNA damage, cells have an arsenal of defense mechanisms, the DNA damage response (DDR). It involves the recruitment and assembly of large complexes of proteins that orchestrate and prioritize a network of responses, which includes DNA repair, activation of cell cycle checkpoints and the decision for cell death (Branzei and Foiani 2008; Derks et al. 2014). Upon DNA damage, the activation of cell cycle checkpoints halts cell proliferation, providing a time to repair damaged DNAs. When damages are beyond repair, cell death or cellular senescence is induced to remove damaged cells from the tissue, avoiding mutations and cancer. Therefore, tightly regulated DDR is of utmost importance. It delicately balances incorrect repair and hyper-activation that maximizes survival and decides on cell fate. Incorrect repair drives carcinogenesis while hyper-activation could induce cell death pathway or senescence.
The current view of DDR is in a complex way by evoking cellular processes that might ultimately lead to DNA repair, damage fixation as mutations or damage elimination by various routes of cell death (Fritz and Kaina 2006; Kaina 2003). As reviewed in Derks et al. (2014), DNA lesions firstly are detected by a class of sensor proteins that then recruit various factors to the damage site, e.g. repair factors. The sensors also transmit a signal to the transducers, the most prominent proteins as ATM and ATR checkpoint kinases. In turn, these transducers amplify the damage signal to the effectors, e.g. p53 or microRNAs, which control the activity of several cellular processes and pathways, such as cell cycle arrest and apoptosis. The sensor and transducer signaling in DDR largely rely on the protein–protein interactions and alterations in protein activity by protein post-translational modifications. Current studies on the transcriptomics, a genome-wide RNA transcript expression level, and proteomics have tremendously expanded our knowledge on the DDR (Daub 2012; Jin et al. 2004). They presented that hundreds of additional proteins are targets of checkpoint kinases and more than a thousand genes are differentially expressed upon DNA damage as a result of transcription factor/microRNA regulation (Derks et al. 2014).
Mammalian DNA polymerase δ (Pol δ), a four-subunit complex enzyme consisting of p125, p50, p68, and p12, plays a crucial and versatile role in eukaryotic DNA replication and is also involved in various DNA repair processes and genetic recombination (Branzei and Foiani 2008; Meng et al. 2010; Rahmeh et al. 2012; Sancar et al. 2004; Zhou et al. 2011, 2012). In the previous studies, we had observed that Pol δ is also regulated in the DDR as reviewed by Lee et al. (2012). The Pol δ smallest subunit p12 is degraded in response to DNA damage that is induced by UV irradiation, alkylating agents, or replication stresses (Chea et al. 2012; Meng et al. 2009; Zhang et al. 2007, 2013). There arises a question of how the sensor and transducer signaling in DDR to be regulated in coordination with the p12 degradation upon DNA damage. In this work, we identified and analyzed protein changes in HU-treated HeLa cells by two-dimensional electrophoresis (2-DE) in combination with mass spectrometric (MS). Comparing with 736 ± 13 proteins in non-treated control group, a total of 741 ± 19 proteins in HU-treated cells were detected, of which 34 proteins exhibited significantly altered protein expression levels. Among them, 17 proteins were up-regulated while 17 proteins were down-regulated. More interestingly, the up-regulation of translationally controlled tumor protein (TCTP) was further identified to be associated with p12 down-regulation by Western blot analysis.
This work provides some useful information for better understanding the involvement of polymerase δ regulation in the network of global cellular stress responses to chemical carcinogens on protein level in DDR, possibly contributing to the etiology of human cancer or other diseases that can result from loss of genomic stability.
Materials and methods
Cell culture and treatments
HeLa cells, purchased from American Type Culture Collection (ATCC, Manassas, VA, USA), were maintained in Dulbecco's modified Eagle’s medium (DMEM, Gibco-Invitrogen, Grand Island, NY, USA) with 10 % fetal bovine serum (FBS, Gibco-Invitrogen, Grand Island, NY, USA) at 37 °C as previously described (Fan et al. 2014; Zhang et al. 2007). Cells were grown in 100-mm culture dishes to about 70 % confluence, treated with genotoxic agents, 4 mM of HU, 150 nM of aphidicolin (APH), or 1.5 mM of methyl methanesulfonate (MMS), respectively, purchased from Sigma-Aldrich, St. Louis, MO, USA, and then analyzed at different time periods of incubation, as previously described (Fan et al. 2014; Zhang et al. 2007). In parallel, non-treated cells were used as the control. For the validation of the up-regulation of TCTP, the p12 degradation was blocked by the addition of proteasome inhibitor MG132 (carbobenzoxy-l-leucyl-l-leucyl-l-leucinal) at a final concentration of 10 μM to the culture medium 30 min prior to the HU treatment.
Western blot analysis and antibodies
Total cell lysates of treated HeLa cells were analyzed, using non-treated cells as the control, by Western blot as described (Fan et al. 2014). Anti-phospho-Chk1 (Ser345) rabbit polyclonal antibody was purchased from Cell Signaling Technology (Beverly, MA, USA). Mouse monoclonal antibody anti-PCNA PC-10 and anti-translationally controlled tumor protein (TCTP), HRF (B-3), were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibody against β-actin was obtained from Beyotime Institute of Biotechnology (Haimen, China). The antibodies for Pol δ subunits were: mouse polyclonal antibody against p50 (ZJM5002), rabbit polyclonal antibodies against p125 (ZJR12501), p68 (ZJR6803), and p12 (ZJR1204), prepared in our laboratory, respectively, as described previously (Fan et al. 2014; Zhou et al. 2011).
Protein sample preparation
The protein samples were prepared as previously described (Yang et al. 2013) and made some modifications. The HeLa cells from 20 culture plates (100-mm) were harvested after treatment with 4 mM HU for 8 h and washed three times with phosphate buffered saline solution. The cell pellet was re-suspended in extraction buffer (7 M urea, 2 M thiourea, 4 % Chaps, 2 % Bio-Lyte pH 3–10, 1 mM PMSF and 1 % DTT (Bio-Rad, Hercules, CA, USA)) on ice for 30 min, followed by sonication using 4 × 15 s bursts with a 15 s cooling period between each burst and centrifugation at 12,500 rpm for 60 min. The supernatant was collected and proteins were precipitated for 2 h on ice by adding cold trichloroacetic acid (TCA) to a final concentration of 10 %. After centrifugation at 12,500 rpm for 20 min at 4 °C, the pellet was washed four times using cold acetone containing 13 mM DTT, followed by a further centrifugation (12,500 rpm, 4 °C, 20 min). The pellet was lyophilized and dissolved in sample buffer (7 M urea, 2 M thiourea, 4 % Chaps, 0.5 % Bio-Lyte pH 3–10, 1 mM PMSF and 1 % DTT) on ice overnight. Following a final centrifugation (12,500 rpm, 4 °C, 20 min), the supernatant was used for 2-DE. In parallel, non-treated cells from 20 culture plates (100-mm) were taken as a control experiment. The total protein concentration was measured with the Bradford assay (Bradford 1976).
