Abstract
Exposure to chemotherapeutic agents has been linked to an increased risk of type 2 diabetes (T2D), a disease characterized by both the peripheral insulin resistance and impaired glucose-stimulated insulin secretion (GSIS) from pancreatic β-cells. Using the rat β-cell line INS-1 832/13 and isolated mouse pancreatic islets, we investigated the effect of the chemotherapeutic drug doxorubicin (Adriamycin) on pancreatic β-cell survival and function. Exposure of INS-1 832/13 cells to doxorubicin caused impairment of GSIS, cellular viability, an increase in cellular toxicity, as soon as 6 h post-exposure. Doxorubicin impaired plasma membrane electron transport (PMET), a pathway dependent on reduced equivalents NADH and NADPH, but failed to redox cycle in INS-1 832/13 cells and with their lysates. Although NADPH/NADP+ content was unaffected, NADH/NAD+ content decreased at 4 h post-exposure to doxorubicin, and was followed by a reduction in ATP content. Previous studies have demonstrated that doxorubicin functions as a topoisomerase II inhibitor via induction of DNA cross-linking, resulting in apoptosis. Doxorubicin induced the expression of mRNA for mdm2, cyclin G1, and fas whereas downregulating p53, and increased the melting temperature of genomic DNA, consistent with DNA damage and induction of apoptosis. Doxorubicin also induced caspase-3 and -7 activity in INS-1 832/13 cells and mouse islets; co-treatment with the pan-caspase inhibitor Z-VAD-FMK temporarily attenuated the doxorubicin-mediated loss of viability in INS-1 832/13 cells. Together, these data suggest that DNA damage, not H2O2 produced via redox cycling, is a major mechanism of doxorubicin toxicity in pancreatic β-cells.
Keywords: insulin secretion, pancreatic β-cell, redox cycling, NAD(P)H, doxorubicin, Adriamycin
Multiple studies have highlighted the association between diabetes and cancer (Garg et al., 2014; Rosta, 2011), and causality has been suggested to be bi-directional. Diabetes has been proposed to be a contributing factor in cancer development, mediated by obesity-associated chronic inflammation, hyperglycemia, and hyperinsulinemia (Sciacca et al., 2013). In a reciprocal fashion, chemotherapeutic cancer treatments have been suggested to lead to the development of diabetes (Feng et al., 2013), and pancreatic β-cell impairment has been implicated in this process (Dispenzieri and Loprinzi, 1997; Geetha et al., 1999). For example, breast cancer survivors have an increased incidence of diabetes, (Juanjuan et al., 2015).
Doxorubicin (trade name Adriamycin, 2015) is a popular anthracycline chemotherapeutic used to treat a variety of cancers including breast cancer, leukemia, and bladder cancer (Singal and Iliskovic, 1998). Cancer treatment is usually administered as a cocktail of chemotherapeutics, making it difficult to dissect which drug is responsible for which specific side-effect, such as impaired glucose homeostasis (a pre-requisite for diabetes development). Although human clinical data linking development of diabetes following exclusive exposure to doxorubicin are not available, recently published rodent studies have demonstrated a strong correlation between doxorubicin treatment and elevated blood triglyceride and glucose levels (Arunachalam et al., 2013), and identified dys-regulation of adipocyte (Biondo et al., 2016) and muscle metabolism (de Lima Junior et al., 2016) as one of the underlying mechanisms. However, the effect of doxorubicin on pancreatic β-cells, which play a critical role in the maintenance of glucose homeostasis and normal blood glucose levels, has not been exhaustively investigated.
Doxorubicin was previously shown to inhibit insulin secretion by rat islets in vitro at doses below those used in chemotherapeutic therapy, suggesting it may be a possible target for chemotherapy-induced diabetes (Deleers and Goormaghtigh, 1985). Although doxorubicin’s mechanism of toxicity has been characterized in various tumor cell types (extensively reviewed in Gewirtz, 1999; Tacar et al., 2013), the mechanisms responsible for doxorubicin toxicity in islets or pancreatic β-cells have never been determined. Doxorubicin can undergo NADPH-dependent redox cycling with cytochrome P450 reductase (Kostrzewa-Nowak et al., 2005) and rat liver microsomes and heart sarcosomes (Bachur et al., 1977), suggesting a role for superoxide (Goldsmith et al., 1978) and its derivative reactive oxygen intermediates such as H2O2 in mediating doxorubicin toxicity in those tissues. The redox cycling of many compounds including the quinone menadione is supported by insulin-secreting rat pancreatic β-cell line INS-1 832/13 and isolated murine islets of Langerhans, as our laboratory has previously shown (Heart et al., 2012). A second significant mechanism of doxorubicin-mediated toxicity is through DNA damage induced by the inhibition of DNA unwinding and topoisomerase activity, resulting in apoptosis (Gewirtz, 1999). The relative contribution of these mechanisms to toxicity in β-cells may differ.
Here we have evaluated mechanisms by which the prototypical DNA damage-inducing chemotherapeutic drug, doxorubicin, affects pancreatic β-cell metabolism and function. We utilized both isolated mouse islets of Langerhans and clonal INS-1 832/13 cells, which retain key parameters (glucose-stimulated-insulin secretion and metabolism) of native β-cells, to determine how doxorubicin affects these functional pathways and induces toxicity. We found that doxorubicin induces toxicity primarily through DNA damage, with almost no contribution of redox cycling to the mechanism of toxicity.
METHODS
Reagents
10-Acetyl-3.7-dihydroxyphenoxazine (AMPLEX-RED) was purchased from Life Technologies (Grand Island, New York). CellTiter-Blue, ApoTox-Glo Triplex Assay, and RealTime-Glo MT Cell Viability Assay were purchased from Promega (Madison, Wisconsin). EvaGreen PCR master mix was purchased from Emperical Bioscience, Grand Rapids, MI). 1-(2-cyano-3,12,28-trioxooleana-1,9(11)-dien-28-yl)-1H-imidazole (CDDO Im) was purchased from Tocris (Minneapolis, Minnesota). All other chemicals were purchased from Sigma Aldrich (St. Louis, Missouri). All chemicals were handled and disposed of in accordance with institutional regulations.
Islets and clonal pancreatic β-cells
To obtain pancreatic islets, male C57BL/6 mice were euthanized by isoflurane. All procedures were performed in accordance with the University of South Florida’s Institutional Guidelines for Animal Care in compliance with United States Public Health Service regulations. Pancreatic islets were isolated by collagenase (Roche, Indianapolis, Indiana) digestion as previously described (Heart et al., 2009). After digestion and separation by histopaque gradient centrifugation, islets were hand-picked and used after an overnight culture in RPMI supplemented with 10% fetal calf serum (Hyclone), penicillin/streptomycin and 5.5 mM glucose in a humidified incubator with 5% CO2. For caspase 3/7 assays, 36 islets were seeded in complete media into white walled, clear bottom, tissue culture treated 96-well plates, and equilibrated for 3 h prior to treatment. Islets were treated with doxorubicin in the presence of complete media for 6 h, followed by analysis for using the Caspase-Glo 3/7 Assay (Promega) according to the protocol of the manufacturer. To determine the effect of doxorubicin on islet gene expression, freshly isolated islets were incubated for 24 h in complete media in the presence of doxorubicin or vehicle control, and stored in RNA later until PCR analysis (described below). Clonal INS-1 832/13 cells were provided by Dr. Christopher Newgard (Duke University) and were maintained and cultured as described previously (Hohmeier and Newgard, 2004). All experiments with INS-1 832/13 cells were performed with cells at ∼90% confluence in 96-well plates (cell viability and toxicity studies) or 24-well plates (insulin secretion studies) in complete cell culture media or KRB buffer (140 mM NaCl, 30 mM HEPES pH 7.4, 4.6 mM KCl, 1 mM MgSO4, 0.15 mM Na2HPO4, 0.4 mM KH2PO4, 5 mM NaHCO3, and 2 mM CaCl2), supplemented with indicated concentrations of glucose.
