Abstract
Mammalian early embryonic development is controlled by a unique program of gene expression, and involves epigenetic reprogramming of histone modifications and DNA methylation. SET and MYND domain-containing protein 3 (SMYD3) is a histone H3 lysine 4 methyltransferase that plays important roles in transcription regulation. The expression of SMYD3 has been studied in some cancer cell lines. However, its expression in oocytes and embryos has not previously been reported. Here, we detected the SMYD3 mRNA and found that it was expressed throughout bovine oocyte in vitro maturation and early embryonic development. Microinjection of SMYD3 siRNA at germinal vesicle stage decreased the transcription level of NANOG, and blocked the development of in vitro fertilization embryos at 4–8 cell stage. Conversely, Microinjection of SMYD3 siRNA at pronuclear stage did not affect early embryonic development. Our findings suggest that SMYD3 regulates the expression of NANOG, and plays an essential role in bovine early embryonic development.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-014-9838-5) contains supplementary material, which is available to authorized users.
Keywords: SMYD3, Maternal-to-zygotic transition, Pre-implantation embryos, SMYD3 splice variants
Introduction
The mammalian early embryonic development begins at the fusion of the highly differentiated egg and sperm. After fusion, the paternal and maternal genomes are reprogrammed to a totipotent state, and the embryonic genome is activated after a period of reliance on maternal factors (Ma et al. 2001; Schultz 1993). Large scale synthesis of mRNA from the diploid embryonic genome occurs at a species-specific time point; it starts at the end of the first cell cycle in murine embryos, at the 4-cell stage in human embryos and at the 8-cell stage in bovine embryos (Braude et al. 1988; Misirlioglu et al. 2006; Telford et al. 1990). This process is called as maternal-to-zygotic transition (MZT) and is regulated by a well-orchestrated gene regulatory network. However, this gene regulatory network that directs early embryonic development has remained elusive. Recently, some researchers have demonstrated that Pou5f1 (Oct3/4) and Nanog affect MZT and pre-implantation embryonic development in mouse and zebrafish (Bachvarova 1985; Lee et al. 2013).
Histone modification functions in the regulation of chromatin structure, as well as transcription activation and repression during embryonic development (Biel et al. 2005). Among the well characterized histone modifications, methylation of lysine residues 4, 9, 27 and 36 on histone H3 and residue 20 on histone H4 (Jenuwein and Allis 2001) are considered to be critical for transcription regulation (Kouzarides 2002; Lachner and Jenuwein 2002). More specifically, current researches suggest that methylation of lysine 9 on histone H3 is involved in transcriptionally repressive heterochromatin formation with heterochromatin associated proteins (Rea et al. 2000), and that lysine 4 methylation on histone H3 (H3-K4) is important for transcriptional activation (Kouzarides 2002; Santos-Rosa et al. 2002; Wang et al. 2001). Interestingly, a functional link between H3-K4 methylation and meiosis-specific gene expression has been demonstrated (Hayashi et al. 2005). PR domain-containing 9 (also known as Meisetz) is an H3-K4 trimethyltransferase that causes transcriptional activation, and is specifically expressed in early meiotic germ cells in testes and ovaries (Hayashi et al. 2005). Thus, other trimethyltransferases should work to maintain the high level of H3-K4me3 in maturing bovine oocytes. We hypothesized that this function is performed by SET and MYND domain-containing protein 3 (SMYD3).
SMYD3 is a histone H3-K4-specific dimethyltransferase and trimethyltransferase. It contains the evolutionarily conserved SET (Su[var]3-9, enhancer-of-zeste, trithorax protein) and MYND (myeloid translocation protein 8, Nervy, DEAF1) domains, both of which have been found in many transcription regulators linked to development, chromatin stability and cancer (Schneider et al. 2002). Because of the histone H3-K4-specific methyltransferase activity of the SET domain and the protein–protein interaction activity of the MYND domain, SMYD3 plays an important role in transcription regulation (Hamamoto et al. 2004). Although the regulatory mechanism of SMYD3 in cancer cells has been extensively investigated, there is an absence of knowledge about its role in oocyte maturation, fertilization and early embryonic development in bovine. Here, we studied the expression of SMYD3 in maturing bovine oocytes and early stage of embryos, and used RNA interference to examine its possible role during oocyte maturation and early embryonic development.
Materials and methods
Oocyte recovery and embryo production
Bovine ovaries were collected at a commercial slaughterhouse and transported to the laboratory in a 0.9 % NaCl aqueous solution containing antimycotic agent. Cumulus-oocyte complexes (COCs) were collected from 3 to 6 mm-diameter follicles and only healthy COCs with at least five layers of cumulus cells were used for in vitro maturation and embryo production, as previously described (Blondin and Sirard 1995).