Two-dimensional electrophoresis (2-DE)
The 2-DE was performed using a Bio-Rad PROTEAN electrophoresis system (Bio-Rad) using a protocol described in our previously studies with some modifications (Qin et al. 2012; Yang et al. 2013). About 1.5 mg protein samples were loaded on isoelectric focusing (IEF) dry gel strips (17 cm, non-linear pH 3–10) during the rehydration step (passive rehydration, room temperature, 12–13 h) and IEF was performed at 20 °C with a voltage gradient of 100 V for 1.5 h, 250 V for 1.5 h, 500 V for 1 h, 1,000 V for 1 h, 10,000 V for 5 h, and then continued for a total of 60 kVh. Subsequently, the strips were equilibrated for 15 min with an equilibration buffer (6 M urea, 0.375 M Tris–HCl pH 8.8, 20 % glycerol, 2 % SDS) containing 2 % DTT and then for another 15 min with equilibration buffer containing 2.5 % iodoacetamide (Bio-Rad, USA). After equilibration steps, the second dimension SDS-PAGE was performed on 12 % polyacrylamide gels. Separated proteins on the gels were detected by staining with 0.116 % Coomassie brilliant blue (CBB, Bio-Rad, USA)) G-250, visualized and scanned on a high precision scanner (ScanMaker 9700XL, Microtek (Hsinchu, Taiwan)) at a resolution of 600 dpi, analyzed with PDQuest software (Bio-Rad, USA) and numbered with Arabic numerals. Triplicate experiments were performed for each pair of tests, HU-treatment and non-treatment. The intensity ratio of the corresponding spots in a pair of gels was calculated. The spots with a ratio of ≥2 or ≤0.5 were defined as quantitative different spots.
In-gel digestion and mass spectrometry (MS) analysis
In-gel digestion and MS analysis were performed as a procedure described previously (Yang et al. 2013) by excising protein spots from the gels, washing with water, distaining and dehydrating with ammonium bicarbonate and acetonitrile, lyophilizing in vacuum. The dried gel spots were rehydrated in 2 μl of digestion buffer containing 25 mM NH4HCO3 and 10 ng of trypsin/μl (Promega, Madison, WI, USA) at 4 °C for 30 min. The digestion was continued by adding 10–15 μl of 25 mM NH4HCO3 at 37 °C overnight. The digested peptides were collected and analyzed on a MALDI-TOF mass spectrometer (Ultraflex-TOF-TOF, Bruker, Bremen, Germany).
Protein identification and bioinformatics analysis of differentially expressed proteins
All of the obtained peptide mass fingerprint data were analyzed using MASCOT (Matrix Science, London, UK) and NCBInr eukaryotic protein sequence database. The parameter was set as follows: missed cleavage was 1; fixed modification was carbamidomethyl (C); variable modification was Glu → pyro-Glu (N-term Q) or oxidation of methionine (M); mass tolerance was ±0.2 Da; mass value was MH+. The protein with a minimum ion score of 83 (P < 0.05) was considered to be reliably identified.
The gene ontology (GO) analysis was performed as described (Ye et al. 2006). The sequences of identified proteins were queried against GO Database (OBO v1.2 format: http://www.geneontology.org/GO.downloads.ontology.shtml) to obtain the GO plot.
The KEGG pathway analysis was carried out as previously described (Arakelyan and Nersisyan 2013; Kanehisa and Goto 2000; Qin et al. 2012). The sequences of identified proteins were queried against KEGG GENES (http://blast.genome.jp/) using BLASTP program with BLOUSM62 scoring matrix. The resultant genes were used to search KEGG reference pathway database (http://www.genome.jp/kegg/tool/search_pathway.html) to obtain reference pathways for those proteins.
Results
Optimal sample conditions corresponding to the p12 subunit degradation
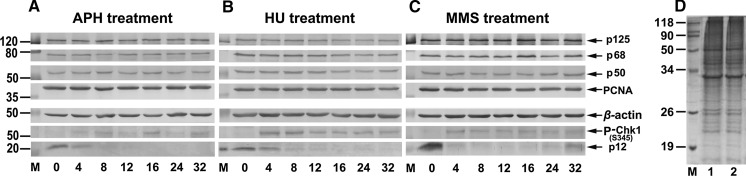
Based on our previous observation that p12 subunit of Pol δ was depleted in cultured human cells by the treatment of alkylating agents or by the induction of replication stress, we re-examined p12 degradation in cultured HeLa cells by treatment with APH, HU, and MMS, respectively, which are known to trigger the intra-S-phase checkpoint via ATR/Chk1 activation. As shown in Fig. 1a, 150 nM of APH treatment caused specific depletion of the p12 subunit in a dose-dependent manner with concomitant phosphorylation of Chk1-S345. While the other three subunits of Pol δ and PCNA were relatively unaffected. The depletion of p12 was reached by 4 h and the p12 levels were depleted below detection levels by 8 h. Similarly, p12 degradation was observed by 4 h, but slightly slower rate of complete degradation of p12, by 16 h, with much more clearly concomitant phosphorylation of Chk1-S345, after treatment with 4 mM of HU (Fig. 1b). For MMS treatment, a concentration of 1.5 mM would rapidly lead to the disappearance of p12 by 4 h, a nearly complete degradation, and p12 levels were recovered after 24 h (Fig. 1c).
Fig. 1.
