Abstract
Recent findings suggest that Alzheimer's disease (AD) is a disconnection syndrome characterized by abnormalities in large-scale networks. However, the alterations that occur in network topology during the prodromal stages of AD, particularly in patients with stable mild cognitive impairment (MCI) and those that show a slow or faster progression to dementia, are still poorly understood. In this study, we used graph theory to assess the organization of structural MRI networks in stable MCI (sMCI) subjects, late MCI converters (lMCIc), early MCI converters (eMCIc), and AD patients from 2 large multicenter cohorts: ADNI and AddNeuroMed. Our findings showed an abnormal global network organization in all patient groups, as reflected by an increased path length, reduced transitivity, and increased modularity compared with controls. In addition, lMCIc, eMCIc, and AD patients showed a decreased path length and mean clustering compared with the sMCI group. At the local level, there were nodal clustering decreases mostly in AD patients, while the nodal closeness centrality detected abnormalities across all patient groups, showing overlapping changes in the hippocampi and amygdala and nonoverlapping changes in parietal, entorhinal, and orbitofrontal regions. These findings suggest that the prodromal and clinical stages of AD are associated with an abnormal network topology.
Keywords: closeness centrality, clustering, modularity, structural covariance networks, transitivity
Introduction
Alzheimer's disease (AD) is a devastating neurodegenerative disorder that slowly deprives individuals of their memories and other essential cognitive functions, including executive, visuospatial abilities and language. There is increasing evidence showing that the progression of these cognitive symptoms occurs in an orderly fashion, which reflects the accumulation of amyloid deposits and the spatial distribution of neurofibrillary tangles (Frisoni et al. 2010). For instance, while memory loss takes place at early disease stages, reflecting the presence of tangles in medial temporal regions (Scahill et al. 2002; Thal et al. 2002; Thompson et al. 2004); aphasia and apraxia typically occur at later stages, reflecting the spread of tangles and plaques into neocortical areas (Braak et al. 2006; McDonald et al. 2009). This incremental spread of pathology between interconnected brain regions suggests that AD might be a disconnection syndrome characterized by abnormalities in brain networks (Pievani et al. 2011).
Graph theory has become a very useful tool in neuroimaging to assess the relationship between different brain regions and their organization into large-scale networks (Filippi et al. 2013). Using this method, previous studies have shown that healthy individuals have an efficient network topology, combining a high level of integration between distant brain regions (short path length) and a good local communication between neighboring areas (high clustering). This small-world configuration (Humphries et al. 2006) is thought to be specially suited for cognitive processing (Bassett and Bullmore 2006) and has been shown in previous functional MRI, structural MRI, electroencephalogram, and diffusion tensor imaging (DTI) studies (Achard and Bullmore 2007; Ferri et al. 2007; Bassett et al. 2008; Iturria-Medina et al. 2008; Gong et al. 2009). The combination of high efficiency and clustering in a small-world architecture is an attractive principle of brain network organization as it could deliver both segregated and integrated information processing (Bullmore and Sporns 2012). For instance, visual processing is a segregated process that would benefit from the clusters of connections between regions that are topologically close, whereas executive processing is an integrated process that would benefit from the long paths that allow transferring information across the whole network (Bullmore and Sporns 2012). In addition, it has been shown that regions showing high clustering tend to be very well connected to areas with the same functional specialization (Sporns and Betzel 2016). In line with this, several studies have shown that the brain is organized into communities or modules of interconnected regions (Hagmann et al. 2008; Meunier et al. 2009), which may correspond to large-scale brain systems such as the executive control, the dorsal attention, and the default-mode network. This network property is commonly referred to as modularity and has been shown in healthy individuals by previous studies (Chen et al. 2008; He et al. 2009).
Graph theory can be applied to different neuroimaging modalities to assess brain networks in vivo in AD, including DTI, structural, and functional MRI (for reviews, see Tijms, Moller, et al. 2013; Tijms, Wink, et al. 2013; Dai and He 2014) as well as positron emission tomography (PET) (Sanabria-Diaz et al. 2013; Sepulcre et al. 2013; Yao et al. 2015). Using structural MRI, a few studies have shown that AD patients present changes in global network organization compared with healthy controls (Dai and He 2014; Phillips et al. 2015); however, there is surprisingly little agreement about the nature of these changes: while some studies showed an increased path length and clustering in the networks of AD patients, others found decreases or no changes at all (Li et al. 2012; Dai and He 2014). Regarding network configuration, some studies using structural MRI have found a preserved small-world organization in AD patients, while others found random or regular network topologies (Dai and He 2014). This inconsistency between studies could be due to the inclusion of small samples of patients with heterogeneous clinical characteristics. Despite this interest in assessing brain connectivity in AD, to date no studies have assessed modularity in the structural MRI networks of AD patients.
Amnestic mild cognitive impairment (MCI) is a transition state between normal aging and AD with a high risk of progression to dementia (Petersen et al. 1999). Although MCI has been associated with reduced white matter integrity and abnormal functional connectivity (Medina et al. 2006; Zhang et al. 2007; Petrella et al. 2011; Binnewijzend et al. 2012), it is not clear whether these patients also present altered brain network topology: Yao et al. (2010) found no differences in the path length or clustering between MCI patients and controls, while Phillips et al. (2015) found differences that varied according to the network construction method. There is increasing evidence showing that MCI patients progress to AD at a rate of approximately 15% per year (Grundman et al. 2004). The assessment of network organization in stable MCI patients and those who show a slow or faster progression to dementia is important as it could provide important clues into which network changes mark the transition to AD and improve our understanding on the effects of disease progression on brain networks.
The aim of the current study was to establish the nature of structural abnormalities in the organization of brain networks in stable MCI (sMCI) subjects, patients who show a slow progression to dementia (late MCI converters, lMCIc), patients who show a fast progression to dementia (early MCI converters, eMCIc), and AD patients using graph theory. To achieve this goal, we assessed over 1000 patients and controls from 2 large multicenter cohorts: the Alzheimer's Disease Neuroimaging Initiative (ADNI) and the AddNeuroMed study. We calculated various global and local network measures, including the characteristic path length, the mean clustering coefficient, the small-worldness, the nodal clustering, and the nodal closeness centrality. In addition, in contrast to previous studies, we calculated for the first time the transitivity and modularity in the structural MRI networks of MCI and AD patients. These graph theory measures reflect how well a region is connected to its neighbouring areas and within brain modules, providing important information on the network's ability for specialized processing to occur within densely interconnected groups of brain regions (Rubinov and Sporns 2010). We hypothesized that global network measures would show abnormalities across all patient groups, with lMCIc, eMCIc, and AD patients showing more severe network changes compared with controls than sMCI patients. In addition, based on previous evidence showing that the sequence of brain abnormalities between regions of the default-mode network is reminiscent of the spread of tangle pathology in AD (Buckner et al. 2005, 2009), we hypothesized that patients would show changes in local network measures in the regions of this network.
Methods
Subjects
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.usc.edu) and the AddNeuroMed study. In total, 1008 subjects were included, consisting of 301 controls, 425 MCI, and 282 AD patients. Regarding MCI patients, 87 converted to AD after 1 year (eMCIc), 71 converted to AD after 3 years (lMCIc), and 110 remained stable after 3 years (sMCI). In addition, 157 MCI patients remained stable after 1 year but had no additional follow-ups after that period. We classified these subjects as sMCI-1y and compared them with the other groups in a supplementary analysis. Eight subjects were excluded from the previous groups due to uncertain diagnosis.
The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. In the ADNI cohort, the inclusion criteria for the control group were Mini-Mental State Examination (MMSE) scores between 24 and 30, a Clinical Dementia Rating-Sum of Boxes (CDR-SB) score of 0, and lack of depression, MCI, or dementia. Inclusion criteria for the MCI group followed the Peterson criteria (Petersen et al. 1999) for amnestic MCI. AD participants met the National Institute for Neurological and Communicative Disorders and Stroke-Alzheimer's Disease and Related Disorder Association (NINDS/ADRDA) criteria for probable AD, had a MMSE score between 18 and 26, and a CDR-SB of 0.5–1.0. Exclusion criteria comprised history of structural brain lesions or head trauma, significant neurological disease other than incipient AD, and the use of psychotropic medications that could affect memory. For up-to-date information about the ADNI study, see www.adni-info.org.
AddNeuroMed is an Integrated Project funded by the European Union Sixth Framework program (Lovestone et al. 2007, 2009). AddNeuroMed aims to develop and validate novel surrogate markers of disease and treatment, based upon in vitro and in vivo models in animals and humans in AD. The neuroimaging part of AddNeuroMed uses MRI and magnetic resonance spectroscopy (MRS) to establish imaging markers for early diagnosis and detection of disease and efficacy of disease modifying therapy in man, as well as translational imaging biomarkers in animal models of AD. Human data were collected from 6 different sites across Europe: University of Kuopio (Finland), University of Perugia (Italy), Aristotle University of Thessaloniki (Greece), King's College London (United Kingdom), University of Lodz (Poland), and University of Toulouse (France) (Lovestone et al. 2009; Simmons et al. 2009, 2011). The inclusion criteria for the control group were Mini-Mental State Examination (MMSE) scores between 24 and 30, a CDR score of 0, 65 years, or above, and lack of depression, dementia, other neurological diseases, unstable systematic illnesses, or organ failure. The inclusion criteria for MCI patients were similar to the control group except for the CDR score of 0.5 and report of memory problems by the patient or informant. AD patients met the NINDS/ADRDA and DSM-IV criteria for probable AD, had a MMSE score between 12 and 28, had 65 years or above, and did not have significant neurological or psychiatric illnesses other than AD, unstable systematic illnesses, or organ failure.
MRI Acquisition
Data acquisition for the AddNeuroMed study was designed to be compatible with ADNI (Jack et al. 2008; Simmons et al. 2009, 2011). In particular, all participants, both from ADNI and AddNeuroMed, were scanned on a 1.5 Tesla MRI system using a sagittal 3D T1-weighted MPRAGE sequence: repetition time (TR) = 9–13 ms; echo time (TE) = 3.0–4.1 ms; inversion time (IT) = 1000 ms; flip angle (FA) = 8°; voxel size = 1.1 × 1.1 × 1.2 mm3. Images from ADNI were acquired in 58 sites, while images from AddNeuroMed were acquired in 5 sites or centers. We have combined these 2 cohorts in several previous studies (Spulber et al. 2013; Falahati et al. 2016), showing that they present similar patterns of atrophy and predictive power in discriminating patients with AD or MCI from controls (Westman et al. 2011).
