Abstract
Medulloblastoma is a highly aggressive brain tumor and one of the leading causes of morbidity and mortality related to childhood cancer. These tumors display differential ability to metastasize and respond to treatment, which reflects their high degree of heterogeneity at the genetic and molecular levels. Such heterogeneity of medulloblastoma brings an additional challenge to the understanding of its physiopathology and impacts the development of new therapeutic strategies. This translational effort has been the focus of most pre-clinical studies which invariably employ experimental models using human tumor cell lines. Nonetheless, compared to other cancers, relatively few cell lines of human medulloblastoma are available in central repositories, partly due to the rarity of these tumors and to the intrinsic difficulties in establishing continuous cell lines from pediatric brain tumors. Here, we report the establishment of a new human medulloblastoma cell line which, in comparison with the commonly used and well-established cell line Daoy, is characterized by enhanced proliferation and invasion capabilities, stem cell properties, increased chemoresistance, tumorigenicity in an orthotopic metastatic model, replication of original medulloblastoma behavior in vivo, strong chromosome structural instability and deregulation of genes involved in neural development. These features are advantageous for designing biologically relevant experimental models in clinically oriented studies, making this novel cell line, named USP-13-Med, instrumental for the study of medulloblastoma biology and treatment.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-015-9914-5) contains supplementary material, which is available to authorized users.
Keywords: Brain tumor, Cancer, Cell line, Medulloblastoma, Stemness
Introduction
Primary tumors in the central nervous system (CNS) are frequent in children and adolescents, who have their quality of life significantly affected due to motor, cognitive, and endocrine defects secondary to the tumors or their treatment (Ward et al. 2014). Medulloblastoma is the most common malignant brain tumor in young children up to 4 years of age, and one of the leading causes of morbidity and mortality related to childhood cancer (Ostrom et al. 2013). It is considered by the World Health Organization (WHO) as an aggressive grade IV tumor, characterized by presence of poorly differentiated cell with high proliferative rate and ability to spread throughout the cerebrospinal axis (Louis et al. 2007).
Approximately 40 % of medulloblastoma patients already have metastasis at diagnosis, which is a predictive factor of poor prognosis (Wu et al. 2012). Other clinical parameters, such as age at diagnosis and size of residual tumor after surgical resection, are employed to stratify medulloblastoma patients according to the risk of tumor recurrence and to guide their treatment. Despite significant therapeutic improvements in recent decades (Ries et al. 1999), about 15 and 40 % of patients with either average or high risk of recurrence, respectively, do not respond properly to current treatment, displaying poor overall survival (Zeltzer et al. 1999; Pizer et al. 2011; Othman et al. 2014).
Improvements in survival rates rely on a better knowledge of the molecular and cellular biology of medulloblastoma, as well as on experimental tools to model this disease and address tumor response to new therapeutic approaches. In the past few years, a significant progress was made in the genetic characterization of medulloblastoma, revealing a high degree of tumor heterogeneity. A new classification proposes to encapsulate such heterogeneity within four medulloblastoma subgroups with distinct molecular/genetic features (Ramaswamy et al. 2011; Kool et al. 2012; Taylor et al. 2012; Northcott et al. 2012; Ramaswamy et al. 2013a; Schroeder and Gururangan 2014; Goschzik et al. 2014; Pietsch et al. 2014). Concurrently, insights into tumor development, evolution, and resistance to therapy were obtained by the seminal isolation and characterization of highly tumorigenic medulloblastoma cells exhibiting stem cell properties (Singh et al. 2003).
Pre-clinical studies are valuable to develop new drugs and treatment protocols for medulloblastoma and they invariably employ cell lines, Daoy being the most commonly used human medulloblatoma cell line (Xu et al. 2015). Compared to other types of cancer, there are relatively few cell lines of medulloblastoma available in central repositories, in part due to intrinsic difficulties in establishing continuous cell lines from pediatric brain tumors, and also because of the significantly lower incidence of these tumors relative to other cancers (Jemal et al. 2010).
Given that medulloblastoma are highly heterogeneous at the molecular, histological, and clinical levels, novel cell lines are instrumental for studying medulloblastoma biology. Here, we report the establishment and characterization of a new cell line derived from a medulloblastoma patient with advantageous features for pre-clinical studies, namely enhanced aggressive traits, stem cell properties, increased chemoresistance, tumorigenicity in an orthotopic metastatic model, and resemblance of original medulloblastoma behavior.
Materials and methods
Cell line establishment
The USP-13-Med cell line was established from a medulloblastoma specimen of classic histological subtype, derived from a 3 year-old boy admitted for treatment in the Medical School Hospital of the University of São Paulo. The procedure was approved by the Internal Review Board (CEP-IB No. 121/2011), and informed consent was obtained from the patient’s parents.
Tumor fragment was mechanically disaggregated with a scalpel, followed by a step of enzymatic disaggregation with collagenase (0.1 %) for 60 min at 37 °C to obtain a cell suspension. Patient-derived cells were cultivated with Dulbecco’s Modified Eagle Media (DMEM, Gibco, Carlsbad, CA USA) supplemented with 10 % Fetal Bovine Serum (FBS, Gibco), 100 U/mL Penicillin, 100 µg/mL Streptomycin and 250 ng/mL Fungizone® (Gibco) at 37 °C at 5 % CO2 atmosphere. Cells were subcultivated at 80 % confluence and cryopreserved in liquid nitrogen. An immortalized clonal cell line was obtained after selection of a tumor cell colony developed in the soft agar assay described below. Downstream characterization was performed side-by-side with Daoy (ATCC, Manassas, VA, USA), a well-studied human medulloblastoma cell line.
Anchorage-independent growth of tumor cells
Patient-derived cells were subjected to the soft agar colony formation assay in order to estimate anchorage-independent growth of tumor cells, which is a proper in vitro assay for detecting transformed cells. Five hundred cells were seeded over a 0.6 % agarose solution and covered with a 0.3 % agarose solution in a well of a six-well plate. Both solutions contained supplemented media at normal concentrations (10 % FBS, 100 U/mL Penicillin, 100 µg/mL Streptomycin and 250 ng/mL Fungizone®). Plates were incubated at 37 °C with 5 % CO2 in a humidified atmosphere. Media was replaced every 3 days.
Population doubling time
In order to evaluate the proliferative profile of USP-13-Med and Daoy cells, a population doubling level (PDL) assay was performed as recommended by WHO (2010) and described by Schaffer (1990). The cumulative PDL over 60 cell generations in vitro was calculated by the formula: PDL = Log10 (N/No) × 3.33 where: N = number of harvested cells after a period of growth and No = number of cells seeded at every new passage. The PDT (population doubling time) was calculated each passage using the following formula recommended by ATCC: DT = t × ln2/ln (N/No), where t is the incubation time (ATCC 2014).
Tumorsphere formation assay
Cells were seeded onto a 96-well ultra-low attachment plate (Corning, Corning, NY, USA) in DMEM/F12 with B-27 supplement, N-2 supplement, 20 ng/mL EGF and 20 ng/mL bFGF (Gibco), at an initial density of 1.25 × 104 cells/mL. Tumorspheres over 50 µm in diameter were counted after 7 days of incubation at 37 °C with 5 % CO2 humidified atmosphere. After 7 days of culture, the tumorspheres were enzymatic dissociated with TrypLE Select (Gibco), fixed for 1 h with 3.7 % paraformaldehyde, and permeabilized with Triton-X100 0.1 % in PBS. CD133/1 antibody conjugated with PE (Miltenyi, Bergisch Gladbach, Germany) and Nestin antibody conjugated with APC (Becton–Dickinson, Franklin Lakes, NJ, USA) were used to detect CD133+ and Nestin+ cells by flow cytometry. Data were collected with the Guava EasyCyte 5HTTM Flow Cytometer (Millipore—Guava Technologies, Billerica, MA, USA), using GuavaSoft 2.1 and FlowJo v10 softwares. Cells that underwent all the procedures but were not stained with CD133/1 or Nestin conjugated antibody were considered negative controls. The experiment was conducted twice, in triplicate. Plots are representatives of the acquired data.