COCs were put through in vitro maturation after three washes with M199 (Gibco, Life Technologies Co., Grand Island, NY, USA) supplemented with 1 % FBS (Sigma-Aldrich Co., St Louis, MO, USA). A group of 80–100 COCs were placed in one well of a 4-well plate with 1 ml maturation medium (M199 supplemented with 10 % FBS, 0.38 mM sodium pyruvate, 0.5 mg/ml follicle-stimulating hormone, 0.5 mg/ml luteinizing hormone, 1 µg/ml estradiol, 0.075 mg/ml penicillin and 0.05 mg/ml streptomycin) covered with mineral oil (M1180; Sigma-Aldrich Co.). The COCs were incubated at 38.5 °C in 5 % CO2.
In vitro fertilization (IVF) was conducted following a standard protocol established in our lab. Briefly, after 10 s of gentle shaking, a straw of semen was thawed for 10 s in a 37 °C water bath. Sperms were then washed twice by centrifugation at 720×g for 5 min in 10 ml of Brackett and Oliphant’s (BO) sperm-washing medium containing 3 mg/ml of bovine serum albumin (BSA, Sigma-Aldrich Co.) and 10 mM caffeine (Sigma-Aldrich Co.) (Brackett and Oliphant 1975). The washed sperm pellet was then resuspended in BO sperm-washing medium at a concentration of 1.0 × 106 sperms/ml for subsequent IVF.
After in vitro maturation, bovine COCs were washed twice and transferred into 50 µl drops of BO medium (30 oocytes/drop) containing 6 mg/ml of BSA (Sigma-Aldrich Co.) and 10 mg/ml of heparin (Sigma-Aldrich Co.), and pre-equilibrated for 2 h at 38.5 °C in 5 % CO2; 50 µl of sperm suspension was then added to each drop of medium. Oocytes were incubated with sperms for 6 h at 38.5 °C in 5 % CO2 incubator. After IVF, embryos were further cultured in CR1 medium supplemented with 6 mg/ml of BSA (Sigma-Aldrich Co.) in a humidified atmosphere at 38.5 °C in 5 % CO2 for 48 h. Cleaved embryos were cultured for an additional 5 days in CR1 medium supplemented with 5 % FBS on cumulus cell monolayers in a humidified atmosphere at 38.5 °C in 5 % CO2.
Oocyte and embryo collection
The bovine oocytes were collected after 0, 8, 12, 18 or 24 h of in vitro maturation. All of the oocytes collected at 18 and 24 h had extruded the first polar body. Following IVF, 2-, 4- and 8-cell embryos, as well as morula- and blastocyst-stage embryos were collected at 1, 1.5, 2, 5 and 8 days post-fertilization, respectively. Immature GV oocytes were collected for quantitative real-time PCR analysis after selection and mechanical removal of cumulus cells. Denuded oocytes were washed with RNase-free PBS to ensure elimination of all cumulus cells and frozen in three pools of 20 oocytes at −80 °C until RNA extraction. Additionally, three pools of 20 embryos of 2-, 4-, 8-cell, as well as morula- and blastocyst-stages were collected for quantitative real-time RT-PCR experiments. All the embryos were washed three times with RNase-free PBS, frozen, and stored at −80 °C until RNA extraction.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using TaKaRa’s RNAiso reagent (TAKARA Biotechnology Co., Shiga, Japan), according to the manufacturer’s instructions. RNA was eluted in 15 µl of RNase free water and frozen at −80 °C until use. For real-time RT-PCR, 10 µl of total RNA from pools of 20 oocytes and embryos was reverse transcribed using PrimeScript RT reagent Kit with gDNA Eraser (TAKARA Biotechnology Co.) to synthesize cDNA. The primers used for real-time RT-PCR are listed in Table 1, and were designed using the Primer 3 web interface (Available at http://frodo.wi.mit.edu/). For each gene examined, a standard curve was included to assess the amplification efficiency. Real-time quantitative PCR reactions were performed at 95 °C for 30 s followed by 40 cycles at 95 °C for 5 s and 58 °C for 31 s using specific primers on an ABI PRISM®7300 PCR cycler (Applied Biosystems, Life Technologies Co.). The averages of three replicate Cycle threshold (Ct) values were used for calculating relative gene expression levels, and were normalized to the reference gene, GAPDH.
Table 1.
