Abstract
Background
Circulating tumor cells (CTCs) are cells that detach from a primary tumor, circulate through the blood stream and lymphatic vessels, and are considered to be the main reason for remote metastasis. Due to their origin, tumor cells have different gene expression levels than the surrounding blood cells. Therefore, they might be detectable in blood samples from breast cancer patients by real-time quantitative polymerase chain reaction (RT-qPCR).
Materials and Methods
Blood samples of healthy donors and adjuvant breast cancer patients were withdrawn and the cell fraction containing white blood cells and tumor cells was enriched by density gradient centrifugation. RNA was isolated and reverse transcribed to cDNA, which was then used in TaqMan real-time PCR against cytokeratin (CK)8, CK18 and CK19. 18S and GAPDH were used as controls.
Results
All 3 CKs were, on average, found to be significantly higher expressed in adjuvant breast cancer samples compared to negative controls, probably due to the presence of CTCs. Unfortunately, gene expression levels could not be correlated to tumor characteristics.
Conclusions
RT-qPCR could make up a new approach for the detection of CTCs from blood samples of breast cancer patients, but a correlation of the PCR data to gold standard methods in CTC detection would help to further improve the informative value of the qPCR results.
Keywords: Circulating tumor cells, RT-qPCR, Cytokeratins, Adjuvant breast cancer
Introduction
Breast cancer, the neoplasm with the highest incidence worldwide, is the most frequent cause of cancer-related death in women. Fortunately, its mortality has been regressive since the 1970s, but still more than half a million women died from breast cancer and its consequences in 2011.
The main reason for death is seldom the primary tumor itself but the overall tumor mass, due to the occurrence of remote metastases in other organs. The so-called circulating tumor cells (CTCs) are considered to be at the root of metastatic lesions. It is well known that CTCs are cells that dissociate from a primary tumor, move into the circulation through invasion of blood or lymphatic vessels, settle down at secondary sites in the body and can there become the origin of metastasis formation [1,2,3,4]. A number of clinical studies were published showing that the presence of CTCs in the blood of cancer patients is correlated with poor outcome of the disease [5,6,7] and a worse prognosis for overall survival in comparison to patients without CTCs [8,9,10]. In light of this, CTCs are already used as tumor markers [11].
Different techniques for CTC detection from blood samples have already been established [12]. It has already been shown, that real-time polymerase chain reaction (PCR) is a high-throughput method for detection and molecular characterization of CTCs [13,14,15], which also allows the monitoring of treatment efficiency [16,17,18,19].
The detection of CTCs via real-time quantitative PCR (RT-qPCR) is based on the fact that tumor cells are of epithelial origin, assuming that blood samples with CTCs would show higher expression levels of typical epithelial genes than blood samples without CTCs [20]. Our research group has already worked on the topic of CTC detection from blood samples [21]. We were able to establish and validate this method in an in vitro model system [22,23]. The present study is now the extension of our former work by moving from the model system towards patient samples, focusing on the possibility to detect CTCs in blood samples of adjuvant breast cancer patients to obtain hints on the clinical significance of this method.
For this purpose, blood samples from healthy donors and breast cancer patients in the adjuvant setting were analyzed by RT-qPCR. For that purpose we used standardized gene expression values of cytokeratin (CK)8, CK18, and CK19. We selected these cytokeratin genes as markers since their proteins are also in use as tumor cell markers in the alkaline phosphatase-antialkaline phosphatase (APAAP) staining [24,25], a routine detection method for tumor cells.
We found an increase in the expression levels of all 3 CK genes in the adjuvant patient group in comparison to the healthy control group, which was statistically significant. Unfortunately, the gene expression values could not be related to tumor characteristics, so that a comparison of the RT-qPCR results with gold standard methods in CTC detection would be necessary to further validate the method.
Materials and Methods
Blood Samples
From each of 20 breast cancer patients, 20 ml blood was withdrawn during primary breast-conserving surgery by using rather fine concave needles, to avoid epithelial cells from the skin to enter the circulation. These patients were treated consecutively with adjuvant breast cancer therapy. Written consent of the patients was obtained prior to the procedure (ethical votum LMU 148–12, in accordance with the Declaration of Helsinki). 20 ml blood was also obtained from each of 20 healthy female donors who had no surgical interventions or previous biopsies before blood withdrawal.