Determination of cisplatin LC50
USP-13-Med and Daoy cultures were serum deprived for 24 h for synchronization. A total of 104 cells/mL were then seeded in a well of a 6-well microplate and allowed to attach overnight to the surface of the culture plate, prior to a 48 h-treatment with increasing doses of cisplatin (Sigma Aldrich, St. Louis, MO, USA). Apoptotic cells were estimated by the Guava Nexin® Annexin V Assay kit (Millipore). Staining was performed according to the manufacturer’s instructions, and samples were analyzed in the Guava EasyCyte 5HT™ Flow Cytometer (Millipore—Guava Technologies), using GuavaSoft 2.1 software. Concentrations lethal to 50 % of cells (LC50) were calculated from dose–response curves.
Cell migration assay
Cell migration was evaluated in vitro using Oris™ Cell Migration Assay Technology (Platypus Techonologies, Madison, WI, USA) according to the manufacturer’s procedure. Cells were seeded in high confluence (density of 106 cells/mL) and allowed to attach overnight. The Oris™ cell seeding stopper was then removed, exposing the cell migration detection zone. The assay was incubated at 37 °C under a 5 % CO2 humidified atmosphere, for 24 h, to allow cell migration. Photomicrographs were taken at times 0, 12 and 24 h and measurement was performed with ImageJ software, subtracting the migrated area after 12 or 24 h from the original area at 0 h.
3-Dimensional (3D) spheroid cell invasion assay
USP-13-Med and Daoy cell invasion capacity were evaluated in vitro using Cultrex® 3D Spheroid Cell Invasion Assay (Trevigen, Gaithersburg, MD, USA) following the manufacturer’s recommendation. Cells were resuspended in Spheroid Formation ECM at density of 8.104/mL, and added to a 96-well round bottom ultra-low attachment microplate (Corning). Spheroids were allowed to form for 24 h before invasion matrix and medium with 10 % FBS were added. Photomicrographs were taken on days 0, 2 and 4 after addition of invasion matrix. The amount and extension of the spindle-like protrusions formed by invading cells from the tumor spheroids were considered to calculate the invaded area using ImageJ software, following the manufacturer’s instructions.
Chromosomal copy number aberrations
DNA from USP-13-Med was extracted using standard cholorophorm/phenol protocol. aCGH was performed using 60 K whole-genome platform (Agilent Technologies, Santa Clara, CA, USA). All procedures were carried out following the manufacturer’s recommendation. Microarray scanned images were processed using the Feature Extraction Software (Agilent Technologies). Copy number aberrations (CNAs) were called using the statistical algorithm ADM-2 (sensitivity threshold: 6.7) in Genomic Workbench 6.9 Software. Chromosome deletions and duplications were considered when log2 ratio of Cy3/Cy5 intensities were detected <−0.3 and >0.3, respectively. Hybridization was gender matched.
Global gene expression analysis
Total RNA of USP-13-Med cells was extracted with the RNeasy kit (Qiagen, Venlo, Netherlands), following the manufacturer’s protocol. Gene expression profiling of USP-13-Med was evaluated against two samples of non-neoplastic human cerebellum RNA (Agilent Technologies—cat# 540007; Clontech, Mountain View, CA, USA—cat# 636535), used as reference samples. Independent microarray hybridizations were carried out for each sample using Affymetrix GeneChip® Human Gene 2.0 ST whole-transcript arrays, measuring both protein coding and long intergenic non-coding RNA transcripts (lincRNA). Hybridization procedures and quality control tests were performed following the manufacturer’s protocol. Microarray chips were scanned by GeneChip® Scanner 3000 7G System and a quality control was processed by Affymetrix® Expression Console Software (Affymetrix, Santa Clara, CA, USA). Differentially expressed genes were identified with the RankProd method, with a p value cutoff of 0.05 adjusted for false discovery rate (Benjamini and Hochberg 1995), using MeV software v.4.9. Functional Annotation of differentially expressed genes was based on an ontology term enrichment analysis using the DAVID v.6.7, as previously described (Huang et al. 2009). Statistical association between being differentially expressed and belonging to a given category was accessed by the Fisher Exact test (p value ≤0.05). The ontologies used were those defined by the KEGG database of metabolic pathways and by the Gene Ontology Consortium. The probe-to-GO and the probe-to-KEGG mapping were established based on the official annotation provided by the manufacturer.
Orthotopic metastatic model
Both USP-13-Med and Daoy cells were suspended at a density of 106 cells/5 μL DMEM and 5 µL of this cell suspension were injected in the right lateral ventricle of BALB/c Nude mice at a ratio of 1 µL/min with a high-precision microsyringe (701RN; Hamilton Company, Reno, NV, USA), by stereotaxic surgery. Control animals were injected with 5 μL of DMEM only. The coordinates used were 1 mm to the right; 0.5 mm posterior of the Bregma and 2.2 mm of depth. The scalp was closed with 2–0 silk suture and the animals were housed under standard controlled conditions (7:00 am to 7:00 pm light/dark cycle; 20–22 °C; 45–55 % humidity) with food and water ad libitum. Animals were euthanized after 50 days of inoculation or after the development of neurological deficits. This metastatic model was adapted from Studebaker et al. (2012). All efforts were made to minimize animal suffering as proposed by the International Ethical Guideline for Biomedical Research (CIOMS/OMS, 1985). The study was approved by the ethics committee for animal research of the University of São Paulo (CEUA protocol no. 132/2011). Brains were collected, formalin-fixed and paraffin-embedded. A total of 10 randomized 7-μm tissue sections were stained with hematoxylin-eosin for histological analysis. Images were taken in a digital inverted microscope (EVOS Cell Imaging System, Life Technologies, Carlsbad, CA, USA).
Statistical analysis
Statistical analysis of cellular assays was performed with Graph Pad Prism 6 using unpaired Student’s t test and statistical significance was established at p < 0.05. Data are presented as the mean ± SEM.
Results
Proliferative profile of USP-13-Med cells
Establishment of an immortalized human medulloblastoma cell line, named USP-13-Med, was only feasible when screening cells based on their 3D growth properties. Previous attempts to cultivate medulloblastoma cells from fresh specimens were not successful. Cells would stop growing after a few passages under standard monolayer culture conditions. Since cells with enhanced tumorigenic capacity usually survive hostile conditions and display anchorage-independent growth, freshly harvested tumor cells were then transferred from 2D to a 3D culture model in soft-agar, which better mimics the physiological environment of tumor cells.
