Abstract
The 14-3-3 gene family identified in all eukaryotic organisms is involved in a wide range of biological processes, particularly in resistance to various abiotic stresses. Here, we performed the first comprehensive study on the molecular characterization, phylogenetics, and responses to various abiotic stresses of the 14-3-3 gene family in Brachypodium distachyon L. A total of seven 14-3-3 genes from B. distachyon and 120 from five main lineages among 12 species were identified, which were divided into five well-conserved subfamilies. The molecular structure analysis showed that the plant 14-3-3 gene family is highly evolutionarily conserved, although certain divergence had occurred in different subfamilies. The duplication event investigation revealed that segmental duplication seemed to be the predominant form by which the 14-3-3 gene family had expanded. Moreover, seven critical amino acids were detected, which may contribute to functional divergence. Expression profiling analysis showed that BdGF14 genes were abundantly expressed in the roots, but showed low expression in the meristems. All seven BdGF14 genes showed significant expression changes under various abiotic stresses, including heavy metal, phytohormone, osmotic, and temperature stresses, which might play important roles in responses to multiple abiotic stresses mainly through participating in ABA-dependent signaling and reactive oxygen species-mediated MAPK cascade signaling pathways. In particular, BdGF14 genes generally showed upregulated expression in response to multiple stresses of high temperature, heavy metal, abscisic acid (ABA), and salicylic acid (SA), but downregulated expression under H2O2, NaCl, and polyethylene glycol (PEG) stresses. Meanwhile, dynamic transcriptional expression analysis of BdGF14 genes under longer treatments with heavy metals (Cd2+, Cr3+, Cu2+, and Zn2+) and phytohormone (ABA) and recovery revealed two main expression trends in both roots and leaves: up-down and up-down-up expression from stress treatments to recovery. This study provides new insights into the structures and functions of plant 14-3-3 genes.
Keywords: 14-3-3 genes, Brachypodium distachyon, gene structures, phylogenetic relationships, gene duplication, abiotic stress, qRT-PCR
Introduction
14-3-3 proteins are a family of highly conserved acidic molecules of ~30 kDa, which were first identified in human brain and named by their particular elution and migration patterns on two-dimensional diethylaminoethyl-cellulose chromatography and starch gel electrophoresis (Moore et al., 1967). Further, studies revealed that, apart from their abundance in mammalian brain, 14-3-3 proteins are present in all eukaryotic organisms (Ferl, 1996). Such proteins are highly helical and generally dimeric in structure, including homo- and hetero-dimers, in which the monomer includes nine antiparallel α-helices, and the dimer creates a large negatively charged channel where the residues conserved among all of the isoforms can be found (Liu et al., 1995). Recent crystal structure analysis of 14-3-3 proteins has shown that the residues in the nine α-helices are highly conserved compared with other isoforms (Benzinger et al., 2005), while the C-terminal is the most variable region within the sequence, necessary for dimer formation and functional diversity (Jones et al., 1995).
Although a few 14-3-3 proteins interact with other proteins without a phosphorylated or even with a phosphorylated sequence, the majority of them specifically bind to phosphoserine- and/or phosphothreonine-containing motifs (Muslin et al., 1996). To date, three types of 14-3-3 unique binding motifs have been identified, including R[S/Φ][+]pSXP (mode I), RX[Φ/S][+]pSXP (mode II), and pS-X1-2-COOH (motif III), where Φ is an aromatic oraliphatic amino acid, + is a basic amino acid, pS is phosphoserine/phosphothreonine, and X is any type of amino acid (Yaffe et al., 1997). The phosphorylation of 14-3-3 proteins themselves on specific residues also has critical effects on their functional execution (Aitken, 2002; Wilson et al., 2016).
The sessile nature of plants results in them having to continuously monitor and adjust critical physiological processes to appropriately respond to changing external environmental conditions (Denison et al., 2011). The character of 14-3-3 proteins, binding other proteins to regulate their functions and/or changing the functional structure themselves, makes them ideally suited to regulate plants' internal physiological processes to respond to changing external environmental conditions (Aitken, 2006). The 14-3-3 protein family is involved in a wide range of biological processes in plants given their divergent functions, such as photosynthesis, primary metabolism, growth and cell division, hormone pathways, and abiotic and biotic stresses (de Boer et al., 2013). In particular, 14-3-3 proteins play key roles in the adverse stress responses of plants. For example, the overexpression of Arabidopsis 14-3-3 λ in cotton directly revealed that 14-3-3 proteins play a major functional role in environmental stress responses (Yan et al., 2004). 14-3-3 proteins also have roles in the responses to biotic stress (Yang et al., 2009). In addition, 14-3-3 proteins have close and complex relationships with phytohormones, such as ABA, gibberellic acid (GA), and H2O2, which participates in hormone signaling pathways (Schoonheim et al., 2007; Cutler et al., 2010).
Brachypodium distachyon L. (2n = 10) is a wild annual grass endemic to the Mediterranean and Middle East, which belongs to the Pooideae subfamily in the grasses (Draper et al., 2001). It is promising as a model system of grasses, especially for economically valuable crops such as wheat and barley, because it has the physical and biological attributes necessary to serve as a model system, including small size, diploid accessions, a small genome (272 Mb), self-fertility, short generation time with a life cycle of < 4 months, a small stature but large seeds, and simple growth requirements (Draper et al., 2001; Vogel and Hill, 2008). The genome sequencing and annotation of B. distachyon 21 (Bd21) were completed in 2010, leading to the functional genome analysis available (International Brachypodium Initiative, 2010). To date, the plant 14-3-3 gene family has been widely investigated in a large number of green plants, such as Arabidopsis (Wu et al., 1997), rice (Chen et al., 2006), soybean (Li and Dhaubhadel, 2011), and Medicago truncatula (Qin et al., 2016). However, the structural features, phylogenetics, and functional properties of the 14-3-3 gene family in B. distachyon have not been reported so far.
In the present work, we performed the first comprehensive study on the molecular characterization, phylogenetic relationships, and abiotic stress responses of the 14-3-3 gene family in B. distachyon L. Our results provide new insights into the structures and functions of plant 14-3-3 genes.
Materials and methods
Sequences retrieval and identification of 14-3-3 genes
14-3-3 gene families were identified from 13 species representing the plant lineage from unicellular green algae to multicellular plants. The search was performed using “14-3-3” as a keyword in TAIR database (https://www.arabidopsis.org/), and the Phytozome v10.3 database (http://www.phytozome.org). Thirteen 14-3-3 genes from Arabidopsis thaliana were first retrieved and then used as query sequences to run BLAST program against phytozome v10.3. The sequences obtained from corresponding plant-genome annotation resources were analyzed, in which the sequences up to E ≤ 1E–10 was selected as candidate sequences, and partial and redundant sequences were excluded manually. Sequences were collected from the following groups and species: the unicellular green algae Chlamydomonas reinhardtii; the moss Physcomitrella patens; the lycophyte Selaginella moellendorffii; the monocotyledonous angiosperms Brachypodium distachyon, Oryza sativa, Triticum aestivum, Setaria italica, Zea mays, and Sorghum bicolor; and the dicotyledonous angiosperms Arabidopsis thaliana, Glycine max, Solanum lycopersicum, and Gossypium raimondii. The 14-3-3 conserved domain of selected candidate sequences were further confirmed by the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/; Letunic et al., 2015) and Pfam (http://pfam.xfam.org/; Finn et al., 2014) database. The proteins up to E ≤ 1E–10, with methionine as the initial amino acid, and with amino acid sequence between 220 and 350 residues were selected as candidate proteins. Furthermore, sequences with partially conserved functional domains were further excluded, which is based on the search results of the SMART and Pfam (PF 00244). The protein sequences and the corresponding coding sequences and genomic sequences of the collected 14-3-3 genes were all obtained from phytozome database.
Chromosomal locations and phylogenetic analysis
Location information of 14-3-3 genes on B. distachyon chromosome maps obtained from Phytozome v10.3 were mapped by MapInspect program and modified manually. Phylogenetic trees were constructed based on Bayesian inference using Markov Chain Monte Carlo (MCMC) method (Hall, 2005). Initially, full proteins were performed multiple sequence alignment based on MUSCLE program (http://www.ebi.ac.uk/Tools/msa/muscle/; Edgar, 2004). Bayesian inference phylogenetic construction was performed with MrBayes v 3.2 using GTR (General Time Reversible) model with Γ distributed rates (gamma-distributed rate variation) (Ronquist et al., 2012). Bayesian analysis included mcmc ngen = 106 and samplefreq = 100. When the average standard deviation of split frequencies was below 0.01, the operation was terminated. After discarding the burn-in samples (first 25% of samples), the remaining data were used to generate a Bayesian tree, which was presented using software FigTree v1.4.2.
Exon-intron structure, conserved motif, chemical character, and promoter analysis
The exon-intron structure and motif analysis of 14-3-3 genes were performed according to the programs described previously (Song et al., 2014) with minimal modifications, in which the maximum number of motifs = 10. The 14-3-3 protein pI/Mw were determined by the Compute pI/Mw tool (http://web.expasy.org/compute_pi/) (Bjellqvist et al., 1994). The 1500 bp upstream sequences as promoter regions were collected from Phytozome database, and they were then submitted to PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al., 2002) to search their putative cis-acting elements.
Analyses of duplication events, positive selection, functional divergence, and coevolution
The duplication events, positive selection, functional divergence, and coevolution analyses of 14-3-3 genes were conducted using multiple methods according to previous description (Song et al., 2014) with minimal modifications, of which the mean synonymous substitution rates (λ) used for the approximate date of the duplication event calculation are following: 6.5 × 10−9 for Brachypodium (Blanc and Wolfe, 2004), rice (Yu et al., 2005), and maize (Fan et al., 2014), 6.1 × 10−9 for soybean (Blanc and Wolfe, 2004), and 1.5 × 10−8 for Arabidopsis (Bowers et al., 2003). However, an accurate λ for Solanum lycopersicum and Gossypium raimondii was not found.
Three-dimensional structure visualization of BdGF14 proteins
The structure of BdGF14 proteins was modeled by searching SWISS-MODEL database (http://swissmodel.expasy.org/) using the amino acid sequence (Biasini et al., 2014), which matched to the rice 14-3-3 protein (OsGF14c, PDB:3AXY), showing 235 from a total of 256 residues (Taoka et al., 2011). The Pymol software (Version 1.7.4 Schrödinger, LLC., http://pymol.org/) was applied to visualize the three-dimensional structure of BdGF14 proteins.
Plant cultures and stress treatments
Before germination, the uniform seeds of standard diploid inbred line of Bd21 were sterilized with 75% alcohol and 15% sodium hypochlorite, and then washed three times with sterile water. After sterilization, the seeds were germinated on filter paper saturated with water in complete darkness condition at 26°C for 3 days. At the 4th day, seedlings were transferred to and grown in dedicated cultivation baskets with full-strength Hoagland solution [5 mM KNO3, 5 mM Ca(NO3)2, 2 mM MgSO4, 1 mM KH2PO4, 50 μM FeNa2(EDTA)2, 50 μM H3BO3, 10 μM MnC12, 0.8 μM ZnSO4, 0.4 μM CuSO4, and 0.02 μM (NH4)6MoO24.] in the greenhouse under a 16/8 h (light/dark) photocycle at 28/26°C (day/night) condition with relative humidity of 70%. The nutrient solution was changed every 3 days. Seedlings at three leaves stage were treated with the following conditions: salinity stress (200 mM NaCl), drought stress (20% (w/v) PEG 6000), heavy metal stress [300 μM Cd2+ (CdCl2), Cr3+ (CrCl3), Cu2+ (CuSO4) and Zn2+ (ZnSO4)], hormone stress (20 mM H2O2, 100μM ABA and SA), hot stress (42°C) and cold stress (4°C). Leaf and root samples of control seedlings were harvested at 0 h. Except for hot and cold stress, samples were collected at 2 h, the corresponding samples of treated seedlings were harvested at 6, 12, 24, 48 h and recover 48 h. Each sample was collected with three biological replicates. All samples were immediately stored at −80°C prior to use.