The white blood cell fraction, potentially containing CTCs, was enriched by density gradient centrifugation with Histopaque (Invitrogen, Darmstadt, Germany) for 30 min at 400 × g. Harvested cell pellets were washed twice at 250 × g, 4 °C for 10 min with phosphate-buffered saline (PBS; Biochrom, Berlin, Germany) and stored at −80 °C until further sample processing.
RNA Isolation
RNA was extracted from the cell pellets by the Trizol (Invitrogen, Darmstadt, Germany)/chloroform (Merck, Darmstadt, Germany) extraction method and resuspended in diethylpyrocarbonate (DEPC)-treated water. RNA concentrations and ratios were determined photometrically (Nanodrop, Implen, Munich, Germany), while RNA integrity was controlled by performing denaturing formaldehyde gel electrophoresis.
Reverse Transcription
Of the isolated total RNA, 4 μg was used for cDNA generation by the SuperScript III First Strand Synthesis Super Mix Kit (Invitrogen, Darmstadt, Germany). Reactions were set up and run according to the manufacturer's instructions; thereafter, samples were kept at −20 °C until use in RT-qPCR.
RT-qPCR
RT-qPCR reactions were performed on 96-well plates (Micro Amp® Fast Optical 96-well reaction plate with barcode; ABI, Foster City, CA, USA). For each reaction, 2 μl of the respective cDNA was used, and a reaction mix containing TaqMan® Fast Universal Mastermix (ABI, Foster City, CA, USA), H2O, and the respective TaqMan® hydrolysis probe (ABI, Foster City, CA, USA; CK8: Hs_02339472_g1, CK18: Hs_01920599_gH, CK19: Hs_00761767_g1, 18S: Hs_03928990_g1, GAPDH: Hs_00266705_g1) was added. Plates were run in the 7500 Fast Real Time PCR system. 18S (coding for the 18S ribosomal protein) and GAPDH (coding for glyceraldehyde 3-phospate dehydrogenase) were included as reference genes. Reactions for every gene were always set up as quadruplicates including non-template and no-reverse transcription controls; the PCR efficiency of the hydrolysis probes used was stated as 100 ± 10% by the provider, and was therefore not controlled again.
Evaluation