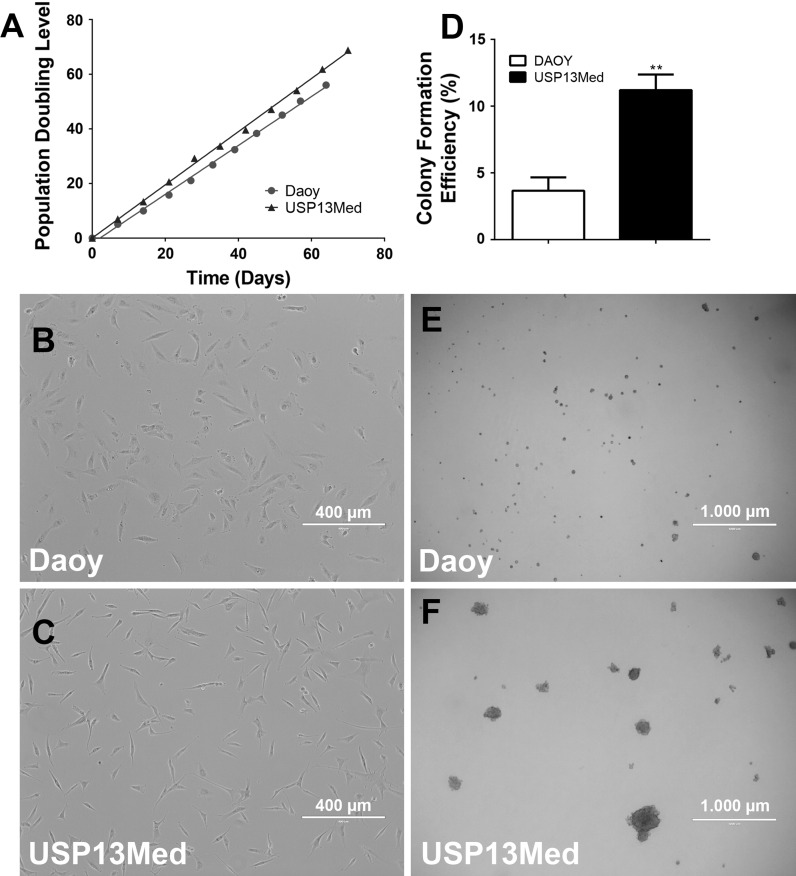
Under such conditions, few small cell colonies could be observed after 35 days of cell seeding. One well developed tumor cell colony was later picked at day 42 and transferred to one well of a 96-well microplate, for expansion under standard culture condition. The colony-derived cells endured clonal expansion for several generations without signs of replicative senescence (Fig. 1a). These cells had adherent properties and fibroblastoid morphology somewhat similar of that of Daoy cells (Fig. 1b, c). This cell morphology was not altered after 30 culture passages. The population doubling time for this cell line was calculated based on a PDL of over 60–70 generations. As observed in Fig. 1a, USP-13-Med cells proliferated faster than Daoy cells, displaying a PDT of 24.5 h, compared to a PDT 29.8 h observed for Daoy cells, under the same experimental conditions.
Fig. 1.
Proliferative rate and colony forming efficiency of USP-13-Med cells. a Population doubling level of USP-13-Med and Daoy cells assayed until at least 60 generations in vitro. USP-13-Med cells did not present signs of senescence under such conditions. Cellular morphology of Daoy (b) and USP-13-Med (c) cells in cultures under 40–50 % of cell confluence. Bar size 400 µm. d USP-13-Med displayed significantly higher colony forming efficiency than Daoy cells. Only colonies higher than 50 µm in diameter were considered. **p < 0.01. Representative images of Daoy (e) and USP-13-Med (f) colonies developed after 15 days of anchorage-independent growth in soft agar. Bar size 1000 µm
When subjected to anchorage-independent growth in soft agar, USP-13-Med cells confirmed their highly proliferative behavior, exhibiting a significantly higher efficiency of colony generation compared with Daoy cells (Fig. 1d). Additionally USP-13-Med colonies were well developed with an average diameter of 150 µm, while Daoy colonies were relatively smaller, inferior of 50 µm in diameter (Fig. 1e, f).
USP-13-Med cells display stem-like features in vitro
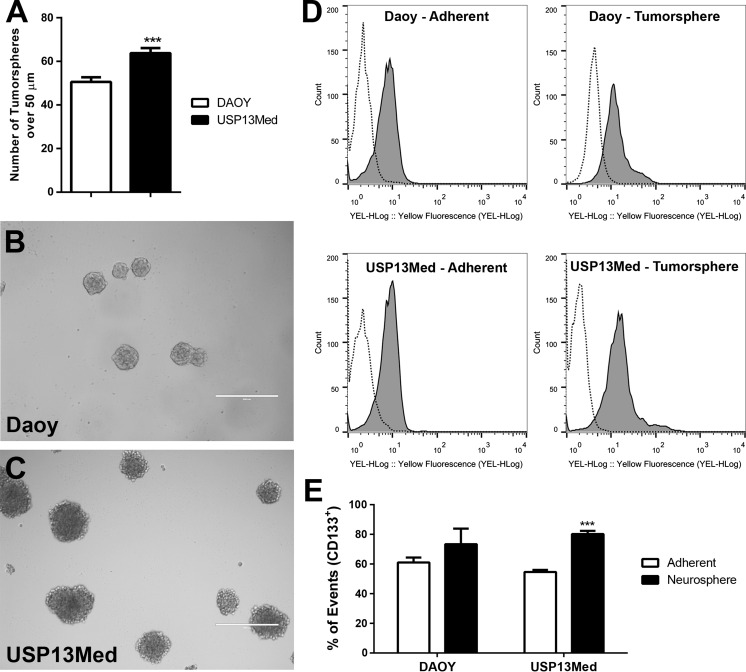
Ability to generate tumorspheres comprised by self-renewable progenitor-like cells, and increased resistance to chemotherapeutic agents are classic hallmarks of cancer stem cells. USP-13-Med cells demonstrated capacity to generate tumorspheres in vitro with efficiency significantly higher than that of Daoy cells (Fig. 2a). Under identical culture conditions, tumorspheres from USP-13-Med cells were also larger than Daoy spheres (Fig. 2b, c). Consistent with what have been reported for medulloblastoma stem cells (Singh et al. 2003), USP-13-Med tumorspheres were significantly enriched in cells expressing the neuroprogenitor cell marker CD133. Under standard monolayer culture conditions, about 50 % of USP-13-Med cells were already positive for CD133. This proportion increased to nearly 80 % when USP-13-Med cells were transferred to tumorsphere generating media. The same physiological response to environmental stimulus was not observed in cultures of Daoy cells, where the proportion of CD133+ cells remained unaltered despite conditioning in neural stem cell media (Fig. 2d, e). A subpopulation of cells highly expressing CD133 appeared in tumorspheres of both cell lines. It suggests that the tumorsphere culture condition also stimulates the expression of this marker. A similar result was observed for Nestin expression, which increased in tumorsphere cells (Supplementary Figure 1).
Fig. 2.
USP-13-Med cells are capable of generating tumorspheres at high efficiency. a Total number of tumorspheres over 50 µm in diameter generated from USP-13-Med and Daoy cells, after 7 days of culture under low-attachment conditions. Representative images of tumorspheres generated by Daoy (b) and USP-13-Med (c) cells are shown. Bar size 400 µm. d, e Population of CD133+ cells in USP-13-Med and Daoy tumorspheres compared with standard culture condition (adherent). The flow cytometry analysis shows an enrichment in CD133+ cells in USP-13-Med tumorspheres. Dashed line Negative Control; Gray Experimental condition. ***p < 0.001
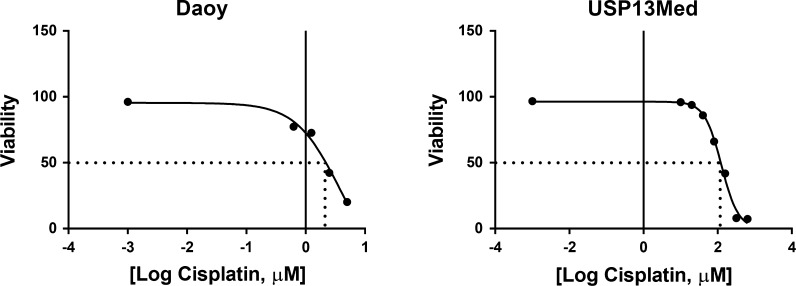
Furthermore, USP-13-Med cells were also more resistant to cisplatin, a common chemotherapeutic agent used in the clinic, than Daoy cells. Based on the amount of apoptotic cell death after 48 h of cisplatin treatment, the LC50 calculated for USP-13-Med cells was 126.4 µM, a value about 34 times higher than the LC50 calculated for Daoy cells (3.7 µM) (Fig. 3), indicating increased chemoresistance.