Degradation of p12 subunit by the treatments with genotoxic agents. a–c Optimal sample conditions corresponding to the p12 degradation. The HeLa cells were treated with 150 nM of aphidicolin (a APH treatment), 4 mM of hydroxyurea (b HU treatment), and 1.5 mM of methyl methanesulfonate (c MMS treatment) and then analyzed by Western blot at indicated time, respectively. The numbers shown on the left indicate the positions in kDa protein markers (M). The four subunits of Pol δ (p125, p68, p50, and p12), PCNA, β-actin (as a loading control), and P-Chk1-S345 (as an indicator for ATR/Chk1 signaling activation) are indicated by arrows. The horizontal axis shows the time after individual treatment. d Comparison of protein sample concentrations for 2-DE. The samples at 8 h after treatment with 4 mM of HU were analyzed by “in-gel” determination of extracted protein concentration. Fifty micrograms of total protein each lane calculated from Bradford assay was loaded. 1 Non-treatment as a control. 2 HU treated sample. M protein markers in kDa
For the best sample conditions used for 2-DE, which should be a clear down-regulation of p12 but not be completely depleted with a clear intra-S-phase checkpoint activation, therefore, we chose 4 mM of HU treatment in sampling at 8 h after treatment according to the comparisons among different treatments shown in Fig. 1. For sample preparation, the harvested cells were lysed and total protein was extracted as described in Materials and Methods. To further confirm the accuracy of protein concentration measured by the Bradford assay, we used “in-gel” determination of extracted protein concentration. Fifty micrograms of total protein per lane was loaded according to the calculation of measured concentration by the Bradford assay (Fig. 1d, column 1: control at 14.6 mg/ml, column 2: HU-treatment at 16.8 mg/ml). As expected, the prepared sample could be suitably used for 2-DE experiments in appropriate condition.
Global identification of differentially expressed proteins
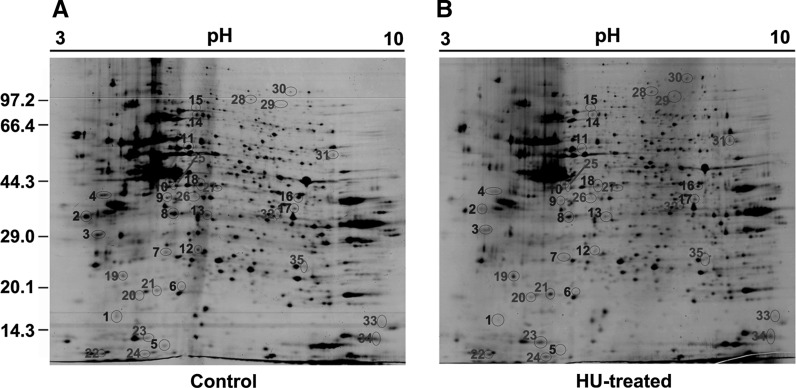
To obtain better resolution and sensitivity, we did a pre-test by loading 500 μg of proteins on a smaller strip (7 cm, linear pH 3–10). Most molecular mass varied within a normal range of approximately 10–120 kDa, suggesting that the protein exaction was properly made. However, there were slightly crowded proteins around the pH 5–7 (data not shown). Therefore, in the present study, the 2-DE was performed using a 17 cm of IEF strip with non-linear pH 3–10 by loading 1.5 mg of protein samples. The gels were analyzed by PDQuest software and numbered with Arabic numerals as shown in Fig. 2. A total of 741 ± 19 protein spots were observed on 2-DE gel for HU-treated cells (Fig. 2b) and 736 ± 13 protein spots for non-treated control (Fig. 2a). Thirty-five different spots were found and analyzed by MS analysis. The down-regulated spots were numbered as 1–18 in blue while up-regulated spots were numbered as 19–35 in red shown in Fig. 2. The results are summarized in Table 1. The spot 2 and 3 are identified as the same protein. It may result from the protein post-translational modifications, such as ubiquitination or phosphorylation, which led to a slight shift of molecular mass on the gels.
Fig. 2.
2-DE of extracted proteins. 2-DE for the extracted proteins from control (a) and HU-treated group (b) were performed as described in “Materials and methods”. The differentially expressed protein spots are indicated by circles and labeled with Arabic numerals in blue for down-regulated proteins and in red for those of up-regulated. The numbers shown on the left indicate the protein markers in kDa
Table 1.
List of the differentially expressed protein spots identified by MALDI-TOF MS
| Spot No. | NCBI accession | Protein name | pIa | MWb | AAc | Seq Cov (%)d | Ratioe Treated:C |
|---|---|---|---|---|---|---|---|
| 1 | gi|7019485 | Programmed cell death protein 6 | 5.16 | 21,912 | 191 | 35 | 0.13 ± 0.010 |
| 2 | gi|338043 | SF2p32 | 4.74 | 31,287 | 278 | 10 | 0.25 ± 0.018 |
| 3 | gi|338043 | SF2p32 | 4.74 | 31,287 | 278 | 10 | 0.48 ± 0.051 |
| 4 | gi|75070716 | Heterogeneous nuclear ribonucleoprotein C | 4.95 | 33,670 | 306 | 2 | 0.33 ± 0.021 |
| 5 | gi|263657 | Cytidine deaminase | 5.84 | 16,556 | 146 | 35 | 0.20 ± 0.018 |
| 6 | gi|4929697 | CGI-114 protein | 5.60 | 23,967 | 205 | 7 | 0.47 ± 0.047 |
| 7 | gi|5453790 | Nicotinamide N-methyltransferase | 5.56 | 30,011 | 264 | 12 | 0.08 ± 0.004 |
| 8 | gi|4506667 | 60S acidic ribosomal protein P0 | 5.71 | 34,423 | 317 | 5 | 0.24 ± 0.013 |
