Abstract
Hydroxyurea, an anticancer agent and potent teratogen, induces oxidative stress and activates a DNA damage response pathway in the gestation day (GD) 9 mouse embryo. To delineate the stress response pathways activated by this drug, we investigated the effect of hydroxyurea exposure on the transcriptome of GD 9 embryos. Timed pregnant CD-1 mice were treated with saline or hydroxyurea (400 mg/kg or 600 mg/kg) on GD 9; embryonic gene and protein expression were examined 3 h later. Microarray analysis revealed that the expression of 1346 probe sets changed significantly in embryos exposed to hydroxyurea compared with controls; the P53 signaling pathway was highly affected. In addition, P53 related family members, P63 and P73, were predicted to be activated and had common and unique downstream targets. Western blot analysis revealed that active phospho-P53 was significantly increased in drug-exposed embryos; confocal microscopy showed that the translocation of phospho-P53 to the nucleus was widespread in the embryo. Furthermore, qRT-PCR showed that the expression of P53-regulated genes (Cdkn1A, Fas, and Trp53inp1) was significantly upregulated in hydroxyurea-exposed embryos; the concentration of the redox sensitive P53INP1 protein was also increased in a hydroxyurea dose-dependent fashion. Thus, hydroxyurea elicits a significant effect on the transcriptome of the organogenesis stage murine embryo, activating several key developmental signaling pathways related to DNA damage and oxidative stress. We propose that the P53 pathway plays a central role in the embryonic stress response and the developmental outcome after teratogen exposure.
Keywords: teratogen, embryonic stress, developmental toxicity, birth defects
During gestation embryos may be exposed to a variety of harmful environmental and therapeutic agents with detrimental effects on development, resulting in birth defects, miscarriage, or even embryonic death. Organogenesis is a particularly vulnerable stage of embryo development (Carney et al., 2004). During this time exposure to a wide variety of teratogens, including thalidomide, ethanol, anticonvulsants, or anticancer drugs, may compromise normal embryo development (Kovacic and Somanathan, 2006). The ability of embryos to respond to the stress triggered by teratogenic exposures is likely to be an important determinant of their developmental outcome. Many such exposures activate specific stress response signaling pathways in the organogenesis-stage conceptus; these include the oxidative stress, DNA damage response, P53, hypoxia, NFκB, and mitogen-activated protein kinases (MAPKs) signaling pathways (Torchinsky and Toder, 2010; Vinson and Hales, 2003; Wells et al., 2009).
Stress responses may play an adaptive role in protecting the embryo from insult, for example by regulating cell proliferation or cycle checkpoints and providing the time to repair damaged DNA, thus ensuring embryo survival and normal development (Aylon and Oren, 2007). Alternatively, activation of these stress response pathways may “tip the balance” in favor of excess cell death and altered cell differentiation pathways, leading to abnormal development, or embryolethality (Brill et al., 1999; Faustman, 2012). Genetic or pharmacological manipulation of a number of these pathways has been shown to alter embryo fate after a teratogen exposure (Nicol et al., 1995; Yan and Hales, 2006). There is a clear need to identify the stress response factors that are at play in the embryo during organogenesis. It is also important to understand how they are coordinated. Does exposure to a teratogen activate distinct stress response signaling pathways or does it disrupt several interactive pathways? The goal of this study was to analyze the effects of exposure to a model teratogen, hydroxyurea, on the organogenesis stage murine embryo transcriptome.
Hydroxyurea, a drug used clinically in the treatment of sickle cell anemia and chronic myeloid leukemia, inhibits ribonucleotide reductase, depleting the endogenous pool of ribonucleotides, and inhibiting DNA synthesis, leading to DNA replication fork stalling and strand breaks (Timson, 1975). Hydroxyurea is a potent teratogen in animal models; it causes severe hind-limb (syndactyly, polydactyly), tail (curled or absent), and craniofacial (cleft palate, exencephaly) defects in murine embryos exposed during organogenesis (DeSesso, 1979; DeSesso et al., 2000; Yan and Hales, 2005). We have shown that exposure to teratogenic doses of hydroxyurea induces oxidative stress (Schlisser et al., 2010; Yan and Hales, 2005), activates the P38 and c-Jun N-terminal kinase (JNK) signaling pathways (Yan and Hales, 2008), and triggers a DNA damage response (Banh and Hales, 2013) in organogenesis-stage murine embryos. Glutathione depletion (Yan and Hales, 2006) or the overexpression of superoxide dismutase (Larouche and Hales, 2009), altering the oxidative stress response, or inhibition of the activation of the P38 or JNK MAPK signaling pathways (Yan and Hales, 2008) will modify the teratogenicity of hydroxyurea.
Our goal is to use a genome wide approach to delineate the major cellular pathways that are activated in the murine embryo in response to a teratogenic dose of hydroxyurea during organogenesis. Our data show that hydroxyurea exposure has a significant impact on the gene expression profile in these embryos. Genome-wide pathway analysis of the genes with changes in expression has revealed that many are downstream of the tumor suppressor protein P53.
MATERIALS AND METHODS
Experimental animals
Timed-pregnant CD1 mice were purchased from Charles River Canada Ltd (St Constant, Quebec, Canada) and housed in the McIntyre Animal Resource Centre (McGill University, Montreal, Quebec, Canada). Animal treatments were conducted in accordance with the guidelines outlined in the Canadian Guide to the Care and Use of Experimental Animals. Mice were mated by the supplier between 8:00 and 10:00 AM on gestation day (GD) 0. Between 8:00 and 10:00 AM on GD 9 pregnant dams were treated with either saline (control) or hydroxyurea (Aldrich Chemical Co., Madison, Wisconsin) at 400 or 600 mg/kg (designated as HU400 and HU600, respectively) by intraperitoneal injection. Previous studies showed a decrease in fetal weights and an increase in the incidence of caudal malformations in the 400 mg/kg treatment group; these effects were more severe and were accompanied by embryolethality in the 600 mg/kg treatment group (Schlisser and Hales, 2013). The dams were euthanized by CO2 asphyxiation and cervical dislocation after 3 h; this time point was chosen based on previous reports that hydroxyurea activation of stress response mechanisms in GD9 embryos, such as P38 MAPK, peaked at 3 h (Yan and Hales, 2008). The uteri were removed and embryos were explanted in Hanks’ balanced salt solution (Invitrogen Canada, Inc., Ontario, Canada). At the time of collection, whole embryos from each litter were separated for future sample processing: 2–3 embryos were stored in 4% paraformaldehyde for immunofluorescence experiments; 6–10 embryos were flash frozen in liquid nitrogen for Western Blot analysis; 2 embryos were stored in RNAlater Stabilization Reagent (Qiagen, Mississauga, Ontario, Canada) for qRT-PCR.
RNA extraction, microarray probe preparation and hybridization
Agilent SurePrint G3 Mouse GE 8x60k Microarrays (Agilent Technologies, Mississauga, Ontario, Canada) were used to probe embryonic gene expression in control and hydroxyurea-treated embryos (HU400) at 3 h post-treatment. Total RNA was extracted using RNeasy Plus Mini Kits (Qiagen, Mississauga, Ontario, Canada). The RNA concentration and purity of each sample were assessed by spectrophotometry using a NanoDrop1000 spectrophotometer (Fisher Scientific, Wilmington, Delaware) and Agilent 2100 BioAnalyzer (Agilent Technologies, Mississauga, Ontario, Canada). For each sample, 600 ng of Cy3 labeled cDNA was used for the single-color microarray. After 17 h of hybridization at 65 °C, the microarray slides were washed as per the recommendations for the Agilent hybridization kit. Arrays were scanned with Agilent DNA Microarray Scanner. Whole litters were pooled for each sample and each experiment was replicated 6 times.
