Abstract
Sarcoidosis is a systemic granulomatous disease of unknown etiology. Granulomatous inflammation in sarcoidosis may affect multiple organs, including the lungs, skin, CNS, and the eyes, leading to severe morbidity and mortality. The underlying mechanisms for sustained inflammation in sarcoidosis are unknown. We hypothesized that metabolic changes play a critical role in perpetuation of inflammation in sarcoidosis. 1H nuclear magnetic resonance (NMR)-based untargeted metabolomic analysis was used to identify circulating molecules in serum to discriminate sarcoidosis patients from healthy controls. Principal component analyses (PCA) were performed to identify different metabolic markers and explore the changes of associated biochemical pathways. Using Chenomx 7.6 NMR Suite software, we identified and quantified metabolites responsible for such separation in the PCA models. Quantitative analysis showed that the levels of metabolites, such as 3-hydroxybutyrate, acetoacetate, carnitine, cystine, homocysteine, pyruvate, and trimethylamine N-oxide were significantly increased in sarcoidosis patients. Interestingly, succinate, a major intermediate metabolite involved in the tricyclic acid cycle was significantly decreased in sarcoidosis patients. Application of integrative pathway analyses identified deregulation of butanoate, ketone bodies, citric cycle metabolisms, and transmethylation. This may be used for development of new drugs or nutritional modification.
Keywords: Sarcoidosis, Metabolomics, 1H NMR, TCA cycle, β-Oxidation
1 Introduction
Sarcoidosis is a multisystem granulomatous disease of unknown etiology (Iannuzzi et al. 2007; Hunninghake et al. 1999) characterized by a heightened immune response to an unknown antigen. The coordination of multiple immune cells including macrophages and dendritic cells, as well as T and B cell differentiation and proliferation contribute to granuloma formation, ultimately leading to organ dysfunction (Rastogi et al. 2011; Iannuzzi et al. 2007; Hunninghake et al. 1999). While the exact mechanisms underlying this disease are still unknown, genes and environmental interplay including metabolic derangements may play a role in the development and progression of the disease. Interconnections between metabolic pathways are particularly complex, yet to identify how cellular metabolism influences the immune response is critical in identifying how to exploit metabolic pathways to alter immune responses and disease outcome. Recent studies have shown that glucose availability and acetyl-CoA production influence epigenetics and drive the T cell differentiation program (Wellen et al. 2009). Additionally, several amino acids, including glutamine and leucine play important roles in T cell function and may directly influence T cell activation through metabolic reprogramming (Wang et al. 2011a; Sinclair et al. 2013). Therefore, detection of metabolic alterations in patients with sarcoidosis may be important in understanding the mechanisms of sustained inflammation in this disease.
Metabolomics is a systematic investigation of the metabolic responses in biological systems, providing a comprehensive and quantitative study of a complete set of intracellular and extracellular metabolites (Trujillo et al. 2006). Given the importance of metabolic cues in the regulation of inflammatory responses both in macrophages as well as T-cells, metabolomics has been used to study such perturbations in numerous diseases (Jayavelu and Bar 2014; Wen et al., 2013; Barallobre-Barreiro et al. 2013; Wang et al. 2010; DeFeo et al. 2011; Lu et al. 2013; Noga et al. 2012). However, the role of metabolomics in sarcoidosis has not been investigated.
In this study, we tested the hypothesis that patients with sarcoidosis may have aberrant metabolomic profiles contributing to granulomatous inflammation. Using NMR-based analysis, we showed that the concentration of certain serum metabolites is altered in patients with sarcoidosis.
2 Research design and methods
2.1 Study population
Patients with sarcoidosis were recruited from the Sarcoidosis and Interstitial Lung Disease Center at Wayne State University School of Medicine, Detroit Michigan. Adult patients (n = 20) diagnosed with sarcoidosis using standard guidelines (Hunninghake et al. 1999) were enrolled in the study. The presence of known extra-pulmonary organ involvement was recorded according to ATS guidelines (Hunninghake et al. 1999). Healthy subjects matched for race, age and BMI from the community (n = 17) with no known lung diseases were enrolled as controls.
Pulmonary function tests were performed following ATS guidelines in a licensed laboratory in all patients. All spirometric studies were performed using a calibrated pneumotachograph and lung volumes were measured in a whole-body plethysmograph (Jaeger Spirometry and Sen-sorMedics Vmax 22, VIASYS Respiratory Care, Inc; Yorba Linda, CA, USA) (Redlich et al. 2014).
Blood samples were collected before initiation of any treatment after written consent was obtained from subjects. Blood was allowed to clot for 30 min at room temperature and centrifuged at 2500 rpm for 10 min using SST vacutainer tubes (BD Bioscience, Franklin Lakes, NJ, USA). Serum was collected and stored at −80 °C until further analysis was performed. The study design and data acquisition were approved by the Institutional Review Board at Wayne State University and Detroit Medical Center, Detroit, MI.
2.2 Serum metabolomic analysis using 1H NMR
Serum samples were analyzed following filtration through a 3-kD nominal cut-off membrane (Amicon Ultra 0.5 mL centrifugal filters, Millipore Corp, Billerica, MA) to remove proteins, lipids, lipoproteins, and protein-bound forms of certain molecules. Each serum sample was transferred to a prewashed filter and centrifuged at 13,800 g for 60 min as previously described (Gregory et al. 2013). The filters were further washed with deuterium oxide (D2O) and the filtrate was mixed with the filtered serum in a 1:1 ratio. A reference buffer solution containing 5 mmol/L disodium-2,2-dimethyl 2-silapentane-5-sulphonate (DSS) and 10 mmol/L imidazole in D2O was added to the samples in a 1:9 ratio. The D2O provided a field-frequency lock, whereas DSS was used as the chemical shift reference and imidazole as pH indicator for the NMR spectra. A total of 600 µL mixture was transferred to a 5 mm NMR tube (Sigma Aldrich, St. Louis, MO). 1H NMR spectra were acquired at 599.773 MHz frequency and a temperature of 298 K on a 600 MHz Agilent spectrophotometer (Agilent Technologies, Santa Clara, CA), using the nuclear Overhauser effect spectroscopy (NOESY) pulse sequence with presaturation. A total of 64 scans and a 10.0 ppm spectral width were obtained. The NMR data were processed using ACD/Spec Manager 7.00 software (Advanced Chemistry Development Inc., Toronto, Canada) using auto-phase and baseline correction. Intelligent binning was used to divide the spectra into 1000 integrated regions of 0.01 ppm width each, corresponding to the region of δ10 to 0 (Chen et al. 2014). The peak ppm values were calibrated with reference to the DSS signal at 0 ppm. Before conducting the analysis, the region of spectra associated with residual water (4.63–5.05 ppm) was removed. The integrated data were normalized to the total integrals of each spectrum, and then the spectra were digitized to a table of common integrals and exported as a non-negative value text file for further analysis.
2.2.1 Multivariate data analysis
Processed NMR data was subjected to multivariate pattern recognition analysis using Soft Independent Modeling of Class Analogy (SIMCA) P+13.0 software package (Umetrics, Umeå, Sweden). Data were mean-centered and each variable was divided by the square root of its standard deviation (Pareto scaled). Spectra were statistically analyzed by principal component analysis (PCA) and partial least square discriminant analysis (PLS-DA). PCA is an unsupervised method of analysis of the spectral data meant to visualize inherent clustering between groups. Data were visualized with the score plots of the first two principal components (PC1 and PC2), in which the data points represent individual samples. The metabolites associated with the group separations were visualized in the corresponding loading plots, where each point represents a single NMR spectral region segment. Supervised analysis statistics PLS-DA was used to maximize identification of the differences among the groups and aid in screening of the metabolites responsible for the class separation by removing systematic variations unrelated to pathological status. To entirely interpret the model, score and loading plots combined with variable importance in the projection (VIP) were used.