The fluorescence signals were evaluated with the Applied Biosystems Sequence Detection Software (SDS v1.3.1), and CT (cycle threshold), ΔCT, ΔΔCT, and RQ (relative expression) values were automatically calculated according to the 2-ΔΔCT method [26]. Non-template and -RT controls did not yield any fluorescent signals.
SDS-generated files were transferred to Microsoft© Excel™ and corresponding graphs were created. Statistical evaluations were done by SPSS v.20, using one-way ANOVA to compare the gene expression levels of sample groups.
Results
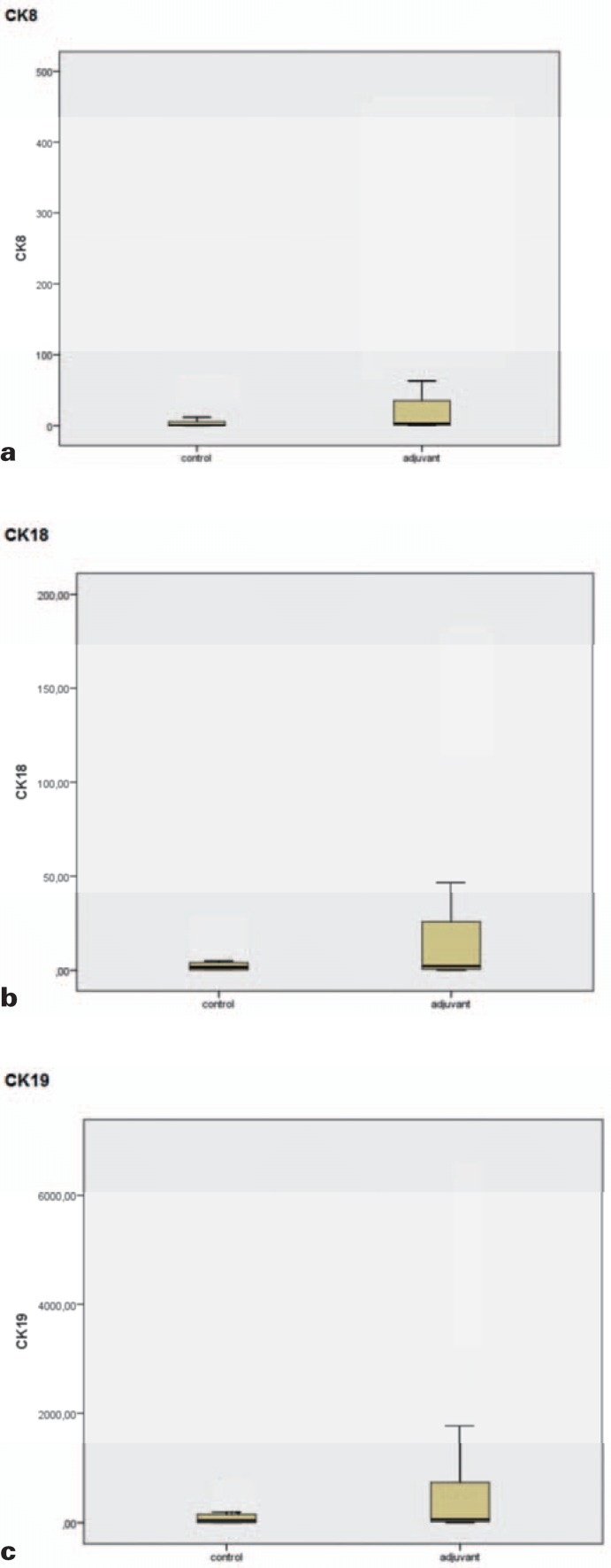
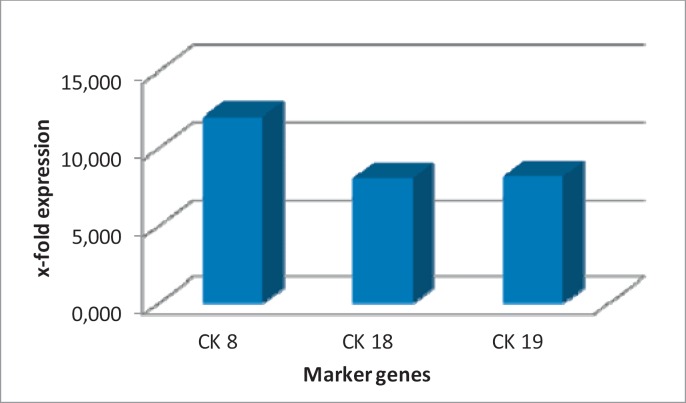
Gene expression values of CK8, CK18 and CK19 were standardized to 18S and GAPDH, and average gene expression values were compared between the healthy control group and the adjuvant breast cancer patients (fig. 1a–c, table 1). For CK8, the average RQ value in the control group was 4.95, while in the group of adjuvant breast cancer patients, the value was 59.607. We detected the same trend for CK18 and CK19: For CK18, the averaged value of the healthy control group was 4.151 and the patient group value was 33.539; and for CK19, the average values were 116.297 and 952.937, respectively. This corresponds to a 12-fold difference in gene expression for CK8 and an 8-fold difference between the 2 sample groups for CK18 and CK19 (fig. 2).
Fig. 1.
Comparison of average RQ values from samples of healthy donors versus adjuvant breast cancer samples. (a) CK8, (b) CK18, (c) CK19. In all three cases, the RQ values are higher for the adjuvant breast cancer group than for the negative control group.
Table 1.
Average RQ values of all 20 negative controls and adjuvant breast cancer samples
| Marker gene | RQ |
p | |
|---|---|---|---|
| Negative controls, average (SD) | Adjuvant breast cancer samples, average (SD) | ||
| CK8 | 4.95 (13.807) | 59.607 (128.609) | 0.047 |
| CK18 | 4.151 (6.918) | 33.539 (66.957) | 0.041 |
| CK19 | 116.297 (176.702) | 952.937 (1826.096) | 0.057 |
For the adjuvant breast cancer samples, the RQ values are significantly higher (indicated by the p-value) than for the negative control samples, indicating the presence of CTCs.
