Abstract
Positive-sense RNA viruses have a small genome with very limited coding capacity and are highly dependent on host components to fulfill their life cycle. Recent studies have suggested that DEAD-box RNA helicases play vital roles in many aspects of RNA metabolism. To explore the possible role of the RNA helicases in viral infection, we used the Turnip mosaic virus (TuMV)-Arabidopsis pathosystem. The Arabidopsis genome encodes more than 100 putative RNA helicases (AtRH). Over 41 Arabidopsis T-DNA insertion mutants carrying genetic lesions in the corresponding 26 AtRH genes were screened for their requirement in TuMV infection. TuMV infection assays revealed that virus accumulation significantly decreased in the Arabidopsis mutants of three genes, AtRH9, AtRH26, and PRH75. In the present work, AtRH9 was further characterized. Yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays showed that AtRH9 interacted with the TuMV NIb protein, the viral RNA-dependent RNA polymerase. Moreover, the subcellular distribution of AtRH9 was altered in the virus-infected cells, and AtRH9 was recruited to the viral replication complex. These results suggest that Arabidopsis AtRH9 is an important component of the TuMV replication complex, possibly recruited via its interaction with NIb.
RNA viruses have small and compact genomes with limited coding capacity and live exclusively in their host cells. During the long-time co-evolution with the hosts, they have acquired the ability to recruit host proteins and reprogram host metabolites to fulfill their life cycle1,2,3,4,5,6. In the last decade, extensive efforts have been made to identify and characterize a number of important host factors recruited for virus infection, since their disruption may provide novel protection strategies for crop plants. These host factors play versatile roles during plant RNA virus replication2,5,7, including (1) assistance in assembly of the viral replication complex (VRC) and cellular membrane remodelling; (2) recruitment of viral proteins and template RNA to the VRC; (3) regulation of the switch from viral genome translation to replication; (4) participation in the intracellular transport of viral proteins and viral RNA; and (5) facilitating folding of viral proteins as protein chaperones.
Potyviruses, belonging to the Potyviridae, represent the largest group of known plant viruses and include many agriculturally and economically important pathogens such as Turnip mosaic virus (TuMV)8,9. As one of the most prevalent viral pathogens, TuMV infects a wide range of plant species, mostly (although not exclusively) in the Brassicaceae10,11. Like other potyviruses, TuMV has a positive-sense, single-stranded RNA genome, which is covalently linked with a viral genome-linked protein VPg at the 5′ end, and contains a 3′-polyadenylated [poly(A)] tail. The viral genomic RNA codes for a long open reading frame (ORF) and another relatively short ORF that results from RNA polymerase slippage in the P3 coding sequence12,13,14,15. The two polyproteins translated from these two ORFs are ultimately cleaved by three viral proteinase domains into 11 mature proteins: P1, helper component protease (HC-Pro), P3, P3N-PIPO, 6K1, cylindrical inclusion (CI) protein, 6K2, VPg, nuclear inclusion a (NIa), nuclear inclusion b (NIb), and capsid protein (CP)16.
The VRC of all positive-sense RNA viruses is intimately associated with intracellular membranous structures17. A variety of host proteins are recruited to form the VRC1,2,4,5. For example, the host proteins of the TuMV VRC include proteins such as eukaryotic translation initiation factors (eIFs)18,19, a cysteine-rich protein PVIP20, heat shock cognate 70-3 (Hsc70-3)21, poly(A) binding protein (PABP)22, eukaryotic elongation factor 1A (eEF1A)23, Arabidopsis RNA helicase 8 (AtRH8)24 and DNA-binding protein phosphatase 1 (DBP1)25.
DEAD-box RNA helicases (DDXs) represent a large family of RNA helicases (RHs) that have been implicated in almost every step of RNA metabolism from transcription, mRNA splicing and translation, RNA modification and transport, ribosome biogenesis and RNP complex assembly to mRNA degradation26. DDXs utilize ATP to catalyze the separation of RNA duplexes and promote the structural rearrangement of RNA-protein complexes27. The name of the family was derived from the highly conserved amino acid sequence D-E-A-D (Asp–Glu–Ala–Asp) in motif II28 of the protein. In a seminal work, Noueiry and colleagues showed that the yeast gene DED1, encoding a human DDX3-like RNA helicase, is indispensable for the translation of Brome mosaic virus (BMV)29. Consistently, downregulation of DED1 negatively affects Tomato bushy stunt virus (TBSV) infection by inhibiting the accumulation of virus-encoded replication proteins30,31. The DED1-encoded protein Ded1p is also required for the replication of Flock house virus (FHV)32. The yeast DDX Ded1 and its plant ortholog AtRH20 suppress viral RNA recombination and maintain viral genome integrity33. Similar to Ded1p, another DDX Dbp2p (the homolog of the human p68 protein) promotes tombusvirus positive-strand synthesis in yeast31. Dbp2p binds to the 3′ end of the TBSV negative-sense RNA and unwinds the local secondary structure31. In plants, AtRH20, an RNA helicase in Arabidopsis sharing high sequence similarity with Dbp2, can stimulate TBSV positive-sense RNA synthesis, suggesting RNA helicases in plants may assist viral replication in a similar manner31. Recently, two additional cellular RNA helicases, e.g., the eIF4AIII-like yeast FAL1 and the DDX5-like Dbp3 and their orthologs in Arabidopsis, AtRH2 and AtRH5, have been shown to be present in the tombusvirus VRCs34. In the case of potyviruses, an Arabidopsis DEAD-box RNA helicase, AtRH8 and a Prunus persica DDX-like protein, PpDDXL, have been identified to interact with potyviral VPg. AtRH8 co-localizes with the viral replication vesicles, suggesting AtRH8 is likely involved in viral genome translation and replication24. These studies demonstrate the essential roles of DDXs in virus infections.
The Arabidopsis genome encodes more than 100 putative DDXs (AtRH)35. To further identify AtRHs required for potyviral infection and explore their possible roles therein, we used Arabidopsis mutant plants generated by T-DNA insertion. Over 41 Arabidopsis T-DNA insertion mutants corresponding to 26 AtRH genes were screened for their resistance to TuMV infection. We found that TuMV accumulation was inhibited in Arabidopsis mutants of three RH genes AtRH9, AtRH26 and PRH75. Herein we describe our characterization of Arabidopsis AtRH9 and present evidence that AtRH9 interacts with TuMV NIb and is recruited to the VRC to promote TuMV infection.