Image Preprocessing
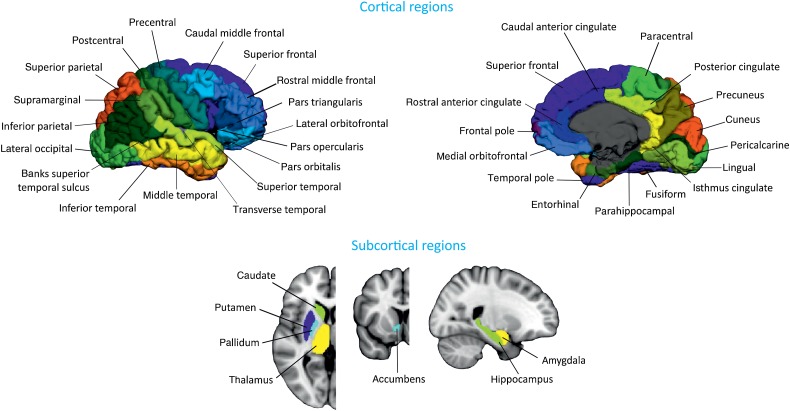
All T1-weighted images were preprocessed using the FreeSurfer software, version 5.1. Briefly, preprocessing included: correction of motion artifacts and spatial distortions due to gradient nonlinearity and B1 field inhomogeneity; removal of nonbrain tissue using a hybrid watershed/surface deformation procedure (Segonne et al. 2004); automated transformation into the Talairach standard space; intensity normalization (Sled et al. 1998); tessellation of the gray/white matter boundary; automated topology correction (Segonne et al. 2007); and surface deformation following intensity gradients to optimally place the gray/white and gray/CSF borders at the location where the greatest shift in intensity defines the transition to the other tissue class (Fischl and Dale 2000). Once the cortical models were complete, registration to a spherical atlas took place, which utilizes individual cortical folding patterns to match cortical geometry across subjects (Fischl et al. 1999). This was followed by parcellation of the cerebral cortex into 68 cortical regions using the atlas by Desikan et al. (2006) (Fig. 1). In addition to these 68 regions, 7 subcortical structures were also included: hippocampus, amygdala, thalamus, caudate, putamen, accumbens, and pallidum (Fig. 1). All data was preprocessed through the HiveDB database system (Muehlboeck et al. 2014). We excluded 8 subjects that were outliers in cortical thickness and subcortical volume measures.
Figure 1.
Brain regions used in network construction and analysis. The cortical thickness and subcortical volumes were extracted from these regions for every subject.
Network Construction and Analysis
For every cortical region, a linear regression was performed to control for the effects of age, gender, and education (He et al. 2007). The same procedure was carried out for subcortical regions including intracranial volume as an additional covariate. To ensure that our results were not influenced by the fact that subjects were scanned at different centers, we included scanning site as an additional covariate in a supplementary analysis.
The residuals of these regressions were used to substitute the raw values and build the structural covariance networks. In these networks, every node corresponded to a brain region and the edges represented the correlations between them (He et al. 2007). In this study, both cortical thicknesses and subcortical volumes were used to build the structural networks due to previous evidence showing the involvement of cortical and subcortical regions in MCI and AD and similarly to previous graph theory studies (Hosseini et al. 2013; Pereira et al. 2015).
For every group, we built an 82 × 82 association matrix, where each entry was defined as the Pearson correlation coefficient between corrected anatomical measures of every pair of regions across participants (Fig. 2). For each association matrix, a binary matrix was computed, where the correlation coefficient was considered 1 if it was above a threshold and 0 if it was below (He et al. 2007). Since thresholding association matrices of different groups may yield networks with a different number of nodes and edges, we thresholded the association matrices at a range of network densities D (D min = 5% to D max = 35%, in steps of 1%) and compared the network topologies across that range. For densities below 5%, the number of edges was inferior to the number of nodes, corresponding to a widely disconnected network. For D above 35%, the networks became similar to random graphs and showed a small-world index close to 1. All self-connections and negative correlations were excluded from the analyses.
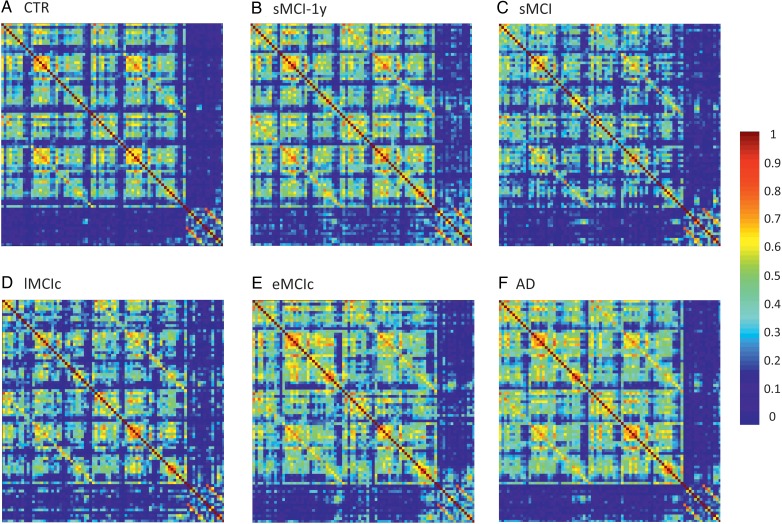
Figure 2.
Structural correlation matrices for (A) controls (CTR), (B) patients with stable mild cognitive impairment after 1 year (sMCI-1y), (C) patients with sMCI (after 3 years), (D) lMCIc, (E) eMCIc, and (F) AD patients. In these matrices, the first rows and columns correspond to the correlations between cortical regions, while the last ones correspond to the correlations between subcortical areas. The color bar indicates the strength of the correlation coefficients: warmer colors represent stronger correlations, while colder colors represent weaker correlations.
To detect differences between groups in global network topology, we calculated the characteristic path length, the mean clustering coefficient, the transitivity, the modularity, and the small-worldness. The characteristic path length is the average shortest path length between all pairs of nodes in the network and indicates how easy it is (on average) to reach a node from any other node in the network (Watts and Strogatz 1998); this measure was calculated only within connected components of the network as implemented in the formulas by Rubinov and Sporns (2010). The clustering coefficient is the average over the whole network of the fraction of a node's neighbors that are also neighbors of each other; it reflects how well the nodes are connected to nearby regions forming clusters (Watts and Strogatz 1998). The transitivity is similar to the clustering coefficient but, instead of being normalized individually by every node, it is normalized by the whole network and is not influenced by nodes with a low degree (Newman 2003). The modularity describes the extent to which a network can be divided into modules or communities of regions with a large number of within-modules connections and a minimal number of between-module connections (Newman 2006). The small-worldness is a measure of how much a network is locally interconnected compared with a random network but still retaining global connectivity between distant brain regions (Watts and Strogatz 1998; Humphries et al. 2006).
To assess differences between groups in regional network topology, we calculated the nodal clustering and the closeness centrality. We selected these 2 nodal network measures, because they are sensitive to different aspects of network topology and remain largely unexplored in MCI and AD. Specifically, the nodal clustering is a measure of segregation, which reflects the ability for specialized information processing to occur within groups of brain regions, while the closeness centrality is a measure of interaction that reflects the ability to combine information from distributed brain areas (Rubinov and Sporns 2010). The nodal clustering is calculated as the mean clustering coefficient but only for a given node. The closeness centrality is the inverse of the average shortest path length from 1 node to all other nodes in the network. To compare the roles of the nodes in each module and their differences between groups, we also calculated the within-module degree and participation coefficient. The within-module degree measures the connectivity of the node within the module compared with the other nodes in the same module. The participation coefficient expresses how strongly a node is connected to other modules and tends to 1 if a node has a homogeneous connection distribution with all the modules and 0 if it does not have any intermodule connections (Guimera and Amaral 2005; Guimera et al. 2005).
The formulas that were used to calculate the global and nodal graph theory measures are provided by Rubinov and Sporns (2010). We used BrainNet Viewer (http://www.nitrc.org/projects/bnv/) for network visualization (Xia et al. 2013).
Comparison of Network Measures Between Groups
We tested the statistical significance of the differences between groups using nonparametric permutation tests with 1000 permutations (Bassett et al. 2008; He et al. 2008). In each permutation, the corrected anatomical values of every subject were randomly reassigned to one of a pair of groups with the same number of subjects as in the original groups. Then, an association matrix was built for each pair of randomized groups, and the binary matrices were calculated at a range of network densities. The network measures were calculated at each density, and the differences between the new randomized groups were computed. This randomization procedure was repeated 1000 times for every density value, and the 95% confidence intervals (CI) of each distribution were used as critical values for a 2-tailed test of the null hypothesis at P < 0.05. To correct the nodal network results for multiple comparisons, we used a false discovery rate (FDR) procedure (Genovese et al. 2002) at a q value of 0.05.
Results
The characteristics of the sample can be found in Table 1. There were significant differences in gender and years of education between the groups (P < 0.001). For this reason, the cortical thickness and subcortical measures were corrected by these variables in the current study. As expected, all patient groups showed lower MMSE scores (P < 0.001) and higher CDR scores (P < 0.001) compared with controls. In addition, lMCIc, eMCIc, and AD patients showed lower MMSE scores compared with sMCI patients (P < 0.05). AD patients showed worse MMSE scores compared with sMCI, lMCIc, eMCIc patients (P < 0.05), and sMCI-1y patients had lower MMSE scores compared with sMCI patients (P < 0.05).
Table 1.