Total mRNA extraction, qRT-PCR, and PCA analysis
Total mRNA was isolated from frozen samples using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. Genomic DNA removal and cDNA synthesis were conducted by using PrimeScript® RT reagent Kit with gDNA Eraser (TaKaRa). Gene-specific primers of each 14-3-3 gene in B. distachyon were designed using on-line tool Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi; Untergasser et al., 2007). The primers were checked by blasting primer sequences in the NCBI database (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome), and all primers were specifically consistent with the respective sequence of its target gene. The primer sequences for the qRT-PCR assays are listed in Table S1. The transcription level of each 14-3-3 gene in Bd21 were quantified with a CFX96 Real-Time PCR Detection System (Bio-Rad) using the intercalating dye SYBR-green following the 2(-Delta Delta C(T)) method (Livak and Schmittgen, 2001). The B. distachyon constitutively expressed S-adenosylmethionine decarboxylase gene (SamDC) was used as a reference for normalization (Hong et al., 2008). qRT-PCR was performed in a 20 μL volume reaction system containing 10 μL 2 × SYBR® Premix ExTaq® (TaKaRa), 2 μL 10-fold diluted cDNA, 0.15 μL of each gene-specific primer and 7.7 μL ddH2O. The PCR conditions were as follows: 95°C for 3 min, 40 cycles at 95°C for 20 s, 61°C for 15 s, and 72°C for 10 s. Triplicates for each PCR reaction and at least three biological replicates were performed for each gene. The qRT-PCR efficiency was determined by five serial 10-fold dilutions of cDNA. The optimal performance was conducted, in which the PCR amplification efficiency (E) of 95–105% and correlation coefficient (R2) of 0.995–0.999 were controlled (Figure S1).
The hierarchical cluster analysis of qRT-PCR results was performed using Cluster 3.0 according to the method described by Eisen et al. (1998) with minor modifications. In particular, the gene expression level at 0 h was defined as 1, the gene expression level at the remaining time periods was changed according to the original ratios. Subsequently, the relative ratios of gene expression were conducted Log2 transforming, and then the Euclidean distance similarity metric was used to define the similarity and the hierarchical clusters were assembled using the complete-linkage clustering method.
Principal Component Analysis (PCA) is a method that focuses on finding the main variations and revealing hidden structures present in the data set (Kristiansen et al., 2010). In the present study, coefficient and KMO and Bartlett's test of sphericity were used for dimension reduction analysis and the results were shown in the loading plot and scatter plot, respectively. PCA was performed using SPSS 19.0 software, and the loading plot and scatter plot of PCA was calculated and displayed with the average expression level of each genes.
Results
Genome-wide identification of the 14-3-3 gene family in B. distachyon and 12 other plant species
To explore the molecular characteristics and phylogenetic relationships of the 14-3-3 gene family in B. distachyon and other main plant species, we initially downloaded 13 known Arabidopsis 14-3-3 protein sequences from the TAIR database (https://www.arabidopsis.org/) and the Phytozome v10.3 database (http://www.phytozome.org), which were subsequently used as queries for searches of the Phytozome v10.3 database. The BLASTP search results were further checked using the online tools SMART and Pfam to confirm the presence of conserved 14-3-3 domains, and redundant and partial sequences were removed manually. Finally, a total of seven 14-3-3 genes from B. distachyon and 120 from 12 other plant species were obtained, including 2 from Chlamydomonas reinhardtii, 10 from Physcomitrella patens, 4 from Selaginella moellendorffii, 7 from Oryza sativa, 10 from Triticum aestivum, 9 from Setaria italica, 12 from Zea mays, 5 from Sorghum bicolor, 13 from Arabidopsis thaliana, 18 from Glycine max, 13 from Solanum lycopersicum, and 17 from Gossypium raimondii (Figure S2). Based on the nomenclature suggestions of rice 14-3-3 genes, each gene except for those from Arabidopsis was named as follows: the first two letters correspond to the species (for example, Os: Oryza sativa), followed by the family designation (GF14) and a letter as shown. Basic information on these 14-3-3 genes (including gene name, loci, protein length, intron number, predicted isoelectric point (PI), and molecular weight) was shown in Table S2, and their conserved motifs identified by SMART and Pfam were listed in Table S3. The encoding proteins of 14-3-3 genes ranged from 230 to 342 amino acids (aa) in length, and the majority of them were about 260 aa. The PI and molecular weight (MW) ranged from 4.61 to 6.26 and 25.85 to 38.13 kDa, respectively.
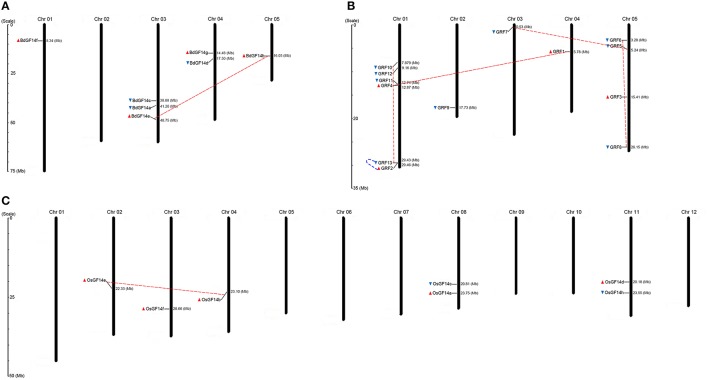
14-3-3 genes of B. distachyon, rice, and Arabidopsis were mapped to their chromosomes using the MapInspect program (Figure 1). In B. distachyon, the predicted seven BdGF14 genes were located on four chromosomes, in which BdGF14f was located on chromosome 01 while three BdGF14 genes (BdGF14a, c, and e) were on chromosome 03. The remaining three genes were distributed on chromosomes 04 (BdGF14d and g) and 05 (BdGF14b). In Arabidopsis, chromosome 01 had six 14-3-3 genes (GRF2, 4, 10, 11, 12, and 13), as the largest group of 14-3-3 genes, followed by chromosome 05 with four genes (GRF3, 5, 6, and 8). The last three genes (GRF9, 7, and 1) were distributed on the chromosome 02, 03, and 04, respectively. Similarly, rice OsGF14a and c were located on chromosome 08 and OsGF14d and h on chromosome 11, while OsGF14e, f and b were located on the chromosome 02, 03, and 04, respectively. These results suggest that 14-3-3 genes have no obvious chromosomal preferences and that they are more likely to be clustered together in certain chromosomal regions.
Figure 1.
Chromosomal distribution map of 14-3-3 genes in B. distachyon, Arabidopsis and rice. (A) B. distachyon. (B) Arabidopsis. (C) Rice. The chromosome numbers are indicated at the top of each bar, while the size of a chromosome is indicated by its relative length. The sea blue triangle and red triangle indicate the transcriptional direction of genes, sea blue (5′ to 3′) and red (3′ to 5′). The dashed lines with sea blue color represent tandem duplicated genes, whereas the dashed lines with red color represent segmentally duplicated genes.
Phylogenetic and structural analyses of 14-3-3 genes
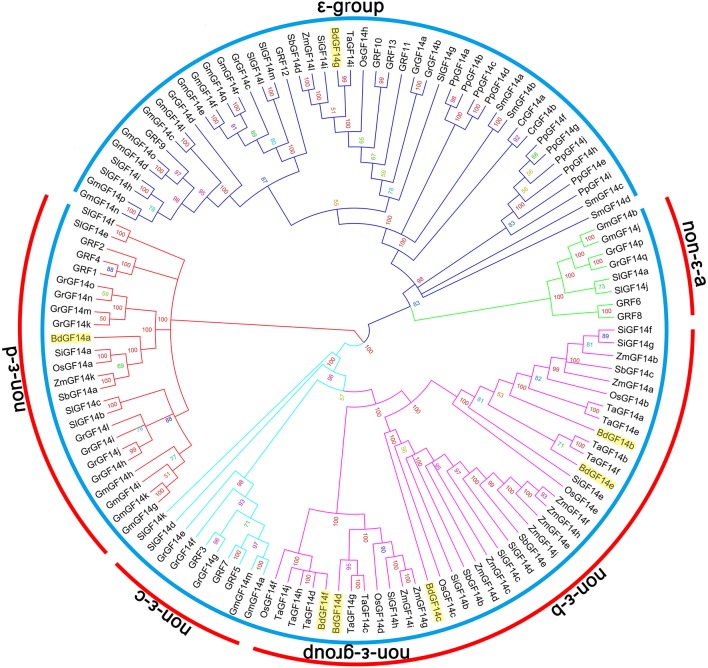
To investigate the evolutionary relationships of 14-3-3 genes within multiple plant species, the full amino acid sequences of 127 identified proteins were used for multiple sequence alignment using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) program. An unrooted phylogenetic tree was constructed using the Markov Chain Monte Carlo (MCMC) method based on Bayesian inference (Hall, 2005), as shown in Figure 2. To further confirm the topology of the phylogenetic tree, an unrooted neighbor-joining (N-J) phylogenetic tree was constructed, which exhibited similar topology to the Bayesian tree with only minor modifications (data not shown). According to the topology, the phylogenetic tree was separated into two distinct subgroups (ε-group and non-ε-group). Similarly, the non-ε-group could be further divided into four subgroups, named the non-ε-a subgroup, non-ε-b subgroup, non-ε-c subgroup, and non-ε-d subgroup. Among these five subgroups, ε-group was the largest, containing 46 14-3-3 gene members, followed by the non-ε-b subgroup (39 members), non-ε-d subgroup (24 members), non-ε-c subgroup (10 members), and non-ε-a subgroup (8 members).
Figure 2.
Phylogenetic relationships of plant 14-3-3 genes. A total of 127 complete protein sequences of the corresponding 14-3-3 genes obtained from 13 species from unicellular green algae to multicellular plants were aligned with MUSCLE program, and the phylogenetic tree was constructed based on Bayesian inference using Markov Chain Monte Carlo (MCMC) methods. The sky blue arcs indicate subgroups of 14-3-3 genes, while the red arcs indicate subfamilies of non-ε-group 14-3-3 genes. The 14-3-3 genes from B. distachyon are indicated by filled yellow rectangle.
