Significance
The essence of the adaptive immune response depends on the specificity of antigen receptors. This report identifies shared alpha–beta T-cell receptor (TCR) pairs from the tissues of HLA-A2+ patients with breast cancer and control donors. Using an emulsion-based RT-PCR assay, we analyzed TCR sequences from tissues ex vivo. We identified multiple TCR pairs shared between tumors, but not control samples. Although recent reports have concluded that anticancer T-cell responses depend on patient-specific mutation-associated neoantigens, this study provides evidence that T cells also recognize shared antigens. This approach has broad application to a variety of research questions where the end goal is to examine T-cell repertoires and/or identify T-cell antigens.
Keywords: T-cell receptors, breast cancer, emulsion RT-PCR, high-throughput sequencing, T-cell repertoire profiling
Abstract
Infiltration of T cells in breast tumors correlates with improved survival of patients with breast cancer, despite relatively few mutations in these tumors. To determine if T-cell specificity can be harnessed to augment immunotherapies of breast cancer, we sought to identify the alpha–beta paired T-cell receptors (TCRs) of tumor-infiltrating lymphocytes shared between multiple patients. Because TCRs function as heterodimeric proteins, we used an emulsion-based RT-PCR assay to link and amplify TCR pairs. Using this assay on engineered T-cell hybridomas, we observed ∼85% accurate pairing fidelity, although TCR recovery frequency varied. When we applied this technique to patient samples, we found that for any given TCR pair, the dominant alpha- or beta-binding partner comprised ∼90% of the total binding partners. Analysis of TCR sequences from primary tumors showed about fourfold more overlap in tumor-involved relative to tumor-free sentinel lymph nodes. Additionally, comparison of sequences from both tumors of a patient with bilateral breast cancer showed 10% overlap. Finally, we identified a panel of unique TCRs shared between patients’ tumors and peripheral blood that were not found in the peripheral blood of controls. These TCRs encoded a range of V, J, and complementarity determining region 3 (CDR3) sequences on the alpha-chain, and displayed restricted V-beta use. The nucleotides encoding these shared TCR CDR3s varied, suggesting immune selection of this response. Harnessing these T cells may provide practical strategies to improve the shared antigen-specific response to breast cancer.
Infiltration of numerous tumors by CD8+ alpha–beta T cells is associated with better outcomes and longer survival times for patients with breast cancer (1–5). Targeted immune-based therapies hold great promise toward improving breast cancer treatments (6, 7). Studies have examined the immune phenotype of breast cancer tumor-infiltrating lymphocytes (TILs), suggesting that activated nonsuppressive T cells are of most benefit (8). Further assessment of the TIL repertoire has broad implications for breast cancer therapies in antigen discovery, cancer vaccines, and adoptive cell therapies (7).
Unique genetic recombination events are required to produce the T-cell receptor (TCR) (reviewed in 9). The alpha- and beta-chains of the TCR undergo V(D)J recombination and heterodimerize in the thymus, resulting in a diverse T-cell repertoire that specifically recognizes peptide–MHC complexes. Many studies have analyzed the alpha- and beta-chains of TIL TCRs separately using high-throughput sequencing to describe the diversity of TILs (10–13). Pairs are readily identified after expansion of T-cell clones, although culture of T cells can lead to substantial skewing of the repertoire (14), which may select for T cells of varied affinity or avidity (15). Single-cell sequencing identifies alpha–beta pairs, but is often laborious and has relatively low throughput. Tumor-reactive TCR pairs can be generated from activated T cells after culture with an autologous tumor using gene capture followed by frequency-based matching (11). Recently, a high-throughput statistical method to determine the matched alpha–beta pairs was published, demonstrating high-fidelity pairing by this combinatorial analysis (16).
We sought to develop a method that was well-suited for limiting numbers of T cells ex vivo. In addition, we reasoned that identified TCRs shared between patients would target shared antigens due to the low incidence of common mutations in breast cancer as well as the low mutational burden (17). Methods using emulsion PCR increase the sensitivity and fidelity of standard PCR reactions (18), and were recently applied to paired TCR repertoire analysis of peripheral blood CD8 T cells (19). This technique facilitates the analysis of large numbers of TCR pairs from single cells. However, the technique, as published, is limited to amplification of a single V-beta gene and a limited number of V-alpha genes. To elucidate the T-cell repertoire responding to breast cancer, we required a method that included primers recognizing all potential TCR V genes.