Preparation of cellular lysates
All procedures were performed at 4 °C: INS-1 832/13 cells, grown in 150 mm dishes, were washed 3 times with PBS, and dislodged using a rubber policeman. Cells, collected by centrifugation at 200g for 5 min, were re-suspended in PBS and sonicated 10× using 1-s pulses at 25% power output. Supernatants, obtained by centrifugation of lysed cells at 3000g were harvested and quantified for protein content using a Micro-BCA Protein Assay kit (Pierce, Rockford, Illinois).
Hydrogen peroxide assay
The Amplex Red/horseradish peroxidase assay was used to quantify the production of extracellular H2O2 both in vivo (intact cells) and in vitro (cellular lysates), as previously described with minor modifications (Gray et al., 2007).
Intact cells
Cells, grown in 96-well plates to ∼80% confluency, were pre-incubated for 1 h in the KRB buffer and then exposed to reaction mixtures containing 25 μM Amplex Red and 1 U/ml horseradish peroxidase in the KRB buffer, supplemented with indicated concentrations of glucose.
Cellular lysates
Cell lysate proteins (0.375 mg/mL) were supplemented with reduced nicotinamides NADPH or NADH (0.5 mM) in KRB buffer and the production of H2O2 was quantified by calculating the rate of fluorescence increase (540 nm excitation and 595 nm emission) due to H2O2-dependent conversion of Amplex Red to fluorescent Resorufin by horseradish peroxidase. Fluorescence was monitored using a SpectraMax M5 multi-mode microplate reader (Molecular Devices, Sunnyvale, California). H2O2 formed by auto-oxidation of NADPH or NADH in the KRB buffer in the absence or presence of doxorubicin or menadione (10 μM) was subtracted from all measurements.
Determination of nucleotides
NAD(P)+, NAD(P)H, and ATP were determined using the NAD+/NADH, NADP+/NADPH, and ATP kits (Abcam, Cambridge, Massachusetts) according to the protocols of the manufacturer.
PMET activity assay
PMET activity was measured as previously described with minor modifications (Gray et al., 2011). Following a 60 min pre-incubation in KRB buffer supplemented with 3 mM glucose, INS-1 832/13 cells were exposed to the doxorubicin (0.01–10 μM) in the presence of glucose (3 mM or 16 mM) for 2 h. Reduction of WST-1 was measured by the increase in absorbance at 450 nm with a reference wavelength of 630 nm. Reduction of ferricyanide was measured by the decrease in absorbance at 420 nm with a reference wavelength at 500 nm.
Oxygen consumption
Consumption of oxygen was measured using the Oxygraph Plus System clark electrode (Hansatech, Norfolk, United Kingdom). A stirred suspension of INS-1 832/13 cells in KRB supplemented with 11 mM glucose was maintained at 37 °C in a temperature-controlled cuvette. After a stable rate of oxygen consumption was achieved, menadione (10 μM) or doxorubicin (10 μM) was sequentially added to the cell suspension, and the change in the oxygen consumption was calculated over a period of 15 min.
Insulin secretion
INS-1 832/13 cells were pre-incubated for 2 h in KRB buffer in the presence of 3 mM glucose, and then subjected to 60 min of static incubation in KRB containing indicated levels of glucose and doxorubicin. The amount of insulin released into the KRB was determined by ELISA kit (Alpco Diagnostics, Salem, New Hampshire). Data were normalized for cellular protein content determined by the Micro-BCA Protein Assay kit (Pierce, Rockford, Illinois). The glucose-dependent insulin secretory index (GSIS16/3), defined as the ratio between the insulin secretion at 16 mM (stimulatory, surrogate to postprandial glucose levels) and 3 mM (basal, surrogate to fasting glucose levels) glucose, was calculated.
Cell viability and toxicity assays
All assays purchased from Promega were performed according to the instructions of the manufacturer using a SpectraMax M5 multi-mode microplate reader (Molecular Devices, Sunnyvale, California). CellTiter-Blue assays (cat #G8081, Promega) were performed by the addition of 16 uL of CellTiter-Blue dye to wells containing 80 μL of cell culture media and measured by the increase in fluorescence (560 nm excitation, 590 nm emission). The ApoTox-Glo Triplex Assay (cat #G6320) was used to assess viability, toxicity, and caspase 3/7 enzymatic activity. The RealTime-Glo MT Viability Assay was used to assess viability in real time over a time course of 24 h. The GSH-Glo Glutathione Assay was used to quantify reduced glutathione.
Genomic DNA crosslinking melt curve analysis
INS-1 832/13 cells were treated with doxorubicin or vehicle control (water) for 24 h. Genomic DNA was isolated using TriReagent (Sigma) according to the instructions of the manufacturer. About 11 mg of DNA were treated with 1× EvaGreen SYBR master mix, incubated at RT for 30 min, and a DNA melt curve was performed, beginning at 55 °C, incrementing by 1 °C every min through 100 °C. Fluorescence of EvaGreen is expressed relative to the fluorescence at 55 °C.
Quantitative real-time RT-PCR
Total RNA was extracted using either TriReagent (for INS-1 832/13 cells, Sigma, St. Louis, MO) or the RNeasy Micro Kit and Qiashredder (islets of Langerhans, Qiagin, Valencia, California), and RNA was reverse-transcribed using either the High Capacity cDNA Reverse Transcription kit (INS-1 832/13 cells, Applied Biosystems) or the RTScript cDNA Synthesis Kit (islets of Langerhans, Empirical Bioscience, Grand Rapids, Michigan) according to the instructions of the manufacturers. Standard curves were generated using serial 2-fold dilutions from pooled cDNA samples to confirm ≥ 90% reaction efficiency for each primer set. Real-time PCR was performed using on a MyiQ2 Real-Time PCR Detection System (Bio-Rad) using Fast Plus EvaGreen qPCR Master Mix (Biotium, Hayward, California). All PCR primers sequences were generated using PrimerQuest (Integrated DNA Technologies, Coralville, Iowa). A minimum of 3 samples was analyzed for each experimental group. INS-1 832/13 cells were controlled by the expression of beta-2 microglobulin. Primer sequences were as follows: NQO1: GGCCATCATTTGGGCAAGTCCATT and ACTGAAAGCAAGCCAGGCAAACTG; HO1: TGCTCGCATGAACACTCTGGAGAT and ATGTTGAGCAGGAAGGCGGTCTTA; SOD: GGTGTGGCCAATGTGTCCATTGAA and CAATCCCAATCACACCACAAGCCA; prdx5: ATGATGCCTTCGTGACTGCAGAGT and TCAGTGCCTTTACTACGCCCTTGT; p53: ACAATGCTAGTCCCTTCACTGCCT and ATTTCACTGTAGGTGCCAGGTCCA;mdm2: GAACTGGCTTCCAGACGATAA and TGCCATCAGGCACATCTAAG; cyclin g1: GGCAGCACATGCCTTTAATC and CTGTCCTGGAACTCACTGTAAA; fas: TTTCCTCAGTCTTCCGCTATTT and GACGCCTCAGTTCACAGTATTA; and b2mg: ACACTGAATTCACACCCACCGAGA and TGATTACATGTCTCGGTCCCAGGT. Islets of Langerhans were controlled by the expression of beta-actin. Primer sequences were as follows: NQO1: CTGCTGGTGGTGACAAGCACATTT and AGCCTCCTTCATGGCGTAGTTGAA; HO1: CTCCCTGTGTTTCCTTTCTCTC and GCTGCTGGTTTCAAAGTTCAG; SOD: GGCAAAGGTGGAAATGAAGAAA and CTCAGACCACACAGGGAATG; Prdx5: ATGGTGATAGACAACGGCATAG and CAGAGTTGAGAGAGGATGTTGG; p53: TGGACCCTGGCACCTACAATGAAA and ATGCAGACAGGCTTTGCAGAATGG; Mdm2: AGCTGACAGAGAATGATGCTAAA and GGAAGTCGATGGTTGGGAATAG; Cyclin g1: GGCAGCACATGCCTTTAATC and CTCTAGCTGTCTTGGAACTCAC; Fas: ACCATGCCAACCTGGTAAA and CATGTACTCCTTCCCTTCTGTG; Beta actin: GAGGTATCCTGACCCTGAAGTA and CACACGCAGCTCATTGTAGA.