Sequences of the real-time PCR primers
| Gene | Sequences |
|---|---|
| SMYD3 | Forward: 5′-ATCCTTTGGCGTACACGGTG-3′ |
| Reverse: 5′-ATTCGGCATTGAGAGCATCG-3′ | |
| OCT4 | Forward: 5′-AGTAGGTTGGGTAGAGGGGTATTAG-3′ |
| Reverse: 5′-AAATTAAAAAAATCTCCTAAAAAAAA-3′ | |
| NANOG | Forward: 5′-ATAATGGTTTTGGTGAGATTGGTAG-3′ |
| Reverse: 5′-ATAAAACTCAACCATACTTAACCCC-3′ | |
| CDX2 | Forward: 5′-AAGACAAATACCGGGTCGTG-3′ |
| Reverse: 5′-CTGCGGTTCTGAAACCAAAT-3′ | |
| GAPDH | Forward: 5′-TTCAACGGCACAGTCAAGG-3′ |
| Reverse: 5′-ACATACTCAGCACCAGCATCAC-3′ |
Immunofluorescence microscopy
Oocytes and embryos were fixed in 3.7 % paraformaldehyde for 30 min at room temperature and permeabilized with 0.5 % Triton X-100 in PBS for 15 min, followed by blocking overnight at 4 °C in 1 % BSA in PBS with 10 % goat serum. The samples were then stained with anti-SMYD3 antibody (ab16027, 1:200; Abcam, Cambridge, MA, USA). The primary antibody binding was detected with an Alexa Fluor 488 donkey anti-rabbit IgG (H+L) secondary antibody (A21206, 1:1,000; Invitrogen, Life Technologies Co.). The nucleus was stained with either PI or DAPI. Images were captured with a Nikon A1 confocal microscope (Nikon Co., Otawara, Tochigi, Japan). Experiments were performed in triplicate, and at least fifteen oocytes or embryos were observed each time.
RNA interference
Three different small interfering RNA (siRNA) oligonucleotides targeting different regions of SMYD3 and one nonsense siRNA (Nos-siRNA) were purchased from Invitrogen (Life Technologies Co.). The three SMYD3 siRNA oligonucleotides were mixed and diluted to a concentration of 20 µM (1:1:1). The Nos-siRNA was also diluted to 20 µM. 10 pl siRNAs were microinjected into a GV stage oocyte or a one-cell embryo at pronuclei stage 7 h post fertilization using an EppendorfFemtoJet® and FemtoJet express Microinjectors (Eppendorf North America, Hauppauge, NY, USA). After injection, we inhibited oocyte maturation for 24–48 h so that siRNAs have time to reduce protein abundance. The oligonucleotides used for SMYD3 and the Nos-siRNA are listed in Table 2.
Table 2.
Sequences of the oligonucleotides used for siRNA microinjection
| Name | Sequences |
|---|---|
| SMYD3 siRNA-1 | Forward: 5′-CCACAAGCGAGAAUGCAAA-3′ |
| Reverse: 5′-UUUGCAUUCUCGCUUGUGG-3′ | |
| SMYD3 siRNA-2 | Forward: 5′-GUGAUCUGCAACUCUUUCA-3′ |
| Reverse: 5′-UGAAAGAGUUGCAGAUCAC-3′ | |
| SMYD3 siRNA-3 | Forward: 5′-GAGGUGUGCAGAUAAUGAA-3′ |
| Reverse: 5′-UUCAUUAUCUGCACACCUC-3′ | |
| Nonsense siRNA | Forward: 5′-UUCUCCGAACGUGUCACGU-3′ |
| Reverse: 5′-ACGUGACACGUUCGGAGAA-3′ |
Statistical analysis
Statistically significant differences between gene expression levels were determined using one-way ANOVA followed by a Newman–Keuls test with the GraphPad Prism 5 software (GraphPad Software, Inc., USA. http://www.graphpad.com/company/). Replicates were included in the statistical model. Differences were considered statistically significant at the 99 % confidence level (P < 0.01). Data are presented as mean ± SD.
