Abstract
Oncology practice increasingly requires the use of molecular profiling of tumors to inform the use of targeted therapeutics. However, many oncologists use third-party laboratories to perform tumor genomic testing, and these laboratories may not have electronic interfaces with the provider’s electronic medical record (EMR) system. The resultant reporting mechanisms, such as plain-paper faxing, can reduce report fidelity, slow down reporting procedures for a physician’s practice, and make reports less accessible. Vanderbilt University Medical Center and its genomic laboratory testing partner have collaborated to create an automated electronic reporting system that incorporates genetic testing results directly into the clinical EMR. This system was iteratively tested, and causes of failure were discovered and addressed. Most errors were attributable to data entry or typographical errors that made reports unable to be linked to the correct patient in the EMR. By providing direct feedback to providers, we were able to significantly decrease the rate of transmission errors (from 6.29% to 3.84%; P < .001). The results and lessons of 1 year of using the system and transmitting 832 tumor genomic testing reports are reported.
INTRODUCTION
Molecular profiling of tumors is becoming the standard of care in an increasing number of cancer types to inform the practice of precision medicine.1,2 However, the implementation of somatic gene testing into clinical practice can be difficult because the results of such testing can be reported asynchronously and separately from other pathology information.3-5 In addition, tumors are often tested at laboratory facilities that are not integrated with oncologists’ offices and their electronic medical records (EMRs). This disconnect can create challenges in reporting important genetic information from the laboratory to the treating oncologist.6
Oncologists at Vanderbilt University Medical Center (VUMC) have partnered with Foundation Medicine Incorporated (FMI, Cambridge, MA), a provider of tumor exome sequencing, to analyze patients’ tumors for genetic variants that may inform clinical decision making.7 This collaboration, like most with third-party laboratories, has relied on the use of faxed documents to report test results. Although faxing has been the standard for third-party laboratory reporting for decades, the use of black and white faxes with subsequent paper copies that are scanned into an EMR leads to a process that requires many manual steps and results in a loss of color information and poor readability in the EMR8 (Appendix Fig A1, online only). This lack of information interoperability has been cited as a target for improving the practice of precision medicine.9 At VUMC, an assessment of faxed molecular profile reports demonstrated poor information fidelity and lack of provider notification; these issues prevented optimal utility of molecular profiling. We hypothesized that automated electronic reporting from FMI directly into the VUMC EMR could be feasibly implemented and could enable provider notification. We describe the design, evaluation, and implementation over 1 year of using this system.
METHODS
The initial evaluation of somatic gene testing at VUMC was to perform a lean-based assessment with a modified value-stream mapping of ordering tests and reporting workflow.10 To address the shortcomings identified in this assessment, we defined a document structure and a data transfer protocol for electronic laboratory reporting. Through an iterative design and testing process that used lean methodology, a postprocessed laboratory report was packaged into a standardized extensible markup language (XML) document that included demographics, ordering information, and the FMI-designated actionable variants that recapitulate the first page of the paper report. A full-color portable document format (PDF) report was attached to the XML file. By using multiple layers of security, these results were transmitted nightly from FMI to VUMC servers. The files were parsed, matched, and incorporated into VUMC patients’ medical records. Provider information was matched to trigger existing notification methods.11 A log of all transmissions and any detected errors was maintained to evaluate the process. Differences in error rates were compared by using the χ2 test. Because of its quality improvement nature, this work was determined to be unrelated to human subjects (VUMC Institutional Review Board #140813).
The multidisciplinary team included a nurse who served as the tissue librarian and helped conduct the workflow analysis. Physician members of the Vanderbilt-Ingram Cancer Center Research Informatics Core helped define the data and transmission structure and perform the evaluation and data analysis. Administrators from the Vanderbilt-Ingram Cancer Center helped define functional and security requirements. Members of the VUMC Health Information Technology department created the XML receiver and parser that would incorporate the report into the EMR and trigger automated notification. Staff from FMI performed the molecular profiling, helped define the data and transmission structure, and worked with VUMC Health Information Technology to implement the transmissions.
RESULTS
Analysis of the ordering and reporting workflow revealed that the faxed and scanned laboratory reports were difficult to read, and providers were not consistently being notified when the reports were returned. The data structure and transmission protocol addressed these issues.