Results
Screening for Arabidopsis AtRH gene mutants resistant to TuMV infection
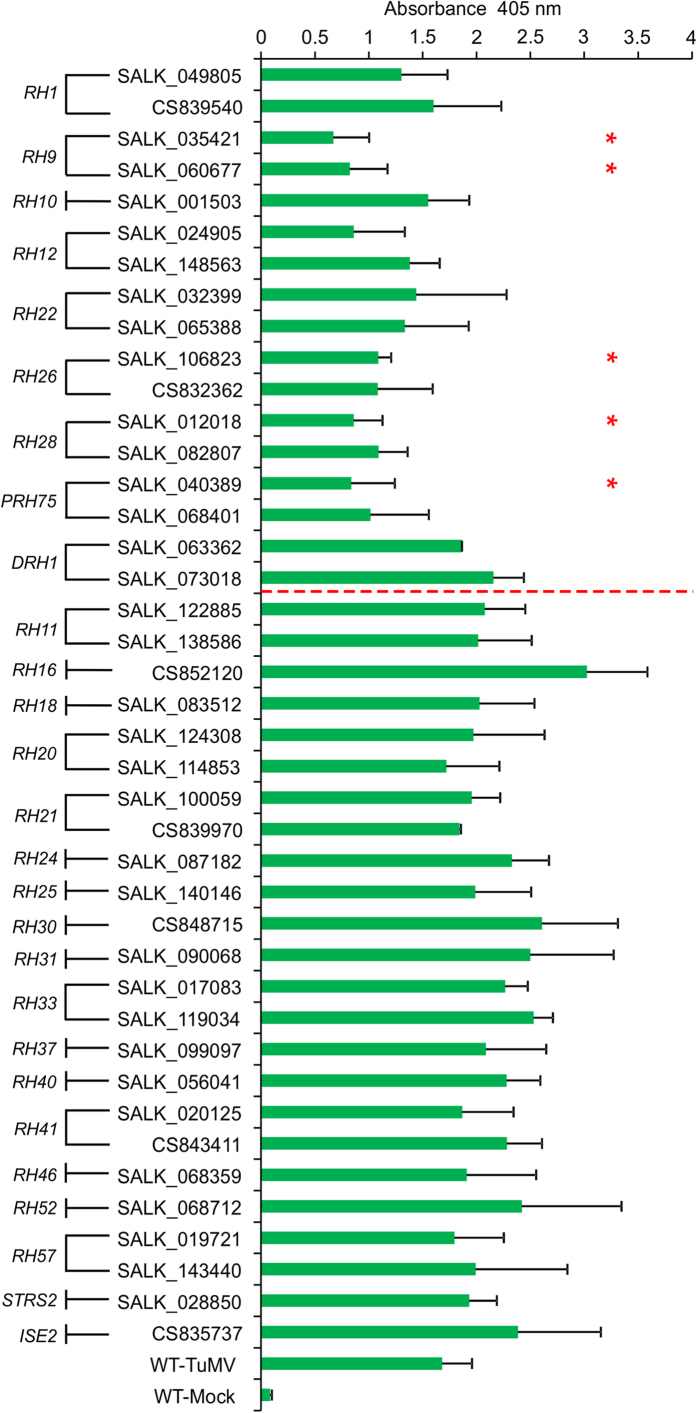
Previous studies predicted that the Arabidopsis genome contained 53 AtRH genes36,37. A relatively more recent study suggested a total of 113 putative helicase genes encoded by the Arabidopsis genome35. The majority of them are ‘computer predicted putative helicases’, and only a few have been experimentally confirmed to have helicase activity. The biological functions of RHs in biological processes are poorly understood in general. To elucidate the role of AtRH genes in viral infection, Arabidopsis T-DNA insertion lines corresponding to 42 AtRH genes were selected from the TAIR database. These genes encode the proteins that are either related to eIF4A24 or have possible functions in the formation of plasmodesmata (PD)38 or stress response regulation39. Seed stocks of 128 Arabidopsis T-DNA insertion lines corresponding to these 42 AtRH genes were obtained from the Arabidopsis Biological Resource Center (ABRC). Mutant and insertion information was obtained from the Salk Institute Genomic Analysis Laboratory website (http://signal.salk.edu/). Genotyping identified a total of 53 homozygous Arabidopsis T-DNA insertion lines corresponding to 26 AtRH genes. Of these, we chose 41 homozygous lines with T-DNA insertions in either an exon or the 5′ UTR region for TuMV infection assays (Table 1). The 41 selected Arabidopsis T-DNA insertion lines and wild-type (WT) control plants were rub-inoculated with TuMV, followed by observation of disease symptoms. Newly-emerged leaves from systemically TuMV-infected T-DNA mutant plants and WT were sampled and assayed for viral CP accumulation at 10 days post inoculation (dpi) by Enzyme-linked Immunosorbent Assay (ELISA) (Fig. 1).
Table 1. List of homozygous Arabidopsis atrh T-DNA insertion lines for TuMV infection assay.
| Gene name | Locus | ArabidopsisT-DNA insertion lines |
|---|---|---|
| RH1 | AT4G15850 | SALK_049805; CS839540 |
| DRH1 | AT3G01540 | SALK_063362; SALK_073018 |
| RH9 | AT3G22310 | SALK_035421; SALK_060677 |
| RH10 | AT5G60990 | SALK_001503 |
| RH11 | AT3G58510 | SALK_122885; SALK_138586 |
| RH12 | AT3G61240 | SALK_024905; SALK_148563 |
| RH16 | AT4G34910 | CS852120 |
| RH18 | AT5G05450 | SALK_083512 |
| RH20 | AT1G55150 | SALK_124308; SALK_114853 |
| RH21 | AT2G33730 | SALK_100059; CS839970 |
| RH22 | AT1G59990 | SALK_032399; SALK_065388 |
| RH24 | AT2G47330 | SALK_087182; SALK_045730 |
| RH26 | AT5G08610 | SALK_106823; CS832362 |
| RH28 | AT4G16630 | SALK_012018; SALK_082807 |
| RH30 | AT5G63120 | CS848715 |
| RH31 | AT5G63630 | SALK_090068 |
| RH33 | AT2G07750 | SALK_017083; SALK_119034 |
| RH37 | AT2G42520 | SALK_099097 |
| RH40 | AT3G06480 | SALK_056041 |
| RH41 | AT3G02065 | SALK_020125; CS843411 |
| RH46 | AT5G14610 | SALK_068359 |
| RH53 | AT3G22330 | SALK_056387 |
| RH57 | AT3G09720 | SALK_019721; SALK_143440 |
| PRH75 | AT5G62190 | SALK_040389; SALK_068401 |
| STRS1 | AT1G31970 | SALK_062509 |
| STRS2 | AT5G08620 | SALK_028850 |
Figure 1. ELISA Analysis of Arabidopsis atrh T-DNA Mutants.
A total of 41 homozygous Arabidopsis T-DNA insertion mutant lines corresponding to 26 AtRH genes were selected for TuMV infection assay. The Arabidopsis atrh T-DNA insertion mutants and WT plants were mechanically inoculated with TuMV. ELISA analysis was used to determine the accumulation of TuMV CP in atrh T-DNA mutants and WT plants 10 days post inoculation (dpi). Extracts from newly-emerged leaves of TuMV-infected individual plants were subjected to ELISA using TuMV CP-specific antibody. The x-axis represents ELISA values. Error bars represent standard deviation (n ≥ 5). Asterisk indicates significantly different from WT plants (unpaired two-tailed Student’s t test, p < 0.05).
Among the 41 T-DNA lines, 18 AtRH gene mutants, corresponding to the genes DRH1, AtRH11, AtRH16, AtRH18, AtRH20, AtRH21, AtRH24, AtRH30, AtRH31, AtRH33, AtRH37, AtRH40, AtRH41, AtRH46, AtRH53, AtRH57, STRS1, and STRS2, displayed higher (albeit not statistically significant) CP accumulation than WT plants. In these mutants, TuMV infection induced severe phenotypes, including stunted growth, yellowish and curled leaves, chlorotic and mosaic lesions on leaves, similar to the symptoms in TuMV-infected WT (data not shown). In contrast, the TuMV CP accumulation level in T-DNA insertion lines of AtRH9, AtRH26 and PRH75 were significantly lower than that observed in WT plants. These three mutants, i.e., atrh9, atrh26 and prh75, exhibited attenuated symptoms in the systemically-infected plants. Apparently, TuMV infection in these mutants was affected. Intriguingly, ELISA results also revealed that TuMV CP levels were reduced in newly-emerged leaves of T-DNA insertion lines of AtRH1, AtRH10, AtRH12, AtRH22 and AtRH28 relative to WT plants. But the reduction did not reach significant levels. Altogether this screen identified the biological relevance of three AtRH genes, i.e., AtRH9, AtRH26 and PRH75, to TuMV infection.