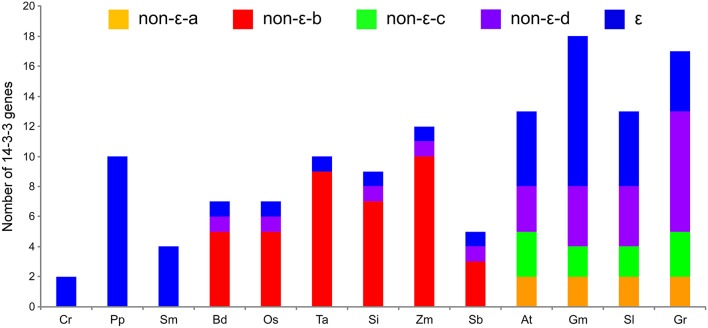
In detail, the distribution of 14-3-3 genes in different individual species had significant differences, as well as unique characters in each subgroup (Figure 3). 14-3-3 genes identified from lower plant species, including the unicellular green algae (Cr), moss (Pp), and lycophyte (Sm), were all present in the ε-group, while the non-ε-b subgroup only contained monocotyledon genes (Bd, Os, Ta, Si, Zm, and Sb), which included the majority of monocot 14-3-3 genes. Similarly, the non-ε-a and non-ε-c subgroups only contained the dicot genes (At, Gm, Sl, and Gr). On the other hand, the distribution among the same lineage showed high consistency. For B. distachyon and rice in the monocots, for example, BdGF14g belonged to the ε-group, while BdGF14a to the non-ε-d subgroup and BdGF14b-f to the non-ε-b subgroup (highlighted by green rectangles in (Figure 2). The phylogenetic tree showed that 14-3-3 genes in rice had almost the same distribution pattern as those in B. distachyon, in which the non-ε-b subgroup contained OsGF14b-f, and OsGF14a was separately placed in the non-ε-d subgroup. Among the dicots, the distribution patterns of 14-3-3 genes were also similar.
Figure 3.
The distribution of 14-3-3 genes based on subgroups in individual species. Cr, Chlamydomonas reinhardtii; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Bd, Brachypodium distachyon; Os, Oryza sativa; Ta, Triticum aestivum; Si, Setaria italica; Zm, Zea mays; Sb, Sorghum bicolor; At, Arabidopsis thaliana; Gm, Glycine max; Sl, Solanum lycopersicum; Gr, Gossypium raimondii.
The exon-intron organization of 14-3-3 genes in all selected plant lineages was analyzed by comparing the predicted coding sequences (CDSs) and their corresponding genomic sequences using the online tool Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). A detailed schematic diagram of the relative length of introns and conservation of the corresponding exon sequences within individual 14-3-3 gene paralog was shown in Figure S3, which was grouped and listed according to the phylogenetic tree construction. The results showed that gene members of the same subgroup possessed similar exon-intron structures. 14-3-3 genes generally had one to six introns, and most of them had three to five (Figure S4). There were no significant differences in the number of introns among individual genes within the same subgroup, while considerable differences were present in different subgroups. The ε-group had more introns than the non-ε-group. In particular, three subgroups, non-ε-a, non-ε-c, and non-ε-d subgroups, had a high percentage of 14-3-3 genes with three introns, up to 62.5, 90.0, and 83.3%, respectively. However, in the non-ε-b subgroup, those with four introns accounted for 89.7%. As the largest subgroup, the ε-group contained 29 genes (63.0%) with five introns, followed by those with three or four introns (15%) (Figure S4A). For the individual species, 14-3-3 genes had multiple introns, but the 14-3-3 genes in the monocots tended to have four introns, while those in the dicots had three or five (Figure S4B).
Conserved protein motif analysis of 14-3-3 genes was conducted using the Multiple Em for Motif Elicitation (MEME) program (http://meme-suite.org/). In total, 10 conserved motifs were detected from 127 14-3-3 genes, as shown in Figure S5; the order and number of motifs in individual genes were shown in Figure S6. The number of motifs contained in 14-3-3 genes was primarily seven or eight (Figure S7A). Similarly to the introns in 14-3-3 genes, the number of conserved motifs was highly consistent within the subgroup, but considerable differences were present in different subgroups. All 14-3-3 genes in the non-ε-a subgroup had seven motifs, while the dominant number of motifs in the non-ε-b, non-ε-c, and non-ε-d subgroups was eight, accounting for 89.7, 90.0, and 79.2%, respectively. The ε-group, different from the other four non-ε groups, primarily contained seven or eight motifs, reaching 39.1 and 56.5% of the total, respectively. For individual species, there were similar patterns in the number of conserved motifs, although there were significant differences in different lineages. Most of the 14-3-3 genes in the lower land plants had seven motifs, but those in monocot and dicot species had eight (Figure S7B). These characteristics were highly consistent with the number of introns.
Duplication events in the 14-3-3 gene family
In this study, we investigated the duplication events in the 14-3-3 gene family. The results showed that 14.3% (1 of 7) of 14-3-3 genes were derived from segmental duplication in B. distachyon, the same as in rice, but this rate was much lower than in Arabidopsis (30.8%, 4 of 13) (Figure 1). Among 13 plant species, soybean (Gm) had been most active in undergoing 14-3-3 gene duplication events, in which 55.6% (10 of 18) were derived from segmental duplication presented in 16 pairs of segmental duplication genes (Table 1). Among the lineages, dicots had 23 genes (37.7%) resulting from segmental duplication, corresponding to 32 pairs of segmental duplication genes, which is a much higher rate than in monocots (10.0%). The lower plant species had not undergone segmental duplication events. In addition, the segmental duplication events in monocots had only occurred in the non-ε-b subgroup, while those in dicots were separated among the remaining four subgroups.
Table 1.
Estimates of the dates for the segmental duplication events of 14-3-3 gene family.
| Gene pairs | KS (means ± s.d.) | Estimated time (mya) | GWD (mya) | References | |
|---|---|---|---|---|---|
| BdGF14b | BdGF14e | 0.593 ± 0.140 | 45.6 | 56–72 | International Brachypodium Initiative, 2010 |
| OsGF14b | OsGF14e | 0.612 ± 0.228 | 47.0 | 30–40 | Vandepoele et al., 2003 |
| ZmGF14a | ZmGF14b | 0.270 ± 0.000 | 20.8 | 5–12, 70 | Schnable et al., 2009 |
| ZmGF14c | ZmGF14d | 0.200 ± 0.020 | 15.4 | ||
| ZmGF14g | ZmGF14i | 0.330 ± 0.120 | 25.4 | ||
| GRF10 | GRF13 | 0.811 ± 0.116 | 27.0 | 28–48 | Ermolaeva et al., 2003 |
| GRF6 | GRF8 | 0.732 ± 0.144 | 24.4 | ||
| GRF5 | GRF7 | 0.656 ± 0.136 | 21.9 | ||
| GRF1 | GRF4 | 0.700 ± 0.000 | 23.3 | ||
| GmGF14a | GmGF14m | 0.182 ± 0.166 | 14.9 | 13, 59 | Schmutz et al., 2010 |
| GmGF14c | GmGF14l | 0.138 ± 0.048 | 11.3 | ||
| GmGF14d | GmGF14o | 0.147 ± 0.049 | 12.0 | ||
| GmGF14e | GmGF14f | 0.124 ± 0.036 | 10.2 | ||
| GmGF14e | GmGF14q | 0.620 ± 0.035 | 50.8 | ||
| GmGF14e | GmGF14r | 0.595 ± 0.060 | 48.8 | ||
| GmGF14f | GmGF14q | 0.638 ± 0.025 | 52.3 | ||
| GmGF14f | GmGF14r | 0.715 ± 0.147 | 58.6 | ||
| GmGF14g | GmGF14i | 0.613 ± 0.203 | 50.2 | ||
| GmGF14g | GmGF14h | 0.618 ± 0.141 | 50.7 | ||
| GmGF14g | GmGF14k | 0.123 ± 0.048 | 10.1 | ||
| GmGF14i | GmGF14h | 0.172 ± 0.055 | 14.1 | ||
| GmGF14h | GmGF14k | 0.450 ± 0.020 | 36.9 | ||
| GmGF14i | GmGF14k | 0.590 ± 0.137 | 48.4 | ||
| GmGF14n | GmGF14p | 0.135 ± 0.060 | 11.1 | ||
| GmGF14q | GmGF14r | 0.143 ± 0.048 | 11.7 | ||
| SlGF14d | SlGF14k | 0.470 ± 0.000 | * | 52–91 | Tomato Genome Consortium, 2012 |
| SlGF14l | SlGF14m | 0.628 ± 0.080 | * | ||
| SlGF14h | SlGF14i | 0.810 ± 0.120 | * | ||
| GrGF14a | GrGF14b | 0.413 ± 0.096 | * | 60 | Paterson et al., 2012 |
| GrGF14h | GrGF14l | 0.593 ± 0.222 | * | ||
| GrGF14h | GrGF14i | 0.607 ± 0.131 | * | ||
| GrGF14k | GrGF14m | 0.445 ± 0.105 | * | ||
| GrGF14k | GrGF14o | 0.455 ± 0.105 | * | ||
| GrGF14m | GrGF14o | 0.570 ± 0.227 | * | ||
| GrGF14m | GrGF14n | 0.630 ± 0.110 | * | ||
| GrGF14n | GrGF14o | 0.460 ± 0.000 | * | ||
| GrGF14p | GrGF14q | 0.745 ± 0.195 | * | ||
represents the unknown data.
Tandem duplication events often generate multiple members of one family within the same or neighboring intergenic regions. Interestingly, the tandem duplication events of 14-3-3 genes were extremely rare compared with the segmental duplication events and only four pairs of tandem duplication genes (PpGF14f/PpGF14g, PpGF14i/PpGF14j, GRF13/GRF2, and GmGF14m/GmGF14n) were found.
To estimate the approximate ages of segmental duplication events, the synonymous base substitution rates (Ks-values) were used as a proxy for time. The results showed that only one pair of segmental duplication genes were found in B. distachyon and rice, while maize, Arabidopsis, soy bean, tomato, and cotton had three, four, sixteen, three, and nine pairs, respectively (Table 1). The segmental duplication genes in B. distachyon were estimated to have originated ~45.6 million years ago (MYA), roughly consistent with the occurrence of large-scale duplications 56–72 MYA (International Brachypodium Initiative, 2010). In rice, the estimated time of emergence of the candidate segmental duplication genes was 47.0 MYA, also roughly consistent with the occurrence of large-scale duplications in rice 30–40 MYA (Vandepoele et al., 2003). Similarly, the estimated times of segmental duplication events for the remaining segmental duplication genes identified, except for those in tomato and cotton, roughly matched their genome-wide duplication(GWD; Table 1).
Functional divergence in the 14-3-3 gene family
The Type I and Type II functional divergences of gene clusters in the 14-3-3 gene family were estimated through a posterior analysis using the DIVERGE v3.0 program to investigate whether amino acid substitutions caused functional diversification, which was based on the N-J trees (data not shown).
For Type I functional divergence (θI), the estimated likelihood ratio statistic (LRT) values of the 10 pairs of 14-3-3 gene subfamilies ranged from 0.000 to 6.480 (Table 2). Especially for the subfamily pairs of non-ε-a/non-ε-c, non-ε-a/non-ε-d, and non-ε-a/non-ε-b, they were ranked in the first three positions among the 10 subfamily pairs, with LRT values up to 6.480, 5.426, and 2.802, respectively. Subsequently, we analyzed whether Type II functional divergence (θII) occurred among the pairs of 14-3-3 gene subfamilies. The results showed that Type II (θII) coefficients between five subfamily pairs, namely, ε/non-ε-a, ε/non-ε-b, ε/non-ε-c, ε/non-ε-d, and non-ε-a/non-ε-d, were all significantly greater than zero, indicating that there were significant changes in amino acid properties among these subfamilies. In contrast, the other five pairs of 14-3-3 gene subfamilies (non-ε-a/non-ε-b, non-ε-a/non-ε-c, non-ε-b/non-ε-c, non-ε-b/non-ε-c, and non-ε-c/non-ε-d) had coefficients of < 0, suggesting that no significant changes had occurred in amino acid properties between these subfamilies (Table 2).
Table 2.