Characteristics of the sample
| CTR (n = 301) | sMCI-1y (n = 157) | sMCI (n = 110) | lMCIc (n = 71) | eMCIc (n = 87) | AD (n = 282) | F or χ2 (P value) | |
|---|---|---|---|---|---|---|---|
| Age | 75.1 (5.7) | 75.0 (6.5) | 74.7 (7.5) | 74.8 (7.0) | 74.1 (6.7) | 75.6 (7.0) | 0.9 (0.475) |
| Gender (m/f)a,b,c,d,e | 156/145 | 88/69 | 74/36 | 43/28 | 54/33 | 130/152 | 19.2 (0.002) |
| Education (y)f,a,g,h,i,j,k,l,c,m,d,e | 14.2 (4.4) | 11.8 (5.2) | 15.7 (3.0) | 16.1 (3.0) | 13.9 (4.2) | 12.2 (4.9) | 21.2 (<0.001) |
| MMSEf,a,g,n,h,i,b,o,l,c,d,e | 29.1 (1.1) | 27.0 (1.6) | 27.6 (1.7) | 26.7 (1.7) | 26.6 (1.8) | 22.4 (3.5) | 272.8 (<0.001) |
| CDRf,a,g,n,h,b,c,d,e | 0 | 0.5 | 0.5 | 0.5 | 0.5 | 0.9 (0.4) | 880.2 (<0.001) |
Note: Means are followed by standard deviations. Differences in age, years of education, and MMSE scores were assessed using an analysis of variance (ANOVA). Differences in CDR scores were assessed using a Kruskal–Wallis test and differences in gender were assessed using a χ2 test. CTR, controls; sMCI, stable mild cognitive impairment after 1 year (sMCI-1y) or 3 years (sMCI); lMCIc, late mild cognitive impairment converters; eMCIc, early mild cognitive impairment converters; AD, Alzheimer's disease; MMSE, mini-mental state examination; CDR, clinical dementia rating scale.
aSignificant differences between CTR and sMCI patients (P < 0.05).
bSignificant differences between sMCI-1y and AD patients (P < 0.05).
cSignificant differences between sMCI and AD patients (P < 0.05).
dSignificant differences between lMCIc and AD patients (P < 0.05).
eSignificant differences between eMCIc and AD patients (P < 0.05).
fSignificant differences between CTR and sMCI-1y patients (P < 0.05).
gSignificant differences between CTR and lMCIc patients (P < 0.05).
hSignificant differences between CTR and AD patients (P < 0.05).
iSignificant differences between sMCI-1y and sMCI patients (P < 0.05).
jSignificant differences between sMCI-1y and lMCIc patients (P < 0.05).
kSignificant differences between sMCI-1y and eMCIc patients (P < 0.05).
lSignificant differences between sMCI and eMCIc patients (P < 0.05).
mSignificant differences between lMCIc and eMCIc patients (P < 0.05).
nSignificant differences between CTR and eMCIc patients (P < 0.05).
oSignificant differences between sMCI and lMCIc patients (P < 0.05).
Global Network Analysis
The weighted correlation matrices for each group are presented in Figure 2. We observed that the correlation patterns of all groups showed strong correlations between bilaterally homologous regions.
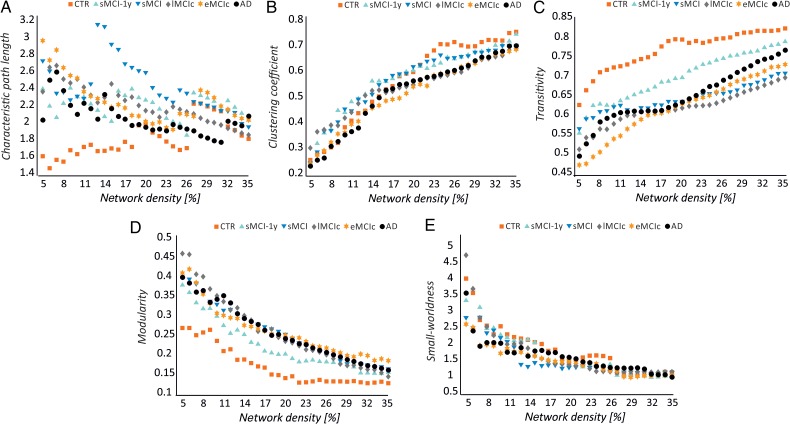
In general, with progressively higher values of network density (D), the characteristic path length and modularity decreased, the mean clustering coefficient and transitivity increased, and a small-world topology was observed across all groups (Fig. 3).
Figure 3.
Changes in global network measures as a function of network density. Characteristic path length (A), clustering coefficient (B), transitivity (C), modularity (D), and small-worldness (E) for controls (CTR), patients with stable mild cognitive impairment after 1 year (sMCI-1y), patients with sMCI (after 3 years), lMCIc, eMCIc, and AD patients.
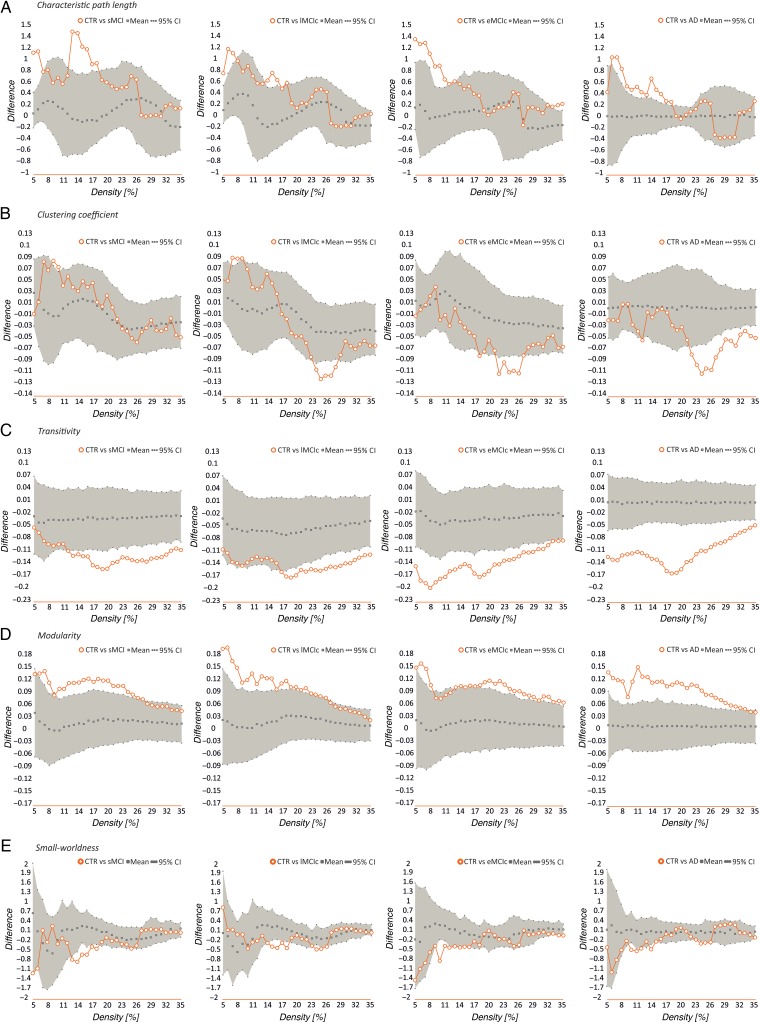
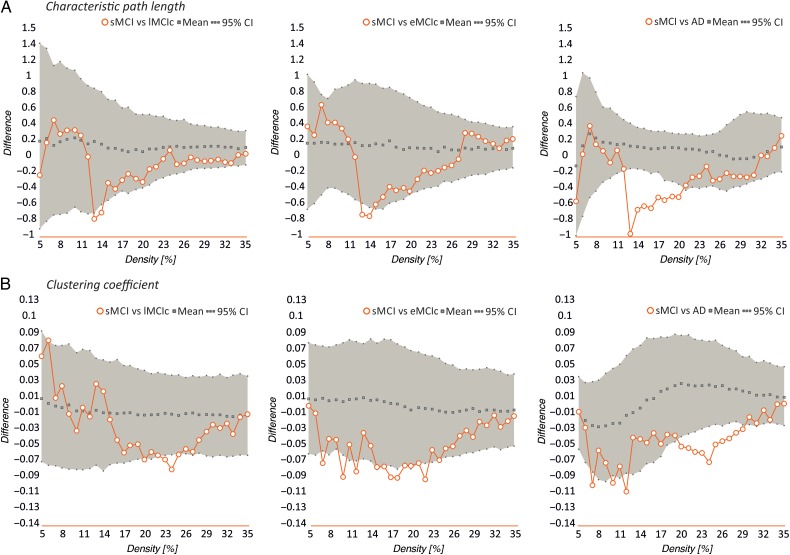
Our statistical analyses showed significant increases in the characteristic path length in the sMCI, lMCIc, eMCIc, and AD groups compared with controls at several network densities (P range, 0.043–0.001) (Fig. 4). The clustering coefficient also showed significant changes, being decreased across different densities in lMCIc, eMCIc, and AD (P range, 0.043–0.001) but not in sMCI patients, compared with controls. The transitivity and modularity showed the greatest differences between patients and controls: the transitivity was significantly decreased (P range, 0.049–0.001) and the modularity was significantly increased (P range, 0.049–0.001) in patients at most network densities (Fig. 4). We also found significant decreases in the small-worldness in the patient groups compared with controls (P range, 0.040–0.010); however, these differences were only observed at a few network thresholds (Fig. 4).
Figure 4.
Differences between controls, sMCI, lMCIc, eMCIc, and AD patients in global network measures. Plots showing the differences between controls (CTR) and sMCI patients; CTR and lMCIc; CTR and eMCIc; CTR and AD patients in the characteristic path length (A), clustering coefficient (B), transitivity (C), modularity (D), and small-worldness (E). The plots show the upper and lower bounds of the 95% confidence intervals (CI) (in gray) and the differences in the network measures between groups (in orange circles) as a function of network density. If these differences fall outside the CIs, there is a statistical significant difference at P < 0.05.
When we compared the different patient groups, we observed that lMCIc, eMCIc, and AD patients had a decreased characteristic path length (P range, 0.048–0.001) and clustering coefficient (P range, 0.042–0.001) compared with sMCI patients (Fig. 5). There were no significant differences in transitivity or modularity between these groups. In addition, there were no differences in global network topology between lMCIc, eMCIc, and AD patients.
Figure 5.
Differences between sMCI, lMCIc, eMCIc, and AD patients in global network measures. Plots showing the differences between sMCI and lMCIc patients; sMCI and eMCIc patients; sMCI and AD patients in the characteristic path length (A) and clustering coefficient (B). The plots show the upper and lower bounds of the 95% confidence intervals (CI) (in gray) and the differences in the network measures between groups (in orange circles) as a function of network density. If these differences fall outside the CIs, there is a statistical significant difference at P < 0.05.
The global network comparisons carried out in the sMCI-1y group can be found in Supplementary Figure 1. These patients showed evidence of larger paths (P range, 0.045–0.004) and changes in the clustering (P range, 0.044–0.018) at a few network thresholds, but no changes in the transitivity or modularity compared with controls, in contrast to the other MCI groups. They also showed higher clustering compared with eMCIc (P range, 0.046–0.018) and AD patients (P range, 0.043–0.016) and higher transitivity (P range, 0.048–0.019) compared with AD patients at several densities. The modularity and small-worldness did not show significant changes in sMCI-1y compared with the other patient groups.
To assess whether our results were influenced by different scanning centers, we repeated the analyses comparing controls to the patient groups after including the centers as an additional covariate. These analyses showed similar differences in the characteristic path length, mean clustering, transitivity, and modularity between patients and controls, suggesting that the differences in scanning sites did not influence our results (see Supplementary Table 1).