The accumulation of TuMV was reduced in atrh9 mutant plants
Of the three gene mutants with resistance to TuMV infection, the two atrh9 T-DNA insertion lines SALK_035421 and SALK_060677 showed the largest reduction in CP accumulation. Based on information from the Arabidopsis database, SALK_035421 contains a T-DNA insertion in the 6th exon, whereas SALK_060677 has a T-DNA in the 3′ UTR region (Supplemental Fig. 1a,b,d). As SALK_035421 was confirmed to be a true knockout mutant40, we focused on this mutant. The AtRH9 gene (also At3g22310) encodes an RH that was previously designated Putative Mitochondrial RNA Helicase1 (PMH1), and SALK_035421 was named pmh1-141. In this study, to be consistent with the gene name AtRH9 in the AtRH family37, we renamed the T-DNA insertion line SALK_035421 as atrh9. A SALK_035421 homozygous insertion line was obtained from ABRC and the homozygous T-DNA insertion was verified using PCR analysis (Supplemental Fig. 1b). Loss of transcripts in atrh9 was confirmed by reverse transcription (RT)-PCR analysis using AtRH9-specific primers (Supplemental Fig. 1c), consistent with the published data40.
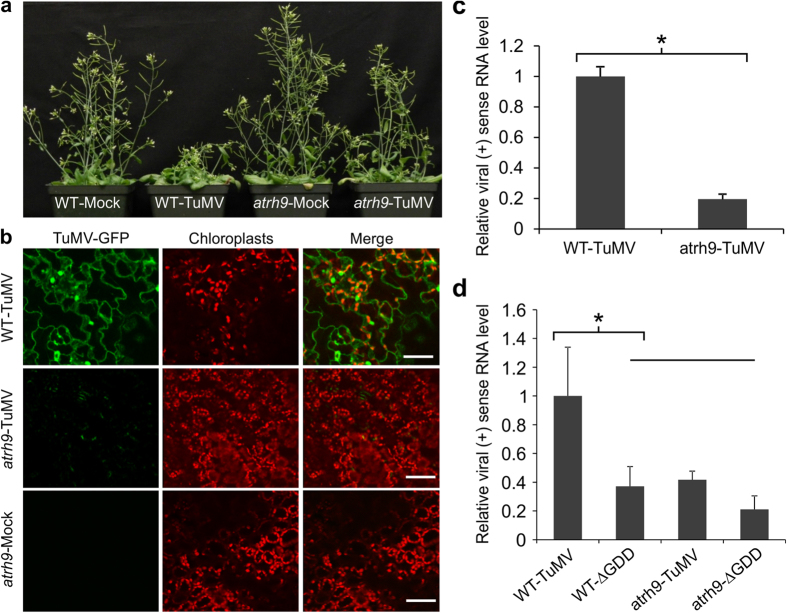
Under standard growth conditions, atrh9 mutant plants displayed no abnormal phenotypes distinguishable from Arabidopsis WT plants (Fig. 2a). In contrast to the severe TuMV symptoms (necrosis, chlorotic leaves and dwarfing) observed in WT plants, atrh9 mutant plants displayed minor symptoms, such as curled bolts and slightly shorter in height (Fig. 2a). To confirm atrh9 was resistant to TuMV, three-week-old atrh9 mutants and WT plants were agroinfiltrated with a GFP-tagged TuMV infectious clone (TuMV-GFP). TuMV infection was monitored by confocal microscopy. In all three independent experiments, strong signals of GFP fluorescence were observed in the newly-emerged leaves of infected WT plants, whereas only weak and scattered GFP fluorescence was detected in atrh9 mutant plants 10 days post agroinfiltration (dpa) (Fig. 2b). Quantitative RT-PCR (qRT-PCR) was carried out to quantify TuMV accumulation. Consistent with the observed reduction in GFP fluorescence, we observed a sharp decline of viral genomic RNA accumulation in atrh9 mutant plants. As shown in Fig. 2c, TuMV viral accumulation in the mutant underwent a substantial decrease by 80% with respect to that in WT plants at 15 dpa.
Figure 2. TuMV Accumulation in atrh9 Mutant Plants.
(a) Three-week-old Arabidopsis atrh9 mutants (SALK_035421) and WT plants were agroinfiltrated with TuMV-GFP infectious clone. Phenotypes of TuMV-infected atrh9 mutants and WT plants. Images were taken 10 days post agroinfiltration (dpa). Mock, inoculated with buffer; TuMV, inoculated with TuMV. (b) Newly-emerged leaves were observed by confocal microscopy 10 dpa, and representative images are shown. Mock, atrh9 mutants and WT plants were agroinfiltrated with buffer. TuMV-GFP, green fluorescence emissions; Chl, chloroplast autofluorescence. Bars, 50 μm. (c) Relative fold changes of TuMV viral genomic RNA in atrh9 mutant plants were determined by real-time RT-PCR at 10 dpa. RNA was extracted from newly-emerged leaves of infected individual plants. Three independent experiments, each consisting of three biological replicates were carried out for quantification analysis. Each value was normalized against Actin2 transcripts in the same sample. The values are presented as means of fold changes relative to WT. Error bars represent standard deviation (n = 9). Statistically significant difference from WT plants, determined by an unpaired two-tailed Student’s t test, is indicated. *p < 0.05. (d) Detection of TuMV viral RNA by qRT-PCR in WT and atrh9 mutant plants agroinfiltrated with a TuMV infectious clone (TuMV) and a replication-defective mutant (∆GDD). Both clones contain a separate expression cassette allowing transcript of mCherry transcripts in plant cells. RNA was purified from agroinfiltrated local leaves 54 hours post agroinfiltration (hpa). mCherry transcript was used as an internal control. The values are presented as means of fold change relative to WT. Error bars denote standard deviation of three biological replicates. Statistically significant differences from WT plants, determined by an unpaired two-tailed Student’s t test, are indicated. *p < 0.05.
To determine if viral replication is affected in the primarily infected cells in atrh9 plants, a TuMV infectious clone containing a separate expression cassette for expression of mCherry and its corresponding replication-defective mutant (∆GDD) were agroinfiltrated into leaves of Arabidopsis WT and atrh9 mutant plants. As potyviral intercellular movement would not occur until 96 hours post agroinfiltration (hpa)42, total RNA was isolated from the agroinfiltrated leaves at 54 hpa, and subjected to qRT-PCR analysis to determine viral RNA accumulation using mCherry as an internal control. It was found that viral genomic RNA in atrh9 mutant plants was significantly lower than that in WT plants (Fig. 2d). Although it was slightly higher than that in both atrh9 and WT Arabidopsis plants agroinfiltrated with the replication-defective TuMV ∆GDD clone, there was no significant difference (Fig. 2d). Taken together, these results suggest that AtRH9 is a host factor promoting TuMV replication.