Results
The expression and localization of SMYD3 in oocytes and pre-implantation embryos
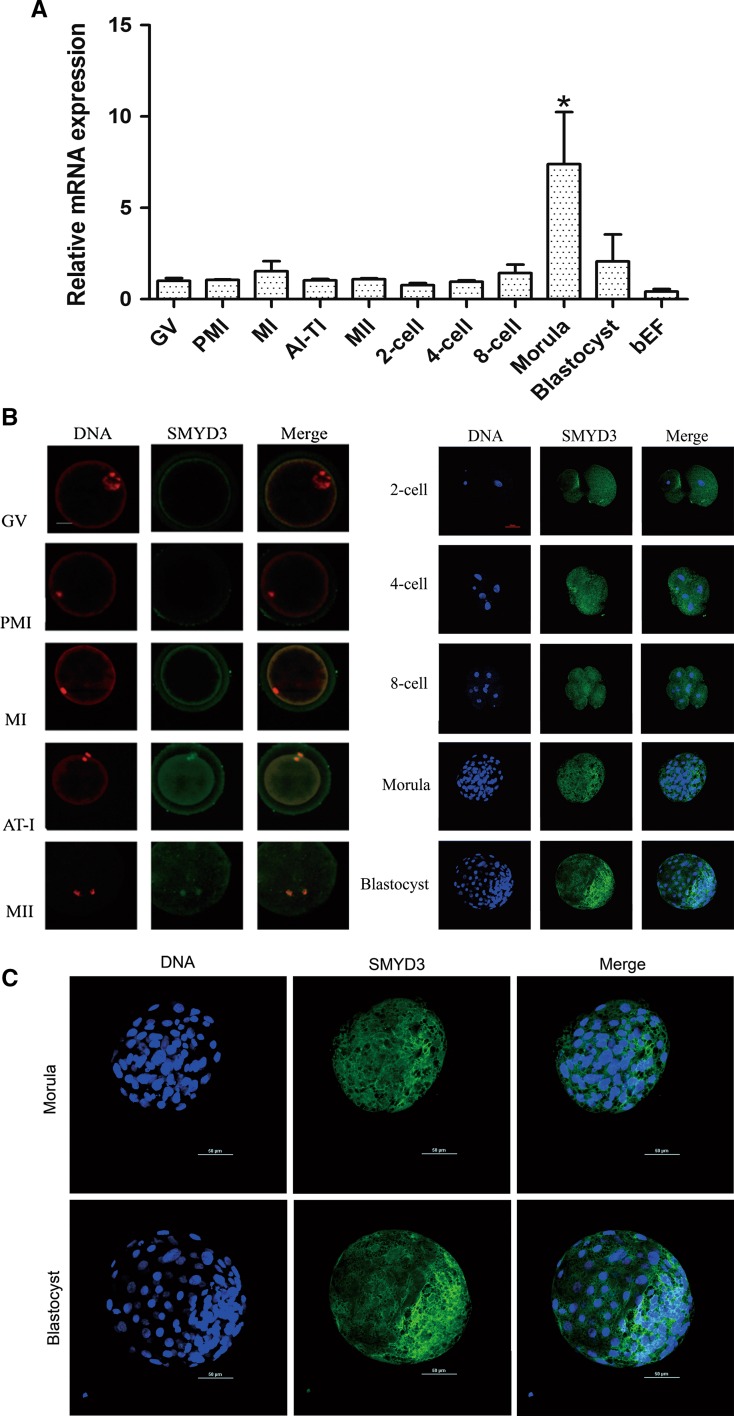
According to the changes in chromosome morphology during meiosis, the maturation of bovine oocytes can be subdivided into GV, GVBD, MI, AI-TI and MII stages, which correspond to the time at 0, 8, 12, 18 and 24 h of in vitro maturation, respectively, as determined by our unpublished data. The transcription levels of SMYD3 during bovine oocyte maturation and pre-implantation development were assessed by RT-qPCR using equal numbers of oocytes or embryos. SMYD3 mRNA levels were maintained constant during oocyte maturation and early embryonic development up to 8-cell stage, at which point SMYD3 mRNA level was significantly increased before returning to baseline at blastocyst stage (Fig. 1a). Next, we assessed SMYD3 protein expression and localization in oocytes and early cleavage stage of bovine embryos by immunofluorescence analysis using anti-SMYD3 antibody. The signals began to be detected in parts of metaphase I stage oocytes (about 22 % showed positive signals). The expression level was increased in the subsequent stages including oocyte maturation (up to 62.5 % at AI-TI and 77 % at MII) and the studied stages of early embryonic development. SMYD3 co-localized with PI-stained chromosomes in AI-TI and MII oocytes, and was subsequently expressed in the cytoplasm and nuclei of developing embryos (Fig. 1b, c).
Fig. 1.
Expression and localization of SMYD3 in bovine maturing oocytes and early developing embryos. a qRT-PCR analysis of SMYD3 transcription in different stages of oocytes and embryos. Data were normalized to GAPDH. Error bars represent standard deviation (SD) (n = 3). *P < 0.01. b Representative images of immunostained oocytes and embryos showing SMYD3 expression and localization (green). DNA was stained with PI (red) or DAPI (blue). c Enlargement of the images of the morula and blastocyst stage embryos (presented in b) showing the SMYD3 localization primarily in cytoplasm. The experiments were performed in triplicate, and at least fifteen oocytes or embryos were observed each time. Scale bar 50 μm
Knockdown of SMYD3 in GV oocyte affected the SMYD3 expression, the rates of 8-cell embryo and further embryo development
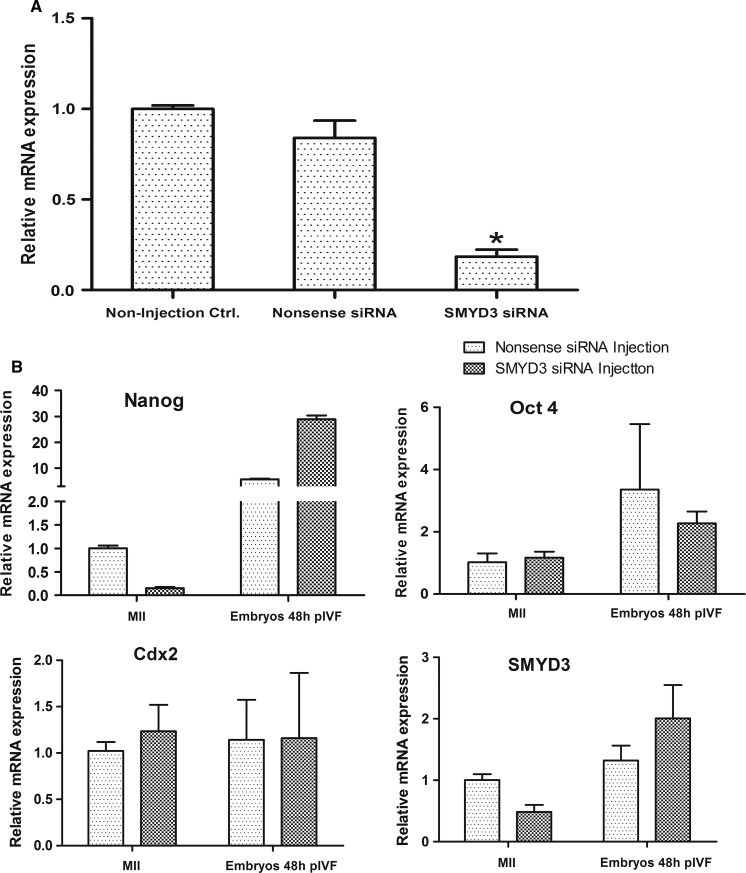
In order to study the role of SMYD3 in bovine oocyte maturation and embryonic development, SMYD3 siRNA was injected into denuded GV stage oocytes (DOs). After cultured in maturation medium containing 0.2 mM 3-isobutyl-1-methyl-xanthin (IBMX) for inhibiting oocyte maturation for 24 h, all oocytes were collected and RT-qPCR was used to analyze the efficiency of mRNA knockdown. Compared with the non-injected control group, the expression of SMYD3 in the Nos-siRNA group was lower, but not significantly decreased, while it was significantly reduced to 18.4 % in experimental group injected with SMYD3 