Abstract
Retaining terminal transferase activity of telomerase, the ribonucleoprotein enzyme which add telomeric repeats on chromosome end is thought to be required to prevent cellular ageing. Additionally, telomerase considered as a marker for cell proliferation and immortalization in eukaryotes. We examined telomerase activity in tissues and lymphoid cell culture of Penaeus monodon. Along with telomerase activity, telomere repeats and an attempt on identification of telomerase reverse transcriptase (PmTERT) were made. Telomeric repeat amplification protocol revealed that telomerase-dependent telomeric lengthening has been taking place in P. monodon and the adult tissues were retaining this capacity throughout their lifespan with the highest activity in ovary, testis and lymphoid organ. However, telomerase activity could not be detected in lymphoid cells in culture. The canonical telomeric repeats added by telomerase of lymphoid tissue extract were identified as TTAGG, but pentameric repeats GGTTA and AGGTT were also added by the telomerase. PmTERT protein sequence (partial) shared 100 % identity with the TERT sequence of Daphnia pulex, 27 % sequence identity with Purple sea urchin and 24–25 % with Zebra fish. Undetectable telomerase activity in lymphoid cell culture supports the hypothesis that the inadequate telomerase activity or gene expression may be a reason that prevents neoplastic transformation and spontaneous immortalization of the cells in vitro. Thus, it is envisaged that telomerase activation in lymphoid cells may surmount cellular ageing for in vitro transformation and cell line establishment.
Keywords: Telomerase, Telomere, PmTERT gene, Telomerase reverse transcriptase, Penaeus monodon, Lymphoid cell culture
Introduction
Ageing is a characteristic of normal cells as a result of their limited proliferative capacity, after attaining finite life span; the normal cells cease to divide and enter into a state of senescence (Bibby 2002). Diploid cells divide only a finite number of times in culture (the Hayflick limit) and undergo replicative senescence, due to low or absence of telomerase activity and subsequent progressive shortening of the telomere (Hayflick and Moorhead 1961; Hayflick 1965; Harley 1991; Allsopp et al. 1992; Schumpert et al. 2015). With each cell division, telomeres shorten due to the inability of DNA polymerases to replicate the ends of linear DNA molecules (Morin 1997; Nakamura et al. 1997) and when telomeres reach a critical length, cells enter a non-dividing stage termed senescence (Harley et al. 1990; Cerni 2000; Lang et al. 2004). Further, the role of telomere shortening in cellular senescence has been observed in different cell lineages (Harley et al. 1990; Allsopp et al. 1992), and confirmed that immortalized cells should have telomerase activity and stable telomeres (Kim et al. 1994; Harley 1991; Olovnikov 1973). Accordingly, germ cells and immortal cell lines express telomerase and maintain telomere length through countless cell divisions (Bibby 2002; Singhapol et al. 2013).
Telomerase is specialized ribonucleoprotein complex required for the synthesis of telomere terminal repeats through telomere terminal transferase activity that have an important role in chromosome structure and function. The essential components required for this activity are telomerase reverse transcriptase (TERT), the catalytic component, and telomerase RNA (TR or TERC: telomerase RNA component) which is the template for DNA repeat synthesis (Bryan and Cech 1999; Blackburn 1991, 1994, 2001).
Eventhough the molecular mechanism of telomerase activation that lead to cell immortalization is not known, the association of telomerase activation in cells during cellular immortalization and expression in tumor growth has been illustrated (Counter et al. 1992; Balasubramanian et al. 1999). Moreover, telomerase is shown to have a correlation with cell cycle progression and the abnormal expression of regulatory molecules such as cyclins, cyclin dependent kinases (cdks), and cyclin dependent kinase inhibitors (cdkis) may cause alterations in cell cycle with uncontrolled cell growth (Balasubramanian et al. 1999). In addition, telomerase reverse transcriptase (TERT) has been considered as the limiting factor for telomerase activity in most normal cells (Jayesh et al. 2012). Further, it has been proven that its ectopic expression or activation has led to the extended replicative lifespan of many cell types (Bodnar et al. 1998; Vaziri and Benchimol 1998; He et al. 2009) and their immortalization as well (Venin et al. 2000).
Like vertebrates (Forsyth et al. 2002) and plants (Fitzgerald et al. 1996), the distribution of telomerase activity has been studied in insects (Sasaki and Fujiwara 2000) and other invertebrates including sponges (Koziol et al. 1998), lobster (Klapper et al. 1998a), shrimps (Lang et al. 2004), molluscs (Owen et al. 2007), sea squirts (Sköld et al. 2011) corals and algae (Zielke and Bodnar 2010). Subsequent to the detection of telomerase activity by Lang et al. (2004) in Penaeus japonicus, there has been no further studies in this direction. Eventhough the enzymatic activity of telomerase has been well studied, very little is known about the telomerase genes and their expression profile in many of the cells including those of shrimps.
The highly sensitive PCR-based telomerase activity assay, telomeric repeat amplification protocol (TRAP, Kim et al. 1994; Kim and Wu 1997) is the widely accepted technique used to evaluate telomerase activity in tissues or cell extracts. The identification of Penaeus monodon telomerase reverse transcriptase (PmTERT) gene and its expression product, may lead to discover telomerase inhibitors (Gomez et al. 2002) and activation of telomerase in shrimp cells through PmTERT induction for immortalization.
P. monodon was selected for the present study due to its commercial importance, and the realization of the resistance of its cells to in vitro transformation and immortalization (Jayesh et al. 2015b). Accordingly, telomerase activity was investigated in its tissues and organs and in its cell cultures aiming at telomerase activation in future for attaining immortalization of the cells.
In this study, a hypothesis was drawn that the existence of shortened telomeres and the inadequate expression of telomerase might be the deficit that diminished the proliferative capability preventing spontaneous neoplastic transformation and immortalization of lymphoid cells. To test this hypothesis we measured telomerase in terms of telomerase terminal transferase activity in various life cycle stages of P. monodon, different tissues, organs and primary lymphoid cell culture. An attempt was also made for the identification (amplification and sequencing) of shrimp telomerase reverse transcriptase (PmTERT) gene as well.
Materials and methods
Methodology was designed to test three objectives 1: Detection of telomerase activity in various tissues and organs, and the lymphoid cell culture; 2: Identification of telomeric repeats; and 3: Identification of the PmTERT gene. These objectives were identified based on the hypothesis that inadequate telomerase activity in shrimp cell culture in vitro might be the reason for preventing continuous proliferation and subsequent spontaneous immortalization. All chemicals used for the experiments, unless specifically stated otherwise, were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Experimental animal
White spot syndrome virus negative P. monodon larvae obtained from a local hatchery were stocked and reared in a recirculation aquaculture system (RAS) for shrimp with 15 g l−1 salinity integrated with nitrifying bioreactors. Detritus in the system was managed by the addition of Detrodigest™ National Centre for Aquatic Animal Health (NCAAH) (Cochin, India) and Enterotrophotic™ (NCAAH) to control vibrios at the rate of 100 ml m−3 of water. The larvae were fed commercially available pelleted feed [Higashimaru Feeds (India) Ltd. (Alappuzha, Kerala, India)]. They were maintained for 3 months and the animals (8–12 g) were confirmed negative to WSSV by nested PCR prior to use as the donor animals for various tissues (Jose et al. 2011; 2012).
