Significance
The genome-wide distribution of histone modifications influences cellular processes that involve access to the DNA. How cells establish and maintain these patterns is currently under debate. Understanding the underlying mechanisms is a prerequisite for predicting the cellular response to epigenetic drugs or programmed epigenetic editing. Here we simulated linear and looping-driven propagation mechanisms and compared the results to the spatiotemporal methylation profiles recently observed in fission yeast. We found that looping-driven spreading, which arises from productive collisions among nucleosomes and chromatin-bound histone modifiers, explains several key observations on engineered methylation domains. These findings point to important roles of chromatin-bound enzymes and chromatin dynamics in controlling modification patterns and epigenetic memory, which might not be restricted to repressive histone methylation.
Keywords: epigenetic memory, heterochromatin, epigenome editing, histone modification, stochastic simulation
Abstract
Histone modifications can redistribute along the genome in a sequence-independent manner, giving rise to chromatin position effects and epigenetic memory. The underlying mechanisms shape the endogenous chromatin landscape and determine its response to ectopically targeted histone modifiers. Here, we simulate linear and looping-driven spreading of histone modifications and compare both models to recent experiments on histone methylation in fission yeast. We find that a generalized nucleation-and-looping mechanism describes key observations on engineered and endogenous methylation domains including intrinsic spatial confinement, independent regulation of domain size and memory, variegation in the absence of antagonists, and coexistence of short- and long-term memory at loci with weak and strong constitutive nucleation. These findings support a straightforward relationship between the biochemical properties of chromatin modifiers and the spatiotemporal modification pattern. The proposed mechanism gives rise to a phase diagram for cellular memory that may be generally applicable to explain epigenetic phenomena across different species.
Histone posttranslational modifications regulate cellular processes including gene expression, DNA replication, and DNA repair (1, 2). Some of these modifications can spread along the genome independently of the underlying DNA sequence, forming extended domains of modified histones. Well-known examples are di/trimethylation of histone 3 at lysine 9 (H3K9me2/3) and at lysine 27 (H3K27me2/3), which are enriched in heterochromatin and play a role in gene silencing (3–5). H3K9me2/3 can spread around centromeres (6) and telomeres (7), and H3K27me2/3 can spread around dedicated response elements within the genome (8, 9), causing so-called chromatin position effects by repressing genes within the methylated domains (7, 10, 11). The enzymes that are responsible for heterochromatic H3K9me2/3 are Clr4 in fission yeast and Suv39h in metazoans, and the PRC2 complex catalyzes H3K27me2/3 (12). By stably tethering these enzymes to chromatin, extended engineered domains enriched for the respective modification can be formed (13–20). Furthermore, both modifications can confer epigenetic memory at least across several cell divisions (14–17, 21).
Aberrations in histone modification patterns and the responsible enzymes are involved in several diseases, including cancer (22, 23). It has recently become possible to recruit histone-modifying enzymes to endogenous target sites, e.g., by using the CRISPR-Cas9 system, thereby eliciting programmed changes to histone modifications and triggering specific position effects (24, 25). Due to the functional relevance of the chromatin landscape, this technique holds promise for future clinical applications (26, 27). Therefore, it is particularly important to understand if and how far modifications can spread and how long engineered domains are maintained after being established by short-term recruitment of a histone modifier.
Because it is currently not feasible to directly follow the spreading process in individual cells in real time, several theoretical models have been proposed and mostly compared with steady-state methylation profiles observed in ensembles of WT or mutant cells (5, 28, 29). Spreading of H3K9me3 is commonly thought to occur linearly along the chromatin fiber (3, 30), whereas spreading of H3K27me3 is thought to involve long-range interactions (29, 31). Both spreading mechanisms are illustrated in Fig. 1. Recently, models involving long-range interactions have also been proposed for H3K9 methylation. One study in mouse cells (32) linked heterochromatic H3K9 levels to the amount of methyltransferases that are stably tethered to chromatin and propagate the modification via long-range interactions. An\ study on heterochromatin in fission yeast (33) showed how long-range interactions in combination with cooperativity can give rise to bistability, which means that chromatin domains delimited by site-specific boundary factors can adopt at least two discrete states with characteristic methylation levels that are stabilized over time. However, recent experiments have shown that engineered H3K9 methylation domains in fission yeast remain confined in the absence of site-specific boundaries and gradually decay over time after the modifier has been removed (15, 16).
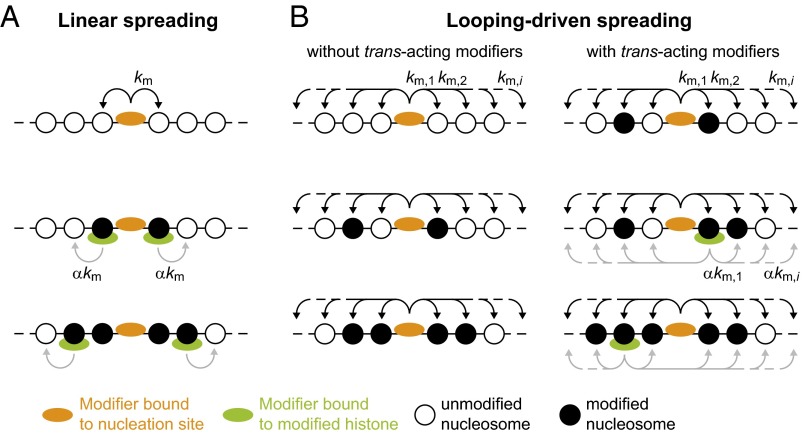
Fig. 1.
Spreading models. (A) According to the linear spreading model, a tethered histone modifier (orange) can modify the histone tails on adjacent nucleosomes. Spreading is mediated by additional modifiers (green) that can bind to modified histones and modify the nearest neighbor while bound. Extended domains are only obtained in the presence of such modifiers. (B) In the looping model, a tethered histone modifier can modify nucleosomes beyond the nearest neighbor according to the contact probability for two chromatin segments separated by a certain distance (Fig. S1). The resulting domain size is intrinsically confined by the maximum loop size that is formed with sufficiently high probability. Spreading can also occur in the absence of trans-acting modifiers that bind to modified histones (Left). The presence of such modifiers (green; Right) can increase the modification level within the domain and can stabilize the domain over time.
Identifying the molecular mechanism that drives spreading of histone modifications is particularly important because it not only governs their distribution in steady state but also defines the prerequisites for the emergence of epigenetic memory and the general rules that determine the effects of epigenome editing. The most obvious difference between linear and looping-driven spreading is that the latter model allows for spreading to distant loci on the same chromosome or on other chromosomes that are located in close spatial proximity. These phenomena have been well characterized for Polycomb loci that exhibit stable long-range interactions among each other, which are specific enough to be picked up in cell ensembles (4, 29). However, both spreading models also make different predictions for domains formed in the immediate vicinity of chromatin-bound histone modifiers on the same chromosome as shown below. These differences are apparent when comparing the relationship among spatial extension, formation kinetics and temporal stability of modified domains but are less pronounced when solely comparing steady states. Recent experiments on engineered histone methylation domains in fission yeast (15, 16) provide information on the spatiotemporal behavior of methylation marks under different conditions and allow for explicitly testing the predictions made by different models.
Here, we show that a generalized nucleation-and-looping model explains key experimental findings on engineered H3K9 methylation domains in fission yeast (15, 16, 18) and epigenome editing experiments in human cells (19, 24, 25). Several features specifically arise from looping interactions beyond the next nucleosome and are therefore not observed in linear spreading simulations. The looping model requires only two parameters, describing the nucleation and feedback strength, which can be readily related to the known biochemical properties of histone modifiers. In a broader context, our findings support a relationship among the biochemical properties and genome-wide binding distribution of histone modifiers, chromatin dynamics, and the spatiotemporal properties of the resulting modification pattern, which lead us to propose a phase diagram for epigenetic memory that may be generally applicable to predict the chromatin landscape.
Results
Simulation Setup for Linear and Looping-Driven Spreading.
We used two different models to simulate spreading of histone modifications. Both models include two types of chromatin binding that correspond to the independent parameters in the models: (i) recruitment to specific nucleation sites, which represent the genomic binding sites of endogenous enzymes or the sites that are targeted by enzymes fused to a heterologous DNA-binding domain such as TetR and dCas9, and (ii) binding to modified histones, which can be mediated by the reader domains that are present in many histone modifiers. According to the linear spreading model, modifiers can only collide productively with neighboring nucleosomes while bound to a nucleation site or another nucleosome (Fig. 1A). According to the looping model, modifiers can productively collide with nucleosomes at more distant genomic positions as long as they reside in close spatial proximity, which is motivated by the fundamental ability of chromatin segments to move by diffusion. This type of motion enables chromatin-bound enzymes to collide with potential substrate nucleosomes beyond the neighboring nucleosome (Fig. 1B and Fig. S1A).
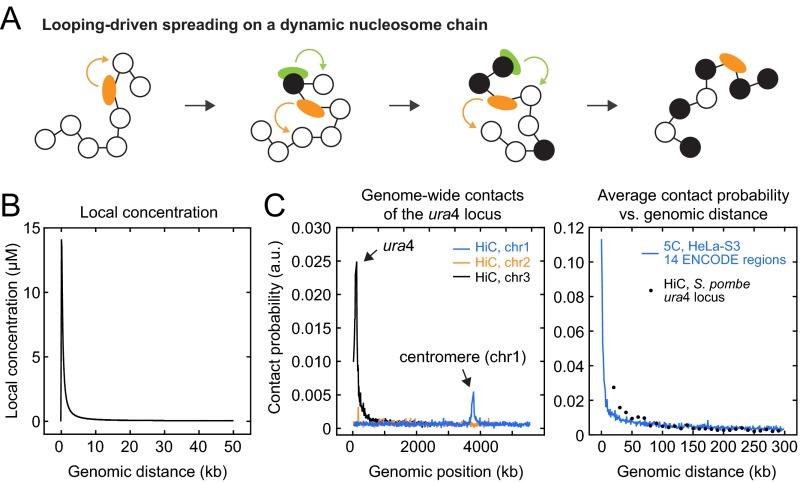
Fig. S1.
Spreading models and contact probabilities. (A) Schematic representation of dynamic intrachromosomal contacts between substrate nucleosomes and histone modifiers that lead to looping-driven spreading. (B) Local concentration for two segments on the same chromosome that are separated by a given genomic distance. For details, see ref. 40. (C) (Left) Intra- and interchromosomal contacts for the ura4 locus observed by HiC in Clr4-deficient fission yeast cells (41). The largest probabilities are found for intrachromosomal contacts with sites adjacent to the ura4 locus on chromosome 3. Interchromosomal contact probabilities are much smaller, with a peak coinciding with the constitutively methylated centromere on chromosome 1. (Right) Average intrachromosomal contacts of the ura4 locus in fission yeast (black, same data as in A) and of 15 ENCODE regions in human HeLa-S3 cells (blue, data from ref. 42) with all loci separated by a given genomic distance. Both datasets were normalized relative to the total contacts within a 500-kb region around the interrogated site. In both species, intrachromosomal contacts between sites that are separated by up to 100 kb are present.