RQ = Relative expression, SD = standard deviation, CK = cytokeratin.
Fig. 2.
Fold differences in RQ values between adjuvant breast cancer samples and negative control samples. The most prominent difference is seen for CK8.
For statistical analysis, a t-test on independent samples was applied to the data. The test yielded statistically significant differences in gene expression levels for CK8 (p = 0.047) and CK18 (p = 0.041). For CK19, a borderline significance was detected (p = 0.057) (table 1).
Combination of the RQ values of each patient sample and the respective patient and tumor data did not result in any significant correlation (table 2, also for standard deviations).
Table 2.
Correlation of tumor characteristics and RQ values for CK8, CK18, and CK19
| No. | T stage | Size, mm | N stage | Histology | Age | ER status | PR status | HER2 status | RQ CK8 | RQ CK18 | RQ CKl9 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | pT1b | 11 | pN1a (1/2) | inv. duct | 58 | neg. | neg. | n.d. | 7.47l | 3.272 | 102.127 |
| 2 | cT2 | 27 | n.d. | inv. duct | 39 | n.d. | n.d. | n.d. | 3.349 | 1.528 | 32.533 |
| 3 | pT1c | l9 | pN0 (0/1) | inv. duct | 52 | neg. | neg. | pos. | 0.90l | 0.32 | 28.47 |
| 4 | pT1c | 12 | pN1a (1/14) | inv. duct | 73 | pos. | pos. | neg. | 3.126 | 2.639 | 185.744 |
| 5 | pT1b | 6 | pN0 (0/3) | inv. duct | 66 | pos. | pos. | pos. | 0.306 | 0.185 | 14 |
| 6 | pT1c | 12 | pN0 (0/2) | inv. duct | 66 | pos. | pos. | neg. | 203.192 | 117.464 | 3338.293 |
| 7 | pT2 | 25 | pN0 (0/1) | inv. duct | 70 | pos. | pos. | pos. | 63.171 | 46.598 | 1269.884 |
| 8 | pT1c | 13 | pN0 (0/2) | inv. duct | 70 | pos. | pos. | pos. | n.d. | n.d. | n.d. |
| 9 | pT2 | 30 | pN0 (0/3) | inv. duct | 46 | pos. | pos. | pos. | 1.432 | 0.806 | 21.22 |
| 10 | pT2 | 22 | pN0 (0/l) | inv. duct | 48 | pos. | pos. | neg. | 5.576 | 3.424 | 103.026 |
| 11 | pT1c | 18 | pN0 (0/2) | inv. duct | 63 | pos. | pos. | neg. | 0.075 | 0.054 | 1.873 |
| 12 | cT3 | 55 | pN0 (0/2) | inv. duct | 53 | neg. | neg. | neg. | 0.105 | 0.121 | 1.259 |
| 13 | pT1c | 12 | pN0 (0/2) | inv. duct | 52 | pos. | pos. | pos. | 1.89 | 1.019 | 72.961 |
| 14 | pT2 | 25 | pN0 (0/7) | inv. lob | 63 | pos. | pos. | pos. | 1.629 | 1.786 | 52.762 |
| 15 | pT1c | 13 | pN0 (0/1) | inv. duct | 70 | pos. | pos. | pos. | 0.407 | 0.634 | 17.851 |
| 16 | pT2 | 40 | pN0 (0/5) | inv. duct | 70 | pos. | pos. | pos. | 155.595 | 135.514 | 4635.474 |
| 17 | pT1c | 20 | pN0 (0/1) | inv. duct | 71 | neg. | neg. | neg. | 6.834 | 4.656 | 106.96 |
| 18 | pT1c | 15 | pN0 (0/1) | inv. duct | 51 | pos. | pos. | neg. | 402.414 | 173.955 | 6310.456 |
| 19 | pT1c | 18 | pN0 (0/4) | inv. duct | 50 | pos. | neg. | pos. | 2.911 | 2.64 | 41.437 |
| 20 | pT1c/pT1b | l2/9 | pN0 (0/1) | inv. duct | 70 | pos./pos. | pos./pos. | pos./neg. | 273.943 | 142.751 | 1769.476 |
No correlation between tumor data and RQ values could be shown. For patients 3, 5, 11, 12, and 15, a downward deviation of the RQ values from the average RQ value is seen; in patients 6, 7, 16, 18, and 20, the RQ values deviate upwards from the average RQ.