Preparation of telomerase extracts from tissues and primary cell culture
Telomerase extract from tissues and cell cultures were prepared by following the methodology of Lang et al. (2004) with slight modification. The extracts were prepared from testis, ovary, lymphoid tissue, heart, hepatopancreas, muscle, eyestalk, nerve cord and post larvae (PL-5) of P. monodon having Artemia nauplii as control. Approximately 50 mg each of the sample was transferred to 200 µl CHAPS lysis buffer (10 mM Tris–HCl (pH 7.5), 1 mM EGTA, 0.1 mM benzamidine, 5 mM 2-mercaptoethanol, 0.5 % CHAPS (3[(3-Cholamidopropyl) dimethylammonio]-propanesulfonic acid), and 10 % glycerol (v/v)), crushed with sterile micropestles and inserted into crushed ice for 30 min. The mixture was centrifuged for 30 min at 20,000×g at 4 °C; the supernatant was transferred into fresh 1.5 ml micro centrifuge tubes and frozen in liquid nitrogen, and stored at −80 °C till used (Lang et al. 2004).
Lymphoid cell cultures along with the control cell line, HeLa (Human cervical cancer cell line) were used to extract telomerase. Lymphoid cell culture was developed in SCCM following the methodology explained by Jayesh et al. (2013, 2015a). HeLa was used as control as it had been widely used for the study on telomerase (Bryan et al. 1995; Kim et al. 1994). HeLa was received from the cell repository of National Centre for Cell Science (NCCS), Pune, India and maintained in minimum essential medium (MEM, Eagle’s) with 2 mM l-glutamine and Earle’s balanced salt solution adjusted to contain 0.35 g l−1 sodium bicarbonate, 0.1 mM non-essential amino acids and 1 mM sodium pyruvate supplemented with 10 % fetal bovine serum and antibiotic mixture containing 100 µg ml−1 streptomycin and 100 IU ml−1 penicillin at 37 °C (Heraeus-BBK6220, Hanau, Germany). The growth media were changed twice in a week. Cultured cells were washed with ice-cold PBS, ice-cold wash buffer [10 mM HEPES–KOH (pH 7.5), 1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol], and the cells were removed with cell scrapers (Greiner Bio-One, Frickenhausen, Germany) and centrifuged for 5 min at 1000×g at 4 °C. Cell pellets were resuspended in ice-cold wash buffer, pelleted again, and resuspended in ice-cold CHAPS lysis buffer. Telomerase extracts were prepared as described above. Protein concentration of each extract was determined by BCA (bicinchoninic acid) protein assay kit (Sigma).
Determination of telomerase activity using telomeric repeat amplification protocol (TRAP)
TRAP assay was performed in accordance with Kim et al. (1994) with modifications, to measure telomere terminal transferase activity of telomerase present in tissues or cell extracts. The TS primer (5′-AATCCGTCGAGCAGAGTT-3′) served as forward primer while CX-ext-Shrimp (5′-GTGTAACCTAACCTAACC-3′, Lang et al. 2004) and CX-ext-Lobster (5′-GTGCCTTCCTTCCTTCCTTCCTTCCTA-3′, Klapper et al. 1998a) as the reverse primers for the PCR of extended telomere (in vitro), by the action of telomerase of shrimp tissues/organs and cell culture extracts while using Artemia extracts as the control. The reverse primer CX-ext-HeLa (5′-CCCTTACCCTTACCCTTACCCTAA-3′, Kim et al. 1994) was used for PCR amplification of the extended telomere repeats (in vitro) on TS primer by the action of telomerase from HeLa.
The addition of telomeric repeats on TS primers by the activity of telomerase takes place as elongation steps in the TRAP assay. The 50 µl reaction mixture for elongation is composed of 20 mM Tris–HCl (pH 8.3), 1.5 mM MgCl2, 63 mM KCl, 0.05 % Tween 20, 1 mM EGTA, 0.01 % BSA, 0.5 mM each dNTP, 1 µM TS primer, and tissue and/or cell extracts that contained 6 µg protein. The reaction mixture was incubated at 30 °C for 60 min followed by incubation at 90 °C for 90 s to inactivate telomerase. After elongation, 5 µl sodium acetate (1 M) and 100 µl ethanol were added to the elongation reaction mixture and kept overnight at 4 °C for precipitation. Each precipitate was suspended in 50 µl PCR mixture consisting 5 µl 10× Taq Buffer, 0.2 mM each dNTP, 1 µM CX-ext primer, and 2 U Taq enzyme (New England Biolabs, Hitchin, Herts, UK). PCR was performed in Mastercycler personal™ (Eppendorf, Wesseling, Germany) and the programme used for the amplification was 95 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, extension at 72 °C for 60 s, followed by final extension at 72 °C for 10 min.
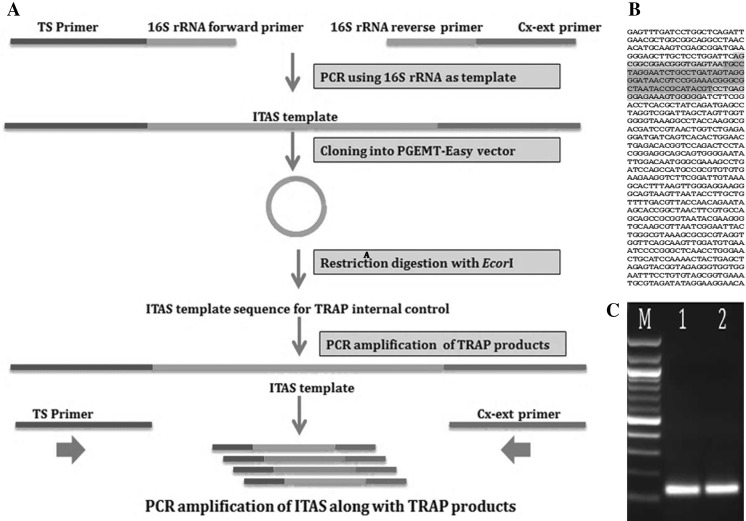
Along with the positive controls internal amplification standard (ITAS) was also used. For ITAS, the primers used were a fusion of TS primer (Bold) and P. aeruginosa MCCB 103 16S rRNA (Accession No: EF053508) gene (underlined) as the forward primer (NP214-F 5′-AAT CCG TCG AGC AGA GTTAGC GGC GGA CGG GTG AGT AA-3′) and CX-ext of shrimp (Bold) and P. aeruginosa MCCB 103 16S rRNA gene (underlined) as the reverse primer (NP214-R5′-GTG TAA CCT AAC CTA ACCCCC CCA CTT TCT CCC TCA GG-3′) which yielded 138-bp PCR product. The ITAS product was cloned into pGEMT, amplified and the purified plasmid was restriction digested with EcoRI, purified products were used as TRAP assay control (Fig. 1; for more details refer Jayesh 2013). Approximately 18 fg template DNA of ITAS was added into every 50 µl telomerase PCR mixture as control (Lang et al. 2004).