Transmission feasibility was demonstrated with an initial test transmission of 524 reports that had been created before September 2014. We alerted providers that they would receive multiple notifications via the EMR as these old reports were delivered. Logs of files transmitted by FMI and received by VUMC were identical; 100% of files were received with no evidence of files being lost in transmission. However, 33 files (6.29%) that failed to be incorporated into the EMR were investigated (Fig 1). All reports that were integrated into the EMR demonstrated text quality identical to other EMR elements, and the PDF contained color information at resolution comparable to publications on the Internet (Appendix Fig A1). The system went live with nightly transmission of new reports in October 2014.
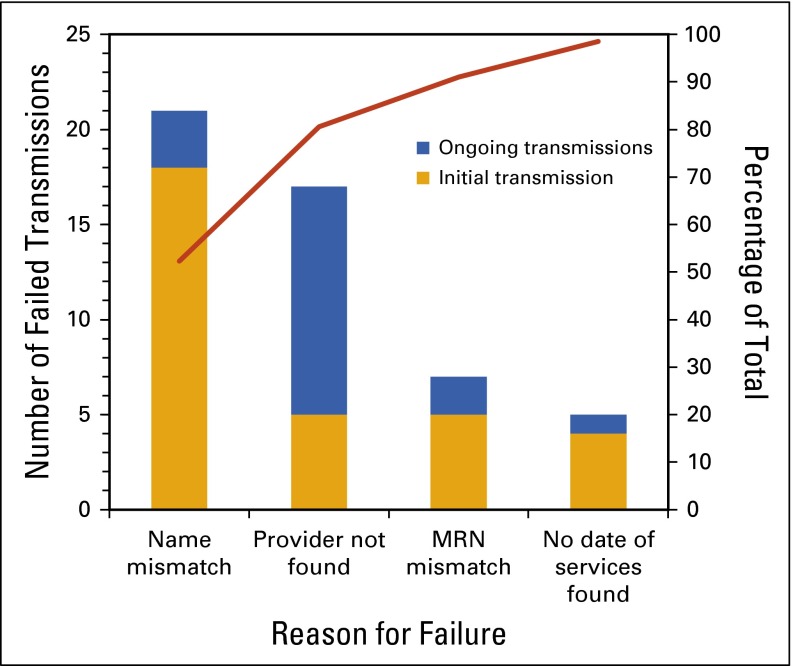
FIG 1.
Pareto diagram for 67 genomic report transmission failures. MRN, medical record number.
Errors during the initial transmission included reports that failed to integrate into the EMR because of misspelled patient names, incorrect medical record numbers (MRNs), missing date of service, and missing provider information (Fig 2). During the initial transmission, there were also two periods of time in which the receiving VUMC server was unable to communicate with a separate system that verifies patient information, resulting in those reports being rejected. Examples of MRN errors that resulted in reports being rejected included using the pathology accession number for MRN, including the “#” symbol, and typographical errors. Providers who were new to the division accounted for the majority of provider errors; however, these were not rejected outright, but rather incorporated into the EMR and routed to a results reporting team for manual provider notification. Taken as a whole, many errors were attributable to mistakes in the manual ordering process when the test was first ordered.
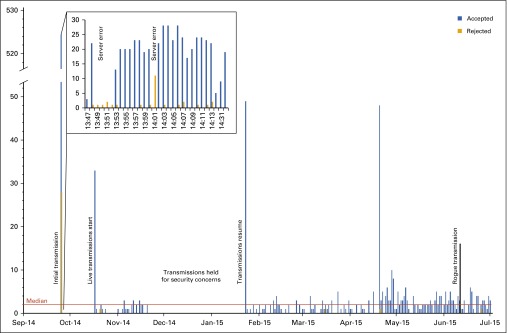
FIG 2.
Run chart demonstrating the failed and successful genomic report transmissions over 1 year.
After reporting this conclusion to the ordering providers, the error log subsequently demonstrated that fewer files were rejected as a result of increased diligence in ordering. Mean rejection rate decreased from 6.29% (95% CI, 4.21% to 8.38%) to 3.84% (95% CI, 1.92% to 5.74%; P < .001). In fall 2014, the transmissions were halted until January 2015 because of security concerns. When transmissions resumed in January 2015, continued low rates of rejected files were observed until June when 16 files for non-VUMC patients were transmitted and rejected by VUMC servers (Fig 1). This rogue transmission also contained the first file that failed for technical reasons; the file had corrupted contents and is under investigation.
