Abstract
Aeromonas hydrophila sodium-driven polar flagellum has a complex stator-motor. Consist of two sets of redundant and non-exchangeable proteins (PomA/PomB and PomA2/PomB2), which are homologs to other sodium-conducting polar flagellum stator motors; and also two essential proteins (MotX and MotY), that they interact with one of those two redundant pairs of proteins and form the T-ring. In this work, we described an essential protein for polar flagellum stability and rotation which is orthologs to Vibrio spp. FlgT and it is encoded outside of the A. hydrophila polar flagellum regions. The flgT was present in all mesophilic Aeromonas strains tested and also in the non-motile Aeromonas salmonicida. The A. hydrophila ΔflgT mutant is able to assemble the polar flagellum but is more unstable and released into the culture supernatant from the cell upon completion assembly. Presence of FlgT in purified polar hook-basal bodies (HBB) of wild-type strain was confirmed by Western blotting and electron microscopy observations showed an outer ring of the T-ring (H-ring) which is not present in the ΔflgT mutant. Anchoring and motility of proton-driven lateral flagella was not affected in the ΔflgT mutant and specific antibodies did not detect FlgT in purified lateral HBB of wild type strain.
Keywords: Aeromonas, flgT, polar and lateral flagella
Introduction
Motility represents an important advantage for bacteria in moving toward favouable conditions, in avoiding of detrimental environments, or in having successful competes with other microorganisms (Frenchel, 2002). The motility organ used by many bacteria to move through liquid or semisolid media is the flagellum, although their number and placement shows differences between species. Flagella are supramolecular reversible rotary complexes anchored in the bacterial surface and made up of many different proteins. A flagellum consists of a filament, a hook and a basal body. The basal body is embedded in the cell envelope and works as a reversible rotary motor, whereas the hook and the filament function as a universal joint and a propeller, respectively (Berg, 2003; Macnab, 2003). The flagella basal body consists in some rings that allow the flagellum rod crossing through the cell envelope, a reversible rotary motor and a protein export apparatus that translocate the flagellar components. In Gam-negative bacteria there are three rings involved: L-, P-, and MS-rings. The L-ring is composed of the FlgH protein and outer membrane-embedded. The P-ring is composed of the FlgI protein, lies in the periplasmic space and is associated with the peptidoglycan layer. Both rings form the LP ring complex that functions as a molecular bushing. The MS-ring is composed of the FliF protein and inner membrane-embedded, being the starting point for motor assembly (Ueno et al., 1992; Macnab, 1996; DeRosier, 1998). The flagellum motor is made of a rotor and about a dozen stator complexes. The rotor is composed of an axial rod and the C-ring, which assemble around the MS-ring and the export apparatus. The C-ring lies in the cytoplasm, is composed of the FliM, FliN, and FliG proteins and is the site of torque generation and switching the direction of flagellum rotation (Khan et al., 1991; Francis et al., 1994; Katayama et al., 1996). Above the C-ring, surrounding the MS-ring in the inner membrane and attached to the peptidoglycan layer are the stators complex. Each stator complex is made up of two membrane proteins with an apparent 4:2 stoichiometry. These membrane proteins constitute an ion channel that transform the flow of proton or sodium ions across the cytoplasmic membrane into the energy required for flagella motor rotation (McCarter, 2001; Yorimitsu and Homma, 2001; Blair, 2003; Terashima et al., 2008). Most bacterial flagella use a single type of stator complex: proton- or sodium-dependent. The proton-dependent stator complex is made up of MotA and MotB, like in Escherichia coli and Salmonella enterica serovar Typhimurium flagella (Blair and Berg, 1990; Stolz and Berg, 1991; Macnab, 1996). The sodium-dependent stator complex is made up of PomA and PomB, as in Vibrio species (Asai et al., 1997; McCarter, 2001; Yorimitsu and Homma, 2001) or MotP and MotS, as in alkaliphilic Bacillus species (Ito et al., 2004). However, the flagella motor of some bacterial species is energized by two different sets of stator complexes. In Bacillus subtilis, MotAB, and MotPS; and in Shewanella oneidensis MR-1, MotAB, and PomAB, supports flagellar rotation by proton and sodium ions flow, respectively (Ito et al., 2004; Paulick et al., 2009). Nevertheless, in Aeromonas hydrophila, PomAB, and PomA2B2 are both sodium-coupled stator complexes with different sensitivity to sodium concentrations (Wilhelms et al., 2009) and in Pseudomonas aeruginosa PAO1, MotAB, and MotCD are both proton-dependent stator complex (Doyle et al., 2004; Toutain et al., 2005). Surrounding the conserved stator structure, different bacterial species display various additional components. The lateral flagella proton-dependent stator of Vibrio parahaemolyticus requires an additional protein, MotY, with a peptidoglycan-binding domain (Stewart and McCarter, 2003). The polar flagellum sodium-dependent stator of Vibrio species, S. oneidensis MR-1 and A. hydrophila contain two additional proteins: MotX and MotY, which make up a beneath structure of P-ring which is named T-ring (Okabe et al., 2002; Yagasaki et al., 2006; Terashima et al., 2008; Koerdt et al., 2009). Furthermore, surrounding the polar-flagellum LP-rings of Vibrio species is the H-ring, which is composed of FlgT protein. The T- and H-rings are required for properly assembly of the PomAB stator complex around the rotor in Vibrio species (Terashima et al., 2006, 2010, 2013).
Aeromonas are found ubiquitously in the environment, but are mainly associated with fresh or estuarine water. They are the causative agent of wide spectrum of diseases in man and animals and some species are becoming food and waterborne pathogens of increasing importance (von Graevenitz, 2007; Ghenghesh et al., 2008). Mesophilic Aeromonas have a single polar flagellum produced constitutively and 50–60% of clinical isolates also have lateral inducible flagella. Fully functional polar and lateral flagella are essential for a proper attachment, biofilms formation, and colonization (Merino et al., 1997; Rabaan et al., 2001; Gavín et al., 2002). Although, both flagella types are structurally similar, they have some differences at the export apparatus and the motor. The FliO protein is only present in the polar flagella export apparatus. The lateral flagella are proton-driven and their stator complex made up of two proteins, LafT and LafU (Canals et al., 2006a; Molero et al., 2011). However, the polar flagellum is sodium-driven and their stator complex consists of two sets of membrane proteins: PomAB and PomA2B2 (Wilhelms et al., 2009), as well as two essential proteins: MotXY, which make up the T-ring (Molero et al., 2011).
In this study, we reported a protein orthologous to FlgT of Vibrio spp., which present in all mesophilic Aeromonas and is encoded outside of the polar flagellum regions, which is involved in the stability and rotation of an unsheathed flagellum sodium-driven with two different stator complex.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown on Luria-Bertani (LB) Miller broth and LB Miller agar at 37°C. Aeromonas strains were grown either in tryptical soy broth (TSB) or agar (TSA) at 25°C. When required ampicillin (100 μg/ml), kanamycin (50 μg/ml), tetracycline (20 μg/ml), chloramphenicol (25 μg/ml), rifampicin (100 μg/ml), and spectinomycin (50 μg/ml) were added to the different media. Media were supplemented with 0.2% (w/v) L-arabinose to induce recombinant proteins expression under the arabinose promoter on pBAD33.
Table 1.
Bacterial strains and plasmid used in this study.