Fig. 1.
Designing internal amplification standard (ITAS). a Schematics of the preparation of ITAS as internal control for TRAP assay. b P. aeruginosa MCCB103 16S rRNA gene sequence (GenBank Accession No: EF053508) used for constructing ITAS. The marked region is the 104-bases from 16S rRNA gene contributed in ITAS. c Gel image of PCR amplified TRAP internal control ITAS designed from the 16S rRNA gene sequence of P. aeruginosa, TS and Cx-ext primer sequence, M: 100-bp molecular marker, 1 and 2: amplified ITAS of 138-bp size
The whole PCR product was (50 µl) mixed with 3 µl gel loading dye and analyzed on 15 % non denaturing acrylamide gels. Electrophoresis was performed in 0.5× Tris–borate EDTA (pH 8.3) buffer at a voltage of 155 V for 5–6 h (PowerPac™ HV, Bio-Rad, Hercules, CA, USA), the gel was stained in ethidium bromide solution and SYBR Green (Invitrogen, Carlsbad, CA, USA) for 30 min and photographed using Gel Doc™ XR+ imaging system. (Bio-Rad). Relative telomerase activity and the histogram of the telomerase expression were measured using Image J software [National Institute of Health (NIH), Bethesda, MD, USA]. The telomerase activity observed from nerve cord was defined as 1 for calculating the relative telomerase activity as it was the lowest telomerase activity recorded.
Identification of telomere repeats of P. monodon
TRAP products of lymphoid tissue extract separated on PAGE with DNA fragments ranging from 100 to 700-bp were recovered from the gel using the ‘crush and soak’ method (Maxam and Gilbert 1980). The extracted TRAP product was stored at −20 °C (SANYO, Osaka, Japan) till used. To get the required DNA concentration for sequencing, PCR amplification was carried out using extracted TRAP product as the template. The 50 µl PCR mixture containing 5 µl 10× buffer, 5 µl dNTP (2.5 mM), 2 µl Taq polymerase (2U), 2 µl extracted TRAP products and 2 µl (1 µM) of each TS primer (forward) and CX-ext-shrimp (reverse) primer and the mixture was made up to 50 µl with MilliQ. The PCR programme used for the amplification was 95 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, extension at 72 °C for 1 min, followed by final extension at 72 °C for 10 min. The whole PCR product was subjected for gel purification using GenElute™ Gel Extraction kit (Sigma) and was subjected for further purification using GenElute™ PCR Clean-Up kit (Sigma) to remove the excess salt, and subsequently sequenced. The sequence of TRAP products was subjected for the identification of telomeric tandem repeats of P. monodon by Fourier transformation using the spectral repeat finder (SRF) (Sharma et al. 2004). Contiguous telomeric sequence and its position were analyzed along with the identification of interstitial occurrence of telomeric repeats and its association with other sequences in P. monodon.
Attempt on identification of telomerase reverse transcriptase gene (PmTERT) in P. monodon
As the genome sequence of P. monodon was not available, the identification of TERT sequences in P. monodon using available TERT protein sequence from various organisms (Bombyx mori, Tribolium castaneum, Danio rerio, Apis mellifera, Ciona intestinalis) was found difficult. To tackle the issue, we hypothesized that the TERT sequence of a crustacean might show some similarity with that of P. monodon and the available genome sequence of a crustacean was selected to test this hypothesis. The only available crustacean whole genome sequence was from Daphnia pulex (Colbourne et al. 2011) which was used as the representative crustacean genome. The available TERT protein sequences of invertebrates and vertebrates were used from the database http://telomerase.asu.edu/ (Podlevsky et al. 2007). With this protein sequences as query the D. pulex genome database (Assembly version Daphnia pulex v1.0) at http://genome.jgi-psf.org was searched using the TBLASTN algorithm. The protein sequence and the GenBank accession numbers used for the TBLASTN search were of C. intestinalis (EF077623), Ciona savignyi (EF514225), A. mellifera (NM001040681), B. mori (DQ467676), Bombyx mandarina (DQ467677), T. castaneum, (NM001040706), Caenorhabditis elegans, (NM059972) and Caenorhabditis remanei (NM 059973). From among them the complimentary TERT gene sequence present in D. pulex genome was used for designing the primer using the software GeneTool Lite™ (version 1.0) to amplify PmTERT from P. monodon.
P. monodon larvae used in this study were obtained from a shrimp hatchery at Cochin, India (Abad shrimp hatchery, Cochin). Approximately 2000 nauplii were used for RNA extraction. The nauplii were collected through a sieve and kept in RNAlater overnight at 4 °C, drained off the RNAlater and stored at −80 °C till use. This sample was directly used for RNA extraction using TRI reagent (Sigma). RNA concentration and quality were determined by optical density (OD 260/280 nm) measurement using a UV–visible spectrophotometer (Hitachi, Tokyo, Japan). An aliquot of 5 µg RNA was subjected to cDNA synthesis. The 20 µl reaction mix containing M-MuLV reverse transcriptase (200 U), RNase inhibitor (8 U), Oligo (dT)12 primer (40 pmol), dNTP mix (1 mM), RTase buffer (1×) and MgCl2 (2 mM) and the synthesis was performed by incubating at 42 °C for 1 h. PmTERT was amplified from P. monodon through PCR using the primers NP593 F-5′ CGC TCG CCA TTA GTG GTC AGA TAA AGG AAA 3′ and NP593R -5′ TGA AGA AAA TTC GTC TGG CAT CCA GTG ATG 3′ designed from the TERT gene of D. pulex. An aliquot of 25 µl PCR mix contained 1 µl cDNA, 0.5 U Taq DNA polymerase, 200 µM dNTP mix, 10 pmol each of forward and reverse primers and 1× PCR buffer. The PCR programme used for the amplification of PmTERT was 95 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 65 °C for 30 s, extension at 72 °C for 1 min, followed by final extension at 72 °C for 10 min. After PCR, the partial sequence of PmTERT gene was cloned into pGEM-T Easy vector (Promega, Madison, WI, USA) and transformed into E. coli DH5α. The cloned plasmid was used for sequencing and the sequence was subjected for BLAST search in various genome/proteome databases viz; European Nucleotide Archive (ENA) http://www.ebi.ac.uk/ena/, D. pulex genome database in http://genome.jgi-psf.org. Protein Knowledge base (UniProtKB) http://www.uniprot.org/uniprot, and the sequence similarity identification was performed (Altschul et al. 1997).