Microarray analysis
Analysis of the scanned microarrays was done using Agilent’s Feature Extraction software version 11.5.1.1. Statistical analysis, clustering and principal component analysis (PCA) were done using GeneSpring Gx version 13.1 (Agilent Technologies). Detected flags were filtered based on expression (20th–100th percentiles) and 1.5-fold change. Pathway statistical analysis and prediction of activated upstream regulators were done using the Ingenuity Pathway Analysis (IPA) software (Qiagen). Pathway Studio (Elsevier) was used to generate the schematic representation of the P53 pathway with highlighted downstream targets. Venny v2.0.2, a free online tool, was used to create the Venn diagrams and lists of P53/P63/P73 overlapping and unique targets (Oliveros, 2007–2015). Several arrays were determined to be of low quality by PCA and quality analysis (Supplementary Figs. 1 and 2) and were not included, reducing the number of replicate experiments for each treatment to 4.
Western blotting
Whole tissue lysates were prepared for detection and quantification of P53, phospho-P53 (S15), P53INP1, and ACTIN protein concentrations. Each sample, consisting of 6-10 embryos, was lysed and processed as previously described (Banh and Hales, 2013). Total protein content from each sample was quantified using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Ltd., Mississauga, Ontario, Canada). Proteins (20 µg) were loaded and separated on 10% SDS-PAGE and transferred at 14 volts overnight at 4°C to PVDF membranes (Bio-Rad Laboratories, Ltd.). Afterwards, membranes were blocked with either 5% BSA or milk in 1X TBS containing 0.1% Tween-20 (TBS-T) for 1 h at room temperature. Membranes were incubated with primary antibodies against P53 (1:1000, cs2524, Cell Signaling Technology, Inc., Danvers, MA), phospho-P53 (S15) (1:1000, cs12571, Cell Signaling Technology), P53INP1 (1:1000, ab202026, Abcam Toronto, Ontario, Canada), or ACTIN (1:1000, sc-1616, Santa Cruz Biotechnology, Texas) overnight at 4°C. Membranes were washed 5 times for 5 min intervals with 1X TBST-T and then incubated for 1 h at room temperature with horseradish peroxidase conjugated anti-mouse (1:5000, cs-7076, Cell Signaling Technology), anti-rabbit (1:5000, NA934V, GE Healthcare, Buckinghamshire, UK), or anti-goat (1:5000, sc-2020, Santa Cruz Biotechnology) secondary antibodies.
Membranes were washed and proteins were detected using the enhanced chemiluminescence technique (ECL Prime, RPN2236, GE Healthcare). Protein bands were quantified using ImageJ software (National Institutes of Health, Bethesda, Maryland) where the area under the curve represents band intensity. All intensities were normalized to that of the ACTIN loading control.
Real-time qRT-PCR
RNA extracted from whole embryos was diluted to a working solution of 2 ng RNA/µl and transcripts were quantified using the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems, Foster City, California) and the StepOnePlus Real-Time PCR System (Applied Biosystems). Each reaction was composed of 10 μl SYBR Green Master Mix, 1–2 μl forward/reverse primer, 0.16 μl Reverse Transcriptase mix, 5 µl sample and completed to 20 µl with RNase-DNase-free water. The PCR reactions were conducted under the following conditions: 48°C for 30 min, 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, 72°C for 30 s and melting curve at 95°C for 15 s, 60°C for 15 s and 95°C for 15 s. Primer sets were purchased from Qiagen: TNF receptor superfamily member 6 (Fas, QT00095333); transformation related protein 53 (Trp53, QT00101906); tumor protein p53 inducible nuclear protein 1 (Trp53inp1, QT00112910); cyclin-dependent kinase inhibitor 1A (Cdkn1a, QT00137053); hypoxanthine phosphoribosyltransferase 1 (Hprt1, QT00166768). Serial dilutions of whole embryo RNA pooled from all treatment groups were used as an internal reference and to create a standard curve for optimizing primer efficiency and concentration. Each reaction was conducted in triplicate, averaged, and normalized to the amounts of Hprt1 RNA transcripts. The levels of Hprt1 were found to be stable in all treatment groups. The relative quantity of each transcript was determined by the StepOnePlus Software (version 2.3).
Preparation of slides for immunofluorescence
Whole embryo sagittal sections were prepared as described previously (Banh and Hales, 2013). Phospho-P53 immunoreactivity was detected as follows: sections were blocked with 10% goat serum (0.5% BSA, 0.1% Triton X-100, 10% goat serum in PBS) for 1 h at room temperature in a dark humidified chamber followed by incubation with the phospho-P53 primary antibody (1:100, cs12571, Cell Signaling Technology) overnight at 4°C. Next, slides were washed 3 times for 5-minute intervals in PBS, followed by a 1 h incubation with AlexaFluor-594 goat anti-rabbit polyclonal secondary antibody (1:200, A-11037, Life Technologies Inc., Burlington, Ontario) at room temperature. Slides were then washed 3 times for 5-minute intervals in PBS, mounted with Vectashield mounting medium supplemented with 4',6-diamidino-2-phenylindole (DAPI) as a counterstain (H-1200, Vector Laboratories Inc., Burlington, Ontario) and covered with glass cover slips. Each slide was mounted with 2–3 embryos from each treatment group. Negative control experiments were done in parallel by blocking the phospho-P53 primary antibody for 1 h at room temperature with phospho-P53 blocking peptide (1:50, SAB 51094, Signalway Antibody LLC, Baltimore, Maryland) then proceeding with the above protocol.
Confocal microscopy
Visualization of phospho-P53 immunoreactivity and cellular localization was done using a Leica TCS 8MP multiphoton confocal microscope. For whole embryo imaging, tile images were scanned at 400 Hz with an optical slice of 5 μm, zoom factor equal to 1, and a pinhole setting of 600 μm using a HC PL APO CS2 20x/0.75 water immersion objective lens. The tiled images were then stitched together using the proprietary Leica Image Analysis software (version 3.2). For cellular localization of phospho-P53, a HC PL APO CS2 63x/1.40 oil immersion lens was used to acquire z-stack images of 2 randomly selected regions within 3 different tissues, the heart (H), the caudal neuroepithelium (CNE), and the rostral neuroepithelium (RNE) of 1–2 embryos per treatment group, from 4 to 5 different litters. All images were captured at a 1024 × 1024 resolution and optical and laser settings were optimized and maintained within each imaging experiment; the 522 nm laser was used for phospho-P53 fluorescence excitation, whereas the multiphoton laser line was set at 730 nm for DAPI excitation.
Quantitative image analysis
Phospho-P53 intensity and nuclear localization were analyzed using Imaris image analysis software (version 8.1.2, Bitplane AG, Zurich, Switzerland). For whole embryo analysis, stitched tile scan images acquired from the Leica software were cropped and imported into Imaris. The mean intensity of the red-channel, representing phospho-P53, was isolated and measured by creating a red-channel specific surface mask, utilizing a region-of-interest approach to neutralize any background signal and to determine the optimal threshold intensity, subsequently applying the optimal parameters to the whole image. Parameters for the surface mask were selected by using the images from the HU600 treatment group. The average mean intensities of phospho-P53 from 4–5 litters per treatment group were compared.