We developed an expanded version of the emulsion RT-PCR protocol and then applied it to the CD8+ TCR repertoires of HLA-A2+ tumors, lymph nodes (LNs), and peripheral blood lymphocytes (PBLs) from patients with breast cancer and PBLs from control donors. Using this method on hybridomas whose alpha-TCR pairs were known, we showed that 85% of the alpha- and beta-chains paired with the expected partner. Within patient samples with an unknown repertoire, we found that for any given TCR pair, a dominant alpha- or beta-binding partner comprised ∼90% of the total binding partners for the analyzed alpha- or beta-TCR. Comparing TIL TCRs with TCRs found in the corresponding sentinel LNs, we found a fourfold increase in repertoire overlap in LNs that were tumor-involved relative to LNs that were not. TIL sequences from a patient with bilateral breast cancer showed 10% overlap between the repertoires of the two tumors. Finally, we identified multiple shared alpha–beta pairs among patient tumors that were not present in control samples.
Results
To identify alpha- and beta-TCR sequences of TILs, we modified the emulsion RT-PCR protocol of Turchaninova et al. (19) to include primers for all V-alpha and V-beta TCR genes (Tables S1 and S2). These primers included overlapping complementary regions to facilitate annealing of the appropriate alpha and beta genes to each other during the initial PCR. Following purification of DNA from the emulsion, a second PCR using nested C-region primers and blocking oligos with noncomplementary 3′ ends (Table S3) was used to amplify alpha–beta joined products and suppress amplification of unpaired genes, respectively. A third nested PCR was performed to add the adaptors and barcodes for Illumina sequencing (Table S3). For analysis of the sequence reads of the paired alpha- and beta-TCRs, we developed a modified version of MiTCR (20), which we have named CompleteTCR.
Table S1.
Primers for TRAV gene fragments
| Conc, nM | TRAV gene | Complement TRBV primer overlap | TRAV-specific | Real-time PCR Ct* |
| 60 | 1 | attgggcaggttgtccctgatt | AGGAGCTCCAGATGAAAGACTC | 29.39 |
| 60 | 2 | attgggcaggttgtccctgatt | GCTCATCCTCCAGGTGCGGGAG | 17.23 |
| 60 | 3 | attgggcaggttgtccctgatt | CCTCCTTCCACCTGAAGAAACC | 29.73 |
| 60 | 4 | attgggcaggttgtccctgatt | CCTGAGCGACACTGCTGTGTAC | 28.94 |
| 60 | 5 | attgggcaggttgtccctgatt | CTGTCTCTGCGCATTGCAGACA | 18.95 |
| 60 | 6 | attgggcaggttgtccctgatt | GACTGAAGGTCACCTTTGATACC | 33.36 |
| 60 | 7 | attgggcaggttgtccctgatt | GCCGTGCAGCCTGAAGATTCA | 33.70 |
| 60 | 8 | attgggcaggttgtccctgatt | CCTGTGATTTCCTATGCCTGTC | 22.31 |