Results
Telomere terminal transferase activity in various tissues and lymphoid cell culture of P. monodon
Results of PCR based telomeric repeat amplification protocol (TRAP) showed the presence/absence of typical DNA ladder formation by telomerase activity confirming the presence/absence of telomere terminal transferase activity in the extracts of various tissues and organs of P. monodon, post larvae, Artemia nauplii, primary lymphoid cell culture, and HeLa, as presented in Fig. 2A1, A2. The internal amplification standard (ITAS) was used to normalize the peak to perform the densitometric analysis and to confirm the amplification of the product with TS and Cx-est primers (Fig. 4). In this experiment, telomerase activity was detected in testis, ovary, lymphoid organ, heart, hepatopancreas, muscle, eyestalk, nerve cord and post larvae of P. monodon, with slight variation in its expression. The histogram of telomerase activity of the extract was measured by densitometric analysis using Image J software (Fig. 2b). Moreover, the relative telomerase activity of various tissues of P. monodon was calculated; for nerve cord 1 ± 0.08, heart 1.23 ± 0.18, eyestalk 1.83 ± 0.21, muscles 2 ± 0.05, post larvae (PL) 2.07 ± 0.15, hepatopancreas 2.57 ± 0.26, ovary 3.5 ± 0.15, testis 3.5 ± 0.14 and lymphoid organ 3.5 ± 0.07 (Fig. 3). As shown in Fig. 3, telomerase was expressed in all tissues analysed, with the highest relative telomere terminal transferase activity in ovary, testis and lymphoid organ and moderate activity in eyestalk, muscles, hepatopancreas and tissues from post larvae. The extracts from HeLa and Artemia nauplii gave rise to typical ladder formation in TRAP assay due to telomere terminal transferase activity. However, telomerase activity was found to be either negative or inadequate in primary lymphoid cell culture. Moreover, results from control cells, tissues and the amplified internal amplification standard (ITAS) confirmed that the experimental protocols were valid without any ambiguity.
Fig. 2.
Telomerase activity in various tissues and lymphoid cell culture along with controls. A1 TRAP assay results: M-100-bp Marker, 1, 2 HeLa cells (+ve control), 3 hepatopancreas, 4 heart, 5 post larvae, 6 Artemia, 7 muscles, 8 nerve cord, 9 ovary, 10 testis, 11 eyestalk, ITAS internal amplification standard, A2 TRAP assay: M-100-bp molecular marker, HeLa cells (+ve control), 1, 2, 3, 4, 5 primary lymphoid cell culture, Lymp-lymphoid organ, PL post larvae. b Telomerase activity scored as histogram from various tissue and lymphoid cell along with controls. 1 HeLa cell line, 2 Artemia, 3 ovary, 4 testis, 5 lymphoid, 6 hepatopancreas, 7 eyestalk, 8 muscle, 9 post larvae, 10 heart, 11 nerve cord, 12 primary lymphoid cell culture
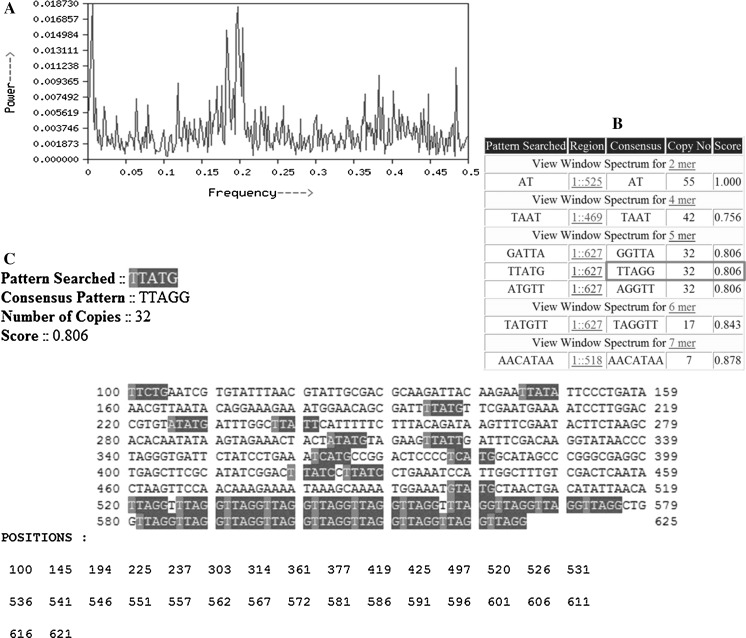
Fig. 4.
Identification of the telomeric repeats added on the artificial chromosome end (TS primer) by lymphoid tissue telomerase extract of P. monodon using Spectral Repeat Finder (SRF). a Fourier Spectrum telomeric sequence designed from SRF, b output from SRF, red box indicate that TTAGG are repeated 32 times in the given telomeric sequence, c position of TTAGG repeats in the telomeric sequence identified from SRF indicating contiguous TTAGG sequences
Fig. 3.
Relative telomerase activity in various tissues of P. monodon and its post larvae. The results represent the mean ± SD (n = 3)
Identification of canonical telomeric repeats added by the telomere terminal transferase (telomerase) of lymphoid tissue from P. monodon
Sequencing results of the telomeric repeats added on to the artificial chromosome end (TS primer) by the action of telomere terminal transferase activity (telomerase) of the enzymatic preparation from lymphoid tissue revealed that the pentameric TTAGG repeats were the canonical telomeric repeats of P. monodon. Meanwhile, the results from spectral repeat finder (SRF, Fig. 4) suggested that in addition to the known TTAGG pentameric repeats of insects and invertebrates, telomere terminal transferase activity of P. monodon added two other pentameric repeats GGTTA and AGGTT (Fig. 4b). Further, the sequence analysis of the PCR products of telomerase activity showed the interstitial occurrence of TTAGG and association with certain unrelated sequences and low-copy repeats. However, among the repeats, the TTAGG was contiguous with 32 repeats (Fig. 4c), (TTAGG)32 suggesting their highest possibility as telomeric repeats of P. monodon.
Identification of PmTERT genes
Direct PCR amplification of PmTERT gene from P. monodon using the primers designed from the TERT gene sequence of various invertebrates and vertebrates including B. mori,T. castaneum, D. rerio, A. mellifera,C. intestinalis and Human telomerase reverse transcriptase (Table 1) were unsuccessful. Meanwhile, the TBLASTN search of D. pulex genome using the C. intestinalis (EF077623) and C. savignyi (EF514225) TERT protein sequences as query successfully identified the putative exons that were homologous with TERT gene sequence of D. pulex. However, TERT protein sequence used from other invertebrate species such as A. mellifera (NM001040681), B. mori (DQ467676), B. mandarina (DQ467677), T. castaneum (NM001040706), C. elegans (NM059972) and C. remanei (NM 059973) did not give any results from the TBLASTN search indicating its divergence with crustacean TERT sequence. Even though C. intestinalis and C. savignyi TERT protein sequences were homologous with D. pulex whole genome at the scaffold #47, C. intestinalis TERT protein sequence was used for further analysis. Moreover, the D. pulex TERT protein was found to be homologous with C. intestinalis TERT located in the scaffold #47 region in whole genome at 332981–333070 and at 333746–333883 DNA base pairs (Fig. 5a), and this region was used for designing gene-specific primers to amplify partial cDNA fragments of P. monodon TERT gene, and designated as PmTERT.
Table 1.