To simulate histone modification domains, we used an array of 1,000 nucleosomes, which corresponds to a genomic region of 150–230 kb depending on the nucleosome repeat length (34). Each nucleosome could reside in a modified (M) or an unmodified (U) state (Fig. 2A). Additional states were considered where indicated. Modification reactions were catalyzed by productive collisions between nucleosomes and histone modifiers either tethered to a nucleation site or bound to a modified histone as indicated. We used different effective rate constants for modifiers bound to a nucleation site (km) and for modifiers bound to a modified histone (α⋅km) because collisions with a nucleation site or a modified histone are only productive if the histone modifier is present. In this manner the effective rate is related to the inherent catalytic activity of the enzyme and the respective saturation degree (SI Materials and Methods). For simplicity, productive collisions with free enzymes that would induce an additional homogenous modification background were not considered explicitly. They are implicitly reflected by the efficiency of the reverse reaction, which was assumed to occur uniformly throughout the domain and represents the activity of soluble antagonists and histone turnover. For H3K9me2/3, the simulated scenario describes the endogenous situation with low H3K9 methylation levels outside heterochromatin in fission yeast, low free concentration and activity of soluble Clr4/Suv39h, higher H3K9me3 loss rates, and weak chromatin-binding of demethylases compared with methyltransferases (16, 32, 35, 36).
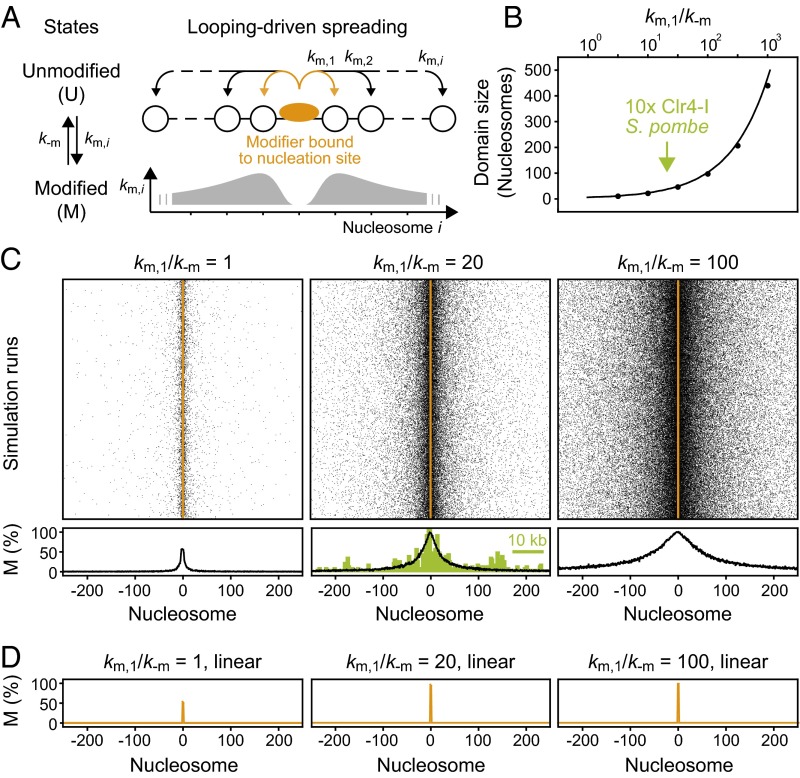
Fig. 2.
Chromatin-bound modifiers can induce confined domains of modified histones with characteristic size. (A) Nucleosomes (white) can be modified via productive collisions with an adjacent chromatin-bound histone modifier (orange). The modification rate km,i at distance i from the modifier scales with the intramolecular contact probability (gray; Fig. S1). (B) Domain sizes for histones with a modification level above 50% at different km,1/k−m ratios. The solid line reflects analytical calculations and the points are results from stochastic simulations. The arrow indicates the size of an engineered H3K9me3 domain in Clr4-deficient fission yeast (16). (C) Spatial distribution of modified nucleosomes around the nucleation site. Each row reflects an independent simulation run. Average profiles are shown below. The ChIP-seq profile for an engineered H3K9me3 domain in Clr4-deficient fission yeast cells (16) is shown for comparison (green). (D) In the absence of interactions beyond the next neighbor, which reflects a linear spreading mechanism without trans-acting modifiers, no extended domain is formed.
For the looping model, we considered only intrachromosomal collisions between histone modifiers and nucleosomes, whose probability scales with the relative distance of both molecules along the chain (Fig. S1 A and B). The contact probabilities used here are based on experiments in human and Drosophila cells (37–39) that are summarized in ref. 40. Qualitatively similar contact probabilities were found in other species (41, 42) and, in particular, around the ura4 locus in fission yeast (Fig. S1C), which was used for recent epigenome engineering experiments (15, 16, 18). Interchromosomal collisions for generic loci such as the ura4 locus are less frequent or less conserved in cell ensembles (Fig. S1C) and were therefore neglected.
Chromatin-Bound Modifiers Can Form Confined Domains via Looping.
To assess the consequence of chromatin-bound histone modifiers, we placed one stably tethered enzyme at the center of the simulated nucleosomal array (Fig. 2A). At this stage, we did not include trans-acting modifiers that could catalyze additional reactions. Due to the distance-dependent contact probability between the tethered modifier and adjacent nucleosomes in the looping model, stable and localized domains of different sizes were formed (Fig. 2 B and C). In contrast, linear spreading did not produce extended domains in the absence of trans-acting modifiers (Fig. 2D). For the looping model, domain sizes increased with the ratio between the modification rate km and the reverse rate k−m (Fig. 2B) because the modification level for each nucleosome is determined by the local balance between both reactions. The two rates are related to the biochemical properties of the corresponding enzymes (SI Materials and Methods), with the modification rate depending on the activity of the modifier in the bound state, the local contact probability and the saturation degree of the nucleation site.
Domain sizes of 20–60 nucleosomes were obtained for ratios km/k−m = 10–40, which corresponds to the range of experimentally observed H3K9me2/3 domain sizes generated by one or several directly or indirectly recruited methyltransferases in fission yeast, mouse, and human (15–19, 25, 43). In principle, very large rate ratios can produce very broad domains (Fig. 2B). In practice, however, the maximum ratio and thus the upper limit for the domain size is intrinsically limited by the basal reverse rate and by the maximum genomic distance at which intramolecular contacts occur frequently enough on the relevant time scale. The minimum basal reverse rate is determined by the histone turnover rate and the activity of antagonists that remove modifications or add mutually exclusive ones (SI Materials and Methods).
These results show that according to the looping model one or several stably tethered histone modifiers can be sufficient to form an extended domain of modified histones even in the absence of trans-acting modifiers, which is in stark contrast to the linear spreading model. The H3K9me3 profile formed around a chromatin-bound Clr4 mutant lacking its chromodomain in fission yeast cells that are deficient for endogenous Clr4 (16), which is considered to be the trans-acting modifier in WT cells, is shown in the center panel of Fig. 2C (green). The looping simulation resembles both the overall geometry and the width of the experimental profile and the rate ratio inferred from this comparison is compatible with expectations from in vitro measurements (Table S1). The linear spreading model predicts that no domain is formed in these cells (Fig. 2D) unless Clr4 functions as a potent trans-acting modifier even in the absence of its chromodomain. This hypothesis is revisited below where we discuss the influence of feedback mediated by trans-acting modifiers on domain size and memory.
Table S1.
Biochemical properties of H3K9 methyltransferases
| Enzyme | kcat (min−1) | KM (µM) | c (µM) | KD, K9me3 (µM)* | km (min−1)† | α | k−m (min−1) |
| hSuv39h1 | 0.1–0.2‡ | 0.3–0.9‡ | 0.08§ | 20¶ | 0.1–0.2 | 0.004# | 0.0001|| |
| hSuv39h2 | 13** | 13–33** | 0.02§ | ND | <4–7 | ND | |
| yClr4 | 0.02†† | 15†† | ND | 0.6–1.5††,‡‡ | 0.01 | ND | 0.006–0.04§§ |
The catalytic rate constant kcat, Michaelis constant KM, free intracellular concentration c, dissociation constant for H3K9me3 binding KD, K9me3, resulting effective reaction rate km, and scale parameter α are listed for different H3K9 methyltransferases. The values of kcat and KM for hSuv39h2 refer to its isolated catalytic domain, with the full-length protein being less active. Reverse rates k−m for H3K9 methylation are also indicated. ND, not determined.
The low micromolar KD values for binding to H3K9me3 compare with the low nanomolar KD values for binding of TetR, GAL4, and Cas9 to their target sites (67–69).
The effective modification rate was calculated according to Eq. S2 using the indicated values for kcat and KM, as well as the local concentration c0 ∼ 11–14 µM of the adjacent nucleosome for the respective nucleosome repeat length (40). The modification rates listed here refer to a single enzyme. They should increase (roughly linearly with the number of recruited enzymes) if multimeric DNA-binding proteins are used for recruitment and/or if nucleation sites contain multiple binding sites, provided that only a relatively small fraction of intramolecular collisions is productive and that nucleation sites are relatively small.
Measured value was taken from ref. 62.
Measured value was taken from ref. 32.
Measured value was taken from ref. 63.
This value was calculated according to Eq. S4 using the indicated values for c and KD,K9me3, as well as KD,1 = 0.2 nM for the TetR-tetO interaction (68) and β = 1.
The decay half-time of 5 d for H3K9me2 in mammalian cells translates into a loss rate of 0.0001 min−1 (19). Small demethylation rates below the detection limit of 0.072 min−1 were also reported for H3K9me3 (66).
Measured value was taken from ref. 64.
Measured value was taken from ref. 35.
Measured value was taken from ref. 65.
Domain Sizes Are Determined by Interplay of Nucleation and Feedback.
Many histone modifiers contain reader domains that bind to the modification they catalyze (12), which leads to positive feedback because increasing modification levels drive increased recruitment of modifiers. Clr4 and Suv39h1/2 contain a chromodomain that binds to methylated H3K9. Although the binding affinity for these modifications is typically low compared with the affinity of TetR or dCas9 for their target sites (Table S1), such enzymes can transiently bind modified histones and modify nucleosomes in close spatial proximity while bound (35, 36, 44). To account for this effect, we assumed that modifiers bound to modified nucleosomes can catalyze reactions at histone tails in their proximity in the same way as modifiers tethered to ectopic nucleation sites, via transient looping (Fig. 3A, Left) or via next neighbor contacts (Fig. 3A, Right). To account for the lower affinity for modified nucleosomes compared with that for nucleation sites, the respective modification rates were multiplied with a scaling factor α. The value of α is related to the concentration of the modifier and its affinity for the respective sites (SI Materials and Methods).