| 60 | 8-1 | attgggcaggttgtccctgatt | TCCTTTAATCTGAGGAAACCCTC | 25.56 |
| 60 | 8-2 | attgggcaggttgtccctgatt | GAATTTAAGAGGAGTCAATCTTCC | 30.28 |
| 60 | 8-3 | attgggcaggttgtccctgatt | CATCAGAGGTTTTGAGGCTGAAT | 19.41 |
| 60 | 8-4 | attgggcaggttgtccctgatt | CATCAACGGTTTTGAGGCTGAAT | 16.37 |
| 60 | 9 | attgggcaggttgtccctgatt | GAAACCACTTCTTTCCACTTGGA | 30.77 |
| 60 | 10 | attgggcaggttgtccctgatt | CTGCACATCACAGCCTCCCAG | 26.79 |
| 60 | 11 | attgggcaggttgtccctgatt | GAATATCGCAGCCTCTCATCTG | 29.34 |
| 60 | 12 | attgggcaggttgtccctgatt | CAGTGATTCAGCCACCTACCTC | 26.95 |
| 60 | 13-1 | attgggcaggttgtccctgatt | GACAGCCAAACATTTCTCCCTG | 27.38 |
| 60 | 13-2 | attgggcaggttgtccctgatt | GACAGTGAAACATCTCTCTCTGC | 24.30 |
| 60 | 14 | attgggcaggttgtccctgatt | CCAACCTTGTCATCTCCGCTTC | 30.14 |
| 60 | 15 | attgggcaggttgtccctgatt | GAATATGCTGGTCTCTCATCCTG | 29.31 |
| 60 | 16 | attgggcaggttgtccctgatt | CCATTTGCTCAAGAGGAAGACTC | 30.20 |
| 60 | 17 | attgggcaggttgtccctgatt | CTTGTTGATCACGGCTTCCCG | 28.25 |
| 60 | 18 | attgggcaggttgtccctgatt | AGAAGCCCTCGGTGCAGCTG | 17.24 |
| 60 | 19 | attgggcaggttgtccctgatt | CCACCAGTTCCTTCAACTTCAC | 29.69 |
| 60 | 20 | attgggcaggttgtccctgatt | CACATCACAGCCCCTAAACCTG | 19.99 |
| 60 | 21 | attgggcaggttgtccctgatt | CATCAGGACGTAGTACTTTATACA | 32.85 |
| 60 | 22 | attgggcaggttgtccctgatt | GTACATTTCCTCTTCCCAGACC | 28.35 |
| 60 | 23 | attgggcaggttgtccctgatt | GCAGTTCTCATTGCATATCATGG | 23.95 |
| 60 | 24 | attgggcaggttgtccctgatt | ATCAAAGGATCCCAGCCTGAAG | 29.36 |
| 60 | 25 | attgggcaggttgtccctgatt | GCACATCACAGCCACCCAGAC | 28.69 |
| 60 | 26-1 | attgggcaggttgtccctgatt | AGTCCAGCACCTTGATCCTGC | 25.82 |
| 60 | 26-2 | attgggcaggttgtccctgatt | ACAGAAAGTCCAGTACCTTGATC | 30.31 |
| 60 | 27 | attgggcaggttgtccctgatt | AGGACAGTTCTCTCCACATCAC | 30.18 |
| 60 | 28 | attgggcaggttgtccctgatt | CTATACATCAGATTCCCAGCCTG | 23.75 |
| 60 | 29 | attgggcaggttgtccctgatt | GAGACTCTGCAGTGTACTTCTG | 28.51 |
| 60 | 30 | attgggcaggttgtccctgatt | GCAAAGCTCCCTGTACCTTACG | 32.32 |
| 60 | 31 | attgggcaggttgtccctgatt | GCTTATCATATCATCATCACAGCC | 28.13 |
| 60 | 32 | attgggcaggttgtccctgatt | CCCAACCAGGAGACTCATTCC | 31.49 |
| 60 | 33 | attgggcaggttgtccctgatt | TCCTTAAAACTGACTCAGCCAAG | 29.19 |
| 60 | 34 | attgggcaggttgtccctgatt | GCAAAGTTCCCTGCATATCACAG | 31.65 |
| 60 | 35 | attgggcaggttgtccctgatt | AGCTTCCTGAATATCTCAGCATC | 27.92 |
| 60 | 36 | attgggcaggttgtccctgatt | CAGCATCCTGAACATCACAGCC | 28.48 |
Conc, concentration; TRAV, TCR variable region alpha.
Real-time PCR Ct values were determined using PBL cDNA from four control donors.
Table S2.