Statistical analysis
Data are expressed as means ± standard deviation. Significance was determined for multiple comparisons using 1- or 2-way Analysis of Variance (ANOVA) followed by the Bonferroni post test and indicated using an asterisk and/or + symbol (Neter et al., 1990).
RESULTS
Doxorubicin was previously demonstrated to inhibit insulin secretion from isolated rat islets in a dose-dependent manner (Deleers and Goormaghtigh, 1985). In order to determine a mechanism of toxicity, we employed the rat insulinoma cell line, INS-1 832/13, which retains many metabolic and functional properties of native pancreatic β-cells (Hohmeier and Newgard, 2004) and isolated islets of Langerhans.
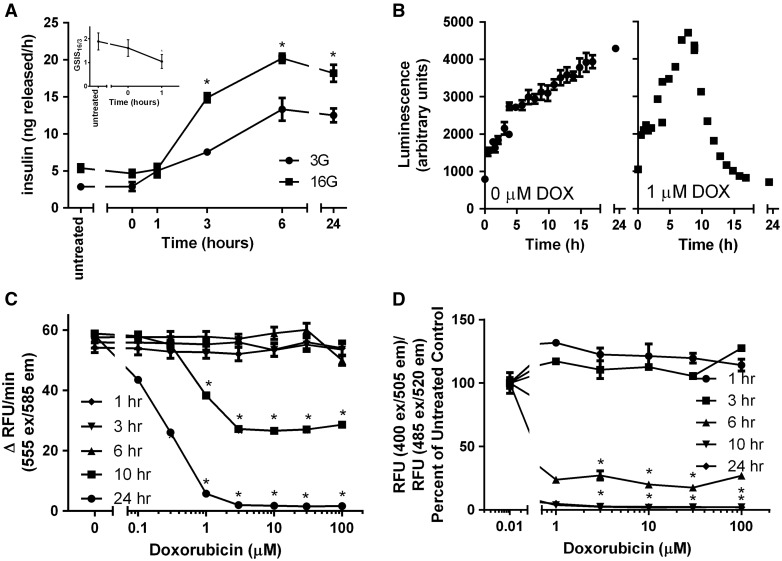
For most insulin secretion studies and toxicity studies, INS-1 832/13 cells were exposed to 1 μM doxorubicin for 1 h, followed by 0, 1, 6, or 24 h recovery in complete culture media. Doxorubicin caused a time-dependent increase in insulin release at both basal glucose (3 mM, represents fasting glucose) and stimulatory glucose (16 mM, represents stimulatory/postprandial glucose) (Figure 1A). However, the glucose-stimulated insulin secretion index (GSIS16/3, the ratio between insulin release at 16 and at 3 mM glucose), a better measure of physiological (metabolic-dependent) insulin release (as opposed to toxicologically induced insulin release due to the necrosis and membrane rupture) was decreased as soon as 1 h following exposure to 1 μM doxorubicin (Figure 1A, inset).
FIG. 1.
Doxorubicin impairs GSIS and induces time- and dose-dependent toxicity in INS-1 832/13 cells. Cells were treated with doxorubicin (1 μM) for 1 h followed by its removal and recovery for 0–24 h in complete growth media. (A) Insulin secretion at basal (3 mM) or stimulatory (16 mM) glucose was measured (A) and expressed as the glucose-stimulated insulin secretion index (GSIS16/3), (A, inset). (B) Cell viability in the presence or absence of doxorubicin (1 mM) was measured in real time using the RealTime-GloTM MT Cell Viability Assay. Cells were treated with varying concentrations of doxorubicin or vehicle as in A and tested for (C) cell viability by CellTiter-Blue and (D) live/dead cell protease activity. Asterisks indicate a significant difference (P < .05) from vehicle-treated controls as determined by one way ANOVA followed by a Bonferroni comparison.
The RealTime Glo MT Cell Viability Assay measures cell viability (determined as cellular reductive potential) in real time. As untreated cells grow and replicate, the signal strengthens over time (Figure 1B, left panel). Doxorubicin (1 μM) caused a reduction in cellular viability beginning at approximately 8 h following treatment, resulting in complete loss of viability by 18–24 h (Figure 1B, right panel). The time of onset of toxicity varied in a dose-dependent manner, with higher doses causing toxicity at earlier time points (not shown). Viability decreased in a dose- and time-dependent manner (Figure 1C). INS-1 832/13 cells experienced a significant increase in toxicity (measured by live and dead cell protease activity) 6 h post-doxorubicin treatment (Figure 1D). Doxorubicin was previously reported to undergo redox cycling both in vitro and in vivo in intact cardiomyocytes and HL60 cells (Davies and Doroshow, 1986; Doroshow and Davies, 1986; Fisher et al., 1983; Gewirtz, 1999; Kostrzewa-Nowak et al., 2005). However, redox cycling was not found to be a significant contributor to doxorubicin-mediated toxicity (Supplementary Figures S1 and S2).