Nodal Network Analysis
We identified several changes in nodal network measures between groups. In summary, the nodal clustering was decreased in patients compared with controls and showed widespread changes only in the AD group. The nodal closeness centrality was decreased in the bilateral hippocampi and amygdala across all patient groups and showed increases in medial parietal, medial temporal, and limbic regions that varied according to the patient group. Below, we describe these changes in greater detail.
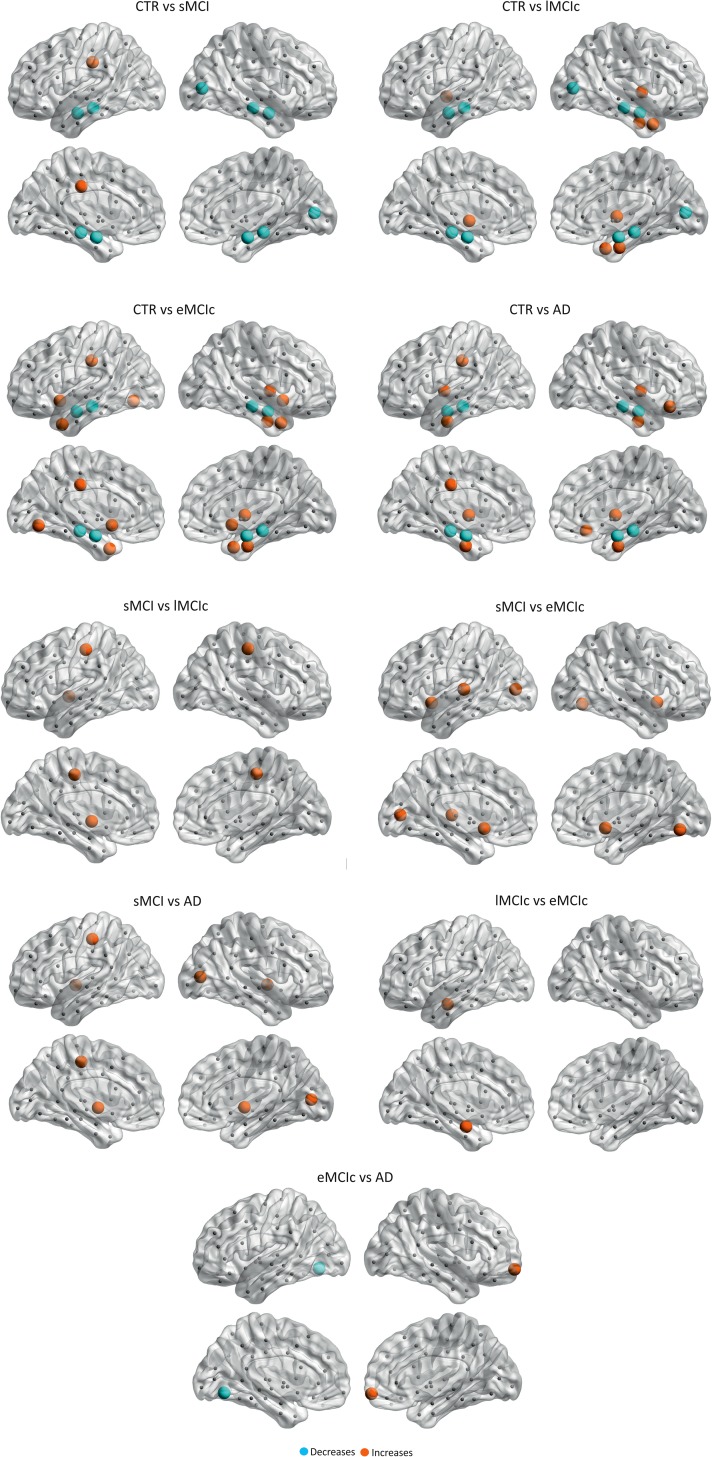
After correcting for multiple comparisons (FDR, q < 0.05), we observed that sMCI patients showed clustering decreases in the left superior frontal gyrus, while eMCIc patients showed decreases in the right postcentral gyrus compared with controls. In AD patients, the nodal clustering decreases involved several regions: the bilateral precuneus, superior frontal gyri, lateral orbitofrontal gyri, middle temporal gyri, inferior temporal gyri, fusiform, hippocampi, and amygdala; the left pars triangularis gyrus, postcentral gyrus; and the right caudal middle frontal gyrus, pars opercularis gyrus, and lateral occipital gyrus (Fig. 6 and Table 2). AD patients also showed significant clustering decreases in the left postcentral gyrus compared with sMCI patients (FDR, q < 0.05).
Figure 6.
Significant decreases in the nodal clustering coefficient in sMCI, eMCIc, and AD patients. CTR, controls; sMCI, stable MCI; eMCIc, early MCI converters, AD, Alzheimer's disease. The regions showing clustering decreases in patients are listed in Table 2.
Table 2.
Significant differences in the nodal clustering coefficient between groups (FDR-corrected)
| Region | CTR | sMCI | P value |
|---|---|---|---|
| Lh Superior frontal G | 0.83 | 0.50 | 0.002 |
| CTR | eMCIc | ||
| Rh Postcentral | 0.94 | 0.56 | 0.001 |
| CTR | AD | ||
| Lh Superior frontal G | 0.83 | 0.57 | 0.001 |
| Lh Lateral orbitofrontal G | 0.81 | 0.51 | 0.001 |
| Lh Pars triangularis G | 0.90 | 0.65 | 0.001 |
| Lh Postcentral G | 0.96 | 0.68 | 0.001 |
| Lh Precuneus | 0.78 | 0.58 | 0.001 |
| Lh Middle temporal G | 0.78 | 0.62 | 0.017 |
| Lh Inferior temporal G | 0.94 | 0.64 | 0.002 |
| Lh Fusiform | 0.79 | 0.56 | 0.004 |
| Lh Hippocampus | 1 | 0.33 | 0.001 |
| Lh Amygdala | 1 | 0.33 | 0.001 |
| Rh Superior frontal G | 0.78 | 0.52 | 0.001 |
| Rh Caudal middle frontal G | 0.92 | 0.77 | 0.009 |
| Rh Lateral orbitofrontal G | 0.71 | 0.45 | 0.003 |
| Rh Pars opercularis G | 0.96 | 0.62 | 0.001 |
| Rh Precuneus | 0.80 | 0.62 | 0.017 |
| Rh Lateral occipital G | 0.74 | 0.60 | 0.007 |
| Rh Middle temporal G | 0.81 | 0.61 | 0.016 |
| Rh Inferior temporal G | 0.90 | 0.66 | 0.003 |
| Rh Fusiform | 0.84 | 0.53 | 0.001 |
| Rh Hippocampus | 1 | 0.33 | 0.001 |
| Rh Amygdala | 1 | 0.33 | 0.001 |
| sMCI | AD | ||
| Lh Postcentral | 0.95 | 0.68 | 0.001 |
Note: CTR, controls; sMCI, stable mild cognitive impairment; lMCIc, late mild cognitive impairment converters; eMCIc, early mild cognitive impairment converters; AD, Alzheimer's disease; Lh, left hemisphere; Rh, right hemisphere; G, gyrus.
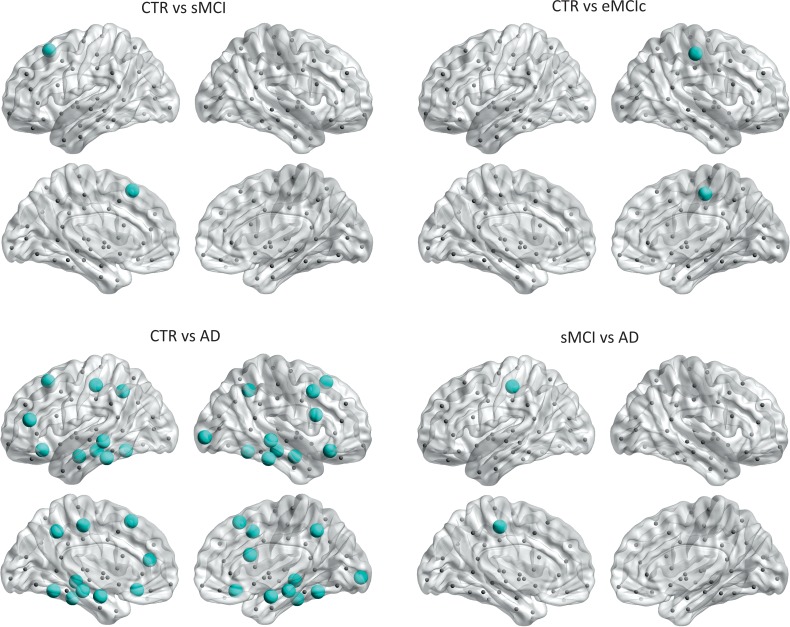
Regarding the closeness centrality, in addition to the significant decreases found in the hippocampi and amygdala in sMCI, lMCIc, eMCI, and AD patients compared with controls, there were additional decreases in the right pericalcarine gyrus in sMCI and lMCIc patients; the right accumbens in eMCIc patients. The increases in closeness centrality were observed in the left posterior cingulate in sMCI patients; the left pallidum, right insula, right temporal pole, right entorhinal in lMCIc patients; the left posterior cingulate, right lateral orbitofrontal gyrus, bilateral insula, bilateral entorhinal in AD patients (Fig. 7 and Table 3).
Figure 7.
Significant differences in the closeness centrality between controls and sMCI, lMCIc, eMCIc, and AD patients. CTR, controls; sMCI, stable mild cognitive impairment; lMCIc, late MCI converters; eMCIc, early MCI converters; AD, Alzheimer's disease. The regions showing significant closeness centrality decreases are colored in blue, while the regions showing closeness centrality increases in patients are colored in orange. These regions are listed in Table 3.
Table 3.