Primers for TRBV gene fragments
| Conc, nM | TRBV gene | Complementary TRAV primer overlap | TRBV-specific | Real-time PCR Ct* |
| 60 | 1 | aatcagggacaacctgcccaat | GACAGCTCTCGCTTATACCTTC | 25.86 |
| 60 | 2 | aatcagggacaacctgcccaat | CAAATTTCACTCTGAAGATCCGGTCC | 29.58 |
| 60 | 3-1 | aatcagggacaacctgcccaat | CTCACTTAAATCTTCACATCAATTC | 25.45 |
| 60 | 4-1 | aatcagggacaacctgcccaat | CCTTCACCTACACGCCCTGC | 27.29 |
| 60 | 4-2/3 | aatcagggacaacctgcccaat | TCCTTCACCTACACACCCTGC | 24.18 |
| 60 | 5-1 | aatcagggacaacctgcccaat | GCTCTGAGATGAATGTGAGCAC | 24.78 |
| 60 | 5-3 | aatcagggacaacctgcccaat | GCTCTGAGATGAATGTGAGTGC | 24.87 |
| 60 | 5-4/5/6/7/8 | aatcagggacaacctgcccaat | GCTCTGAGCTGAATGTGAACGC | 26.50 |
| 60 | 6-1 | aatcagggacaacctgcccaat | TCGCTCAGGCTGGAGTCGGC | 24.67 |
| 60 | 6-2/3 | aatcagggacaacctgcccaat | GCTGGGGTTGGAGTCGGCTG | 25.62 |
| 60 | 6-4 | aatcagggacaacctgcccaat | CCTCACGTTGGCGTCTGCTG | 25.96 |
| 60 | 6-5 | aatcagggacaacctgcccaat | GCTCAGGCTGCTGTCGGCTG | 24.60 |
| 60 | 6-6 | aatcagggacaacctgcccaat | GCTCAGGCTGGAGTTGGCTG | 24.76 |
| 60 | 6-7 | aatcagggacaacctgcccaat | CCCTCAAGCTGGAGTCAGCTG | 27.94 |
| 60 | 6-8 | aatcagggacaacctgcccaat | ACTCAGGCTGGTGTCGGCTG | 25.87 |
| 60 | 6-9 | aatcagggacaacctgcccaat | GCTCAGGCTGGAGTCAGCTG | 25.59 |
| 60 | 7-1 | aatcagggacaacctgcccaat | CACTCTGAAGTTCCAGCGCAC | 24.87 |
| 60 | 7-2 | aatcagggacaacctgcccaat | CACTCTGACGATCCAGCGCAC | 24.20 |
| 60 | 7-3 | aatcagggacaacctgcccaat | ACTCTGAAGATCCAGCGCACAG | 24.14 |
| 60 | 7-4 | aatcagggacaacctgcccaat | ACTCTGAAGATCCAGCGCACAG | 24.22 |
| 60 | 7-6 | aatcagggacaacctgcccaat | CACTCTGACGATCCAGCGCAC | 23.90 |
| 60 | 7-7 | aatcagggacaacctgcccaat | CACTCTGACGATTCAGCGCAC | 24.80 |
| 60 | 7-8 | aatcagggacaacctgcccaat | CACTCTGAAGATCCAGCGCAC | 24.05 |
| 60 | 7-9 | aatcagggacaacctgcccaat | CACCTTGGAGATCCAGCGCAC | 24.17 |
| 60 | 9 | aatcagggacaacctgcccaat | GCACTCTGAACTAAACCTGAGC | 26.77 |
| 60 | 10-1 | aatcagggacaacctgcccaat | CCCTCACTCTGGAGTCTGCTG | 26.44 |
| 60 | 10-2 | aatcagggacaacctgcccaat | CCCTCACTCTGGAGTCAGCTA | 27.16 |
| 60 | 10-3 | aatcagggacaacctgcccaat | CTCCTCACTCTGGAGTCCGC | 25.23 |
| 60 | 11 | aatcagggacaacctgcccaat | CACTCTCAAGATCCAGCCTGCA | 25.57 |
| 60 | 12-3/4/5 | aatcagggacaacctgcccaat | CACTCTGAAGATCCAGCCCTC | 24.41 |
| 60 | 13 | aatcagggacaacctgcccaat | CATTCTGAACTGAACATGAGCTCC | 26.92 |
| 60 | 14 | aatcagggacaacctgcccaat | CTACTCTGAAGGTGCAGCCTG | 24.93 |
| 60 | 15 | aatcagggacaacctgcccaat | GATAACTTCCAATCCAGGAGGC | 26.07 |