| Strain or plasmid | Genotype and/or phenotypea | Reference |
|---|---|---|
| Strains | ||
| Aeromonas hydrophila | ||
| AH-3 | A. hydrophila wild type, serogroup O :34 | Merino et al., 1991 |
| ATCC7966T | A. hydrophila wild type | Seshadri et al., 2006 |
| AH-405 | AH-3, spontaneous Rifr | Altarriba et al., 2003 |
| ATCC7966-Rif | ATCC7966T, spontaneous Rifr | This work |
| AH-3ΔflgT | AH-405; ΔflgT | This work |
| ATCCΔAHA1089 | ATCC7966-Rif; ΔAHA_1089 | This work |
| AH-3::flaAΔflaB | AH-405; flaA::Kmr; ΔflaB | Canals et al., 2006b |
| AH-3:: flhA | AH-405; flhA::Kmr | Canals et al., 2006b |
| AH-3ΔlafA | AH-405; ΔlafA | Wilhelms et al., 2013 |
| AH-3::flaAΔflaBflgT | AH-3::flaA::Kmr; ΔflaB; ΔflgT | This work |
| AH-3ΔlafAflgT | AH-3ΔlafA; ΔflgT | This work |
| AH-3::flrA | AH-405; flrA::Kmr | Wilhelms et al., 2011 |
| AH-3::flrBC | AH-405; flrB::pSF, Kmr | Wilhelms et al., 2011 |
| AH-3::fliAP | AH-405; fliAP:: Kmr | Canals et al., 2006b |
| AH-3::lafK | AH-405; lafK::Kmr | Canals et al., 2006a |
| AH-3::lafS | AH-405; lafS::Kmr | Wilhelms et al., 2013 |
| Escherichia coli | ||
| DH5α | F- endA hdsR17(rk- mk+) supE44 thi-1 recA1 gyr-A96ϕ80lacZ | Hanahan, 1983 |
| MC1061λpir | thi thr1 leu6 proA2 his4 argE2 lacY1 galK2 ara14 xyl5 supE44λ pir | Rubires et al., 1997 |
| Plasmids | ||
| pLA2917 | Cosmid vector, Tcr Kmr | Allen and Hanson, 1985 |
| pLA-FLGT | pLA2917 with AH-3 flgT, Tcr. | This work |
| pRK2073 | Helper plasmid, Spr | Rubires et al., 1997 |
| pGEMT | Cloning vector, Apr. | Promega |
| pDM4 | Suicide plasmid, pir dependent with sacAB genes, oriR6K, Cmr. | Milton et al., 1996 |
| pDM-AHA1089 | pDM4ΔAHA_1089 of ATCC7966T, Cmr. | This work |
| pDM-FLGT | pDM4ΔflgT of AH-3, Cmr. | This work |
| pET-30 Xa/LIC | IPTG inducible expression vector KmR | Novagen |
| pET-30-FlgT | pET-30 Xa/LIC with A. hydrophila AH-3 flgT | This study |
| pBAD33 | pBAD33 arabinose-induced expression vector with Cmr | Guzman et al., 1995 |
| pBAD33-FLGT | pBAD33 with AH-3 flgT gen, Cmr | This work |
a Kmr, kanamycin resistant; Apr, ampicillin resistant; Rifr, rifampicin resistant; Cmr, chloramphenicol resistant; Spr, spectinomycin resistant; Tcr, tetracycline resistant.
Motility Assays (Swarming and Swimming)
Fresh bacterial grown colonies were transferred with a sterile toothpick onto the center of a soft agar plate (1% tryptone, 0.5% NaCl, 0.25% agar). Plates were incubated face up for 24–48 h. at 25°C and motility was assessed by examining the migration of bacteria through the agar from the center toward the periphery of the plate. Moreover, swimming motility was assessed by light microscopy observations in liquid media.
Transmission Electron Microscopy (TEM)
Bacterial suspensions were placed on Formvar-coated grids and negative stained with a 2% solution of uranyl acetate pH 4.1. Preparations were observed on a Jeol JEM 1010 transmission electron microscope.
DNA Techniques
DNA manipulations were carried out according to standard procedures (Sambrook et al., 1989). DNA restriction endonucleases were obtained from Promega. T4 DNA ligase and alkaline phosphatise were obtained from Invitrogen and GE Healthcare, respectively. PCR was performed using the BioTaq DNA polymerase (Ecogen) in a Gene Amplifier PCR System 2400 Perkin Elmer Thermal Cycler. Colony hybridizations were carried out by colony transfer onto positive nylon membranes (Roche) and then lysed according to the manufacturer’s instructions. Probe labeling with digoxigenin, hybridization and detection (GE Healthcare) were carried out as recommended by the suppliers.
Nucleotide Sequencing and Computer Sequence Analysis
Plasmid DNA for sequencing was isolated by Qiagen plasmid purification kit (Qiagen, Inc. Ltd.) as recommended by the suppliers. Double-strand DNA sequencing was performed by using the Sanger dideoxy-chain termination method (Sanger et al., 1977) with the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystem). Custom-designed primers used for DNA sequencing were purchased from Sigma–Aldrich.
DNA sequence was translated in all six frames, and their deduced amino acid sequences were inspected in the GenBank, EMBL, and SwissProt databases by using the BLASTX, BLASTP, or PSI-BLAST network service at the National Center for Biotechnology Information (NCBI) (Altschul et al., 1997). Protein family profile was performed using the Protein Family Database Pfam at the Sanger Center (Bateman et al., 2002).
RT-PCR
Total RNA was isolated from A. hydrophila AH-3, AH-3::flrA, AH-3::flrBC, AH-3::fliAp, AH-3::lafK, and AH-3::lafS which were grown at 25°C in liquid media (TSB) or plates (TSA) by RNA Protect Bacteria Reagent (Qiagen) and RNeasy Mini kit (Qiagen). To ensure that RNA was devoid of contaminating DNA, the preparation was treated with RNase-free TurboDNase I (Ambion). First-strand cDNA synthesis was carried out using the Thermoscript RT-PCR system (Invitrogen) and random primers on 5 μg of total RNA DNase-digested, according to the manufacturer’s instructions. PCR without reverse transcriptase was also performed to confirm the absence of contaminating DNA in the RNA sample. The second strand synthesis and subsequent DNA amplification of flgT internal fragment was carried out using the BioTaqDNA polymerase (Bioline) and the pair of oligonucleotides5′-CAGTGGCTGG ACGAGAAC-3′ and 5′- TTCCAATACTGCCAGATCC-3′ designed using the Prime program from the Genetics Computer Group package (Madison, Wisconsin). Amplicons were visualized by agarose gel electrophoresis with ethidium bromide staining. A. hydrophila ribosomal 16S primers were used as a control of cDNA template.
Constructions of Defined Mutants
The single defined insertion ATCCΔAHA1089 and AH-3ΔflgT were obtained by allelic exchange as described by Milton et al. (1996). Briefly, DNA regions upstream and downstream of AHA_1089 of A. hydrophila ATCC7966T were ampli-fied using the primer pairs A1 (5′-CGCGGATCCAATCTTGACCACCACCACT-3′) and B1 (5′-CCCATCCACTAAACT TAAACAGGCGTAGACCTCGTCTGT-3′), and C1 (5′-TGTTTAAGTTTAGTGGAT GGGGATCAGTTCCGCATCCAG-3′) and D1 (5′-CGCGGATCCCTCGATGGTCCA ATCCAT-3′) in two sets of asymmetric PCRs to amplify DNA fragments of 610 (A1B1) and 674 (C1D1) bp, respectively. Regions upstream and downstream of flgT of A. hydrophila AH-3 were amplified using the primer pairs A1 and B2 (5′-CCCATCCA CTAAACTTAAACACTGTTCACGGGCATAGAC-3′), and C2 (5′-TGTTTAAGTTT AGTGGATGGGGTGATAGGCCAGAACGAAC-3′) and D2 (5′- CGCGGATCCTGT CAGCTGTTTGGTTACG-3′) in two sets of asymmetric PCRs to amplify DNA fragments of 617 (A1B2) and 619 (C2D2) bp, respectively. DNA fragment A1B1 and C1D1 or A1B2 and C2D2 were annealed at their overlapping regions (underlined letters in primers B and C) and amplified as a single fragment using primers A1 and D1 or A1 and D2. The AD fusion products were purified, BamHI digested (the BamHI site is double-underlined in primer A1, D1, and D2), ligated into BglII-digested and phosphatase-treated pDM4 vector (Milton et al., 1996) and electroporated into E. coli MC1061(λpir) and plated on chloramphenicol plates at 30°C to obtain pDM-AHA1089 and pDM-FLGT plasmids, respectively. Introduction of the plasmids into A. hydrophila ATCC7966-Rif or AH-405 rifampicin-resistant (Rifr), was performed by triparental matings using the E. coli MC1061 (λpir) containing the insertion constructs and the mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing chloramphenicol and rifampicin. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. After sucrose treatment, transformants that were rifampicin-resistant (Rifr) and chloramphenicol sensitive (CmS) were chosen and confirmed by PCR.
The mutants AH-3::flaAΔflaBflgT and AH-3ΔlafAΔflgT were obtained by introduction of the pDM-FLGT plasmid into A. hydrophila AH-3::flaAΔflaB and AH-3::lafA, respectively, by triparental mating using the E. coli MC1061 (λpir) containing the plasmid and the mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing chloramphenicol, kanamycin and rifampicin or chloramphenicol and rifampicin, respectively. After sucrose treatment, transformants that were rifampicin and kamycine-resistant or rifampicin-resistent and chloramphenicol sensitive, respectively, were chosen, and confirmed by PCR.
Plasmid Constructions
Plasmid pBAD33-FLGT containing the complete flgT gene from A. hydrophila AH-3 under the arabinose promoter (pBAD) on pBAD33 (Guzman et al., 1995) was obtained by PCR amplification of genomic DNA. Oligonucleotides 5′-TCTAGACACGGTTCTGTGGTCTGTA-3′ and 5′-GTCGACGG GACCGCTCTATCCTACT-3′ generated a band of 1319bp containing the flgT gene (the XbaI site is underlined and the SalI site double-underlined). The amplified band containing the flgT gen was ligated into pGEM-Teasy (Promega) and transformed into E. coli XL1-Blue. The DNA insert was recovered by XbaI and SalI restriction digestion and ligated into XbaI-SalI digested pBAD33 vector to construct the pBAD33-FLGT plasmid. Recombinant plasmid was introduced by electroporation into the E. coli DH5α (Hanahan, 1983) and was sequenced. For complementation assay, the recombinant plasmid was introduced into the AH-3ΔflgT mutant (Rifr) by triparental mating using the E. coli DH5α containing the pBAD33-FLGT plasmid and the mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing chloramphenicol and rifampicin.