| 9 | gi|3646128 | Thioredoxin-like protein | 5.25 | 37,751 | 335 | 8 | 0.32 ± 0.028 |
| 10 | gi|61212932 | ATP-dependent RNA helicase DDX39A | 5.46 | 49,129 | 427 | 10 | 0.27 ± 0.0211 |
| 11 | gi|48762932 | T-complex protein 1 subunit theta | 5.78 | 54,207 | 497 | 9 | 0.19 ± 0.009 |
| 12 | gi|1008915 | Proteasome activator hPA28 subunit beta | 5.44 | 27,502 | 239 | 24 | 0.27 ± 0.023 |
| 13 | gi|1060907 | Quinolinate phosphoribosyltransferase | 5.81 | 31,232 | 297 | 7 | 0.39 ± 0.025 |
| 14 | gi|332356380 | Albumin | 5.73 | 66,531 | 585 | 13 | 0.43 ± 0.034 |
| 15 | gi|49256408 | Eukaryotic translation initiation factor 4B | 5.54 | 69,166 | 611 | 9 | 0.37 ± 0.031 |
| 16 | gi|312137 | Fructose bisphosphate aldolase | 6.41 | 39,816 | 364 | 18 | 0.46 ± 0.056 |
| 17 | gi|728777 | Replication factor C subunit 5 | 6.72 | 38,496 | 340 | 6 | 0.46 ± 0.021 |
| 18 | gi|18379349 | Synaptic vesicle membrane protein VAT-1 homolog | 5.88 | 42,122 | 483 | 12 | 0.49 ± 0.036 |
| 19 | gi|1172837 | Ran-specific GTPase-activating protein | 5.19 | 23,310 | 201 | 3 | 3.61 ± 0.059 |
| 20 | gi|310943005 | Chain A, Crystal Structure Analysis of R29mE81 M Double Mutant of Human CLIC1 | 5.07 | 26,899 | 241 | 10 | 2.76 ± 0.054 |
| 21 | gi|5453559 | ATP synthase subunit d, mitochondrial isoform a | 5.21 | 18,537 | 161 | 34 | 2.97 ± 0.031 |
| 22 | gi|4503545 | Eukaryotic translation initiation factor 5A-1 isoform B | 5.08 | 17,049 | 154 | 14 | 4.22 ± 0.086 |
| 23 | gi|4507669 | Translationally-controlled tumor protein | 4.84 | 19,697 | 172 | 22 | 2.94 ± 0.011 |
| 24 | gi|3041660 | Cyclin-dependent kinase 4 inhibitor A | 5.52 | 16,532 | 156 | 14 | 4.66 ± 0.071 |
| 25 | gi|28201890 | RuvB-like 2 | 5.49 | 51,156 | 463 | 11 | 2..56 ± 0.011 |
| 26 | gi|189617 | Protein PP4-X | 5.65 | 36,262 | 321 | 17 | 2.14 ± 0.013 |
| 27 | gi|62897507 | Isocitrate dehydrogenase 3(NAD +) alpha precursor variant | 6.47 | 40,050 | 366 | 10 | 2.19 ± 0.006 |
| 28 | gi|125962 | Lamin-A/C | 6.57 | 74,139 | 664 | 23 | 2.83 ± 0.023 |
| 29 | gi|313104306 | Far upstream element-binding Protein 2 | 6.26 | 73,146 | 711 | 8 | 3.02 ± 0.009 |
| 30 | gi|115311883 | Methyltransferase-like protein 19 | 6.98 | 84,628 | 757 | 17 | 2.32 ± 0.011 |
| 31 | gi|38455427 | T-complex protein 1 subunit delta | 7.96 | 58,401 | 539 | 9 | 2.41 ± 0.013 |
| 32 | gi|7387554 | Mitotic checkpoint protein BUB3 | 6.36 | 37,154 | 328 | 3 | 2.12 ± 0.024 |
| 33 | gi|119597758 | dUTP pyrophosphatase, isoform CRA_d | 9.88 | 23,042 | 218 | 13 | 3.08 ± 0.071 |
| 34 | gi|4506517 | Regulator of G-protein signaling 2 | 9.05 | 24,381 | 211 | 5 | 2.76 ± 0.019 |
| 35 | gi|4758788 | NADH dehydrogenase[ubiquinone] iron-sulfur protein 3, mitochondrial precursor | 6.89 | 30241 | 264 | 35 | 2.32 ± 0.010 |
a Isoelectric point. b Molecular weight. c Amino acid. d Sequence coverage (%). e The spot intensity ratios were calculated for HU-treated (treated) to non-treated control (C)
Thus, a total of 34 proteins (17 up-regulated and 17 down-regulated) were identified as response proteins to HU-trigged DNA damage in HeLa cells corresponding to the p12 subunit degradation.
Bioinformatics analysis of differentially expressed proteins
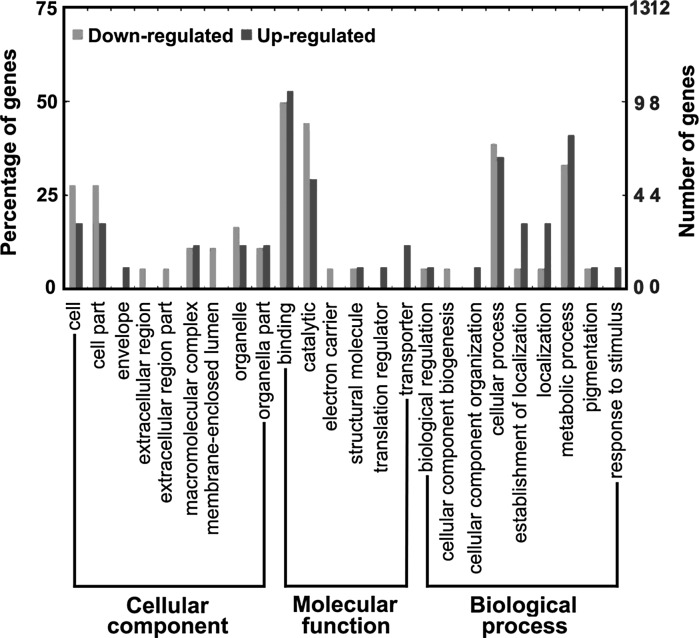
In order to annotate the physiological roles of these differentially expressed proteins identified by 2-DE combining with MS, GO analysis was performed by querying protein sequence against GO Database (Gene Ontology Annotation Plot, WEGO). All of them are classified in terms of cellular component, molecular function, and biological process as shown in Fig. 3. Most of the identified proteins associated with cellular component are involved in cell, cell part, macromolecular complex, membrane-enclosed lumen, organelle, and organelle parts, except for the involvement in envelope, extracellular region, and extracellular region part. These include proteins associated with molecular functions are mainly involved in binding and catalysis, and in addition, three up-regulated proteins that are involved in translation regulation and transport. On the association of biological process, they are largely involved in cellular process and metabolic process of which 5 down-regulated and 6 up-regulated proteins are involved in metabolic process while 6 down-regulated and 5 up-regulated proteins are associated with cellular process. And besides, they are also involved in biological regulation, cellular component biogenesis, cellular component organization, establishment of localization, localization, pigmentation, and response to stimulus.
Fig. 3.