Fig. 3.
Increased chemoresistance of USP-13-Med cells. Dose–response curves of USP-13-Med and Daoy cells treated with cisplatin in vitro. Data are presented as viable (non-apoptotic) cells after 48 h of treatment with increasing log-transformed concentrations of cisplatin. The estimated LC50 values for USP-13-Med and Daoy cells were 126.4 and 3.7 µM, respectively
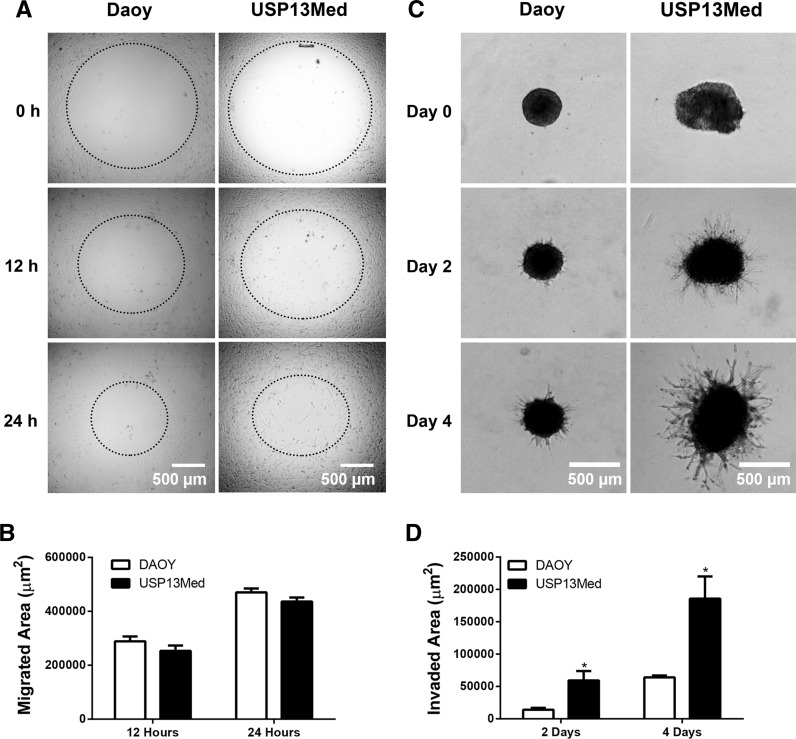
Another intrinsic feature of cancer stem-like cells is their high ability to invade tissues. Although USP-13-Med and Daoy cells displayed similar migration properties in a wound healing-like assay (Fig. 4a, b), when grown as tumor spheroids surrounded by a biological matrix, a condition that better reproduce tumor behavior in vivo, USP-13-Med cells exhibited a significantly higher invasion capacity than Daoy cells (Fig. 4c, d), revealing a more aggressive phenotype.
Fig. 4.
Migration and invasion capabilities of USP-13-Med cells. a Representative images from the wound healing-like migration assay with USP-13-Med and Daoy cells after 0, 12 and 24 h of assay. Bar size 500 µm. b Quantification of migrated area after 12 and 24 h. c Representative images of USP-13-Med and Daoy tumor spheroids embedded in invasion matrix at days 0, 2 and 4. The spindle-like protrusions observed are formed by tumor cells invading the matrix. The higher the cell invasion, the higher the length and amount of such protrusions. d Measurement of area invaded by USP-13-Med and Daoy cells after 2 and 4 days of assay. Bar size 500 µm. *p < 0.05
USP-13-Med cell behavior in vivo recapitulates main features of metastatic medulloblastoma
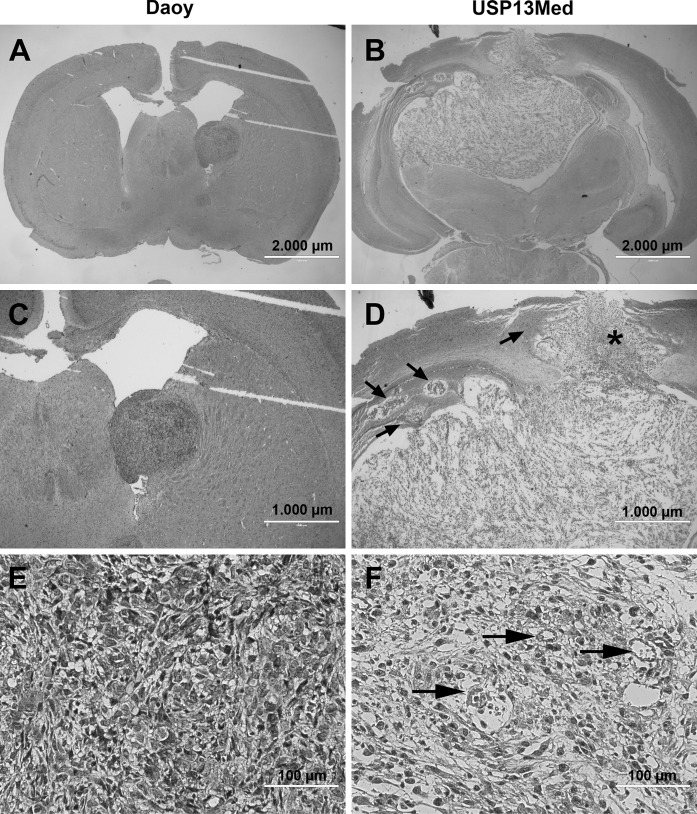
Besides being a fast growing tumor, medulloblastoma is also characterized by its infiltrative behavior and ability to spread. When orthotopically injected in the right lateral ventricle of nude mice, mimicking a M1 metastasis stage (cells localized in the cerebrospinal fluid), all animals of both groups developed tumor (Daoy, n = 3; USP-13-Med, n = 4). Three out of four animals injected with USP-13-Med cells developed clinical symptoms (weight loss and gait abnormalities) while, during the experimental period set, animals injected with Daoy cells did not display clinical symptoms. USP-13-Med cells were able to engraft in the brain tissue and generate M2 metastatic tumors characterized by scattered necrotic areas intercalated by high cell density zones. Tumor mass was well developed, well vascularized, and had an irregular shape with several regions of evident intraparenchymal invasion. Although Daoy cells were also able to colonize the brain when injected in the cerebrospinal fluid, the ensuing tumors were more homogeneous, displaying well defined borders. Necrotic zones and vasculature were not evident in the tumor mass and cells had a uniform phenotype (Fig. 5).
Fig. 5.