ER = Estrogen receptor, PR = progesterone receptor, HER2 = human epidermal growth factor receptor, RQ = relative expression, CK = cytokeratin, n.d. = not detectable, inv. duct = invasive ductal, inv. lob = invasive lobular, neg. = negative, pos. = positive.
Discussion
RT-qPCR seems to be a promising method for CTC detection in blood samples [5,6,7,13,14,15] of patients suffering from different types of cancer, such as breast cancer, for example. But still a lot of work needs to be done. As the on-hand preliminary study shows, the presence of tumor cells in blood samples can indeed be detected but the number of CTCs [27] cannot yet be quantified with reference to the used technique. To overcome this limitation, standard curves have to be generated to be able to correlate gene expression levels to the number of CTCs contained in a certain blood sample. However, as different types of breast cancer express different genes at different levels, the creation of reliable standard curves will be a challenging task. Another drawback of the methodology is that it cannot be completely avoided that, by blood withdrawal or previous therapeutical interventions, single epithelial cells will enter the circulation and are later detected as CTCs, as it was shown in a number of publications [28,29,30], resulting in false-positive outcomes. Although using similar detection methods, other research groups did not find epithelial cells in blood samples of healthy control persons [31,32]. Therefore it could be generally concluded that sample withdrawal has to be carried out with care, so that the number of such false-positive cells is kept rather small, e.g. by using fine butterfly needles for blood extraction. An analysis of the samples by ancillary methods would help to overcome this obstacle [33].
In the following, a simultaneous analysis of blood samples by RT-qPCR and a gold standard method, the CellSearch™ system, could on the one hand help to generate these standard curves and on the other hand might be useful for the correlation of patient/tumor data and RQ values. Also, an analysis of blood from metastatic patients could improve PCR-based detection of CTCs as the CTC incidence in this patient group is significantly higher, but bears the disadvantage that material from metastatic patients is rather rare. Another important and interesting point would be to decide by the RT-qPCR results whether a certain blood sample came from a cancer patient or was withdrawn from a healthy donor. But for this purpose, a lot of work still needs to be done, especially in defining a set of marker genes on which this decision could be made with high reliability [34,35,36]. The analysis of more marker genes like Her-2 [37], MMP13 [38], UBE2Q2 [39], or Nectin-4 [40] for their use in CTC detection could help in this point, allowing a more sensitive detection with simultaneous characterization of the tumor cells as well. With the help of such a marker gene panel, an epithelial cell adhesion molecule (EPCAM)-independent CTC detection method could be developed, meaning that the epithelial-to-mesenchymal transition (EMT; [41,42]) would no longer influence the CTC detection. The latter could be regarded as a drawback of the present study as well. Thereby new roads towards a more individualized treatment with increasing treatment efficiency and reduced therapeutic side effects could eventually be opened.
Disclosure Statement
The authors declare no conflicts of interest.
Acknowledgement
This work was supported by a grant from the Heuer-Stiftung für medizinische Forschung.