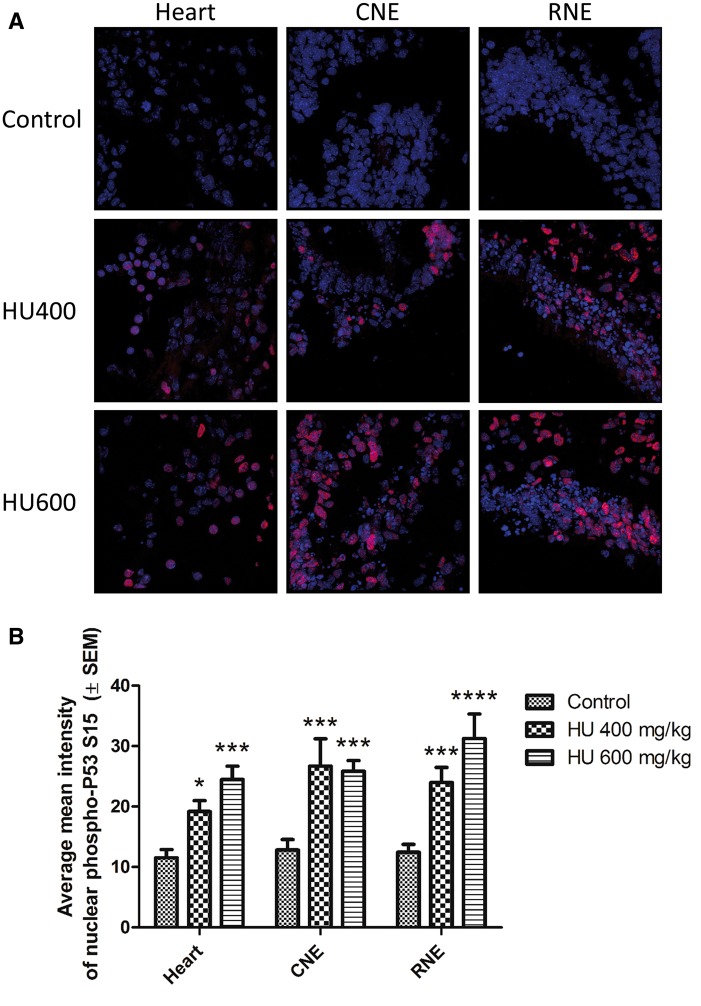
Nuclear localization of phospho-P53 was quantified as follows: z-stack images taken at 63x magnification were quantified by applying a whole image surface mask specific to the blue-channel, representing the DAPI counterstain. The mean intensity of the red-channel within the mask was quantified, reflecting the content of phospho-P53 immunoreactivity within the nuclear staining, as shown in the movie file (Supplementary movie file, movie1.mpg). The nuclear localization of phospho-P53 was compared between 3 different tissues within each embryo and across the treatment groups. These regions were selected based on earlier studies that showed significant levels of phospho-P38 MAPK intensity as a result of hydroxyurea exposure (Banh and Hales, 2013). The average mean nuclear intensity was determined from 1 to 2 embryos per litter from each treatment group.
Statistical analyses
All data were statistically analyzed using the GraphPad Prism Software (version 5, Graph Pad Software Inc., La Jolla, California) except for the microarray and pathway analysis results. The microarray data were analyzed in GeneSpring using an unpaired t-test with a Benjamini–Hochberg multiple comparison correction to compare differences between the control and hydroxyurea treatment group. Pathway analysis was conducted using the Fischer Exact test in the IPA™ software. Data from all other experiments were log transformed then tested using 1-way or 2-way ANOVA, followed by a Bonferroni post-hoc multiple comparison correction to detect statistical differences among the control and hydroxyurea treated groups. The 2-way ANOVA test was used to analyze statistical differences in phospho-P53 nuclear translocation among the control and hydroxyurea treated groups as well as differences among the 3 selected tissues, and any statistically significant interaction between the treatment and tissue variables. The level of significance for all statistical tests was set to P < .05.
RESULTS
Hydroxyurea Exposure Activates the P53 Pathway
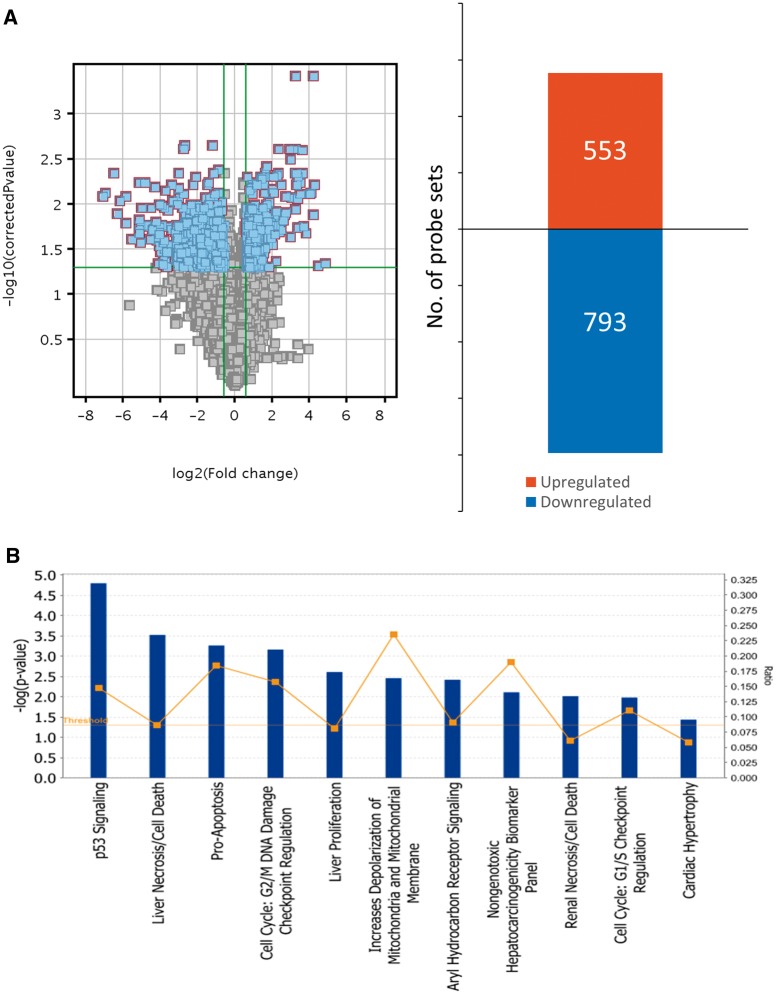
Microarray experiments were conducted using GD 9 embryos exposed to saline or hydroxyurea (400 mg/kg) for 3 h. All raw data are deposited in the GEO repository, under study number GSE54579 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54579). A total of 28 080 probe sets were detected under both conditions; principal component analysis revealed a clear separation between the control and the hydroxyurea treated group (Supplementary Figure 2). Data filtration and analysis of statistical differences revealed that the expression of 1346 transcripts was significantly changed by at least 1.5-fold in hydroxyurea-treated embryos compared with control; 553 were upregulated whereas 793 were downregulated (Figure 1A). Upregulated transcripts with the greatest fold change included cell cycle checkpoint regulators (Ptprv, Cdkn1a and Ccng1), programmed cell death factors (Pmaip1, Fas, Tp53inp1, Phlda3, and Anxa8) and DNA damage response factors (Zfp365 and Ddit4l) (Table 1 and Supplementary Table 1). Thus, hydroxyurea exposure alters the expression of genes that regulate cell cycle arrest and cell death by apoptosis, in addition to inducing a DNA damage response.
FIG. 1.
Hydroxyurea significantly impacts gene expression profiles in GD 9 embryos and activates several cell cycle and cell death related pathways. A, Left: Volcano plot of all detected entities in the microarray. Blue dots indicate transcripts that were significantly changed (-Log P-value = 1.3; P < .05) by at least 1.5-fold. Right: Bar graph of detected probes that were significantly upregulated or downregulated by at least 1.5-fold in response to hydroxyurea treatment in GD 9 embryos. B, Data collected from GeneSpring were analyzed using the Ingenuity Pathway Analysis (IPA) software to predict and examine the molecular pathways that were most affected by hydroxyurea exposure. Vertical bars indicate the level of significance of each pathway indicated by –Log P-value; boxes (▪) indicate the ratio between detected genes in the microarray and total number of known genes in the database for that pathway. Statistical analysis was conducted with the IPA software. N = 4 for each treatment group.
Table 1.