| 60 | 16 | aatcagggacaacctgcccaat | GCCTTGAGATCCAGGCTACGA | 29.83 |
| 60 | 17 | aatcagggacaacctgcccaat | CTTCCACGCTGAAGATCCATCC | 28.85 |
| 60 | 18 | aatcagggacaacctgcccaat | GCATCCTGAGGATCCAGCAGG | 19.87 |
| 60 | 19 | aatcagggacaacctgcccaat | CCTCTCACTGTGACATCGGCC | 24.69 |
| 60 | 20-1 | aatcagggacaacctgcccaat | CCACTCTGACAGTGACCAGTG | 25.47 |
| 60 | 23-1 | aatcagggacaacctgcccaat | CAGCCTGGCAATCCTGTCCTC | 25.95 |
| 60 | 24-1 | aatcagggacaacctgcccaat | CTCCCTGTCCCTAGAGTCTGC | 26.85 |
| 60 | 25-1 | aatcagggacaacctgcccaat | CCCTGACCCTGGAGTCTGCC | 25.46 |
| 60 | 27 | aatcagggacaacctgcccaat | CCTGATCCTGGAGTCGCCCA | 23.24 |
| 60 | 28 | aatcagggacaacctgcccaat | CTCCCTGATTCTGGAGTCCGC | 25.35 |
| 60 | 29-1 | aatcagggacaacctgcccaat | CTAACATTCTCAACTCTGACTGTG | 25.64 |
| 60 | 30 | aatcagggacaacctgcccaat | CAGTTCATCCTGAGTTCTAAGAAG | 30.24 |
Conc, concentration; TRBV, TCR variable region beta.
Real-time PCR Ct values were determined using PBL cDNA from four control donors.
Table S3.
Other primers
| Conc, nM | Name | Sequence (5′ to 3′) |
| Initial PCR | ||
| 600 | Calpha Rev | ACACATCAGAATCCTTACTTTGTG |
| 600 | Cbeta Rev | GCAGTATCTGGAGTCATTGAGG |
| 600 | Stepout | ATTGGGCAGGTTGTCCCTGATT |
| First nested PCR | ||
| 200 | Calpha nest 1 | AATCGGTGAATAGGCAGACAGAC |
| 200 | Cbeta nest 1 | TGATGGCTCAAACACAGCG |
| 1,550 | Valpha block | TTTTTTTATTGGGCAGGTTGTCCCTGATTTTT/3PHOS/ |
| 1,550 | Vbeta block | TTTTTTTAATCAGGGACAACCTGCCCAATTTT/3PHOS/ |
| Second nested PCR | ||
| 400 | Calpha nest2 | CAAGCAGAAGACGGCATACGAGATINDEXXGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNTCAGGGTTCTGGATAT |
| 400 | Cbeta nest2 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNXXXXCCTCGGGTGGGAACAC |
INDEXX, internal indices used to differentiate among samples; NNNNN, zero to eight randomized nucleotides to provide cluster diversification for the MiSeq analysis; XXXX, a second set of internal indices used to differentiate among samples.
To confirm that all 92 V-alpha and V-beta primers amplified a corresponding V-alpha or V-beta gene, we performed real-time PCR reactions using each of these primers and pooled cDNA derived from the PBL T cells of four donors. All primer reactions produced a quantifiable product, which crossed cycle threshold (Ct) between 15 and 35 cycles (Tables S1 and S2), demonstrating that this primer set binds and amplifies all potential alpha- and beta-TCR genes.