List of primers designed from various species to amplify PmTERT gene from P. monodon
| S. No | Species | Primer sequence |
|---|---|---|
| 1 | Bombyx mori | F-5′-TTTATCGAAATATGAATACCCCC-3′ |
| R-5′-CCATTCCATATATTCCAGTGAA-3′ | ||
| F-5′-GTGAAACGTCGATTTCTAGCTTAA-3′ | ||
| R-5′ GGTATTAAATTGGAACATTTCCATGTT-3′ | ||
| 2 | Tribolium castaneum | F-5′-ATGGTCCACTACTATCGC-3′ |
| R-5′-AAATAACTCGCATCCACCTC-3′ | ||
| 3 | Danio rerio | F-5′CCCCAAGCACGCGCACAGATGTC-3′ |
| R-5′ATGCTGTGTTTACGAGTGTGTGT-3′ | ||
| 4 | Apis mellifera | F-5′TAGCTTAAGGGTTGTTGTTTT-3′ |
| R-5′ATGAAAAAAATGTGATAAAATA-3′ | ||
| 5 | Ciona intestinalis | F-5′TTCGGTCGGTTTTGTATCTCCA-3′ |
| R-5′CACAGGGGAGGCAGGGATAGT-3′ | ||
| 6 | Homo sapiens | F-5′GCCGAATTCTGCCGTTGCCCAAGAGG-3′ |
| R-5′GCGTGGATCCCAAGCAGCTCCAGA-3′ |
Fig. 5.
Identification of PmTERT gene from P. monodon. a TBLASTN results suggesting that the C. intestinalis TERT sequence is complimentary with the sequence of Daphnia pulex genome scaffold 47, confirming the complimentary region of Daphnia pulex genome containing putative TERT gene sequence. b DNA sequence along with translated amino acid sequence (partial) of PmTERT gene. c BLAST result of PmTERT partial gene sequence and its identity with water flea, zebra fish and sea urchin
The primers NP593F-5′ CGC TCG CCATTA GTG GTC AGA TAA AGG AAA 3′ and NP593R-5′ TGA AGA AAA TTC GTC TGG CAT CCA GTG ATG 3′ were designed from the region of scaffold #47 and PCR amplification was successfully carried out; a 68-bp sized product (PmTERT) was obtained from P. monodon, and sequenced. PmTERT gene sequence was further converted to amino acid sequence and used for BLAST analysis (Fig. 5b). The BLAST search in European Nucleotide Archive (ENA) resulted in 100 % identity with EMBL-CDS: EFX76361.1: D. pulex telomerase reverse transcriptase, and the scaffold #47 of the D. pulex genome database in http://genome.jgi-psf.org. Further to this identity, the sequence similarity identification revealed that PmTERT protein sequence (partial) shared 100 % identity with the TERT sequence of D. pulex (GenBank accession no. E9GVZ2), 27 % sequence identity with Strongylocentrotus purpuratus (Purple sea urchin) and only 24–25 % with D. rerio (Zebra fish) confirming its divergence (Fig. 5c).
Discussion
The results from telomerase activity demonstrate that telomerase is constitutively active in all tissues tested from P. monodon. The typical DNA ladder formation on using the tissue extract during TRAP assay was confirmed by the extension of TTAGG oligonucleotide repeats added by the presence of complimentary sequence on RNA components of telomerase. This implies that telomerase-dependent telomeric lengthening has been taking place in P. monodon and the adult tissues have been retaining this capacity throughout their lifespan. The need for telomerase activity may be explained from the active cell proliferation required in various stages of its life cycle for attaining growth including regeneration of exoskeleton during molting, suggesting maintenance of stem cellness throughout life (Vogt 2012). Such constitutive activity of telomerase has been reported from lobsters (Klapper et al. 1998a), bivalves (Owen et al. 2007), shrimps (Lang et al. 2004), fishes (Klapper et al. 1998b; Anchelin et al. 2011) and insects (Sasaki and Fujiwara 2000; Mohan et al. 2011). Moreover, telomerase activity has been considered as cell proliferation and cell aging marker (Vogt 2012; Belair et al. 1997). Contradictory to this, most of the human somatic tissues lack telomerase activity (Lang et al. 2004) and has been detected only in germline and tumor cells (Kim et al. 1994; Wright et al. 1996).
Lang et al. (2004) suggested that among various tissues tested from the shrimp P. japonicus, consistent telomerase expression was observed in ovary and testis, and explained that this was due to the presence of germline and low level of differentiated cells along with active cell division. In the same line, the present study support the findings of Lang et al. (2004) with the highest (relative) telomerase activity detected in ovary, testis and lymphoid organ of P. monodon. Meanwhile, Klapper et al. (1998a) reported highest telomerase activity in hepatopancreas and heart tissue of the lobster Homarus americanus. Contradictory to the findings of Lang et al. (2004), in the present study, telomerase activity was not detected in lymphoid cell culture in vitro, despite its activity in tissue (in vivo) counterpart. This leads to the hypothesis that inadequate telomerase activity may be a factor that prevents neoplastic transformation and spontaneous immortalization of shrimp cells in vitro. However, the reasons for the loss of telomerase activity in shrimp cells in vitro comparing to its continued activity in vivo could not be explained at this stage, and it is open for further research.
Sequencing results showed that the pentameric repeats, TTAGG, were getting added on to the TS primers by the telomeric terminal transferase (telomerase) present in the enzymatic preparation from lymphoid tissue of P. monodon. These repeats were added corresponding to the complimentary RNA sequence present in telomerase of the animal, suggesting that the TTAGG repeats were synthesized based on telomerase of the shrimp. Moreover, the sequence analysis of added telomere repeats on to TS primers and the results from spectral repeat finder revealed the interstitial occurrence of the telomere repeats. This supported the finding of Mohan et al. (2011) on the interstitial occurrence of telomeric repeats and its association with certain unrelated low-copy repeats in mealy bug, Planococcus lilacinus.
The synthesis of canonical TTAGG telomeric sequence by telomerase has been reported from many species of lobsters (Klapper et al. 1998a), shrimps (Lang et al. 2004), and insects (Sasaki and Fujiwara 2000; Vitkova et al. 2005; Monti et al. 2011), and Frydrychová et al. (2004) investigated the phylogenetic distribution of TTAGG telomeric repeats in insects (Frydrychová et al. 2004). Besides, the telomerase-independent mechanism of telomere maintenance, such as the alternative lengthening of telomeres (ALT) has been reported in Drosophila (Pardue et al. 1996), yeasts (Lundblad and Blackburn 1993) and human (Bryan et al. 1995, 1997; Henson et al. 2002). In this context the telomerase-dependent telomere maintenance has been considered as conserved mechanism for maintaining long-term cell proliferation capability (Klapper et al. 1998a). The telomere sequence (TTAGGG)n is conserved among many phyla in Animalia, including Vertebrata (Meyne et al. 1989), Mollusca (Wang and Guo 2001; Gallardo-Escarate et al. 2005; Sakai et al. 2005) Annelida (Jha et al. 1995), Platyhelminthes, (Joffe et al. 1996) Onychophora, (Vitkova et al. 2005) and Echinodermata (Okazaki et al. 1993). However, another telomere type, (TTAGGC)n, is found in Nematoda (Muller et al. 1991), and (TTAGG)n in Arthropoda (Okazaki et al. 1993; Sahara et al. 1999; Frydrychová et al. 2004; Vitkova et al. 2005; Schumpert et al. 2015) including shrimps (Lang et al. 2004) and lobsters (Klapper et al. 1998a) in the kingdom Animalia (Sakai et al. 2007). More recently, Gomes et al. (2010) concluded that with the exception of Nematodes and Arthropods, the (TTAGGG)n sequence was conserved in most Metazoa. Precisely, the results from the present study reinstated the fact that the distribution patterns of TTAGG repeats were essentially consistent with telomerase-dependent telomeric maintenance in the Phylum Arthropoda (Okazaki et al. 1993; Klapper et al. 1998a; Sahara et al. 1999; Sasaki and Fujiwara 2000; Lang et al. 2004; Vitkova et al. 2005; Monti et al. 2011).