Functional divergence between subfamilies of the 14-3-3 gene family.
| Group1 | Group2 | Type-I | Type-II | |||
|---|---|---|---|---|---|---|
| θI ± s.e. | LRT | Qk > 0.99 | θII ± s.e. | Qk > 0.99 | ||
| ε | non-ε-a | 0.27 ± 0.022 | 0.000 | 0 | 0.202 ± 0.137 | 45 |
| ε | non-ε-b | 0.001 ± 0.022 | 0.000 | 0 | 0.270 ± 0.134 | 0 |
| ε | non-ε-c | 0.001 ± 0.022 | 0.000 | 0 | 0.194 ± 0.136 | 46 |
| ε | non-ε-d | 0.060 ± 0.061 | 0.957 | 0 | 0.284 ± 0.126 | 0 |
| non-ε-a | non-ε-b | 0.186 ± 0.111 | 2.802 | 0 | 0.052 ± 0.064 | 5 |
| non-ε-a | non-ε-c | 0.278 ± 0.110 | 6.480* | 0 | 0.045 ± 0.050 | 4 |
| non-ε-a | non-ε-d | 0.150 ± 0.065 | 5.426* | 0 | 0.063 ± 0.058 | 5 |
| non-ε-b | non-ε-c | 0.121 ± 0.118 | 1.063 | 0 | −0.010 ± 0.058 | 0 |
| non−ε−b | non−ε−d | 0.042 ± 0.073 | 0.337 | 0 | −0.010 ± 0.065 | 0 |
| non−ε−c | non−ε−d | 0.006 ± 0.082 | 0.005 | 0 | −0.022 ± 0.053 | 0 |
θI and θII, the coefficients of Type-I and Type-II functional divergence; LRT, Likelihood Ratio Statistic, for
p < 0.05; Qk, posterior probability.
To further identify the critical amino acid sites for functional divergence in 14-3-3 gene subfamilies, the posterior probability (Qk) of divergence was calculated for each site. Type I and Type II functional divergence-related residues between the specified subgroups with Qk < 0.99 were excluded to reduce false positives. No critical amino acid sites were found for Type I functional divergence in any 14-3-3 subfamily pairs, as shown in Table 2 and Table S4. In contrast, Type II functional divergence-related residues were much more numerous than those of Type I. In particular, 45, 46, and 5 amino acid sites critical for Type II functional divergence were identified in ε/non-ε-a, ε/non-ε-c, and non-ε-a/non-ε-d subfamily pairs, respectively. These results suggested that functional divergence in these groups was primarily attributable to rapid changes in amino acid physiochemical properties, not to changes in evolutionary rate. The non-ε-a/non-ε-b and non-ε-a/non-ε-c subfamily pairs had five and four critical amino acid sites, respectively. However, these two subfamily pairs had a θII coefficient < 0, suggesting that both subfamily pairs may also have undergone functional divergence during evolutionary history, but not at a significant level. For the remaining five gene subfamily pairs (ε/non-ε-b, ε/non-ε-d, non-ε-b/non-ε-c, non-ε-b/non-ε-d, and non-ε-c/non-ε-d), no critical amino acid sites were identified, indicating that functional divergence among these subfamily pairs was not significant.
Positive selection and coevolution analysis of the 14-3-3 gene family
To assume variable selective pressure among sites but no variation among branches in the phylogeny, a site-specific likelihood model was employed to the 14-3-3 gene family of the selected 13 species. Two pairs of models were applied in this analysis, forming two LRTs: M0 (one ratio) and M3 (discrete), and M7 (beta) and M8 (beta and ω). However, in the results, no amino acid sites were identified to be under positive selection (Table S5), suggesting the lack of selective pressure among the 14-3-3 gene family.
According to the negative results from the site-specific likelihood model, we further conducted a branch-site model analysis of the 14-3-3 gene family to test the positive selection effect at individual sites in different subfamilies. From the results, two critical positive selection sites were identified in the ε subfamily (1M and 148S, P < 0.01) (Table 3). Apart from the ε subfamily, two critical positive selection sites 2S and 95S, and 96K and 108H, were also found in each of the non-ε-a and non-ε-d subfamilies, respectively. The remaining two subfamilies (non-ε-b and non-ε-c) had no positive selection sites and had undergone less selective pressure. Furthermore, protein sequences (CAPS) were used for coevolution analysis, and three amino acids showed coevolution among the 13 plant species, namely, 236L, 237T, and 238E (Table S6).
Table 3.
Parameters estimation and likelihood ratio tests for the branch-site and free-ratio models among 14-3-3 genes.
| Cluster | Model | npa | lnL | 2⊿l | Positive selected sitesb |
|---|---|---|---|---|---|
| ε | Model A-null | 255 | −27619.537638 | Not allowed | |
| Model A | 256 | −27613.788734 | 11.50** | 1M, 148S | |
| non-ε-a | Model A-null | 255 | −27621.004255 | Not allowed | |
| Model A | 256 | −27619.359293 | 3.39 | , 95S | |
| non-ε-b | Model A-null | 255 | −27627.122669 | Not allowed | |
| Model A | 256 | −27627.122670 | 0.00 | none | |
| non-ε-c | Model A-null | 255 | −27627.122670 | Not allowed | |
| Model A | 256 | −27627.122670 | 0.00 | none | |
| non-ε-d | Model A-null | 255 | −27617.430951 | Not allowed | |
| Model A | 256 | −27616.779011 | 1.30 | , 108H |
p < 0.01 are marked by **.
Number of parameters in the ω distribution.
The numbers of Positive-selection sites are inferred at posterior probabilities >95%.
Red color marked amino acids were identified in both Type-II functional divergence analysis and positive selection analysis.
Three-dimensional structure prediction of 14-3-3 proteins and identification of critical amino acid sites
The three-dimensional structures of seven BdGF14 proteins from B. distachyon were predicted by searching the SWISS-MODEL database and visualized using Pymol software. All 14-3-3 proteins displayed similar structural features; the representative BdGF14c is shown in Figure 4. The results showed that BdGF14c well-matched OsGF14c (PDB: 3AXY), with 97.02% identity (Figure 4A), in which the monomer consists of a bundle of nine anti-parallel α-helices (αA-αI, Figure 4B), and the dimer usually presents a characteristic cup-like shape with a large central channel containing two amphipathic binding grooves (Figures 4C,D). Four critical sites (1S, 2M, 96K, and 148S) were identified and shown to be associated with positive selection, as well as Type II functional divergence (Table S4). These sites are located on the N-terminal, αD- and αF-helices of the three-dimensional structure, respectively (Figure 4E). Multiple sequence alignment was performed to further investigate the location of the four critical sites in the protein sequences (Figure S8). Interestingly, all of these four sites were presented on the outer convex surface (Figure 4E). Further analysis showed that a large number of the identified sites from the Type II functional divergence (Table S4) were also located on the outer convex surface (data not shown).
Figure 4.
Three-dimensional structure of B. distachyon 14-3-3 protein BdGF14c. (A) Structural comparison of BdGF14c (green) and OsGF14c (PDB: 2o98B, marine). The N- and C-terminals are indicated, the helices are labeled αA-αI. (B) Structure of BdGF14c monomer. (C) Structure of BdGF14c homodimer. Monomer 01 labeled green color, monomer 02 labeled cyan color. (D) Electrostatic surface representation of BdGF14c. The protein surface is colored according to its electrostatic potential: blue for positive and red for negative charge. (E) The precise position of the four critical amino acid identified by both Type-II functional divergence and positive selection. Each structure is rotated 90° to show different view sides of the protein.
Analysis of promoter regions
The binding of transcription factors (TFs) to corresponding TF binding sites (TFBSs) upstream of target genes can regulate gene expression. Thus, the cis-acting elements in the 1500-bp region upstream of the translation initiation codon of the 14-3-3 gene family were identified using PlantCARE. In total, seven types of regulatory elements were found, four of which are strongly related to crucial physiological process, including light periods, developmental regulation, hormonal responses, and environmental responses (Table S7).
Among these elements, Sp1, G-box (Menkens et al., 1995), G-Box, Box 4 (Lois et al., 1989), and Box I (Arguello-Astorga and Herrera-Estrella, 1996) belonged to the light-responsive elements. Sp1 was shown to be the most abundant light-responsive element in the 14-3-3 gene family, with a mean number of 3.921 copies per gene, and it was much more abundant in lower plants (5.250) and monocots (6.780) than in dicots (1.229). The G-box was ranked as the second most abundant light-responsive element, followed by Box 4, G-Box, and Box I. These results suggest that 14-3-3 genes may function in photosynthesis and/or carbohydrate metabolism. Development-related elements were also identified, but their abundance showed no significant differences, except for CCGTCC-box (meristem development-related element), the abundance of which in lower plants and monocots was ~17.2- and 11.6-fold that in dicots, respectively. Phytohormones and other abiotic stress-responsive mechanisms also play crucial roles for defense against environmental stresses in plants. ABRE (Simpson et al., 2003), GARE-motif (Ogawa et al., 2003), TCA-element (Pastuglia et al., 1997), and TGA-element (Guilfoyle et al., 1993) are hormone-responsive elements. Among these elements, the ABRE element was found to be the most abundant (2.937), being related to the ABA signaling pathway, indicating that the expression of several 14-3-3 genes was induced by ABA-mediated signal transduction. Similarly, several elements were considered to be related to environmental stress, including MBS (Yamaguchi-Shinozaki and Shinozaki, 1993), ARE (Klotz and Lagrimini, 1996), HSE (Freitas and Bertolini, 2004), and TC-rich elements (Klotz and Lagrimini, 1996). Promoter analysis showed that the diversity of cis-acting regulatory elements in the upstream regions of 14-3-3 genes provided further support for the diversified functional roles of 14-3-3 genes in response to plant growth, development, and phytohormone and abiotic stresses.
Expression profiles of 14-3-3 genes across different tissues and abiotic stresses in B. distachyon
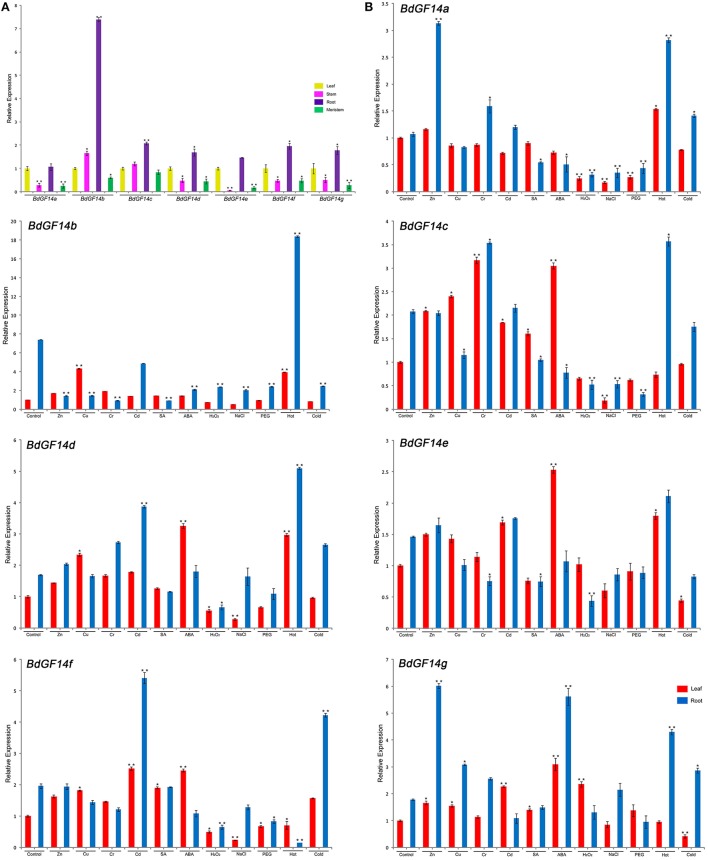
The transcriptional expression profiles of seven BdGF14 genes from Bd21 in different organs and tissues (leaf, stem, root, and meristem) and abiotic stresses (salinity, drought, temperature, heavy metal, and phytohormone stresses) were analyzed by qRT-PCR. The results showed that all BdGF14 genes were expressed in leaf, stem, root, and meristem, but had higher expression levels in root, particularly for BdGF14b, with 7.4-, 4.5-, and 12.3-fold higher expression than in leaf, stem, and meristem, respectively. In contrast, BdGF14a and BdGF14e in stem and meristem, and BdGF14g in meristem had lower expression (Figure 5A and Table S8).