GO analysis for specific proteins in the control and treated groups. The right coordinate axis indicates the number of proteins for each GO annotation, and the left one represents the proportion of proteins for every GO annotation
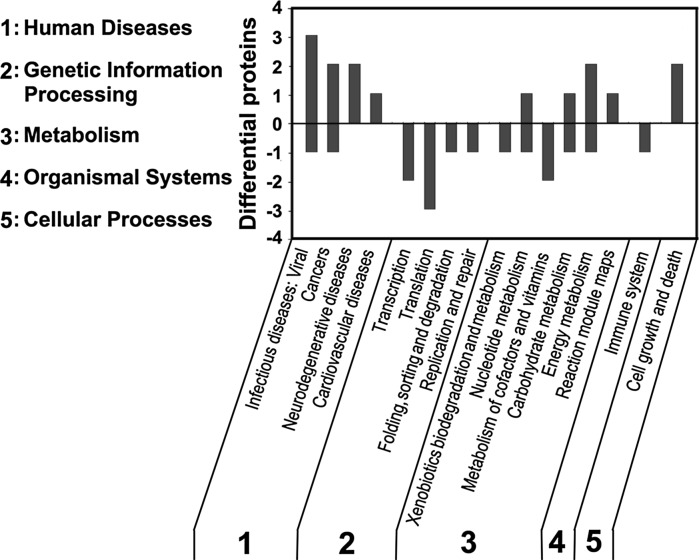
For further characterization of those protein functions that were used for GO analysis, KEGG pathway analysis was performed to systematically analyze the gene functions that link genomic information with higher order functional information (Kanehisa and Goto 2000). Among these 34 proteins, 21 proteins could be mapped to KEGG pathways while the rest cannot be mapped, which may be due to lack of available annotations for primary and secondary metabolic pathways. The differentially expressed proteins were mainly involved in 5 primary pathways, being assigned as human diseases, genetic information processing, metabolisms, organismal systems, and cellular processes, with the subsets that were involved in 16 secondary pathways as described in Fig. 4. As expected, a number of response proteins to HU-trigged DNA damage in cells corresponding to the degradation of p12 were associated with human disease-dependent pathway with the sets that were involved in infectious diseases (viral), cancers, neurodegenerative diseases, and cardiovascular disease, etc. Partially, they were also involved in other four primary pathways concerning transcription, translation, folding, sorting and degradation, replication and repair, xenobiotics biodegradation and metabolism, nucleotide metabolism, metabolism of cofactors and vitamins, carbohydrate metabolism, energy metabolism, immune system, cell growth and death, etc.
Fig. 4.
KEGG pathway analysis. The differentially expressed proteins were analyzed for their KEGG pathways. The up-regulated proteins are shown above the X axis and those for down regulated are shown below the X axis. The resultant pathways belong to five categories: 1 human diseases, 2 genetic information processing, 3 metabolism, 4 organismal systems, 5 cellular processes
Validation of the up-regulation of TCTP versus down-regulation of p12 subunit
Because of its critical role in tumorigenesis, one of these response proteins, TCTP, was selected for further validation to see if its up-regulation is indeed associated with the depletion of p12 upon DNA damage in living cells. Cells grown to ~70 % confluence were treated with 4 mM of HU and harvested by 8 h after incubation. Then Western blot analysis was performed. As shown in Fig. 5b, in concomitancy with p12 subunit depletion, TCTP was significantly up-regulated at 8 h by HU-treatment (Fig. 5b, column 2), compared with the control without HU-treatment (Fig. 5b, column 1). Furthermore, this TCTP up-regulation was effectively abolished corresponding to the blockage of p12 degradation by the treatment of MG132 (Fig. 5b, column 3). The other three subunits p125, p68, p50, and PCNA were relatively unaffected, which is consistent with the observations in Fig. 1.
Fig. 5.
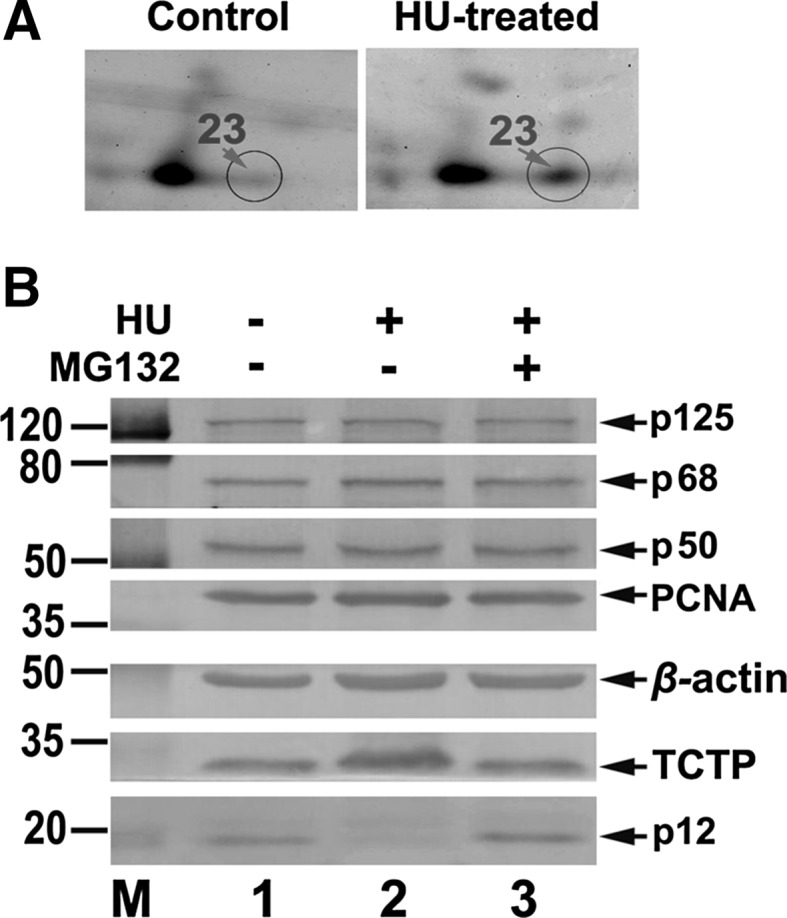
Validation of TCTP up-regulation versus p12 degradation. a An enlarged spot 23 in 2-DE gel. Left panel shows the 2-DE gel around spot 23 for non-treated group and right panel shows the 2-DE gel around spot 23 for HU-treated group. Circled spots 23 are indicated by arrows in red. b HeLa cells grown to ~70 % confluence were treated with 4 mM of HU, harvested by 8 h after incubation, and analyzed by Western blot. Group 1: non-treated cells as a control. Group 2: 4 mM of HU treated cells. Group 3: by the addition of 10 μM proteasome inhibitor MG132 into the culture medium 30 min before HU treatment. The numbers shown on the left indicate the positions in kDa protein markers (M). The four subunits of Pol δ (p125, p68, p50, and p12), PCNA, β-actin, and TCTP are indicated by arrows
Combining with the judgment on the enlarged 2-DE gels around sport 23 (Fig. 5a) in which the protein level was significantly up-regulated nearly threefold in HU-treated HeLa cells based on the identification by MS analysis (Table 1), these observations strongly support that the up-regulation of TCTP is associated with p12 depletion in response to HU-trigged DNA damage.