Telomerase reverse transcriptase (TERT) is the catalytic subunit of telomerase, that synthesizes telomeric DNA (TTAGG)n in the case of Arthropods or (TTAGGG)n in the case of most of the Metazoa (excepting Arthropods) directly onto chromosome ends. Various studies have proven that the ectopic expressions of TERT in cells induce the telomerase activity and lengthen telomeres in human (Bodnar et al. 1998; Counter et al. 1998; Vaziri and Benchimol 1998; Perrem et al. 2001), dog (Techangamsuwan et al. 2009) and mouse (Armstrong et al., 2000). Meanwhile, immortalization of cells was successfully performed with the ectopic expression of TERT in epithelial cells of human (Morales et al. 1999; Piao et al. 2005) and goat (He et al. 2009). As a corollary, identification of gene encoding TERT in P. monodon (PmTERT) is of great importance as the envisaged immortalization of shrimp cells could be possible with the ectopic expression of PmTERT. The TERT cDNA has been isolated and characterized from yeasts (Counter et al. 1997) and vertebrates such as fish (Rao et al. 2011), frog (Kuramoto et al. 2001), chicken (Delany and Daniels 2004), mouse (Greenberg et al. 1998), hamster (Guo et al. 2001), dog (Nasir et al. 2004) and human (Meyerson et al. 1997); invertebrates such as ascidians (Li et al. 2007), protozoan (Bryan et al. 1998), and insects (Gillis et al. 2008; Osanai et al. 2006). However, the telomerase reverse transcriptase has not yet been identified from P. monodon. In the present study, partial sequence of P. monodonTERT (PmTERT) could be identified and characterized. Moreover, the BLAST search analysis suggested its similarity with TERT of Daphnia, zebra fish and sea squirt. In the context of the non-availability of TERT gene sequence of P. monodon, the partial gene sequence identified from this study is vital for characterization of PmTERT of the shrimp.
In conclusion, telomerase activity was detected in all tissues tested and the TTAGGn repeats were identified as the canonical telomeric repeats from P. monodon. However, telomerase activity was not detected from the primary lymphoid cell culture indicating the inactivation/non expression of telomerase gene or inadequate telomerase in cells in vitro. Additionally, partial putative TERT region of P. monodon (PmTERT) was identified which could be used for gene mining and eventual transfer of the active telomerase reverse transcriptase to lymphoid cells in vitro leading to expression of the enzyme and subsequent proliferation and immortalization of the cells. The reason for loss of telomerase activity in shrimp cell in vitro has to be addressed which might lead to the discovery of telomerase inhibitors in P. monodon cell culture and removal of the blocks, necessarily required for shrimp cell line development.
Acknowledgments
This research was supported by Department of Biotechnology, Government of India (BT/PR8050/AAQ/03/289/2006 and BT/PR5126/AAQ/3/591/2012). The first author thanks DBT for Fellowship.
References
- Allsopp RC, Vaziri H, Pattersont C, Goldsteint S, Younglai EV, Futcher AB, Greidert CW, Harley CB. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchelin M, Murcia L, Alcaraz-Pérez F, García-Navarro EM, Cayuela ML. Behaviour of telomere and telomerase during aging and regeneration in zebrafish. PLoS One. 2011;6:e16955. doi: 10.1371/journal.pone.0016955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Lako M, Lincoln J, Cairns PM, Hole N. mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mech Dev. 2000;97:109–116. doi: 10.1016/S0925-4773(00)00423-8. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Kim KH, Ahmad N, Mukhtar H. Activation of telomerase and its association with G1-phase of the cell cycle during UVB-induced skin tumorigenesis in SKH-1 hairless mouse. Oncogene. 1999;18:1297–1302. doi: 10.1038/sj.onc.1202417. [DOI] [PubMed] [Google Scholar]
- Belair CD, Yeager TR, Lopez PM, Reznikoff CR. Telomerase activity: a biomarker of cell proliferation, not malignant transformation. Proc Natl Acad Sci USA. 1997;94:13677–13682. doi: 10.1073/pnas.94.25.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby MC. Introduction to telomeres and telomerase. In: Double JA, Thompson MJ, editors. Telomeres and telomerase: methods and protocols. Totowa: Humana Press; 2002. [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Telomeres: no end in sight. Cell. 1994;77:621–623. doi: 10.1016/0092-8674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/S0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt ShE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddell RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Sperger JM, Chapman KB, Cech TR. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerni C. Telomeres, telomerase, and myc: an update. Mutat Res. 2000;462:31–47. doi: 10.1016/S1383-5742(99)00091-5. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Cáceres CE, Carmel L, Casola C, Choi JH, Detter JC, Dong Q, Dusheyko S, Eads BD, Fröhlich T, Geiler-Samerotte KA, Gerlach D, Hatcher P, Jogdeo S, Krijgsveld J, Kriventseva EV, Kültz D, Laforsch C, Lindquist E, Lopez J, Manak JR, Muller J, Pangilinan J, Patwardhan RP, Pitluck S, Pritham EJ, Rechtsteiner A, Rho M, Rogozin IB, Sakarya O, Salamov A, Schaack S, Shapiro H, Shiga Y, Skalitzky C, Smith Z, Souvorov A, Sung W, Tang Z, Tsuchiya D, Tu H, Vos H, Wang M, Wolf YI, Yamagata H, Yamada T, Ye Y, Shaw JR, Andrews J, Crease TJ, Tang H, Lucas SM, Robertson HM, Bork P, Koonin EV, Zdobnov EM, Grigoriev IV, Lynch M, Boore JL (2011) The ecoresponsive genome of Daphnia pulex. Science 331:555–561 [DOI] [PMC free article] [PubMed]
- Counter CM, Avilion AA, LeFeuvrel CE, Stewart NG, Greider CW, Harley CB, Bacchettil S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Ellisen Leif W, Caddle SD, Haber DA, Weinberg RA. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- Delany ME, Daniels LM. The chicken telomerase reverse transcriptase (chTERT): molecular and cytogenetic characterization with a comparative analysis. Gene. 2004;339:61–69. doi: 10.1016/j.gene.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, McKnight TD, Shippen DE. Characterization and developmental patterns of telomerase expression in plants. Proc Natl Acad Sci USA. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth NR, Wright WE, Shay JW. Telomerase and differentiation inmulticellular organisms: turn it off, turn it on, turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- Frydrychová R, Grossmann P, Trubac P, Vítková M, Marec F. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome. 2004;47:163–178. doi: 10.1139/g03-100. [DOI] [PubMed] [Google Scholar]