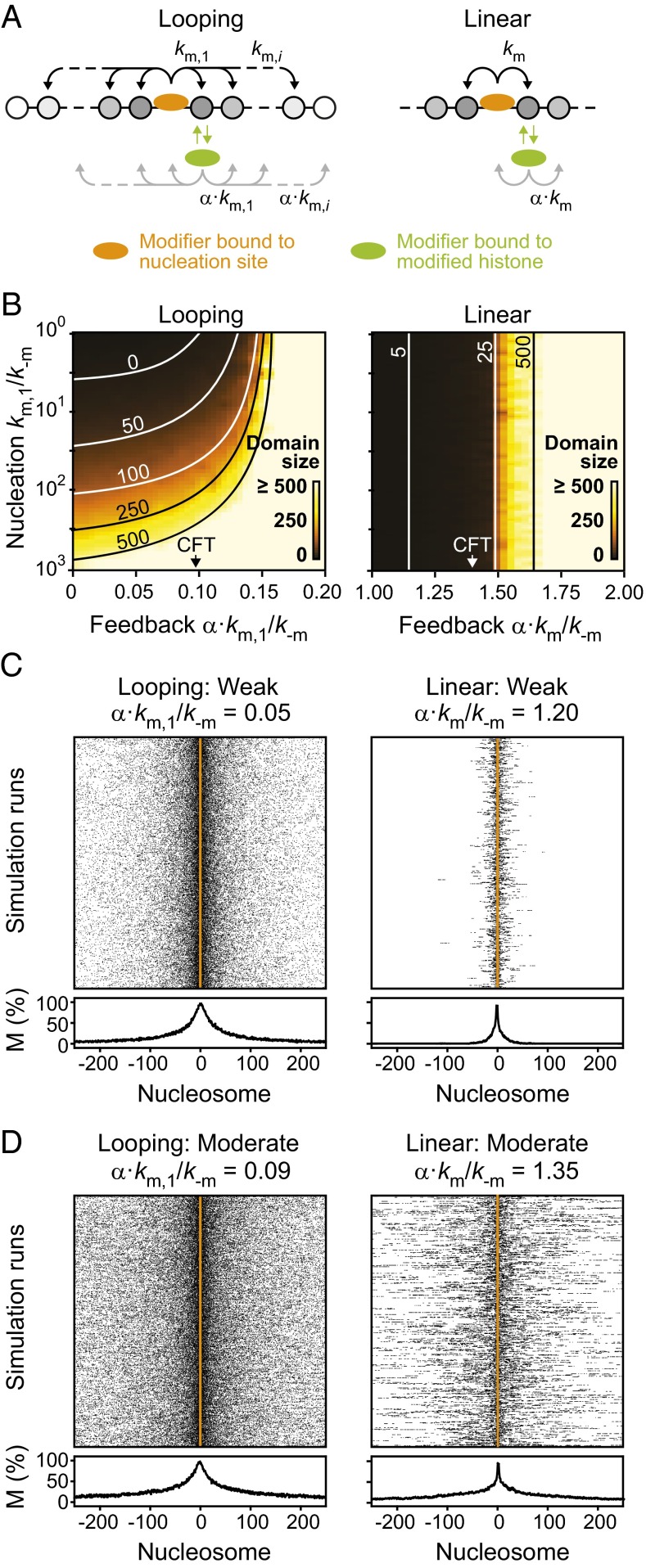
Fig. 3.
Influence of nucleation and feedback strength on domain size. (A) Trans-acting modifiers can propagate the modification via positive feedback. Due to the different affinity for modified nucleosomes compared with nucleation sites, the rates were scaled with a factor α. In the looping model (Left), bound modifiers can modify nucleosomes beyond the next neighbor, whereas only next neighbor contacts are considered in the linear spreading model (Right). (B) Resulting domain sizes as a function of nucleation and feedback strength. Lines indicate parameter combinations that yield the indicated domain size. (C and D) Spatial distribution of modified nucleosomes across the simulated domain for weak (C) and moderate (D) feedback strengths. See Fig. S2 for strong feedback.
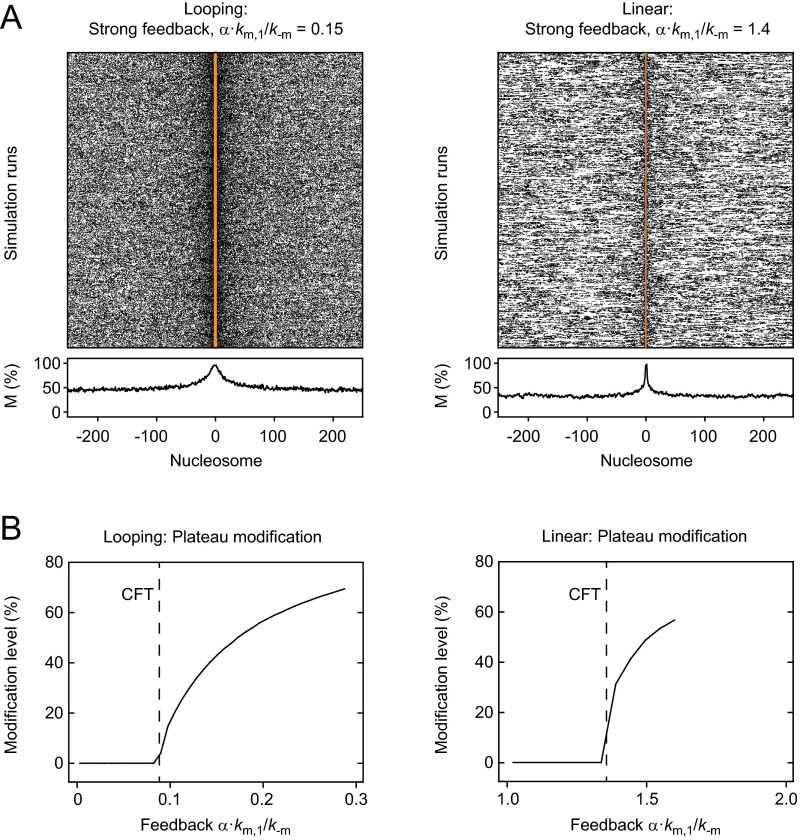
The resulting domain sizes in the presence of these additional transiently bound modifiers are shown in Fig. 3B. Both models produced extended domains, which remained spatially confined for weak feedback and became unconfined for strong feedback (Fig. S2A). To characterize this transition, we found it useful to define a critical feedback threshold (CFT) for each model. It reflects the feedback strength at which confinement was lost and a uniform background modification level was established in addition to the localized domain (Fig. S2 A and B). Simulations above the CFT that yielded a confined domain with a background modification level below 50% received a finite domain size in Fig. 3B because we used a cutoff modification level of 50% to measure the domain size. In the regime above the CFT cells would require boundary factors or other mechanisms to confine modification domains and prevent uncontrolled spreading initiated by spurious nucleation events. Domain sizes for the linear spreading model were solely determined by the feedback strength (Fig. 3B, Right). In contrast, domain sizes for the looping model depended on both nucleation and feedback strength (Fig. 3B, Left), with the largest contribution of feedback near the CFT. Spatial distributions of modified nucleosomes for the nucleation strength estimated for engineered domains in fission yeast (km/k−m = 20; Fig. 2B), and different feedback strengths are shown in Fig. 3 C and D and Fig. S2A.
Fig. S2.
Unconfined spreading for strong feedback. (A) Spatial distribution of modified nucleosomes in the presence of strong feedback above the CFT for looping-driven (Left) and linear (Right) spreading. In addition to localized domains, a uniform modification background was present. (B) The background modification level is plotted as a function of the feedback strength for looping-driven (Left) and linear (Right) spreading.
These simulations show that both linear and looping-driven spreading can produce confined domains of similar size and shape, which agree with experimentally observed domains in steady state (15–18). This finding is in line with previous simulations of linear spreading (30). The prediction from the looping model that the domain size can be decoupled from the feedback strength provides a straightforward explanation for the finding that engineered H3K9 methylation domains in fission yeast do not significantly change their size in the absence or presence of endogenous Clr4 (16) and suggests that Clr4-mediated feedback in fission yeast is well below the CFT. In contrast, the linear spreading model predicts that relatively strong feedback close to the CFT is required to produce domains that match the experimentally observed size and predicts that modulation of the feedback strength by changing the abundance of endogenous Clr4 translates into changes of the domain size.
Feedback Enables Short-Term Memory of Confined Domains.
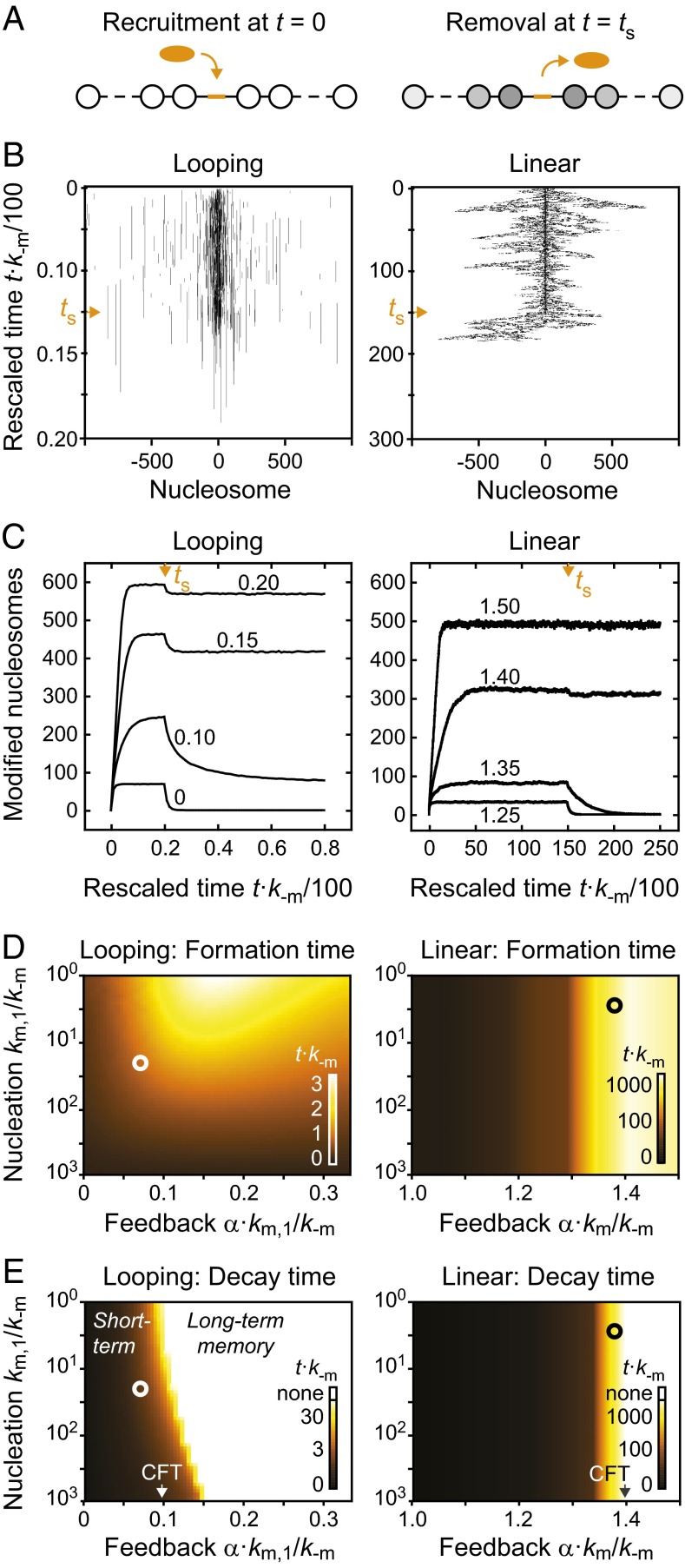
To monitor the formation and stability of induced domains in the absence and presence of trans-acting modifiers that induce feedback, we performed simulations in which the modifier was removed when a steady state had been reached (Fig. 4A). The time evolution for individual nucleosomal arrays is shown in Fig. 4B. The left panel shows a looping-driven spreading simulation in the absence of feedback, which corresponds to the steady-state profile that is shown and compared with experiments in Fig. 2C. The right panel shows linear spreading with the feedback strength α⋅km/k−m = 1.35, which produced similar steady-state modification levels to the looping simulation in the left panel. The time evolution of the number of modified nucleosomes is shown in Fig. 4C. Simulations of looping-driven spreading reached 50% of their steady state level in less than 3/k−m (Fig. 4D), whereas linear spreading was much slower. For the feedback strength α⋅km/k−m = 1.35, it took more than 250/k−m to reach 50% of the steady-state modification level. This time period is two orders of magnitude longer than for looping-driven spreading. For inverse demethylation rates of ∼20 min in fission yeast (Table S1), these values translate into 1 h for looping-driven spreading and more than 80 h for linear spreading, which compares to the generation time of 2–3 h (45). Thus, linear spreading cannot reach its steady state within one or a few cell cycles. The drastic difference observed here is caused by the presence of long-range interactions in the looping model, which significantly accelerate domain formation. Linear spreading requires ∼1/km or more to extend the domain by one nucleosome on each side and is therefore much slower for a given set of rate constants.