The system currently receives a median of two reports per day and has successfully integrated more than 900 test reports into patients’ medical records. Providers appreciate the automatic EMR-based notification and the better readability of the color PDF. In addition, providers were able use the EMR’s text search function to quickly find a patient’s genomic results within their medical record, a feature previously unavailable with scanned documents.
DISCUSSION
We have evaluated this system, which has demonstrated the feasibility of an automated, secure electronic reporting solution for molecular profiling reports being transferred from a third-party laboratory into an EMR. This system improves information fidelity by preserving text quality and report color and allows use of an existing physician notification system. The accuracy of incorporating results into the correct medical record was significantly improved by informing ordering providers about the importance of providing accurate information during the ordering process.
Although there were concerns that prompted multiple reviews of the system, we demonstrated that these concerns can be surmounted. There was a possibility that files lost in transmission would not be logged; however, the initial test was accurate, and monitoring medical records for undiscovered errors in the system has not revealed any missing files. Many failures of the system were attributable to propagated ordering errors, so the system can be improved by increasing awareness of the importance of providing accurate information during ordering.
This system uses common document types (XML and PDF) that could be extended to other practices. The importance and challenges of reporting cancer genetic testing was highlighted recently by the College of American Pathology, which proposed an XML-based electronic cancer checklist that could be implemented with a system similar to the one we described here.6 However, the XML document that was developed would not necessarily be extensible to other third-party laboratories or clinical sites. One potential solution would be to use the interoperable Fast Healthcare Interoperability Resources (FHIR) standard from Health Level Seven International, which uses the SMART platform (SMART on FHIR Genomics).12 With increasing attention to the importance of standards in the interoperability of cancer-specific data, it is likely that the SMART on FHIR Genomics framework will be a solution for future exchanges of genomic information.13 Although using such a standard would not necessarily resolve the process issues encountered during this implementation, it would facilitate implementation with vendor EMRs that support such data standards.14-16
Future efforts to extend this system at VUMC could include using computerized physician order entry (CPOE) to order the molecular profiling test. Closed-loop systems such as that offered by Syapse (Palo Alto, CA) incorporate CPOE into the physician’s workflow on certain EMR platforms. However, CPOE can require prohibitively costly interfaces between EMR, laboratory information systems, and third-party laboratories. In an ideal setting, CPOE can automatically populate names, MRNs, and other identifying information, thus eliminating typographical errors in ordering. In addition, CPOE can provide an opportunity for clinical decision support in the increasingly complex practice of precision oncology. As molecular profiling of tumors increases, solutions for integrating results reporting into clinical workflows will allow greater utility from these tests.
Appendix
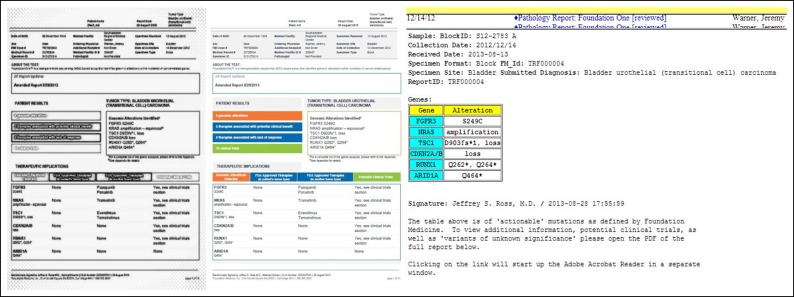
FIG A1.
Faxing and scanning of tumor molecular profiling reports (left) results in loss of color information and reduced readability compared with the original color PDF report (center). In addition, parsed XML information can be displayed directly in the electronic medical record (right) thus not impeding the practitioner's workflow when finding results of genetic testing. Data shown are dummy data created for quality assurance purposes; dates and accession numbers do not correspond to any patient protected health information.