Genes upregulated in embryos exposed to 400 mg/kg hydroxyurea compared with control, filtered by fold change >9.0 and P < .05, sorted by descending order.
| Gene symbol | Description | Fold change | P-value |
|---|---|---|---|
| Agtr1b | Angiotensin II receptor, type 1b | 18.87 | .0002 |
| Ptprv | Protein tyrosine phosphatase, receptor type, V | 18.49 | .0007 |
| 1700007K13Rik | RIKEN cDNA 1700007K13 gene | 17.71 | .0000 |
| Tap1 | Transporter 1, ATP-binding cassette, sub-family B | 16.46 | .0003 |
| Svop | SV2 related protein | 16.06 | .0003 |
| Eda2r | Ectodysplasin A2 receptor, transcript variant 3 | 14.11 | .0011 |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 12.70 | .0009 |
| Pmaip1 | Phorbol-12-myristate-13-acetate-induced protein 1 | 12.17 | .0002 |
| Zfp365 | Zinc finger protein 365 | 12.08 | .0001 |
| Prrg4 | Proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) | 11.32 | .0003 |
| Fas | Fas (TNF receptor superfamily member 6) | 10.99 | .0002 |
| Ddit4l | DNA-damage-inducible transcript 4-like | 10.26 | .0002 |
| Zfp750 | Zinc finger protein 750 | 10.00 | .0004 |
| P2ry6 | Pyrimidinergic receptor P2Y, G-protein coupled, 6 | 9.70 | .0002 |
| Ltb4r1 | Leukotriene B4 receptor 1 | 9.54 | .0002 |
| Eva1c | Eva-1 homolog C (C. elegans) | 9.43 | .0002 |
| D630023F18Rik | RIKEN cDNA D630023F18 gene | 9.33 | .0006 |
| Trp53inp1 | Transformation related protein 53 inducible nuclear protein 1 | 9.32 | .0001 |
| Ccng1 | Cyclin G1 | 9.17 | .0000 |
To delineate the cellular pathways and upstream regulators that are activated during organogenesis in response to a teratogenic dose of hydroxyurea, we ran a pathway analysis on the microarray data. Of the 1346 transcripts detected in the microarray, only 743 transcripts were assigned a functional pathway by the IPA software. The most significantly activated pathway in GD 9 embryos exposed to hydroxyurea was the P53 pathway (P = 1.6e−5) (Figure 1B, Supplementary Table 2); 2 MAPK pathways, the P38 and JNK pathways, were also predicted to be activated (Supplementary Table 2). As anticipated based on the individual genes that were highly upregulated, pathway analysis revealed that hydroxyurea exposure activated cell death/pro-apoptotic pathways and cell cycle checkpoint pathways; in addition, it affected mitochondrial function and aryl hydrocarbon receptor signaling (Figure 1B, Tables 2 and 3).
Table 2.
G1/S cell cycle checkpoint regulation pathway genes that were upregulated by >1.5-fold in embryos exposed to 400 mg/kg hydroxyurea.
| Gene symbol | Gene name | Fold change | P-value |
|---|---|---|---|
| Cdkn1a | Cyclin-dependent kinase inhibitor 1a (p21, cip1) | 12.70 | .0004 |
| Cdkn2b | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 2.55 | .0026 |
| Rb1 | Retinoblastoma 1 | 2.40 | .0008 |
| Ccne1 | Cyclin E1 | 1.87 | .0017 |
| Foxo1 | Forkhead box O1 | 1.62 | .0027 |
| Atm | ATM serine/threonine kinase | 1.57 | .0010 |
Table 3.
Pro-apoptotic pathway genes that were upregulated by >1.5 fold in embryos exposed to 400 mg/kg hydroxyurea.
| Gene symbol | Gene name | Fold change | P-value |
|---|---|---|---|
| Fas | Fas cell surface death receptor | 10.99 | .0002 |
| Tnfrsf10a | Tumor necrosis factor receptor superfamily, member 10a | 4.99 | .0002 |
| Apaf1 | Apoptotic peptidase activating factor 1 | 3.90 | .0003 |
| Bbc3 | BCL2 binding component 3 | 2.73 | .0007 |
| Casp7 | Caspase 7, apoptosis-related cysteine peptidase | 2.26 | .0008 |
| Bik | BCL2-interacting killer (apoptosis-inducing) | 1.92 | .0011 |
| Dapk1 | Death-associated protein kinase 1 | 1.89 | .0037 |
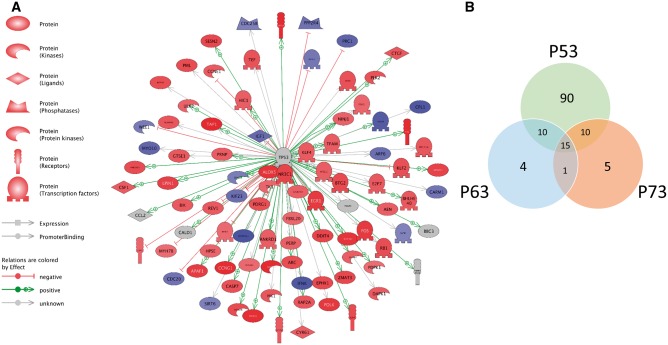
The p53 pathway was the highest predicted activated upstream regulator after hydroxyurea treatment; indeed, many of the most affected genes were downstream targets of P53 transcriptional regulation (Figure 2A, Table 4, and Supplementary Table 3). Other predicted upstream regulators related to the P53 pathway and protein family included BRCA1, P63, and P73 (Table 5). A comparative analysis of genes predicted to be regulated by p53 family members showed that 90 genes were downstream targets of P53; there were 4 and 5 unique genes that were targets of P63 and P73, respectively. Interestingly, there were a number of genes that were shared or common targets of all 3 proteins, P53, P63, and P73, as depicted in the overlapping regions of the Venn diagram (Figure 2B). A complete list of the common and unique targets of these 3 upstream regulators is available in the Supplementary Table 4. Together, these data show that the tumor-suppressor protein P53 and its related family members, P63 and P73 play a major role in the response of embryos to exposure to a teratogenic dose of hydroxyurea during organogenesis.
FIG. 2.
Pathway analysis predicts P53 as the most activated upstream regulator in response to hydroxyurea exposure in GD 9 embryos. A, Schematic representation of interactions between P53 and downstream targets detected in the microarray. Colors indicate the level of transcript expression as determined by the Log fold change extracted from the microarray data using GeneSpring; red denotes upregulation, blue denotes downregulation, whereas grey denotes no effect. B, Venn diagram of common and unique genes associated with the transcription factors P53, P63, and P73. Data were extracted from analysis of predicted activated upstream regulators as determined by the IPA software.
Table 4.
P53 signaling pathway genes that were upregulated by >1.5-fold in embryos exposed to 400 mg/kg hydroxyurea.
| Gene symbol | Gene name | Fold change | P-value |
|---|---|---|---|
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 12.70 | .0009 |
| Fas | Fas cell surface death receptor | 10.99 | .0002 |
| Tp53inp1 | Tumor protein p53 inducible nuclear protein 1 | 9.32 | .0001 |
| Ccng1 | Cyclin G1 | 9.17 | .0000 |
| Tnfrsf10a | Tumor necrosis factor receptor superfamily, member 10a | 4.99 | .0002 |
| Apaf1 | Apoptotic peptidase activating factor 1 | 3.90 | .0003 |
| Pidd1 | p53-induced death domain protein 1 | 3.35 | .0010 |
| Bbc3 | BCL2 binding component 3 | 2.73 | .0007 |
| Pml | Promyelocytic leukemia | 2.50 | .0011 |
| Rb1 | Retinoblastoma 1 | 2.40 | .0008 |
| Perp | PERP, TP53 apoptosis effector | 1.77 | .0008 |
| Gadd45a | Growth arrest and DNA-damage-inducible, alpha | 1.76 | .0017 |
| Atm | ATM serine/threonine kinase | 1.57 | .0010 |
| Kat2b | K(lysine) acetyltransferase 2B | 1.56 | .0034 |
Table 5.