Fig. 4.
Temporal stability of modification domains. (A) Modification domains were established on recruitment of a modifier to the center of the nucleosomal array. After a steady state had been reached, the modifier was removed. (B) Time evolution for individual simulations of looping-driven (Left) and linear spreading (Right). Vertical axes indicate the elapsed time, which was multiplied with the reverse rate k−m to make the results independent of the actual rate constant. Orange arrowheads reflect the time point at which the modifier was removed. (C) Time evolution of the number of modified nucleosomes within the entire domain for looping-driven (Left) and linear spreading (Right). In both cases, the nucleation strength km/k−m = 20 was used. Orange arrowheads reflect the time point at which the modifier was removed. Numbers within the panels indicate the feedback strength α⋅km/k−m. (D and E) Time required for reaching 50% of the steady-state modification level after the modifier was recruited (D) or removed (E). Calibration bars in the bottom right corners show the assignment between color code and rescaled time t⋅k−m. White circles in the left panels denote nucleation strength km/k−m = 20 and feedback strength α⋅km/k−m = 0.07, which yield domains that have similar properties to those found in WT fission yeast cells (Fig. 5). Black circles in the right panels denote feedback strength α⋅km/k−m = 1.38, which produces a similar number of modified nucleosomes in linear spreading simulations. White regions in E indicate parameter combinations for which domains did not decay below the 50% level.
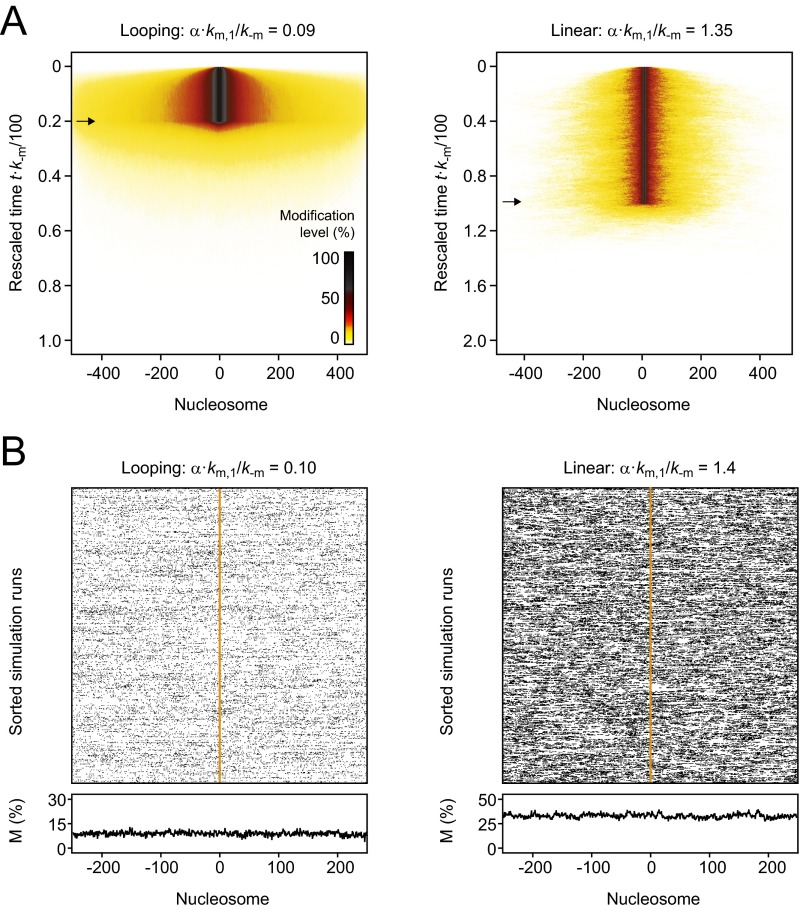
Both models predict that for weak and moderate feedback strength below the CFT domains decay after the modifier that initiated the domain is removed (Fig. 4C). In both models, the decay rates decreased with increasing feedback strength (Fig. 4E), giving rise to short-term memory that can be sufficient to maintain the modified domain for time periods corresponding to several cell divisions. For sufficiently strong feedback below but close to the CFT, domains that persisted after removal of the modifier were not necessarily localized around the nucleation site in each simulation (Fig. 4B). The observation that individual domains can bifurcate and detach from the nucleation site is in agreement with previous linear spreading simulations (30). However, domains averaged over several independent simulation runs, corresponding to an ensemble of cells, remained localized for both models (Fig. S3A). For feedback strengths above the CFT, modification levels did not fully decay after the modifier had been released (Fig. 4E). In this regime, domains did not remain intrinsically confined (Fig. S3B), showing that for both models long-term memory at engineered domains coincides with uncontrolled spreading.
Fig. S3.
Memory at engineered domains. (A) Average time evolution of the spatial distribution of modified nucleosomes for looping-driven (Left) and linear (Right) spreading in the presence of moderate feedback. The modification level is represented by the color code shown in the bottom right corner of the left panel. Arrows indicate removal of the modifier. In both cases, domains remained localized around the nucleation site at the center of the array and ultimately decayed after the modifier had been removed. (B) Spatial distribution of modified nucleosomes long after removal of the modifier (t = 3,000/k−m) for looping-driven (Left) and linear (Right) spreading in the presence of strong feedback. Although modification levels are maintained, the position of the initiating stimulus is not remembered.
Looping-Driven Spreading Describes Engineered H3K9 Methylation Domains.
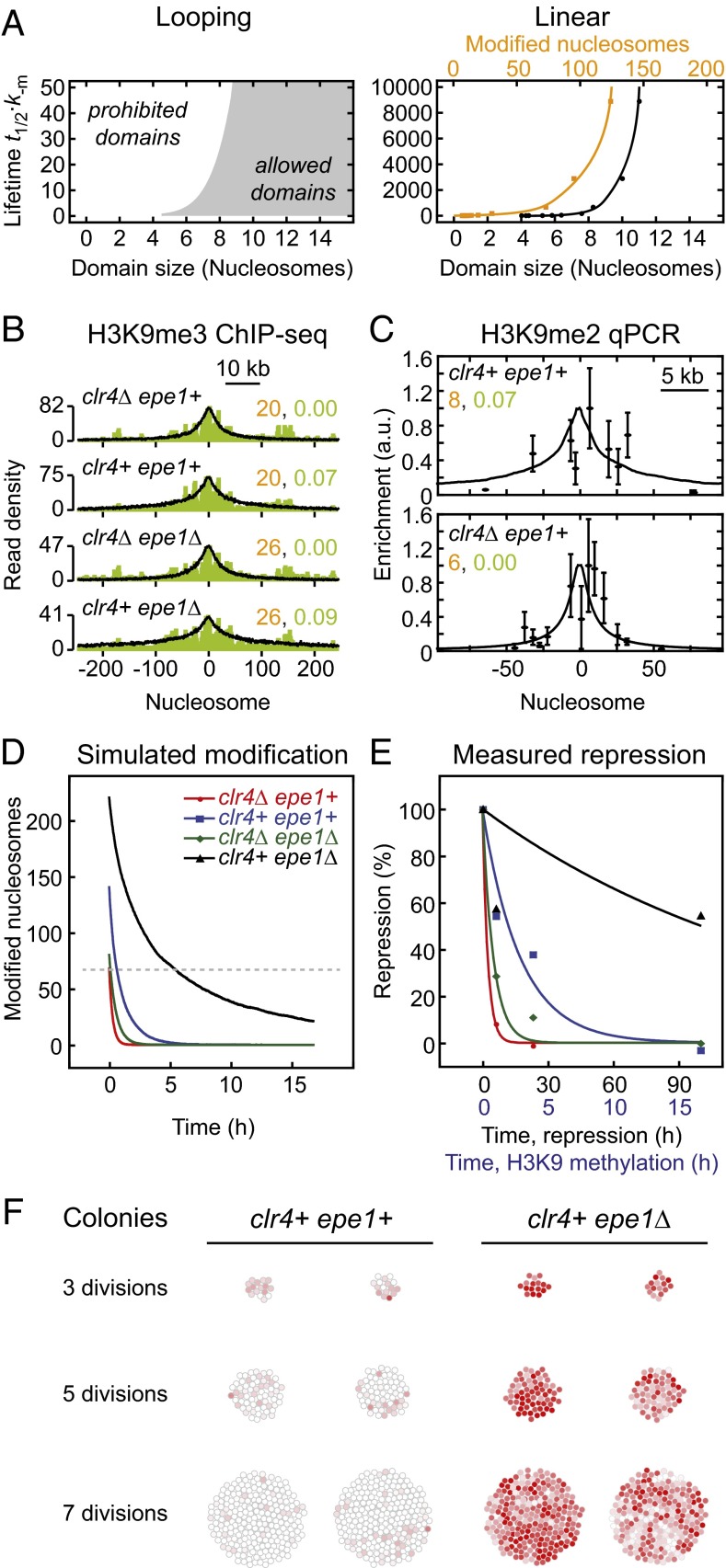
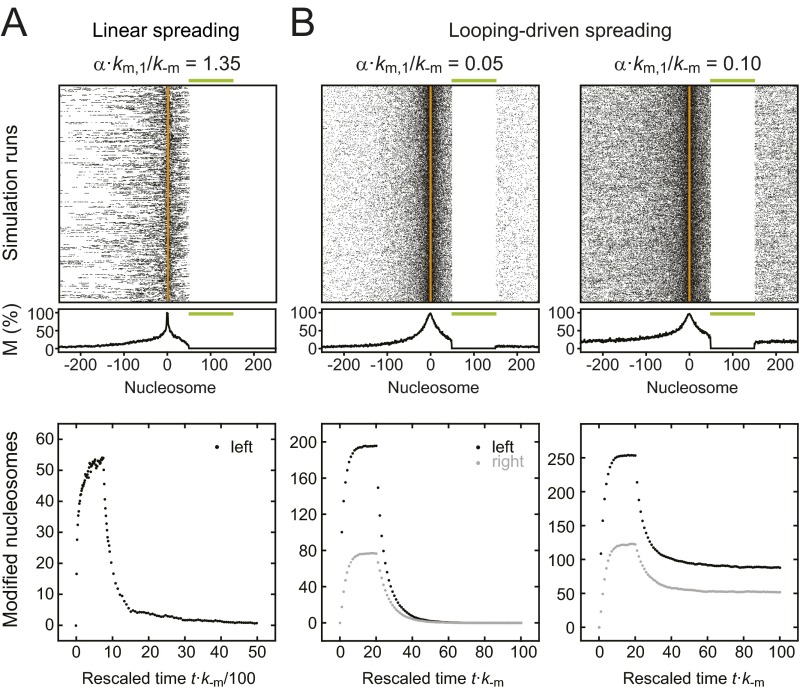
As shown above, linear and looping-driven spreading models make different predictions for the relationship among feedback and nucleation strength, domain size, spreading rate, and temporal stability (Figs. 3 and 4). In particular, domain size and decay time in the linear spreading model are coupled because both are solely determined by the feedback strength. Therefore, each decay time can be uniquely assigned to a defined domain size and modification level (Fig. 5A, Right). The broadest domain with finite decay time had a domain size of less than 15 nucleosomes (using the 50% cutoff definition for domain size above), which corresponds to a total of ∼130 modified nucleosomes within the simulated array. The total number of modified nucleosomes was considerably larger than the domain size because modified domains contained extended regions with modification levels below 50%. Broader domains only formed above the CFT, where intrinsic confinement was lost and domains did not decay. In contrast, the looping model relates each decay time to a minimum domain size and therefore allows for a range of differently sized domains for a given lifetime (gray region in the left panel of Fig. 5A). Notably, these differences between both models are independent of the molecular mechanism that mediates feedback, which means that it is equivalent if Clr4/Suv39h is recruited to methylated histones by its chromodomain or by scaffold proteins like Swi6/HP1.