Figure 5.
Expression profiles of B. distachyon 14-3-3 genes in tissues and under short time stress treatments. (A) The expression profiles of seven B. distachyon 14-3-3 genes in different tissues, including leaf, stem, root and meristem. (B) The remaining seven subfigures were expression profiles of seven B. distachyon 14-3-3 genes under various abiotic stresses, including Zn2+, Cu2+, Cr3+, Cd2+, SA, ABA, H2O2, NaCl, PEG, Hot (42°C) and Cold (4°C). SA, salicylic acid; ABA, abscisic acid; PEG, polyethylene glycol. The expression levels of 14-3-3 genes in the control group were defined as 1.0. The leaf and root are separately analyzed, red color bar presents leaf, sky blue color bar presents root. Statistically significant differences between control group and treatment group were calculated by an independent Student's t-tests: *P < 0.05, **P < 0.01.
The transcriptional expression changes of seven BdGF14 genes in Bd21 seedling roots and leaves under various abiotic stresses were investigated, including osmotic [NaCl and polyethylene glycol (PEG)], temperature (hot: 42°C and cold: 4°C), heavy metal (Zn2+, Cu2+, Cr3+, and Cd2+), and phytohormone stresses [ABA, salicylic acid (SA), and H2O2]. Initially, we analyzed the expression profiles of BdGF14 genes by applying 6 h treatments with salinity, drought, heavy metals, and phytohormones and 2 h treatments at the set temperatures. The morphological appearance under these treatments did not change significantly, except for H2O2 treatment, in which the seedling roots became brown after treatment (Table S9, Figure S9, S10).
In general, Seven GdGF14 genes showed significant transcriptional expression changes under all stress treatments in both roots and leaves (Figure 5). In roots, GdGF14 genes under different heavy metal stresses displayed upregulation or downregulation. Specifically, five genes (GdGF14a, GdGF14c, GdGF14d, GdGF14e, and GdGF14f) under Cd2+ stress, four (GdGF14a, GdGF14d, GdGF14e, and GdGF14g) under Zn2+ stress, and four (GdGF14a, GdGF14c, GdGF14d, and GdGF14g) under Cr3+ stress were upregulated. Under Cu2+ stress, only BdGF14g showed upregulated expression. Phytohormone and osmotic generally induced downregulated expression of GdGF14 genes, but BdGF14d and BdGF14g under ABA stress and BdGF14g under NaCl stress were upregulated. Except for GdGF14f, which was downregulated, the other six genes were significantly upregulated under high-temperature stress. Four genes (GdGF14a, GdGF14d, GdGF14f, and GdGF14g) and three (GdGF14b, GdGF14c, and GdGF14e) under low-temperature stress were upregulated and downregulated, respectively.
BdGF14 genes in leaves generally showed greater expression differences in response to abiotic stress. Except for BdGF14a showing downregulation under Cu2+, Cr3+, and Cd2+ stresses, the other genes were all upregulated under heavy metal stress. Compared with those in roots, more genes showed upregulated expression under phytohormone treatments, while similar expression patterns were found under NaCl and PEG treatments. In response to temperature stresses, four genes (BdGF14a, BdGF14b, BdGF14d, and BdGF14e) under a high temperature and only one gene (BdGF14f) under a low temperature were upregulated (Figure 5 and Table S8). Our results demonstrated that multiple abiotic stresses could induce the upregulated expression of individual BdGF14 genes, including BdGF14g under seven treatments in roots and eight treatments in leaves, and four genes (BdGF14b, BdGF14d, BdGF14e, and BdGF14f) under seven treatments in leaves.
Based on the results above, five treatments (Cd2+, Cr3+, Cu2+, Zn2+, and ABA) generally induced significant upregulation of GdGF14 genes. Thus, the dynamic transcriptional expression changes of seven GdGF14 genes under long-term treatments with six time points (0, 6, 12, 24, and 48 h, and recovery 48 h) were further detected by qRT-PCR. The morphological observation results showed that, except for Zn2+ treatment, plant growth under stress treatments was significantly inhibited (Table S9 and Figure S10). After recovery for 48 h, almost no morphological changes were observed, except for Cd2+ treatment with minor changes. Interestingly, plants in the Zn2+ treatment group grew at a speed equivalent to those in the control group (Table S9 and Figure S10).
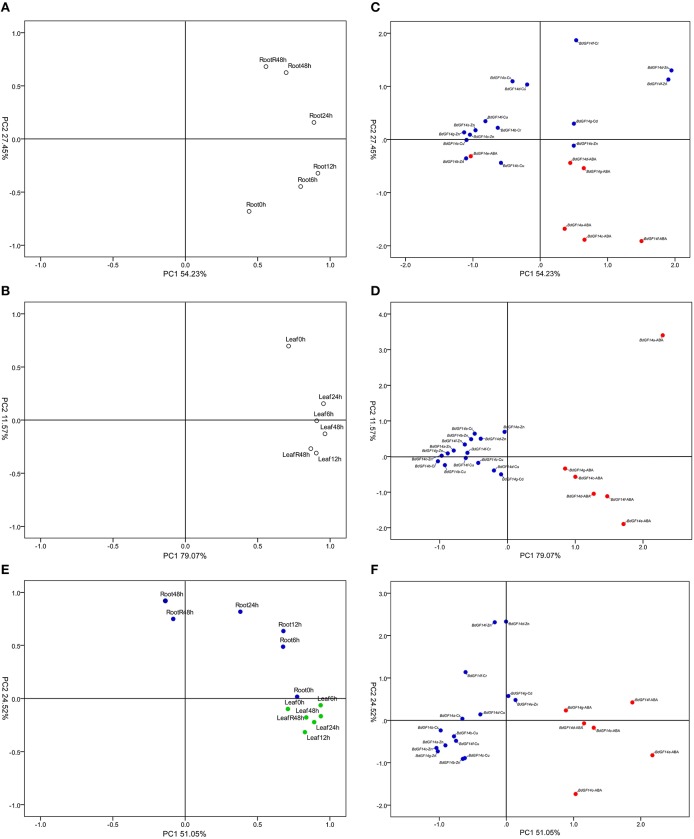
Principal component analysis (PCA) showed that the first 6 h induced larger expression changes of BdGF14 genes than the subsequent periods in leaves (Figure 6A), while two periods (0–6 and 24–48 h) showed greater expression differences than the other three periods in roots (Figure 6B). Heavy metal (Cd2+, Cr3+, Cu2+, Zn2+) and phytohormone (ABA) treatments were clearly separated, indicating their distinct expression differences. The pattern in roots featured less close clustering than that in leaves (Figures 6C,D), indicating that roots had stronger responses against abiotic stresses than leaves. The integrative analysis results of roots and leaves showed similar patterns to the separate analyses mentioned above (Figures 6E,F).
Figure 6.
PCA of B. distachyon 14-3-3 genes in response to abiotic stresses. (A) PCA of each treatment time point in the root. (B) PCA of each treatment time point in the leaf. (C) PCA of each treatment type in the root. (D) PCA of each treatment type in the leaf. (E) Integrative PCA of each treatment time point. (F) Integrative PCA of each treatment type. In the subfigure (C,D,F), blue color solid dots indicate heavy metal treatments, red color solid dots indicate ABA treatment. In the subfigure (E), blue color solid dots indicate treatment in the root, green color solid dots indicate treatment in the leaf.
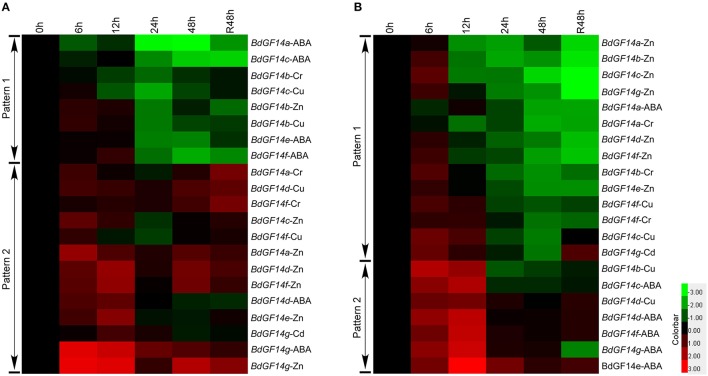
Gene expression profiles revealed that all BdGF14 genes in both roots and leaves showed similar expression patterns under five stress treatments, in which two distinct expression patterns, I and II, were present, corresponding to up-down and up-down-up expression trends from stress treatments to recovery, respectively (Figure 7). At 6 and 12 h after the five treatments, more genes showed significant upregulation in both roots and leaves, but all genes were downregulated at 24 h, and the lowest expression levels occurred at 48 h in leaves. However, the BdGF14 genes at 48 h and recovery in roots displayed more significant upregulation than those in leaves.
Figure 7.
Expression profiles of B. distachyon 14-3-3 genes under long stress treatments. (A) Gene expression profiles in the root. (B) Gene expression profiles in the leaf. Samples were harvested at 0, 6, 12, 24, 48, and recovery 48 h. The expression levels of 14-3-3 genes at 0 h were defined as 1.0 in both tissues.
Discussion
In this paper, we present the first comprehensive survey on the molecular characterization, phylogenetic relationships, and abiotic stress responses of the 14-3-3 gene family in B. distachyon. Here, we focus on the structure, evolution, and function of plant 14-3-3 genes, combining our results and previous reports.
Molecular characterization, phylogenetics, and evolutionary conservation of the 14-3-3 gene family in B. distachyon
Our results demonstrated that BdGF14 genes, as well as other plant 14-3-3 genes, can be divided into five well-conserved subfamilies, but clear divergences occurred in different subfamilies, which is highly consistent with the results from Arabidopsis and rice (Chen et al., 2006). The genes in the same subfamily shared similar exon-intron organization and motif compositions, but greater structural differences were present between subfamilies (Figures S2–S6), suggesting the high structural conservation of plant 14-3-3 genes within a subfamily. The motif analysis results using the MEME online tool provide further evidence for the evolutionary conservation of the 14-3-3 genes in B. distachyon and other plant species, in which the former seven motifs from 5′ to 3′ were identical among 127 genes (Figures S5–S7).