USP-13-Med cells generate aggressive brain tumors with an invasive and necrotic phenotype. Representative images of brain tumors developed after intracerebroventricular injection of USP-13-Med and Daoy cells in BALB/c Nude mice. a, b USP-13-Med cells developed a larger tumoral mass invading both brain hemispheres and characterized by an extensive necrotic area, when compared to Daoy cells. Bar size 2000 µm. c, d Several tumor cell islands (arrow) and/or bud-like protrusions (asterisk) invading the brain parenchyma were observed in USP-13-Med tumors, while Daoy tumors tended to be more circumscribed and less developed in size. Bar size 1000 µm. e, f USP-13-Med tumors displayed a more complex histopathology, with extensive necrotic areas intercalated with areas with high cellularity typically located nearby blood vessels (arrows). Daoy tumors, on the other hand, displayed a homogenous morphology characterized by high cell density. Bar size 100 µm
Subchromosomal aberrations in USP-13-Med cells
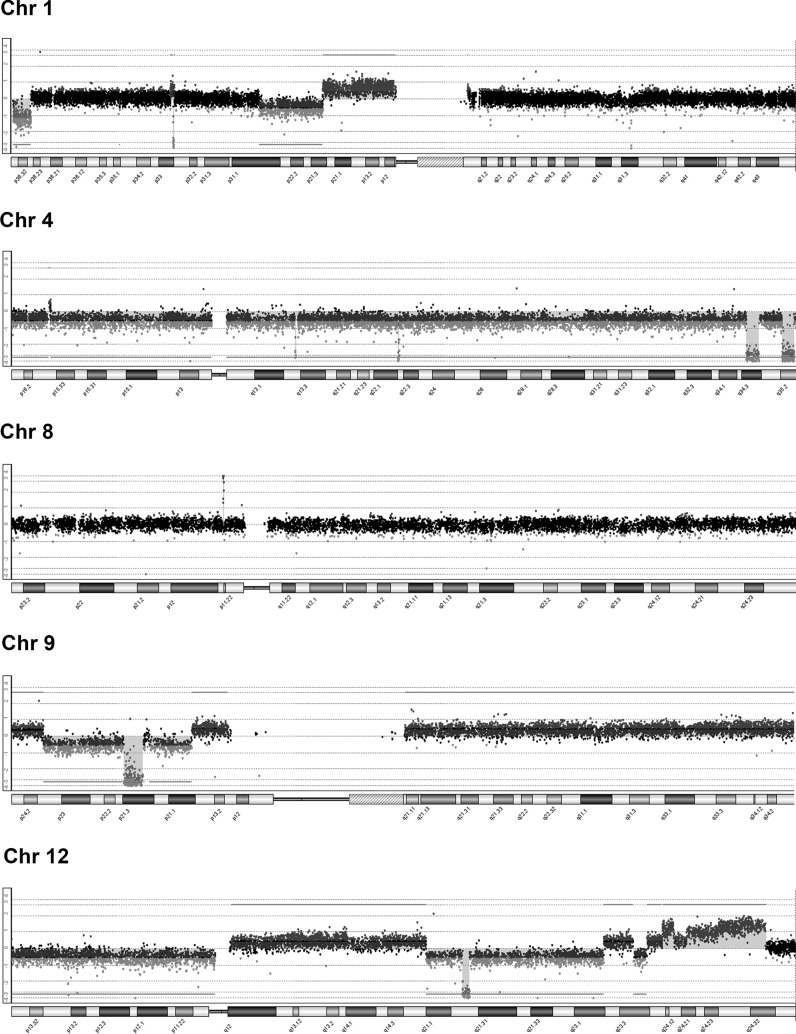
USP-13-Med cells displayed extensive chromosomal copy number aberration. Most chromosomes showed some kind of low level alteration. About 55 losses and 65 gains were detected (Supplementary Table 1), reflecting a very strong chromosome structural instability.
Notably, USP-13-Med cells have focused and very high amplifications in chromosome 8, cytoband 8p11.22, harboring ADAM5P and ADAM3A pseudogenes, and in chromosome 3q26.1, in a region containing no genes. Duplications and higher copy number gains in chromosome 2, 6, 7, 11, 12 and 20 were also found. The chromosome 6p21.1 duplication harbors the VEGFA gene functioning in angiogenesis, while the MSI1 gene implicated in medulloblastoma development is located in the duplicated q24.11–q24.32 region of chromosome 12. Of note, the duplicated segments of chromosome 12 also harbor several non-coding RNAs of unknown function.
Marked apparently homozygous deletions in chromosomes 1 (1p32.3), 4 (q13.2, q22.2, q34.3, q35.2), 9 (p21.3) and 12 (q21.2) are also characteristic of this cell line. Deletion of 1p32.3 results in loss of the tumor suppressor gene CDKN2C, while deletion of 9p21.3 implicates loss of two tumor suppressor genes, CDKN2A and CDKN2B. The deleted regions of chromosome 4 (q22.2, q34.3, q35.2) encompass both long and small non-coding RNAs of unknown function. The deleted 9p21.3 is also enriched in long non-coding RNAs (FOCAD-AS1, MIR31HG, CDKN2A-AS1, CDKN2B-AS1, LINC01239, LOC101929563), and microRNAs (miR-4473, miR-491 and miR-31). Finally, deleted 12q21.2 harbors the NAV3 gene, which is frequently deleted in tumors. Major CNA in USP-13-Med are shown in Fig. 6 and Table 1.
Fig. 6.
Major chromosomal copy number aberration found in USP-13-Med cells. Ideograms of chromosomes displaying major alterations in copy number are shown. Homozygous deletions were found on chromosomes 1, 4, 9 and 12, while an amplification was observed in chromosome 8. Blue or red dots in the aCGH profiles of these chromosomes represent gains or losses, respectively. (Color figure online)
Table 1.
Major chromosomal copy number aberration and respective affected genes found in USP-13-Med cells
| Chr | Cytoband | Amplification | Deletion | Gene names |
|---|---|---|---|---|
| chr1 | p32.3 | 0 | −2.518147 | FAF1, CDKN2C |
| chr4 | q22.2 | 0 | −3.07152 | GRID2 |
| chr4 | q34.3 | 0 | −3.045391 | AK094945, LINC01098, LINC01099, JC172092, L13714, JC172136, DQ587130, miR-544, DQ593458 |
| chr4 | q35.2 | 0 | −2.892707 | F11-AS1, MTNR1A, DQ578736, FAT1, DQ598830, DQ587130, DQ593458, RP11-138B4.1, DQ596939, DQ601017, ZFP42, DQ591044, TRIML2, TRIML1, LINC01060, L13714 |
| chr4 | q13.2 | 0 | −2.411487 | UGT2B17 |
| chr8 | p11.22 | 2.882072 | 0 | ADAM5P, ADAM3A |
| chr8 | p11.22 | 4.377834 | 0 | ADAM5P, ADAM3A |
| chr9 | p21.3 | 0 | −3.18852 | CDKN2BAS1, DMRTA1, miR-384 |
| chr9 | p21.3 | 0 | −2.747862 | MLLT3, miR-4473, FOCAD, FOCAD-AS1, miR-491, HACD4, IFNB1, IFNW1, IFNA21, IFNA4, IFNA7, IFNA10, IFNA16, IFNA17, IFNA14, IFA22P, IFNA5, KLHL9, IFNA6, IFNA13, IFNA2, IFNA8, IFNA1, miR-31HG, IFNE, miR-31, MTAP, CDKN2A-AS1, CDKN2A, CDKN2B-AS1, CDKN2B, DMRTA1, LINC01239, LOC01929563 |
| chr12 | q21.2 | 0 | −3.090438 | E2F7, NAV3 |
No significant copy number aberrations were detected in cytoband locations relative to TP53 (17p13.1), nor in APC (5q22.2), MYC (8q24.21), MYCN (2p24.3), CTNNB1 (3p22.1), GLI2 (2q14.2), and CDK6 (7q21.2), which have been found in specific molecular subtypes of medulloblastoma. A deletion, possibly in mosaic, was detected in 13q14.2, encompassing RB1, and low copy gains were detected in 9q22.32, 12q12–q21.1, and 14q22.1–q32.33, harboring, PTCH1, GLI1, WNT1, and OTX2, respectively.