Isolation of the A. hydrophila Polar Flagellar Hook-basal Bodies
Isolation of the A. hydrophila polar flagella HBBs was carried out from an overnight culture in T.S.B. (1000 ml) at 25°C as described by Terashima et al. (2006). Briefly, after cultivation, the cells were harvested in a sucrose solution (0.5 M sucrose, 50 mM Tris-HCl at pH 8.0) and converted into spheroplasts by adding lysozyme and EDTA to final concentrations of 0.1 mg/ml and 2 mM, respectively. After lysis of spheroplasts with 1% (w/v) Triton X-100, 5 mM MgSO4, and 0.1 mg/ml DNase I were added to reduce viscosity and then, 5 mM EDTA was added. Unlysed cells and cellular debris were recovered by centrifugation at 17.000 × g for 20 min. Polyethylene glycol 6000 and NaCl were added to the lysate to final concentrations of 2% and 100 mM, respectively, and flagella were collected by centrifugation at 27000 g for 30 min. The pellet was suspended in TET buffer [10 mM Tris-HCl at pH 8.0, 5 mM EDTA, 0.1% (w/v) Triton X-100]. To remove cellular debris, the suspension was centrifuged at 1.000 × g for 15 min at 4°C and the supernatant was centrifuged at 100.000 × g for 30 min. To dissociate the flagella into monomeric flagellin, the pellet was suspended in TET buffer and diluted 30-fold in 50 mM glycine-HCl (pH 3.5) containing 0.1% (w/v) Triton X-100 and shaken for 60 min at room temperature. After treatment, the mixture was centrifuged at 1.000 × g for 15 min at 4°C and supernatant centrifuged 150.000 × g for 40 min and pellet suspended in TET buffer.
Anti-FlgT Polyclonal Serum
To obtain the A. hydrophila AH-3 FlgT we overexpressed A. hydrophila AH-3 flgT in E. coli using pET-30 Xa/LIC vector (Novagen). The A. hydrophila AH-3 flgT was amplified from AH-3 genomic DNA using primers PETflgTfor 5′-GGTATTGAGGGTCGCATGAAATTACCGCTGCTG-3′ and PETflgTrev 5′-AGAGGAGAGTTAGAGCCGCGGGCATTATACAAGAAG-3′. The PCR product was ligated into pET-30 Xa/LIC (Novagen) by their overlapping regions (underlined letters in primers) and electroporated into E. coli BL21(λDE3). The His6-FlgT protein was overexpressed and cell lysates obtained as previously reported for other proteins (Canals et al., 2007; Jiménez et al., 2009). The total membrane fraction was obtained by ultracentrifugation (200.000 × g 30 min at 10°C), the His6-FlgT protein was solubilized and purified with a Ni2+-NTA agarose (Quiagen) as previously reported (Al-Dabbagh et al., 2008). Approximately 200 μg of purified AH-3 FlgT was emulsified with 1 ml of Freunds complete adjuvant and inoculated subcutaneously into adult New Zealand rabbits. Booster injections of the flagellin protein were administered 4 and 6 weeks later. Antibodies were obtained by bleeding 10 days after the booster injection.
Immunological Methods
Western blot of whole cell proteins and supernatants from Aeromonas strains grown in T.S.B. at 25°C or purified polar and lateral flagella basal bodies, was performed as briefly described. Whole cells and supernatants came from equivalent numbers of cells harvested by centrifugation. The cell pellet was suspended in 50–200 μl of SDS PAGE loading buffer and boiled for 5 min.
After SDS-PAGE and transfer to nitrocellulose membrane at 1.3 A for 1 h, the membranes were blocked with bovine serum albumin (3 mg/ml), and probed with polyclonal rabbit anti-FlgT antibodies (1:1000). The unbound antibody was removed by three washes in PBS, and a goat anti-rabbit immunoglobulin G alkaline phosphatase conjugated secondary antibody (1:1000) was added. The unbound secondary antibody was removed by three washes in PBS. The bound conjugate was then detected by the addition of 5-bromo-4-chloroindolylphosphate disodium-nitroblue tetrazolium. Incubations were carried out for 1 h, and washing steps with 0.05% Tween 20 in phosphate-buffered saline were included after each incubation step.
Adherence Assay to HEp-2 Cell
Adherence assay was conducted as a slight modification of that described by Carrello et al. (1988). Bacteria were grown statically in brain heart infusion broth (BHIB) at 25°C, harvested by gentle centrifugation (1,600 × g, 5 min), and resuspended in PBS (pH 7.2) at approximately 107 CFU/ml (A600 = 0.07). The HEp-2 cell monolayer was infected with 1 ml of the bacterial suspension for 90 min at 37°C in 5% CO2. Following infection, the non-adherent bacteria were removed from the monolayer by three washes with PBS. The remaining adherent bacteria and the monolayers were then fixed in 100% methanol for 5 min. Methanol was removed by washing them with PBS, and the HEp-2 cells with the adherent bacteria were stained for 45 min in 10% (vol/vol) Giemsa stain (BDH) prepared in Giemsa buffer. The coverslips were air dried, mounted, and viewed by oil immersion under a light microscope. Twenty HEp-2 cells/coverslips were randomly chosen, and the number of bacteria adhering/HEp-2 cell was recorded. Assays were carried out in duplicates or triplicates.
Biofilm Formation
Quantitative biofilm formation was performed in a microtiter plate as described previously (Pratt and Kotler, 1998), with minor modifications. Briefly, bacteria were grown on TSA and several colonies were gently resuspended in TSB (with or without the appropriated antibiotic); 100 μl aliquots were place in a microtiter plate (polystyrene) and incubated 48 h at 30°C without shaking. After the bacterial cultures were poured out, the plate was washed extensively with water, fixed with 2.5% glutaraldehyde, washed once with water and stained with 0.4% crystal violet solution. After solubilisation of the crystal violet with ethanol-acetone (80/20, v/v) the absorbance was determined at 570 nm.
Results
Identification of a New Aeromonas spp. Protein Essential for Motility
Mesophilic Aeromonas have a constitutive unsheathed polar flagellum energized by sodium ions. The stator complex of Aeromonas polar flagellum is composed of two redundant pairs of membrane proteins: PomAB and PomA2B2, with different sensitivity to sodium concentrations; and two motility essential proteins (MotXY) which make up the T-ring (Wilhelms et al., 2009; Molero et al., 2011). In Vibrio spp. the sodium-driven polar flagellum shows a ring (H-ring) surrounding the LP-rings, which is composed of the FlgT protein and may be involved in the assembly of MotXY to the basal body (Cameron et al., 2008; Terashima et al., 2010). The analysis of A. hydrophila ATCC7966T, A. salmonicida subsp. salmonicida A449, A. veronii B565 and A. caviae Ae398 genome sequences (Seshadri et al., 2006; Reith et al., 2008; Beatson et al., 2011; Li et al., 2011) revealed an open reading frame (AHA_1089, ASA_3241, B565_3123, and AcavA_05659, respectively), annotated as hypothetical protein which deduced amino acid sequences exhibit 27–28% identity, 46–48% similarity and E-value of 1e-34 to 6e-36 to Vibrio spp. FlgT (Figure 1). The A. hydrophila AH-3 genomic library was screened by colony blotting using an AHA_1089 DNA probe leading to the identification of clone pLA-FLGT (Nogueras et al., 2000), which carries the entire flgT gene. A. hydrophila AH-3 FlgT is predicted to be 386 amino acids in length and exhibits 96% identity/98% similarity to A. hydrophila ATCC7966T AHA_1089. Furthermore, Aeromonas FlgT harbor a signal peptide for secretion with a cleavage site between Ala18 and Glu19 (Figure 1), which suggest it is translocated to the periplasmic space like MotX and MotY. As described in Vibrio FlgT, the Aeromonas FlgT show two conserved cysteine residues that might form a disulfide bond for protein stabilization (Figure 1).
FIGURE 1.
Alignment of FlgT amino acid sequences of Vibrio cholerae (VC_2208), Vibrio parahaemolyticus (VP_0767), and Vibrio vulnificus FlgT (VV_0952); and the hypothetical protein of Aeromonas hydrophila ATCC7966T (AHA_1089), A. salmonicida subsp. salmonicida A449 (ASA_3241), A. veronii B565 (B565_3123), A. caviae Ae398 (AcavA_05659), and A. hydrophila AH-3 (AH-3). Black letters in light gray boxes indicate residues that matched in at least two of the three Vibrio sequences. White letters in dark gray boxes indicate residues that matched in at least three of the five Aeromonas sequences. White letters in black boxes indicate residues that are present in Vibrio and Aeromonas sequences. The arrowhead shows the site of signal sequence cleavage. The stars show the conserved cysteine residues.