Fig. 5.
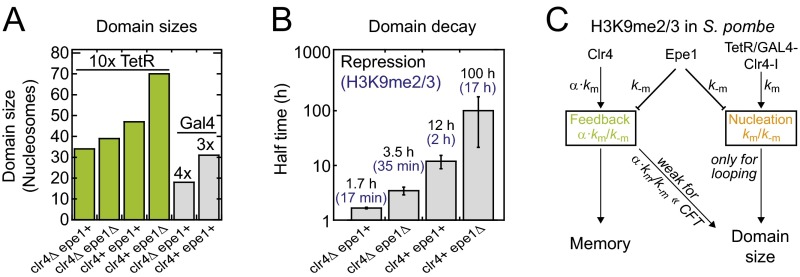
Quantitative comparison between simulations and experiments. (A) Relationship between domain size and lifetime for looping-driven (Left) and linear spreading (Right). The domain size reflects the region around the nucleation site that exhibits a modification level above 50%, whereas the number of modified nucleosomes (orange; Right) indicates the total number of modified nucleosomes within the simulated array. (B) Overlay of H3K9me3 ChIP-seq profiles (green, data from ref. 16) with looping-driven spreading simulations. The nucleation and feedback strengths used for the simulations are indicated in orange and green, respectively. (C) Overlay of qPCR measurements (Upper, data from ref. 15; Lower, data from ref. 18) with looping-driven spreading simulations. Parameters from B were adjusted to reflect the reduced number of nucleation sites (10 sites in B, 3/4 sites in clr4Δ/clr4+ cells in C). (D) Simulated decay of engineered domains for the parameters indicated in C. The dashed line indicates the initial modification level for clr4Δ epe1+ cells, which is sufficient for repression. (E) Time evolution of the repression level observed in different fission yeast strains. Points are measurements from ref. 16; lines are monoexponential fit functions. The same color code as in D was used. See Fig. S4B for fit parameters. (F) Simulated colonies originating from a single cell that contained an engineered modification domain. The modifier was released at the first cell division. The color of each cell reflects its modification level (red, modified; white, unmodified).
To test both models for H3K9 methylation in fission yeast, we compared our simulation results to recent experimental data (15, 16, 18). Engineered H3K9 methylation domains were broad yet intrinsically confined (Fig. 5 B and C) and decayed after the tethered modifier had been removed (Fig. 5E). This behavior is inconsistent with the prediction from linear spreading that only much smaller domains exhibit a finite lifetime (Fig. 5A, Right). Furthermore, domains with similar sizes (Fig. 5B and Fig. S4A) led to very different decay kinetics for the repression level that is a proxy for the methylation level (Fig. 5E and Fig. S4B), suggesting that domain size and temporal stability are not strongly coupled. Finally, to match the lifetime of ∼2 h that was experimentally determined for an engineered domain in WT fission yeast (15) linear spreading predicts a modification rate α⋅km ∼ 33 min−1 if a domain with 100 modified nucleosomes is considered. This rate is more than 1,000 times larger than the catalytic rate kcat ∼ 0.02 min−1 that has been measured for Clr4 in vitro (Table S1).
Fig. S4.
Parameters for engineered H3K9 methylation domains in fission yeast. (A) Summary of engineered H3K9me2/3 domain sizes at the ura4 locus in different fission yeast strains. Plotted domain sizes correspond to the full width at half maximum of the simulated profiles in Fig. 5 B and C. According to the simulations, each tetO binding site is associated with a nucleation strength km/k−m = 2, which translates into km/k−m = 20 for the 10× tetO array. The feedback strength in WT cells is α⋅km/k−m = 0.7, and deletion of Epe1 decreases k−m by a factor of 1.3, which increases the nucleation and feedback strength accordingly (see C). (B) Fit results for the decay curves shown in Fig. 5E (black). Error bars correspond to fit errors. Although the precise relationship between the (spatially modulated) modification level and the repression level is unknown, the ratios among repression decay rates can be used as a proxy for ratios among methylation decay rates (blue). The decay rate for the methylation level in clr4+epe1+ cells was taken from ref. 15. To obtain repression levels, the average GFP intensity in ref. 16 was converted into a repression level R according to R = 1 − [I(t) − IOFF]/(ION − IOFF), where IOFF is the average intensity at t = 0 h, which corresponds to the methylated domain in steady state, and ION is the average intensity of the respective clr4Δ cells at t = 100 h, which corresponds to the intensity for the fully activated domain. (C) Simplified model for the regulation of the size and temporal stability of engineered H3K9 methylation domains in fission yeast. Indicated proteins regulate the respective rate.
In contrast to linear spreading, the looping model is in good agreement with the experimental findings on engineered H3K9 methylation domains. A single parameter set with nucleation strength km/k−m = 2 per ectopic binding site and feedback strength α⋅km/k−m = 0.07 in WT cells as well as a reduction of the k−m rate by a factor of 1.3 on deletion of the antagonist Epe1 is consistent with the domain size and geometry observed around different nucleation sites in four different fission yeast strains (Fig. 5 B and C). Furthermore, it is in qualitative agreement with the decay kinetics of the repression level (Fig. 5 D and E). The reduction of the reverse rate upon deletion of Epe1 is expected (Fig. S4C) because the protein increases histone turnover and is a putative demethylase (16, 46). According to the looping model the lifetime of 2 h found in WT cells translates into modification rates km ∼ 0.04 min−1 (per ectopic binding site) and α⋅km ∼ 0.001 min−1, which is in good agreement with the in vitro value of kcat ∼ 0.02 min−1 for Clr4 (Table S1). Therefore, we focus in the following section on looping-driven spreading.
Variegation in the Presence of Feedback.
To assess how different modification levels are spatially distributed within a colony, we conducted looping simulations similar to those shown in Fig. 4 using the parameters for WT and Epe1-deficient cells listed in Fig. 5B. We assigned simulation trajectories to individual cells and started the simulation with a cell that contained an engineered domain. At the first cell division, we removed the modifier and replaced the cell with two daughter cells that inherited the identical domain and evolved independently thereafter. This scenario is motivated by the experimental observation that the decay of engineered domains is not dominated by replication-dependent dilution (15). In every time step, cells were colored according to their current modification level, and cells were spatially shifted apart to prevent overlap. As shown in Fig. 5F and Movie S1, modification levels decayed rapidly in WT cells, whereas moderate feedback and the reduced reverse rate in Epe1-deficient cells led to variegated colonies that contained spatial clusters of cells with similar modification levels. Distributions of modification levels for cell ensembles in the presence of different feedback strengths are shown in Fig. S5. These simulations reproduce the phenotypic heterogeneity observed among Epe1-deficient fission yeast cells (16).
Fig. S5.
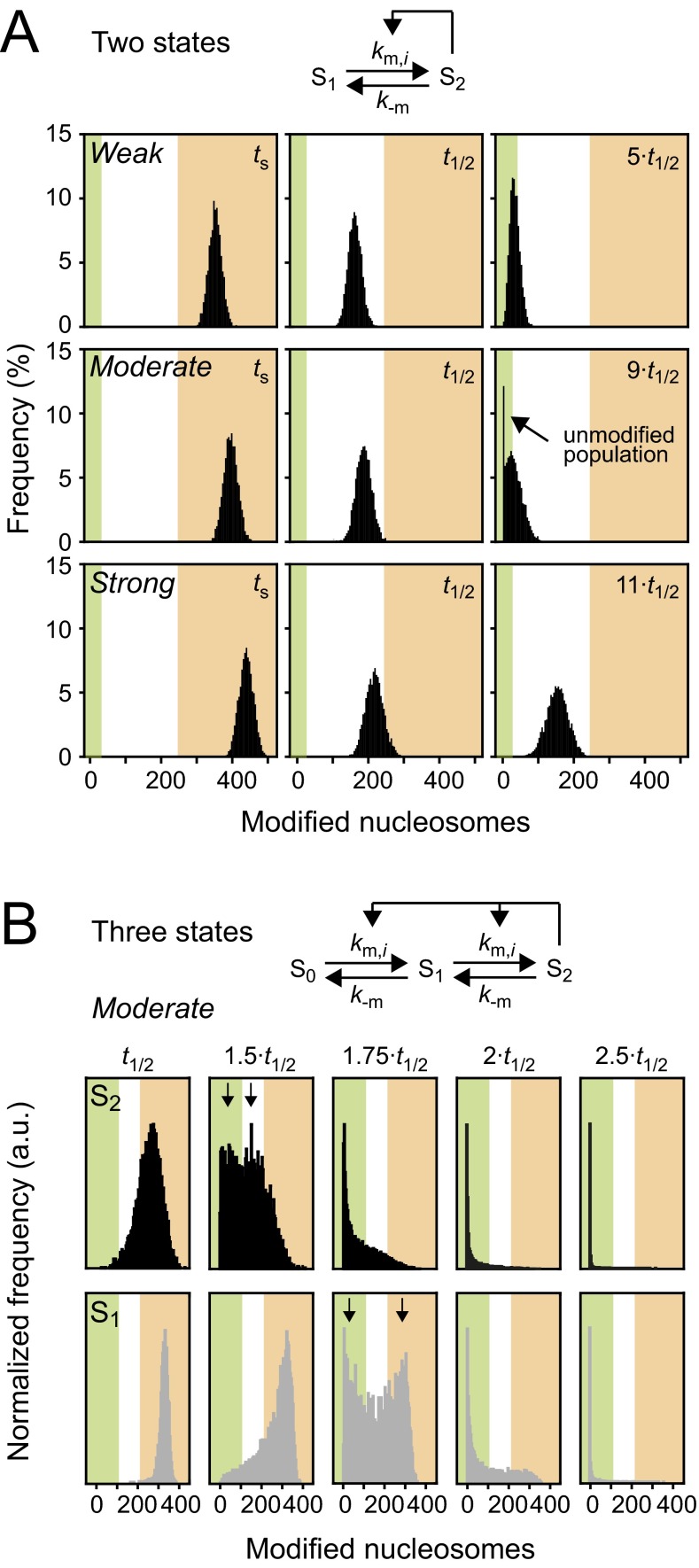
Distribution of modification levels for looping-driven spreading. (A) The time evolution of modification levels was followed after the modifier that had initiated an engineered domain was removed at ts. Simulations for weak (Top), moderate (Middle), and strong (Bottom) feedback are shown. Moderate feedback led to an unmodified population that was separated from the rest of the cell ensemble. Green and orange regions reflect unmodified and modified states. ts indicates the initial time point that corresponds to steady state modification levels; t1/2 indicates the time point at which the modification level had decayed to 50% of the steady-state level. (B) If nucleosomes were allowed to adopt an additional modification state, moderate feedback led to more broadly distributed modification levels. For intermediate time points, bimodal distributions were observed (see arrows that indicate the individual populations). For H3K9 methylation, the states S1 and S2 can be identified with H3K9me2 and H3K9me3, which are both catalyzed by Clr4/Suv39h and are both repressive. Similar histograms have been experimentally observed for the repression level of a reporter gene within an engineered H3K9me2/3 domain (16). Time points are indicated relative to t1/2 (see A for definition).