2.2.2 Identification and quantification metabolites
The pre-processed 1H NMR-spectra were imported into Chenomx NMR Suite 7.6 software (Chenomx Inc., Edmonton, Canada). The reference spectral library was calibrated based on the position, intensity, and line-width of the reference peak. Essentially, a Lorentzian peak shape model of each reference compound was generated from the database and superimposed upon the actual spectrum. Spectra were randomly ordered for profiling and compounds were profiled in the order of decreasing concentration.
2.2.3 Metabolic pathway and network analyses
To identify the relevant metabolic pathways and networks involved, we applied two programs: (1) MetaboAnalyst 3.0 software, a web-based metabolomics data processing tool (Xia and Wishart 2011b) and, (2) the Kyoto Encyclopedia of Genes and Genomes (KEGG), a self-sufficient, integrated resource consisting of genomic, chemical, and network information (Altermann and Klaenhammer 2005).
2.3 Statistical analysis
All data are reported as mean ± SD using SPSS 22.0 software (IBM, Armonk, NY, USA). An independent samples t test was applied to detect significant distinction between the concentrations of metabolites of the two groups. A Pearson correlation test was applied to identify any association between the metabolites found to be altered in sarcoidosis patients, as compared with healthy controls. A p value of <0.05 was considered statistically significant.
3 Results
3.1 Separation of metabolomic profiles of sarcoidosis patients and healthy controls using unsupervised and supervised analysis
Patients were recruited at the time of diagnosis in our center and before starting any treatment. Sera were collected at the same day of bronchoscopy. Demographics and disease characteristics of the sarcoidosis cohort and healthy controls are summarized in Table 1. There was no significant difference in age, race and BMI between patients and healthy controls (p > 0.05). As shown in Table 1, none of the patients had evidence of decreased oxygen saturation or diminished lung function.
Table 1.
Subject demographics, organ involvements, pulmonary function tests
| Patients | Controls | |
|---|---|---|
| Age years | 44.4 ± 11 | 40.6 ± 11.8 |
| BMI (kg m−2) | 31.9 ± 5.2 | 31.0 ± 7.1 |
| Gender [N (%)] | ||
| Male | 6 (30) | 5 (30) |
| Female | 14 (70) | 12 (70) |
| Race [N (%)] | ||
| African American | 20 (100) | 17 (100) |
| Smoking status | ||
| Yes | 3 (15) | 0 (0) |
| No | 17 (85) | 17 (100) |
| Organ involvement | ||
| Lung | 20 (100) | N/A |
| Skin | 12 (60) | |
| Eye | 7 (35) | |
| Multi-organ | 19 (99) | |
| PFT values | ||
| FEV1 % | 88 (5.7) | N/A |
| FVC % | 95 (4.5) | N/A |
| Baseline O2 saturation | 98 (0.37) | N/A |
Results are reported as mean ± SEM
N (%) number of subjects and percent given in parentheses, BMI body mass index, PFT pulmonary function test, FEV1 % percent predicted forced expiratory volume, FVC % percent predicted forced vital capacity
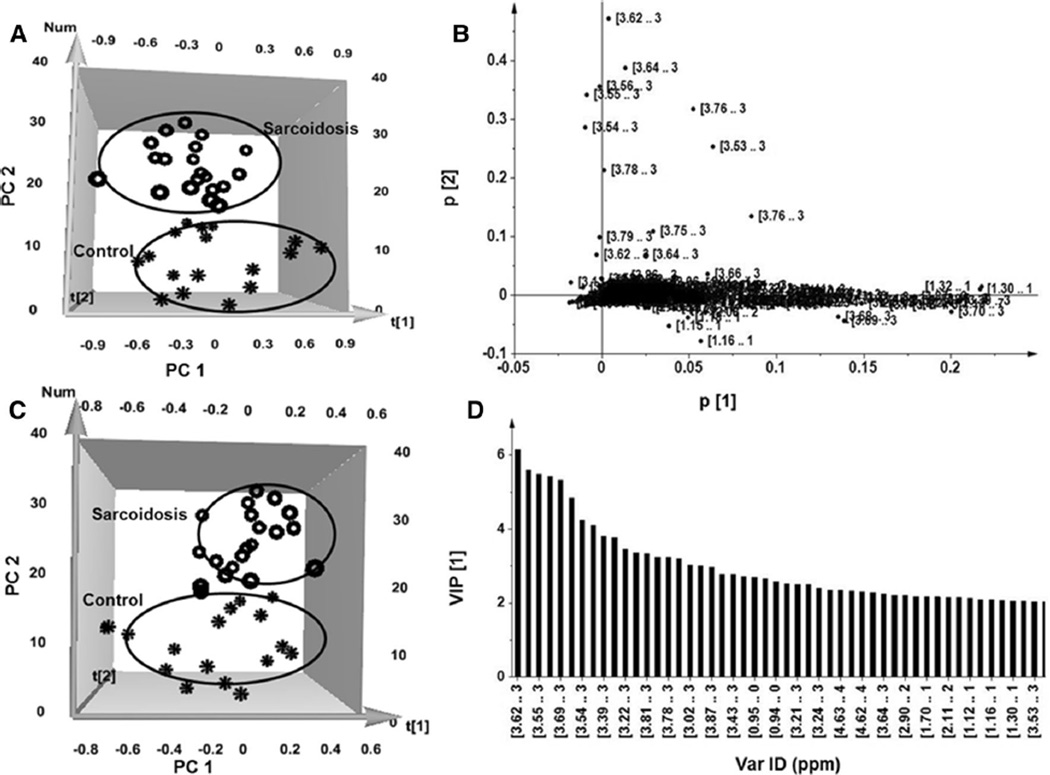
Unsupervised principal component analysis (PCA) was carried out to determine whether it is possible to distinguish healthy controls from sarcoidosis patients. Figure 1a, shows a distinct separation of the NMR spectra acquired from samples of patients and healthy controls as indicated in a 3-dimensional PCA score plot. The first component (PC1) accounts for the greatest variability in the data set, and the succeeding component (PC2) accounts for the second most variability in the data set. The plot revealed a distinct discrimination along the PC2 direction, representing 27 and 34 % variance, respectively (R2X = 0.73, Q2 = 0.66). The loading plot in Fig. 1b shows a distinct distribution of variables across PC1 and PC2 that provide information about the significance of the contribution of each variable to the pattern in the score plots. The cluster closer to the origin of the plot represents the metabolites that are similar in both groups, whereas the regions distant from the origin represent the metabolites that separate the two groups.
Fig. 1.
Characterization of the serum metabolomic changes in sarcoidosis patients and healthy controls. a 3D_NOESY_PCA score plot. Each circle represents one patient spectrum with varying concentrations of metabolites, whereas each star represents one healthy control. b PCA loading plot revealing important regions (3.5–3.75, 1.29, 3.2–3.4, and 2.4 ppm) in the spectra containing metabolites responsible for the clustering shown by the score plot. These metabolites strongly contribute to discrimination between sarcoidosis patients and healthy controls. c The partial least-square-discriminant analysis (3D_NOESY_PLS-DA) score plots based on metabolic fingerprints. d Variable importance in the projection (VIP) plot provides a ranked list of the 40 most important spectra regions (ppm) which lead to the distinction between the sarcoidosis patients and healthy control groups
Next, we applied supervised partial least squares-discriminant analysis (PLS-DA) to the data set to remove factors unrelated to group characteristics and to maximize the group separation and identify discriminating metabolites. The PLS-DA score plot clearly shows class separation of spectra of healthy controls and the sarcoidosis group (Fig. 1c). The PLS-DA score plot provided a stronger clustering for the sarcoidosis group (Fig. 1c), which was similar to PCA score plot (Fig. 1a).