Significant differences in the nodal closeness centrality between groups (FDR-corrected)
| Region | CTR | sMCI | P value |
|---|---|---|---|
| Lh Posterior cingulate | 0.30 | 0.48 | 0.001 |
| Lh Hippocampus | 1 | 0.29 | 0.001 |
| Lh Amygdala | 1 | 0.34 | 0.001 |
| Rh Pericalcarine | 0.38 | 0.28 | 0.001 |
| Rh Hippocampus | 1 | 0.38 | 0.001 |
| Rh Amygdala | 1 | 0.33 | 0.001 |
| CTR | lMCIc | ||
| Lh Pallidum | 0.39 | 1 | 0.001 |
| Lh Hippocampus | 1 | 0.39 | 0.001 |
| Lh Amygdala | 1 | 0.34 | 0.001 |
| Rh Insula | 0.28 | 0.53 | 0.001 |
| Rh Pericalcarine | 0.38 | 0.30 | 0.001 |
| Rh Temporal pole | 0.29 | 0.46 | 0.001 |
| Rh Entorhinal | 0.22 | 0.45 | 0.001 |
| Rh Hippocampus | 1 | 0.44 | 0.001 |
| Rh Amygdala | 1 | 0.33 | 0.001 |
| CTR | eMCIc | ||
| Lh Posterior cingulate | 0.30 | 0.54 | 0.001 |
| Lh Lingual G | 0.43 | 0.56 | 0.001 |
| Lh Temporal pole | 0.39 | 0.49 | 0.001 |
| Lh Hippocampus | 1 | 0.26 | 0.001 |
| Lh Amygdala | 1 | 0.35 | 0.001 |
| Lh Accumbens | 0.36 | 1 | 0.001 |
| Rh Insula | 0.28 | 0.49 | 0.001 |
| Rh Temporal pole | 0.29 | 0.45 | 0.001 |
| Rh Entorhinal | 0.22 | 0.46 | 0.001 |
| Rh Hippocampus | 1 | 0.32 | 0.001 |
| Rh Amygdala | 1 | 0.32 | 0.001 |
| Rh Accumbens | 0.54 | 1 | 0.001 |
| CTR | AD | ||
| Lh Insula | 0.38 | 0.51 | 0.001 |
| Lh Posterior cingulate | 0.30 | 0.55 | 0.001 |
| Lh Entorhinal | 0.29 | 0.44 | 0.001 |
| Lh Hippocampus | 1 | 0.31 | 0.001 |
| Lh Amygdala | 1 | 0.31 | 0.001 |
| Rh Lateral orbitofrontal G | 0.47 | 0.58 | 0.006 |
| Rh Insula | 0.28 | 0.53 | 0.001 |
| Rh Entorhinal | 0.22 | 0.44 | 0.001 |
| Rh Hippocampus | 1 | 0.31 | 0.001 |
| Rh Amygdala | 1 | 0.31 | 0.001 |
| sMCI | lMCIc | ||
| Lh Postcentral G | 0.41 | 0.58 | 0.005 |
| Lh Pallidum | 0.21 | 1 | 0.001 |
| Rh Postcentral | 0.41 | 0.61 | 0.001 |
| sMCI | eMCIc | ||
| Lh Pericalcarine | 0.26 | 0.41 | 0.001 |
| Lh Transverse temporal G | 0.40 | 0.54 | 0.001 |
| Lh Accumbens | 0.21 | 1 | 0.001 |
| Rh Lingual G | 0.32 | 0.50 | 0.001 |
| Rh Accumbens | 0.21 | 1 | 0.001 |
| sMCI | AD | ||
| Lh Postcentral G | 0.41 | 0.60 | 0.001 |
| Lh Pallidum | 0.21 | 1 | 0.001 |
| Rh Pericalcarine | 0.28 | 0.37 | 0.001 |
| Rh Pallidum | 0.21 | 1 | 0.001 |
| lMCIc | eMCIc | ||
| Lh Accumbens | 0.30 | 1 | 0.001 |
| eMCIc | AD | ||
| Lh Lingual | 0.55 | 0.43 | 0.001 |
| Rh Frontal pole | 0.34 | 0.45 | 0.001 |
Note: CTR, controls; sMCI, stable mild cognitive impairment; lMCIc, late mild cognitive impairment converters; eMCIc, early mild cognitive impairment converters; AD, Alzheimer's disease; Lh, left hemisphere; Rh, right hemisphere; G, gyrus.
There were also significant differences in closeness centrality between the patient groups, mostly in temporal, occipital, and subcortical regions (for further details, see Table 3).
In Table 4, we present a summary of the most relevant global and nodal network results found in the current study.
Table 4.
Summary of the most relevant global and nodal network results
| Measures | CTR vs. sMCI | CTR vs. lMCIc | CTR vs. eMCIc | CTR vs. AD | sMCI vs. lMCIc | sMCI vs. eMCIc | sMCI vs. AD |
|---|---|---|---|---|---|---|---|
| Characteristic path length | ↑ | ↑ | ↑ | ↑ | ↓ | ↓ | ↓ |
| Clustering coefficient | — | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ |
| Transitivity | ↓ | ↓ | ↓ | ↓ | — | — | — |
| Modularity | ↑ | ↑ | ↑ | ↑ | — | — | — |
| Small-worldness | ↓ | ↓ | ↓ | ↓ | — | — | — |
| Nodal clustering | ↓ 1 region |
— | ↓ 1 region |
↓ 21 regions |
— | — | ↓ 1 region |
| Nodal closeness centrality | ↓ 5 regions |
↓ 5 regions |
↓ 4 regions |
↓ 4 regions |
— | — | — |
| ↑ 1 region |
↑ 4 regions |
↑ 8 regions |
↑ 6 regions |
↑ 3 regions |
↑ 5 regions |
↑ 4 regions |
Note: Compared with controls, all patient groups showed an increased path length and modularity as well as changes in the nodal closeness centrality. The mean clustering coefficient was decreased only in lMCIc, eMCIc, and AD groups, while the nodal clustering showed the most prominent changes in AD patients by being decreased in a total of 21 regions compared with controls. Compared with sMCI patients, the other patient groups showed a decreased path length, mean clustering coefficient, and increased closeness centrality. There were also nodal clustering decreases in 1 region in AD patients compared with sMCI patients.
Brain Modules
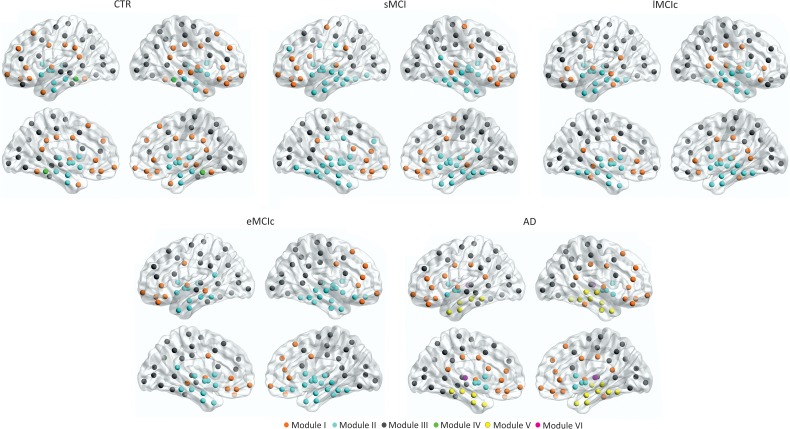
We identified 4 modules in controls; 3 modules in sMCI, lMCIc, and eMCIc patients; and 5 modules in AD patients (Fig. 8). For a full list of the regions belonging to each module, see Table 5.
Figure 8.
Brain modules in controls and sMCI, lMCIc, eMCIc, and AD patients. CTR, controls; sMCI, stable mild cognitive impairment; lMCIc, late MCI converters; eMCIc, early MCI converters; AD, Alzheimer's disease. Four modules were identified in the networks of CTR; 3 modules were identified in sMCI, lMCIc, and eMCIc patients; 5 modules were identified in the networks of AD patients. For each group, the left and right lateral (top) and medial (bottom) brain views are shown.
Table 5.
Brain modules in controls, sMCI, lMCIc, eMCIc, and AD patients
| Hemisphere | Brain region | Modules CTR | Modules sMCI | Modules lMCIc | Modules eMCIc | Modules AD |
|---|---|---|---|---|---|---|
| Left | Superiorfrontal | I | III | III | III | III |
| Left | Frontalpole | I | I | III | I | I |
| Left | Rostralmiddlefrontal | I | II | III | III | III |
| Left | Caudalmiddlefrontal | III | III | III | III | III |
| Left | Parsorbitalis | III | I | III | I | I |
| Left | Lateralorbitofrontal | I | I | I | I | I |
| Left | Parstriangularis | III | I | III | III | I |
| Left | Parsopercularis | III | I | III | III | I |
| Left | Medialorbitofrontal | I | I | I | I | I |
| Left | Rostralanteriorcingulate | I | I | I | I | I |
| Left | Caudalanteriorcingulate | I | II | I | I | I |
| Left | Insula | I | II | I | I | I |
| Left | Precentral | III | I | III | III | III |
| Left | Postcentral | III | III | III | III | III |
| Left | Supramarginal | I | I | III | III | III |
| Left | Superiorparietal | III | III | III | III | III |
| Left | Inferiorparietal | III | III | III | III | III |
| Left | Paracentral | III | III | III | III | III |
| Left | Posteriorcingulate | I | II | III | III | I |
| Left | Isthmuscingulate | I | II | I | III | I |
| Left | Precuneus | III | III | III | III | III |
| Left | Cuneus | III | III | III | III | III |
| Left | Pericalcarine | III | III | III | III | III |
| Left | Lingual | III | II | III | III | III |
| Left | Lateraloccipital | III | III | III | III | III |
| Left | Transversetemporal | III | I | III | III | III |
| Left | Bankssts | III | III | III | III | III |
| Left | Superiortemporal | III | II | II | III | III |
| Left | Middletemporal | III | II | I | I | III |
| Left | Inferiortemporal | III | II | I | III | III |
| Left | Temporalpole | I | II | II | II | V |
| Left | Entorhinal | II | II | II | II | V |
| Left | Parahippocampal | IV | II | II | II | V |
| Left | Fusiform | I | II | II | III | III |
| Left | Thalamus | II | II | II | II | VI |
| Left | Caudate | II | II | II | II | II |
| Left | Putamen | II | II | II | II | II |
| Left | Pallidum | II | II | II | II | II |
| Left | Hippocampus | II | II | II | II | V |
| Left | Amygdala | II | II | II | II | V |
| Left | Accumbens | II | II | II | II | II |
| Right | Superiorfrontal | I | III | III | I | I |
| Right | Frontalpole | I | I | III | I | I |
| Right | rostralmiddlefrontal | I | III | III | I | I |
| Right | Caudalmiddlefrontal | III | III | III | III | III |
| Right | Parsorbitalis | I | I | III | I | I |
| Right | Lateralorbitofrontal | I | I | I | II | I |
| Right | Parstriangularis | III | I | III | I | I |
| Right | Parsopercularis | III | III | III | III | III |
| Right | Medialorbitofrontal | I | I | III | II | I |
| Right | Rostralanteriorcingulate | I | I | I | I | I |
| Right | Caudalanteriorcingulate | I | I | I | I | I |
| Right | Insula | I | II | I | II | I |
| Right | Precentral | III | I | III | III | III |
| Right | Postcentral | III | III | III | III | III |
| Right | Supramarginal | I | III | III | III | III |
| Right | Superiorparietal | III | III | III | III | III |
| Right | Inferiorparietal | III | III | III | III | III |
| Right | Paracentral | III | III | III | III | III |
| Right | Posteriorcingulate | I | III | I | III | I |
| Right | Isthmuscingulate | I | III | I | II | I |
| Right | Precuneus | III | III | III | III | III |
| Right | Cuneus | III | III | III | III | III |
| Right | Pericalcarine | III | III | III | III | III |
| Right | Lingual | III | III | III | III | III |
| Right | Lateraloccipital | III | III | III | III | III |
| Right | Transversetemporal | III | I | I | III | III |
| Right | Bankssts | III | III | I | III | III |
| Right | Superiortemporal | I | II | II | II | V |
| Right | Middletemporal | I | I | II | II | V |
| Right | Inferiortemporal | III | II | I | II | I |
| Right | Temporalpole | I | II | II | II | V |
| Right | Entorhinal | II | II | II | II | V |
| Right | Parahippocampal | IV | II | II | II | V |
| Right | Fusiform | I | II | II | II | V |
| Right | Thalamus | II | II | II | II | VI |
| Right | Caudate | II | II | II | II | II |
| Right | Putamen | II | II | II | II | II |
| Right | Pallidum | II | II | II | II | II |
| Right | Hippocampus | II | II | II | II | V |
| Right | Amygdala | II | II | II | II | V |
| Right | Accumbens | II | II | II | II | II |
Note: CTR, controls; sMCI, stable mild cognitive impairment; lMCIc, late mild cognitive impairment converters; eMCIc, early mild cognitive impairment converters; AD, Alzheimer's disease.