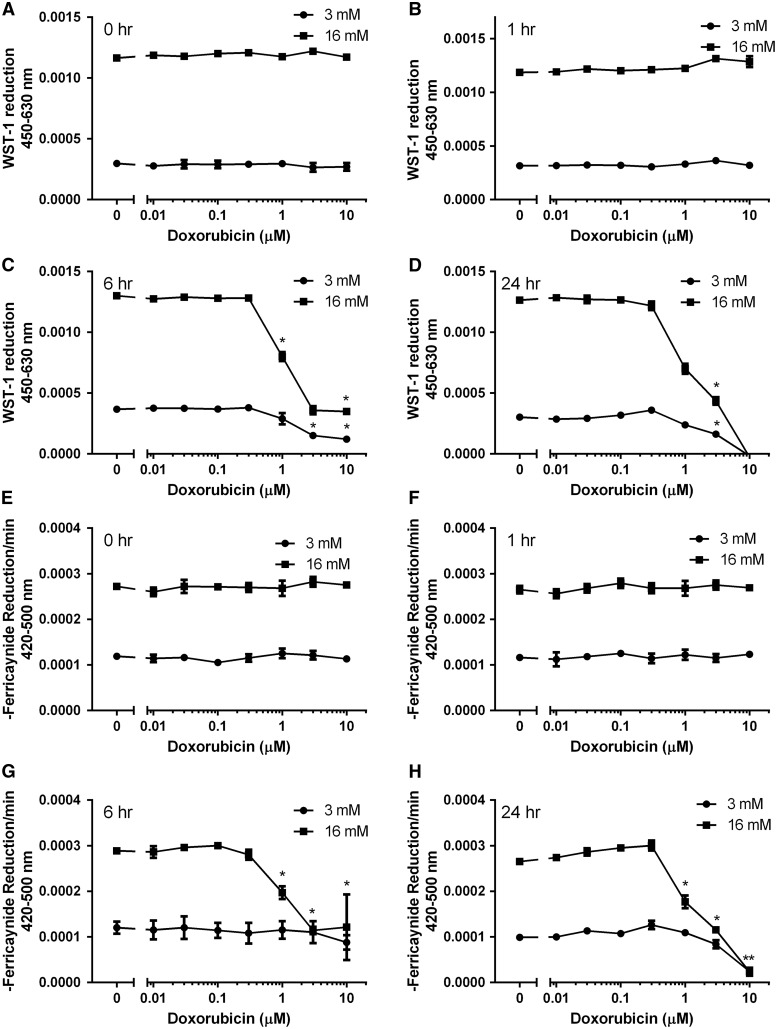
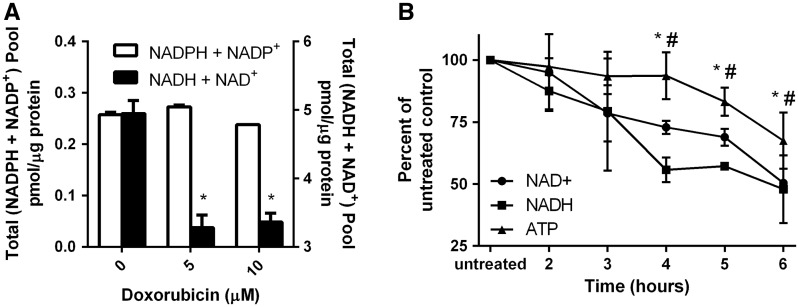
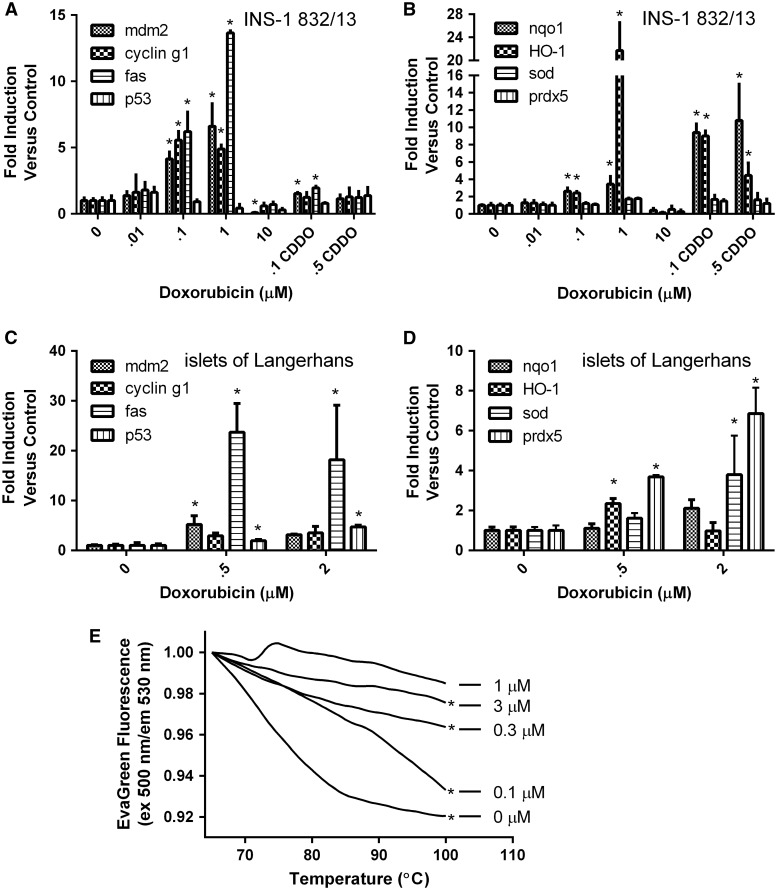
Plasma membrane electron transport (PMET), an extra-mitochondrial pathway which utilizes cytosolic reduced equivalents (NADH and NADPH), was previously shown to operate in a glucose-dependent fashion and be important for GSIS in INS-1 832/13 cells (Gray et al., 2011). Doxorubicin induced a time- and dose-dependent inhibition of PMET activity as measured by reduction of WST-1 (Figure 2A–D) and ferricyanide (Figure 2E–H) in live cells. Doxorubicin-mediated inhibition of PMET was first observed at 6 h. Because PMET activity is contingent upon the availability of NADH and NADPH, we examined if doxorubicin affected levels of reduced and oxidized forms of NADH and NADPH. Although the NADPH + NADP+ pool (the sum of reduced plus oxidized forms of nicotinamide adenine dinucleotide phosphate) was unaffected by doxorubicin, the total NADH + NAD+ pool (the sum of reduced plus oxidized forms of nicotinamide adenine dinucleotide) was significantly reduced by doxorubicin treatment (Figure 3A). Consistent with the doxorubicin-dependent depletion of NAD+, the levels of NADH (formed by the enzymatic reduction of available NAD+) decrease in parallel (Figure 3B). As NADH is a source of protons for ATP synthesis, the doxorubicin-dependent decrease in the ATP levels follows the decrease in the NAD+ and NADH levels (Figure 3B). Consistent with the doxorubicin-dependent induction of DNA damage which triggers the apoptotic pathway (Tacar et al., 2013), doxorubicin treatment for 24 h was found to induce the expression of DNA damage (fas, mdm2, and p53) and cell cycle (cyclin g1) genes in a dose-dependent manner in INS-1 832/13 cells (Figure 4A) and in islets of Langerhans (Figure 4C). In INS-1 832/13 cells, doxorubicin also induced the expression of antioxidant genes regulated by nrf2, NQO1, and HO-1, but did not induce the expression of genes regulated independently of nrf2 – SOD or prdx5 (Figure 4B). This is consistent with the activation of the antioxidant response element by the transcription factor nrf2. Indeed, treatment of INS-1 832/13 cells with CDDO Imidazole, a well-characterized inducer of nrf2-dependent gene expression, induced the expression of NQO1 and HO-1. Interestingly, isolated islets of Langerhans failed to respond to doxorubicin treatment with strong induction of antioxidant genes, suggesting a difference between islets and the tumor cell line model (Figure 4D). Doxorubicin also caused a dose-dependent increase in total cellular DNA melting temperature, consistent with the induction of DNA cross-linking (Figure 4E).
FIG. 2.
Doxorubicin inhibits plasma membrane electron transport (PMET) in a time- and dose-dependent manner. INS-1 832/13 cells, exposed to basal (3 mM) or stimulatory (16 mM) glucose, were treated with doxorubicin for 1 h and analyzed for WST-1 and ferricyanide reduction immediately (A and E) or after 1 h (B and F), 6 h (C and G), or 24 h (D and H) following doxorubicin removal. Asterisks indicate a significant difference (P < .05) from vehicle-treated controls as determined by one-way ANOVA followed by a Bonferroni comparison.