To further identify the variables accounting for the separation between the two groups, variable importance in projection (VIP) statistics were calculated based on the PLS weights and the variability explained by the PLS-DA. A VIP score >1 is considered sufficient to discriminate between study groups (Ni et al. 2008). Using a VIP > 1, we initially identified a total of 60 variables. Increasing the threshold of VIP from 1 to 2, to reach a more stringent analysis, we considered the first 40 variables as optimal discriminating metabolites for the clustering of sarcoidosis and healthy subjects. Figure 1d shows that the most relevant regions of the spectra identified by the VIP plot (VIP > 2) include 0.9–1.3, 2.9, 3.2, and 3.4–3.8 ppm, similar to those depicted by the PCA loading plot, validating the consistency of the data analyses using two different methodologies.
3.2 Identification and quantification of metabolites altered in sarcoidosis patients
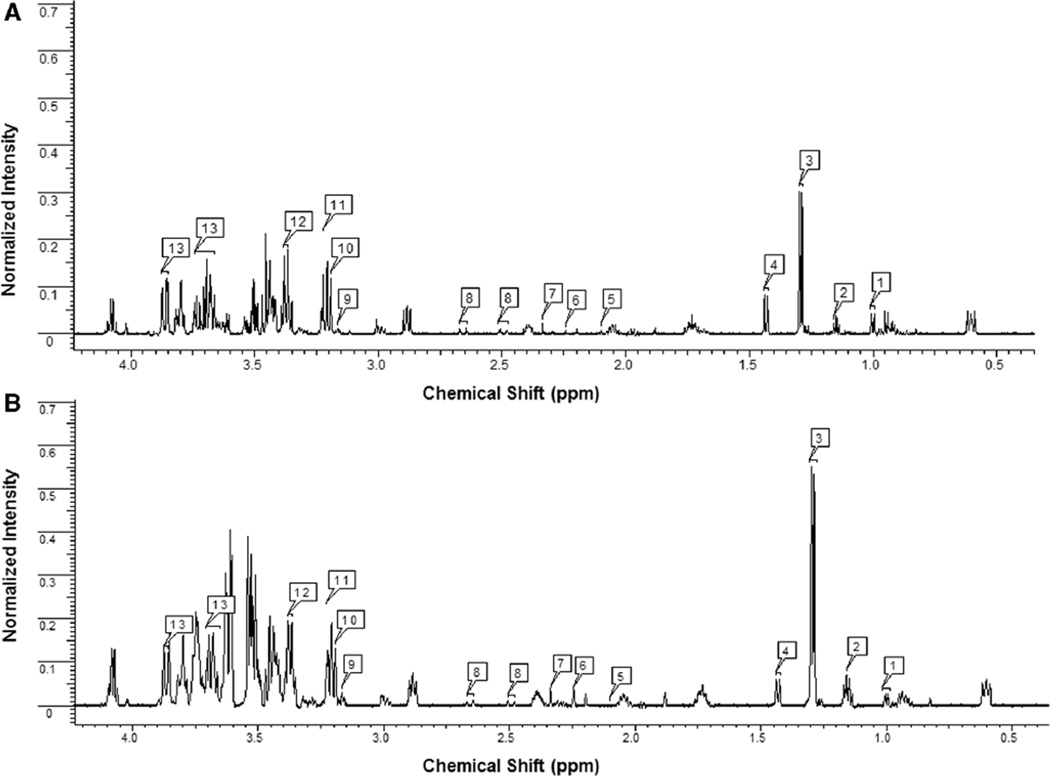
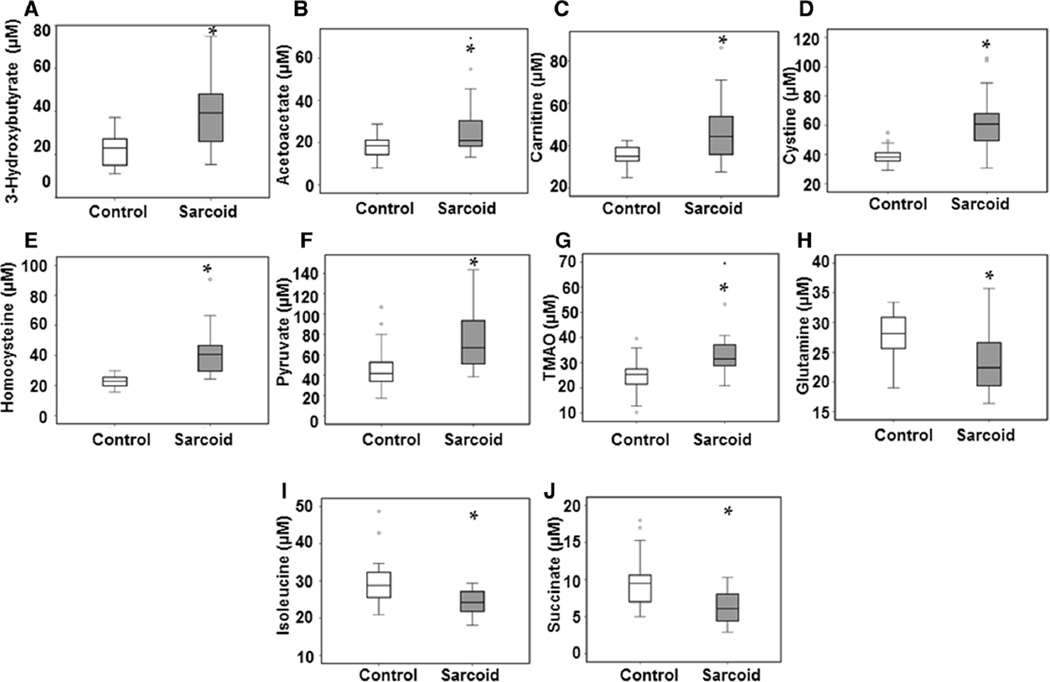
To further identify the complete signature of metabolites of sarcoidosis patients, Chenomx 7.6 Suite NMR software was used to scan the metabolomic profiles of study subjects. 1H NMR spectra of sera provided well resolved peaks, allowing us to detect over 50 metabolites, including amino acids, intermediates of the tricarboxylic acid cycle (TCA), organic acids, and phospholipid-associated molecules. Representative NOESY 1H NMR spectra of major metabolites detected in serum samples from a healthy control and a sarcoidosis patient are shown in Fig. 2a, b, respectively. There was a clear difference in the chemical shift of spectra between the two individuals. After quantification of each metabolite, values were subjected to an independent t-test analysis. Among all the metabolites analyzed, we identified numerous metabolites that were significantly increased (p < 0.05) in sarcoidosis patients as compared to healthy controls (Table 2A). Metabolites that showed a higher concentration included 3-hydroxybutyrate (3-HB, p = 0.002), acetoacetate (p = 0.015), carnitine (p = 0.019), cystine (p = 0.000), homocysteine (p = 0.023), pyruvate (p = 0.029), and trimethylamine N-oxide (TMAO, p = 0.003). In contrast, a number of metabolites showed decreased concentrations in serum of sarcoidosis patients, such as glutamine (p = 0.013), isoleucine (p = 0.019), and succinate (p = 0.003) (Table 2B). Although, succinate and pyruvate appeared in close proximity in the NMR spectrum (around 2.4 ppm), they were easy to distinguish owing to their characteristic peaks. All these metabolites were identified to MSI level 1. Using box plots, Fig. 3 depicts the relative concentration of altered metabolites between controls and sarcoidosis patients.