Briefly, in controls, Module I included the superior frontal gyri, posterior cingulate, and supramarginal gyri, which are part of the default-mode network. Module II included the entorhinal gyri and subcortical regions. Module III was the largest, including several lateral frontal, parietal, and occipital regions. Module IV included the parahippocampal gyri.
In sMCI, lMCIc, and eMCIc patients, the modules were similar to those in controls. However, Module I did not include the superior frontal, lateral parietal, or posterior parietal regions and Module II included additional areas in the patient groups. In AD patients, Module II lost several regions that formed 2 new modules, which were not present in the other groups: one composed of the caudate, putamen, accumbens, and pallidum (Module IV) and the other composed of the bilateral thalami (Module V).
To assess differences between groups in the previous modules, we measured the within-module degree and participation coefficient. We found significant increases in the within-module degree and decreases in the participation coefficient in AD patients compared with controls, after FDR corrections (Table 6). The within-module degree increases were observed in the left postcentral, left superior parietal, right pars opercularis gyri, and right insula, which were part of Module I and Module III. The participation coefficient decreases were observed in the left pars orbitalis and bilateral cuneus, which were part of Module III. Although they did not survive correction for multiple comparisons, lMCIc and eMCIc patients also showed within-module degree increases and participation coefficient decreases in some of these regions, compared with controls (Table 6).
Table 6.
Differences in the within-module degree and participation coefficient between groups
| Within-module degree | CTR | lMCIc | P value |
|---|---|---|---|
| Region | |||
| Rh Pars opercularis G | −3.29 | 0.53 | 0.002 |
| Within-module degree | CTR | eMCIc | P value |
| Region | |||
| Lh Postcentral G | −0.35 | 1.22 | 0.004 |
| Lh Superior Parietal G | −0.35 | 0.99 | 0.017 |
| Within-module degree | CTR | AD | P value |
| Region | |||
| Lh Postcentral G | −1.12 | 0.70 | 0.001 |
| Lh Superior Parietal G | −1.12 | 0.89 | 0.002 |
| Lh Superior Temporal G | 0.39 | 1.86 | 0.003 |
| Rh Pars Opercularis G | −2.81 | 0.42 | 0.001 |
| Participation coefficient | CTR | AD | P value |
| Region | |||
| Lh Lateral Occipital G | 0.66 | 0.23 | 0.004 |
| Rh Postcentral G | 0.66 | 0.24 | 0.008 |
| Rh Lateral Occipital G | 0.66 | 0.32 | 0.001 |
Note: CTR, controls; sMCI, stable mild cognitive impairment; lMCIc, late mild cognitive impairment converters; eMCIc, early mild cognitive impairment converters; AD, Alzheimer's disease; Lh, left hemisphere; Rh, right hemisphere; G, gyrus. Differences between controls and AD patients survived corrections for multiple comparisons with FDR, while the other differences between groups were significant at an uncorrected level (P < 0.05).
Discussion
This study is the largest to date to assess network topology in MCI patients that remain stable, show a slow or fast progression to dementia as well as AD patients. Our findings revealed an abnormal organization in the networks of all patient groups as reflected by an increased path length, reduced transitivity, and increased modularity, compared with controls. The clustering coefficient showed a different pattern, being decreased in lMCIc, eMCIc, and AD but not in sMCI patients. Altogether, these findings suggest that the prodromal and clinical stages of AD are associated with a reduced ability to integrate information across distributed brain regions and an altered communication between neighboring areas and modules.
In the current study, we found a larger path length across all patient groups compared with controls, indicating an abnormal global network organization. Previous evidence suggests that a short path length ensures rapid information transmission across remote brain regions that are thought to be the basis of cognitive functioning (Sporns and Zwi 2004). The path length increases found in our study show that the distance between any 2 regions was greater in the patient's networks, making the communication between them less efficient. This finding is in line with those of previous graph theory studies in AD, which found an increased path length in the structural and functional networks of these patients (He et al. 2008; Lo et al. 2010; Sanz-Arigita et al. 2010; Yao et al. 2010; Shu et al. 2012). In one of these studies using DTI to build individual white matter networks, the path length increases were associated with worse MMSE performance (Shu et al. 2012), suggesting that this measure might indeed be a marker of cognitive dysfunction in AD.
Our analyses also showed decreases in the mean clustering coefficient in lMCIc, eMCIc, and AD patients relative to controls, indicating that there were fewer connections between neighboring areas in their networks. This result is in line with some (Tijms, Moller, et al. 2013; Tijms, Wink, et al. 2013) but not all previous studies (He et al. 2008; Yao et al. 2010), suggesting that differences in methodology, sample sizes, and patient characteristics might lead to different network findings. In a previous study, Li et al. (2012) found that MCI converters presented longitudinal decreases of the clustering coefficient suggesting that reductions in this network measure are associated with conversion to dementia in AD.
Furthermore, our study is the first to assess transitivity and modularity in the structural networks of MCI and AD patients. We found that these measures identified greater abnormalities in the networks of all patient groups compared with the path length or clustering, reaching significance across most network densities. Similarly to the clustering coefficient, the transitivity is a measure that reflects how well a region is integrated within its local cluster. However, in contrast to the clustering, the transitivity is less influenced by nodes with fewer connections (Rubinov and Sporns 2010), being a superior measure in networks with poorly connected nodes. Hence, we recommend the use of this measure in future studies assessing structural networks in amnestic MCI and AD as it offers greater sensitivity to the effects of the disease. The modularity is a more sophisticated measure that describes the existence of communities of regions within the network (Newman 2004). This network measure increases when brain regions are well connected within their module but are poorly connected with regions belonging to other modules. In the current study, we found significant modularity increases in sMCI, lMCIc, eMCIc, and AD patients compared with controls, indicating higher intramodule connectivity and lower connectivity between modules. This finding indicates that there is a worse communication between modules in patients, suggesting that their whole-brain networks were fragmented into a few large, isolated components. The within-module degree increases and participation coefficient decreases we found in frontal, parietal, and occipital regions in the patient groups compared with controls further confirm that the modules were well connected within themselves but not between each other in patients. In a previous fMRI study, significant modularity increases were also found in patients with Parkinson's disease with mild cognitive impairment, who have a higher risk of developing dementia (Baggio et al. 2014). These increases in modularity can be interpreted as an abnormal process by which the connections between brain areas belonging to a certain module increase, leaving the other modules relatively isolated. In that study, the abnormal modularity increases were associated with worse memory and visuospatial performance in Parkinson's patients, confirming they were pathological and related to greater clinical decline (Baggio et al. 2014). In our study, we also observed that, despite having similar modules to controls, the regions belonging to each module changed across the patient groups, with AD patients showing 2 modules that were not present in the other groups. Hence, our findings suggest that there is a reorganization of the modules in sMCI, lMCIc, eMCIc, and AD patients. Similarly to previous studies assessing modularity in structural MRI networks (Chen et al. 2008, 2011; Wu et al. 2012), we did not find an exact correspondence between the brain modules and previously reported resting-state fMRI networks (Greicius et al. 2004).
In addition to global network changes, we also observed alterations in the topology of specific brain regions. We found there were decreases in the nodal clustering of several areas in AD patients compared with controls, indicating worse local communication between neighboring areas. Some of these areas belonged to the default-mode network and included the precuneus and superior frontal gyri (Greicius et al. 2004), in line with our initial hypothesis and with previous studies showing pathological changes within this network in AD (Buckner et al. 2005, 2009; Li et al. 2013). In addition, we also observed clustering decreases in other frontal and temporal regions in AD patients, suggesting that the regional clustering changes were quite widespread, in line with evidence showing extended brain atrophy at advanced stages of AD (Lehmann et al. 2011). The MCI patient groups only showed clustering decreases in a few frontal and parietal regions, in contrast with the widespread changes observed in AD patients.
The nodal closeness centrality showed both increases and decreases in sMCI, lMCIc, eMCIc, and AD patients compared with controls. The decreases were mainly observed in the bilateral hippocampi and amygdala across all patient groups, indicating a loss of number of connections between these structures and other regions of the network. This could be related to white matter integrity loss or disruption of white matter fibers connecting these brain areas, which has been previously observed in MCI and AD patients in DTI studies (for a review, see Chua et al. 2008). The increases of closeness centrality were mainly observed in the posterior cingulate, temporal pole, entorhinal cortex, insula, and orbitofrontal regions. The closeness centrality is a measure of interaction between regions; the fact that it is increased in regions showing pathological changes in AD (Braak and Braak 1991; Thal et al. 2002; Frisoni et al. 2010) could be related to shared mechanisms in neurodegeneration (Zhu et al. 2012). Within the graph theory framework, 2 regions might correlate with each other not only if they are structurally or functionally connected but also if they become atrophied at the same rate (Alexander-Bloch et al. 2013). Thus, since medial temporal, medial parietal, and limbic regions show atrophy since early stages of AD, it seems natural that they might strongly interact with other regions in the network that become atrophied with disease progression.