Silencing AtRH9 in Arabidopsis results in partial resistance to TuMV infection
To further confirm the involvement of AtRH9 in TuMV infection, a Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) system was employed to silence AtRH9 expression in Arabidopsis. A cDNA fragment of AtRH9 was cloned into a pTRV2-derived vector to produce pTRV2-AtRH9. Arabidopsis WT seedlings were co-agroinfiltrated with the vectors pTRV2-AtRH9 and pTRV1. A bleaching phenotype was observed in control plants 12 days after co-agroinfiltration with pTRV2-PDS and pTRV1, indicating VIGS was established. At this time point, the TuMV infection assay was applied on AtRH9-downregulated plants (treated with pTRV2-AtRH9 and pTRV1) as well as WT plants (treated with buffer) and negative control plants (treated with empty pTRV vectors). qRT-PCR was performed to evaluate AtRH9 expression and TuMV accumulation at 15 dpi (Supplemental Fig. 2). The severely reduced level of the AtRH9 transcript in AtRH9-downregulated plants was coupled with partial resistance to TuMV infection (Supplemental Fig. 2). Consistent with the results from TuMV infection assays on the atrh9 mutant plants, these data reaffirm our hypothesis that downregulation of AtRH9 effectively inhibits TuMV infection in Arabidopsis.
AtRH9 is differentially distributed in TuMV-infected in Nicotiana benthamiana leaf cells, and co-localized with the VRCs
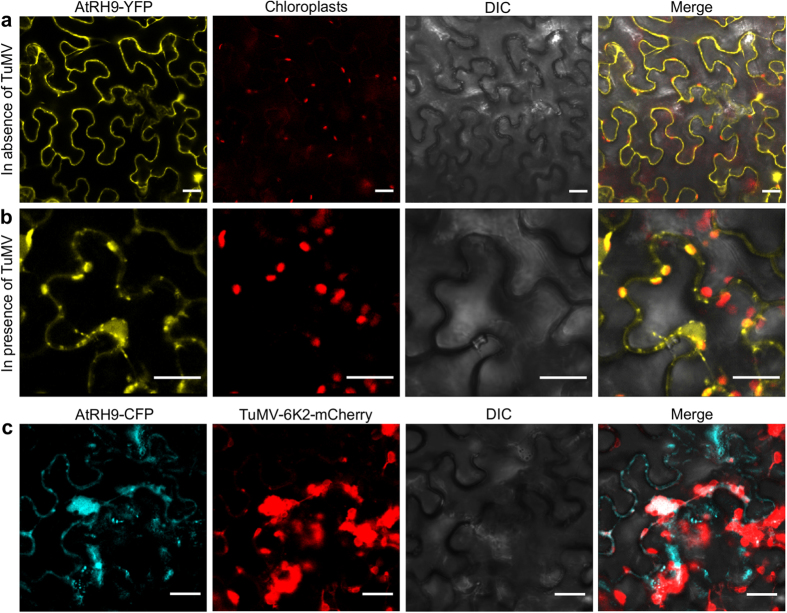
To gain insight into the molecular function of AtRH9 in TuMV infection, subcellular localization of AtRH9 was performed. A translational fusion of AtRH9 with YFP controlled by the CaMV 35S promoter was transiently expressed in N. benthamiana leaf epidermal cells via agroinfiltration. Subcellular localization of fusion proteins was monitored using a Leica TCS SP2 inverted confocal microscopy at 48 hpa. Consistent with a previous study41, most of the AtRH9-YFP was evenly distributed in the cytoplasm of N. benthamiana leaf cells without TuMV infection (Fig. 3a). In contrast, in TuMV-infected cells, AtRH9-YFP was found to form punctate and irregular-shaped aggregations, many of which were associated with chloroplasts (Fig. 3b). Thus, the distribution pattern of AtRH9 in TuMV-infected leaf cells was clearly different from that in the leaf cells free from TuMV infection.
Figure 3. Altered Localization of AtRH9 in Arabidopsis during TuMV Infection.
(a) Transient expression of AtRH9-YFP in N. benthamiana leaf epidermal cells. YFP fluorescence was observed using a confocal microscope 48 hours post agroinfiltration (hpa). (b) AtRH9-YFP was expressed in N. benthamiana leaves infected by TuMV::6K2-mCherry. Localization of AtRH9 is associated with chloroplasts in the course of viral infection at 72 hpa. Bars, 20 μm. (c) Transient expression of AtRH9-CFP in N. benthamiana leaves infected by TuMV::6K2-mCherry. CFP fluorescence was observed using a confocal microscope 72 hpa. AtRH9-CFP was observed mainly with irregularly-shaped globular-like aggregations from red fluorescence from TuMV VRCs (6K2-mCherry). DIC, differential interference contrast. Bars, 20 μm.
Our previous studies have shown that 6K2-induced chloroplast-associated vesicles are the sites for potyviral replication43,44. These 6K2-labelled structures contain double-stranded replicative RNA intermediates, viral replicase proteins and many host factor proteins43,45,46,47,48. To determine if AtRH9 co-localizes with the chloroplast-associated 6K2 vesicles during TuMV infection, AtRH9-CFP was transiently expressed in N. benthamiana leaf cells infected by a TuMV infectious clone (TuMV::6K2-mCherry), which produces red-fluoresce labelled 6K2 vesicles during infection. AtRH9-CFP co-localized with 6K2-induced irregular-shaped aggregations in the cytoplasm of TuMV-infected leaf cells (Fig. 3c). These results indicated that AtRH9 was recruited to the VRC during TuMV replication, suggesting a functional role of AtRH9 in viral proliferation.
AtRH9 interacts with TuMV RNA-dependent RNA polymerase
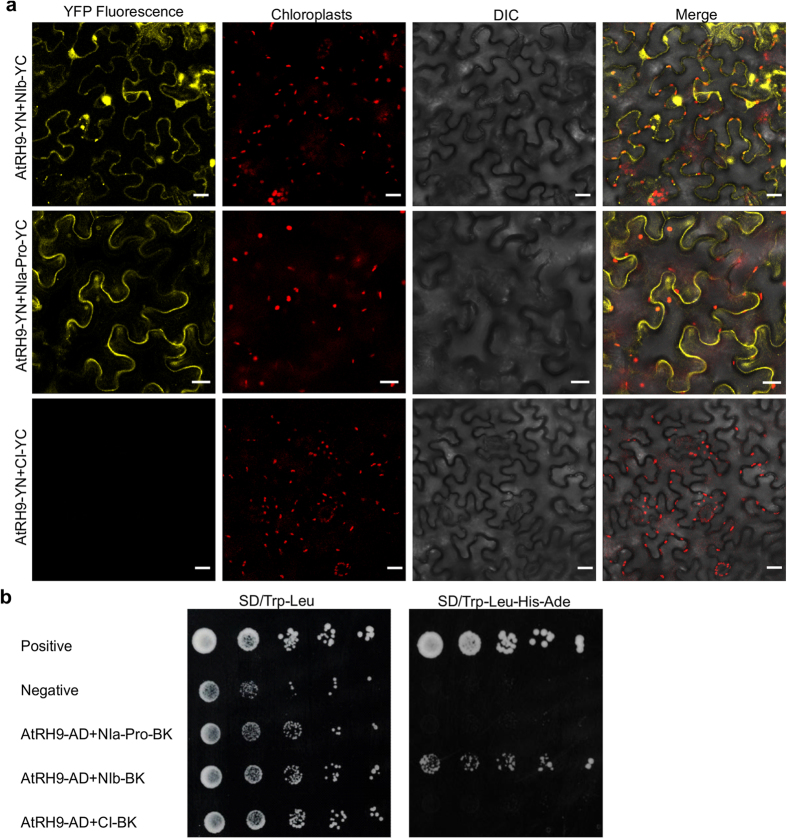
To investigate how AtRH9 is recruited to the VRC, we examined if AtRH9 interacts with TuMV viral proteins in vivo using the BiFC assay. The AtRH9 gene and the coding sequence for each of the 11 TuMV viral proteins were introduced into BiFC vectors containing the DNA fragments encoding the N- or C- terminal moiety of YFP. The resulting plasmids were transiently co-expressed in N. benthamiana epidermal cells. The YFP signal would be reconstituted when split fluorescent protein segments were brought together as a result of positive interactions between two tested proteins. Yellow fluorescence was visualized when AtRH9 and NIb or NIa-Pro were co-expressed, suggesting AtRH9 interacts with NIa-Pro and NIb (Fig. 4a). No positive interactions were found between AtRH9 and other viral proteins such as CI (Fig. 4a) or in negative controls (Supplemental Fig. 3).