Duplication events play critical roles in gene evolution, which is not only one of the primary driving forces in the evolution of genomic and genetic systems (Moore and Purugganan, 2003) but also a major mechanism for the establishment of new functions (Long and Langley, 1993). Segmental duplication was shown to be the predominant form by which the 14-3-3 gene family has expanded, which occurred about 10.1–58.6 MYA (Table 1). In B. distachyon, only one pair of segmental genes (BdGF14e/BdGF14b) was found, which was estimated to have arisen 45.6 MYA. The results from genome sequencing indicated that the divergence time of B. distachyon from wheat was 32–39 MYA, and 40–60 MYA from rice and sorghum (International Brachypodium Initiative, 2010). Thus, the segmental duplication event in the B. distachyon 14-3-3 gene family occurred before the divergence from wheat and almost concurrently with the divergence from rice and sorghum.
Great variations can lead to gene diversification in biological functions (Zhu et al., 2015). The functional diversity of proteins was highly correlated with amino acid properties. We identified seven amino acid sites (1M, 2S, 96K, 148S, 236L, 237T, and 238E) as making important contributions to the functional diversity of 14-3-3 genes (Table 3, Tables S4, S6). Interestingly, the former four amino acids were mapped onto the outer convex surface of the three-dimensional structure of BdGF14c (Figure 4), and the remaining three were on the C-terminal (Figure S8). It is known that the inner walls of the central channel and the dimer interface in 14-3-3 proteins are formed by conserved residues, whereas the less conserved residues are located on the outer convex surface (Obsilova et al., 2014). In addition, the most flexible region of the 14-3-3 protein molecule is the C-terminal segment, which exhibits the highest sequence variability among 14-3-3 isoforms (Truong et al., 2002). Thus, we speculate that the diversity of amino acids at these sites may play roles in determining the functional divergence of 14-3-3 proteins.
Expression and function of 14-3-3 genes in B. distachyon
14-3-3 proteins show great functional diversity, being involved in a wide range of biological processes in plants, such as photosynthesis, primary metabolism, growth and cell division, hormone pathways, and abiotic and biotic stresses (de Boer et al., 2013). Results from cotton revealed that 14-3-3 proteins performed functions in cell elongation during the elongation phase of fiber development (Zhang et al., 2010). GmGF14 genes abundantly expressed in embryos during early seed germination showed functions in soybean seed development (Li and Dhaubhadel, 2011), while 14-3-3 genes in Medicago truncatula were expressed almost equally in roots, stems, leaves, and flowers (Qin et al., 2016). The results from the present study indicated that the 14-3-3 genes are preferentially expressed in the roots and leaves (Figure 5A). During seedling growth phases, roots, and leaves are the most important organs necessary for nutrient absorption and photosynthesis, respectively. The abundant expression of 14-3-3 genes may contribute to the rapid vegetative growth of plant seedlings.
Considerable work has revealed that the 14-3-3 genes are involved in various abiotic stresses in different plant species, such as rice (Chen et al., 2006), Populus (Tian et al., 2015), common bean (Li et al., 2015), and Hevea brasiliensis (Yang et al., 2014). In particular, the presence of stress-related cis-elements in the promoter region of 14-3-3 genes could play important roles in responses to abiotic stresses. In the current study, significant expression changes of BdGF14 genes in response to various abiotic stresses indicated their important roles in resisting such stresses. Particularly, high temperature induced the significant upregulation of most BdGF14 genes, among which BdGF14a, b, d, and e were upregulated in both roots and leaves. However, BdGF14 genes showed greater expression differences in these two organs under low-temperature stress: generally upregulation in roots and downregulation in leaves, but BdGF14f showed upregulated expression in both roots and leaves (Figure 5). At present, the mechanism of plant 14-3-3 gene function in high-temperature stress defense is still unclear. Considerable work has demonstrated that ABA/SA and Ca2+, representing ABA-dependent and calcium-dependent protein kinase (CDPK) signaling pathways, are involved in plant high-temperature stress responses (Mosser et al., 1990; Talanova and Titov, 1994; Lopez-Delgado et al., 1998). We speculate that 14-3-3 genes might also participate in these signaling pathways to defend against high-temperature stress. Except for the ABA-dependent signaling pathway that is considered as a critical signaling pathway involved in the low-temperature responses of plants (Roberts et al., 2002), functional analysis revealed that RAC1A (corresponding to GRF3 in Figure 2) functions as a negative regulator in controlling the biosynthesis of ethylene to modulate constitutive freezing tolerance and cold acclimation in Arabidopsis (Catalá et al., 2014). However, investigation in maize roots found that 14-3-3 proteins participated in low-temperature and osmotic stress responses through binding to the plasma membrane H+-ATPase to activate the proton pump, leading to K+ influx and water uptake (Shanko et al., 2003).
ABA, a critical plant hormone, participates in a wide range of abiotic stresses, and regulates abiotic stress tolerance to cope with environmental stresses (Dong et al., 2015). Functional studies have indicated that 14-3-3 proteins interact with many TFs, such as the ABF family (Jakoby et al., 2002), WRKY (Rushton et al., 2010), and basic leucine zipper (bZIP; Sornaraj et al., 2016). These downstream TFs are involved in the ABA-dependent signaling pathway, important for plants in response to many abiotic stresses, such as phytohormone (ABA and SA), osmotic (salinity and drought), and temperature stresses (Rock, 2000; Dong et al., 2015). In the current study, we found that exogenous ABA and SA generally induced upregulation of BdGF14 genes in leaves and downregulation in roots (Figures 5, 7). We speculate that plant 14-3-3 genes mainly participate in the ABA-dependent signaling pathway to cope with phytohormone stress by interacting with the downstream TFs.
Heavy metal contamination is a widespread and increasingly serious problem globally. The over-accumulation of reactive oxygen species (ROS) in cells is one of the most common consequences of heavy metal toxicity in plants, resulting from the interaction of heavy metals with electron transport activities (Shahid et al., 2014). We found that the majority of BdGF14 genes were upregulated in leaves and downregulated in roots under heavy metal stresses (Figures 6, 7). One of the most common strategies for plants against heavy metal stresses is to activate the ROS-mediated mitogen-activated protein kinase (MAPK) cascade signaling pathway to scavenge extra ROS for detoxification (Jalmi and Sinha, 2015). 14-3-3 proteins, apart from binding to the downstream TFs, also interact with MAPK (Fritz et al., 2006). Thus, 14-3-3 genes perform their functions in response to heavy metal stress mainly through the ROS-mediated MAPK cascade signaling pathway.
NaCl, PEG, and H2O2 stresses generally induced significant downregulation of BdGF14 genes (Figure 5). Previous reports showed that PvGF14e and PvGF14h genes in common bean and ZmGF14-6 gene in maize (corresponding to ZmGFc in Figure 2) were downregulated under saline conditions and drought stress (Campo et al., 2012; Li et al., 2015), consistent with our results (Figure 5). A recent proteomic analysis revealed that BdGF14b and BdGF14d were downregulated under H2O2 stress in both roots and leaves (Bian et al., 2015). These 14-3-3 genes may be part of an adjustment in the perception of salinity and drought stresses (Talanova and Titov, 1994). Moreover, although phytohormone, osmotic, and temperature stresses mainly activate the ABA-dependent signaling pathway, these stresses can also induce the accumulation of ROS in plant cells plus the common downstream TFs. Thus, 14-3-3 genes could participate in different signaling pathways to cope with frequent and complex crosstalk caused by various abiotic stresses.
Putative metabolic pathways of 14-3-3 genes involved in abiotic stresses
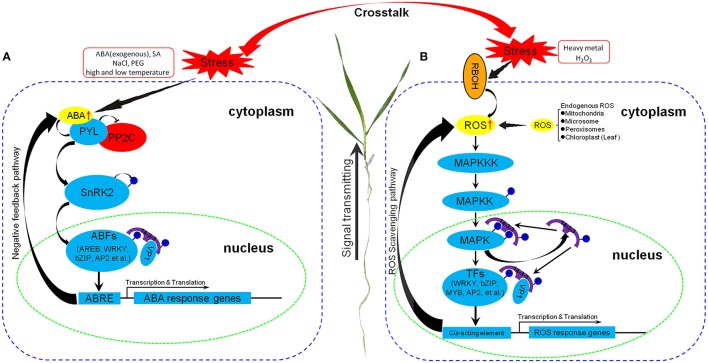
According to our results in combination with previous reports, we proposed the putative metabolic pathways of plant 14-3-3 genes in response to abiotic stresses, mainly involved in the ABA-dependent signaling pathway and/or ROS-mediated MAPK cascade signaling pathway (Figure 8). Abiotic stresses such as exogenous ABA, SA, NaCl, PEG, and high- and low-temperature stresses can induce the accumulation of ABA in cells, which subsequently triggers activation of the ABA-dependent signaling pathway and regulates the expression of downstream ABA-responsive genes against the stresses (Ng et al., 2014; Figure 8A). Meanwhile, the level of ROS in cells was increased by heavy metal stress, which triggered the MAPK cascade signaling pathway, and then initiated the expression of heavy metal stress response genes to scavenge the extra ROS for defense (Jalmi and Sinha, 2015) (Figure 8B). Under various abiotic stresses, plant 14-3-3 genes were upregulated and participated in these signaling pathways in response to abiotic stresses through interacting with TFs and regulating the expression of downstream stress response genes. Furthermore, various stresses result in frequent and complex crosstalk, and thus 14-3-3 genes are able to cope with multiple abiotic stresses through participating in different signaling pathways.
Figure 8.
Schematic representation of putative metabolic pathways of plant 14-3-3 genes involved in abiotic stresses. (A) Metabolic pathway through ABA-dependent signaling pathway. (B) Metabolic pathway through ROS-mediated MAPK cascade signaling pathway. PYL, Pyrabactin resistance 1-like family; PP2C, type 2C protein phosphatase; SnRK2, Snf1-related protein kinase 2; ABF, ABRE-binding factor; AREB, ABRE-binding proteins; bZIP, Basic region/leucine zipper motif (bZIP) transcription factor; AP2, APETALA 2; VP1, Abscisic acid-VIVIPAROUS1; ABRE, ABA-responsive element; RBOH, Respiratory burst oxidase homolog; MAPK, Mitogen activated protein kinase; MAPKK, MAPK kinase; MAPKKK, MAPKK kinase; MYB, myeloblastosis family of transcription factor. Positive regulators are shown in sky-blue, negative regulators are shown in red, signal molecules are shown in yellow.
Conclusion
The current study has demonstrated that plant 14-3-3 genes are highly conserved in individual subfamilies, but are divergent between different subfamilies. Segmental duplication seems to be the predominant form by which the 14-3-3 gene family has expanded, and all of the putative duplicated genes may postdate the monocot-dicot split. Functional divergence analysis revealed that the 14-3-3 gene family experienced radical shifts in cluster-specific amino acid properties, without significant changes in the evolutionary rate at certain amino acid sites. Transcriptional expression analysis of the BdGF14 gene indicated that the 14-3-3 genes are significantly regulated by various abiotic stresses. Through participating in different signaling pathways and employing different strategies, BdGF14 genes play important roles in the responses to multiple abiotic stresses. Meanwhile, frequent and complex crosstalk between different signaling pathways could occur under different adverse stresses. Individual 14-3-3 genes can cope with multiple abiotic stresses mainly through functioning in different signaling pathways. These results not only provide a better understanding of the evolutionary processes of 14-3-3 genes in plants but also offer a new insight into the molecular mechanisms of 14-3-3 genes in response to abiotic stresses in plants.