Fig. 2.
Representative 1H NMR spectroscopy of serum samples obtained from a healthy control (a) and a patient with biopsy proven sarcoidosis (b). Metabolites were identified using the Chenomx NMR Suite 7.6 software. Major metabolites recognized include 1 Valine; 2 3-Hydroxybutyrate; 3 Lactate; 4 Alanine; 5 Homocysteine; 6 Acetoacetate; 7 Pyruvate; 8 Citrate; 9 Choline; 10 Carnitine; 11 Trimethylamine-N-oxide; 12 Cystine; 13 Glucose
Table 2.
Serum metabolites quantified to be altered in sarcoidosis patients as compared with healthy controls
| Metabolite | Peak regions (ppm) | Change | p value | KEGG No. |
|---|---|---|---|---|
| (A) | ||||
| 3-Hydroxybutyrate | 1.2, 2.3, 2.4, 4.1 | ↑ | 0.005 | C01089 |
| Acetoacetate | 2.3,3.4 | ↑ | 0.011 | C00164 |
| Carnitine | 2.4, 3.2, 3.4, 4.6 | ↑↑ | 0.002 | C00318 |
| Cystine | 3.2, 3.4, 4.1 | ↑↑ | 0.000 | C00491 |
| Homocysteine | 2.1,2.2,2.6,2.7,3.9 | ↑↑ | 0.000 | C00155 |
| Pyruvate | 2.4 | ↑ | 0.006 | C00022 |
| Trimethylamine-N-oxide | 3.3 | ↑↑ | 0.001 | C01104 |
| (B) | ||||
| Glutamine | 2.1,2.2,2.4,2.5,3.8,6.9,7.6 | ↓↓ | 0.002 | C00064 |
| Isoleucine | 0.9,1.0,1.2, 1.5, 2.0,3.7 | ↓↓ | 0.003 | C00407 |
| Succinate | 2.4 | ↓↓ | 0.002 | C00042 |
Serum metabolites found to be significantly higher (A) or lower (B) in sarcoidosis patients, as compared with healthy controls. The metabolites were quantified using the Chenomx 7.6 NMR Suite database and significance was obtained using the Independent Sample T-test, SPSS, p < 0.05
Fig. 3.
Relative levels of metabolites found to be altered in sarcoidosis patients as compared with healthy controls, represented by box plots. a 3-Hydroxybutyrate; b Acetoacetate; c Carnitine; d Cystine; e Homocysteine; f Pyruvate; g Trimethylamine N-oxide; h Glutamine; i Isoleucine; j Succinate. For the box plots, the bottom and top of the boxes represent the 25th and 75th percentile, respectively. The top and bottom bars represent the entire stretch of the data points for the subjects, except the extreme points, which are indicated with circles (o). The hyphen indicates the median value. The y axis represents the relative concentrations of the metabolites
3.3 Concordance between the altered metabolites within a pathway
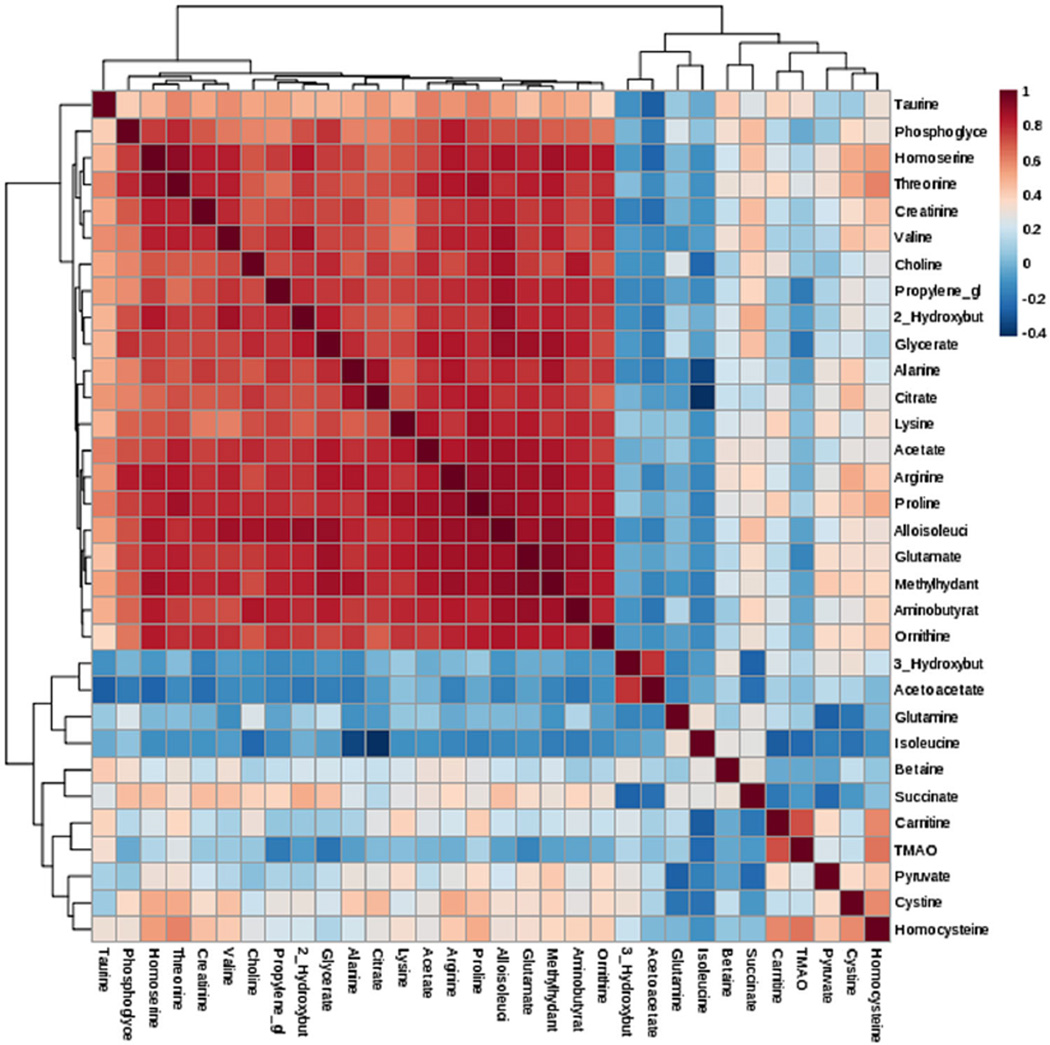
Correlations, as observed between the concentrations of metabolites in a biological sample, may be used to gain additional information about the physiological or patho-physiological state of the organism (Steuer et al. 2003). Next, we asked whether there is a correlation among metabolites within a pathway. Figure 4 shows the correlation heatmap among thirty-two metabolites in sarcoidosis subjects using the Pearson’s correlation and Spearman’s rank correlation along with hierarchical clustering using hclust function. Table 3 shows Pearson correlations between the metabolites, which were significantly expressed. We identified a positive correlation between 3-HB and acetoacetate (R = 0.6), these metabolites being important in the fatty acid β-oxidation pathway. Homocysteine strongly correlated with cystine (R = 0.7) and carnitine (R = 0.6), indicating a connection between the homocysteine pathway, and carnitine production (Table 3). Similarly, pyruvate positively correlated with carnitine (R = 0.4), TMAO (R = 0.4), and homocysteine (R = 0.4). The highest positive correlation was found between TMAO and carnitine (R = 0.8) and TMAO and homocysteine (R = 0.8). It has been shown that certain gut microbiota metabolize l-carnitine into trimethylamine (TMA), which can be further oxidized to TMAO, a highly atherogenic metabolite (Koeth et al. 2013). In addition, even though taurine was not significantly different between the two groups, it positively correlated with carnitine (R = 0.5) and TMAO (R = 0.5). It is important to note that taurine is an intermediate in the methylamine/homocysteine pathway.