In the current study, we also compared global and local network topology between the patient groups. We observed that sMCI patients had a larger path length and reduced nodal closeness centrality in several regions compared with the other patients, indicating that they presented greater abnormalities in the communication or interaction between distant brain areas. Previous evidence suggests that the initial pathological changes occurring in AD do not target regions that are close to each other but rather distant brain areas (Zhou et al. 2012), which are often connected by long and poorly myelinated axons. Hence, it is possible that these changes are more prominent in sMCI patients, which are potentially at earlier stages of AD. In contrast to the path length, the clustering coefficient was reduced in lMCIc, eMCIc, and AD patients compared with sMCI patients, suggesting that the loss of connections between neighboring areas reflects better the changes occurring in patients that are on the path to develop AD or already have dementia.
In the past few years, there has been increasing evidence showing that there is substantial heterogeneity among MCI patients. For instance, many MCI subjects remain stable for several years, while others show a fast progression to dementia and some can even fully reverse to normal cognition (Koepsell and Monsell 2012). Moreover, there are several non-AD pathologies that may produce amnestic MCI such as frontotemporal dementia (Yaffe et al. 2006), vascular dementia (Zanetti et al. 2006), and hippocampal sclerosis (Dickson et al. 1994). In the current study, we observed heterogeneity in the network topology abnormalities between the MCI groups. Specifically, sMCI-1y patients showed evidence of increased clustering and almost no changes in the transitivity and modularity compared with controls (see Supplementary Fig. 1), in contrast to the other MCI patients. It is possible that the sMCI-1y group included a mixture of subjects who remained stable, converted to dementia after a few years, had a non-AD related disorder or simply did not have any neurodegenerative disease (the cognitive deficits they presented were due to a transient medical condition). This heterogeneity might account for the lack of changes in the transitivity and modularity in sMCI-1y, in contrast to the other groups that were more homogeneous. Studies assessing network topology in MCI subjects should consider their results with respect to this important heterogeneity.
Although brain networks are sparse, current neuroimaging analyses build network representations that are continuous association matrices (Fornito et al. 2013). For this reason, many studies apply a threshold to these matrices in an attempt to retain the true brain connections and remove the potentially spurious ones. One way of applying a threshold is to retain the connections that overcome a level of significance. However, this approach will result in different groups of subjects having different numbers of edges or connections. In the current study, we applied a threshold to the connectivity matrices of each group by retaining the most significant connections, while ensuring an equal number of connections across groups. Although this step would ideally consist of applying a single threshold to the connectivity matrices of different groups, there is currently no absolute way of determining which threshold is best (Fornito et al. 2013). For this reason, we decided to test for group differences across a range of densities, similarly to previous studies (He et al. 2008; Yao et al. 2010). Since it does not make sense to compute topological measures in networks that have a random configuration, in the current study we defined the higher bound of this range using the small-world index, which indicates whether the networks are meaningfully organized. Our results showed that there were significant differences between groups at different densities, suggesting they were consistent.
We would like to highlight that the present study has some limitations. First, despite providing useful information, the analysis of structural covariance networks does not allow correlation analyses to be performed with clinical measures since there are no individual networks but only a network per group. Nevertheless, Tijms et al. (2012, 2016), Tijms, Moller, et al. (2013), and Tijms, Wink, et al. (2013) have overcome this limitation by providing a method that can create single-subject structural networks using structural MRI; this method could be considered in future graph theory studies assessing structural networks in large cohorts of AD and MCI patients. Secondly, we had limited longitudinal data regarding the clinical diagnosis of patients of only up to 3 years. Hence, it is possible that many of the individuals included in the sMCI group converted to dementia shortly after this period.
In conclusion, our study is the largest to date to assess structural network topology in stable MCI, progressive MCI, and AD by including 1008 patients and controls from 2 large multicenter cohorts. Our findings show, for the first time, that the transitivity and modularity are important graph theory measures that offer greater sensitivity to MCI and AD compared with the path length and clustering coefficient, which have been used more frequently in graph theory studies in AD. In addition, in contrast to previous studies, we provide a detailed description of nodal network changes in sMCI, lMCIc, eMCIc, and AD patients. Specifically, we show that while the nodal clustering showed widespread changes in AD patients, the closeness centrality detected alterations in several regions in all groups, showing overlapping changes in the hippocampi and amygdala and nonoverlapping changes in medial parietal and limbic areas in sMCI, lMCIc, eMCIc, and AD patients. These results offer an important glimpse into how AD progresses across different brain regions and ultimately leads to changes in global network organization.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This study was supported by InnoMed, (Innovative Medicines in Europe) an Integrated Project funded by the European Union of the Sixth Framework program priority FP6-2004-LIFESCIHEALTH-5, Life Sciences, Genomics and Biotechnology for Health. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. We would also like to thank the Swedish Foundation for Strategic Research (SSF), the Strategic Research Programme in Neuroscience at Karolinska Institutet (StratNeuro), Hjärnfonden, and Birgitta och Sten Westerberg for additional financial support. J.B.P. was funded by a Marie Curie fellowship for postdoctoral researchers (grant No. FP7-PEOPLE-2012-IEF-328758). Funding to pay the Open Access publication charges for this article was provided by the Swedish Foundation for Strategic Research (SSF).
Supplementary Material
Notes
Conflict of Interest: None declared.
References
- Achard S, Bullmore E. 2007. Efficiency and cost of economical brain functional networks. PLoS Comput Biol. 3(2):e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Giedd JN, Bullmore E. 2013. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 14(5):322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggio HC, Sala-Llonch R, Segura B, Marti MJ, Valldeoriola F, Compta Y, Tolosa E, Junque C. 2014. Functional brain networks and cognitive deficits in Parkinson's disease. Hum Brain Mapp. 35(9):4620–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E. 2006. Small-world brain networks. Neuroscientist. 12(6):512–523. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. 2008. Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 28(37):9239–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewijzend MA, Schoonheim MM, Sanz-Arigita E, Wink AM, van der Flier WM, Tolboom N, Adriaanse SM, Damoiseaux JS, Scheltens P, van Berckel BN et al. 2012. Resting-state fMRI changes in Alzheimer's disease and mild cognitive impairment. Neurobiol Aging. 33(9):2018–2028. [DOI] [PubMed] [Google Scholar]
- Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K. 2006. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 112(4):389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. 1991. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82(4):239–259. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. 2009. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci. 29(6):1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC et al. 2005. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 25(34):7709–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2012. The economy of brain network organization. Nat Rev Neurosci. 13(5):336–349. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Germann J, Evans AC. 2008. Revealing modular architecture of human brain structural networks by using cortical thickness from MRI. Cereb Cortex. 18(10):2374–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Gong G, Evans AC. 2011. Age-related alterations in the modular organization of structural cortical network by using cortical thickness from MRI. Neuroimage. 56(1):235–245. [DOI] [PubMed] [Google Scholar]
- Chua TC, Wen W, Slavin MJ, Sachdev PS. 2008. Diffusion tensor imaging in mild cognitive impairment and Alzheimer's disease: a review. Curr Opinion Neurol. 21(1):83–92. [DOI] [PubMed] [Google Scholar]
- Dai Z, He Y. 2014. Disrupted structural and functional brain connectomes in mild cognitive impairment and Alzheimer's disease. Neurosci Bull. 30(2):217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT et al. 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 31(3):968–980. [DOI] [PubMed] [Google Scholar]
- Dickson DW, Davies P, Bevona C, Van Hoeven KH, Factor SM, Grober E, Aronson MK, Crystal HA. 1994. Hippocampal sclerosis: a common pathological feature of dementia in very old (≥ 80 years of age) humans. Acta Neuropathol. 88:212–221. [DOI] [PubMed] [Google Scholar]
- Falahati F, Ferreira D, Soininen H, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, Lovestone S, Eriksdotter M, Wahlund LO et al. 2016. The effect of age correction on multivariate classification in Alzheimer's disease, with a focus on the characteristics of incorrectly and correctly classified subjects. Brain Topogr. 29:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri R, Rundo F, Bruni O, Terzano MG, Stam CJ. 2007. Small-world network organization of functional connectivity of EEG slow-wave activity during sleep. Clin Neurophysiol. 118:449–456. [DOI] [PubMed] [Google Scholar]
- Filippi M, van den Heuvel MP, Fornito A, He Y, Hulshoff Pol HE, Agosta F, Comi G, Rocca MA. 2013. Assessment of system dysfunction in the brain through MRI-based connectomics. Lancet Neurol. 12(12):1189–1199. [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM. 2000. Measuring the thickness of the human cerebral cortex from magnetic resonance images. PNAS. 97(20):11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. 1999. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 9(2):195–207. [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M. 2013. Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage. 80:426–444. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Fox NC, Jack CR Jr, Scheltens P, Thompson PM. 2010. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 6(2):67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. 2002. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 15(4):870–878. [DOI] [PubMed] [Google Scholar]
- Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, Beaulieu C. 2009. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb Cortex. 19:524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. 2004. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. PNAS. 101(13):4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundman M, Petersen RC, Ferris SH, Thomas RG, Aisen PS, Bennett DA, Foster NL, Jack CR Jr, Galasko DR, Doody R et al. 2004. Mild cognitive impairment can be distinguished from Alzheimer disease and normal aging for clinical trials. Arch Neurol. 61(1):59–66. [DOI] [PubMed] [Google Scholar]