Figure 4. Protein-Protein Interactions between AtRH9 and TuMV Viral Proteins.
(a) BiFC assay for interactions between AtRH9 and TuMV NIb, NIa-Pro. N. benthamiana leaves were co-agroinfiltrated with constructs expressing AtRH9 with TuMV NIb, NIa-Pro and CI fused to the N- and C- terminal half of YFP, respectively. The reconstructed YFP fluorescence was recorded 48 hours post agroinfiltration (hpa) by confocal microscopy. DIC, differential interference contrast. Bars, 20 μm. (b) Yeast two-hybrid assay for protein-protein interaction between AtRH9 and NIb. Growth of yeast cells co-transformed with pGAD-AtRH9 and pGBK-NIb, pGBK-NIa-Pro or pGBK-CI were placed on synthetic medium lacking Tryptophan and Leucine (SD/Trp-Leu) to confirm the correct transformation. Different dilutions of yeast transformants were spotted onto a high-stringency selective medium (SD/Trp-Leu-His-Ade) to detect positive interactions, respectively. Co-transformation of pGAD-VPg with pGBK-eIF(iso)4E or pGAD-AtRH9 with pGBK empty vector were used as the positive or negative controls, respectively.
To confirm the interaction of AtRH9 with TuMV NIb and NIa-Pro, yeast two hybrid (Y2H) assays were performed. Positive interaction was detected between AtRH9 and NIb using a high-stringency selective medium (SD/Trp-Leu-His-Ade) (Fig. 4b). However, no interaction was observed between AtRH9 and NIa-Pro (Fig. 4b). Therefore, it is very likely that Arabidopsis AtRH9 is recruited to the VRC via interactions with the viral NIb protein.
Discussion
In this study, a reverse genetic screening was performed to identify AtRHs that are required for TuMV infection. Of the 41 homozygous T-DNA insertion lines disrupting 26 AtRH genes, three genes AtRH9, AtRH26 and PRH75 were identified as their mutants showed less susceptibility to TuMV infection than WT controls. As shown in Fig. 1, the amount of TuMV CP was significantly reduced and the symptoms were less severe in the atrh9, atrh26 and prh75 T-DNA mutants in comparison with WT plants. In recent years, several AtRHs including AtRH234, AtRH534, and AtRH2031, have been documented to function in infection by tombusviruses, whereas AtRH8 is implicated in potyvirus infection. As all these RHs belong to the family of DDXs that have both RNA-binding and helicase activities, it is reasonable for us to suggest that some members in the DDX family are indeed recruited to assist virus infection in plants. However, so far none of these AtRHs has been found to be involved in infections by two different groups of viruses. As indicated above, AtRH20 is implicated in tombusvirus infection31. In this study, TuMV infection was not significantly affected in the atrh20 mutant (Fig. 1), suggesting AtRH20 is not involved in TuMV infection. Therefore, different viruses may recruit different DDXs to support their infections.
To explore the functional role of the identified RHs in TuMV infection, we focused on AtRH9, since it showed the least accumulation of CP in our screen. qRT-PCR analysis of TuMV accumulation in newly-emerged leaves of atrh9 mutants or in primarily infected cells revealed that viral RNA was drastically reduced in comparison with that in TuMV-infected WT plants (Fig. 2). To further exclude the possibility that other unknown mutations in the atrh9 mutant are responsible for reduced TuMV replication, AtRH9 expression was knockdown in WT Arabidopsis seedlings using a VIGS vector and then mechanically inoculated with TuMV. TuMV accumulation was also significantly reduced in the newly-emerged systemic leaves (Supplemental Fig. 2). These results strongly suggest that AtRH9 is a host factor for TuMV replication.
AtRH9 (or PMH1) is hypothesized to be a mitochondrial protein and might be involved in RNA metabolism in mitochondria as an RNA chaperon40. However, only small amounts of AtRH9 are present in mitochondrial high molecular weight complexes40. This is in contrast to the PHH1 homolog PMH2, which is abundantly present in mitochondrial high molecular weight complexes, acts as a posttranscriptional RNA chaperon, and is required for efficient group II intron splicing. AtRH9 does not influence group II intron splicing efficiency40. AtRH9 mRNA level is enhanced in response to biotic stress caused by different pathogens41. The subcellular localization of AtRH9 was visualized as punctate particles in the cytoplasm, consistent with the assumed mitochondrial distribution41. During TuMV infection, however, the distribution pattern of AtRH9 changed and some AtRH9 was apparent in close proximity with chloroplasts (Fig. 3). This observation raised the possibility that AtRH9 is recruited to TuMV VRCs that are associated with chloroplasts44. We thus conducted a co-localization study, and demonstrated that AtRH9 co-localizes with 6K2-induced VRCs. These data strongly suggest that AtRH9 is involved in viral replication.
To understand how AtRH9 is recruited for TuMV infection, the BiFC assay and Y2H assay were conducted to test protein-protein interactions between AtRH9 and TuMV viral proteins. The analysis revealed that AtRH9 interacts with NIb, the viral RNA-dependent RNA polymerase in yeast and plant cells and NIa-Pro, the virus-encoded protease in plant cells (Fig. 4). TuMV NIb catalyzes the synthesis of new viral genomic RNA. The NIb protein accumulates predominantly in the nucleus as nuclear inclusions and is also recruited into the cytoplasmic membrane-bound vesicles that house the VRC during viral infection21,23,49. The interaction of AtRH9 with NIb was confirmed by both BiFC and Y2H assays. Therefore, it is possible that AtRH9 participates in viral replication by associating with viral RdRp. TuMV NIa-Pro is also present in the VRCs and is also responsible for catalyzing the cleavage of P3/6K1, 6K1/CI, CI/6K2, 6K2/NIa, NIa/NIb, NIb/CP and the cleavage site between VPg and NIa-Pro domains in NIa protein44,50. However, no interaction between AtRH9 and NIa-Pro was detected by the Y2H assay. It is possible that the interaction between AtRH9 and NIa-Pro found in the BiFC assay was either transient or mediated by one more host protein that binds to both AtRH9 and NIa-Pro and thus serves as a bridging interactor. Further study is needed to understand how AtRH9 interacts with NIa-Pro in plants.