Fig. 4.
Correlation and Clustering result of 32 metabolites in form of a Heatmap. Clustering was performed using the hclust function in package stat, MetaboAnalyst 3.0 software. Hierarchical cluster analysis, each sample begins as a separate cluster and the algorithm proceeds to combine them until all samples belong to one cluster. Two parameters need to be considered when performing hierarchical clustering. The first one is similarity measure-Euclidean distance, Pearson’s correlation, Spearman’s rank correlation. The other parameter is clustering algorithms, including average linkage (clustering uses the centroids of the observations), complete linkage (clustering uses the farthest pair of observations between the two groups), single linkage (clustering uses the closest pair of observations) and Ward’s linkage (clustering to minimize the sum of squares of any two clusters)
Table 3.
Correlation of metabolites found to be altered in patients with sarcoidosis as compared with healthy controls
| 3-HB | Carnitine | TMAO | Homocysteine | |
|---|---|---|---|---|
| Acetoacetate | 0.78** | 0.1 | 0.3 | 0.0 |
| Pyruvate | 0.29 | 0.37* | 0.27 | 0.43* |
| Isoleucine | −0.2 | −0.44* | −0.28 | −0.38* |
| Carnitine | 0.25 | 1 | 0.74** | 0.59** |
| Cystine | 0.32 | 0.18 | 0.22 | 0.63** |
| Homocysteine | 0.16 | 0.59** | 0.7** | 1 |
| Taurine | −0.05 | 0.52** | 0.5** | 0.28 |
Values represent Pearson correlation, N = 15.
p < 0.05;
p < 0.01.
SPSS software
3-HB 3-Hydroxybutyrate, TMAO trimethylamine N-oxide
Isoleucine was negatively correlated with carnitine (R = −0.4), homocysteine (R = −0.4), and TMAO (R = −0.4). Similarly, succinate was negatively correlated with 3-HB (R = −0.5) and pyruvate (R = −0.5), indicating an alteration in energy production and the TCA cycle in sarcoidosis (Table 3). Moreover, succinate was positively correlated with decreased levels of isoleucine (R = 0.4), an amino acid that after deamination and decarboxylation yields succinyl-CoA to feed into the TCA cycle (Nelson and Cox 2012). A significant correlation among the metabolites within these pathways suggests concordance among metabolites measured in the sera of sarcoidosis subjects.
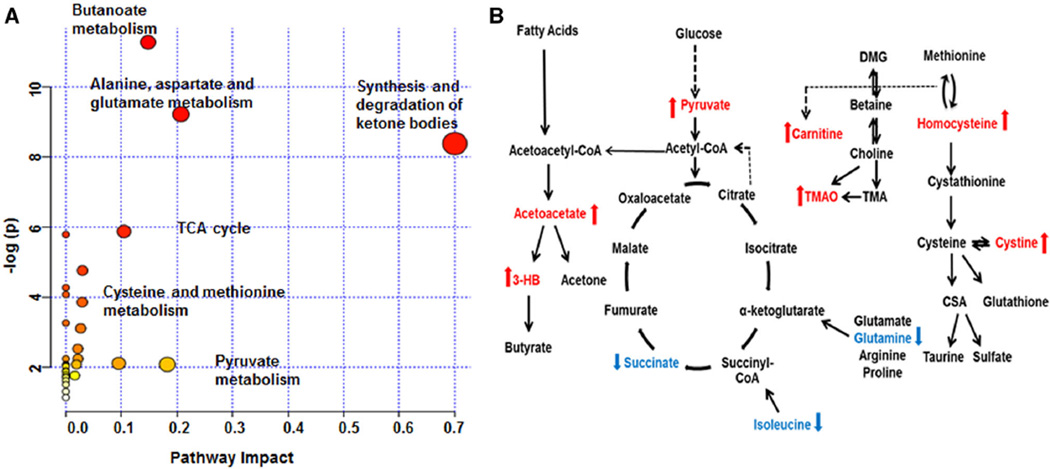
To identify metabolic network(s) and biological relevance of the identified metabolic de-arrangements in sarcoidosis, the MetaboAnalyst 3.0 software (Xia and Wishart 2011b) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used. The significant pathways identified by MetaboAnalyst 3.0 include butanoate metabolism, alanine, aspartame and glutamate metabolism, and the synthesis and degradation of ketone bodies. In addition, the TCA cycle, tyrosine, propanoate, and phenylalanine metabolism, cysteine and methionine metabolism, and pyruvate metabolism are involved (Table 4; Fig. 5a). The impact of the metabolic pathways is determined by the number of hits, false discovery rate (FDR), and p-value (Table 4). Similar, using KEGG database, we identified three major pathways altered in this disease: (1) fatty acid β-oxidation, (2) glycolysis/TCA cycle, and (3) homocysteine/methylamine (Fig. 5b).
Table 4.
MetaboAnalyt 3.0 identified highly significant pathways
| Pathway name | Hits | p-value | −log(p) | FDR |
|---|---|---|---|---|
| Butanoate metabolism | 4 | 1.2797E–5 | 11.266 | 0.0010237 |
| Alanine, aspartate and glutamate metabolism | 3 | 9.9926E–5 | 9.2111 | 0.0039971 |
| Synthesis and degradation of ketone bodies | 2 | 2.3105E–4 | 8.3729 | 0.0061613 |
| Citrate (TCA) cycle | 2 | 0.002837 | 5.865 | 0.049516 |
| Tyrosine metabolism | 3 | 0.0030948 | 5.778 | 0.049516 |
| Phenylalanine metabolism | 2 | 0.013984 | 4.2699 | 0.15981 |
| Cysteine and methionine metabolism | 2 | 0.021227 | 3.8525 | 0.02938 |
| Arginine and proline metabolism | 2 | 0.038491 | 3.2573 | 0.30793 |
Pathway name, hits, significance, −log(p) as well as false discovery rate (FDR)
Fig. 5.
Schematic networks detected in sarcoidosis patients illustrating the most predominant altered metabolic pathways and the biochemical linkage among the metabolites detected in sarcoidosis. a MetaboAnalyst 3.0 output. NMR data were uploaded into MetaboAnalyst 3.0 software and significant metabolic pathways arranged according to the scores from the enrichment analysis (y axis) and from the topology analysis (x axis). b The three major pathways include fatty acid metabolism, glycolysis/TCA cycle, and homocysteine/methylamine pathway. The metabolites that are significantly increased in sarcoidosis patients are depicted in red, whereas the metabolites that are reduced are in blue (p < 0.05). Dashed arrows indicate multiple step processes
4 Discussion
The “omics” technologies are primarily aimed at the universal detection of genomics, transcriptomics, proteomics, and recently metabolomics in a specific biological sample. Metabolites are the end products of cellular processes and considered as the ultimate response of an organism to genetic and environmental factors, representing the actual biological phenotype (Urbanczyk-Wochniak et al. 2003). Metabolomics is a rapidly evolving technique that simultaneously studies large numbers of small molecules and can aid to complete the knowledge gap in understanding the physiology of an organism. Evaluation of metabolomics in human chronic inflammatory diseases is still in the early stage. To our knowledge this study is the first to evaluate metabolomics in sarcoidosis using 1H NMR spectroscopy.