AUTHOR CONTRIBUTIONS
Conception and design: Matthew J. Rioth, Lauren Hackett, Mia Levy, Jeremy Warner
Administrative support: Lauren Hackett
Provision of study materials or patients: Jeremy Warner
Collection and assembly of data: Matthew J. Rioth, David B. Staggs, Erich Haberman, Mike Tod, Jeremy Warner
Data analysis and interpretation: Matthew J. Rioth, Jeremy Warner
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Implementing and Improving Automated Electronic Tumor Molecular Profiling
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jop.ascopubs.org/site/misc/ifc.xhtml.
Matthew J. Rioth
No relationship to disclose
David B. Staggs
Patents, Royalties, Other Intellectual Property: StarPanel
Lauren Hackett
No relationship to disclose
Erich Haberman
No relationship to disclose
Mike Tod
Employment: Foundation Medicine
Stock or Other Ownership: Foundation Medicine
Travel, Accommodations, Expenses: Foundation Medicine
Mia Levy
Leadership: Personalis
Consulting or Advisory Role: Personalis, GenomOncology
Jeremy Warner
No relationship to disclose
References
- 1.Tsimberidou AM, Eggermont AM, Schilsky RL. Precision cancer medicine: The future is now, only better. 2014 . Am Soc Clin Oncol Educ Book 61-69. [DOI] [PubMed]
- 2.Van Allen EM, Wagle N, Levy MA. Clinical analysis and interpretation of cancer genome data. J Clin Oncol. 2013;31:1825–1833. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronquillo JG, Li C, Lester WT. Genetic testing behavior and reporting patterns in electronic medical records for physicians trained in a primary care specialty or subspecialty. J Am Med Inform Assoc. 2012;19:570–574. doi: 10.1136/amiajnl-2011-000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasser S. Kurdolgu AA, Izatt T, et al: An integrated framework for reporting clinically relevant biomarkers from paired tumor/normal genomic and transcriptomic sequencing data in support of clinical trials in personalized medicine. Biocomputing 2015, Proceedings of the Pacific Symposium, Kohala Coast, HI, January 4-8, 2015, pp 56-67. [PubMed]
- 5.Rubin MA. Health: Make precision medicine work for cancer care. Nature. 2015;520:290–291. doi: 10.1038/520290a. [DOI] [PubMed] [Google Scholar]
- 6.Simpson RW, Berman MA, Foulis PR, et al. Cancer biomarkers: The role of structured data reporting. Arch Pathol Lab Med. 2015;139:587–593. doi: 10.5858/arpa.2014-0082-RA. [DOI] [PubMed] [Google Scholar]
- 7.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGowan GK. The signing of laboratory reports. J Clin Pathol. 1974;27:427–429. doi: 10.1136/jcp.27.5.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. The White House, Office of the Press Secretary: Fact Sheet: President Obama’s Precision Medicine Initiative. https://www.whitehouse.gov/the-press-office/2015/01/30/fact-sheet-president-obama-s-precision-medicine-initiative.
- 10.Toussaint JS, Berry LL. The promise of Lean in health care. Mayo Clin Proc. 2013;88:74–82. doi: 10.1016/j.mayocp.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Jirjis J, Weiss JB, Giuse D, et al. A framework for clinical communication supporting healthcare delivery. AMIA Annu Symp Proc. 2005:375–379. [PMC free article] [PubMed] [Google Scholar]
- 12.Alterovitz G, Warner J, Zhang P, et al. SMART on FHIR Genomics: Facilitating standardized clinico-genomic apps. J Am Med Inform Assoc. 2015;22:1173–1178. doi: 10.1093/jamia/ocv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warner JL, Maddux SE, Hughes KS, et al. Development, implementation, and initial evaluation of a foundational open interoperability standard for oncology treatment planning and summarization. J Am Med Inform Assoc. 2015;22:577–586. doi: 10.1093/jamia/ocu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Open Epic: Ancillary systems. https://open.epic.com/Ancillary.
- 15. Cerner: Laboratory. http://www.cerner.com/solutions/Hospitals_and_Health_Systems/Laboratory/
- 16. GE Healthcare: Centricity perinatal standard interfaces. https://www2.gehealthcare.com/us/maintenance_support/maintenance_offerings/ch.standard-interfaces.octonary#COLD Feed.