Top predicted activated upstream regulators in GD 9 embryos exposed to 400 mg/kg hydroxyurea by descending P-value order.
| Upstream regulator | Molecule type | Predicted activation state | Activation z-score | P-value of overlap |
|---|---|---|---|---|
| TP53 | Transcription regulator | Activated | 5.74 | 3.12E−23 |
| BRCA1 | Transcription regulator | Activated | 3.17 | 5.49E−10 |
| CCND1 | Transcription regulator | – | 0.63 | 5.09E−09 |
| CDKN1A | Kinase | – | 0.11 | 2.81E−08 |
| TP63 | Transcription regulator | Activated | 3.49 | 4.78E−08 |
| TOPBP1 | Other | – | −1.89 | 1.54E−07 |
| TP73 | Transcription regulator | Activated | 3.01 | 1.69E−07 |
| IGF1 | Growth factor | – | 1.68 | 2.77E−07 |
| PTH | Other | – | 0.69 | 3.75E−07 |
P53 and phospho-P53 Protein Concentrations Are Significantly Increased After Hydroxyurea Exposure
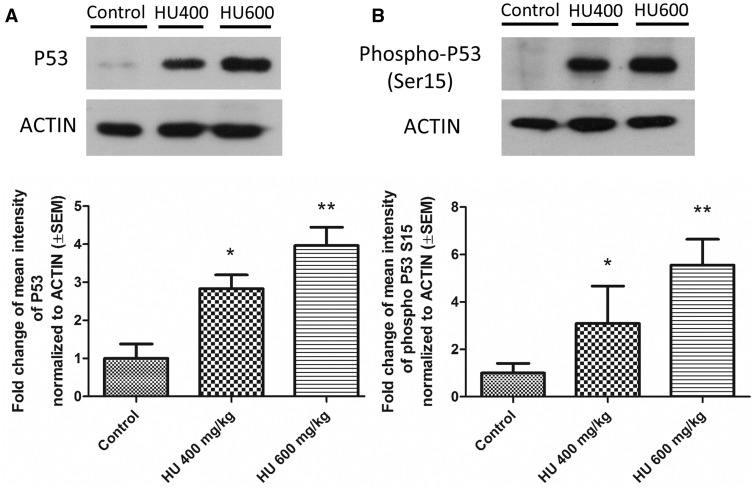
Because P53 activity is largely regulated post-transcriptionally the next question was whether hydroxyurea exposure affected the protein concentrations of P53 and phospho-P53 in treated embryos. Using Western blots P53 and phospho-P53 (Ser15) protein levels were found to be significantly increased in both hydroxyurea treatment groups compared with the control group (Figs. 3A and B). Quantitative analysis showed that in the HU400 and HU600 treated embryos P53 concentrations were increased by 3- and 4-fold, respectively, whereas phospho-P53 levels were increased 3- and 5.5-fold. These results indicate that phosphorylated P53 accumulates in murine embryos in response to a teratogenic dose of hydroxyurea.
FIG. 3.
P53 and Phospho-P53 protein levels increased in a dose-dependent fashion after hydroxyurea exposure. A, Top: Representative blots of P53 protein expression. Bottom: Quantification of P53 levels in embryos from the 400 mg/kg hydroxyurea treatment group (HU400) (Fold change [FC] = 2.83, SEM = ±0.36) and the 600 mg/kg group (HU600) (FC = 3.96, SEM = ±0.48). B, Top: Representative blots of phospho-P53 protein expression. Bottom: Quantification of phospho-P53 levels in HU400 (FC = 3.1, SEM = ±1.5) and HU600 treated embryos (FC = 5.55, SEM = ±1.2). P53 and phospho-P53 levels were significantly upregulated in both hydroxyurea treated groups compared with control. All protein levels were normalized to ACTIN protein expression. N = 4–5 for each treatment group. *P < .05, **P < .01, one-way ANOVA with Bonferroni post-hoc test.
Hydroxyurea Treatment Induces Phospho-P53 Immunoreactivity and Nuclear Localization in Various Embryonic Tissues
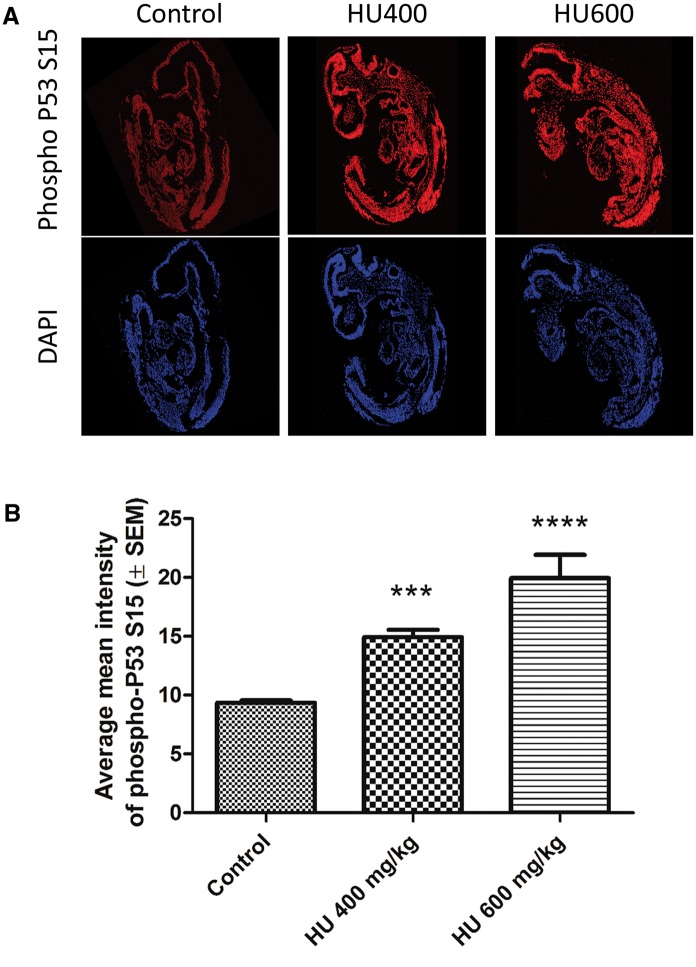
To determine whether hydroxyurea activates P53 in a tissue-specific manner phospho-P53 immunoreactivity was visualized in paraffin-embedded whole embryo sections using confocal microscopy. Phospho-P53 immunoreactivity significantly increased in a dose-dependent fashion throughout the embryo (Figure 4A), with a 2-fold increase in the highest hydroxyurea dose group, HU600 (Figure 4B). The extent to which phospho-P53 was translocated to the nucleus was determined by quantifying the co-localization of phospho-P53 and DAPI. The nuclear concentrations of phospho-P53 in 3 regions, the heart, the caudal neuroepithelium (CNE) and the rostral neuroepithelium (RNE), were compared (Figure 5A; these embryo regions are depicted in Supplementary Figure 3). Nuclear concentrations of phospho-P53 were significantly elevated in all 3 regions of the embryos exposed to hydroxyurea treatment, compared with controls (Figure 5B). There were no significant differences in phospho-P53 nuclear translocation among the 3 different tissues within each treatment group.
FIG. 4.
Hydroxyurea exposure induced a widespread increase in phospho-P53 immunoreactivity. A, Representative multiphoton and confocal microscopy images taken at × 20 of whole embryo sections showing increasing phospho-P53 immunoreactivity (red, top panel) and the DAPI nuclear counterstain (blue, bottom panel). B, Average mean intensity of phospho-P53 immunoreactivity in whole embryo sections. A significant increase in phospho-P53 intensity was detected in the HU400 and HU600 groups. N = 4–5 for each treatment group. ***P < .001, ****P < .0001, one-way ANOVA with Bonferroni post-hoc test.
FIG. 5.
Nuclear translocation of phospho-P53 increased significantly with hydroxyurea treatment. A, Representative multiphoton and confocal images of phospho-P53 immunoreactivity taken at × 63 magnification. Three tissues were analyzed per embryo across all treatment groups (Heart, CNE: caudal neuroepithelium, RNE: rostral neuroepithelium). B, Quantification of phospho-P53 colocalization with DAPI. N = 4–5 for each tissue and treatment group. *P < .05, ***P < .001, ****P < .0001 compared with control, 2-way ANOVA with Bonferroni post-hoc test. There were no statistically significant differences in phospho-P53 nuclear translocation between tissues in the embryo, and no statistically significant interaction between the treatment and tissue variables.