References
- 1.Mocellin S, Keilholz U, Rossi CR, Nitti D. Circulating tumor cells: the ‘leukemic phase’ of solid cancers. Trends Mol Med. 2006;12:130–139. doi: 10.1016/j.molmed.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 3.Ring A, Smith IE, Dowsett M. Circulating tumour cells in breast cancer. Lancet Oncol. 2004;5:79–88. doi: 10.1016/S1470-2045(04)01381-6. [DOI] [PubMed] [Google Scholar]
- 4.Smerage JB, Hayes DF. The measurement and therapeutic implications of circulating tumour cells in breast cancer. Br J Cancer. 2006;94:8–12. doi: 10.1038/sj.bjc.6602871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molloy TJ, Bosma AJ, van't Veer LJ. Towards an optimized platform for the detection, enrichment, and semi-quantitation of circulating tumor cells. Breast Cancer Res Treat. 2008;112:297–307. doi: 10.1007/s10549-007-9872-5. [DOI] [PubMed] [Google Scholar]
- 6.Molloy TJ, Devriese LA, Helgason HH, Bosma AJ, Hauptmann M, Voest EE, Schellens JH, van't Veer LJ. A multimarker qPCR-based platform for the detection of circulating tumour cells in patients with early-stage breast cancer. Br J Cancer. 2011;104:1913–1919. doi: 10.1038/bjc.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xenidis N, Ignatiadis M, Apostolaki S, Perraki M, Kalbakis K, Agelaki S, Stathopoulos EN, Chlouverakis G, Lianidou E, Kakolyris S, Georgoulias V, Mavroudis D. Cytokeratin-19 mRNA-positive circulating tumor cells after adjuvant chemotherapy in patients with early breast cancer. J Clin Oncol. 2009;27:2177–2184. doi: 10.1200/JCO.2008.18.0497. [DOI] [PubMed] [Google Scholar]
- 8.Braun S, Pantel K, Muller P, Janni W, Hepp F, Kentenich CR, Gastroph S, Wischnik A, Dimpfl T, Kindermann G, Riethmuller G, Schlimok G. Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N Engl J Med. 2000;342:525–533. doi: 10.1056/NEJM200002243420801. [DOI] [PubMed] [Google Scholar]
- 9.Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, Pierga JY, Marth C, Oruzio D, Wiedswang G, Solomayer EF, Kundt G, Strobl B, Fehm T, Wong GY, Bliss J, Vincent-Salomon A, Pantel K. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 10.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 11.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 12.Van der Auwera I, Peeters D, Benoy IH, Elst HJ, Van Laere SJ, Prove A, Maes H, Huget P, van Dam P, Vermeulen PB, Dirix LY. Circulating tumour cell detection: a direct comparison between the CellSearch system, the AdnaTest and CK-19/mammaglobin RT-PCR in patients with metastatic breast cancer. Br J Cancer. 2010;102:276–284. doi: 10.1038/sj.bjc.6605472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sieuwerts AM, Kraan J, Bolt-de Vries J, van der Spoel P, Mostert B, Martens JW, Gratama JW, Sleijfer S, Foekens JA. Molecular characterization of circulating tumor cells in large quantities of contaminating leukocytes by a multiplex real-time PCR. Breast Cancer Res Treat. 2009;118:455–468. doi: 10.1007/s10549-008-0290-0. [DOI] [PubMed] [Google Scholar]
- 14.Sieuwerts AM, Mostert B, Bolt-de Vries J, Peeters D, de Jongh FE, Stouthard JM, Dirix LY, van Dam PA, Van Galen A, de Weerd V, Kraan J, van der Spoel P, Ramirez-Moreno R, van Deurzen CH, Smid M, Yu JX, Jiang J, Wang Y, Gratama JW, Sleijfer S, Foekens JA, Martens JW. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin Cancer Res. 2011;17:3600–3618. doi: 10.1158/1078-0432.CCR-11-0255. [DOI] [PubMed] [Google Scholar]
- 15.Stathopoulou A, Gizi A, Perraki M, Apostolaki S, Malamos N, Mavroudis D, Georgoulias V, Lianidou ES. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the LightCycler system. Clin Cancer Res. 2003;9:5145–5151. [PubMed] [Google Scholar]