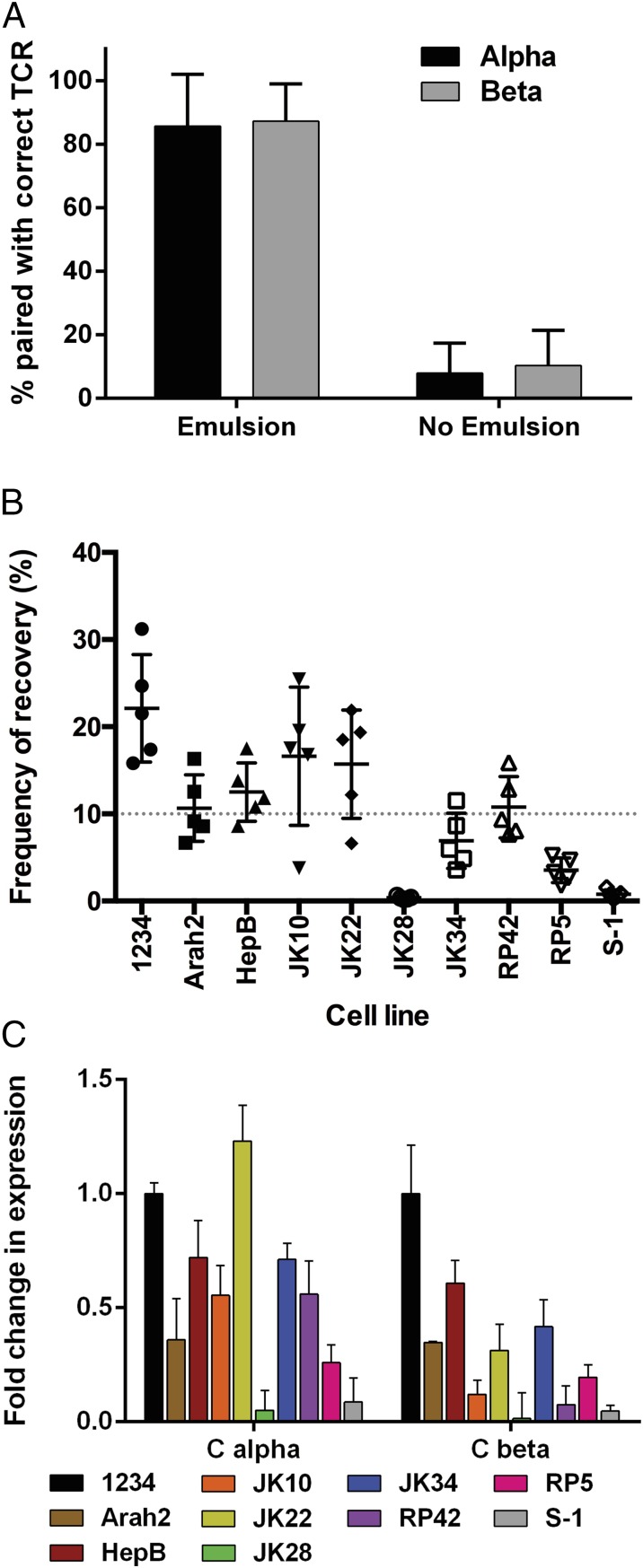
To determine the robustness of this protocol and the fidelity of alpha–beta pairing, we mixed 10 hybridomas expressing different, known alpha- and beta-TCRs before adding them to the emulsion RT-PCR protocol (Table S4). Replicate samples were processed and sequenced without the emulsion, using the same cycling parameters for all steps to determine pairing fidelity and efficiency (Fig. 1A). The frequency of TCRs paired with the correct input partner was ∼85% when the emulsion was used, and was reduced to 5–10% in the absence of the emulsion. Analysis of the incorrect pairs showed no single alpha-chain was dominantly paired with any given beta-chain, or vice versa, suggesting random pairing in the absence of the emulsion. Although the hybridomas were added in equal numbers, the recovered read frequencies rarely reflected the input frequencies (Fig. 1B). Real-time PCR of the TCR transcripts from the hybridomas before adding them to the emulsion showed the ratio of the alpha- to beta-transcript, and the amount of transcript between clones varied greatly, likely skewing the resulting sequences (Fig. 1C).
Table S4.