siRNA (Fig. 2a). Next, we investigated oocyte maturation and embryonic development after injecting SMYD3 siRNA into DOs, using non-injected COCs, DOs, and Nos-siRNA injected DOs as controls (Tables 3, 4). Down-regulation of SMYD3 did not affect oocyte maturation and cleavage, but influence the formation of 8-cell embryos and further embryonic development. Incidentally, we found in this experiment that removal of the cumulus cells from in vitro matured COCs had no effect on oocyte maturation, but significantly decreased cleavage rate.
Fig. 2.
Effect of SMYD3 knockdown on OCT4, NANOG and CDX2 expression. a Effect of SMYD3 knockdown in bovine oocytes by SMYD3 siRNA interference. Suppression of SMYD3 was examined at 24 h after GV oocytes injected with SMYD3 siRNA. The non-injection control and nonsence siRNA injection group were used as control. Error bars represent standard deviation (SD) (n = 3). *P < 0.01. b Graphs showing the relative mRNA transcript level of OCT4, NANOG, CDX2 and SMYD3 relative to GAPDH are shown for metaphase II oocytes and embryos 48 h after in vitro fertilization (pIVF) which were treated with siRNA. Error bars represent standard deviation (SD) (n = 3)
Table 3.
The effect of SMYD3 knockdown in bovine GV oocytes on oocyte maturation
| Non-injection groups | Injection groups | |||
|---|---|---|---|---|
| COCs (%) | Dos (%) | Nos-siRNA (%) | SMYD3-siRNA (%) | |
| No. of cultured oocytes | 184 | 168 | 140 | 277 |
| MII oocytes | 139 (75.54 ± 0.1)a | 128 (76.19 ± 0.4)a | 94 (67.14 ± 0.5)b | 183 (66.06 ± 0.7)b |
The experimental replicates at least three times, and the result shown as mean ± SD
a,bMean the groups have significant difference (P < 0.01)
Table 4.
The effect of SMYD3 knockdown in bovine GV oocytes on embryonic development
| Non-injection groups | Injection groups | |||
|---|---|---|---|---|
| COCs (%) | DOs (%) | Nos-siRNA (%) | SMYD3-siRNA (%) | |
| Total No. | 233 | 257 | 156 | 152 |
| 2-Cell | 156 (66.95 ± 2.1)a | 110 (42.80 ± 2.0)b | 67 (42.95 ± 2.2)b | 67 (44.07 ± 2.5)b |
| 8-Cell | 95 (40.34 ± 2.2)a | 45 (17.51 ± 1.9)b | 20 (12.82 ± 2.0)b | 5 (3.29 ± 2.6)c |
| Blastocyst | 71 (30.47 ± 1.9)a | 28 (10.89 ± 2.1)b | 13 (8.33 ± 1.7)b | 0 (0 ± 0)c |
The experimental replicates at least three times, and the result shown as mean ± SD
a,b,cMean the groups have significant difference (P < 0.01)
Knockdown of SMYD3 in zygote had no effect on early embryonic development
To define the exact time of SMYD3 affecting the embryo development, we also injected SMYD3 siRNA into one cell embryos at 7 h post-fertilization. The result showed that there were no significant differences in cleavage rates, 8-cell rates and blastocyst rates among the non-injected zygotes, Nos-siRNA injected zygotes and SMYD3-siRNA injected zygotes (Table 5). This result suggested the deletion of SMYD3 mainly affected the substance that accumulated at maturation stage and resulted in blocking the embryonic development ultimately.