Discussion
Both environmental chemicals and spontaneous cellular events can cause serious DNA damage, threatening the integrity of the genome. As a consequence, mutations will be generated which can lead to development of numerous human diseases such as cancer. Upon DNA damage, the DDR is triggered by a perfect coordination of an intricate interplay of sensor, signal transducer, and effecter proteins to ensure faithful DNA repair. This process is typically associated with a temporary cell-cycle arrest. If genetic material cannot be restored, cell death or cellular senescence pathway is triggered to prevent tumorigenesis (Ciccia and Elledge 2010). Our previous studies showed that the p12 subunit is rapidly degraded in cultured living cells by DNA damage or replication stress induced by UV, alkylating agents, oxidative, or replication stresses (Zhang et al. 2007). The depletion of p12 results in a conversion of Pol δ heterotetramer (Pol δ4) to a heterotrimer (Pol δ3) with altered Pol δ properties in vivo (Meng et al. 2009, 2010). It is now generally accepted that the display of p12 depletion is a common response to DNA damage. The p12 subunit is rapidly degraded by DNA damage or replication stress brought about by treatments with UV, MMS, HU, or APH not only in cultured neoplastic cells, such as HeLa, human lung carcinoma H1299, human colorectal carcinoma HCT116, etc., but also in cultured derivatives from modified normal cells, such as HEK293T, GM00637, H38-5 or ts20TGR, mouse cells derived from BALB/c 3T3 A31, etc. (Zhang et al. 2007). The apparent p12 degradation and consequent transformation of Pol δ by modification of its quaternary structure represent a novel DDR mechanism in mammalian cells (Lee et al. 2012). Our recent study indicates that p12 subunit is also degraded in calcium-triggered apoptotic HeLa cells (Fan et al. 2014). This suggests a novel potential role of p12 involved in cellular apoptosis by altered Pol δ function(s) due to the loss of p12. Therefore, in the present study, we used HeLa cells as study model. The major focus is to understand the cellular responses, correspondingly the degradation of p12 subunit, to DNA damage in tumor cells using mass spectrometry-based proteomics.
HU is a common chemical antimetabolite. It is cytostatic by inhibiting ribonucleotide reductase, an enzyme important in creating deoxynucleosides for DNA, inducing the stalling of replication forks that leads to the generation of aberrant DNA structures containing single-stranded DNA arising from replication fork collapse, inappropriate homologous recombination (Andreassen et al. 2006), or the uncoupling of the replication helicase from Pol α (Byun et al. 2005). For this purpose, HU was selected to be used in the treatment of cells for the creation of replication stress. As expected, p12 was degraded in a time-dependent manner by the treatment of cells with 4 mM HU (Fig. 1b). The p12 degradation was dependent on the activation of ATR/Chk1 that triggers the intra-S-phase checkpoint (Fig. 1b, P-Chk1-S345).
In recent years, MS-based proteomics has been widely used in cellular biochemistry due to the fact that enormous progress has been made on its reproducibility, higher sensitivity, specificity, accuracy, and software developments (Daub 2012; Derks et al. 2014). After protein complex isolation, MS has been instrumental to identify novel protein–protein interactions and modifications in DDR research. Specialized proteomics screens dramatically expanded the components and repertoire of post-translational modification (PTM) events such as phosphorylation, acetylation, (poly)ADP-ribosylation, ubiquitination, sumoylation and neddylation within DDR (Harper and Elledge 2007; Jackson and Bartek 2009). As indicated in a preview by Daub (2012), such multipronged PTM analysis could be combined with spatiotemporal proteomics, to capture subcellular distribution and localization changes and record the temporal dynamics of protein abundance and modifications. Here, we used mass spectrometry-based proteomics to investigate the global cellular response to DNA damage in HU-treated HeLa cells. In order to meet the requirement for greater sampling amounts for 2-DE, the total proteins of each group (HU-treated and non-treated) were isolated from at least 20 culture plates (100-mm) following a standard protocol as stated in “Materials and methods” section. In such case, the collected cells at the same time in each treatment could stay in the same status. To ensure the constant results in each treatment, triplicate experiments were performed for each pair of tests. About 736 ± 13 proteins in the control group and 741 ± 19 proteins in the HU-treated group were detected. However, only 34 spots were identified as differentially expressed proteins in this content (Table 1). A poorer identification rate may be due to the methodology we currently used. Therefore, more improvements in the protocol for MS-based proteomics are needed in our future studies, such as sample preparation, gel staining, faster mass spectrometers of higher sensitivity and accuracy, and software developments allowing for automated data processing. There is lack of information about some important factors, for example, Y-family polymerases (e.g., Pols η, ι, κ, and REV1) that are generally considered to act as the main translesion polymerases that effectively bypass a variety of DNA lesions, and those main DNA damage recognition factors MRN and the PI3 kinases ATM, ATR and DNA-PK, which phosphorylate a multitude of proteins and thus induce DDR (Roos and Kaina 2013), etc. Although lacking available information about the factors mentioned above, 34 differentially expressed proteins still provide us some key clues. Corresponding to the p12 degradation, 17 up- and 17 down-regulated proteins were identified (Table 1). Go analysis indicates their physiological roles that mainly associate with cellular components, molecular functions, and biological processes (Fig. 3), which are involved in 5 primary pathways with the subsets involving 16 secondary pathways by further KEGG analysis (Fig. 4). Among them, some important factors should be noticed. For example, one of down-regulated protein, programmed cell death protein 6 (PDCD6), also called apoptosis-linked gene 2 protein (ALG2), functions as calcium-binding protein required for T cell receptor-, Fas-, and glucocorticoid-induced cell death (Suzuki et al. 2012; Yoon et al. 2012). Besides their multifunction, four proteins, ATP-dependent RNA helicase DDX39A, complement component 1 Q subcomponent-binding protein (SF2p32), Lamin-A/C, and far upstream element-binding Protein 2 are involved in pre-mRNA splicing or as splicing factors to play an important role in DNA damage and repair (Dittmer and Misteli 2011; Filippov et al. 2007; Marengo and Wassarman 2008; Singh et al. 2013). Replication factor C (RFC) subunit 5 is down regulated, suggesting that RFC and Pol δ may be synergistically involved in DNA repair events on lagging or leading strand with their altered functions by the down-regulation of their smallest subunits.