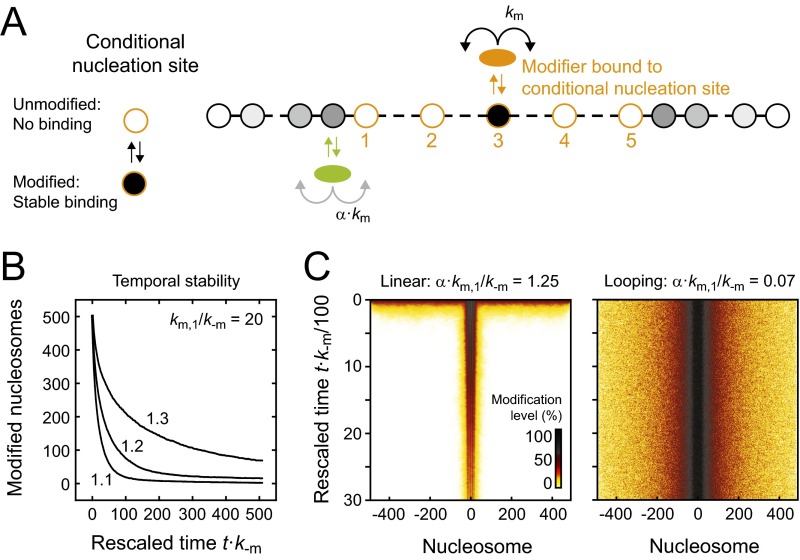
Conditional Nucleation Sites Can Facilitate Stable Epigenetic Memory.
The simulations above show that stable memory at engineered domains requires strong feedback above the critical threshold, which in both models is accompanied with unconfined spreading. Therefore, intrinsically confined modification domains cannot be stably maintained. For the case of H3K9 methylation, Clr4/Suv39h-mediated feedback is indeed too weak to induce stable memory at engineered domains (15, 16, 19). How can cells selectively enable stable epigenetic memory at endogenous heterochromatin but exhibit only short-term memory at engineered domains?
To answer this question, we considered the following scenario that recapitulates the situation in pericentric heterochromatin: We placed five modification-dependent conditional nucleation sites at the center of the simulated domain, which behave as fully functional high-affinity binding sites for modifiers if modified and are not bound at all if unmodified (Fig. 6A). Adjacent conditional nucleation sites were separated by seven nucleosomes to match the density previously estimated in mouse cells (32). Conditional nucleation sites are motivated by the experimental finding that Clr4/Suv39h requires H3K9 methylation for stable recruitment to heterochromatin (28, 32, 47, 48). The strong methylation dependence is probably not caused by direct interaction of Clr4/Suv39h with methylated H3K9 alone but rather by the methylation-dependent presence of several other factors like Swi6/HP1, which form a nucleation complex that is required for stable and specific recruitment of the enzyme (32).
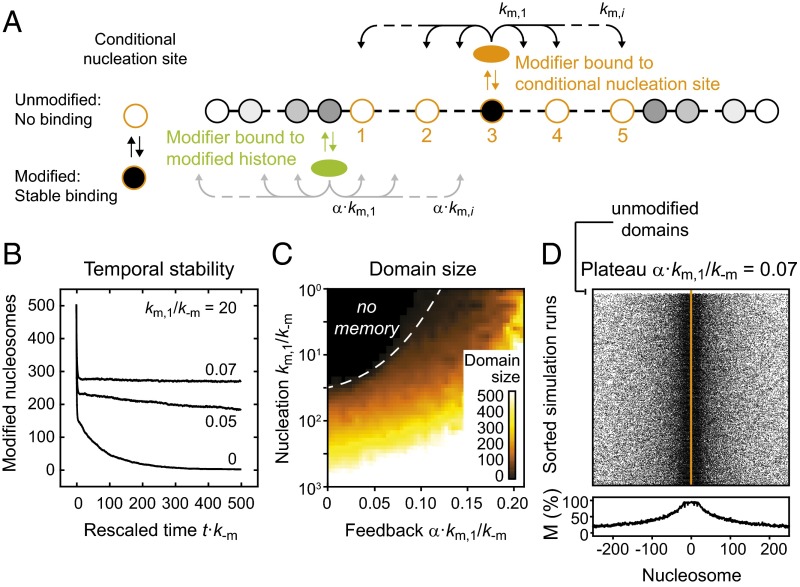
Fig. 6.
Conditional nucleation sites can confer epigenetic memory. (A) Conditional nucleation sites act as regular nucleation sites if modified, but do not bind modifiers if unmodified. For our simulations we considered looping-driven spreading at an array of five conditional nucleation sites (orange circles, numbered). (B) In the presence of conditional nucleation sites and moderate feedback, modified domains were stably maintained for many simulation steps. Numbers within the panel indicate the feedback strength that was used. (C) Sizes of maintained domains. (D) Spatial distribution of modified nucleosomes after time t = 500/k−m. Domains remained localized and confined. The white top rows represent the small fraction of simulations that lost the domain because they started with all conditional nucleation sites being unmodified. In the other simulations, the domain was stably maintained.
We then sought to determine in which situations the presence of these conditional nucleation sites is sufficient to maintain a modified domain. We conducted looping-driven spreading simulations with an initial modification level of 50%, which is similar to the H3K9me2/3 level for pericentric heterochromatin in mouse fibroblasts (see ref. 32 and references therein). To this end, 50% of all nucleosomes across the entire simulation array were randomly modified before the simulation was started. The time evolution of the average modification level for different feedback strengths is shown in Fig. 6B. Remarkably, modification levels decayed in the absence of feedback but were stably maintained for more than 500/k−m in the presence of weak or moderate feedback, which corresponds to several days in fission yeast and several weeks in human cells and is much longer than the lifetimes of engineered domains (Fig. 4E, Left).
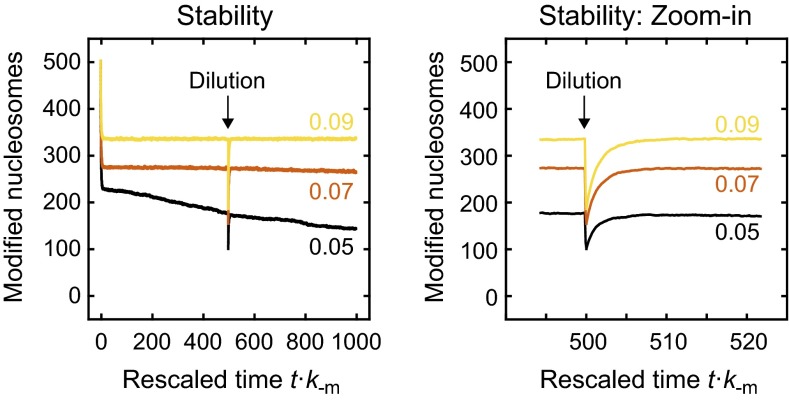
The domain sizes for different parameter combinations are shown in Fig. 6C. Stable memory required both sufficient nucleation and feedback strength. For large nucleation strengths, domains persisted even in the absence of feedback. The spatial distribution of modified nucleosomes for km/k−m = 20 is shown in Fig. 6D. In contrast to the long-term memory at engineered domains for α⋅km/k−m ≥ 0.10, which was accompanied by uncontrolled spreading (Fig. S3B), domains remained confined and localized around the center of the conditional nucleation site array for α⋅km/k−m < 0.10 (Fig. 6D). Furthermore, domains persisted if 50% of the nucleosomes were randomly selected and replaced with unmodified nucleosomes (Fig. S6), which might reflect the situation during replication where modified nucleosomes are transiently diluted. This type of stable epigenetic memory was not observed in linear spreading simulations conducted with the same configuration of conditional nucleation sites (Fig. S7A), in which average modification levels visibly decayed within 500/k−m for the feedback strengths tested (Fig. S7B) and in which after 3,000/k−m residual modifications were restricted to nucleation sites (Fig. S7C).
Fig. S6.
Stability of domains around conditional nucleation sites. Looping-driven spreading simulations were conducted in the presence of an array of conditional nucleation sites like in Fig. 6 with nucleation strength km/k−m = 20 and the indicated feedback strengths (color-coded). Modified nucleosomes were diluted at time t·k−m = 500 by randomly selecting 50% of the nucleosomes across the entire simulated array and replacing them with unmodified ones. For the parameter values tested, modification levels rapidly recovered after dilution.
Fig. S7.
Linear spreading around conditional nucleation sites. (A) Linear spreading simulations were conducted in the presence of an array of conditional nucleation sites like in Fig. 6. Conditional nucleation sites were either fully active if modified (km/k−m = 20) or completely inactive if unmodified. (B) Initially, 50% of the nucleosomes within the domain were randomly selected and modified. Subsequently, the simulation was started and the modification level was followed over time. Modification levels decayed for weak and moderate feedback strengths (values in the panel indicate the feedback strength α⋅km/k−m). (C) Average time evolution of the spatial distribution of modified nucleosomes for linear (Left) and looping-driven (Right) spreading with the indicated feedback strength and nucleation strength km/k−m = 20. The modification level is represented by the color code shown in the bottom right corner of the left panel. In the case of linear spreading, only nucleosomes next to the nucleation sites retained a moderate modification level for prolonged time, which eventually decayed completely. In contrast, looping-driven spreading could maintain an extended domain without visible reduction of modification levels.
These simulations illustrate how cells can simultaneously implement short-term memory at ectopic sites and long-term memory at conditional nucleation sites. For increasing constitutive recruitment in the absence of modifications, which was set to zero in the simulations shown here, conditional nucleation sites convert into the regular nucleation sites discussed above and induce constitutively modified domains that are formed independently of preexisting modifications (see Fig. 3B for threshold values). Therefore, emergence of epigenetic memory in our simulations is a consequence of sufficiently weak basal recruitment in the absence of modifications and sufficiently strong recruitment in their presence. Basal recruitment to conditional nucleation sites might involve methylated cytosines and the factors that bind to it in mammalian cells (see ref. 32 and references therein) and components of the RNAi machinery and Clr3 in fission yeast (47, 49, 50).