FIG. 3.
Doxorubicin causes a significant dose- and time-dependent reduction in the total NADH/NAD+ pool and ATP content. INS-1 832/13 cells were treated with doxorubicin for 1 h. Following doxorubicin removal and 6 h recovery in growth media, cells were analyzed for (A) (NADPH + NADP+) and (NADH + NAD+) Asterisks indicate a significant difference (P < .05) from vehicle-treated controls as determined by one-way ANOVA followed by a Bonferroni comparison. (B) INS-1 832/13 cells were treated with doxorubicin (5 mM) for 1 h and analyzed for NAD+, NADH, and ATP at 2-6 h post-removal of doxorubicin. * and # indicate a significant difference (P < .05) between NAD+ and ATP and NADH and ATP, respectively.
FIG. 4.
Effects of doxorubicin on gene expression and DNA damage. INS-1 832/13 cells were incubated with doxorubicin or the nrf2 activator CDDO for 24 h and analyzed for the expression of (A) cell-cycle/DNA damage and (B) oxidative stress genes. Islets of Langerhans were incubated with doxorubicin for 24 h and analyzed for the expression of (C) cell-cycle/DNA damage and (D) oxidative stress genes. (E) INS-1 832/13 cells were incubated with doxorubicin for 24 h. Isolated genomic DNA was treated with EvaGreen and the melting temperature of the genomic DNA was determined. Asterisks indicate a significant difference (P < .05) from vehicle-treated controls as determined by one-way ANOVA followed by a Bonferroni comparison (A–D) or linear regression analysis (P < .05) (E).
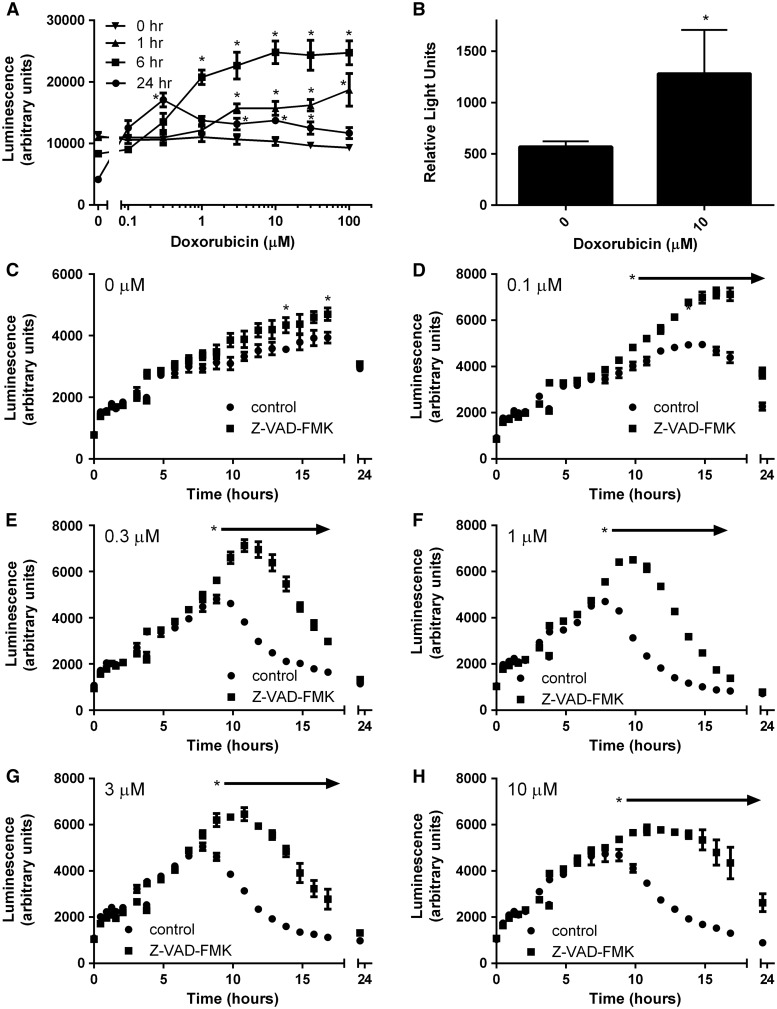
Because doxorubicin was found to cause a delayed toxicity starting at approximately 10 h post-exposure to 1 μM, we next tested whether doxorubicin induced caspase 3/7 activity in INS-1 832/13 cells. Most doses caused an increase in caspase 3/7 activity by 6 h, with higher doses inducing caspase 3/7 within 1 h (Figure 5A). At the 24 h time point, caspase 3/7 activity was depressed, which together with the drastic decrease in cell viability (Figure 1C), was consistent with cell death. Caspase 3/7 activity was similarly induced by doxorubicin treatment in islets of Langerhans (Figure 5B). To test whether cellular death was caspase dependent, INS-1 832/13 cells were treated with doxorubicin in the absence or presence of the pan-caspase inhibitor Z-VAD-FMK. The time of onset of loss of viability was dose dependent, with higher doses causing loss of viability at earlier time points (Figure 5C–H). Z-VAD-FMK temporarily attenuated loss of viability at all doses tested. Taken together, these data suggest that DNA damage followed by induction of apoptosis is an important mechanism of toxicity in pancreatic β-cells.
FIG. 5.
Doxorubicin induces the caspase pathway. (A) INS-1 832/13 cells were treated with doxorubicin or vehicle control and analyzed for induction of caspase 3 and 7 at 4 timepoints post-exposure. (B) Islets of Langerhans were treated with doxorubicin or vehicle control and analyzed for induction of caspases 3 and 7. (C–H) INS-1 832/13 cells were treated with doxorubicin at the indicated concentration or vehicle control for 24 h in the absence or presence of the pan-specific caspase inhibitor Z-VAD-FMK (100 mM) and analyzed for viability in real time using the RealTime-Glo MT Cell Viability Assay. Asterisks indicate a significant difference (P < .05) from vehicle-treated controls as determined by one-way ANOVA followed by a Bonferroni comparison.
DISCUSSION
A single study in 1985 that investigated the effects of doxorubicin on rat pancreatic islets of Langerhans found that doxorubicin inhibited insulin secretion. However, the mechanism of doxorubicin-induced islet dysfunction was not elucidated in this study (Deleers and Goormaghtigh, 1985). In the current work, we investigated mechanisms of doxorubicin toxicity in the well-characterized and widely used insulin-secreting cell line INS-1 832/13 and in isolated murine islets of Langerhans. Doxorubicin induced cytotoxicity in a time- and dose-dependent manner (Figs. 1 and 5C–H); toxicity was paralleled by decreased reductive capacity of the cell (Figure 1B–C), increased cellular permeability (Figure 1D), and decreased plasma membrane electron transport (PMET) activity (Figure 2). Doxorubicin was previously shown to mediate toxicity through 2 primary pathways: redox cycling and induction of DNA damage (Tacar et al., 2013). In this paper, we investigated both mechanisms to determine the relative contribution of each pathway to doxorubicin-induced toxicity in β-cells and demonstrated that DNA damage is the primary mechanism of toxicity in these cells.