P53 Downstream Transcription Targets Are Significantly Upregulated by Hydroxyurea
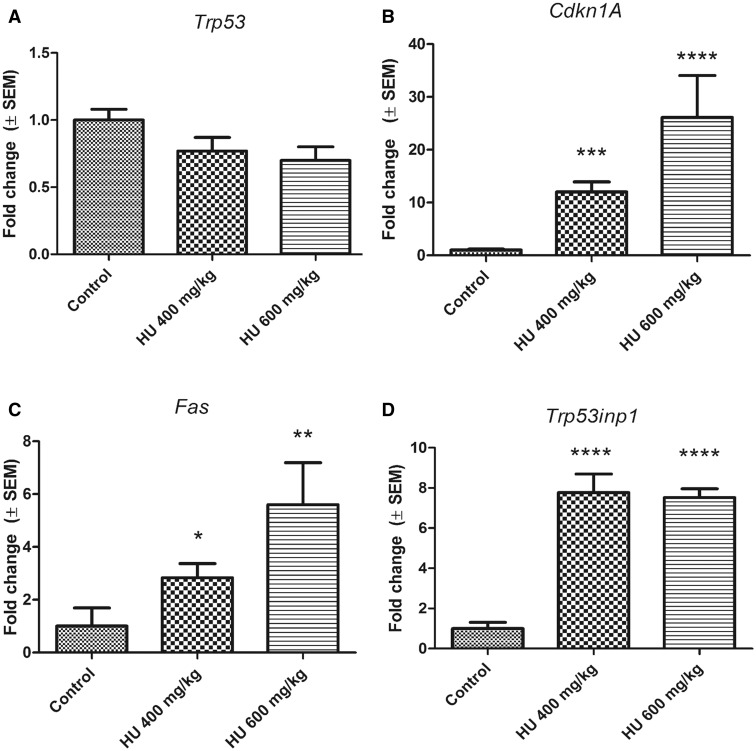
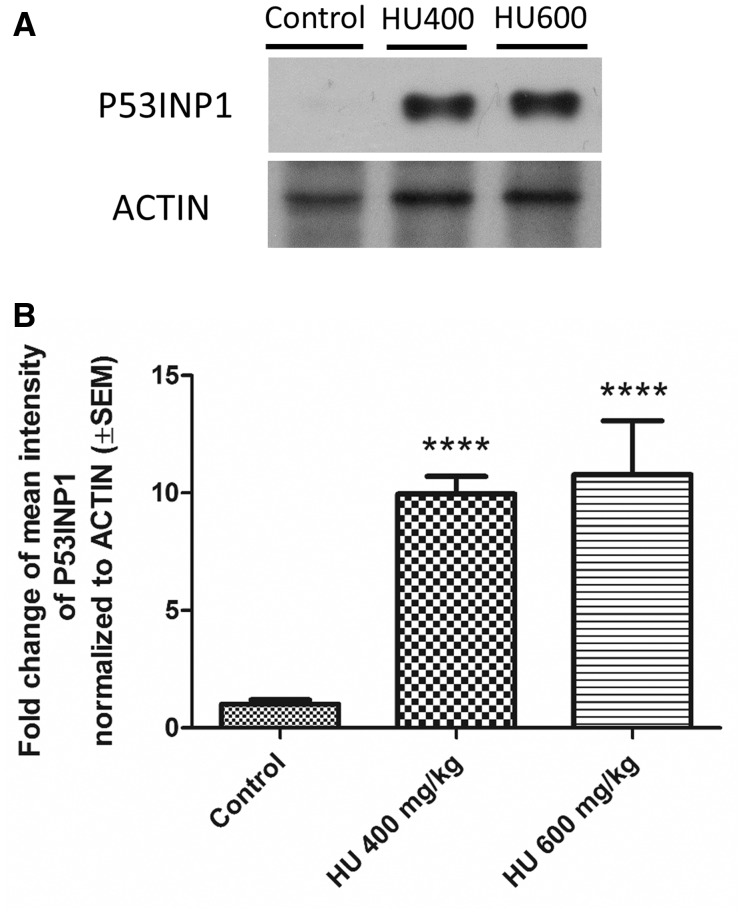
The induction of specific P53 regulated genes was quantified using qRT-PCR to assess P53 pathway activation and to validate the microarray data. Although Trp53 transcript levels did not change with hydroxyurea exposure compared with control (Figure 6A), Cdkn1a (Cyclin-Dependent Kinase Inhibitor 1A [also known as P21, CIP1]), Fas (Fas Cell Surface Death Receptor) and Trp53inp1 (Tumor protein p53-inducible nuclear protein 1) were significantly upregulated in both the HU400 and HU600 treatment groups (Figs. 6B–D). We examined the protein expression level of P53INP1 in response to hydroxyurea exposure as a marker for P53 downstream pro-apoptotic effects. Whereas P53INP1 was almost undetected in control embryos, expression increased significantly in response to hydroxyurea exposure in both the HU400 and HU600 treatment groups (Figure 7).
FIG. 6.
Transcription levels of P53 downstream targets in response to hydroxyurea exposure. A, Trp53 transcript levels after hydroxyurea treatment were not significantly different compared with control, N = 3. B–D, Hydroxyurea exposure significantly induced transcript levels of Cdkn1a (HU400 FC = 12.02, SEM = ±1.90; HU600 FC = 26.11, SEM = ±7.95, N = 5), Fas (HU400 FC = 2.83, SEM = ±0.54; HU600 FC = 5.6, SEM = ±1.59, N = 4–5), and Trp53inp1 (HU400 FC = 7.77, SEM = ±0.93; HU600 FC = 7.52, SEM = ±0.44, N = 5). Each bar represents the fold change of the mean quantity of the transcript relative to Hprt1. *P < .05, **P < .01, ***P < .001, ****P < .0001, one-way ANOVA followed by a Bonferroni post-hoc test.
FIG. 7.
Upregulation of P53INP1 in response to hydroxyurea treatment. A, Representative blot of P53INP1 protein expression levels in embryos exposed to hydroxyurea compared with controls. B, Quantification of immunoblots showed that hydroxyurea treatment significantly upregulated P53INP1 in the HU400 (FC = 9.95, SEM = ±0.75) and HU600 (FC = 10.78, SEM = 2.29) groups. N = 4–5 for each treatment group. ****P < .0001, one-way ANOVA followed by a Bonferroni post-hoc test.
DISCUSSION
Hydroxyurea exposure has a significant impact on the gene expression profile of organogenesis-stage mouse embryos. Genome-wide pathway analysis reveals that many significantly changed genes are downstream targets of the tumor suppressor protein P53. Indeed, P53 is the hub in the regulation of a wide gene network in these embryos. We show here that hydroxyurea exposure significantly increases overall P53 protein levels in the embryo and specifically increases the concentrations of phosphorylated P53. Phospho-P53 immunoreactivity was widespread throughout hydroxyurea exposed embryos.
P53, the “guardian of the genome”, is a transcription factor that regulates the expression of numerous annotated genes in response to stress (Chang et al., 2014); P53 activates the expression of genes that are important in cell cycle regulation, the DNA damage response, apoptosis, senescence, cellular metabolism, mitochondrial function, and the oxidative stress response (Ashcroft et al., 1999; Aylon and Oren, 2007). P53 has important roles in murine embryonic development in addition to its role as a tumor suppressor gene; P53 has been shown to regulate the differentiation and modeling of embryonic progenitor nephrons in the kidney, of osteoblasts during bone formation, and of the interdigital tissue of developing limbs (Aboudehen et al., 2012; Armstrong et al., 1995; Lorda-Diez et al., 2015; Schmid et al., 1991; Wang et al., 2006). In situ hybridization studies showed that P53 transcripts are widely expressed in organogenesis-stage murine embryos up to GD 10 (Schmid et al., 1991). Using mice in which the p53-dependent promoter of Mdm2 was tagged with lacZ, Gottlieb et al. (1997) demonstrated that the pattern of P53 activation in response to ionizing radiation-induced DNA damage became progressively more restricted with embryo age. P53 expression was strongly activated throughout the embryo on GD 8.5 but by GD 10.5 positive staining was observed in the brain regions, the branchial arches, the maxillary area and the limb buds; no staining was observed in the heart. Here we report that immunoreactive phospho-P53 is widespread throughout the embryo after hydroxyurea exposure; however, the translocation of phospho-P53 to the nucleus appears to be treatment and dose-dependent but not embryo region-specific. A previous study from our lab showed that phospho-P38α expression was increased in the neuroepithelium and neural tube, but not in the somites or the heart, after hydroxyurea exposure (Banh and Hales, 2013); P53 phosphorylation (serine 18 in the mouse is equivalent to serine 15 in human P53) is catalyzed by the P38 MAPK kinase in response to DNA strand breaks (Sluss et al., 2004).