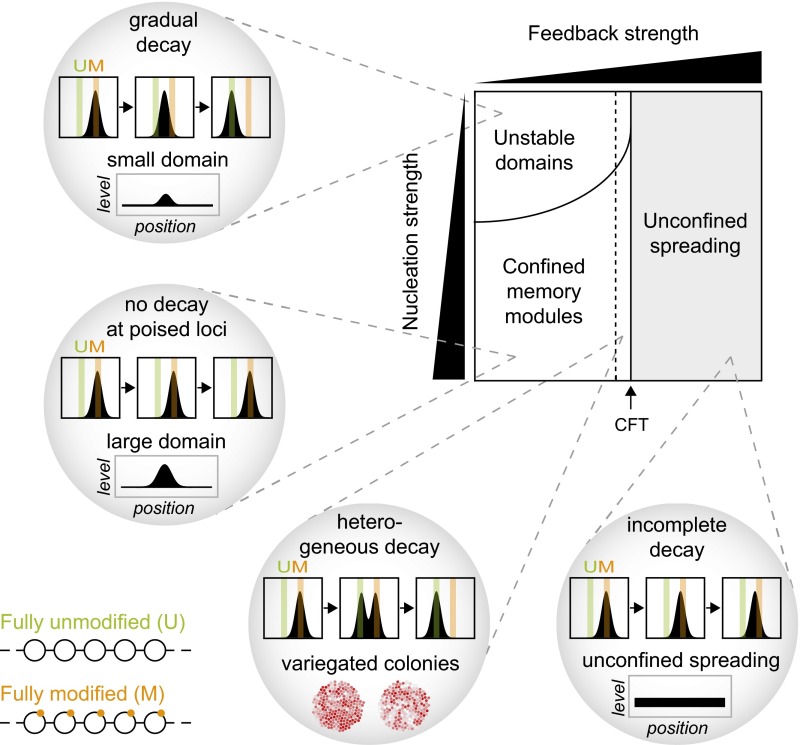
A Phase Diagram for Epigenetic Memory.
Based on our simulations, we constructed a phase diagram that illustrates the key features of different types of memory in relation to the two parameters used in our simulations (Fig. 7). Above a CFT (black arrow), each modified nucleosome can nucleate an unconfined domain of modified nucleosomes, leading to uncontrolled spreading that has to be restricted by boundary factors and opposing modifiers. In this regime, domains do not fully decay after removal of the modifier that initiated the domain (Fig. 4), which is a hallmark of stable epigenetic memory. However, due to loss of intrinsic confinement in this regime, cells can only remember the position of the initiating stimulus with the help of boundary factors that prevent spreading across the whole genome.
Fig. 7.
Phase diagram for epigenetic memory. Cells can implement different types of memory depending on the activity of histone modifiers bound to localized nucleation sites (vertical axis) and modified histones (horizontal axis). For weak and moderate feedback below the CFT (black arrow), modified domains remain intrinsically confined and decay after removal of the tethered modifier unless they are maintained by conditional nucleation sites. Moderate feedback strength close to the CFT increases heterogeneity (Lower Left), leading to variegated colonies composed of distinct subpopulations that are spatially clustered. For strong feedback above the CFT, tethered modifiers can induce domains that are not intrinsically confined (Lower Right). In this regime, cells require mechanisms to restrict uncontrolled spreading.
Below the CFT, chromatin-bound modifiers can establish confined domains whose size in the case of the looping model is intrinsically limited by the maximum range of intramolecular contacts that can form between the modifier and potential substrate nucleosomes. In the presence of moderate feedback, linear spreading can also produce confined domains (Fig. 3B, Right). These domains, however, can transiently detach from the nucleation site and therefore affect more distant genomic regions (Fig. 4B, Right). In this regime, modifiers are not active enough to initiate a domain around each modified nucleosome. Engineered domains fully decay below the CFT unless they are maintained by the presence of conditional nucleation sites that are only functional when modified and do not require an additional persistent stimulus (Fig. 6). Stable epigenetic memory below the CFT was facilitated by looping interactions and was not observed in linear spreading simulations under the conditions used here (Fig. S7). Conditional nucleation sites must be strong enough and have to reside closely enough to each other to allow for epigenetic memory. The repetitive sequences at pericentric heterochromatin, interspersed repeats, rDNA loci, and telomeres might represent poised loci that meet these requirements. They are likely to provide a sufficiently high density of identical binding sites for both nucleosomes and site-specific chromatin proteins that can interact with histone methyltransferases. Furthermore, conditional nucleation sites have to be weak enough in the absence of modifications to prevent formation of a constitutive domain instead of an epigenetic domain.
In the absence of conditional nucleation sites, moderate feedback near the CFT can induce transient heterogeneity among modification levels in individual cells (Fig. 7, Lower Left). Furthermore, it may lead to variegated colonies containing patches of cells with similar modification levels (Fig. 5F and Movie S1). In the absence of feedback, domains are rapidly lost, with lifetimes that are shorter than one generation time. In the presence of moderate feedback, lifetimes increase up to 30/k−m (several days; see Table S1 for k−m rates), reflecting short-term memory that can be sufficient for transmitting information through several cell divisions. For very weak nucleation and feedback strengths, no significant domains are formed.
SI Materials and Methods
Reaction Rates.
Modification reactions were assumed to occur on productive collision of chromatin-bound modifiers with substrate nucleosomes. The intramolecular contact probability was described with a polymer model that is consistent with experiments involving several techniques (40). The effective reaction rate was modeled according to Eq. S1
| [S1] |
Here, c(x) is the local concentration that describes the contact probability between two sites separated by distance x, c0 is the local concentration at the next nucleosome, and km is the effective rate for the reaction at the adjacent nucleosome. The effective rate can be related to the biochemical properties of the enzyme via
| [S2] |
Here, kcat is the catalytic rate, and KM is the Michaelis constant. These equations assume that the reaction rate is proportional to the local concentration between two sites and that the rate-limiting step is the modification reaction, not the intramolecular collision, which justifies the conversion of the collision frequency into a local concentration (40). For sequentially occurring reactions, such as reversal of a mutually exclusive modification followed by a modification reaction, or several consecutive modification reactions (such as sequential attachment of methylation groups to the same residue), kcat can be regarded to represent the rate-limiting step. A sequential scheme was explicitly considered in Fig. S5B. In the case of small KM values the local concentration for short separation distances might be larger than KM and it might be more suitable to define the effective rate constant according to
| [S3] |
This equation accounts for saturation in the local vicinity of the tethered modifier, which changes the local geometry of the resulting domain but does not lead to significant changes in the distance dependence of the reaction rate unless KM values that are much smaller than those measured for most histone modifiers are considered.
To distinguish between modifiers bound to nucleation sites and modifiers bound to modified histones, the modification rate for the latter ones was multiplied with a scaling factor α. This factor accounts for the different saturation degrees of both types of binding sites and for potential allosteric effects for both species. In particular, the relationship between α and the biochemical properties of the enzyme reads
| [S4] |
Here, KD,1 is the dissociation constant for binding to the nucleation site, KD,2 is the dissociation constant for binding to the modified histone, c is the concentration of free modifiers, and β is the allosteric activation/inhibition at modified histones compared with nucleation sites.
The reverse rate k−m is given by the sum of all rates that describe processes antagonizing the modification reaction, including enzymatic removal of the modification and histone turnover.
Domain Sizes in the Presence of Mutually Exclusive Modifications.
The modification level of a nucleosome in steady state in the presence of an antagonizing modification reaction is given by
| [S5] |
Here, km and k−m are the modification and reverse rate, and ka and k−a are the modification and reverse rate for the antagonizing modification, respectively. The apparent reverse rate was defined as
Eq. S5 shows that antagonizing reactions that occur with constant rate ka throughout the domain simply increase the apparent reverse rate, leading to smaller domains. In the general case, in which ka may depend on the position of the substrate nucleosome within the array, a modification level of 50% is reached at the point at which the following relationship among rate constants holds
| [S6] |
Here, km(x) and ka(x) can vary across the array, and k−m and k−a are considered to be constant. The modification reaction is assumed to be strong enough to produce modification levels above 50%, in which case all rate constants are positive. As can easily be seen from Eq. S6, larger modification rates km(x) are needed in the presence of an enzyme that catalyzes the antagonizing modification [ka(x) > 0] to reach a modification level of 50%, which means that the resulting domain becomes smaller.
Discussion
Inspired by recent epigenome editing experiments in yeast and mammalian cells, we simulated a generalized nucleation-and-looping model and compared it to linear spreading. Our results show how chromatin-bound and soluble histone modifiers can produce confined domains with characteristic size and temporal stability. The looping model explains many key experimental observations on heterochromatin and predicts the spatiotemporal behavior of histone modification patterns based on the binding distribution and the biochemical properties of the histone modifiers involved.
Comparison Between Linear and Looping-Driven Spreading.
The simulations presented here reveal similarities and important differences between looping-driven and linear spreading of histone modifications. The differences affect both the spatiotemporal behavior of modification domains formed around chromatin-bound modifiers as summarized below and the relationship between genome organization and histone modification patterns.
(i) Looping-driven spreading does not require feedback, which means that modifications that are not propagated by enzymes that bind to them can also form extended domains around a chromatin-bound modifier. In contrast, linear spreading strictly relies on the presence of feedback. (ii) For any set of catalytic rates, looping-driven spreading occurs much faster than linear spreading. The reason is that long-range interactions in the looping model efficiently propagate the modification although they occur at much lower frequency than next neighbor interactions. (iii) Memory and spreading are independently regulated in the looping model because memory is caused by feedback and spreading can be caused by both feedback and nucleation. Therefore, the looping model allows for large domains that are not inherited or small domains that are inherited. According to the linear spreading model large domains require relatively strong feedback, which inevitably induces memory. (iv) Looping-driven spreading facilitates stable epigenetic memory at conditional nucleation sites, for strong nucleation even in the absence of feedback. Although linear spreading might be able to create similar effects with other configurations of conditional nucleation sites, it would require stronger sites at higher concentration than the looping model and would strictly depend on feedback to generate memory.
The Role of the Reader Domain in Looping-Driven Spreading.
Based on the conclusions above, we propose that the chromodomain of Clr4/Suv39h that recognizes H3K9 methylation has two functions. On the one hand, it enables short-term memory at methylated domains independently of their genomic position by transiently binding to methylated histones and locally propagating the methylation mark (Fig. 4). On the other hand, it stabilizes the enzyme at its endogenous nucleation sites in heterochromatin by converting weak into strong binding (Fig. 6).
In this view, the reduction of the domain size observed at endogenous loci on deletion (47) or mutation (49) of the Clr4 chromodomain is primarily caused by reduced nucleation strength rather than by reduced feedback strength. In line with this model, replacement of the Clr4 chromodomain with that of Swi6/HP1 has a similar effect because Clr4 is redirected to other sites that contain H3K9me2, leading to reduced Clr4 levels at the nucleation site that is marked by H3K9me3 (35). This result reconciles the experimental findings that the chromodomain is required for heterochromatin spreading at endogenous loci (35, 47, 49) but not at ectopic sites where its function in nucleation is bypassed by TetR or Gal4, leading to stably nucleated modifiers that produce broad domains with very similar domain sizes either in the presence or absence of the Clr4 chromodomain (15, 16, 18).
Looping-Driven Spreading and Genome Folding.