More interestingly, TCTP, also called histamine releasing factor (HRF), tumor protein translationally controlled 1 (Tpt1), p23 or fortilin, is found to be up-regulated by ~threefold (Table 1). TCTP is a highly conserved protein and abundantly expressed in eukaryotes. It is reported as multifunctional to participate in numerous cellular processes including cell growth, survival, development, protein synthesis, immune response, tumor reversion, transcription regulation, and induction of pluripotent stem cells or apoptosis (Bommer and Thiele 2004). Comparing to normal tissue, the level of TCTP is increased in many human cancer tissues, such as breast, colorectal, prostate and lung cancer (Arcuri et al. 2004; Deng et al. 2006; Kim et al. 2008; Slaby et al. 2009).Therefore, in recent years, more and more attention is paid to the involvement of TCTP on tumor reversion because of its critical role in tumorigenesis (Miao et al. 2013; Nagano-Ito and Ichikawa 2012). It is now recognized as a therapeutic target in several cancers (Acunzo et al. 2014). In this study, the over-expressed TCTP protein could be recovered to a normal level following the blockage of p12 degradation by the proteasome inhibitor MG132 in HU-treated HeLa cells, suggesting that the up-regulation of TCTP is tightly associated with the p12 degradation in response to DNA damage (Fig. 5b). The fact of linkage of up-regulation of TCTP to the p12 degradation in response to DNA damage should be taken into account in understanding of DDR mechanisms. The further studies of how alterations in Pol δ function and the synergistic action of TCTP could contribute to tumorigenesis and how they may facilitate the communication with other replication and repair proteins should be more deeply investigated.
As stated in the "introduction" section, mammalian cells respond to DNA damage by a host of defense mechanisms, which involves the recruitment and assembly of large complexes of proteins that orchestrate and prioritize a network of responses. Upon encounter of replisomes with damaged DNA, cells are particularly vulnerable during S-phase. If the replicative DNA polymerases perform translesion (TLS) synthesis and introduce errors, or the polymerases are unable to bypass the lesions and lead to replication fork stalling, the mutations would be introduced. Such events that lead to prolonged stalling of replication forks result in replication fork collapse, formation of aberrant fork structures, chromosome damage or loss and ultimately cell death. In order to facilitate our understanding of carcinogens on cells, it is therefore important to shed light on the how protein complexes are recruited and assembled in cells upon DNA damage, and how these processes are taking place. Thus, this present study may enlarge and broaden our view for deeply understanding how global cellular stress responds to DNA damage, which could contribute to the etiology of human cancer or other diseases that can result from loss of genomic stability.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31370790, 30970612, 31100118), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Natural Science Foundation of Jiangsu Province (BK2011495).
Footnotes
Chao You, Yanhua Yang and Lei Zhang have contributed equally to this work.
References
- Acunzo J, Baylot V, So A, Rocchi P. TCTP as therapeutic target in cancers. Cancer Treat Rev. 2014;40:760–769. doi: 10.1016/j.ctrv.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Al-Rubeai M, Singh RP. Apoptosis in cell culture. Curr Opin Biotechnol. 1998;9:152–156. doi: 10.1016/S0958-1669(98)80108-0. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, Ho GP, D’Andrea AD. DNA damage responses and their many interactions with the replication fork. Carcinogenesis. 2006;27:883–892. doi: 10.1093/carcin/bgi319. [DOI] [PubMed] [Google Scholar]
- Arakelyan A, Nersisyan L. KEGGParser: parsing and editing KEGG pathway maps in Matlab. Bioinformatics. 2013;29:518–519. doi: 10.1093/bioinformatics/bts730. [DOI] [PubMed] [Google Scholar]
- Arcuri F, Papa S, Carducci A, Romagnoli R, Liberatori S, Riparbelli MG, Sanchez JC, Tosi P, del Vecchio MT (2004) Translationally controlled tumor protein (TCTP) in the human prostate and prostate cancer cells: expression, distribution, and calcium binding activity. Prostate 60:130–140. doi:10.1002/pros.20054 [DOI] [PubMed]
- Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP) Int J Biochem Cell Biol. 2004;36:379–385. doi: 10.1016/S1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chea J, Zhang S, Zhao H, Zhang Z, Lee EY, Darzynkiewicz Z, Lee MY. Spatiotemporal recruitment of human DNA polymerase delta to sites of UV damage. Cell Cycle. 2012;11:2885–2895. doi: 10.4161/cc.21280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H. DNA damage response: multilevel proteomics gains momentum. Mol Cell. 2012;46:113–114. doi: 10.1016/j.molcel.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Deng SS, Xing TY, Zhou HY, Xiong RH, Lu YG, Wen B, Liu SQ, Yang HJ (2006) Comparative proteome analysis of breast cancer and adjacent normal breast tissues in human. Genomics Proteomics Bioinform 4:165–172. doi:10.1016/S1672-0229(06)60029-6 [DOI] [PMC free article] [PubMed]
- Derks KW, Hoeijmakers JH, Pothof J. The DNA damage response: the omics era and its impact. DNA Repair (Amst) 2014;19:214–220. doi: 10.1016/j.dnarep.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Misteli T. The lamin protein family. Genome Biol. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Zhang Q, You C, Qian Y, Gao J, Liu P, Chen H, Song H, Chen Y, Chen K, Zhou Y (2014) Proteolysis of the human DNA polymerase delta smallest subunit p12 by mu-calpain in calcium-triggered apoptotic HeLa cells. PLoS ONE 9:e93642. doi:10.1371/journal.pone.0093642 [DOI] [PMC free article] [PubMed]