Pathway analysis revealed that cell cycle checkpoint pathways and pro-apoptotic factors are activated in embryos in response to hydroxyurea exposure; these include Cdkn1A (P21), a cyclin-dependent kinase inhibitor that binds and deactivates CDK1, a major driver of the cell cycle (Harper et al., 1993), Fas, which is involved in the extrinsic apoptotic pathway, and the autophagy-related factor Trp53inp1 (Adachi et al., 1997; Seillier et al., 2012). Whether these downstream factors elicit their effects in a pattern that mimics the embryo-wide expression and activation of P53 is yet to be determined. The expression of other genes is also highly upregulated in response to hydroxyurea. These include Ptprv, which transcribes a G1/S cell cycle checkpoint regulator that acts downstream of P53, Pmaip1, which transcribes the protein NOXA that induces mitochondrial mediated apoptosis, Phdla3, which represses the Akt1 pathway and thereby induces apoptosis, and Zfp365, which transcribes a zinc-finger protein that is involved in the DNA damage repair of stalled replication forks (Doumont et al., 2005; Kurata et al., 2008; Zhang et al., 2013). Thus, hydroxyurea induces DNA damage repair factors as well as apoptotic and cell-cycle regulators.
Other P53 family members, namely P63 and P73, may play a role in the response of the embryo to teratogenic doses of hydroxyurea as they have also been shown to respond to DNA damage from other anti-cancer agents (Lin et al., 2009; Wilhelm et al., 2010). These transcription factors have overlapping as well as distinct roles from P53 and are expressed in a region-specific manner during organogenesis (Levrero et al., 2000); when P63 is mutated or knocked out in mice, the offspring have developmental defects that are similar to the malformations that are observed after hydroxyurea exposure (e.g., truncated or absent fore- and hind-limbs) (Duijf et al., 2003; Mills et al., 1999). Our microarray data analysis revealed that several transcripts are common targets of 2 or all of these tumor suppressor family members but each of these upstream regulators is also associated with its unique set of downstream targets. The possible roles of these upstream regulators in the embryonic response to teratogenic stress and the functions of their unique downstream targets are not clear.
In utero exposure to hydroxyurea activates MAPK signaling, inducing the phosphorylation and activation of P38 and JNK (Yan and Hales, 2008), which are known to respond to both DNA damage and oxidative stress. Our pathway analysis and qRT-PCR data revealed a DNA damage response but did not detect an increase in the oxidative stress response at the transcript level. Previous studies have shown an increase in 4-hydroxynonenal (4-HNE) protein adducts after hydroxyurea exposure, suggesting that oxidative stress is induced by this drug (Schlisser et al., 2010). However, we did observe a significant upregulation of P53INP1 at both the transcript and protein levels in this study. P53INP1, a P53 downstream effector that is known to respond to oxidative stress, interacts with P53 via a specific binding domain to regulate its pro-apoptotic function (Peuget et al., 2014; Seillier, 2012). P53 itself is also redox sensitive (Forsberg and Di Giovanni, 2014). Thus, any oxidative stress response in the embryo after 3 h of exposure to hydroxyurea may be best detected at the post-translational level.
P53 is upregulated in embryos exposed to several known teratogens (eg, ionizing radiation, cyclophosphamide, valproic acid) (Hosako et al., 2007; MacCallum et al., 1996; Paradis and Hales, 2015). A number of labs have investigated the role of P53 in mediating the response of embryos to teratogen exposures. Trp53 deficient embryos exhibit more malformations than their wild-type counterparts when exposed to teratogenic doses of benzo[α]pyrene or ionizing radiation, suggesting that p53 plays a protective role (Nicol et al., 1995; Norimura et al., 1996). Other labs have reported that Trp53 null embryos are more resistant than their wild type counterparts to teratogen exposures that included 2-chloro-2′-deoxyadenosine and cyclophosphamide (Pekar et al., 2007; Wubah et al., 1996). Whereas each of these teratogens may damage DNA, it is important to note that they activate different pathways in the embryo; whereas benzo[α]pyrene induces oxidative stress, ionizing radiation, 2-chloro-2′-deoxyadenosine and cyclophosphamide activate a strong apoptotic response (Torchinsky and Toder, 2010). In addition, the timing and duration of exposure to a teratogen during development may be an important factor in determining developmental outcome. Thus, it is possible that P53 plays a role as both a teratological inducer and suppressor depending on the type and timing of the insult.
Our data demonstrate that organogenesis-stage embryos respond to exposure to a potent teratogen and genotoxic agent by activating the P53 signaling pathway. P53 may also play a central role in cross-talk with other stress response pathways, such as the oxidative stress and DNA damage response pathways in the developing embryo after a teratogenic insult.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfrodjounrals.org.
ACKNOWLEDGMENTS
We thank Wolfgang Reintsch (McGill University, Montreal) for his assistance and guidance in using the IMARIS software and with confocal microscopy, and Yaned Gaitan (McGill University, Montreal) for her assistance with extracting the microarray data using the Agilent Feature Extraction software.
FUNDING
This work was supported by funding from the Canadian Institutes of Health Research [grant number: MOP-57867]. NELH is the recipient of a student stipend award from the CIHR Training Program in Reproduction, Early Development, and the Impact on Health (CIHR-REDIH), and the Graduate Excellence Fellowship from McGill University. BFH is a James McGill Professor.
REFERENCES
- Aboudehen K., Hilliard S., Saifudeen Z., El-Dahr S. S. (2012). Mechanisms of p53 activation and physiological relevance in the developing kidney. Am. J. Physiol. Renal Physiol. 302, F928–F940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi S., Cross A. R., Babior B. M., Gottlieb R. A. (1997). Bcl-2 and the outer mitochondrial membrane in the inactivation of cytochrome c during Fas-mediated apoptosis. J. Biol. Chem. 272, 21878–21882. [DOI] [PubMed] [Google Scholar]
- Armstrong J. F., Kaufman M. H., Harrison D. J., Clarke A. R. (1995). High-frequency developmental abnormalities in p53-deficient mice. Curr. Biol. 5, 931–936. [DOI] [PubMed] [Google Scholar]
- Ashcroft M., Kubbutat M. H., Vousden K. H. (1999). Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol. 19, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y., Oren M. (2007). Living with p53, dying of p53. Cell 130, 597–600. [DOI] [PubMed] [Google Scholar]
- Banh S., Hales B. F. (2013). Hydroxyurea exposure triggers tissue-specific activation of p38 mitogen-activated protein kinase signaling and the DNA damage response in organogenesis-stage mouse embryos. Toxicol. Sci. 133, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill A., Torchinsky A., Carp H., Toder V. (1999). The role of apoptosis in normal and abnormal embryonic development. J. Assist. Reprod. Genet. 16, 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney E. W., Scialli A. R., Watson R. E., DeSesso J. M. (2004). Mechanisms regulating toxicant disposition to the embryo during early pregnancy: an interspecies comparison. Birth Defects Res. C Embryo Today 72, 345–360. [DOI] [PubMed] [Google Scholar]
- Chang G. S., Chen X. A., Park B., Rhee H. S., Li P., Han K. H., Mishra T., Chan-Salis K. Y., Li Y., Hardison R. C., et al. (2014). A comprehensive and high-resolution genome-wide response of p53 to stress. Cell Rep. 8, 514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSesso J. M. (1979). Cell death and free radicals: a mechanism for hydroxyurea teratogenesis. Med. Hypotheses 5, 937–951. [DOI] [PubMed] [Google Scholar]
- DeSesso J. M., Jacobson C. F., Scialli A. R., Goeringer G. C. (2000). Hydroxylamine moiety of developmental toxicants is associated with early cell death: A structure-activity analysis. Teratology 62, 346–355. [DOI] [PubMed] [Google Scholar]