Table 5.
The effect of SMYD3 knockdown in zygotes on bovine embryonic development
| Non-injected zygotes | Nos-siRNA injected | SMYD3 siRNA injected | |
|---|---|---|---|
| Total No. | 269 | 279 | 256 |
| 2-cell | 153 (56.88 ± 3.3)a | 163 (58.43 ± 4.1)a | 152 (55.88 ± 4.4)a |
| 8-cell | 94 (34.94 ± 2.8)a | 89 (31.90 ± 3.6)a | 88 (32.35 ± 4.0)a |
| Blastocyst | 78 (29.00 ± 3.0)a | 67 (24.01 ± 4.0)a | 61 (23.83 ± 4.1)a |
The experimental replicates at least three times, and the result shown as mean ± SD
aDifferent superscript letters mean the groups are significantly different (P < 0.01)
Knockdown of SMYD3 in GV oocyte affected the transcription factor expression
To better understand how SMYD3 knockdown results in embryonic development arrest, we surveyed the transcription levels of SMYD3 and three embryonic transcription factors (OCT4, NANOG and CDX2) in MII oocytes and embryos 48 h post-fertilization. Results showed that SMYD3 expression was dramatically reduced in MII stage oocytes after injection of SMYD3 siRNA in GV stage oocytes, while SMYD3 expression was increased in embryos 48 h post-fertilization compared with Nos-siRNA injection group. We observed no significant changes in OCT4 and CDX2 mRNA at either stage; However, NANOG transcription level was significantly decreased in MII oocytes injected with SMYD3 siRNA. In contrast to the changes observed during oocyte maturation, SMYD3 siRNA treatment was associated with a five-fold increase in NANOG expression in the embryos 48 h post IVF. These results show that experimental down-regulation of SMYD3 expression can alter the expression of some key embryonic transcription factors, especially the pluripotency gene NANOG (Fig. 2b).
SMYD3 had at least four splice variants in bovine
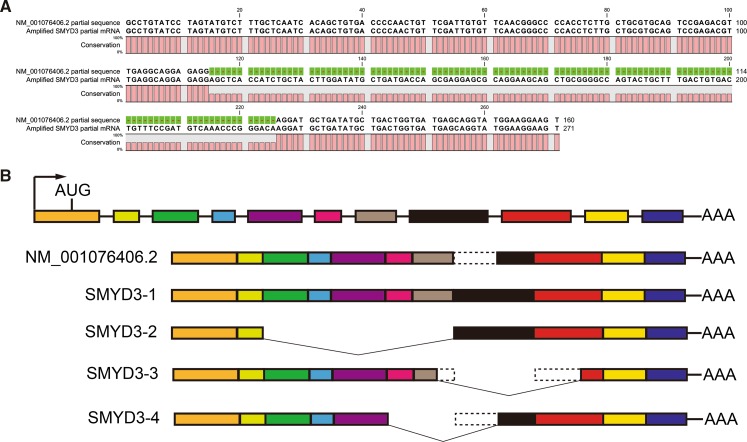
In order to assess the expression of SMYD3, we designed partial-length primers (5′-GCCTGTATCCTAGTATGTCTTTG-3′, 5′-ACTTCCTTCCATACCTGCTC-3′) to amplify the SMYD3 partial mRNA. The size of the predicted product for the RT-PCR was 160 bp; however, the actual amplified product was 111 bp longer than the sequence published by NCBI (NM_001076406.2) (Fig. 3a). To further investigate this phenomenon, we designed full-length primers (5′-GAGGTGTCCTATCTCGGCAA-3′, 5′-AGTAAAGGTACAAGCTGGCTCT-3′) for SMYD3 mRNA and analyzed the total RNA extracted from COCs by RT-PCR. When the products from this extraction were separated by an agarose gel, different sizes of reaction products were observed. These bands were sequenced (Online Resource) and were found to contain four unique SMYD3 splice variants (called SMYD3-1, SMYD3-2, SMYD3-3, and SMYD3-4), which did not align with the NCBI sequence (Fig. 3b).
Fig. 3.