- 16.Androulakis N, Agelaki S, Perraki M, Apostolaki S, Bozionelou V, Pallis A, Kalbakis K, Xyrafas A, Mavroudis D, Georgoulias V. Clinical relevance of circulating CK-19 mRNA-positive tumour cells before front-line treatment in patients with metastatic breast cancer. Br J Cancer. 2012;106:1917–1925. doi: 10.1038/bjc.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daskalaki A, Agelaki S, Perraki M, Apostolaki S, Xenidis N, Stathopoulos E, Kontopodis E, Hatzidaki D, Mavroudis D, Georgoulias V. Detection of cytokeratin-19 mRNA-positive cells in the peripheral blood and bone marrow of patients with operable breast cancer. Br J Cancer. 2009;101:589–597. doi: 10.1038/sj.bjc.6605183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ignatiadis M, Kallergi G, Ntoulia M, Perraki M, Apostolaki S, Kafousi M, Chlouverakis G, Stathopoulos E, Lianidou E, Georgoulias V, Mavroudis D. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, mammaglobin A, and Her2 in early breast cancer. Clin Cancer Res. 2008;14:2593–2600. doi: 10.1158/1078-0432.CCR-07-4758. [DOI] [PubMed] [Google Scholar]
- 19.Wang HY, Ahn S, Kim S, Park S, Park S, Han H, Sohn JH, Kim S, Lee H. Detection of circulating tumor cells in patients with breast cancer using the quantitative RT-PCR assay for monitoring of therapy efficacy. Exp Mol Pathol. 2014;97:445–452. doi: 10.1016/j.yexmp.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 21.Andergassen U, Kolbl AC, Hutter S, Friese K, Jeschke U. Detection of circulating tumour cells from blood of breast cancer patients via RT-qPCR. Cancers. 2013;5:1212–1220. doi: 10.3390/cancers5041212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zebisch M, Kolbl AC, Schindlbeck C, Neugebauer J, Heublein S, Ilmer M, Rack B, Friese K, Jeschke U, Andergassen U. Quantification of breast cancer cells in peripheral blood samples by real-time RT-PCR. Anticancer Res. 2012;32:5387–5391. [PubMed] [Google Scholar]
- 23.Zebisch M, Kolbl AC, Andergassen U, Hutter S, Neugebauer J, Engelstadter V, Gunthner-Biller M, Jeschke U, Friese K, Rack B. Detection of circulating tumour cells on mRNA levels with established breast cancer cell lines. Biomed Rep. 2013;1:231–234. doi: 10.3892/br.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurec AS, Baltrucki L, Mason DY, Davey FR. Use of the APAAP method in the classification and diagnosis of hematologic disorders. Clin Lab Med. 1988;8:223–236. [PubMed] [Google Scholar]
- 25.Noack F, Schmitt M, Bauer J, Helmecke D, Kruger W, Thorban S, Sandherr M, Kuhn W, Graeff H, Harbeck N. A new approach to phenotyping disseminated tumor cells: methodological advances and clinical implications. Int J Biol Markers. 2000;15:100–104. doi: 10.1177/172460080001500119. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Mostert B, Sleijfer S, Foekens JA, Gratama JW. Circulating tumor cells (CTCs): detection methods and their clinical relevance in breast cancer. Cancer Treat Rev. 2009;35:463–474. doi: 10.1016/j.ctrv.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Bleiweiss IJ, Nagi CS, Jaffer S. Axillary sentinel lymph nodes can be falsely positive due to iatrogenic displacement and transport of benign epithelial cells in patients with breast carcinoma. J Clin Oncol. 2006;24:2013–2018. doi: 10.1200/JCO.2005.04.7076. [DOI] [PubMed] [Google Scholar]
- 29.Diaz NM, Vrcel V, Centeno BA, Muro-Cacho C. Modes of benign mechanical transport of breast epithelial cells to axillary lymph nodes. Adv Anat Pathol. 2005;12:7–9. doi: 10.1097/01.pap.0000151267.34438.a1. [DOI] [PubMed] [Google Scholar]
- 30.Winkens T, Pachmann K, Freesmeyer M. Circulating epithelial cells in patients with thyroid carcinoma. Can they be identified in the blood? Nuklearmedizin. 2013;52:7–13. doi: 10.3413/Nukmed-0524-12-08. [DOI] [PubMed] [Google Scholar]