- Doumont G., Martoriati A., Beekman C., Bogaerts S., Mee P. J., Bureau F., Colombo E., Alcalay M., Bellefroid E., Marchesi F., et al. (2005). G1 checkpoint failure and increased tumor susceptibility in mice lacking the novel p53 target Ptprv. EMBO J. 24, 3093–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijf P. H., van Bokhoven H., Brunner H. G. (2003). Pathogenesis of split-hand/split-foot malformation. Hum. Mol. Genet. 12, R51–R60. [DOI] [PubMed] [Google Scholar]
- Faustman E. M. (2012). Experimental approaches to evaluate mechanisms of developmental toxicity In Developmental and Reproductive Toxicology: A Practical Approach (Hood, Ed R. D.), 3rd edn., pp. 29–33. Informa Healthcare, New York. [Google Scholar]
- Forsberg K., Di Giovanni S. (2014). Cross talk between cellular redox status, metabolism, and p53 in neural stem cell biology. Neuroscientist 20, 326–342. [DOI] [PubMed] [Google Scholar]
- Gottlieb E., Haffner R., King A., Asher G., Gruss P., Lonai P., Oren M. (1997). Transgenic mouse model for studying the transcriptional activity of the p53 protein: Age- and tissue-dependent changes in radiation-induced activation during embryogenesis. EMBO J. 16, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hosako H., Little S. A., Barrier M., Mirkes P. E. (2007). Teratogen-induced activation of p53 in early postimplantation mouse embryos. Toxicol. Sci. 95, 257–269. [DOI] [PubMed] [Google Scholar]
- Kovacic P., Somanathan R. (2006). Mechanism of teratogenesis: Electron transfer, reactive oxygen species, and antioxidants. Birth Defects Res. C Embryo Today 78, 308–325. [DOI] [PubMed] [Google Scholar]
- Kurata K., Yanagisawa R., Ohira M., Kitagawa M., Nakagawara A., Kamijo T. (2008). Stress via p53 pathway causes apoptosis by mitochondrial Noxa upregulation in doxorubicin-treated neuroblastoma cells. Oncogene 27, 741–754. [DOI] [PubMed] [Google Scholar]
- Larouche G., Hales B. F. (2009). The impact of human superoxide dismutase 1 expression in a mouse model on the embryotoxicity of hydroxyurea. Birth Defects Res. A Clin. Mol. Teratol. 85, 800–807. [DOI] [PubMed] [Google Scholar]
- Levrero M., De Laurenzi V., Costanzo A., Gong J., Wang J. Y., Melino G. (2000). The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci. 113 (Pt 10), 1661–1670. [DOI] [PubMed] [Google Scholar]
- Lin Y. L., Sengupta S., Gurdziel K., Bell G. W., Jacks T., Flores E. R. (2009). p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genetics 5, e1000680.. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lorda-Diez C. I., Garcia-Riart B., Montero J. A., Rodriguez-Leon J., Garcia-Porrero J. A., Hurle J. M. (2015). Apoptosis during embryonic tissue remodeling is accompanied by cell senescence. Aging 7, 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum D. E., Hupp T. R., Midgley C. A., Stuart D., Campbell S. J., Harper A., Walsh F. S., Wright E. G., Balmain A., Lane D. P., and., et al. (1996). The p53 response to ionising radiation in adult and developing murine tissues. Oncogene 13, 2575–2587. [PubMed] [Google Scholar]
- Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999). p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713. [DOI] [PubMed] [Google Scholar]
- Nicol C. J., Harrison M. L., Laposa R. R., Gimelshtein I. L., Wells P. G. (1995). A teratologic suppressor role for p53 in benzo[a]pyrene-treated transgenic p53-deficient mice. Nat. Genet. 10, 181–187. [DOI] [PubMed] [Google Scholar]
- Norimura T., Nomoto S., Katsuki M., Gondo Y., Kondo S. (1996). p53-dependent apoptosis suppresses radiation-induced teratogenesis. Nat. Med. 2, 577–580. [DOI] [PubMed] [Google Scholar]
- Oliveros J. C. (2007–2015). Venny. An interactive tool for comparing lists with Venn's diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Paradis F. H., Hales B. F. (2015). Valproic acid induces the hyperacetylation of P53, expression of P53 target genes, and markers of the intrinsic apoptotic pathway in midorganogenesis murine limbs. Birth Defects Res. B Dev. Reprod. Toxicol. 104, 177–183. [DOI] [PubMed] [Google Scholar]
- Pekar O., Molotski N., Savion S., Fein A., Toder V., Torchinsky A. (2007). p53 regulates cyclophosphamide teratogenesis by controlling caspases 3, 8, 9 activation and NF-kappaB DNA binding. Reproduction 134, 379–388. [DOI] [PubMed] [Google Scholar]
- Peuget S., Bonacci T., Soubeyran P., Iovanna J., Dusetti N. J. (2014). Oxidative stress-induced p53 activity is enhanced by a redox-sensitive TP53INP1 SUMOylation. Cell Death Differ. 21, 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlisser A. E., Hales B. F. (2013). Deprenyl enhances the teratogenicity of hydroxyurea in organogenesis stage mouse embryos. Toxicol. Sci. 134, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlisser A. E., Yan J., Hales B. F. (2010). Teratogen-induced oxidative stress targets glyceraldehyde-3-phosphate dehydrogenase in the organogenesis stage mouse embryo. Toxicol. Sci. 118, 686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid P., Lorenz A., Hameister H., Montenarh M. (1991). Expression of p53 during mouse embryogenesis. Development 113, 857–865. [DOI] [PubMed] [Google Scholar]
- Seillier M. (2012). Antioxidant Role of p53 and of Its Target TP53INP1 In Antioxidant Enzyme (El-Missiry, Ed M. A.), 1st edn, pp. 122–134. INTECH, Croatia. [Google Scholar]
- Seillier M., Peuget S., Gayet O., Gauthier C., N'Guessan P., Monte M., Carrier A., Iovanna J. L., Dusetti N. J. (2012). TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell Death Differ. 19, 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluss H. K., Armata H., Gallant J., Jones S. N. (2004). Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol. Cell. Biol. 24, 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timson J. (1975). Hydroxyurea. Mutat. Res. 32, 115–132. [DOI] [PubMed] [Google Scholar]
- Torchinsky A., Toder V. (2010). Mechanisms of the embryo's response to embryopathic stressors: A focus on p53. J. Reprod. Immunol. 85, 76–80. [DOI] [PubMed] [Google Scholar]
- Vinson R. K., Hales B. F. (2003). Genotoxic stress response gene expression in the mid-organogenesis rat conceptus. Toxicol. Sci. 74, 157–164. [DOI] [PubMed] [Google Scholar]
- Wang X., Kua H. Y., Hu Y., Guo K., Zeng Q., Wu Q., Ng H. H., Karsenty G., de Crombrugghe B., Yeh J., and., et al. (2006). p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J. Cell Biol. 172, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells P. G., McCallum G. P., Chen C. S., Henderson J. T., Lee C. J., Perstin J., Preston T. J., Wiley M. J., Wong A. W. (2009). Oxidative stress in developmental origins of disease: Teratogenesis, neurodevelopmental deficits, and cancer. Toxicol. Sci. 108, 4–18. [DOI] [PubMed] [Google Scholar]
- Wilhelm M. T., Rufini A., Wetzel M. K., Tsuchihara K., Inoue S., Tomasini R., Itie-Youten A., Wakeham A., Arsenian-Henriksson M., Melino G., et al. (2010). Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 24, 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubah J. A., Ibrahim M. M., Gao X., Nguyen D., Pisano M. M., Knudsen T. B. (1996). Teratogen-induced eye defects mediated by p53-dependent apoptosis. Curr. Biol. 6, 60–69. [DOI] [PubMed] [Google Scholar]
- Yan J., Hales B. F. (2005). Activator protein-1 (AP-1) DNA binding activity is induced by hydroxyurea in organogenesis stage mouse embryos. Toxicol. Sci. 85, 1013–1023. [DOI] [PubMed] [Google Scholar]
- Yan J., Hales B. F. (2006). Depletion of glutathione induces 4-hydroxynonenal protein adducts and hydroxyurea teratogenicity in the organogenesis stage mouse embryo. J. Pharmacol. Exp. Ther. 319, 613–621. [DOI] [PubMed] [Google Scholar]
- Yan J., Hales B. F. (2008). p38 and c-Jun N-terminal kinase mitogen-activated protein kinase signaling pathways play distinct roles in the response of organogenesis-stage embryos to a teratogen. J. Pharmacol. Exp. Ther. 326, 764–772. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Park E., Kim C. S., Paik J. H. (2013). ZNF365 promotes stalled replication forks recovery to maintain genome stability. Cell Cycle 12, 2817–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.