Global gene expression profiling of USP-13-Med cells
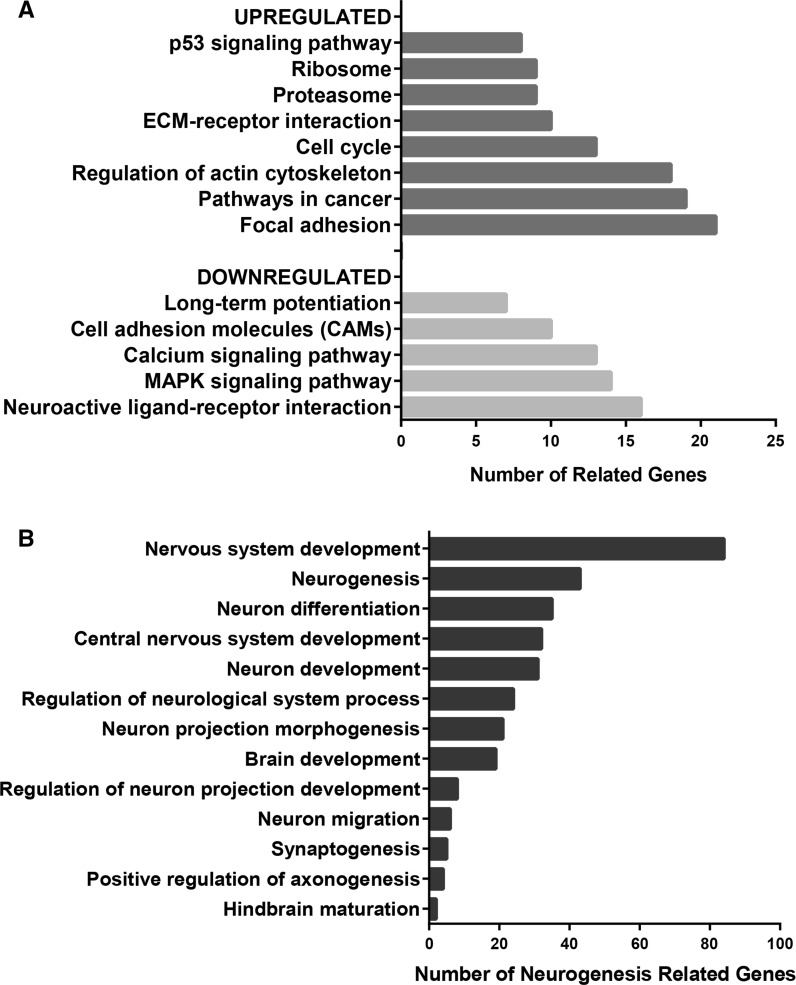
A total of 984 genes were found differentially expressed in USP-13-Med cells compared with normal cerebellum, with a fold change value of at least 2. As listed in Supplementary Table 2, there were 448 upregulated genes and 536 downregulated genes. The list of upregulated genes was significantly enriched in genes functioning in cell cycle regulation, cytoskeleton, extracellular matrix, protein metabolism, and p53 signaling, whereas the list of downregulated genes was enriched in genes involved in cell adhesion, neuronal signaling, MAPK and calcium signaling pathways (Fig. 7a).
Fig. 7.
Enrichment and Functional Annotation Analysis of genes differentially expressed in USP-13-Med cells. a Main functional categories and signaling pathways significantly associated with the subset of either upregulated or downregulated genes in USP-13-Med cells, relative to normal cerebellum. b Biological processes significantly associated with the subset of downregulated genes in USP-13-Med, showing enrichment in genes regulating neurodevelopment. p values ≤0.05 for all comparisons, according to the Fisher Exact Test for ontology enrichment analysis
Although, in both gene lists, no significant enrichment in genes related to the WNT or SHH signaling was detected, roughly 20 % of the downregulated genes encode proteins involved in neural system development, encompassing genes with specific functions in neurogenesis, neural differentiation, neuron migration, synaptogenesis, and other closely related functional categories, all of which being significantly overrepresented in the list of differentially expressed genes (Fig. 7b).
Additionally, classic oncogenes such as NRAS, RRAS, MET, and AURKB, as well as genes encoding proteins related with cancer stem cell properties, including the kinase MELK, the multidrug resistance transporter ABCC4, the radioresistance protein EZH2, the DNA repair enzyme RAD50, and the antiapoptotic protein BIRC5, were found overexpressed in USP-13-Med cells.
Discussion
Tumor cell lines are very important tools to address fundamental questions in cancer biology and develop new therapies. In the case of pediatric brain tumors such as medulloblastoma, only a few cell lines have been more extensively used in research (Xu et al. 2012, 2015). Medulloblastoma, however, comprise a group of highly heterogeneous tumors, as revealed by recent gene expression profiling and exome sequencing studies (Kool et al. 2012; Baron 2012; Mardis 2012; Williams et al. 2013). Current molecular subtypes of medulloblastoma are differentially associated with treatment response, frequency of metastasis, and survival rate (Kool et al. 2012; Northcott et al. 2012; Ramaswamy et al. 2013b). Thus, a diversity of cell lines displaying different characteristics and molecular/genetic anomalies is required to properly cover the heterogeneity of medulloblastoma.
The USP-13-Med cell line, obtained from a 3 year old boy with classic medulloblastoma, display typical cancer hallmarks that contribute to tumor maintenance and aggressiveness (Hanahan and Weinberg 2000, 2011). In particular, this cell line demonstrated an aggressive tumor behavior in vitro, revealed by a significantly higher cell proliferation rate compared to Daoy cells. The population doubling time of USP-13-Med (24.5 h) is also shorter than those reported for other commonly used medulloblastoma cell lines, such D283-Med (52.5 h) (Friedman et al. 1985), CHLA-259 (76 h) (Xu et al. 2012), and MHH-MED-2 (119 h) (Pietsch et al. 1994). Under selective culture conditions, USP-13-Med cells still demonstrated significantly higher capacity to form colonies in soft agar, as well as tumorspheres in neural stem cell medium, compared to Daoy cells. Notably, contrary to Daoy, USP-13-Med line was able to respond to environmental stimuli, enriching the proportion of cells expressing the neural stem cell marker CD133 under tumorsphere-generating conditions.
Some of the chromosomal abnormalities found in USP-13-Med cells are in concert with these observations. While gain in 14q22.1–q32.33, harboring the OTX2 oncogene and deletion of the tumor suppressors CDKN2A and CDKN2B in chromosome 9p21.3, CDKN2C in chromosome 1p32.3, and of the cell cycle repressor E2F7 (Di Stefano et al. 2003) in chromosome 12q21.2 would favor increased cell proliferation rates, additional copies of the MSI1 gene due to gains in the 12q24.11– q24.32 would favor a more primitive, neural stem-like cell phenotype. Besides being a neural stem cell marker (Glazer et al. 2012), the MSI1 protein also promotes cell proliferation by suppressing p21, p27 and p53 translation (Liu et al. 2014). Upregulation of MELK, encoding a serine/threonine kinase with critical role in cancer stem cell proliferation and survival (Ganguly et al. 2014), was also detected in USP-13-Med cells.
In brain tumor cells, both MSI1 (Vo et al. 2012) and MELK (Marie et al. 2008) upregulation, as well as capacity to generate tumorspheres in vitro (Panosyan et al. 2010), have been correlated with poor prognosis. Self-renewable CD133+ medulloblastoma stem cells are also known to be more efficient in initiating tumors in vivo (Singh et al. 2004). Furthermore, cancer stem cells tend to form tumors that better resemble the original anatomopathological features of the human disease, displaying aggressive features such as extensive necrotic areas, invasion to adjacent tissue, evident angiogenesis, and cellular heterogeneity (Galli et al. 2004). Consistently, when orthotopically inoculated in the lateral brain ventricle, USP-13-Med cells were able to generate large tumors characterized by presence of necrotic areas, evident vasculature, and irregular board plenty of cells invading the brain parechyma. This high degree of intraparenchymal invasion is supported by the significantly enhanced 3D invasion capacity observed for USP-13-Med spheroids, compared to Daoy. This is an important USP-13-Med trait, since cell motility is also implicated in tumor aggressiveness and ability to metastasize (van Zijl et al. 2011; Polyak and Weinberg 2009; Dave et al. 2012). Several small tumor islands could be observed in some brains of animals injected with USP-13-Med cells. These structures could represent either new metastatic lesions or protrusions of the main tumor. In any case, these are features indicative of more invasive and aggressive tumors.