Sarcoidosis is an inflammatory disease of unknown etiology and is characterized by granuloma formation in different organs with an increase in T-helper type 1 (Th1)—mediated cytokines. The presence of activated macrophages and the expansion of oligoclonal T and B cells suggest a sustained activation of inflammatory pathways in this disease (Iannuzzi et al. 2007; Rastogi et al. 2011). Recently, it has been recognized that metabolic reprogramming is a prerequisite for inflammation and immunological responses to diverse stimuli, both in macrophages and lymphocytes (Tannahill et al. 2013; Wang et al. 2011a; Sinclair et al. 2013). However, most of these studies were done in animal models or isolated macrophages and lymphocytes (Tannahill et al. 2013). How metabolic reprogramming might relate to sustained inflammation in chronic inflammatory diseases or respiratory diseases, including sarcoidosis, has not been studied. To decrease confounding factors such as chronic respiratory diseases, we only enrolled subjects, who were recently diagnosed with symptomatic sarcoidosis, not on any treatment and had only minimal abnormalities in pulmonary function testing.
We investigated the metabolomic profiles of patients with sarcoidosis compared to healthy controls by analyzing serum metabolites using 1H NMR spectroscopy. Using an untargeted approach, a complete scan of the metabolomic profile was performed using NMR Suite Chenomx software. As a cross-validation, the chemical shift of the altered metabolites matched the regions identified by the PCA loading plot and VIP analysis, confirming that these metabolites represent a novel signature and might be associated with the overall pathobiology of this disease. Applying a web based metabolomics data analysis tool, MetaboAnalyst 3.0 (Xia and Wishart 2011a) to enrich and identify metabolic pathways and relevant networks, several significant pathways were detected (Fig. 5; Table 4). Major aberrant pathways with high impact were as following, butanoate metabolisms, synthesis and degradation of ketone bodies, glutamate metabolism, TCA cycle and cysteine metabolism. Several pathways shared compounds, for instance network analysis identified several significant amino acid pathways as indicated by low p-value and low FDR, such as alanine, phenylalanine, tyrosine as well as arginine, glutamate and proline. We summarized the alterations in three critical pathways: (1) fatty acid metabolism (β-oxidation), (2) glycolysis/TCA cycle, and (3) the homocysteine/methylamine pathways in sarcoidosis patients (Fig. 5b). Furthermore, both MetaboAnalyst 3.0 and the KEGG data base analyses of our data pointed towards aberrant microbiota and colon microbiome, identifying abnormalities in butanoate metabolism and methylamine (TMAO). Gut microbiota, especially anaerobic bacteria are the major producer of butyrate (Louis et al. 2004). Interestingly, butyrate has been shown to be important in the modulation of T cell immunity and it affects histone acetylation (Furusawa et al. 2013, Donohoe et al. 2012). In addition, metabolism of l-carnitine by intestinal microbiota has been recently linked to increased levels of inflammation and cardiovascular risk (Koeth et al. 2013). Wang et al. have shown that metabolism of choline and phosphatidylcholine, as well as dietary l-carnitine (abundant in red meat) by gut microbiota leads to production of the hepatic metabolite trimethylamine (TMA), which is further oxidized to trimethylamine N-oxide (TMAO). That study showed that generation of TMAO from dietary TMA is dependent on the composition of intestinal microbiota, as suppression of gut microbiota with oral broad-spectrum antibiotics led to a decrease in plasma TMAO (19). Both TMA and TMAO are found to be directly linked to the composition of the intestinal microbiota and are considered biomarkers for inflammation and atherosclerosis (Koeth et al. 2013, Wang et al. 2011b). Taken together, our untargeted metabolomic analysis revealed elevated TMAO levels and abnormalities in butanoate metabolism in patients with sarcoidosis raising concern for an association of abnormal gut microflora and chronic inflammation in these patients.
Several metabolites associated with the fatty acid β–oxidation pathway were increased in patients with sarcoidosis, including 3-HB, acetoacetate, and carnitine. Initial steps of β–oxidation occur in the cytoplasm, where fatty acid oxidation is catalyzed through acyl CoA ligase (Nelson and Cox 2012), and then transported into the mitochondria via an acyl-carnitine intermediate (McClelland 2004). In the mitochondria, saturated acyl-CoA is degraded by oxidation at the β-carbon. An increase in ketone bodies (3-HB, acetoacetate) suggests activation of mitochondrial β-oxidation as a marker for lipolysis (Fig. 5b) in sarcoidosis. Further, it is known that acylcarnitines originate as intermediates during transport of substrates across mitochondrial membranes and increased concentrations could indicate an increased flux into the β-oxidation pathway (Bhuiyan et al. 1992). Additionally, there is a possibility that the butanoate pathway through aberrant gut microbiota contributes to increased ketone body production, as it has been shown that acetate is normally regarded as an end product of anaerobic fermentation and is an integral part of the butyrate pathway (Duncan et al. 2004).
In addition, the level of carnitine was significantly increased (p < 0.05) in sarcoidosis patients, indicating that its higher concentration is not the limiting factor of transfer of acetyl-CoA or its analogs into the mitochondria for further utilization via the TCA cycle or β-oxidation. Recently O’Sullivan et al., using an isolated murine T cell system, have shown that differentiation of memory T cells requires fatty acid oxidation to generate ATP for development and survival (Weinberg and Chandel 2014; O’Sullivan et al. 2014). In their system, murine memory T cells display a distinct metabolic profile as compared to effector T cells. Sarcoidosis is characterized by increased local and circulating inflammatory cytokines, the presence of non-caseating granuloma and a lack of cutaneous reaction to tuberculin skin testing (anergy) (Iannuzzi et al. 2007). Anergy in sarcoidosis is associated with lymphatic proliferation, as these patients often present with generalized lymphoadenopathy and splenomegaly (Iannuzzi et al. 2007). How the metabolism, including β-oxidation, contributes to this immunological paradox is not known.
Likewise, choline and its metabolite, betaine, are methyl donors, and are metabolically linked to transmethylation pathways, including synthesis of the cardiovascular disease (CVD) risk factor homocysteine. Our results did not show a significant difference in choline and betaine concentrations, but homocysteine and cystine were significantly increased in sarcoidosis patients, indicating an alteration in the transmethylation pathway. High levels of homocysteine were also previously associated with alteration in immune cell function, perhaps inducing T cell and monocyte activation, resulting in T cell differentiation into a Th1 phenotype (Dawson et al. 2004).