Author contributions
YY and YH designed the study and provide guidance on the whole study. HC carried out the bioinformatic analysis, plant materials culture, qRT-PCR analysis, and drafted the manuscript. YX and LY participated in the qRT-PCR analysis. YB took parted in the plant materials culture. LW and SZ coordinated the study and elaborated on manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was financially supported by grants from the Ministry of Science and Technology China (2016YFD0100500), the National Natural Science Foundation of China (31471485), and the Natural Science Foundation of Beijing City/the Key Developmental Project of Science Technology, Beijing Municipal Commission of Education (KZ201410028031). The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/index/QO9n4B.
Glossary
Abbreviations
- ABA
abscisic acid
- ABRE
ABA-responsive element
- ABF
ABRE-binding factor
- AREB
ABRE-binding proteins
- AP2
APETALA 2
- bZip
Basic region/leucine zipper motif (bZIP) transcription factor
- LRT
Likelihood ratio statistic
- MW
Molecular weight
- MAPK
Mitogen activated protein kinase
- MAPKK
MAPK kinase
- MAPKKK
MAPKK kinase
- MYB
myeloblastosis family of transcription factor
- PEG
polyethylene glycol
- PYL
Pyrabactin resistance 1-like family
- PP2C
type 2C protein phosphatase
- PI
Isoelectric points
- Qk
Posterior probability
- RBOH
Respiratory burst oxidase homolog
- SA
salicylic acid
- SnRK2
Snf1-related protein kinase 2
- TFBS
Transcription factor binding site
- TF
Transcription factor
- VP1
Abscisic acid-VIVIPAROUS1
- WRKY
WRKY transcription factor.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01099
Gene-specific primers for qRT-PCR analysis.
The nomenclature, characteristics of 14-3-3 genes and their deduced proteins in 13 representative species.
The parameters of conserved 14-3-3 domain from SMART and Pfam. (A) The parameters of conserved 14-3-3 domain from SMART. (B) The parameters of conserved 14-3-3 domain from Pfam.
Type-II functional divergence sites identified between different subgroup pairs of 14-3-3 gene family. Red color marked amino acids were identified in both Type-II functional divergence analysis and positive selection analysis. Amino acids listed in the table were matched to BdGF14c.
Tests for positive selection among codons of 14-3-3 genes using free-ratio and site models. aNumber of parameters in the ω distribution.
Critical coevolution amino acids identified from coevolution analysis.
Cis-element analysis of 1500-bp nucleotide sequences upstream of the translation initiation codon of 14-3-3 genes.
Data of the relative expression of B. distachyon 14-3-3 genes under abiotic stress treatments.
Parameters of B. distachyon root length and plant size under control and abiotic stress treatments. Dashes indicate no parameters collected.
Double standard curves and dissolution curves of qRT-PCR. The red standard curves represent the reference gene (S-adenosylmethionine decarboxylase gene), the blue standard curves represent the target genes. The dissolution curves of different genes are indicated.
The number of 14-3-3 genes in individual species. Cr, Chlamydomonas reinhardtii; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Bd, Brachypodium distachyon; Os, Oryza sativa; Ta, Triticum aestivum; Si, Setaria italic; Zm, Zea mays; Sb, Sorghum bicolor; At, Arabidopsis thaliana; Gm, Glycine max; Sl, Solanum lycopersicum; Gr, Gossypium raimondii.
Exon-intron organization of 14-3-3 genes. The yellow boxes and gray lines represent exons and introns, respectively. The bold dark blue lines in the top indicate the 5′ upstream region (left) and the 3′ downstream region (right). The 14-3-3 genes were lined according to Phylogenetic tree construction.
Statistic analysis in intron number. (A) Statistic analysis based on subgroups. (B) Statistic analysis based on individual species.
Schematic representation of conserved 14-3-3 protein motifs. The schematic diagram was derived from MEME. The order of motifs in the Schematic representation was automatically generated by MEME according to scores.
Schematic representation of amino acid motifs of 14-3-3 proteins. Motif analysis was performed using MEME as described in the Methods. The gray solid lines represent the corresponding 14-3-3 proteins and their length. The different-colored boxes represent different motifs and their position and order in individual 14-3-3 protein sequence.
Statistic analysis in motif number. (A) Statistic analysis based on subgroups. (B) Statistic analysis based on individual species.
Multiple sequence alignment of B. distachyon 14-3-3 protein sequences. Eleven 14-3-3 proteins consist of 7 B. distachyon 14-3-3 proteins, 2 Arabidopsis 14-3-3 proteins, and 2 rice 14-3-3 proteins. Nine α helices are marked with red-colored solid rectangle, while N-terminal and C-terminal are marked with light blue-colored solid rectangle, respectively. Red boxes indicate the critical amino acids identified from functional divergence and positive selection, light blue box indicates the critical amino acids identified from coevolution, while green boxes indicate the critical amino acids identified only from positive selection.
Images of B. distachyon plants under abiotic stress treatments for 6 or 2 h. (A) Mild drought stress (PEG 6000). (B) Phytohormone stress (H2O2). (C) Phytohormone stress (SA). (D) Salinity stress (NaCl). (E) High temperature stress (42°C). (F) Low temperature stress (4°C). Bar scale is 5.0 cm.
Images of Bd21 plants under abiotic stress treatments for 48 and 48 h recovery. (A) Control group. (B) Heavy metal stress (Cu2+). (C) Heavy metal stress (Cr3+). (D) Heavy metal stress (Cd2+). (E) Heavy metal stress (Zn2+). (F) Phytohormone stress (ABA). Bar scale is 5.0 cm.
References
- Aitken A. (2002). Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Plant Mol. Biol. 50, 993–1010. 10.1023/A:1021261931561 [DOI] [PubMed] [Google Scholar]
- Aitken A. (2006). 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 16, 162–172. 10.1016/j.semcancer.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Arguello-Astorga G. R., Herrera-Estrella L. R. (1996). Ancestral multipartite units in light-responsive plant promoters have structural features correlating with specific phototransduction pathways. Plant Physiol. 112, 1151–1166. 10.1104/pp.112.3.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzinger A., Popowicz G. M., Joy J. K., Majumdar S., Holak T. A., Hermeking H. (2005). The crystal structure of the non-liganded 14-3-3 sigma protein: insights into determinants of isoform specific ligand binding and dimerization. Cell Res. 15, 219–227. 10.1038/sj.cr.7290290 [DOI] [PubMed] [Google Scholar]
- Bian Y. W., Lv D. W., Cheng Z. W., Gu A. Q., Cao H., Yan Y. M. (2015). Integrative proteome analysis of Brachypodium distachyon roots and leaves reveals a synergetic responsive network under H2O2 stress. J. Proteomics 128, 388–402. 10.1016/j.jprot.2015.08.020 [DOI] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, 252–258. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjellqvist B., Basse B., Olsen E., Celis J. E. (1994). Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15, 529–539. 10.1002/elps.1150150171 [DOI] [PubMed] [Google Scholar]
- Blanc G., Wolfe K. H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. 10.1105/tpc.021345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J. E., Chapman B. A., Rong J., Paterson A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. 10.1038/nature01521 [DOI] [PubMed] [Google Scholar]
- Campo S., Peris-Peris C., Montesinos L., Peñas G., Messeguer J., San Segundo B. (2012). Expression of the maize ZmGF14-6 gene in rice confers tolerance to drought stress while enhancing susceptibility to pathogen infection. J. Exp. Bot. 63, 983–999. 10.1093/jxb/err328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R., López-Cobollo R., Mar Castellano M., Angosto T., Alonso J. M., Ecker J. R., et al. (2014). The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26, 3326–3342. 10.1105/tpc.114.127605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Li Q., Sun L., He Z. (2006). The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 13, 53–63. 10.1093/dnares/dsl001 [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- de Boer A. H., van Kleeff P. J., Gao J. (2013). Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma 250, 425–440. 10.1007/s00709-012-0437-z [DOI] [PubMed] [Google Scholar]
- Denison F. C., Paul A. L., Zupanska A. K., Ferl R. J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22, 720–727. 10.1016/j.semcdb.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Dong T., Park Y., Hwang I. (2015). Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 58, 29–48. 10.1042/bse0580029 [DOI] [PubMed] [Google Scholar]
- Draper J., Mur L. A., Jenkins G., Ghosh-Biswas G. C., Bablak P., Hasterok R., et al. (2001). Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555. 10.1104/pp.010196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868. 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva M. D., Wu M., Eisen J. A., Salzberg S. L. (2003). The age of the Arabidopsis thaliana genome duplication. Plant Mol. Biol. 51, 859–866. 10.1023/A:1023001130337 [DOI] [PubMed] [Google Scholar]
- Fan K., Wang M., Miao Y., Ni M., Bibi N., Yuan S., et al. (2014). Molecular evolution and expansion analysis of the NAC transcription factor in Zea mays. PLoS ONE 9:e111837. 10.1371/journal.pone.0111837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl R. J. (1996). 14-3-3 proteins and signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 49–73. 10.1146/annurev.arplant.47.1.49 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, 222–230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas F. Z., Bertolini M. C. (2004). Genomic organization of the Neurospora crassa gsn gene: possible involvement of the STRE and HSE elements in the modulation of transcription during heat shock. Mol. Genet. Genomics 272, 550–561. 10.1007/s00438-004-1086-5 [DOI] [PubMed] [Google Scholar]
- Fritz A., Brayer K. J., McCormick N., Adams D. G., Wadzinski B. E., Vaillancourt R. R. (2006). Phosphorylation of serine 526 is required for MEKK3 activity, and association with 14-3-3 blocks dephosphorylation. J. Biol. Chem. 281, 6236–6245. 10.1074/jbc.M509249200 [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. J., Hagen G., Li Y., Ulmasov T., Liu Z. B., Strabala T., et al. (1993). Auxin-regulated transcription. Funct. Plant Biol. 20, 489–502. 10.1071/pp9930489 [DOI] [Google Scholar]
- Hall B. G. (2005). Comparison of the accuracies of several phylogenetic methods using Protein and DNA sequences. Mol. Biol. Evol. 22, 792–802. 10.1093/molbev/msi066 [DOI] [PubMed] [Google Scholar]
- Hong S., Seo P. J., Yang M., Xiang F., Park C. M. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8:112. 10.1186/1471-2229-8-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. 10.1038/nature08747 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. 10.1016/S1360-1385(01)02223-3 [DOI] [PubMed] [Google Scholar]
- Jalmi S. K., Sinha A. K. (2015). ROS mediated MAPK signaling in abiotic and biotic stress-striking similarities and differences. Front. Plant Sci. 6:769. 10.3389/fpls.2015.00769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. H., Ley S., Aitken A. (1995). Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 368, 55–58. 10.1016/0014-5793(95)00598-4 [DOI] [PubMed] [Google Scholar]
- Klotz K. L., Lagrimini L. M. (1996). Phytohormone control of the tobacco anionic peroxidase promoter. Plant Mol. Biol. 31, 565–573. 10.1007/BF00042229 [DOI] [PubMed] [Google Scholar]
- Kristiansen L. C., Jacobsen S., Jessen F., Jørgensen B. M. (2010). Using a cross-model loadings plot to identify protein spots causing 2-DE gels to become outliers in PCA. Proteomics 10, 1721–1723. 10.1002/pmic.200900318 [DOI] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, 257–260. 10.1093/nar/gku949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Jiang X., Jin D., Dhaubhadel S., Bian S., Li X. (2015). Identification of 14-3-3 family in common bean and their response to abiotic stress. PLoS ONE 10:e0143280. 10.1371/journal.pone.0143280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Dhaubhadel S. (2011). Soybean 14-3-3 gene family: identification and molecular characterization. Planta 233, 569–582. 10.1007/s00425-010-1315-6 [DOI] [PubMed] [Google Scholar]