In conclusion, by screening and analyzing the susceptibility of Arabidopsis DDX T-DNA insertion mutants using a TuMV infection assay, we have provided evidence that DDXs AtRH9, AtRH26 and PRH75 are required for TuMV infection. Further, we have demonstrated that AtRH9 is recruited to the TuMV VRC through interactions with NIb and/or NIa-Pro to promote TuMV replication. However, how AtRH9 affects TuMV replication remains to be elucidated. It is possible that AtRH9 may participate in the separation of RNA duplexes during viral genome replication. Alternatively, it is also possible that AtRH9 facilitates the proteolytic cleavage of TuMV polyprotein via its interaction with NIa-Pro. Future experiments are directed towards a better understanding of the exact biological functions of the identified DDXs in the viral infection cycle.
Materials and Methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 and Nicotiana benthamiana plants were utilized in this study. Plants were maintained in a growth chamber under constant conditions of 60% relative humidity and a day/night regime of 16 h in the light at 22 °C followed by 8 h at 18 °C in the dark. Seeds for Arabidopsis T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University, Columbus, Ohio, USA. T-DNA insertion information was obtained from the Salk Institute Genomic Analysis Laboratory website (http://signal.salk.edu/). The Arabidopsis atrh9 mutants have been described previously40. The T-DNA insertion sites of Arabidopsis atrh9 T-DNA insertion lines in the Col-0 background SALK_035421 and SALK_060677 were confirmed by PCR using the primer pairs (SALK_ 035421-LP: TCATAAATGGAAGTGGCGAAG and SALK_ 035421-RP: TCTTGTTGCAACTGATGTTGC or SALK_ 060677-LP: TTCTCATCCACGGTCAAGATC and SALK_ 060677-RP: TGTACAAGAACCCGTTCTTGG).
Virus materials, inoculation and TuMV infection assay
The pCambiaTunos/GFP plasmid containing the full-length cDNA of the TuMV genome and pCambiaTunos/6K-mCherry having an additional copy of the 6K2-coding sequence tagged with fluorescent protein mCherry between P1 and HC-Pro were as described previously51.
TuMV infection assay was carried out to test the susceptibility of Arabidopsis T-DNA insertion lines to TuMV infection. The seedlings of Arabidopsis WT plants and selected homozygous mutants were inoculated with TuMV either by mechanical inoculation or using agroinfiltration. Plants were inoculated when at the five to six leaf stage of development. Virus was applied to the two oldest leaves by mechanical inoculation. Approximately 1 g TuMV-infected leaf tissue of N. benthamiana was harvested as the source of virus material. The tissue was homogenized using a mortar and pestle in 10 mL inoculation buffer (50 mM potassium phosphate buffer, pH 7.5]. Carborundum powder was lightly dusted on plant leaves, followed by a gentle rubbing of the TuMV-containing inoculum over the leave surface to facilitate virus entry. The negative control plants were rubbed with inoculation buffer also as mock inoculations. The TuMV infection assay was repeated at least three times for each T-DNA insertion line. Systemically infected Arabidopsis leaves were harvested for ELISA analysis at 10 dpi.
After mechanical inoculation or agroinfiltration24, triple antibody sandwich enzyme-linked immunosorbent assay (TAS-ELISA) was performed to quantify virus accumulation level of WT and Arabidopsis T-DNA insertion lines at the days indicated. The newly emerged leaves of Arabidopsis mutants and WT plants inoculated with TuMV were harvested for ELISA analysis. Leaf tissue was weighed and ground in ELISA sample extraction buffer. TAS-ELISA was conducted with an ELISA kit (Agdia, Elkhart, IN, USA), following the manufacturer’s instructions. Absorbance values were recorded at 405 nm with an iMark microplate reader (Bio-Rad, Mississauga, Ontario, Canada).
To determine TuMV replication in primarily infected cells, Arabidopsis atrh9 mutants and WT plants, at the 5–7 leaf growth stage, were agroinfiltrated with a TuMV infectious clone containing an independent cases to express mCherry (TuMV-GFP//mCherry) and a TuMV replication-defective clone (TuMV-∆GDD-GFP//mCherry) (OD600, 0.1), respectively52. Agroinfiltrated leaf tissues were harvested at 54 hpa. At this time point, viral intercellular movement did not occur42. The harvested tissues were subjected to qRT-PCR analysis.
Plasmid construction and plant transformation
Gateway technology (Invitrogen, Burlington, Ontario, Canada) was used to generate all the plasmid constructs used in this study, unless otherwise stated. Gene sequences were amplified by polymerase chain reaction (PCR) using Phusion High-Fidelity DNA Polymerase (New England Biolabs, Pickering, ON, Canada) for cloning purposes. GoTaq Flexi DNA Polymerase (Promega, Madison, WI, USA) was employed for genotyping and other analysis.
The full-length P1, HC-Pro, P3, 6K1, CI, 6K2, VPg, NIa-Pro, NIb and CP coding regions of TuMV (GenBank accession NC_002509) were obtained by PCR amplification from the pCambiaTunos/GFP infectious clone51,53. AtRH9 (At3g22310) coding sequence was generated from cDNA derived from mRNAs isolated from Arabidopsis leaves. The resulting DNA fragments were purified and transferred into the entry vector pDONR221 or pENTR (Invitrogen) by recombination using BP Clonase (Invitrogen) following the standard conditions and procedures recommended by the supplier54. Insertions in the resulting entry clones were verified by DNA sequencing.
For bimolecular fluorescence complementation (BiFC) assay, the full-length coding sequence of Arabidopsis AtRH9 and the coding sequences of TuMV NIb, NIa-Pro and CI cistrons were introduced into the BiFC vectors pEarleygate201-YN or pEarleygate201-YC55 to produce AtRH9-YN, NIb-YC, NIa-Pro-YC, and CI-YC respectively.
For transient expression analysis in plant cells, the full-length coding sequence of Arabidopsis AtRH9 was transferred by recombination into the binary destination vectors pEarleyGate10156 to generate plant expression vector for transient expression of AtRH9-YFP (yellow fluorescence protein) or pEarleyGate102 to produce AtRH9-CFP (cyan fluorescence protein), respectively.
For the targeted yeast two-hybrid assay (Y2H) assay, inserts of the resulting intermediate entry clones were further transferred into modified Gateway-compatible vectors pGADT7-DEST (prey) or pGBKT7-DEST (bait)56 by recombination using LR Clonase (Invitrogen) to yield pGAD-AtRH9, pGAD-VPg, and pGBK-NIb, pGBK-NIa-Pro, pGBK-CI, pGBK-eIF(iso)4E, respectively.
For Tobacco rattle virus (TRV)-based virus induced gene silencing (VIGS), a 110 bp fragment of AtRH9 was amplified from Arabidopsis cDNA using the primer pair that contained an EcoRI and BamHI site specific to the 5′ end and 3′ end of the fragments, respectively (AtRH9-EcoRI-F: 5′- CCGGAATTCTGATGTTGCTGCCCGTGGACT-3′ and AtRH9-BamHI-R: 5′- CGCGGATCCCACGACCAGTTCGCCCCGTT-3′). The amplified fragment was digested with EcoRI and BamHI and cloned into the corresponding sites of pTRV2 vector57 to generate the vector pTRV2-AtRH9.