Currently, treatment of medulloblastoma is multimodal, consisting in tumor resection followed by chemotherapy, with cisplatin being one of the main chemotherapeutic agents used (Ramaswamy et al. 2015). Radiotherapy may also be applied depending on the age of the patient, with doses varying according to clinical staging (Gerber et al. 2014; Ramaswamy et al. 2015). The 5-year disease-free survival range from about 85 % in average risk patients to 60–65 % in high risk patients (Crawford et al. 2007; Remke et al. 2013). Nonetheless, up to 40 % of patients do not respond to treatment and present tumor recurrence, which often leads to a poor survival outcome (Taylor et al. 2003; Lannering et al. 2012; Tarbell et al. 2013; Othman et al. 2014). The enhanced resistance of USP-13-Med cells to cisplatin is, therefore, another interesting trait of this cell line that could be explored to study chemoresistance and to screen new drug candidates.
This increased resistance to apoptosis induced by chemotherapy may be explained, at least in part, by some of the chromosomal abnormalities found in USP-13-Med cells. The loss of chromosome 12q21.2 encompasses NAV3, whose silencing was recently reported to inhibit apoptosis of breast cancer cells (Cohen-Dvashi et al. 2015). USP-13-Med cells also seem to have dysfunctional p53 signaling. Despite the native p53 status of these cells, multiple deletions affecting genes encoding upstream and downstream mediators of p53 signaling were identified. For instance, the deletion of CDKN2A, which encodes the tumor suppressors p14 and p16 by alternative splicing, may affect the p14-mediated accumulation of p53 and ensuing activation (Midgley et al. 2000). Noteworthy, deletion of 9p21.3 harbors the mirR-491, which was recently reported to have pro-apoptotic activity by targeting both BCL-XL and MCL1 proteins in chemoresistant ovarian cancer cell lines (Denoyelle et al. 2014), as well as miR-31, whose genomic deletion has been correlated with defects in the p53 pathway in cancer cells (Creighton et al. 2010). Decreased miR-31 expression was also reported to enhance chemoresistance of ovarian cancer cells due to up-regulation of its target, receptor tyrosine kinase MET (Mitamura et al. 2013). Finally, gains in 12q12–q21.1 encompassing MDM2 would favor p53 degradation (Pant and Lozano 2014), while gains in 12q24.11–q24.32, as mentioned above, would favor inhibition of p53 translation by MSI1 (Liu et al. 2014).
Interestingly, USP-13-Med cells have few genetic aberrations affecting proteins of both WNT (WNT1) and SHH (PTCH1 and GLI1) signaling pathways. Nonetheless, low copy gain of OTX2 was also detected in USP-13-Med cells. Previous studies reported that OTX2 copy number gain was not associated with WNT or SHH medulloblastoma subtypes (Adamson et al. 2009), but rather with either group 3 or 4 tumors (Taylor et al. 2012). The metastatic behavior USP-13-Med cells is also more coherent with groups 3 and 4, since tumors of the WNT and SHH subgroups are rarely metastatic (Taylor et al. 2012). Since neither amplification of MYC, typical in group 3 tumors, nor of MYCN were found in USP-13-Med cells, the CNA profile of these cells are more consistent with that of group 4 tumors. In agreement with the CNA status, the global gene expression profiling of USP-13-Med cells revealed enrichment in differentially expressed genes involved in neural development and other neurogenesis-related functions which, again, have been correlated with group 4 medulloblastomas (Taylor et al. 2012). Notably, genes involved in WNT or SHH pathways were not significantly overrepresented in the list of differentially expressed genes.
Compared with other subgroups, medulloblastomas categorized in group 4 are more heterogeneous and the molecular pathogenesis of such tumors is uncertain. Confirmation of this molecular classification would add another important value to USP-13-Med cell line as a biologically relevant model since, from the 15 available medulloblastoma cell lines with known molecular classification, only one (CHLA-01-MED) belongs to group 4 (Xu et al. 2015).
In summary, we established a new human medulloblastoma cell line characterized by a high proliferative activity, high colony formation efficiency, enhanced ability to generate tumorspheres enriched in CD133+ cells, as well as increased invasion capacity and chemoresistance, when compared to the well-established cell line Daoy. USP-13-Med was also tumorigenic in vivo in an orthotopic metastatic model, generating well developed tumors with extensive necrosis, evident vascularization and intraparenchymal invasion, typical of aggressive tumors. Numerous chromosomal gains and losses were detected, affecting oncogenes, tumor suppressors, neurogenesis genes, microRNAs, as well as long non-coding RNAs of unknown function. Overexpression of classic oncogenes and cancer stem cell-related genes were also found. Cell behavior, chromosomal aberrations, and global gene expression profile more closely related to those of group 4 medulloblastomas were identified in this cell line. All these features are highly advantageous for pre-clinical studies aiming at improving our understanding of medulloblastoma development and testing new therapeutic strategies. The use of this novel cell line by the scientific community in future studies shall also refine its molecular classification.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Dr. Carla Rosenberg for supervising the CNA analysis and for her helpful comments on the manuscript. This study was supported by funds from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Grants FAPESP-CEPID (2013/08028-1), FAPESP (2010/52686-5); funds from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Grants CNPq (309206/2011-1; 444722/2014-9), INCT-CETGEN (573633/2008-8); and funds from Financiadora de Estudos e Projetos (FINEP), Grant FINEP-CTC (0108057900).
Compliance with ethical standards
Conflict of interest
The authors disclose no potential conflicts of interest.