- 31.Pantel K, Deneve E, Nocca D, Coffy A, Vendrell JP, Maudelonde T, Riethdorf S, Alix-Panabieres C. Circulating epithelial cells in patients with benign colon diseases. Clin Chem. 2012;58:936–940. doi: 10.1373/clinchem.2011.175570. [DOI] [PubMed] [Google Scholar]
- 32.Rhim AD, Thege FI, Santana SM, Lannin TB, Saha TN, Tsai S, Maggs LR, Kochman ML, Ginsberg GG, Lieb JG, Chandrasekhara V, Drebin JA, Ahmad N, Yang YX, Kirby BJ, Stanger BZ. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology. 2014;146:647–651. doi: 10.1053/j.gastro.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofman VJ, Ilie MI, Bonnetaud C, Selva E, Long E, Molina T, Vignaud JM, Flejou JF, Lantuejoul S, Piaton E, Butori C, Mourad N, Poudenx M, Bahadoran P, Sibon S, Guevara N, Santini J, Venissac N, Mouroux J, Vielh P, Hofman PM. Cytopathologic detection of circulating tumor cells using the isolation by size of epithelial tumor cell method: promises and pitfalls. Am J Clin Pathol. 2011;135:146–156. doi: 10.1309/AJCP9X8OZBEIQVVI. [DOI] [PubMed] [Google Scholar]
- 34.Andergassen U, Hofmann S, Kolbl AC, Schindlbeck C, Neugebauer J, Hutter S, Engelstadter V, Ilmer M, Friese K, Jeschke U. Detection of tumor cell-specific mRNA in the peripheral blood of patients with breast cancer - evaluation of several markers with real-time reverse transcription-PCR. Int J Mol Sci. 2013;14:1093–1104. doi: 10.3390/ijms14011093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo M, Li X, Zhang S, Song H, Zhang W, Shang X, Zheng Y, Jiang H, Lv Q, Jiang Y, Hao H. Real-time quantitative RT-PCR detection of circulating tumor cells from breast cancer patients. Int J Oncol. 2015;46:281–289. doi: 10.3892/ijo.2014.2732. [DOI] [PubMed] [Google Scholar]
- 36.Smirnov DA, Zweitzig DR, Foulk BW, Miller MC, Doyle GV, Pienta KJ, Meropol NJ, Weiner LM, Cohen SJ, Moreno JG, Connelly MC, Terstappen LW, O'Hara SM. Global gene expression profiling of circulating tumor cells. Cancer Res. 2005;65:4993–4997. doi: 10.1158/0008-5472.CAN-04-4330. [DOI] [PubMed] [Google Scholar]
- 37.Hayes DF, Walker TM, Singh B, Vitetta ES, Uhr JW, Gross S, Rao C, Doyle GV, Terstappen LW. Monitoring expression of Her-2 on circulating epithelial cells in patients with advanced breast cancer. Int J Oncol. 2002;21:1111–1117. doi: 10.3892/ijo.21.5.1111. [DOI] [PubMed] [Google Scholar]
- 38.Chang HJ, Yang MJ, Yang YH, Hou MF, Hsueh EJ, Lin SR. MMP13 is potentially a new tumor marker for breast cancer diagnosis. Oncol Rep. 2009;22:1119–1127. doi: 10.3892/or_00000544. [DOI] [PubMed] [Google Scholar]
- 39.Nikseresht M, Seghatoleslam A, Monabati A, Talei A, Ghalati FB, Owji AA. Overexpression of the novel human gene, UBE2Q2, in breast cancer. Cancer Genet Cytogenet. 2010;197:101–106. doi: 10.1016/j.cancergencyto.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 40.Fabre-Lafay S, Garrido-Urbani S, Reymond N, Goncalves A, Dubreuil P, Lopez M. Nectin-4, a new serological breast cancer marker, is a substrate for tumor necrosis factor-alpha-converting enzyme (TACE)/ADAM-17. J Biol Chem. 2005;280:19543–19550. doi: 10.1074/jbc.M410943200. [DOI] [PubMed] [Google Scholar]
- 41.Joosse SA, Hannemann J, Spotter J, Bauche A, Andreas A, Muller V, Pantel K. Changes in keratin expression during metastatic progression of breast cancer: impact on the detection of circulating tumor cells. Clin Cancer Res. 2012;18:993–1003. doi: 10.1158/1078-0432.CCR-11-2100. [DOI] [PubMed] [Google Scholar]
- 42.Joosse SA, Pantel K. Biologic challenges in the detection of circulating tumor cells. Cancer Res. 2013;73:8–11. doi: 10.1158/0008-5472.CAN-12-3422. [DOI] [PubMed] [Google Scholar]