As shown above, the looping model is consistent with experiments on engineered H3K9 methylation domains (Fig. 5) and can explain epigenetic memory in the presence of conditional nucleation sites (Fig. 6). Because looping-driven spreading depends on spatial rather than genomic proximity, the model makes distinct predictions for the function of boundary elements that we did not consider in the context of the simulations above. In particular, it can reproduce phenomena like discontinuous heterochromatin spreading and domain skipping (Fig. S8), which have previously been observed in experiments (51–53). Furthermore, the fact that proteins involved in the regulation of genome folding act as boundary factors for spreading (54–56) are fully consistent with the expectations for a looping-driven spreading mechanism involving long-range interactions, whereas linear spreading should not be sensitive to higher-order chromatin structure. The fact that loci enriched for H3K9 methylation tend to form spatial clusters and preferentially contact each other (41, 54, 57) is consistent with the idea that the modification can also spread to distant loci on the same or a different chromosome if contacts are sufficiently frequent.
Fig. S8.
Boundary elements. (A) If a stretch of nucleosomes is kept unmodified (green bar, Upper), linear spreading stops at the boundary, which is reflected by the steady-state distribution after recruitment of the modifier (Upper). The time evolution of the average modification level left from the boundary is shown in Lower. The modifier was released at t·k−m = 1,000. (B) According to the looping model, boundaries do not necessarily stop spreading. Rather, chromatin-bound modifiers in the vicinity of a gap have the ability to form modified domains on both sides of the gap if the contact probability is high enough and the modifier is strong enough (Upper). On removal of the modifier, the modification level on both sides of the gap decays if feedback is below the critical threshold (Lower Left) or persists if feedback is above the critical threshold (Lower Right). The skipping efficiency depends on the activity of the modifier, its distance to the boundary, and the size of the boundary. Boundary factors that reduce the contact probability between the modifier and the substrate nucleosomes on the other side of the boundary, e.g., by altering higher-order chromatin structure, are expected to block looping-driven spreading.
Conclusions
Although the current work was motivated by experiments on engineered H3K9 methylation domains, the results might have implications for other histone modifications. Recent experiments show that targeted recruitment of an acetyltransferase can also lead to the formation of a discontinuous extended domain (24), suggesting that spreading phenomena might not be restricted to heterochromatic histone methylation. Indeed, most modifications form domains that at least in cell ensembles extend beyond a single nucleosome (58, 59), and it is unclear if local spreading contributes to their genomic distribution. The reason why H3K9 di/trimethylation might spread more efficiently than other modifications in the endogenous situation could be the unusually strong recruitment of histone methyltransferases to their unusually high number of binding sites in native heterochromatin. It will be interesting to see in future epigenome editing experiments which modifications can spread around chromatin-bound modifiers and which modifications require local feedback and confer epigenetic memory. We hope that our work will stimulate experiments designed to probe the interplay among chromatin-bound histone modifiers, chromatin structure and the spatiotemporal response of the chromatin landscape.
Materials and Methods
Stochastic Simulations.
Stochastic simulations were carried out according to the Gillespie algorithm (60) implemented in Java. At least 500 independent simulation runs were performed to determine steady-state profiles and time evolutions. For histograms containing heterogeneous populations, 5,000 simulation runs were used. For further details, see SI Materials and Methods.
Simulation of Cell Colonies.
For the simulation of cell colonies, we assigned individual simulation trajectories to cells. Cell divisions were carried out after multiples of the inverse reverse rate 1/k−m by replacing each cell with two daughter cells carrying the distribution of modified nucleosomes from the parent cell. In case of spatial overlap, cells were shifted apart from each other. The direction of motion was calculated as the weighted sum of connecting lines between a cell and each overlapping cell. The distances between each pair of overlapping cells were used as weighting factors.
Conversion Between Base Pairs and Nucleosomes.
To compare simulations with experimentally observed H3K9 methylation profiles from fission yeast, base pairs were converted into nucleosomes using the experimentally determined nucleosome repeat length of 154 bp (61).
Supplementary Material
Acknowledgments
We thank Songtao Jia, Danesh Moazed, Geeta Narlikar, Oliver Rando, Karsten Rippe, Dan Duzdevich, Corentin Moevus, Johannes Stigler, and Tsuyoshi Terakawa for help and discussions. Part of the simulations were conducted with computational resources provided via the German Federal Ministry of Education and Research (BMBF) Grant 01IG07015G of the Services@MediGRID program. F.E. is the recipient of a long-term fellowship from the European Molecular Biology Organization (EMBO LTF 187-2014). This work was supported by NIH Grants R01GM082848 and R01GM074739 (to E.C.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605862113/-/DCSupplemental.
References
- 1.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 2.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011;25(5):409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 4.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 5.Talbert PB, Henikoff S. Spreading of silent chromatin: Inaction at a distance. Nat Rev Genet. 2006;7(10):793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 6.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol. 2013;5(8):a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292(5524):2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- 8.Jermann P, Hoerner L, Burger L, Schübeler D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci USA. 2014;111(33):E3415–E3421. doi: 10.1073/pnas.1400672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13(11):2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akhtar W, et al. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154(4):914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Muller HJ. Types of visible variations induced by X-rays in Drosophila. J Genet. 1930;22(3):299–334. [Google Scholar]
- 12.Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: The properties and functions of repressive Lys methyltransferases. Nat Rev Mol Cell Biol. 2015;16(8):499–513. doi: 10.1038/nrm4029. [DOI] [PubMed] [Google Scholar]
- 13.Schuettengruber B, Cavalli G. Polycomb domain formation depends on short and long distance regulatory cues. PLoS One. 2013;8(2):e56531. doi: 10.1371/journal.pone.0056531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10(11):1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 15.Audergon PNCB, et al. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science. 2015;348(6230):132–135. doi: 10.1126/science.1260638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragunathan K, Jih G, Moazed D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348(6230):1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149(7):1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagansky A, et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324(5935):1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kungulovski G, et al. Targeted epigenome editing of an endogenous locus with chromatin modifiers is not stably maintained. Epigenetics Chromatin. 2015;8:12. doi: 10.1186/s13072-015-0002-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12(24):2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- 21.Bintu L, et al. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351(6274):720–724. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geutjes EJ, Bajpe PK, Bernards R. Targeting the epigenome for treatment of cancer. Oncogene. 2012;31(34):3827–3844. doi: 10.1038/onc.2011.552. [DOI] [PubMed] [Google Scholar]
- 23.Plass C, et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14(11):765–780. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 24.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakore PI, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12(12):1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falahi F, Sgro A, Blancafort P. Epigenome engineering in cancer: Fairytale or a realistic path to the clinic? Front Oncol. 2015;5:22. doi: 10.3389/fonc.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Köeferle A, Stricker SH, Beck S. Brave new epigenomes: The dawn of epigenetic engineering. Genome Med. 2015;7(1):59. doi: 10.1186/s13073-015-0185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146(4):510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bantignies F, Cavalli G. Polycomb group proteins: Repression in 3D. Trends Genet. 2011;27(11):454–464. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Hodges C, Crabtree GR. Dynamics of inherently bounded histone modification domains. Proc Natl Acad Sci USA. 2012;109(33):13296–13301. doi: 10.1073/pnas.1211172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476(7358):105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 32.Müller-Ott K, et al. Specificity, propagation, and memory of pericentric heterochromatin. Mol Syst Biol. 2014;10(8):746. doi: 10.15252/msb.20145377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129(4):813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 34.van Holde KE. Chromatin. Springer; New York: 1989. [Google Scholar]
- 35.Al-Sady B, Madhani HD, Narlikar GJ. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol Cell. 2013;51(1):80–91. doi: 10.1016/j.molcel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller MM, Fierz B, Bittova L, Liszczak G, Muir TW. A two-state activation mechanism controls the histone methyltransferase Suv39h1. Nat Chem Biol. 2016;12(3):188–193. doi: 10.1038/nchembio.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dostie J, et al. Chromosome conformation capture carbon copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ringrose L, Chabanis S, Angrand PO, Woodroofe C, Stewart AF. Quantitative comparison of DNA looping in vitro and in vivo: Chromatin increases effective DNA flexibility at short distances. EMBO J. 1999;18(23):6630–6641. doi: 10.1093/emboj/18.23.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18(4):424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 40.Erdel F, Müller-Ott K, Rippe K. Establishing epigenetic domains via chromatin-bound histone modifiers. Ann N Y Acad Sci. 2013;1305:29–43. doi: 10.1111/nyas.12262. [DOI] [PubMed] [Google Scholar]
- 41.Mizuguchi T, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516(7531):432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA. 2014;111(25):9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan W, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337(6097):971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 45.Fantes PA. Control of cell size and cycle time in Schizosaccharomyces pombe. J Cell Sci. 1977;24:51–67. doi: 10.1242/jcs.24.1.51. [DOI] [PubMed] [Google Scholar]
- 46.Aygün O, Mehta S, Grewal SI. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol. 2013;20(5):547–554. doi: 10.1038/nsmb.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15(4):381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 48.Melcher M, et al. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol. 2000;20(10):3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noma K, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36(11):1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 50.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20(2):173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Belyaeva ES, Zhimulev IF. Cytogenetic and molecular aspects of position effect variegation in Drosophila. III. Continuous and discontinuous compaction of chromosomal material as a result of position effect variegation. Chromosoma. 1991;100(7):453–466. doi: 10.1007/BF00364556. [DOI] [PubMed] [Google Scholar]
- 52.Talbert PB, Henikoff S. A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics. 2000;154(1):259–272. doi: 10.1093/genetics/154.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogel MJ, et al. High-resolution mapping of heterochromatin redistribution in a Drosophila position-effect variegation model. Epigenetics Chromatin. 2009;2(1):1. doi: 10.1186/1756-8935-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamakaka RT. Chromatin: A connection between loops and barriers? Curr Biol. 2002;12(15):R535–R537. doi: 10.1016/s0960-9822(02)01032-1. [DOI] [PubMed] [Google Scholar]
- 56.Yang J, Corces VG. Insulators, long-range interactions, and genome function. Curr Opin Genet Dev. 2012;22(2):86–92. doi: 10.1016/j.gde.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148(3):458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Yue F, et al. Mouse ENCODE Consortium A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillespie DT. Exact stochastic simulation of coupled chemical-reactions. J Phys Chem. 1977;81(25):2340–2361. [Google Scholar]
- 61.Lantermann AB, et al. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17(2):251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 62.Chin HG, Patnaik D, Estève PO, Jacobsen SE, Pradhan S. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: Participation of the chromodomain in enzymatic catalysis. Biochemistry. 2006;45(10):3272–3284. doi: 10.1021/bi051997r. [DOI] [PubMed] [Google Scholar]
- 63.Wang T, et al. Crystal structure of the human SUV39H1 chromodomain and its recognition of histone H3K9me2/3. PLoS One. 2012;7(12):e52977. doi: 10.1371/journal.pone.0052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuhmacher MK, Kudithipudi S, Kusevic D, Weirich S, Jeltsch A. Activity and specificity of the human SUV39H2 protein lysine methyltransferase. Biochim Biophys Acta. 2015;1849(1):55–63. doi: 10.1016/j.bbagrm.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Schalch T, et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34(1):36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zee BM, et al. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285(5):3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamionka A, Bogdanska-Urbaniak J, Scholz O, Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004;32(2):842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parthun MR, Jaehning JA. Purification and characterization of the yeast transcriptional activator GAL4. J Biol Chem. 1990;265(1):209–213. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.