To investigate the role of this protein in the Aeromonas motility, defined insertion mutants were created in two different A. hydrophila strains: ATCC7966T, which only possess polar flagellum (ATCCΔAHA1089), and AH-3, which possess constitutive polar flagellum and inducible lateral flagella (AH-3ΔflgT). Motility assays in liquid media by light microscopy showed that AHA_1089 and flgT mutations abolish swimming motility in ATCC7966T and AH-3, respectively. However, whereas motility in soft agar was abolished in the ATCCΔAHA1089 mutant, in the AH-3ΔflgT mutant it causes a highly decrease of radial expansion (68% reduction), in relation to the wild-type. The radial expansion of AH-3ΔflgT mutant was similar to those observed in mutants without polar flagella as AH-3ΔflaAB mutant (Canals et al., 2006b) (Figure 2).
FIGURE 2.
Motility of A. hydrophila strains: AH-3, ATCC7966T, ATCCΔAHA1089, AH-3ΔflgT, AH-3::flaAΔflaB (AH-3 non-polar flagellum mutant), AH-3::flaAΔflaBflgT, AH-3ΔlafAΔflgT, and AH-3ΔflgT harboring plasmid pBAD33-FLGT grown 20 h at 25°C on soft agar. The mutant complemented with pBAD33 plasmid was grown with 0.2% L-arabinose.
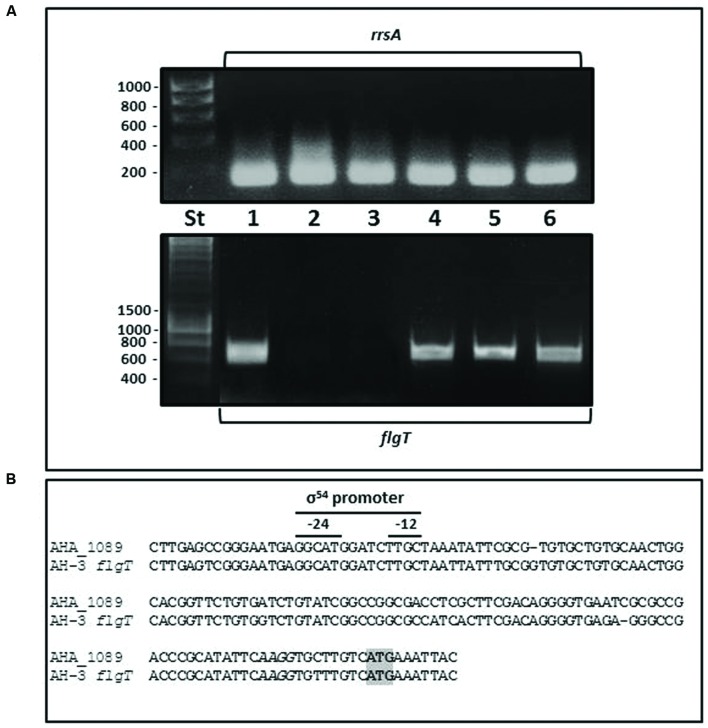
Although the Aeromonas flgT is located outside the polar and lateral flagella chromosomal regions it is involved in flagella motility, therefore we analyze whether flgT is under the control of some flagella regulator. By RT-PCR, we analyzed the flgT transcription in the wild-type AH-3; the non-polar flagella mutants AH-3::flrA, AH-3::flrBC, and AH-3::fliAp; and the non-lateral flagella mutants AH-3::lafK and AH-3::lafS. Data show flgT is not transcribed in AH.3::flrA and AH-3::flrBC, mutants, being transcribed in AH-3::fliAp, AH-3::lafK, and AH-3::lafS mutants (Figure 3). Therefore, Aeromonas flgT is transcribed from a polar-flagellum class III promoter. Furthermore, in silico analysis of DNA sequences upstream of AHA_1089 and AH-3 flgT show putative σ54 promoter sequences (Figure 3).
FIGURE 3.
(A) RT-PCR amplification of A. hydrophila flgT internal fragments from cDNA of A. hydrophila AH-3 (1), AH3::flrA (2), AH-3::flrBC (3), AH-3::fliAP (4), AH-3::lafK (5), and AH-3::lafS (6) mutants. DNA molecular marker (St). A. hydrophila ribosomal 16S (rrsA) amplification was used as a control for cDNA template. RT-PCR amplifications were performed at least twice with total RNA preparations obtained from a minimum of two independent extractions. (B) Promoter sequences of A. hydrophila ATCC7966T AHA_1089 and AH-3 flgT determined in silico. Italic letters indicates Shine-Dalgarno sequences upstream of AHA_1089 and AH-3 flgT start codon ATG (gray box). The -12 and -24 show sequences for the σ54 binding.
Complementation assays of AH-3ΔflgT with pLA-FLGT cosmid or pBAD33-FLGT plasmid induced with 0.2% L-arabinose showed that transconjugants are able to swim in liquid media and have a radial expansion in semi-solid plates identical to that of the wild-type AH-3 (Figure 2).
Role of FlgT in Polar and Lateral Motility
In order to analyze whether flgT is also involved in lateral flagella motility we performed two double mutants: a non-polar flagellated and FlgT mutants (AH-3::flaAΔflaBflgT) and a non-lateral flagellated and FlgT mutants (AH-3ΔlafAΔflgT). Both double mutants are unable to swim in liquid media but whereas motility in soft agar was abolished in the AH-3ΔlafAΔflgT mutant, the AH-3::flaAΔflaBflgT show a highly reduction in relation to observed in the wild-type AH-3 and similar to AH-3ΔflgT and AH-3::flaAΔflaB mutants (Figure 2). These data suggest that FlgT is only involved in polar flagellum motility and do not affect lateral flagella. Furthermore, AH-3::flrA mutant having mutated the polar flagellum master regulator, is unable to transcribe the polar flagellum genes, as well as flgT and shows identical motility phenotype as AH-3::flaAΔflaB and AH-3::flaAΔflaBflgT.
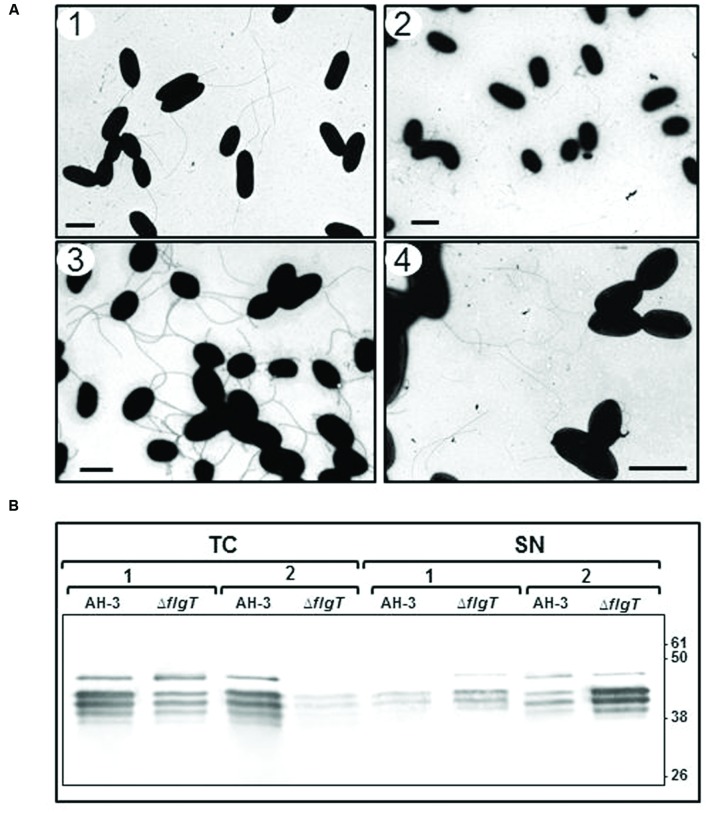
TEM of AH-3ΔflgT mutant, grown overnight at 25°C in liquid medium, showed many broken polar flagella not assembled on the bacterial surface. However, grown in soft agar showed the lateral flagella assembled on it (Figure 4). Using TEM and western-blot assays, we assessed whether the AH-3ΔflgT mutant has a defect in polar flagellum assembly or anchorage. We analyzed in 100 cells of the wild-type AH-3 and the flgT mutant, by TEM, the proportion of polar flagellated bacteria at different times of bacterial growth. In the wild-type, AH-3, the number of polar flagellated cells increase over time into the population; however, the number of polar flagellated cells shown a strong reduction in the flgT mutant over time. Thus, while in the mid-log phase growth (OD600 ≈ 0.5) the 58% of flgT mutant population shows an anchored polar flagellum, in the late-log phase growth (OD600 ≈ 2) the proportion of polar flagellated cells decreased to 12% (Figure 4). Furthermore, to quantify the amount of attached and unattached polar flagellum during growth, we analyzed whole-cells and supernatants of the wild-type AH-3 and the flgT mutant in the mid- and late-log phase growth, by western-blot using specific antiserum against purified polar flagellins (Gavín et al., 2002). These assays showed that most polar flagellins are detected in whole-cells of wild-type, because polar flagellum is anchored in the bacterial surface and only a small amount is released in the supernatant, both in mid- and late-log phase growth (Figure 4). However, in the flgT mutant, the amount of polar flagellins in supernatant, increase during bacterial growth, since the amount of not anchored polar flagellum increases, being higher in the late-log phase than in the mid-log phase (Figure 4). Complementation of AH-3ΔflgT mutant with pBAD33-FLGT plasmid, under induced conditions (0.2% L-arabinose), restore the anchorage of polar flagellum in the late-log phase growth and reduce the amount of polar flagellum in the supernatant. These data suggest that the reduced number of flagellated bacteria in the flgT mutant population was due to a defect in their ability to anchor the polar flagellum to surface.