Apoptosis Due to DNA Damage Is the Primary Mechanism of Doxorubicin Toxicity in Pancreatic β-Cells
Doxorubicin is also known to induce DNA damage through inhibition of topoisomerase II and the crosslinking of DNA (Tacar et al., 2013). This results in activation of p53 and, if damage is sufficient, subsequent activation of the caspase pathway and induction of apoptosis. Our work showed that doxorubicin induced the expression of DNA damage-related genes both in INS-1 832/13 cells and in isolated islets of Langerhans (Figure 4A and C). Doxorubicin also caused an increase in the temperature necessary to melt double-stranded whole cell DNA (Figure 4E), consistent with the induction of DNA cross-links and damage. Our work also showed that caspase 3/7 were induced by doxorubicin treatment in both INS-1 832/13 cells and islets of Langerhans (Figure 5A and B), with higher doses inducing caspases earlier (Figure 5A). Inhibition of caspase enzymatic activity was previously shown to temporarily halt apoptosis in CHO and 293 cells (Sauerwald et al., 2003) and a myeloma cell line (McKenna and Cotter, 2000). Similarly, the pan-specific caspase inhibitor Z-VAD-FMK temporarily protected β-cells from cytotoxicity induced by doxorubicin at a range of doses (Figure 5C–H), but ultimately was unable to block cellular death due to doxorubicin, suggesting that other caspase-independent apoptotic pathways likely play a role. Together, these data provide compelling evidence that DNA damage is a major contributor to toxicity in β-cells, consistent with previous reports in other cell types (Tacar et al., 2013).
Effect of Doxorubicin on Insulin Secretion
Glucose-stimulated insulin secretion (GSIS) is a hallmark of pancreatic β-cell function. Doxorubicin was previously shown to reduce insulin secretion from isolated rat islets of Langerhans when coincubated with glucose (11.1 mM and 16.7 mM glucose) in a dose-dependent manner for 90 min (Deleers and Goormaghtigh, 1985). At 1 μM doxorubicin, insulin secretion was reduced to ∼90% at 16.7 mM glucose and ∼80% at 11.1 mM glucose. In contrast, we found that doxorubicin induced insulin release at both 3 mM and 16 mM glucose reaching a maximum at 6 h, but it decreased the glucose stimulatory index (the ratio between secretion at high stimulatory and low basal glucose), which determines the β-cells secretory responsiveness to glucose and is the true measure of β-cell functionality. Other toxicants are known to induce insulin release from β-cells by, including the well-characterized streptozotocin (reviewed in Lenzen, 2008). This release is thought to be caused by damage to the cell membrane, resulting in non-physiological release of insulin granules ultimately followed by cellular death. This idea is consistent with detection of loss of membrane integrity in response to doxorubicin at 1 μM (Figure 1D).
Lessons Learned From Doxorubicin Toxicity in Other Tissues
Doxorubicin is a widely used chemotherapeutic drug that induces cellular toxicity via oxidative stress, inhibition of DNA replication, and other mechanisms, dependent upon cell type and dose (reviewed in Gewirtz, 1999). Although it is an effective chemotherapy, doxorubicin has significant off-target toxicity, particularly in cardiomyocytes. The relative contribution and impact of individual mechanisms of doxorubicin toxicity is tissue and dose dependent. At submicromolar doses, doxorubicin interferes with DNA unwinding and topoisomerase activity, whereas at higher doses such as those found after intravenous administration, formation of reactive oxygen intermediates (ROI) and DNA damage may occur (reviewed in Gewirtz, 1999).
Doxorubicin generates ROI via redox cycling; a process enabled in part by its favorable reduction potential of −328 mV (Land et al., 1983). Doxorubicin can be enzymatically reduced by NADH dehydrogenase (Davies and Doroshow, 1986), complex I of the mitochondrial respiratory chain (Goormaghtigh et al., 1987), carbonyl reductase (Olson et al., 1988), and cytochrome P450 reductase (Kostrzewa-Nowak et al., 2005). Through bioreductive alkylation of DNA (Skladanowski and Konopa, 1994a, b), inhibition of topoisomerase II (Tacar et al., 2013; Zhang et al., 2012), intercalation between DNA base pairs (Bodley et al., 1989), and/or production of ROI (Gewirtz, 1999; Minotti et al., 2004; Olson et al., 1988), doxorubicin introduces DNA damage and nicks in the DNA backbone. Studies with topoisomerase IIb knockout animals demonstrated protection from doxorubicin-mediated cardiotoxicity (Zhang et al., 2012), implicating this enzyme as a primary target for doxorubicin toxicity in cardiomyocytes. Still, tocopherol pretreatment has been shown to protect against doxorubicin damage, suggesting that ROI may also play a role in cardiotoxicity (Myers et al., 1977; Tanigawa et al., 1986). Because of the exceptional sensitivity of cardiomyocytes to doxorubicin, its administration is limited to a cumulative lifetime dose (Singal and Iliskovic, 1998). Both redox cycling and inhibition of DNA replication due to DNA damage appear to be important in cardiomyocyte toxicity. However, redox cycling was not found to be a significant contributor to toxicity in INS-1 832/13 cells (Supplementary Data).
Organ toxicity due to doxorubicin treatment is not limited to the heart. Indeed, nearly 40% of patients treated with doxorubicin experience some form of liver injury (Tacar et al., 2013). In isolated rat hepatocytes, doxorubicin first causes lipid peroxidation and depletion of mitochondrial glutathione, followed by increased lactate dehydrogenase leakage (Meredith and Reed, 1983). Rats treated for 3 d with a cumulative dose of 25 mg/kg body weight show increased malondialdehyde formation, decreased glutathione, and decreased expression of antioxidant enzyme activity in liver supernatants (Singla et al., 2014). In mice, doxorubicin-dependent lipid peroxidation has been shown to be reduced by pretreatment with a water soluble precursor form of vitamin E (Nagata et al., 1999), implicating doxorubicin-dependent ROI as a major mechanism of liver toxicity. In vitro, rat liver microsomes readily support redox cycling, which can be increased by pretreatment with phenobarbital (Bachur et al., 1977), a known inducer of cytochrome P450 reductase in the liver (Utley and Mehendale, 1989). The semiquinone form of doxorubicin undergoes oxidation by molecular oxygen resulting in superoxide formation (Kalyanaraman et al., 1980; Kovacic and Osuna, 2000) and the parent compound, completing the redox cycle.
Capacity of Doxorubicin to Contribute to the Onset of Diabetes
The frequency of diabetes mellitus is higher among cancer survivors than among the general U.S. population (Stava et al., 2007). Cancer patients who have received chemotherapeutics are at greater risk for the development of diabetes than patients who have not received adjuvant therapy (Stava et al., 2007). Indeed, chemotherapy-dependent development of insulin-dependent diabetes has been documented (Dispenzieri and Loprinzi, 1997; Geetha et al., 1999). Although the β-cell population can be regenerated in adults following injury (Bonner-Weir et al., 2010), the rate of replication is extremely low (Butler et al., 2003; Meier et al., 2008; Tyrberg et al., 2001), and the capacity for replication driven by replicative aging may play an important role in humans (Yanagiya et al., 1993), limiting the potential for replacement of damaged β-cells. However, studies aimed to evaluate diabetic potential of individual chemotherapeutics are lacking (Juanjuan et al., 2015).
Interestingly, we found that although INS-1 832/13 cells responded to doxorubicin treatment with robust increased expression of both NQO1 and HO-1, genes involved in antioxidant response, this pathway was significantly less induced in native islets of Langerhans (Figure 4B and D). These differences might reflect the acquired defense pathways in the cell line, which was derived from a rat insulinoma (Hohmeier and Newgard, 2004). They might also reflect a lower capacity for oxidative defense in native islets of Langerhans, a theme consistently observed throughout the literature (Gray and Heart, 2010). Further work is required to characterize this difference.