- Guimera R, Amaral LA. 2005. Cartography of complex networks: modules and universal roles. J Stat Mech. (P02001):P02001-1–P02001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera R, Mossa S, Turtschi A, Amaral LA. 2005. The worldwide air transportation network: Anomalous centrality, community structure, and cities’ global roles. PNAS. 102(22):7794–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. 2008. Mapping the structural core of human cerebral cortex. PLoS Biol. 6:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. 2007. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 17(10):2407–2419. [DOI] [PubMed] [Google Scholar]
- He Y, Chen Z, Evans A. 2008. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J Neurosci. 28(18):4756–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, Tang H, Zhu C, Gong Q, Zang Y et al. 2009. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PLoS ONE. 4:e5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SM, Black JM, Soriano T, Bugescu N, Martinez R, Raman MM, Kesler SR, Hoeft F. 2013. Topological properties of large-scale structural brain networks in children with familial risk for reading difficulties. Neuroimage. 71:260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MD, Gurney K, Prescott TJ. 2006. The brainstem reticular formation is a small-world, not scale-free, network. Proc Royal Soc B Biol Sci. 273:503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Canales-Rodriguez EJ, Aleman-Gomez Y, Melie-Garcia L. 2008. Studying the human brain anatomical network via diffusion-weighted MRI and graph theory. Neuroimage. 40:1064–1076. [DOI] [PubMed] [Google Scholar]
- Jack CR Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell J, Ward C et al. 2008. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging. 27:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsell TD, Monsell SE. 2012. Reversion from mild cognitive impairment to normal or near-normal cognition Risk factors and prognosis. Neurology. 79(15):1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Crutch SJ, Ridgway GR, Ridha BH, Barnes J, Warrington EK, Rossor MN, Fox NC. 2011. Cortical thickness and voxel-based morphometry in posterior cortical atrophy and typical Alzheimer's disease. Neurobiol Aging. 32(8):1466–1476. [DOI] [PubMed] [Google Scholar]
- Li X, Li TQ, Andreasen N, Wiberg MK, Westman E, Wahlund LO. 2013. Ratio of Aβ42/P-tau181p in CSF is associated with aberrant default mode network in AD. Sci Reports. 3(1339):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Y, Wu G, Shi F, Zhou L, Lin W, Shen D; Alzheimer's Disease Neuroimaging Initiative. 2012. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer's disease using dynamic and network features. Neurobiol Aging. 33(2):427.e15–427.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CY, Wang PN, Chou KH, Wang J, He Y, Lin CP. 2010. Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer's disease. J Neurosci. 30(50):16876–16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovestone S, Francis P, Kloszewska I, Mecocci P, Simmons A, Soininen H, Spenger C, Tsolaki M, Vellas B, Wahlund LO et al. 2009. AddNeuroMed—the European collaboration for the discovery of novel biomarkers for Alzheimer's disease. Ann NY Acad Sci. 1180:36–46. [DOI] [PubMed] [Google Scholar]
- Lovestone S, Francis P, Strandgaard K. 2007. Biomarkers for disease modification trials—the innovative medicines initiative and AddNeuroMed. J Nutrition Health Aging. 11(4):359–361. [PubMed] [Google Scholar]
- McDonald CR, McEvoy LK, Gharapetian L, Fennema-Notestine C, Hagler DJ Jr, Holland D, Koyama A, Brewer JB, Dale AM; Alzheimer's Disease Neuroimaging Initiative. 2009. Regional rates of neocortical atrophy from normal aging to early Alzheimer disease. Neurology. 73(6):457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D, DeToledo-Morrell L, Urresta F, Gabrieli JD, Moseley M, Fleischman D, Bennett DA, Leurgans S, Turner DA, Stebbins GT. 2006. White matter changes in mild cognitive impairment and AD: A diffusion tensor imaging study. Neurobiol Aging. 27(5):663–672. [DOI] [PubMed] [Google Scholar]
- Meunier D, Achard S, Morcom A, Bullmore E. 2009. Age-related changes in modular organization of human brain functional networks. Neuroimage. 44:715–723. [DOI] [PubMed] [Google Scholar]
- Muehlboeck JS, Westman E, Simmons A. 2014. TheHiveDB image data management and analysis framework. Front Neuroinformatics. 7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. 2003. The structure and function of complex networks. SIAM Rev. 45:167–256. [Google Scholar]
- Newman MEJ. 2004. Fast algorithm for detecting community structure in networks. Phys Rev E. 69:066133. [DOI] [PubMed] [Google Scholar]
- Newman ME. 2006. Modularity and community structure in networks. PNAS. 103(23):8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pievani M, de Haan W, Wu T, Seeley WW, Frisoni GB. 2011. Functional network disruption in the degenerative dementias. Lancet Neurol. 10(9):829–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JB, Aarsland D, Ginestet CE, Lebedev AV, Wahlund LO, Simmons A, Volpe G, Westman E. 2015. Aberrant cerebral network topology and mild cognitive impairment in early Parkinson's disease. Hum Brain Mapp. 36(8):2980–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. 1999. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 56(3):303–308. [DOI] [PubMed] [Google Scholar]
- Petrella JR, Sheldon FC, Prince SE, Calhoun VD, Doraiswamy PM. 2011. Default mode network connectivity in stable vs progressive mild cognitive impairment. Neurology. 76(6):511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DJ, McGlaughlin A, Ruth D, Jager LR, Soldan A; Alzheimer's Disease Neuroimaging Initiative. 2015. Graph theoretic analysis of structural connectivity across the spectrum of Alzheimer's disease: the importance of graph creation methods. NeuroImage Clin. 7:377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 52(3):1059–1069. [DOI] [PubMed] [Google Scholar]
- Sanabria-Diaz G, Martinez-Montes E, Melie-Garcia L; Alzheimer's Disease Neuroimaging Initiative. 2013. Glucose metabolism during resting state reveals abnormal brain networks organization in the Alzheimer's disease and mild cognitive impairment. PLoS ONE. 8(7):e68860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Arigita EJ, Schoonheim MM, Damoiseaux JS, Rombouts SA, Maris E, Barkhof F, Scheltens P, Stam CJ. 2010. Loss of ‘small-world’ networks in Alzheimer's disease: graph analysis of FMRI resting-state functional connectivity. PloS one. 5(11):e13788, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. 2002. Mapping the evolution of regional atrophy in Alzheimer's disease: unbiased analysis of fluid-registered serial MRI. PNAS. 99(7):4703–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. 2004. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 22(3):1060–1075. [DOI] [PubMed] [Google Scholar]
- Segonne F, Pacheco J, Fischl B. 2007. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans Med Imaging. 26(4):518–529. [DOI] [PubMed] [Google Scholar]
- Sepulcre J, Sabuncu MR, Becker A, Sperling R, Johnson KA. 2013. In vivo characterization of the early states of the amyloid-beta network. Brain. 136(Pt 7):2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu N, Liang Y, Li H, Zhang J, Li X, Wang L, He Y, Wang Y, Zhang Z. 2012. Disrupted topological organization in white matter structural networks in amnestic mild cognitive impairment: relationship to subtype. Radiology. 265(2):518–527. [DOI] [PubMed] [Google Scholar]
- Simmons A, Westman E, Muehlboeck S, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Wahlund LO, Soininen H, Lovestone S et al. 2011. The AddNeuroMed framework for multi-centre MRI assessment of Alzheimer's disease: experience from the first 24 months. Int J Ger Psychiatry. 26(1):75–82. [DOI] [PubMed] [Google Scholar]
- Simmons A, Westman E, Muehlboeck S, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Wahlund LO, Soininen H, Lovestone S et al. 2009. MRI measures of Alzheimer's disease and the AddNeuroMed study. Ann NY Acad Sci. 1180:47–55. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. 1998. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 17(1):87–97. [DOI] [PubMed] [Google Scholar]
- Sporns O, Betzel RF. 2016. Modular brain networks. Ann Rev Psychol. 67:613–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Zwi JD. 2004. The small world of the cerebral cortex. Neuroinformatics. 2(2):145–162. [DOI] [PubMed] [Google Scholar]
- Spulber G, Simmons A, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, Soininen H, Spenger C, Lovestone S et al. 2013. An MRI-based index to measure the severity of Alzheimer's disease-like structural pattern in subjects with mild cognitive impairment. J Intern Med. 273:396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal DR, Rub U, Orantes M, Braak H. 2002. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology. 58(12):1791–1800. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, De Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM et al. 2004. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 22(4):1754–1766. [DOI] [PubMed] [Google Scholar]
- Tijms BM, ten Kate M, Wink AM, Visser PJ, Ecay M, Clerigue M, Estanga A, Sebastian MG, Izagirre A, Villanua J, Lage PM. 2016. Gray matter network disruptions and amyloid beta in cognitively normal adults. Neurobiol Aging. 37:154–160. [DOI] [PubMed] [Google Scholar]
- Tijms BM, Moller C, Vrenken H, Wink AM, de Haan W, van der Flier WM, Stam CJ, Scheltens P, Barkhof F. 2013. Single-subject grey matter graphs in Alzheimer's disease. PLoS ONE. 8(3):e58921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms BM, Series P, Willshaw DJ, Lawrie SM. 2012. Similarity-based extraction of individual networks from gray matter MRI scans. Cereb Cortex (New York, NY: 1991). 22(7):1530–1541. [DOI] [PubMed] [Google Scholar]
- Tijms BM, Wink AM, de Haan W, van der Flier WM, Stam CJ, Scheltens P, Barkhof F. 2013. Alzheimer's disease: connecting findings from graph theoretical studies of brain networks. Neurobiol Aging. 34(8):2023–2036. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. 1998. Collective dynamics of ‘small-world’ networks. Nature. 393(6684):440–442. [DOI] [PubMed] [Google Scholar]
- Westman E, Simmons A, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, Soininen H, Weiner MW, Lovestone S et al. 2011. AddNeuroMed and ADNI: similar patterns of Alzheimer's atrophy and automated MRI classification accuracy in Europe and North America. Neuroimage. 58(3):818–828. [DOI] [PubMed] [Google Scholar]
- Wu K, Taki Y, Sato K, Kinomura S, Goto R, Okada K, Kawashima R, He Y, Evans AC, Fukuda H. 2012. Age-related changes in topological organization of structural brain networks in healthy individuals. Hum Brain Mapp. 33(3):552–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Wang J, He Y. 2013. BrainNet viewer: a network visualization tool for human brain connectomics. PLoS ONE. 8(7):e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe K, Petersen RC, Lindquist K, Kramer J, Miller B. 2006. Subtype of mild cognitive impairment and progression to dementia and death. Dem Ger Cog Disord. 22(4):312–319. [DOI] [PubMed] [Google Scholar]
- Yao Z, Hu B, Zheng J, Zheng W, Chen X, Gao X, Xie Y, Fang L; Alzheimer's Disease Neuroimaging Initiative. 2015. A FDG-PET study of metabolic networks in apolipoprotein e epsilon4 allele carriers. PLoS ONE. 10(7):e0132300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Zhang Y, Lin L, Zhou Y, Xu C, Jiang T; Alzheimer's Disease Neuroimaging Initiative. 2010. Abnormal cortical networks in mild cognitive impairment and Alzheimer's disease. PLoS Comp Biol. 6(11):e1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M, Ballabio C, Abbate C, Cutaia C, Vergani C, Bergamaschini L. 2006. Mild cognitive impairment subtypes and vascular dementia in community-dwelling elderly people: a 3-year follow-up study. J Am Geriatr Soc. 54:580–586. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schuff N, Jahng GH, Bayne W, Mori S, Schad L, Mueller S, Du AT, Kramer JH, Yaffe K et al. 2007. Diffusion tensor imaging of cingulum fibers in mild cognitive impairment and Alzheimer disease. Neurology. 68(1):13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. 2012. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 73(6):1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Wen W, He Y, Xia A, Anstey KJ, Sachdev P. 2012. Changing topological patterns in normal aging using large-scale structural networks. Neurobiol Aging. 33(5):899–913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.