FIGURE 4.
(A) Transmission electron microscopy of AH-3ΔflgT mutant during the mid-log-phase (OD600 ≈ 0.5) (1) and the late-log-phase (OD600 ≈ 2) (2) growth at 25°C on liquid media and at the late-log-phase growth in soft agar plates (3). A. hydrophila AH-3 during the late-log-phase growth at 25°C on liquid media (4). Bacteria were gently placed onto Formvar-coated copper grids and negatively stained using 2% uranyl acetate. Bar = 2 μm. (B) Western-blot of total bacterial cells (TC) and supernatants (SN) of A. hydrophila AH-3 and AH-3ΔflgT mutant during the mid-log-phase (1) and the late-log-phase (2) growth at 25°C on liquid media, using specific antiserum against purified polar flagellins.
Location of FlgT in the Polar Flagella
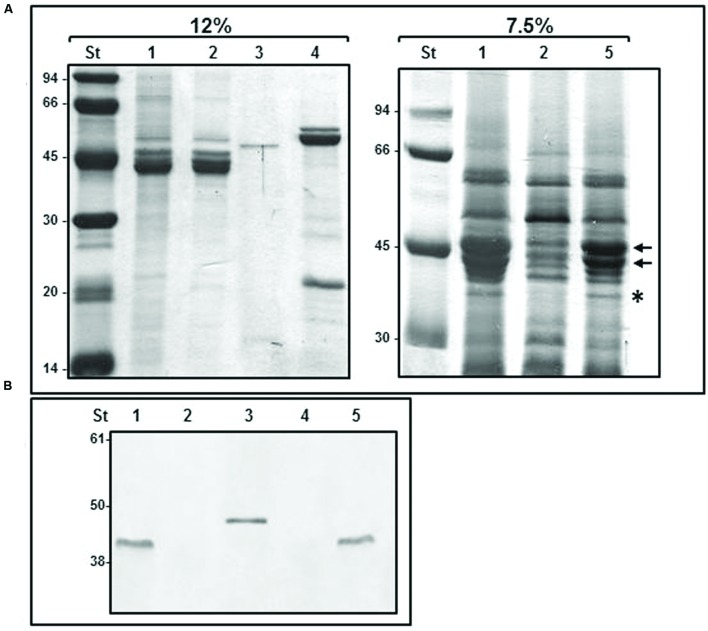
The evidences that FlgT plays a role in the anchoring of Aeromonas polar flagellum to the cell surface prompted us to search its location. In Vibrio spp. the orthologs protein has been detected in the periplasmic space and constitutes the H-ring, which is associated with the polar flagellum basal-body (Terashima et al., 2010). In order to locate the Aeromonas FlgT, we purified polar flagellum HBB of A. hydrophila AH-3 and AH-3ΔflgT mutant growth in liquid media at 25°C and analyzed them by SDS-PAGE and Coomasie-blue stained. In a 12% SDS-PAGE, the bands profile of the wild-type and the mutant were similar; however, in a 7.5% SDS-PAGE they showed some differences. The wild-type shows two intense bands around 40 KDa, which correlate with the molecular weight of polar flagellins (FlaA and FlaB) present in the HBBs fraction as a result of the resistance to despolymerization that have the highly glycosylated polar flagellum of Aeromonas AH-3. These two bands are strongly reduced in the AH-3ΔflgT and also present in the mutant complemented with pBAD33-FLGT grown under inducer conditions (Figure 5). Furthermore, the 7.5% SDS-PAGE showed some bands which are absent in the AH-3ΔflgT mutant, being one of them correlated with the molecular weight of MotY and MotX proteins that constitute the T-ring of the flagellum basal body. In order to known if one of these absent band correspond to FlgT, we make a transductional fusion of AH-3 FlgT with six histidine residues by cloning the A. hydrophila AH-3 flgT in the pET-30 Xa/LIC vector. The His6-FlgT was overexpressed in E. coli and purified protein was used to obtain specific A. hydrophila AH-3 FlgT antiserum. Polar flagellum HBBs of A. hydrophila AH-3 and AH-3ΔflgT mutant were analyzed by western-blot assays using specific A. hydrophila AH-3 FlgT antiserum. We only found positive reaction with the purified His6-FlgT and with a band of 42 KDa present in the polar flagellum HBB of A. hydrophila AH-3 (Figure 5). We also obtained the lateral flagella HBB of AH-3::flhA mutant, which do not have the FlhA protein of the polar flagellum export-apparatus and is unable to constitute the polar flagella basal body. Western-blot assays using AH-3 FlgT antiserum do not had positive reaction with the lateral HBB (Figure 5). Data suggest that FlgT is a component of the polar HBB of Aeromonas as previously described in Vibrio ssp.
FIGURE 5.
Purified HBBs of polar and lateral flagella. (A) 12 and 7.5% SDS-PAGE of purified flagella HBB. The black arrows show FlaA and FlaB polar flagellins. The asterisk shows a band which molecular weight correlate to MotY and MotX proteins. (B) Western blot analysis using A. hydrophila AH-3 FlgT antiserum (1:1,000). Size standard (St); polar flagella HBB of AH-3 (1), AH-3ΔflgT mutant (2); purified His6-FlgT protein (3); lateral flagella HBB of AH-3::flhA (4); and AH-3ΔflgT mutant complemented with pBAD33-FLGT grown under inducer condition (5).
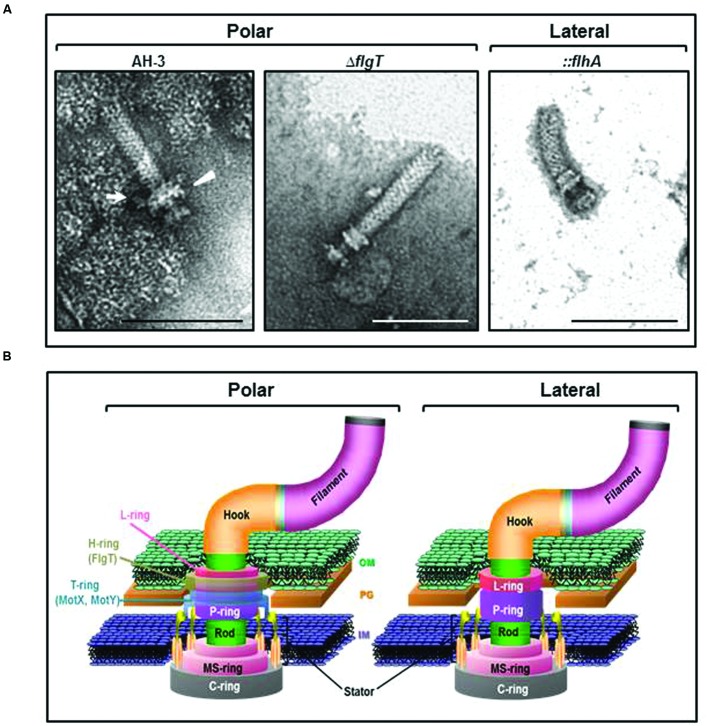
To investigate if FlgT constitute a ring around the LP-ring, we performed TEM of purified polar flagella HBBs from AH-3 and the AH-3ΔflgT mutant. The HBBs of the wild-type AH-3 have a LP-ring with a protuberance which is not present in the LP-ring of the AH-3ΔflgT mutant. Furthermore, the HBBs of AH-3ΔflgT mutant also lost the T-ring, consisting for the MotX and MotY proteins. The lateral flagella HBBs of the wild-type AH-3 were structurally similar to the polar HBBs of the AH-3ΔflgT mutant (Figure 6).
FIGURE 6.