In the current work, we focused on doxorubicin as a prototypical anti-cancer agent with broad usage in the United States and its action on pancreatic β-cell survival and function. We have shown that doxorubicin is highly toxic to these cells at doses below those administered intravenously to cancer patients, providing mechanistic evidence for earlier work demonstrating changes in insulin secretion in isolated rat islets (Deleers and Goormaghtigh, 1985). Our in vitro findings provide rationale for future work investigating the effects of doxorubicin on β-cell survival and function in vivo. Future studies are necessary to analyze islet function ex vivo following intravenous doxorubicin administration to mice or rats and to determine if other chemotherapeutic agents increase the risk of type II diabetes development due in part to toxicity in pancreatic β-cells.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr Neil Copes for help with oxygen consumption measurements and Carolyn Smith for her help in the laboratory. Shpetim Karandrea was supported by the Graduate Student Success Fellowship (USF). Christopher Benton and Malcolm Johns are thanked for donation of a PCR machine and other equipment via the Department of Homeland Security’s Homeland Defense Equipment Reuse (HDER) program to support Cadet research at the U.S. Coast Guard Academy (J.G.). The Coast Guard Alumni Association is thanked for its purchase of the SpectraMax M5 plate reader. The contents of the work presented here do not necessarily represent those of the United States Coast Guard or the federal government.
FUNDING
This work was supported by the National Institutes of Health (Grant no. R01DK097847) to E.A.H. and the American Diabetes Association (Grant no. 7-12-BS-073) to E.A.H.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Arunachalam S., Tirupathi Pichiah P. B., Achiraman S. (2013). Doxorubicin treatment inhibits PPARgamma and may induce lipotoxicity by mimicking a type 2 diabetes-like condition in rodent models. FEBS Lett. 587, 105–110. [DOI] [PubMed] [Google Scholar]
- Bachur N. R., Gordon S. L., Gee M. V. (1977). Anthracycline antibiotic augmentation of microsomal electron transport and free radical formation. Mol. Pharmacol. 13, 901–910. [PubMed] [Google Scholar]
- Biondo L. A., Lima Junior E. A., Souza C. O., Cruz M. M., Cunha R. D., Alonso-Vale M. I., Oyama L. M., Nascimento C. M., Pimentel G. D., Dos Santos R. V., et al. (2016). Impact of doxorubicin treatment on the physiological functions of white adipose tissue. PLoS One 11, e0151548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodley A., Liu L. F., Israel M., Seshadri R., Koseki Y., Giuliani F. C., Kirschenbaum S., Silber R., Potmesil M. (1989). DNA topoisomerase II-mediated interaction of doxorubicin and daunorubicin congeners with DNA. Cancer Res. 49, 5969–5978. [PubMed] [Google Scholar]
- Bonner-Weir S., Li W. C., Ouziel-Yahalom L., Guo L., Weir G. C., Sharma A. (2010). Cell growth and regeneration: replication is only part of the story. Diabetes 59, 2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., Butler P. C. (2003). Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes 52, 102–110. [DOI] [PubMed] [Google Scholar]
- Davies K. J., Doroshow J. H. (1986). Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J. Biol. Chem. 261, 3060–3067. [PubMed] [Google Scholar]
- de Lima Junior E. A., Yamashita A. S., Pimentel G. D., De Sousa L. G. O., Santos R. V. T., Gonçalves C. L., Streck E. L, de Lira F. S., Rosa Neto J. C., (2016). Doxorubicin caused severe hyperglycaemia and insulin resistance, mediated by inhibition in AMPk signalling in skeletal muscle. J. Cachexia Sarcopenia Muscle doi: 10.1002/jcsm.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleers M., Goormaghtigh E. (1985). Adriamycin effects on insulin secretion, Ca2+ movements and glucose oxidation in pancreatic islet cells. Pharmacol. Res. Commun. 17, 227–232. [DOI] [PubMed] [Google Scholar]
- Dispenzieri A., Loprinzi C. L. (1997). Chemotherapy-induced insulin-dependent diabetes mellitus. J. Clin. Oncol. 15, 1287. [DOI] [PubMed] [Google Scholar]
- Doroshow J. H., Davies K. J. (1986). Redox cycling of anthracyclines by cardiac mitochondria. II. Formation of superoxide anion, hydrogen peroxide, and hydroxyl radical. J. Biol. Chem. 261, 3068–3074. [PubMed] [Google Scholar]
- Doxorubicin (doxorubicin hydrochloride). Physician’s Desk Reference website http://www.pdr.net/drug-summary/doxorubicin-hydrochloride?druglabelid=1876&id=2972. Accessed October 21, 2015.
- Feng J. P., Yuan X. L., Li M., Fang J., Xie T., Zhou Y., Zhu Y. M., Luo M., Lin M., Ye D. W. (2013). Secondary diabetes associated with 5-fluorouracil-based chemotherapy regimens in non-diabetic patients with colorectal cancer: results from a single-centre cohort study. Colorectal Dis. 15, 27–33. [DOI] [PubMed] [Google Scholar]
- Fisher J., Ramakrishnan K., Becvar J. E. (1983). Direct enzyme-catalyzed reduction of anthracyclines by reduced nicotinamide adenine dinucleotide. Biochemistry 22, 1347–1355. [DOI] [PubMed] [Google Scholar]
- Garg S. K., Maurer H., Reed K., Selagamsetty R. (2014). Diabetes and cancer: two diseases with obesity as a common risk factor. Diabetes Obes. Metab. 16, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha N., Lali V. S., Hussain B. M., Nair M. K. (1999). Insulin dependent diabetes mellitus induced by chemotherapy and granulocyte, macrophage–colony stimulating factor. J. Assoc. Physicians India 47, 835. [PubMed] [Google Scholar]
- Gewirtz D. A. (1999). A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 57, 727–741. [DOI] [PubMed] [Google Scholar]
- Goldsmith M., Koutcher J. A., Damadian R. (1978). NMR in cancer, XIII: application of the NMR malignancy index to human mammary tumours. Br. J. Cancer 38, 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormaghtigh E., Brasseur R., Huart P., Ruysschaert J. M. (1987). Study of the adriamycin-cardiolipin complex structure using attenuated total reflection infrared spectroscopy. Biochemistry 26, 1789–1794. [DOI] [PubMed] [Google Scholar]
- Gray J. P., Eisen T., Cline G. W., Smith P. J., Heart E. (2011). Plasma membrane electron transport in pancreatic beta-cells is mediated in part by NQO1. Am. J. Physiol. Endocrinol. Metab. 301(1), E113–E121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. P., Heart E. (2010). Usurping the mitochondrial supremacy: extramitochondrial sources of reactive oxygen intermediates and their role in beta cell metabolism and insulin secretion. Toxicol. Mech. Methods 20, 167–174. [DOI] [PubMed] [Google Scholar]
- Gray J. P., Heck D. E., Mishin V., Smith P. J., Hong J. Y., Thiruchelvam M., Cory-Slechta D. A., Laskin D. L., Laskin J. D. (2007). Paraquat increases cyanide-insensitive respiration in murine lung epithelial cells by activating an NAD(P)H:paraquat oxidoreductase: identification of the enzyme as thioredoxin reductase. J. Biol. Chem. 282, 7939–7949. [DOI] [PubMed] [Google Scholar]