Schematic diagram of the structure of the SMYD3 splice variants. a Alignment of the SMYD3 partial sequence we amplified with the NCBI-published sequence (NM_001076406.2, partial sequence) by the CLC Sequence Viewer Software. b A graph of the SMYD3 splice variants we identified is shown alongside the NCBI-published sequence (NM_001076406.2). The color boxes represent exons and the dotted lines mean deletion parts of exons
Discussion
Oocytes have a remarkable and complex life history, and they finally acquire full capacity to support fertilization and embryogenesis after the completion of a set of intracellular changes. During oocyte growth, the cytoplasm accumulates large amounts of mRNA and proteins (Masui and Clarke 1979; Smith and Richter 1985). There is a short phase of intracellular reprogramming which confers developmental competence on oocytes. Genetic screens have identified numerous maternal-effect genes (Bellotto et al. 2002; Perrimon et al. 1989; Schupbach and Wieschaus 1989), and molecular characterization of the maternal gene products supports the notion that embryogenesis begins during oogenesis (Wolpert et al. 2002).
Our studies showed that SMYD3 mRNA level was maintained constant during oocytes maturation and early embryonic development. SMYD3 may trigger epigenetic reprogramming and regulate gene expression in early embryos. Because SMYD3 is a SET domain-containing H3K4 methyltransferase (Hamamoto et al. 2004, 2006), it could regulate downstream genes as a transcription factor containing histone methyltransferase activity or binding with RNA helicase to form a transcriptional complex with RNA polymerase II (Hamamoto et al. 2004).
Immunofluorescent staining showed a location transition of SMYD3, from being located around chromosomes during oocyte maturation to being located in cytoplasm and nucleus of embryonic development stage. Interestingly, Hamamoto et al. previously found that SMYD3 expression was primarily located in cytoplasm during the G0/G1 phase, and in nuclei in the S and G2/M phase in Huh7 cells (Hamamoto et al. 2004). The expression and localization results suggested that SMYD3 was a maternal gene, which combined with chromosomes at maturation stage might affect the chromosome structure and regulate the gene expression as a histone H3K4me3 tri-methyltransferase.
In order to study the role of SMYD3 in bovine oocyte maturation and early embryonic development, we down-regulated the SMYD3 mRNA level by microinjection of SMYD3 siRNA into GV oocytes and fertilized eggs. The results showed that down-regulation of SMYD3 did not affect oocyte maturation and cleavage, but blocked embryonic development at 8-cell stage. In contrast, the embryonic development rate was not affected when injected SMYD3 siRNA at one cell embryo stage 7 h after fertilization. Embryonic genome activation (EGA) entails a dramatic gene expression that must be faithfully executed for development to proceed. Although EGA was previously thought to be relatively promiscuous due to the extensive chromatin remodeling, results of microarray experiments have demonstrated that the genes involved in transcription, RNA processing and cell cycle regulation are preferentially expressed (Zeng et al. 2004). EGA, by definition, must be initiated by maternally derived proteins and/or mRNAs. SMYD3 may act as a transcription activator regulating a set of maternally inherited factors expressed during oogenesis, or as a transcription factor playing a role in EGA.
The mRNA content of oocytes changes during maturation. This switch in mRNA populations is brought about by both general termination of transcription at GVBD, and degradation of a selective subset of transcripts (Smith and Richter 1985). After fertilization, embryos must clear the previous transcriptional program (Lee et al. 2013) and SMYD3 mRNA may be one of those needs to be degraded. Our SMYD3 expression data also showed that SMYD3 had a certain degree of degradation after fertilization. Therefore, injection of SMYD3 siRNA into fertilized eggs had no effect on subsequent embryonic development.
We further studied how SMYD3 affected the embryonic development by detecting the expression change of the transcription factors, OCT4, NANOG and CDX2. The transcription factors OCT4 and NANOG have important roles in early embryonic development (Khan et al. 2012; Ovitt and Scholer 1998), and are essential for maintenance of the undifferentiated state of ES cells in culture (Boyer et al. 2005; Loh et al. 2006). NANOG is also known to be specifically expressed in the inner cell mass (Khan et al. 2012). Recently, some researches have demonstrated that OCT4, NANOG and other transcription factors play important roles in regulating maternal-embryonic transition and pre-implantation development (Foygel et al. 2008; Lee et al. 2013; Tan et al. 2013). They function as maternal factors and directly regulate hundreds of mRNAs that constitute the first wave of zygotic transcription. Our research showed that down-regulation of SMYD3 altered the transcription level of NANOG. A previous study showed that SMYD3 bound to the motif, 5′-CCCTCC-3′ or 5′-GGAGGG-3′, present in the promoter region of downstream genes (Hamamoto et al. 2004). We also found these two motifs in NANOG promoter region (−2000 and −246 of the NANOG promoter, Chromosome: 5; AC_000162.1 (101869150–101871310), NCBI), and speculate that SMYD3 can bind to NANOG promoter and regulate NANOG transcription directly. This might affect the NANOG regulation network and result in developmental arrest.