(A) Transmission electron microscopy of A. hydrophila HBB. Polar flagellum HBB of AH-3 (AH-3) and AH-3ΔflgT (ΔflgT); and lateral flagella HBB of AH-3::flhA (::flhA). White arrow points the H-ring and white triangle points the T-ring. The HBB were gently placed onto Formvar-coated copper grids and negatively stained using 2% uranyl molibdate. Bar = 100 nm. (B) Diagram of the polar and lateral flagellar basal body of A. AH-3. H- and T-rings surround the LP-ring in the polar flagellum basal body of AH-3. OM, outer membrane; PG, peptidoglycan layer; and IM, inner membrane.
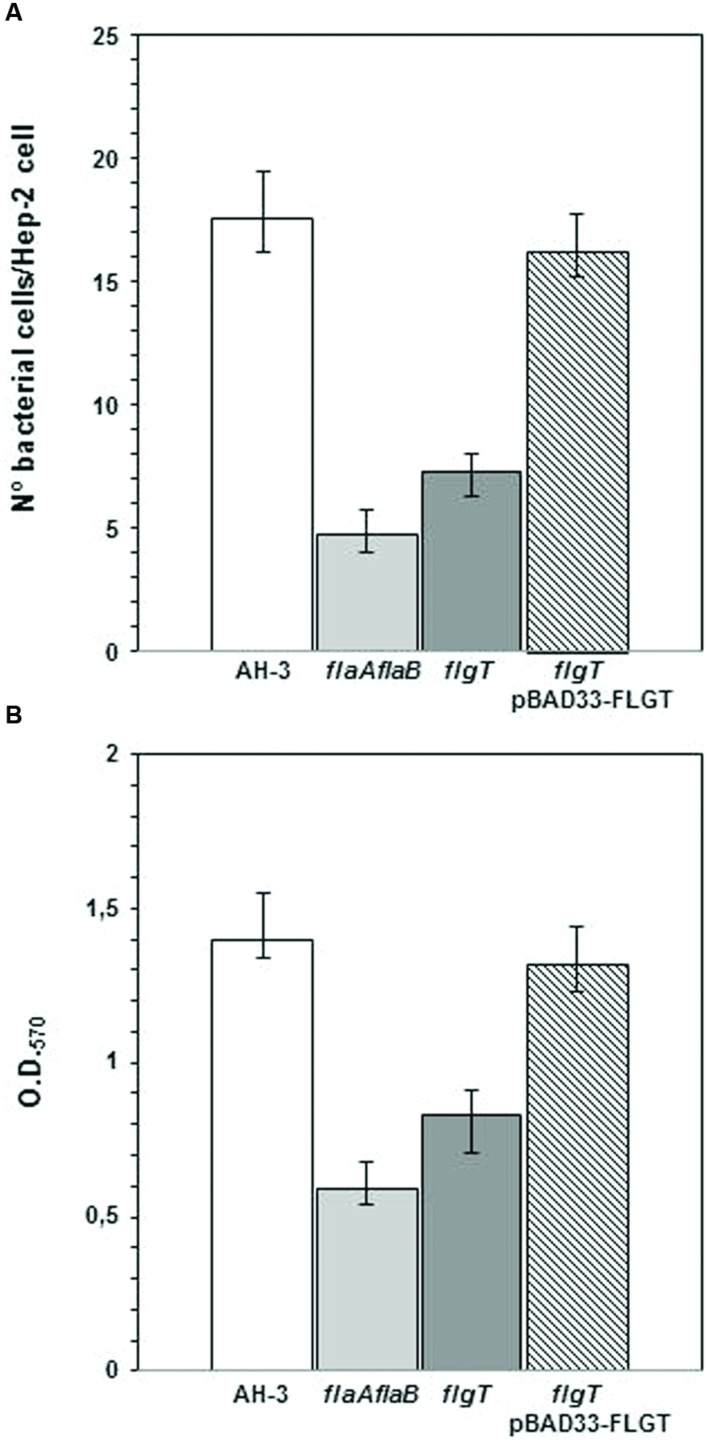
Adhesion to HEp-2 Cells and Biofilm Formation
In order to correlate polar flagella stability and motility with adherence to mammalian cells, we examined the interaction of flgT mutant with cultured monolayers of HEp-2 cells. Differences in adherence were calculated by determining the average number of bacteria adhering to HEp-2 cells (Figure 7). We also compared the ability of the wild type and the flgT mutant to form biofilms in microtiter plates (Figure 7). The A. hydrophila wild type strain, AH-3, exhibited an adhesion value of 17.6 (17.6 ± 1.9) bacteria adhered per HEp-2 cell and a biofilm formation ability with an OD570 value of 1.43 (1.43 ± 0.15). The mutant lacking FlgT showed a 58.5% reduction in HEp-2 cell adhesion, which is slightly higher than that determined in the non-polar flagellated mutant AH-3::flaAΔflaB (72%). The results obtained in biofilm formation (Figure 7) show a similar overall pattern to the adhesion, when comparing the characteristics of wild-type and mutant strains. The effects observed in biofilms formation are less marked. Mutants lacking FlgT showed a 40.7% reduction and the non-polar flagellated AH-3::flaAΔflaB have a 57.8% reduction (Figure 7). Both, adhesion to HEp2-cells and biofilm formation were fully rescued in the flgT mutants by the introduction of the wild-type gene.
FIGURE 7.
(A) Adhesion of A. hydrophila AH-3, non-polar flagellum mutant AH-3::flaAΔflaB, AH-3ΔflgT mutant, and AH-3ΔflgT complemented with pBAD33-FLGT to HEp-2 cells. Mean number of adherent bacteria per HEp-2 cells. (B) Biofilm formation ability of A. hydrophila AH-3, non-polar flagellum mutant AH-3::flaAΔflaB, AH-3ΔflgT mutant, and AH-3ΔflgT complemented with pBAD33-FLGT. OD570 quantifies the amount of crystal violet retained by the biofilm on the microtiter plates after staining. The complemented mutant was grown under induced conditions (0.2% L-arabinose). The average of three independent experiments (each experiment performed in duplicate) is show. Error bars represent standard deviations.
Discussion
Mesophilic Aeromonas possess a constitutive glycosylated polar flagellum energized by an electrochemical potential of sodium ions. In previous study we described that polar flagellum stator complex is composed of two redundant pairs of membrane proteins: PomAB and PomA2B2, with different sensitivity to sodium concentrations; and two essential motility proteins (MotXY) which make up the T-ring (Wilhelms et al., 2009; Molero et al., 2011). The analysis of A. hydrophila ATCC7966T, A. salmonicida subsp. salmonicida A449, A. veronii B565, and A. caviae Ae398 genome sequences (Seshadri et al., 2006; Reith et al., 2008; Beatson et al., 2011; Li et al., 2011) revealed an open reading frame which deduced amino acid sequences exhibit 27–28% identity, 46–48% similarity to Vibrio spp. FlgT. Hybridization assays using an AHA_1089 DNA probe led to identify an homologous gene in A. hydrophila AH-3. As in Vibrio spp., upstream of Aeromonas flgT we found two open reading frames which encode amino acid sequences orthologs to flgO and flgP; however, the chromosomal location is different in Vibrio spp, Shewanella oneidensis, and A. hydrophila. In Aeromonas these genes are outside the polar flagella chromosomal regions and flgT transcribed under the control of a σ54 promoter FlrC-dependent, as determined by RT-PCR analysis in polar flagella transcriptional regulators mutants (AH.3::flrA, AH-3::flrBC, and AH-3::fliAp). Lateral flagella regulators as LafK and LafS do not control flgT transcription (Figure 3). As described in Vibrio (Terashima et al., 2010), the Aeromonas FlgT shows an N-terminal signal peptide for secretion with a cleavage site between Ala18 and Glu19, which suggest is translocated to the periplasmic space like MotX and MotY, and two conserved cysteine residues that might form a disulfide bond for protein stabilization (Figure 1). By constructing specific flgT mutants in the wild-type (AH-3ΔflgT), a non-polar flagella mutant (AH-3::flaAΔflaBflgT) and a non-lateral flagella mutant (AH-3ΔlafAΔflgT) we demonstrated that Aeromonas FlgT is only involved in polar flagella motility. Single and double mutants are unable to swim in liquid medium; however, motility in soft-agar plates was only abolished in the double mutant unable to form lateral flagella and FlgT (AH-3ΔlafAΔflgT). The double mutant unable to produce polar flagella and FlgT (AH-3::flaAΔflaBflgT), as well as the single mutants for polar flagella (AH-3::flaAΔflaB) or FlgT (AH-3ΔflgT) only show reduction of their radial expansion in soft-agar plates, since lateral flagella are able to rotate (Figure 2). The swimming phenotype of wild-type was restored when mutants were complemented using the pLA-FLGT cosmid or pBAD33-FLGT plasmid in presence of L-arabinose.