- Heart E., Cline G. W., Collis L. P., Pongratz R. L., Gray J. P., Smith P. J. (2009). Role for malic enzyme, pyruvate carboxylation, and mitochondrial malate import in glucose-stimulated insulin secretion. Am. J. Physiol. Endocrinol. Metab. 296, E1354–E1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heart E., Palo M., Womack T., Smith P. J., Gray J. P. (2012). The level of menadione redox-cycling in pancreatic beta-cells is proportional to the glucose concentration: role of NADH and consequences for insulin secretion. Toxicol. Appl. Pharmacol. 258, 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmeier H. E., Newgard C. B. (2004). Cell lines derived from pancreatic islets. Mol. Cell. Endocrinol. 228, 121–128. [DOI] [PubMed] [Google Scholar]
- Juanjuan L., Wen W., Zhongfen L., Chuang C., Jing C., Yiping G., Changhua W., Dehua Y., Shengrong S. (2015). Clinical pathological characteristics of breast cancer patients with secondary diabetes after systemic therapy: a retrospective multicenter study. Tumour Biol. 36, 6939–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman B., Perez-Reyes E., Mason R. P. (1980). Spin-trapping and direct electron spin resonance investigations of the redox metabolism of quinone anticancer drugs. Biochim. Biophys. Acta 630, 119–130. [DOI] [PubMed] [Google Scholar]
- Kostrzewa-Nowak D., Paine M. J., Wolf C. R., Tarasiuk J. (2005). The role of bioreductive activation of doxorubicin in cytotoxic activity against leukaemia HL60-sensitive cell line and its multidrug-resistant sublines. Br. J. Cancer 93, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic P., Osuna J. A., Jr. (2000). Mechanisms of anti-cancer agents: emphasis on oxidative stress and electron transfer. Curr. Pharm. Des. 6(3), 277-309. [DOI] [PubMed] [Google Scholar]
- Land E. J., Mukherjee T., Swallow A. J., Bruce J. M. (1983). One-electron reduction of adriamycin: properties of the semiquinone. Arch. Biochem. Biophys. 225, 116–121. [DOI] [PubMed] [Google Scholar]
- Lenzen S. (2008). The mechanisms of alloxan- and streptozotocin-induced diabetes. Diabetologia 51, 216–226. [DOI] [PubMed] [Google Scholar]
- McKenna S. L., Cotter T. G. (2000). Inhibition of caspase activity delays apoptosis in a transfected NS/0 myeloma cell line. Biotechnol. Bioeng. 67, 165–176. [PubMed] [Google Scholar]
- Meier J. J., Butler A. E., Saisho Y., Monchamp T., Galasso R., Bhushan A., Rizza R. A., Butler P. C. (2008). Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes 57, 1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith M. J., Reed D. J. (1983). Depletion in vitro of mitochondrial glutathione in rat hepatocytes and enhancement of lipid peroxidation by adriamycin and 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU). Biochem. Pharmacol. 32, 1383–1388. [DOI] [PubMed] [Google Scholar]
- Minotti G., Menna P., Salvatorelli E., Cairo G., Gianni L. (2004). Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 56, 185–229. [DOI] [PubMed] [Google Scholar]
- Myers C. E., McGuire W. P., Liss R. H., Ifrim I., Grotzinger K., Young R. C. (1977). Adriamycin: the role of lipid peroxidation in cardiac toxicity and tumor response. Science 197, 165–167. [DOI] [PubMed] [Google Scholar]
- Nagata Y., Takata J., Karube Y., Matsushima Y. (1999). Effects of a water-soluble prodrug of vitamin E on doxorubicin-induced toxicity in mice. Biol. Pharm. Bull. 22, 698–702. [DOI] [PubMed] [Google Scholar]
- Neter J., Wasserman W., Kutner M. (1990). Applied Linear Statistical Models. Chicago: Irwin. [Google Scholar]
- Olson R. D., Mushlin P. S., Brenner D. E., Fleischer S., Cusack B. J., Chang B. K., Boucek R. J., Jr. (1988). Doxorubicin cardiotoxicity may be caused by its metabolite, doxorubicinol. Proc. Natl. Acad. Sci. U S A. 85, 3585–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosta A. (2011). Diabetes and cancer risk: oncologic considerations. Orv Hetil. 152, 1144–1155. [DOI] [PubMed] [Google Scholar]
- Sauerwald T. M., Oyler G. A., Betenbaugh M. J. (2003). Study of caspase inhibitors for limiting death in mammalian cell culture. Biotechnol. Bioeng. 81, 329–340. [DOI] [PubMed] [Google Scholar]
- Sciacca L., Vigneri R., Tumminia A., Frasca F., Squatrito S., Frittitta L., Vigneri P. (2013). Clinical and molecular mechanisms favoring cancer initiation and progression in diabetic patients. Nutr. Metab. Cardiovasc. Dis. 23, 808–815. [DOI] [PubMed] [Google Scholar]
- Singal P. K., Iliskovic N. (1998). Doxorubicin-induced cardiomyopathy. N. Engl. J. Med. 339, 900–905. [DOI] [PubMed] [Google Scholar]
- Singla S., Kumar N. R., Kaur J. (2014). In vivo studies on the protective effect of propolis on doxorubicin-induced toxicity in liver of male rats. Toxicol. Int. 21, 191–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skladanowski A., Konopa J. (1994a). Interstrand DNA crosslinking induced by anthracyclines in tumour cells. Biochem. Pharmacol. 47, 2269–2278. [DOI] [PubMed] [Google Scholar]
- Skladanowski A., Konopa J. (1994b). Relevance of interstrand DNA crosslinking induced by anthracyclines for their biological activity. Biochem. Pharmacol. 47, 2279–2287. [DOI] [PubMed] [Google Scholar]
- Stava C. J., Beck M. L., Feng L., Lopez A., Busaidy N., Vassilopoulou-Sellin R. (2007). Diabetes mellitus among cancer survivors. J. Cancer Surviv. 1, 108–115. [DOI] [PubMed] [Google Scholar]
- Tacar O., Sriamornsak P., Dass C. R. (2013). Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 65, 157–170. [DOI] [PubMed] [Google Scholar]
- Tanigawa N., Katoh H., Kan N., Mizuno Y., Tanimura H., Satomura K., Hikasa Y. (1986). Effect of vitamin E on toxicity and antitumor activity of adriamycin in mice. Jpn. J. Cancer Res. 77, 1249–1255. [PubMed] [Google Scholar]
- Tyrberg B., Ustinov J., Otonkoski T., Andersson A. (2001). Stimulated endocrine cell proliferation and differentiation in transplanted human pancreatic islets: effects of the ob gene and compensatory growth of the implantation organ. Diabetes 50, 301–307. [DOI] [PubMed] [Google Scholar]
- Utley W. S., Mehendale H. M. (1989). Phenobarbital-induced cytosolic cytoprotective mechanisms that offset increases in NADPH cytochrome P450 reductase activity in menadione-mediated cytotoxicity. Toxicol. Appl. Pharmacol. 99, 323–333. [DOI] [PubMed] [Google Scholar]
- Yanagiya K., Takato T., Akagawa T., Harii K. (1993). Reconstruction of large defects that include the mandible with scapular osteocutaneous and forearm flaps: report of cases. J. Oral. Maxillofac. Surg. 51, 439–444. [DOI] [PubMed] [Google Scholar]
- Zhang S., Liu X., Bawa-Khalfe T., Lu L. S., Lyu Y. L., Liu L. F., Yeh E. T. (2012). Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 18, 1639–1642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.