- Liu D., Bienkowska J., Petosa C., Collier R. J., Fu H., Liddington R. (1995). Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376, 191–194. 10.1038/376191a0 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lois R., Dietrich A., Hahlbrock K., Schulz W. (1989). A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 8, 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M., Langley C. H. (1993). Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260, 91–95. 10.1126/science.7682012 [DOI] [PubMed] [Google Scholar]
- Lopez-Delgado H., Dat J. F., Foyer C. H., Scott I. M. (1998). Induction of thermotolerance in potato microplants by acetylsalicylic acid and H2O2. J. Exp. Bot. 49, 713–720. 10.1093/jxb/49.321.713 [DOI] [Google Scholar]
- Menkens A. E., Schindler U., Cashmore A. R. (1995). The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. 10.1016/S0968-0004(00)89118-5 [DOI] [PubMed] [Google Scholar]
- Moore B. E., Perez V. J., Carlson F. D. (ed.). (1967). Physiological and Biochemical Aspects of Nervous Integration. Englewood Cliffs, NJ: Prentice-Hall. [Google Scholar]
- Moore R. C., Purugganan M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 15682–15687. 10.1073/pnas.2535513100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser D. D., Kotzbauer P. T., Sarge K. D., Morimoto R. I. (1990). In vitro activation of heat shock transcription factor DNA-binding by calcium and biochemical conditions that affect protein conformation. Proc. Natl. Acad. Sci. U.S.A. 87, 3748–3752. 10.1073/pnas.87.10.3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. (1996). Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897. 10.1016/S0092-8674(00)81067-3 [DOI] [PubMed] [Google Scholar]
- Ng L. M., Melcher K., The B. T., Xu H. E. (2014). Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol. Sin. 35, 567–584. 10.1038/aps.2014.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsilova V., Kopecka M., Kosek D., Kacirova M., Kylarova S., Rezabkova L., et al. (2014). Mechanisms of the 14-3-3 protein function: regulation of protein function through conformational modulation. Physiol. Res. 63(Suppl. 1), 155–164. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604. 10.1105/tpc.011650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M., Roby D., Dumas C., Cock J. M. (1997). Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell 9, 49–60. 10.1105/tpc.9.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J., Jin D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- Qin C., Cheng L., Shen J., Zhang Y., Cao H., Lu D., et al. (2016). Genome-wide identification and expression analysis of the 14-3-3 family genes in Medicago truncatula. Front. Plant Sci. 7:320. 10.3389/fpls.2016.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. R., Salinas J., Collinge D. B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50, 1031–1039. 10.1023/A:1021261614491 [DOI] [PubMed] [Google Scholar]
- Rock C. (2000). Pathways to abscisic acid-regulated gene expression. New Phytol. 148, 357–396. 10.1046/j.1469-8137.2000.00769.x [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Hőhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Schoonheim P. J., Sinnige M. P., Casaretto J. A., Veiga H., Bunney T. D., Quatrano R. S., et al. (2007). 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J. 49, 289–301. 10.1111/j.1365-313X.2006.02955.x [DOI] [PubMed] [Google Scholar]
- Shahid M., Pourrut B., Dumat C., Nadeem M., Aslam M., Pinelli E. (2014). Heavy-metal-induced reactive oxygen species: phytotoxicity and physicochemical changes in plants. Rev. Environ. Contam. Toxicol. 232, 1–44. 10.1007/978-3-319-06746-9_1 [DOI] [PubMed] [Google Scholar]
- Shanko A. V., Mesenko M. M., Klychnikov O. I., Nosov A. V., Ivanov V. B. (2003). Proton pumping in growing part of maize root: its correlation with 14-3-3 protein content and changes in response to osmotic stress. Biochemistry 68, 1320–1326. 10.1023/B:BIRY.0000011653.46422.c3 [DOI] [PubMed] [Google Scholar]
- Simpson S. D., Nakashima K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozak K., et al. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 33, 259–270. 10.1046/j.1365-313X.2003.01624.x [DOI] [PubMed] [Google Scholar]
- Song W., Qin Y., Zhu Y., Yin G., Wu N., Li Y., et al. (2014). Delineation of plant caleosin residues critical for functional divergence, positive selection and coevolution. BMC Evol. Biol. 14:124. 10.1186/1471-2148-14-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sornaraj P., Luang S., Lopato S., Hrmova M. (2016). Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta 1860, 46–56. 10.1016/j.bbagen.2015.10.014 [DOI] [PubMed] [Google Scholar]
- Talanova V. V., Titov A. F. (1994). Endogenous abscisic acid content in cucumber leaves under the influence of unfavourable temperatures and salinity. J. Exp. Bot. 45, 1031–1033. 10.1093/jxb/45.7.1031 [DOI] [Google Scholar]
- Taoka K., Ohki I., Tsuji H., Furuita K., Hayashi K., Yanase T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. 10.1038/nature10272 [DOI] [PubMed] [Google Scholar]
- Tian F., Wang T., Xie Y., Zhang J., Hu J. (2015). Genome-wide identification, classification, and expression analysis of 14-3-3 gene family in Populus. PLoS ONE 10:e0123225. 10.1371/journal.pone.0123225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong A. B., Masters S. C., Yang H., Fu H. (2002). Role of the 14-3-3 C-terminal loop in ligand interaction. Proteins 49, 321–325. 10.1002/prot.10210 [DOI] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. A. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, 71–74. 10.1093/nar/gkm306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Simillion C., Van de Peer Y. (2003). Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15, 2192–2202. 10.1105/tpc.014019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Hill T. (2008). High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 27, 471–478. 10.1007/s00299-007-0472-y [DOI] [PubMed] [Google Scholar]
- Wilson R. S., Swatek K. N., Thelen J. J. (2016). Regulation of the regulators: post-translational modifications, subcellular, and spatiotemporal distribution of Plant 14-3-3 proteins. Front. Plant Sci. 7:611. 10.3389/fpls.2016.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Rooney M. F., Ferl R. J. (1997). The arabidopsis 14-3-3 multigene family. Plant Physiol. 114, 1421–1431. 10.1104/pp.114.4.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., et al. (1997). The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 91, 961–971. 10.1016/S0092-8674(00)80487-0 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1993). Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol. 101, 1119–1120. 10.1104/pp.101.3.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., He C., Wang J., Mao Z., Holaday S. A., Allen R. D., et al. (2004). Overexpression of the Arabidopsis 14-3-3 protein GF14 lambda in cotton leads to a “staygreen” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol. 45, 1007–1014. 10.1093/pcp/pch115 [DOI] [PubMed] [Google Scholar]
- Yang X., Wang W., Coleman M., Orgil U., Feng J., Ma X., et al. (2009). Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 60, 539–550. 10.1111/j.1365-313X.2009.03978.x [DOI] [PubMed] [Google Scholar]
- Yang Z., Li H., Guo D., Tang X., Peng S. (2014). Identification and characterization of the 14-3-3 gene family in Hevea brasiliensis. Plant Physiol. Biochem. 80, 121–127. 10.1016/j.plaphy.2014.03.034 [DOI] [PubMed] [Google Scholar]
- Yu J., Wang J., Lin W., Li S., Li H., Zhou J., et al. (2005). The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38. 10.1371/journal.pbio.0030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. T., Zhou Y., Li Y., Shao S. Q., Li B. Y., Shi H. Y., et al. (2010). Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J. Exp. Bot. 61, 3331–3344. 10.1093/jxb/erq155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Chen G., Zhu J., Zhu Y., Lu X., Li X., et al. (2015). Molecular characterization and expression profiling of NAC transcription factors in Brachypodium distachyon L. PLoS ONE 10:e0139794. 10.1371/journal.pone.0139794 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene-specific primers for qRT-PCR analysis.
The nomenclature, characteristics of 14-3-3 genes and their deduced proteins in 13 representative species.
The parameters of conserved 14-3-3 domain from SMART and Pfam. (A) The parameters of conserved 14-3-3 domain from SMART. (B) The parameters of conserved 14-3-3 domain from Pfam.
Type-II functional divergence sites identified between different subgroup pairs of 14-3-3 gene family. Red color marked amino acids were identified in both Type-II functional divergence analysis and positive selection analysis. Amino acids listed in the table were matched to BdGF14c.
Tests for positive selection among codons of 14-3-3 genes using free-ratio and site models. aNumber of parameters in the ω distribution.
Critical coevolution amino acids identified from coevolution analysis.
Cis-element analysis of 1500-bp nucleotide sequences upstream of the translation initiation codon of 14-3-3 genes.
Data of the relative expression of B. distachyon 14-3-3 genes under abiotic stress treatments.
Parameters of B. distachyon root length and plant size under control and abiotic stress treatments. Dashes indicate no parameters collected.
Double standard curves and dissolution curves of qRT-PCR. The red standard curves represent the reference gene (S-adenosylmethionine decarboxylase gene), the blue standard curves represent the target genes. The dissolution curves of different genes are indicated.
The number of 14-3-3 genes in individual species. Cr, Chlamydomonas reinhardtii; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Bd, Brachypodium distachyon; Os, Oryza sativa; Ta, Triticum aestivum; Si, Setaria italic; Zm, Zea mays; Sb, Sorghum bicolor; At, Arabidopsis thaliana; Gm, Glycine max; Sl, Solanum lycopersicum; Gr, Gossypium raimondii.
Exon-intron organization of 14-3-3 genes. The yellow boxes and gray lines represent exons and introns, respectively. The bold dark blue lines in the top indicate the 5′ upstream region (left) and the 3′ downstream region (right). The 14-3-3 genes were lined according to Phylogenetic tree construction.
Statistic analysis in intron number. (A) Statistic analysis based on subgroups. (B) Statistic analysis based on individual species.
Schematic representation of conserved 14-3-3 protein motifs. The schematic diagram was derived from MEME. The order of motifs in the Schematic representation was automatically generated by MEME according to scores.
Schematic representation of amino acid motifs of 14-3-3 proteins. Motif analysis was performed using MEME as described in the Methods. The gray solid lines represent the corresponding 14-3-3 proteins and their length. The different-colored boxes represent different motifs and their position and order in individual 14-3-3 protein sequence.
Statistic analysis in motif number. (A) Statistic analysis based on subgroups. (B) Statistic analysis based on individual species.
Multiple sequence alignment of B. distachyon 14-3-3 protein sequences. Eleven 14-3-3 proteins consist of 7 B. distachyon 14-3-3 proteins, 2 Arabidopsis 14-3-3 proteins, and 2 rice 14-3-3 proteins. Nine α helices are marked with red-colored solid rectangle, while N-terminal and C-terminal are marked with light blue-colored solid rectangle, respectively. Red boxes indicate the critical amino acids identified from functional divergence and positive selection, light blue box indicates the critical amino acids identified from coevolution, while green boxes indicate the critical amino acids identified only from positive selection.
Images of B. distachyon plants under abiotic stress treatments for 6 or 2 h. (A) Mild drought stress (PEG 6000). (B) Phytohormone stress (H2O2). (C) Phytohormone stress (SA). (D) Salinity stress (NaCl). (E) High temperature stress (42°C). (F) Low temperature stress (4°C). Bar scale is 5.0 cm.
Images of Bd21 plants under abiotic stress treatments for 48 and 48 h recovery. (A) Control group. (B) Heavy metal stress (Cu2+). (C) Heavy metal stress (Cr3+). (D) Heavy metal stress (Cd2+). (E) Heavy metal stress (Zn2+). (F) Phytohormone stress (ABA). Bar scale is 5.0 cm.