In order to known whether inability to swim was produced by an unassembled polar flagellum or a flagellum unable to rotate, the AH-3ΔflgT was analyzed by TEM after grown overnight at 25°C in liquid media. The flgT mutant shows many broken polar flagella not assembled on the bacterial surface (Figure 4). Analysis of attached and unattached polar flagellum at different times of bacterial growth show that the amount of unattached flagella increases over the phase growth, as reported in Vibrio cholerae flgT mutant (Martinez et al., 2010). Thus, in the mid-log phase, more than half of bacterial cells (58%) show attached the polar flagellum, and the amount of polar flagellins is similar in whole cells and supernatant. Nevertheless, in the late-log phase, only a reduced number of cells (12%) show polar flagella attached in its surface, being mostly aflagellate or with broken flagella and the amount of polar flagellins in the supernatant were strongly higher than in whole cells (Figure 4). Although the polar flagellum was assembled in the mid log-phase and probably rotates, their rotation in absence of FlgT makes the flagella structure to be unstable and break. Therefore, the more rotate, more unstable is the flagellum structure and the number of aflageladas cells increase in the late log-phase. These results suggest that Aeromonas flgT mutant is able to assemble the polar flagellum but probably, it is instable, being its rotation responsible of disbanding from the cell surface. Furthermore, the abolishment of FlgT not affect transcription of class IV polar flagella genes, as was reported in Vibrio spp. (Martinez et al., 2010), since the two Aeromonas polar flagellines, FlaA and FlaB, which are transcribed from class IV promoters, have detected by specific antiserum in the AH-3Δ flgT mutant. Differences in lateral flagella assembly were not detected in the wild-type, AH-3, and the Aeromonas flgT mutant after grown in soft-agar plates (Figure 4).
Evidences that FlgT plays a role in the stability and anchoring of Aeromonas polar flagellum to the cell surface and that Vibrio spp. orthologs protein has been associated to the polar flagellum LP-ring (Terashima et al., 2010), in the periplasmic space, prompted us to search the location of FlgT in Aeromonas. Purified polar HBB of the wild-type and the flgT mutant were analyzed by SDS-PAGE stained with Coomasie-blue and by western-blot, using specific A. hydrophila AH-3 FlgT antiserum. The bands profile of the wild-type and the mutant was similar in a 12% SDS-PAGE, however, some differences were visualized in a 7.5% SDS-PAGE (Figure 5). The HBBs fraction of wild-type shows two intense bands (around 40 KDa) whose molecular weight correlates with those of polar flagellins (FlaA and FlaB). The high presences of flagellins are a result of the resistance to despolymerization that have the glycosylated polar flagellum of Aeromonas AH-3. These two bands are strongly reduced in the AH-3ΔflgTbecause HBB were purified after overnight grown and most polar flagella are released to the supernatant in the mutant. Furthermore, some bands around 32 KDa are absent in the AH-3ΔflgT mutant, which correlated with the molecular weight of MotY and MotX proteins that constitute the T-ring of the polar flagellum HBB (Molero et al., 2011). Western-blot assays with specific anti FlgT antiserum shows the presence of FlgT in the polar HBB of wild-type, but absent in the flgT mutant. FlgT was not detected in A. hydrophila lateral flagella HBBs (Figure 5).
Analysis of HBBs by TEM showed polar flagellum HBBs of the flgT mutant were similar to lateral flagella HBBs of the flhA mutant (polar aflagellated mutant) and did not show protuberances associated to the LP-ring, corresponding to the H- and T-rings (Figure 6). As described in Vibrio spp (Terashima et al., 2006, 2010), the data suggest that Aeromonas FlgT constitute the H-ring associated to the LP-ring and probably anchor the T-ting, whose components are MotX and MotY (Molero et al., 2011). However, in contrast to described in Vibrio spp. (Martinez et al., 2010) the loss of the T-ring is not produced by the non-transcription of polar flagellum class IV genes in the flgT mutant, but rather probably for its inability to anchor or stabilize the T-ring in absence of H-ring. The absence of T-ring could correlate with the loss of ≈32 KDa bands in the HBB of flgT mutant analyzed in 7.5% SDS-PAGE, which may correspond to the lost MotX and MotY (Figures 5 and 6)
In our previous research we described that adhesion and biofilms formation of Aeromonas is affected for the loss of polar flagellum, as well as for its inability to rotate, since bacterial do not make sufficient contact with the epithelial cells (Canals et al., 2006b). The loss of FlgT reduces progressively during the grown the amount of bacterial cells with an anchored polar flagellum and therefore, the number of motile bacteria. This phenotype leads to a strong reduction of adherence ability and biofilm formation in relation to wild-type, which is somewhat higher than the quantified in a non-polar flagella mutant (Figure 7).
Then, our data in A. hydrophila suggests that FlgT is present in the HBB of the unsheathed polar flagellum, which is sodium-driven by two different stator complexes. This protein constitutes a substructure in the polar HBB, the H-ring, associated to the LP-ring and it is probably essential for anchorage and stability of the T-ring but is not involved in the transcription of polar flagella genes. Therefore, FlgT is essential for polar flagellum stability and rotation. Furthermore, FlgT is not present in HBB of lateral flagella.
Author Contributions
SM and JT conceived the study and analyzed the data. SM drafted the manuscript and JT critically commented and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Plan Nacional de I + D (Ministerio de Economía y Competitividad, Spain) and from Generalitat de Catalunya (Centre de Referència en Biotecnologia). We thank Maite Polo for her technical assistance and the Servicios Científico-Técnicos from University of Barcelona.
References
- Al-Dabbagh B., Mengin-Lecreux D., Bouhss A. (2008). Purification and characterization of the bacterial UDP-GlcNAC:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J. Bacteriol. 190 7141–7146. 10.1128/JB.00676-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen L. N., Hanson R. S. (1985). Construction of broad-host-range cosmid cloning vector: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J. Bacteriol. 161k955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarriba A., Merino S., Gavín R., Canals R., Rabaan A., Shaw J. G., et al. (2003). A polar flagella operon (flg) of Aeromonas hydrophila contains genes required for lateral flagella expression. Microb. Pathog. 34 249–259. 10.1016/S0882-4010(03)00047-0 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai Y., Kojima S., Kato H., Nishioka N., Kawagishi I., Homma M. (1997). Putative channel component for the fast-rotating sodium-driven flagella motor of a marine bacterium. J. Bacteriol. 179 5104–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S. R., et al. (2002). The pfam protein families database. Nucleic Acids Res. 30 276–280. 10.1093/nar/30.1.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatson S. A., das Graças de Luna M., Bachmann N. L., Alikhan N. F., Hanks K. R., Sullivan M. J., et al. (2011). Genome sequence of the emerging pathogen Aeromonas caviae. J. Bacteriol. 193 1286–1287. 10.1128/JB.01337-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C. (2003). The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72 19–54. 10.1146/annurev.biochem.72.121801.161737 [DOI] [PubMed] [Google Scholar]
- Blair D. F. (2003). Flagellar movement driven by proton translocation. FEBS Lett. 545 86–95. 10.1016/S0014-5793(03)00397-1 [DOI] [PubMed] [Google Scholar]
- Blair D. F., Berg H. C. (1990). The MotA protein of E. coli is a proton conducting component of the flagellar motor. Cell 60 439–449. 10.1016/0092-8674(90)90595-6 [DOI] [PubMed] [Google Scholar]
- Cameron D. E., Urbach J. M., Mekalanos J. J. (2008). A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 105 8736–8741. 10.1073/pnas.0803281105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals R., Altarriba M., Vilches S., Horsburgh G., Shaw J. G., Tomás J. M., et al. (2006a). Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 188 852–862. 10.1128/JB.188.3.852-862.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals R., Jiménez N., Vilches S., Regué M., Merino S., Tomás J. M. (2007). The role of Gne and GalE in the virulence of Aeromonas hydrophila serotype O34. J. Bacteriol. 189 540–550. 10.1128/JB.01260-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals R., Ramirez S., Vilches S., Horsburgh G., Shaw J. G., Tomás J. M., et al. (2006b). Polar flagellum biogenesis in Aeromonas hydrophila. J. Bacteriol. 188 542–555. 10.1128/JB.188.2.542-555.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrello A., Silburn K. A., Budden J. R., Chang B. J. (1988). Adhesion of clinical and environmental Aeromonas isolates to Hep-2 cells. J. Med. Microbiol. 26 19–27. 10.1099/00222615-26-1-19 [DOI] [PubMed] [Google Scholar]