- Filippov V, Filippova M, Duerksen-Hughes PJ. The early response to DNA damage can lead to activation of alternative splicing activity resulting in CD44 splice pattern changes. Cancer Res. 2007;67:7621–7630. doi: 10.1158/0008-5472.CAN-07-0145. [DOI] [PubMed] [Google Scholar]
- Fritz G, Kaina B. Rho GTPases: promising cellular targets for novel anticancer drugs. Curr Cancer Drug Targets. 2006;6:1–14. [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Yang J, Gao Z, Yu Y. Proteomic analysis of cellular responses to low concentration N-methyl-N′-nitro-N-nitrosoguanidine in human amnion FL cells. Environ Mol Mutagen. 2004;43:93–99. doi: 10.1002/em.20001. [DOI] [PubMed] [Google Scholar]
- Kaina B. DNA damage-triggered apoptosis: critical role of DNA repair, double-strand breaks, cell proliferation and signaling. Biochem Pharmacol. 2003;66:1547–1554. doi: 10.1016/S0006-2952(03)00510-0. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Koo KH, Kim YH, Sohn J, Park YG. Identification of potential lung cancer biomarkers using an in vitro carcinogenesis model. Exp Mol Med. 2008;40:709–720. doi: 10.3858/emm.2008.40.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Zhang S, Lin SH, Chea J, Wang X, LeRoy C, Wong A, Zhang Z, Lee EY (2012) Regulation of human DNA polymerase delta in the cellular responses to DNA damage. Environ Mol Mutagen 53:683–698. doi:10.1002/em.21743 [DOI] [PubMed]
- Marengo MS, Wassarman DA. A DNA damage signal activates and derepresses exon inclusion in Drosophila TAF1 alternative splicing. RNA. 2008;14:1681–1695. doi: 10.1261/rna.1048808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhou Y, Zhang S, Lee EY, Frick DN, Lee MY. DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucl Acids Res. 2009;37:647–657. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhou Y, Lee EY, Lee MY, Frick DN. The p12 subunit of human polymerase delta modulates the rate and fidelity of DNA synthesis. Biochemistry. 2010;49:3545–3554. doi: 10.1021/bi100042b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercille S, Massie B. Induction of apoptosis in nutrient-deprived cultures of hybridoma and myeloma cells. Biotechnol Bioeng. 1994;44:1140–1154. doi: 10.1002/bit.260440916. [DOI] [PubMed] [Google Scholar]
- Miao X, Chen YB, Xu SL, Zhao T, Liu JY, Li YR, Wang J, Zhang J, Guo GZ (2013) TCTP overexpression is associated with the development and progression of glioma. Tumour Biol 34:3357–3361. doi:10.1007/s13277-013-0906-9 [DOI] [PubMed]
- Nagano-Ito M, Ichikawa S. Biological effects of Mammalian translationally controlled tumor protein (TCTP) on cell death, proliferation, and tumorigenesis. Biochem Res Int. 2012;2012:204960. doi: 10.1155/2012/204960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Xia H, Shi H, Zhou Y, Chen L, Yao Q, Liu X, Feng F, Yuan Y, Chen K (2012) Comparative proteomic analysis reveals that caspase-1 and serine protease may be involved in silkworm resistance to Bombyx mori nuclear polyhedrosis virus. J Proteomics 75:3630–3638. doi:10.1016/j.jprot.2012.04.015 [DOI] [PubMed]
- Rahmeh AA, Zhou Y, Xie B, Li H, Lee EY, Lee MY. Phosphorylation of the p68 subunit of Pol delta acts as a molecular switch to regulate its interaction with PCNA. Biochemistry. 2012;51:416–424. doi: 10.1021/bi201638e. [DOI] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Ann Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Singh M, Hunt CR, Pandita RK, Kumar R, Yang CR, Horikoshi N, Bachoo R, Serag S, Story MD, Shay JW, Powell SN, Gupta A, Jeffery J, Pandita S, Chen BP, Deckbar D, Löbrich M, Yang Q, Khanna KK, Worman HJ, Pandita TK (2013) Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol Cell Biol 33:1210–1222. doi:10.1128/MCB.01676-12 [DOI] [PMC free article] [PubMed]
- Slaby O, Sobkova K, Svoboda M, Garajova I, Fabian P, Hrstka R, Nenutil R, Sachlova M, Kocakova I, Michalek J, Smerdova T, Knoflickova D, Vyzula R (2009) Significant overexpression of Hsp110 gene during colorectal cancer progression. Oncol Rep 21:1235–1241 [DOI] [PubMed]
- Suzuki K, Dashzeveg N, Lu ZG, Taira N, Miki Y, Yoshida K. Programmed cell death 6, a novel p53-responsive gene, targets to the nucleus in the apoptotic response to DNA damage. Cancer Sci. 2012;103:1788–1794. doi: 10.1111/j.1349-7006.2012.02362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Dai L, Xia H, Zhu K, Liu H, Chen K. Protein profile of rice (Oryza sativa) seeds. Genet Mol Biol. 2013;36:87–92. doi: 10.1590/S1415-47572013000100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucl Acids Res 34:W293–W297. doi:10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed]
- Yoon JH, Choi YJ, Kim SG, Nam SW, Lee JY, Park WS. Programmed cell death 6 (PDCD6) as a prognostic marker for gastric cancers. Tumour Biol. 2012;33:485–494. doi: 10.1007/s13277-011-0280-4. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY. A novel DNA damage response: rapid degradation of the p12 subunit of dna polymerase delta. J Biol Chem. 2007;282:15330–15340. doi: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhou Y, Sarkeshik A, Yates JR 3rd, Thomson TM, Zhang Z, Lee EY, Lee MY (2013) Identification of RNF8 as a ubiquitin ligase involved in targeting the p12 subunit of DNA polymerase delta for degradation in response to DNA damage. J Biol Chem 288:2941–2950. doi:10.1074/jbc.M112.423392 [DOI] [PMC free article] [PubMed]
- Zhou Y, Chen H, Li X, Wang Y, Chen K, Zhang S, Meng X, Lee EY, Lee MY (2011) Production of recombinant human DNA polymerase delta in a Bombyx mori bioreactor. PLoS ONE 6:e22224. doi:10.1371/journal.pone.0022224 [DOI] [PMC free article] [PubMed]
- Zhou Y, Meng X, Zhang S, Lee EY, Lee MY. Characterization of human DNA polymerase delta and its subassemblies reconstituted by expression in the MultiBac system. PLoS ONE. 2012;7:e39156. doi: 10.1371/journal.pone.0039156. [DOI] [PMC free article] [PubMed] [Google Scholar]