Yeast two-hybrid assay
Yeast two-hybrid assay (Y2H) was performed following the Clontech yeast protocol handbook using the small-scale lithium acetate yeast transformation method53. In brief, yeast cells (strain AH109) were plated onto a selective medium lacking tryptophan and leucine (SD-Trp-Leu) to confirm the right transformation and a high-stringency selective medium lacking tryptophan, leucine, histidine, and adenine (SD-Trp-Leu-His-Ade) to detect positive interactions.
TRV-based virus-induced gene silencing (VIGS)
To suppress AtRH9 expression by VIGS, a Tobacco rattle virus-based vector was used. pTRV1 and pTRV2-AtRH9 were introduced into Agrobacterium tumefaciens (A. tumefaciens) and co-agroinfiltrated into Arabidopsis seedlings at the four-leaf stage. Plants co-agroinfiltrated with pTRV1 and pTRV2 empty vector or pTRV2-PDS were used as controls. Treated plants were mechanically inoculated with TuMV at 12 dpa.
RT-PCR and Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from newly emerged leaves of TuMV-infected Arabidopsis mutants and WT plants using the RNeasy Plant Mini Kit (Qiagen) and treated with DNase I following the manufacturer’s instructions. Except otherwise indicated, cDNA synthesized by reverse transcription of RNA samples with Superscript III reverse transcriptase (Invitrogen) and an oligo(dT)12–18 primer (Invitrogen) was used to determine the mRNA expression levels of target genes as well as for quantifying TuMV accumulation levels at specific time points indicated. Arabidopsis Actin II was used as an internal control.
qRT-PCR was conducted using SsoFast Evagreen supermix (Bio-Rad) and analyzed with the CFX96 Real-Time PCR Detection System (Bio-Rad) following the manufacturer’s instructions. Relative transcript abundances were calculated using Bio-Rad CFX Manager software. The expression of the CP gene of TuMV was determined to reflect the virus accumulation level using primer sets TuMV-CP-F (5′-TGGCTGATTACGAACTGACG-3′) and TuMV-CP-R (5′-CTGCCTAAATGTGGGTTTGG-3′). Gene specific primers (AtRH9-F: 5′-TCGTGCTGGAAAGAAAGGAAGCG-3′ and AtRH9-R: 5′-TTCCACAGCAATGCTAGGCAGCTC-3′) were used quantify AtRH9 expression. Arabidopsis Actin II expression level determined with the primer pair (At-Actin2-F: 5′-CACCACAACAGCAGAGCGGGA-3′ and At-Actin2-R: 5′-TCCCACAAACGAGGGCTGGA-3′) was used as a reference to normalize the data. For detection of viral RNA in WT plants and atrh9 mutants at primarily infected cells, total RNA was purified at 54 hpa and treated with DNase I (Invitrogen) to remove contaminated DNA. The mCherry transcript level determined with the primers mCherry-F (5′-CACTACGACGCTGAGGTCAA-3′) and mCherry-R (5′-TGGTGTAGTCCTCGTTGTGG-3′) was used as an internal control.
For all qRT-PCR analyses, three biological replicates were included and for each biological replicate, three technical repeats were carried out. All results are shown as means of biological replicates with corresponding standard errors.
Transient expression in N. benthamiana
For transient expression analysis in N. benthamiana leaves, constructs were generated in Gateway-compatible binary vectors and transformed into A. tumefaciens strain GV3101 via electroporation. Four-week-old N. benthamiana plants were used for Agrobacterium-mediated transient expression. Agrobacterium cultures were grown overnight in LB with appropriate antibiotic selection at 28 °C. The Agrobacterium cells were harvested by centrifugation, and then resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.7 and 150 μM acetosyringone). After incubation for 2 h at room temperature, the culture was diluted to an optical density of 0.5–1.0 at 600 nm (OD600) and agroinfiltrated into leaf tissue under gentle pressure using a syringe barrel58. After agroinfiltration, the plants were maintained under normal conditions for 2–4 days before observation. For Bimolecular Fluorescence Complementation (BiFC) assay, Agrobacterium carrying the N-terminal and C-terminal of YFP fusion constructs was co-agroinfiltrated into N. benthamiana leaves. The reconstitution of YFP signals were monitored at 2–4 dpa as described46.
Confocal microscopy
Confocal microscopy was performed essentially as described previously59,60,61. Fluorescence was visualized at 2–4 dpa using a Leica TCS SP2 inverted confocal microscope (http://www.leica.com/) with an Argon-Krypton laser. Sections from agroinfiltrated leaves were excised and placed between two microscope cover slides with a drop of water. YFP signals were imaged using a 63× water immersion objective at an excitation wavelength of 514 nm, and emissions were collected between 525 and 575 nm. Images of CFP fluorescence were obtained using the same microscope at an excitation wavelength of 458 nm and emissions were collected between 470 and 500 nm. GFP signal was excited at 488 nm and the emitted light was captured at 505 to 525 nm. mCherry fluorescence was excited at 543 nm and the emitted light was captured at 590–630 nm. Light emitted at 630–680 nm was used to record chlorophyll autofluorescence. Data for the different color channels were collected simultaneously. The samples were scanned at a resolution of 512 × 512 pixels. Images were collected with a charge-coupled device camera and analyzed by Leica confocal software.
Additional Information
How to cite this article: Li, Y. et al. Recruitment of Arabidopsis RNA Helicase AtRH9 to the Viral Replication Complex by Viral Replicase to Promote Turnip Mosaic Virus Replication. Sci. Rep. 6, 30297; doi: 10.1038/srep30297 (2016).
Supplementary Material
Acknowledgments
We are very grateful to Jamie McNeil (AAFC) for technical support, Alex Molnar (AAFC) for photography, and Zhaoji Dai (AAFC) for assistance. This work was supported in part by the Plum Pox Management and Monitoring Program (PPMMP), the Government of Canada, and a discovery grant to AW from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Footnotes
Author Contributions Y.L., M.B. and A.W. designed experiments. Y.L. and R.X. performed experiments. All authors interpreted results. Y.L. and A.W. wrote the manuscript. All authors reviewed the manuscript.