Our metabolomic analysis revealed a significant increase in the level of pyruvate suggesting activation of the glycolytic pathway. Because it is known that macro-phage activation leads to glycolysis and enhanced anaerobic metabolism, we expected to have increased levels of lactate in our patients. Although, the lactate levels were increased in our patients, they did not reach statistical significance (p > 0.05). Surprisingly, we found significantly decreased levels of succinate in sarcoidosis patients compared to healthy controls (p = 0.003). The levels of other TCA intermediates were not significantly different in the two groups. Pyruvate plays a central role in human metabolism. Two key enzymes regulate pyruvate metabolism (1) pyruvate dehydrogenase and (2) pyruvate car-boxylase, by which pyruvate is transformed to acetyl-CoA or through carboxylation of pyruvate to form oxaloacetate (Nelson and Cox 2012). Both can be shuttled through the TCA cycle. It was reported that during hypoxia or inflammation, citrate plays an important feature of the metabolic switch regulating mitochondrial dysfunction (Tannahill et al. 2013). Citrate can be converted into cytosolic acetyl-CoA without passing through the conventional clockwise steps of the TCA cycle (O’Neill and Hardie 2013). Acetyl-CoA generated by this pathway can function as a precursor for fatty acid synthesis (Pearce et al. 2013; O’Neill and Hardie 2013). These alternative pathways may contribute to restoration of other TCA intermediates (malate, oxaloacetate and citrate) and could explain their normal levels compared to succinate. Additionally, isoleucine and glutamine which feed into TCA cycle to generate succinate were similarly decreased (Table 2B).
It has been shown that macrophages in response to Tolllike receptor 4 (TLR4) activation switch their metabolism from oxidative phosphorylation to glycolysis, leading to rapid accumulation of succinate (Tannahill et al. 2013). The same study showed that such increased succinate is dependent on glutamine. We found increased glycolysis as indicated by increased pyruvate levels, but this was not associated with an increase in succinate. In contrast, succinate and important branched amino acids such as isoleucine and glutamine were decreased. These findings in sarcoidosis patients indicate a disturbance in the amino acid and energy metabolism and might provide a connection with the overall decreased energy levels and elevated fatigue noted at clinical diagnosis (Lower et al. 2013).
Glutamine is an abundant free amino acid in the circulation and in the intracellular amino acid pool of most tissues. Previously, it has been shown that glutamine is beneficial in severe metabolic stress, by decreasing inflammation and improving the gut barrier function (De-Souza and Greene 2005; Singleton et al. 2005b; Wischmeyer et al. 2003). It has been proposed that the anti-inflammatory effects of glutamine are due to a decrease in cytokine release and nuclear factor-kB activation (Singleton et al. 2005a). In addition, glutamine is an important amino acid that is known to be a key nutrient for T lymphocytes (Nakaya et al. 2014). Recently, it has been reported that the ASC amino acid transporter 2 (ASCT2) is a key regulator of glutamine uptake in CD4+ Th cells and influences the development of pro-inflammatory Th1 and Th 17 responses in vitro and in vivo (Nakaya et al. 2014). The cell-mediated immune-response depends on the availability of glutamine, which can directly impact several metabolic programs required for T cell proliferation and function (Nakaya et al. 2014). Interestingly, one previous study showed increased glutamine levels in subjects with chronic obstructive pulmonary disease (Ubhi et al. 2012), in contrast patients with sarcoidosis exhibited low levels of glutamine, in addition to increased TMAO levels possibly due to altered intestinal microbiota, we speculate that these metabolites may contribute to the high inflammatory state and disturbed energy metabolism found in these patients. These data augment our findings that sarcoidosis patients exhibit an increase in β-oxidation pathway along with decreased intermediates of the TCA cycle, such as succinate.
There is a paucity of the studies addressing the role of metabolomics in respiratory diseases. There are only a few studies examining the metabolomics in human samples in chronic respiratory diseases (Ubhi et al. 2012). The current study is a novel approach in the field of sarcoidosis and unravels metabolic pathways affected in these patients. Several metabolites found to be altered in the present study have been previously associated with atherosclerosis and CVD, indicating a connection between these inflammatory diseases and sarcoidosis. These findings support previous studies that reported the presence of proatherogenic lipoprotein particles in sarcoidosis patients (Vekic et al. 2013; Ivanisevic et al. 2012). Future work should include a larger population sample and other chronic inflammatory diseases, such as lupus and cystic fibrosis, to confirm the specificity of the metabolites identified in this study.
5 Conclusion
In summary, the metabolomic profiling of patients with sarcoidosis revealed changes in the energy production, homocysteine, and amino acid metabolism, as well as a potential role of aberrant gut microbiota. The number of potential pathways involved underscores the complexity of the metabolic reactions to systemic inflammatory response. We view the observed changes as global perturbations and the interpretation of the resulting pattern of correlations between metabolites as a global fingerprint of the physiological or pathophysiological state. This altered pattern of correlations, in addition to changes in average metabolite levels may indicate changes in the underlying state of the system (Steuer et al. 2003). Therefore, metabolomics has great potential to become a critical tool used to gain more insight into the molecular cascades that lead to the patho-genesis of sarcoidosis. Ongoing studies are set to validate and complement the initial discoveries, leading to a novel and non-invasive methodology for the study of sarcoidosis and other chronic diseases.
Acknowledgments
This work was supported by the Department of Medicine and the Center for Molecular Medicine and Genetics, Wayne State University School of Medicine (LS) and National Institute of Health R01HL113508 (LS).
Abbreviations
- ASCT2
Amino acid transporter 2
- BMI
Body mass index
- CD4+
Cluster of differentiation 4
- D2O
Deuterium oxide
- DSS
Disodium-2,2-dimethyl 2-silapentane-5-sulphonate
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NMR
Nuclear magnetic resonance
- NOESY
Nuclear Overhauser effect spectroscopy
- PC
Principal component
- PCA
Principal component analysis
- PLS-DA
Partial least square discriminant analysis
- SIMCA
Soft independent modeling of class analogy
- TCA
Tricarboxylic acid cycle
- Th1
T-helper type 1
- TLR4
Toll-like receptor 4
- TMA
Trimethylamine
- TMAO
Trimethylamine N-oxide
- VIP
Variable importance in the projection
Footnotes
Author contribution
AG contributed to the study design, participated in sample collection, conducted the analysis, interpreted the data and drafted the manuscript. SVG provided access and contributed to multivariate data analysis, as well as assisted with the preparation of the manuscript. CB participated in data interpretation and preparation of the manuscript. LS designed the study, and participated in all areas of the research, data analysis and writing of the manuscript. All authors have read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest AG, SVG, CB, and LS declare that they have no conflict of interest.