References
- Adamson DC, Shi Q, Wortham M, et al. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 2009;70:181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ATCC (2014) ATCC® animal cell culture guide. http://www.atcc.org/guides/animal_cell_culture_guide.aspx. Accessed 16 Jul 2014
- Baron JA. Screening for cancer with molecular markers: progress comes with potential problems. Nat Rev Cancer. 2012;12:368–371. doi: 10.1038/nrc3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing on JSTOR. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- Cohen-Dvashi H, Ben-Chetrit N, Russell R, et al. Navigator-3, a modulator of cell migration, may act as a suppressor of breast cancer progression. EMBO Mol Med. 2015;7:299–314. doi: 10.15252/emmm.201404134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JR, MacDonald TJ, Packer RJ. Medulloblastoma in childhood: new biological advances. Lancet Neurol. 2007;6:1073–1085. doi: 10.1016/S1474-4422(07)70289-2. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Fountain MD, Yu Z, et al. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave B, Mittal V, Tan NM, Chang JC. Epithelial-mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res. 2012;14:202. doi: 10.1186/bcr2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyelle C, Lambert B, Meryet-Figuière M, et al. miR-491-5p-induced apoptosis in ovarian carcinoma depends on the direct inhibition of both BCL-XL and EGFR leading to BIM activation. Cell Death Dis. 2014;5:e1445. doi: 10.1038/cddis.2014.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman HS, Burger PC, Bigner SH, et al. Establishment and characterization of the human medulloblastoma cell line and transplantable xenograft D283 Med. J Neuropathol Exp Neurol. 1985;44:592–605. doi: 10.1097/00005072-198511000-00005. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Ganguly R, Hong CS, Smith LGF, et al. Maternal embryonic leucine zipper kinase: key kinase for stem cell phenotype in glioma and other cancers. Mol Cancer Ther. 2014;13:1393–1398. doi: 10.1158/1535-7163.MCT-13-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber NU, Mynarek M, von Hoff K, et al. Recent developments and current concepts in medulloblastoma. Cancer Treat Rev. 2014;40:356–365. doi: 10.1016/j.ctrv.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Glazer RI, Vo DT, Penalva LOF. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front Biosci. 2012;17:54–64. doi: 10.2741/3915. [DOI] [PubMed] [Google Scholar]
- Goschzik T, Zur Mühlen A, Kristiansen G, et al. Molecular stratification of medulloblastoma: comparison of histological and genetic methods to detect Wnt activated tumors. Neuropathol Appl Neurobiol. 2014 doi: 10.1111/nan.12161. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laks DR, Masterman-Smith M, Visnyei K, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27:980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannering B, Rutkowski S, Doz F, et al. Hyperfractionated versus conventional radiotherapy followed by chemotherapy in standard-risk medulloblastoma: results from the randomized multicenter HIT-SIOP PNET 4 trial. J Clin Oncol. 2012;30:3187–3193. doi: 10.1200/JCO.2011.39.8719. [DOI] [PubMed] [Google Scholar]
- Liu X, Yang W-T, Zheng P-S. Msi1 promotes tumor growth and cell proliferation by targeting cell cycle checkpoint proteins p21, p27 and p53 in cervical carcinomas. Oncotarget. 2014;5:10870–10885. doi: 10.18632/oncotarget.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Genome sequencing and cancer. Curr Opin Genet Dev. 2012;22:245–250. doi: 10.1016/j.gde.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie SKN, Okamoto OK, Uno M, et al. Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int J Cancer. 2008;122:807–815. doi: 10.1002/ijc.23189. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Desterro JM, Saville MK, et al. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- Mitamura T, Watari H, Wang L, et al. Downregulation of miRNA-31 induces taxane resistance in ovarian cancer cells through increase of receptor tyrosine kinase MET. Oncogenesis. 2013;2:e40. doi: 10.1038/oncsis.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Pfister SM, Taylor MD. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8:340–351. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15:ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman RT, Kimishi I, Bradshaw TD, et al. Overcoming multiple drug resistance mechanisms in medulloblastoma. Acta Neuropathol Commun. 2014;2:57. doi: 10.1186/2051-5960-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panosyan EH, Laks DR, Masterman-Smith M, et al. Clinical outcome in pediatric glial and embryonal brain tumors correlates with in vitro multi-passageable neurosphere formation. Pediatr Blood Cancer. 2010;55:644–651. doi: 10.1002/pbc.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Lozano G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev. 2014;28:1739–1751. doi: 10.1101/gad.247452.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch T, Scharmann T, Fonatsch C, et al. Characterization of five new cell lines derived from human primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1994;54:3278–3287. [PubMed] [Google Scholar]
- Pietsch T, Schmidt R, Remke M, et al. Prognostic significance of clinical, histopathological, and molecular characteristics of medulloblastomas in the prospective HIT2000 multicenter clinical trial cohort. Acta Neuropathol. 2014;128:137–149. doi: 10.1007/s00401-014-1276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer B, Donachie PHJ, Robinson K, et al. Treatment of recurrent central nervous system primitive neuroectodermal tumours in children and adolescents: results of a Children’s Cancer and Leukaemia Group study. Eur J Cancer. 2011;47:1389–1397. doi: 10.1016/j.ejca.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Ramaswamy V, Northcott PA, Taylor MD. FISH and chips: the recipe for improved prognostication and outcomes for children with medulloblastoma. Cancer Genet. 2011;204:577–588. doi: 10.1016/j.cancergen.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Ramaswamy V, Remke M, Bouffet E, et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 2013;14:1200–1207. doi: 10.1016/S1470-2045(13)70449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy V, Remke M, Bouffet E, et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 2013;14:1200–1207. doi: 10.1016/S1470-2045(13)70449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy V, Remke M, Adamski J et al (2015) Medulloblastoma subgroup-specific outcomes in irradiated children: who are the true high-risk patients? Neuro Oncol nou357. doi:10.1093/neuonc/nou357 [DOI] [PMC free article] [PubMed]
- Remke M, Ramaswamy V, Taylor MD. Medulloblastoma molecular dissection: the way toward targeted therapy. Curr Opin Oncol. 2013;25:674–681. doi: 10.1097/CCO.0000000000000008. [DOI] [PubMed] [Google Scholar]
- Ries LAG, Smith MA, Gurney JG et al (1999) Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975-1995. In: National Cancer Institute, SEER Program. NIH Pub. http://seer.cancer.gov/archive/publications/childhood/childhood-monograph.pdf. Accessed 4 Aug 2014
- Schaffer WI. Terminology associated with cell, tissue and organ culture, molecular biology and molecular genetics. Vitro Cell Dev Biol. 1990;26:97–101. doi: 10.1007/BF02624162. [DOI] [PubMed] [Google Scholar]
- Schroeder K, Gururangan S. Molecular variants and mutations in medulloblastoma. Pharm Pers Med. 2014;7:43–51. doi: 10.2147/PGPM.S38698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Studebaker AW, Hutzen B, Pierson CR, et al. Oncolytic measles virus prolongs survival in a murine model of cerebral spinal fluid-disseminated medulloblastoma. Neuro Oncol. 2012;14:459–470. doi: 10.1093/neuonc/nor231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbell NJ, Friedman H, Polkinghorn WR, et al. High-risk medulloblastoma: a pediatric oncology group randomized trial of chemotherapy before or after radiation therapy (POG 9031) J Clin Oncol. 2013;31:2936–2941. doi: 10.1200/JCO.2012.43.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RE, Bailey CC, Robinson K, et al. Results of a randomized study of preradiation chemotherapy versus radiotherapy alone for nonmetastatic medulloblastoma: the International Society of Paediatric Oncology/United Kingdom Children’s Cancer Study Group PNET-3 Study. J Clin Oncol. 2003;21:1581–1591. doi: 10.1200/JCO.2003.05.116. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zijl F, Krupitza G, Mikulits W. Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat Res. 2011;728:23–34. doi: 10.1016/j.mrrev.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal C, Li N, Wang X, et al. Bmi1 marks intermediate precursors during differentiation of human brain tumor initiating cells. Stem Cell Res. 2012;8:141–153. doi: 10.1016/j.scr.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Vo DT, Subramaniam D, Remke M, et al. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am J Pathol. 2012;181:1762–1772. doi: 10.1016/j.ajpath.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E, DeSantis C, Robbins A, et al. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- WHO (2010) Recommendations for the evaluation of animal cell cultures as substrates for the manufacture of biological medicinal products and for the characterization of cell banks. http://www.who.int/biologicals/Cell_Substrates_clean_version_18_April.pdf. Accessed 16 July 2014
- Williams SA, Anderson WC, Santaguida MT, Dylla SJ. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab Invest. 2013;93:970–982. doi: 10.1038/labinvest.2013.92. [DOI] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Dubuc A, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Erdreich-Epstein A, Gonzalez-Gomez I, et al. Novel cell lines established from pediatric brain tumors. J Neurooncol. 2012;107:269–280. doi: 10.1007/s11060-011-0756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Margol A, Asgharzadeh S, Erdreich-Epstein A. Pediatric brain tumor cell lines. J Cell Biochem. 2015;116:218–224. doi: 10.1002/jcb.24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- Zeltzer PM, Boyett JM, Finlay JL, et al. Metastasis stage, adjuvant treatment, and residual tumor are prognostic factors for medulloblastoma in children: conclusions from the Children’s Cancer Group 921 randomized phase III study. J Clin Oncol. 1999;17:832. doi: 10.1200/JCO.1999.17.3.832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.