- Gallardo-Escarate C, Alvarez-Borrego J, Del Rio-Portilla MA, Von Brand-Skopnik E, Cross I, Merlo A, Rebordinos L. Karyotype analysis and chromosomal localization by FISH of ribosomal DNA, telomeric (TTAGGG)n and (GATA)n repeats in Haliotis fulgens and H. corrugata (Archeogastropoda: Haliotidae) J Shellfish Res. 2005;24:1153–1159. doi: 10.2983/0730-8000(2005)24[1153:KAACLB]2.0.CO;2. [DOI] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–638. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Gomes NMV, Shay JW, Wright WE. Telomere biology in Metazoa. FEBS Lett. 2010;584:3741–3751. doi: 10.1016/j.febslet.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D, Mergny JL, Riou JF. Detection of telomerase inhibitors based on G-quadruplex ligands by a modified telomeric repeat amplification protocol assay. Cancer Res. 2002;62:3365–3368. [PubMed] [Google Scholar]
- Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- Guo W, Okamoto M, Park NH, Lee YM. Cloning and expression of hamster telomerase catalytic subunit cDNA. Int J Mol Med. 2001;8:73–78. doi: 10.3892/ijmm.8.1.73. [DOI] [PubMed] [Google Scholar]
- Harley CB. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Cancer Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- He YL, Wu YH, He XN, Liu FJ, He XY, Zhang Y. An immortalized goat mammary epithelial cell line induced with human telomerase reverse transcriptase (hTERT) gene transfer. Theriogenology. 2009;71:1417–1424. doi: 10.1016/j.theriogenology.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- Jayesh P (2013) Development of lymphoid cell culture system from Penaeus monodon and molecular approaches for its transformation. Ph.D. Thesis. Cochin University of Science and Technology, India
- Jayesh P, Seena J, Singh ISB. Establishment of shrimp cell lines: perception and orientation. Ind J Virol. 2012;23:244–251. doi: 10.1007/s13337-012-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayesh P, Seena J, Philip R, Singh ISB. A novel medium for the development of in vitro cell culture system from Penaeus monodon. Cytotechnology. 2013;65:307–322. doi: 10.1007/s10616-012-9491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayesh P, Philip R, Singh ISB. Multifactorial interaction of growth factors on Penaeus monodon lymphoid cells and the impact of IGFs in DNA synthesis and metabolic activity in vitro. Cytotechnology. 2015;67:559–571. doi: 10.1007/s10616-014-9697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayesh P, Philip R, Singh ISB. Transgene expression in Penaeus monodon cells: evaluation of recombinant baculoviral vectors with shrimp specific hybrid promoters. Cytotechnology. 2015 doi: 10.1007/s10616-015-9872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AN, Dominquez I, Balajee AS, Hutchinson TH, Dixon DR, Natarajan AT. Localization of a vertebrate telomeric sequence in the chromosomes of two marine worms (phylum Annelida: class Polychaeta) Chromosome Res. 1995;3:507–508. doi: 10.1007/BF00713966. [DOI] [PubMed] [Google Scholar]
- Joffe BI, Solovei IV, Macgregor HC. Ends of chromosomes in Polycelis tenuis (Platyhelminthes) have telomere repeat TTAGGG. Chromosome Res. 1996;4:323–324. doi: 10.1007/BF02263686. [DOI] [PubMed] [Google Scholar]
- Jose S, Jayesh P, Mohandas A, Philip R, Singh ISB. Application of primary haemocyte culture of Penaeus monodon in the assessment of cytotoxicity and genotoxicity of heavy metals and pesticides. Mar Environ Res. 2011;71:169–177. doi: 10.1016/j.marenvres.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Jose S, Jayesh P, Sudheer NS, Poulose G, Mohandas A, Philip R, Bright ISB. Lymphoid organ cell culture system from Penaeus monodon (Fabricius) as a platform for white spot syndrome virus and shrimp immune-related gene expression. J Fish Dis. 2012;35:321–334. doi: 10.1111/j.1365-2761.2012.01348.x. [DOI] [PubMed] [Google Scholar]
- Kim NW, Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP) Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Klapper W, Kuëhne K, Singh KK, Heidorn K, Parwaresch R, Krupp G. Longevity of lobsters is linked to ubiquitous telomerase expression. FEBS Lett. 1998;439:143–146. doi: 10.1016/S0014-5793(98)01357-X. [DOI] [PubMed] [Google Scholar]
- Klapper W, Heidorn W, Kuëhne K, Parwaresch R, Krupp G. Telomerase activity in ‘immortal fish’. FEBS Lett. 1998;434:409–412. doi: 10.1016/S0014-5793(98)01020-5. [DOI] [PubMed] [Google Scholar]
- Koziol C, Borojevic R, Steffen R, Müller WEG. Sponges (Porifera) model systems to study the shift from immortal to senescent somatic cells: the telomerase activity in somatic cells. Mech Ageing Devt. 1998;100:107–120. doi: 10.1016/S0047-6374(97)00120-6. [DOI] [PubMed] [Google Scholar]
- Kuramoto M, Ohsumi K, Kishimoto T, Ishikawa F. Identification and analyses of the Xenopus TERT gene that encodes the catalytic subunit of telomerase. Gene. 2001;277:101–110. doi: 10.1016/S0378-1119(01)00684-9. [DOI] [PubMed] [Google Scholar]
- Lang GH, Wang Y, Nomura N, Matsumura M. Detection of telomerase activity in tissues and primary cultured lymphoid cells of Penaeus japonicus. Mar Biotechnol. 2004;6:347–354. doi: 10.1007/s10126-003-0038-0. [DOI] [PubMed] [Google Scholar]
- Li Y, Yates JA, Chen JJ-L. Identification and characterization of sea squirt telomerase reverse transcriptase. Gene. 2007;400:16–24. doi: 10.1016/j.gene.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-H. [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled dNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/S0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785–795 [DOI] [PubMed]
- Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan KN, Rani BS, Kulashreshta PS, Kadandale JS. Characterization of TTAGG telomeric repeats, their interstitial occurrence and constitutively active telomerase in the mealybug Planococcus lilacinus (Homoptera; Coccoidea) Chromosoma. 2011;120:165–175. doi: 10.1007/s00412-010-0299-0. [DOI] [PubMed] [Google Scholar]
- Monti V, Giusti M, Bizzaro D, Manicardi GC, Mandrioli M. Presence of a functional (TTAGG)n telomere-telomerase system in aphids. Chromosome Res. 2011;19:625–633. doi: 10.1007/s10577-011-9222-7. [DOI] [PubMed] [Google Scholar]
- Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y, Wilson KS, White MA, Wright WE, Shay JW. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- Morin GB. The implications of telomerase biochemistry for human disease. Eur J Cancer. 1997;33:750–760. doi: 10.1016/S0959-8049(97)00061-0. [DOI] [PubMed] [Google Scholar]
- Muller F, Wicky C, Spicher A, Tobler H. New telomere formation after developmentally regulated chromosomal breakage during the process of chromatin diminution in Ascaris lumbricoides. Cell. 1991;67:815–822. doi: 10.1016/0092-8674(91)90076-B. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nasir L, Gault E, Campbell S, Veeramalai M, Gilbert D, McFarlane R, Munro A, Argyle DJ. Isolation and expression of the reverse transcriptase component of the Canis familiaris telomerase ribonucleoprotein (dogTERT) Gene. 2004;336:105–113. doi: 10.1016/j.gene.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Okazaki S, Tsuchida K, Maekawa H, Ishikawa H, Fujiwara H. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm Bombyx mori and in other insects. Mol Cell Biol. 1993;13:1424–1432. doi: 10.1128/MCB.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov AM. A theory of marginotomy: the incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Osanai M, Kojima KK, Futahashi R, Yaguchi S, Fujiwara H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle) Gene. 2006;376:281–289. doi: 10.1016/j.gene.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Owen R, Sarkis S, Bodnar A. Developmental pattern of telomerase expression in the sand scallop, Euvola ziczac. Invertebr Biol. 2007;126:40–45. doi: 10.1111/j.1744-7410.2007.00074.x. [DOI] [Google Scholar]
- Pardue ML, Danilevskaya ON, Lowenhaupt K, Slot F, Traverse KL. Drosophila telomeres: new views on chromosome evolution. Trends Genet. 1996;12:48–52. doi: 10.1016/0168-9525(96)81399-0. [DOI] [PubMed] [Google Scholar]
- Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol Cell Biol. 2001;21:3862–3875. doi: 10.1128/MCB.21.12.3862-3875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao CQ, Liu L, Zhao YL, Balajee AS, Suzuki M, Hei TK. Immortalization of human small airway epithelial cells by ectopic expression of telomerase. Carcinogenesis. 2005;26:725–731. doi: 10.1093/carcin/bgi016. [DOI] [PubMed] [Google Scholar]
- Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen J. The telomerase database. Nucleic Acids Res. 2007;36:339–343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, Wang T, Li M, Li Z, Hong N, Zhao H, Yan Y, Lu W, Chen T, Wang W, Lim M, Yuan Y, Liu L, Zeng L, Wei Q, Guan G, Li C, Hong Y. Medaka tert produces multiple variants with differential expression during differentiation in vitro and in vivo. Int J Biol Sci. 2011;7:426–439. doi: 10.7150/ijbs.7.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara K, Marec F, Traut W. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999;7:449–460. doi: 10.1023/A:1009297729547. [DOI] [PubMed] [Google Scholar]
- Sakai M, Okumura S, Yamamori K. Telomere analysis of pacific abalone Haliotis discus hannai chromosomes by fluorescence in situ hybridization. J Shellfish Res. 2005;24:1149–1151. doi: 10.2983/0730-8000(2005)24[1149:TAOPAH]2.0.CO;2. [DOI] [Google Scholar]
- Sakai M, Okumura S, Onuma K, Senbokuya H, Yamamori K. Identification of a telomere sequence type in three sponge species (Porifera) by fluorescence in situ hybridization analysis. Fish Sci. 2007;73:77–80. doi: 10.1111/j.1444-2906.2007.01304.x. [DOI] [Google Scholar]
- Sasaki T, Fujiwara H. Detection and distribution patterns of telomerase activity in insects. Eur J Biochem. 2000;267:3025–3031. doi: 10.1046/j.1432-1033.2000.01323.x. [DOI] [PubMed] [Google Scholar]
- Schumpert C, Nelson J, Kim E, Dudycha JL, Patel RC. Telomerase activity and telomere length in Daphnia. PLoS One. 2015;10:e0127196. doi: 10.1371/journal.pone.0127196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Issac B, Raghava GP, Ramaswamy R. Spectral Repeat Finder (SRF): identification of repetitive sequences using Fourier transformation. Bioinformatics. 2004;20:1405–1412. doi: 10.1093/bioinformatics/bth103. [DOI] [PubMed] [Google Scholar]
- Singhapol C, Pal D, Czapiewski R, Porika M, Nelson G, Saretzki GC. Mitochondrial telomerase protects cancer cells from nuclear DNA damage and apoptosis. PLoS One. 2013;8:e52989. doi: 10.1371/journal.pone.0052989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sköld NH, Asplund MA, Wood CA, Bishop JDD. Telomerase deficiency in a colonial ascidian after prolonged asexual propagation. J Exp Zool B Mol Dev Evol. 2011;316:276–283. doi: 10.1002/jez.b.21399. [DOI] [PubMed] [Google Scholar]
- Techangamsuwan S, Kreutzer R, Kreutzer M, Imbschweiler I, Rohn K, Wewetzer K, Baumgärtner W. Transfection of adult canine Schwann cells and olfactory ensheathing cells at early and late passage with human TERT differentially affects growth factor responsiveness and in vitro growth. J Neurosci Methods. 2009;176:112–120. doi: 10.1016/j.jneumeth.2008.08.030. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replication life span. Curr Biol. 1998;8:279–282. doi: 10.1016/S0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- Venin SS, Bernard V, Tremblay JP. Telomerase allows the immortalization of T antigen positive DMD myoblasts: a new source of cells for gene transfer application. Gene Ther. 2000;7:619–623. doi: 10.1038/sj.gt.3301132. [DOI] [PubMed] [Google Scholar]
- Vitkova M, Krai J, Traut W, Zrzavy J, Marec F. The evolutionary origin of insect telomeric repeats, (TTAGG)n. Chromosome Res. 2005;13:145–156. doi: 10.1007/s10577-005-7721-0. [DOI] [PubMed] [Google Scholar]
- Vogt G. Hidden treasures in stem cells of indeterminately growing bilaterian invertebrates. Stem Cell Rev. 2012;8:305–317. doi: 10.1007/s12015-011-9303-1. [DOI] [PubMed] [Google Scholar]
- Vogt G. Ageing and longevity in the Decapoda (Crustacea): a review. Zool Anz. 2012;251:1–25. doi: 10.1016/j.jcz.2011.05.003. [DOI] [Google Scholar]
- Wang Y, Guo X. Chromosomal mapping of the vertebrate telomeric sequence (TTAGGG)n in four bivalve molluscs by fluorescence in situ hybridization. J Shellfish Res. 2001;20:1187–1190. [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Zielke S, Bodnar A. Telomeres and telomerase activity in scleractinian corals and Symbiodinium spp. Biol Bull. 2010;218:113–121. doi: 10.1086/BBLv218n2p113. [DOI] [PubMed] [Google Scholar]