References
- Altermann E, Klaenhammer TR. PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics. 2005;6:60. doi: 10.1186/1471-2164-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barallobre-Barreiro J, Chung YL, Mayr M. Proteomics and metabolomics for mechanistic insights and biomarker discovery in cardiovascular disease. Revista Española de Cardiología (English Edition) 2013;66:657–661. doi: 10.1016/j.rec.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Bhuiyan AK, Jackson S, Turnbull DM, Aynsley-Green A, Leonard JV, Bartlett K. The measurement of carnitine and acyl-carnitines: application to the investigation of patients with suspected inherited disorders of mitochondrial fatty acid oxidation. Clinica Chimica Acta. 1992;207:185–204. doi: 10.1016/0009-8981(92)90118-a. [DOI] [PubMed] [Google Scholar]
- Chen G, Li D, Jin Y, Zhang W, Teng L, Bunt C, Wen J. Deformable liposomes by reverse-phase evaporation method for an enhanced skin delivery of (+)-catechin. Drug Development and Industrial Pharmacy. 2014;40:260–265. doi: 10.3109/03639045.2012.756512. [DOI] [PubMed] [Google Scholar]
- Dawson H, Collins G, Pyle R, Deep-Dixit V, Taub DD. The immunoregulatory effects of homocysteine and its intermediates on T-lymphocyte function. Mechanisms of Ageing and Development. 2004;125:107–110. doi: 10.1016/j.mad.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Defeo EM, Wu CL, McDougal WS, Cheng LL. A decade in prostate cancer: from NMR to metabolomics. Nature Reviews Urology. 2011;8:301–311. doi: 10.1038/nrurol.2011.53. [DOI] [PubMed] [Google Scholar]
- De-Souza DA, Greene LJ. Intestinal permeability and systemic infections in critically ill patients: effect of glutamine. Critical Care Medicine. 2005;33:1125–1135. doi: 10.1097/01.ccm.0000162680.52397.97. [DOI] [PubMed] [Google Scholar]
- Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Molecular Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, Flint HJ. Contribution of acetate to butyrate formation by human faecal bacteria. British Journal of Nutrition. 2004;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gregory JF, III, Park Y, Lamers Y, Bandyopadhyay N, Chi YY, Lee K, et al. Metabolomic analysis reveals extended metabolic consequences of marginal vitamin B-6 deficiency in healthy human subjects. PLoS ONE. 2013;8:e63544. doi: 10.1371/journal.pone.0063544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunninghake GW, Costabel U, Ando M, Baughman R, Cordier JF, du Bois R, et al. ATS/ERS/WASOG statement on sarcoidosis. American Thoracic Society/European Respiratory Society/World Association of Sarcoidosis and other Granulomatous Disorders. Sarcoidosis, Vasculitis, and Diffuse Lung Diseases. 1999;16:149–173. [PubMed] [Google Scholar]
- Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. New England Journal of Medicine. 2007;357:2153–2165. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- Ivanisevic J, Kotur-Stevuljevic J, Stefanovic A, Jelic-Ivanovic Z, Spasic S, Videnovic-Ivanov J, et al. Dyslipidemia and oxidative stress in sarcoidosis patients. Clinical Biochemistry. 2012;45:677–682. doi: 10.1016/j.clinbiochem.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Jayavelu ND, Bar NS. Metabolomic studies of human gastric cancer: Review. World Journal of Gastroenterology. 2014;20:8092–8101. doi: 10.3748/wjg.v20.i25.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature Medicine. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. Journal of Bacteriology. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower EE, Malhotra A, Sudurlescu V, Baughman RP. Sarcoidosis, fatigue, and sleep apnea. Chest. 2013;144:1976–1977. doi: 10.1378/chest.13-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Xie G, Jia W, Jia W. Metabolomics in human type 2 diabetes research. Frontiers of Medicine. 2013;7:4–13. doi: 10.1007/s11684-013-0248-4. [DOI] [PubMed] [Google Scholar]
- McClelland GB. Fat to the fire: The regulation of lipid oxidation with exercise and environmental stress. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2004;139:443–460. doi: 10.1016/j.cbpc.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger principles of biochemistry. New York: W.H. Freeman; 2012. [Google Scholar]
- Ni Y, Su M, Lin J, Wang X, Qiu Y, Zhao A, et al. Metabolic profiling reveals disorder of amino acid metabolism in four brain regions from a rat model of chronic unpredictable mild stress. FEBS Letters. 2008;582:2627–2636. doi: 10.1016/j.febslet.2008.06.040. [DOI] [PubMed] [Google Scholar]
- Noga MJ, Dane A, Shi S, Attali A, van Aken H, Suidgeest E, et al. Metabolomics of cerebrospinal fluid reveals changes in the central nervous system metabolism in a rat model of multiple sclerosis. Metabolomics. 2012;8:253–263. doi: 10.1007/s11306-011-0306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- O’Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: Insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi R, Du W, Ju D, Pirockinaite G, Liu Y, Nunez G, Samavati L. Dysregulation of p38 and MKP-1 in response to NOD1/TLR4 stimulation in sarcoid bronchoalveolar cells. American Journal of Respiratory and Critical Care Medicine. 2011;183:500–510. doi: 10.1164/rccm.201005-0792OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlich CA, Tarlo SM, Hankinson JL, Townsend MC, Eschenbacher WL, von Essen SG, et al. Official American Thoracic Society technical standards: Spirometry in the occupational setting. American Journal of Respiratory and Critical Care Medicine. 2014;189:983–993. doi: 10.1164/rccm.201402-0337ST. [DOI] [PubMed] [Google Scholar]
- Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nature Immunology. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton KD, Beckey VE, Wischmeyer PE. Glutamine prevents activation of NF-kappaB and stress kinase pathways, attenuates inflammatory cytokine release, and prevents acute respiratory distress syndrome (ARDS) following sepsis. Shock. 2005a;24:583–589. doi: 10.1097/01.shk.0000185795.96964.71. [DOI] [PubMed] [Google Scholar]
- Singleton KD, Serkova N, Banerjee A, Meng X, Gamboni-Robertson F, Wischmeyer PE. Glutamine attenuates endotoxin-induced lung metabolic dysfunction: Potential role of enhanced heat shock protein 70. Nutrition. 2005b;21:214–223. doi: 10.1016/j.nut.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Steuer R, Kurths J, Fiehn O, Weckwerth W. Interpreting correlations in metabolomic networks. Biochemical Society Transactions. 2003;31:1476–1478. doi: 10.1042/bst0311476. [DOI] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-Mcdermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo E, Davis C, Milner J. Nutrigenomics, proteomics, metabolomics, and the practice of dietetics. Journal of the American Dietetic Association. 2006;106:403–413. doi: 10.1016/j.jada.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Ubhi BK, Riley JH, Shaw PA, Lomas DA, Tal-Singer R, Macnee W, et al. Metabolic profiling detects biomarkers of protein degradation in COPD patients. European Respiratory Journal. 2012;40:345–355. doi: 10.1183/09031936.00112411. [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Luedemann A, Kopka J, Selbig J, Roessner-Tunali U, Willmitzer L, Fernie AR. Parallel analysis of transcript and metabolic profiles: A new approach in systems biology. EMBO Reports. 2003;4:989–993. doi: 10.1038/sj.embor.embor944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekic J, Zeljkovic A, Jelic-Ivanovic Z, Spasojevic-Kalimanovska V, Spasic S, Videnovic-Ivanov J, et al. Distribution of low-density lipoprotein and high-density lipoprotein subclasses in patients with sarcoidosis. Archives of Pathology and Laboratory Medicine. 2013;137:1780–1787. doi: 10.5858/arpa.2012-0299-OA. [DOI] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011a;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011b;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tso VK, Slupsky CM, Fedorak RN. Metabolomics and detection of colorectal cancer in humans: A systematic review. Future Oncology. 2010;6:1395–1406. doi: 10.2217/fon.10.107. [DOI] [PubMed] [Google Scholar]
- Weinberg SE, Chandel NS. Futility sustains memory T cells. Immunity. 2014;41:1–3. doi: 10.1016/j.immuni.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Yang HJ, An YJ, Kim JM, Lee DH, Jin X, et al. Enhanced phase II detoxification contributes to beneficial effects of dietary restriction as revealed by multi-platform metabolomics studies. Molecular and Cellular Proteomics. 2013;12:575–586. doi: 10.1074/mcp.M112.021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischmeyer PE, Jayakar D, Williams U, Singleton KD, Riehm J, Bacha EA, et al. Single dose of glutamine enhances myocardial tissue metabolism, glutathione content, and improves myocardial function after ischemia-reperfusion injury. JPEN. Journal of Parenteral and Enteral Nutrition. 2003;27:396–403. doi: 10.1177/0148607103027006396. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Current Protocols in Bioinformatics, Chapter. 2011a;14 doi: 10.1002/0471250953.bi1410s34. Unit 14 10. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nature Protocols. 2011b;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]