Conclusion
Our data provide the first evidence concerning the function of endogenous SMYD3 during bovine pre-implantation embryonic development. The results show that SMYD3 influences blastocyst formation and NANOG expression, indicating that SMYD3 plays an important role in early embryonic development. Yet the precise function of SMYD3 in regulating NANOG expression during early embryonic development remains to be dissected.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by a Grant from Major Program of the Inner Mongolia Natural Science Foundation (No. 2012ZD04), a Grant from the National Key Basic Research Program of China (No. 2010CB134410) and a Grant from NSFC (No. 30860185).
Conflict of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Bachvarova R. Gene expression during oogenesis and oocyte development in mammals. Dev Biol. 1985;1:453–524. doi: 10.1007/978-1-4615-6814-8_11. [DOI] [PubMed] [Google Scholar]
- Bellotto M, Bopp D, Senti KA, Burke R, Deak P, Maroy P, Dickson B, Basler K, Hafen E (2002) Maternal-effect loci involved in Drosophila oogenesis and embryogenesis: P element-induced mutations on the third chromosome. Int J Dev Biol 46:149–157 [PubMed]
- Biel M, Wascholowski V, Giannis A. Epigenetics—an epicenter of gene regulation: histones and histone-modifying enzymes. Angew Chem Int Ed. 2005;44:3186–3216. doi: 10.1002/anie.200461346. [DOI] [PubMed] [Google Scholar]
- Blondin P, Sirard MA. Oocyte and follicular morphology as determining characteristics for developmental competence in bovine oocytes. Mol Reprod Dev. 1995;41:54–62. doi: 10.1002/mrd.1080410109. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122:947–956. doi:10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed]
- Brackett BG, Oliphant G. Capacitation of rabbit spermatozoa in vitro. Biol Reprod. 1975;12:260–274. doi: 10.1095/biolreprod12.2.260. [DOI] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four-and eight-cell stages of preimplantation development. Nature. 1988;332:459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- Foygel K, Choi B, Jun S, Leong DE, Lee A, Wong CC, Zuo E, Eckart M, Reijo Pera RA, Wong WH, Yao MW (2008) A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS ONE 3:e4109 [DOI] [PMC free article] [PubMed]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y (2004) SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol 6:731–740. doi:10.1038/ncb1151 [DOI] [PubMed]
- Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Khan DR, Dubé D, Gall L, Peynot N, Ruffini S, Laffont L, Le Bourhis D, Degrelle S, Jouneau A, Duranthon V (2012) Expression of pluripotency master regulators during two key developmental transitions: EGA and early lineage specification in the bovine embryo. PLoS ONE 7:e34110. doi:10.1371/journal.pone.0034110 [DOI] [PMC free article] [PubMed]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/S0959-437X(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/S0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38:431–440. doi:10.1038/ng1760 [DOI] [PubMed]
- Ma J, Svoboda P, Schultz RM, Stein P. Regulation of zygotic gene activation in the preimplantation mouse embryo: global activation and repression of gene expression. Biol Reprod. 2001;64:1713–1721. doi: 10.1095/biolreprod64.6.1713. [DOI] [PubMed] [Google Scholar]
- Masui Y, Clarke HJ. Oocyte maturation. Int Rev Cytol. 1979;57:185–282. doi: 10.1016/S0074-7696(08)61464-3. [DOI] [PubMed] [Google Scholar]
- Misirlioglu M, Page GP, Sagirkaya H, Kaya A, Parrish JJ, First NL, Memili E. Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc Natl Acad Sci USA. 2006;103:18905–18910. doi: 10.1073/pnas.0608247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovitt CE, Scholer HR. The molecular biology of Oct-4 in the early mouse embryo. Mol Hum Reprod. 1998;4:1021–1031. doi: 10.1093/molehr/4.11.1021. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Engstrom L, Mahowald AP. Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on the X chromosome. Genetics. 1989;121:333–352. doi: 10.1093/genetics/121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599. doi:10.1038/35020506 [DOI] [PubMed]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411 [DOI] [PubMed]
- Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27:396–402. doi: 10.1016/S0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- Schultz RM. Regulation of zygotic gene activation in the mouse. BioEssays News Rev Mol Cell Dev Biol. 1993;15:531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121:101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LD, Richter JD. Synthesis, accumulation, and utilization of maternal macromolecules during oogenesis and oocyte maturation. Biol Fertil. 1985;1:141–178. doi: 10.1016/B978-0-12-492601-1.50013-3. [DOI] [Google Scholar]
- Tan MH, Au KF, Leong DE, Foygel K, Wong WH, Yao MW. An Oct4-Sall4-Nanog network controls developmental progression in the pre-implantation mouse embryo. Mol Syst Biol. 2013;9:632. doi: 10.1038/msb.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/S1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wolpert L, Beddington R, Jessell T, Lawrence P, Meyerowitz E, Smith J (2002) Principles of development, vol 3. Oxford University Press, Oxford
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.