- DeRosier D. J. (1998). The turn of the screw: the bacterial flagellar motor. Cell 93 17–20. 10.1016/S0092-8674(00)81141-1 [DOI] [PubMed] [Google Scholar]
- Doyle T. B., Hawkins A. C., McCarter L. L. (2004). The complex flagellar torque generator of Pseudomonas aeruginosa. J. Bacteriol. 186 6341–6350. 10.1128/JB.186.19.6341-6350.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N. R., Sosinsky G. E., Thomas D., Derosier D. J. (1994). Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235 1261–1270. 10.1006/jmbi.1994.1079 [DOI] [PubMed] [Google Scholar]
- Frenchel T. (2002). Microbial behavior in a heterogeneous world. Sciences 296 1068–1071. 10.1126/science.1070118 [DOI] [PubMed] [Google Scholar]
- Gavín R., Rabaan A. A., Merino S., Tomás J. M., Gryllos I., Shaw J. G. (2002). Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43 383–397. 10.1046/j.1365-2958.2002.02750.x [DOI] [PubMed] [Google Scholar]
- Ghenghesh K. S., Ahmed S. F., El-Khalek R. A., Al-Gendy A., Klena J. (2008). Aeromonas-associated infections in developing countries. J. Infect. Dev. Ctries 2 81–98. 10.3855/T2.2.81 [DOI] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166 557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- Ito M., Hicks D. B., Henkin T. M., Guffanti A. A., Powers B. D., Zvi L., et al. (2004). MotPS is the stator-force generator for motility of alkaliphilic Bacillus, and its homologue is a second functional Mot in Bacillus subtilis. Mol. Microbiol. 53 1365–2958. 10.1111/j.1365-2958.2004.04173.x [DOI] [PubMed] [Google Scholar]
- Jiménez N., Vilches S., Lacasta A., Regué M., Merino S., Tomás J. M. (2009). A bifunctional enzyme in a single gene catalyzes the incorporation of GlcN into the Aeromonas core LPS. J. Biol. Chem. 284 32995–33005. 10.1074/jbc.M109.038828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama E., Shiraishi T., Oosawa K., Baba N., Aizawa S. (1996). Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J. Mol. Biol. 255 458–475. 10.1006/jmbi.1996.0038 [DOI] [PubMed] [Google Scholar]
- Khan S., Khan I. H., Reese T. S. (1991). New structural features of the flagellar base in Salmonella typhimurium revealed by rapid-freeze electron microscopy. J. Bacteriol. 173 2888–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerdt A., Paulick A., Mock M., Jost K., Thormann K. M. (2009). MotX and MotY are required for flagellar rotation in Shewanella oneidensis MR-1. J. Bacteriol. 191 5085–5093. 10.1128/JB.00206-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu Y., Zhou Z., Huang H., Ren Y., Zhang Y., et al. (2011). Complete genome sequence of Aeromonas veronii strain B565. J. Bacteriol. 193 3389–3390. 10.1128/JB.00347-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. (1996). “Flagella and motility,” in Escherichia coli and Salmonella, ed. Neidhardt F. C. (Washington, DC: American Society for Microbiology; ), 123–145. [Google Scholar]
- Macnab R. M. (2003). How bacteria assemble flagella? Annu. Rev. Microbiol. 57 77–100. 10.1146/annurev.micro.57.030502.090832 [DOI] [PubMed] [Google Scholar]
- Martinez R. M., Jude B. A., Kirn T. J., Skorupski K., Taylor R. K. (2010). Role of FlgT in anchoring the flagellum of Vibrio cholerae. J. Bacteriol. 192 2085–2092. 10.1128/JB.01562-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. L. (2001). Polar flagella motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65 445–462. 10.1128/MMBR.65.3.445-462.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino S., Camprubi S., Tomas J. M. (1991). The role of lipopolysaccharide in complement-killing of Aeromonas hydrophila strains of serotype O:34. J. Gen. Microbiol. 137 1583–1590. 10.1099/00221287-137-7-1583 [DOI] [PubMed] [Google Scholar]
- Merino S., Rubires X., Aguilar A., Tomás J. M. (1997). The role of flagella and motility in the adherence and invasion to fish cell lines by Aeromonas hydrophila serogroup O:34 strains. FEMS Microbiol. Lett. 151 213–217. 10.1111/j.1574-6968.1997.tb12572.x [DOI] [PubMed] [Google Scholar]
- Milton D. L., O’Toole R., Horstedt P., Wolf-Watz H. (1996). Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero R., Wilhelms M., Infanzon B., Tomás J. M., Merino S. (2011). Aeromonas hydrophila motY is essential for polar flagellum function, and requires coordinate expression of MotX and Pom proteins. Microbiology 157 2772–2784 10.1099/mic.0.049544-0 [DOI] [PubMed] [Google Scholar]
- Nogueras M. M., Merino S., Aguilar A., Benedi V. J., Tomás J. M. (2000). Cloning, sequencing, and role in serum susceptibility of porin II from mesophilic Aeromonas hydrophila. Infect. Immun. 68 1849–1854. 10.1128/IAI.68.4.1849-1854.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M., Yakushi T., Kojima M., Homma M. (2002). MotX and MotY, specific components of the sodium-driven flagellar motor, colocalize to the outer membrane in Vibrio alginolyticus. Mol. Microbiol. 46 125–134. 10.1046/j.1365-2958.2002.03142.x [DOI] [PubMed] [Google Scholar]
- Paulick A., Koerdt A., Lassak J., Huntley S., Wilms I., Narberhaus F., et al. (2009). Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol. Microbiol. 71 836–850. 10.1111/j.1365-2958.2008.06570.x [DOI] [PubMed] [Google Scholar]
- Pratt L. A., Kotler R. (1998). Genetic analysis of Escherichia coli biofilm formation: role of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30 285–293. 10.1046/j.1365-2958.1998.01061.x [DOI] [PubMed] [Google Scholar]
- Rabaan A. A., Gryllos I., Tomas J. M., Shaw J. G. (2001). Motility and the polar flagellum are required for Aeromonas caviae adherence to Hep-2 cells. Infect. Immun. 69 4257–4267. 10.1128/IAI.69.7.4257-4267.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith M. E., Singh R. K., Curtis B., Boyd J. M., Bouevitch A., Kimball J., et al. (2008). The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427 10.1186/1471-2164-9-427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubires X., Saigi F., Piqué N., Climent N., Merino S., Albertí S., et al. (1997). A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179 7581–7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74 5463–5467. 10.1073/pnas.74.12.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R., Joseph S. W., Chopra A. K., Sha J., Shaw J. G., Graf J., et al. (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188 8272–8282. 10.1128/JB.00621-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B. J., McCarter L. L. (2003). Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185 4508–4518. 10.1128/JB.185.15.4508-4518.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz B., Berg H. C. (1991). Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173 7033–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H., Fukuoka H., Yakushi T., Kojima S., Homma M. (2006). The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol. Microbiol. 62 1170–1180. 10.1111/j.1365-2958.2006.05435.x [DOI] [PubMed] [Google Scholar]
- Terashima H., Koike M., Kojima S., Homma M. (2010). The Flagellar basal body-associated protein FlgT es essential for a novel ring structure in the sodium-driven Vibrio motor. J. Bacteriol. 192 5609–5615. 10.1128/JB.00720-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H., Kojima S., Homma M. (2008). Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270 39–85. 10.1016/S1937-6448(08)01402-0 [DOI] [PubMed] [Google Scholar]
- Terashima H., Li N., Sakuma M., Koike M., Kojima S., Homma M., et al. (2013). Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc. Natl. Acad. Sci. U.S.A. 110 6133–6138. 10.1073/pnas.1222655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toutain C. M., Zegans M. E., O’Toole G. A. (2005). Evidence fortwo flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187 771–777. 10.1128/JB.187.2.771-777.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T., Oosawa K., Aizawa S. (1992). M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227 672–677. 10.1016/0022-2836(92)90216-7 [DOI] [PubMed] [Google Scholar]
- von Graevenitz A. (2007). The role of Aeromonas in diarrhea: a review. Infection 35 59–64. 10.1007/s15010-007-6243-4 [DOI] [PubMed] [Google Scholar]
- Wilhelms M., Gonzalez V., Tomás J. M., Merino S. (2013). Aeromonas hydrophila lateral flagellar gene transcriptional hierarchy. J. Bacteriol. 195 1436–1445. 10.1128/JB.01994-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelms M., Molero R., Shaw J. G., Tomás J. M., Merino S. (2011). Transcriptional hierarchy of Aeromonas hydrophila polar-flagellum genes. J. Bacteriol. 193 5179–5190. 10.1128/JB.05355-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelms M., Vilches S., Molero R., Shaw J. G., Tomás J. M., Merino S. (2009). Two redundant sodium-driven stator motor proteins are involved in Aeromonas hydrophila polar flagellum rotation. J. Bacteriol. 191 2206–2217. 10.1128/JB.01526-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagasaki J., Okabe M., Kurebayashi R., Yakushi T., Homma M. (2006). Roles of the intramolecular disulfide bridge in MotX and MotY, the specific proteins for sodium driven motors in Vibrio spp. J. Bacteriol. 188 5308–5314. 10.1128/JB.00187-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Homma M. (2001). Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 1505 82–93. 10.1016/S0005-2728(00)00279-6 [DOI] [PubMed] [Google Scholar]