References
- den Boon J. A., Diaz A. & Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe 8, 77–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. D. & Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10, 137–149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. & Krishnaswamy S. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol Plant Pathol 13, 795–803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- García J. A. & Pallás V. Viral factors involved in plant pathogenesis. Curr Opin Virol 11, 21–30 (2015). [DOI] [PubMed] [Google Scholar]
- Wang A. Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol 53, 45–66 (2015). [DOI] [PubMed] [Google Scholar]
- Li Y., Cui H., Cui X. & Wang A. The altered photosynthetic machinery during compatible virus infection. Curr Opin Virol 17, 19–24 (2016). [DOI] [PubMed] [Google Scholar]
- Hyodo K. & Okuno T. Host factors used by positive-strand RNA plant viruses for genome replication. J Gen Plant Pathol 80, 123–135 (2014). [Google Scholar]
- Ivanov K. I., Eskelin K., Lõhmus A. & Mäkinen K. Molecular and cellular mechanisms underlying potyvirus infection. J Gen Virol 95, 1415–1429 (2014). [DOI] [PubMed] [Google Scholar]
- Revers F. & García J. A. Molecular Biology of Potyviruses. Adv Virus Res 92, 101–199 (2015). [DOI] [PubMed] [Google Scholar]
- Sánchez F., Wang X., Jenner C. E., Walsh J. A. & Ponz F. Strains of turnip mosaic potyvirus as defined by the molecular analysis of the coat protein gene of the virus. Virus Res 94, 33–43 (2003). [DOI] [PubMed] [Google Scholar]
- Walsh J. A. & Jenner C. E. Turnip mosaic virus and the quest for durable resistance. Mol Plant Pathol 3, 289–300 (2002). [DOI] [PubMed] [Google Scholar]
- Chung B. Y.-W., Miller W. A., Atkins J. F. & Firth A. E. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci USA 105, 5897–5902 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olspert A., Chung B. Y. W., Atkins J. F., Carr J. P. & Firth A. E. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep 16, 995–1004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodamilans B. et al. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J Virol 89, 6965–6967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. A. The polymerase slips and PIPO exists. EMBO Rep 16, 885–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcuqui-Inchima S., Haenni A.-L. & Bernardi F. Potyvirus proteins: a wealth of functions. Virus Res 74, 157–175 (2001). [DOI] [PubMed] [Google Scholar]
- Laliberté J.-F. & Sanfaçon H. Cellular remodeling during plant virus infection. Annu Rev Phytopathol 48, 69–91 (2010). [DOI] [PubMed] [Google Scholar]
- Wittmann S., Chatel H., Fortin M. G. & Laliberté J. F. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 234, 84–92 (1997). [DOI] [PubMed] [Google Scholar]
- Schaad M. C., Anderberg R. J. & Carrington J. C. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273, 300–306 (2000). [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Thomas C., Harrison S., Revers F. & Maule A. A cysteine-rich plant protein potentiates potyvirus movement through an interaction with the virus genome-linked protein VPg. J Virol 78, 2301–2309 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne P. J. et al. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374, 217–227 (2008). [DOI] [PubMed] [Google Scholar]
- Dufresne P. J., Ubalijoro E., Fortin M. G. & Laliberté J.-F. Arabidopsis thaliana class II poly (A)-binding proteins are required for efficient multiplication of Turnip mosaic virus. J Gen Virol 89, 2339–2348 (2008). [DOI] [PubMed] [Google Scholar]
- Thivierge K. et al. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377, 216–225 (2008). [DOI] [PubMed] [Google Scholar]
- Huang T. S., Wei T., Laliberté J. F. & Wang A. A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol 152, 255–266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelló M. J. et al. A plant small polypeptide is a novel component of DNA-binding protein phosphatase 1-mediated resistance to Plum pox virus in Arabidopsis. Plant Physiol 157, 2206–2215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N. K. & Linder P. The DEAD-box protein family of RNA helicases. Gene 367, 17–37 (2006). [DOI] [PubMed] [Google Scholar]
- Pyle A. M. RNA helicases and remodeling proteins. Curr Opin Chem Biol 15, 636–642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. & Jankowsky E. From unwinding to clamping- the DEAD box RNA helicase family. Nat Rev Mol Cell Bio 12, 505–516 (2011). [DOI] [PubMed] [Google Scholar]
- Noueiry A. O., Chen J. & Ahlquist P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc Natl Acad Sci USA 97, 12985–12990 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Serviene E., Gal J., Panavas T. & Nagy P. D. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J Virol 80, 7394–7404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N., Barajas D. & Nagy P. D. Similar roles for yeast Dbp2 and Arabidopsis RH20 DEAD-box RNA helicases to Ded1 helicase in tombusvirus plus-strand synthesis. Virology 432, 470–484 (2012). [DOI] [PubMed] [Google Scholar]
- Kovalev N., Pogany J. & Nagy P. D. A co-opted DEAD-box RNA helicase enhances tombusvirus plus-strand synthesis. PLoS Pathog 8, e1002537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C., Prasanth K. R. & Nagy P. D. Coordinated function of cellular DEAD-box helicases in suppression of viral RNA recombination and maintenance of viral genome integrity. PLoS Pathog 11, e1004680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N. & Nagy P. D. The Expanding Functions of Cellular Helicases: The tombusvirus RNA replication enhancer co-opts the plant eIF4AIII-Like AtRH2 and the DDX5-Like AtRH5 DEAD-Box RNA helicases to promote viral asymmetric RNA replication. PLoS Pathog 10, e1004051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umate P., Tuteja R. & Tuteja N. Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol 73, 449–465 (2010). [DOI] [PubMed] [Google Scholar]
- Boudet N., Aubourg S., Toffano-Nioche C., Kreis M. & Lecharny A. Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res 11, 2101–2114 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg S., Kreis M. & Lecharny A. The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res 27, 628–636 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebloom S. et al. Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc Natl Acad Sci USA 106, 17229–17234 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant P., Kant S., Gordon M., Shaked R. & Barak S. STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol 145, 814–830 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler D., Schmidt-Gattung S. & Binder S. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol Biol 72, 459–467 (2010). [DOI] [PubMed] [Google Scholar]
- Matthes A. et al. Two DEAD-box proteins may be part of RNA-dependent high-molecular-mass protein complexes in Arabidopsis mitochondria. Plant Physiol 145, 1637–1646 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H. & Wang A. Plum pox virus 6K1 protein is required for viral replication and targets the viral replication complex at the early stage of infection. J Virol 90, 5119–5131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T. & Wang A. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J Virol 82, 12252–12264 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T. et al. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J Virol 84, 799–809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R., Cotton S. & Laliberté J.-F. A model for the biogenesis of Turnip mosaic virus replication factories. Commun Integr Biol 3, 363–365 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T. et al. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6, e1000962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Deng P., Cui H. & Wang A. Visualizing double-stranded RNA distribution and dynamics in living cells by dsRNA binding-dependent fluorescence complementation. Virology 485, 439–451 (2015). [DOI] [PubMed] [Google Scholar]
- Wan J. et al. Ultrastructural characterization of Turnip mosaic virus-induced cellular rearrangements reveals membrane-bound viral particles accumulating in vacuoles. J Virol 89, 12441–12456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo M. A., Freed D. D. & Carrington J. C. Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. H., Valdez P., Olvera R. E. & Carrington J. C. Functions of the tobacco etch virus RNA polymerase (NIb): subcellular transport and protein-protein interaction with VPg/proteinase (NIa). J Virol 71, 1598–1607 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton S. et al. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J Virol 83, 10460–10471 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P., Wu Z. & Wang A. The multifunctional protein CI of potyviruses plays interlinked and distinct roles in viral genome replication and intercellular movement. Virol J 12, 141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong R. & Wang A. SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of Turnip mosaic virus is required for viral infection. J Virol 87, 4704–4715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D. & Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- Lu Q. et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: Components and anchoring nucleoporin. Plant J 61, 259–270 (2010). [DOI] [PubMed] [Google Scholar]
- Earley K. W. et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45, 616–629 (2006). [DOI] [PubMed] [Google Scholar]
- Burch-Smith T. M., Schiff M., Liu Y. & Dinesh-Kumar S. P. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142, 21–27 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I. A., Runions J., Kearns A. & Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1, 2019–2025 (2006). [DOI] [PubMed] [Google Scholar]
- Wei T., Zhang C., Hou X., Sanfaçon H. & Wang A. The SNARE protein Syp71 is essential for Turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog 9, e1003378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen H., Brandizzi F., Verchot J. & Wang A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet 11, e1005164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang C. & Wang A. Divergence and conservation of the major UPR branch IRE1-bZIP signaling pathway across eukaryotes. Sci Rep 6, 27362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.