TRAV, TRAJ, TRBV, and TRBJ use and CDR3 sequences for control T-cell hybridomas
| Clone | TRAV | CDR3α | TRAJ | TRBV | CDR3β | TRBJ |
| S-1 | 12-1 | CVVMGGPGSNYQLIW | 33 | 5-1 | CASSPGQRVGANVLTF | 2-6 |
| JK28 | 9-2 | CALSLYSGAGSYQLTF | 28 | 5-1 | CASSLAQGGETQYF | 2-5 |
| RP42 | 8-6 | CAVSRVSGGYNKLIF | 4 | 28 | CATGGRARGTGELFF | 2-2 |
| HepB | 26-1 | CIAWNSGGSNYKLTF | 53 | 20-1 | CSAPGGGVYNSPLHF | 1-6 |
| 1234 | 26-1 | CIVRVWDAGNNRKLIW | 38 | 18 | CASSPNGQAHEQYF | 2-7 |
| JK10 | 2 | CAVEDQNQGGKLIF | 23 | 6-2/3 | CASRSQGTCNQPQHF | 1-5 |
| Arah2 | 17 | CATDTSAVEYGNKLVF | 47 | 4-1 | CASSQEPSGSYYEQYF | 2-7 |
| RP5 | 9-2 | CALSDNTGFQKLVF | 8 | 28 | CASSLGDQPQHF | 1-5 |
| JK34 | 2 | CAVHNNAGNMLTF | 39 | 2 | CASRPPGLGSINEQFF | 2-1 |
| JK22 | 13-1 | CAASSYSGAGSYQLTF | 28 | 5-1 | CASSLAQGSEKLFF | 1-4 |
TRAJ, TCR joining region alpha; TRBJ, TCR joining region beta.
Fig. 1.
Recovery of TCR pairs by emulsion RT-PCR from 5KC hybridomas. (A) Ten 5KC T-cell hybridomas were mixed in equal numbers, and emulsion RT-PCR was performed either with or without emulsion phase. Sequences were analyzed for pairing with the correct alpha-TCR (black) or beta-TCR (gray). Percentages represent the number of reads that paired with the input alpha or beta out of the total number of reads for the analyzed alpha or beta (n = 5 replicate experiments). (B) Frequency that a given TCR pair was recovered after being added in equivalent numbers is shown as a percentage of the total reads recovered (n = 5 replicate experiments). (C) Real-time quantitative PCR for the transcript abundance in the various 5KC hybridomas before addition to the emulsion RT-PCR normalized to the amount in the 1234 hybridomas (n = 3 replicate experiments). Error bars represent SD from the mean. The average read count was 670,000 across five experiments.
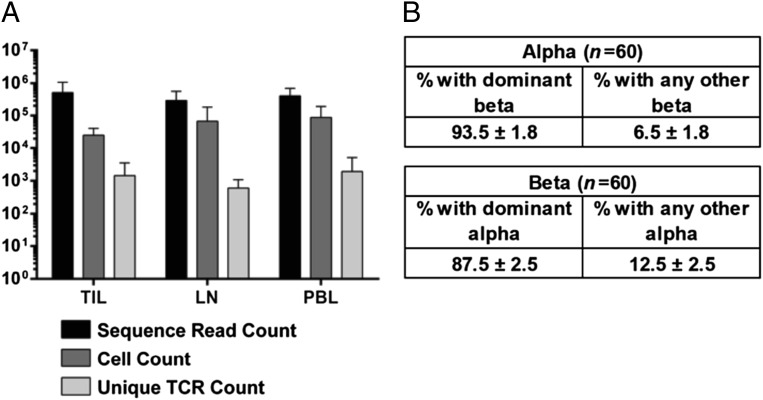
We next applied this technology to identify shared T-cell clonotypes in TILs of patients with breast cancer. Patients provided consent and were HLA-typed, and their tissue samples were collected (Table 1). CD8+ T cells were positively selected from digested tumors, LNs, and PBLs; incubated overnight in IL-2 to increase the concentration of intracellular TCR transcript; and subjected to emulsion RT-PCR (Fig. 2A). Sixty frequently identified TCR pairs across all repertoires were analyzed for the diversity of binding partners of the alpha- and beta-TCR chains. When analyzing all recovered binding partners for the alpha-chains, 90% were a single TCR beta-sequence. For the beta-chain, the percentage dropped to 87%, which we presume is due to the presence of complete allelic exclusion at the TCR-alpha-chain locus or perhaps to the thymic proliferation of the beta-chain–expressing pre-T cell before alpha-chain rearrangements (Fig. 2B).
Table 1.
Patient information
| Patient ID | Age, y | Tumor stage | Tumor grade | Sentinal LN status | ER* | PR* | Her2* | HLA-A alleles | |
| p1 | 59 | IIIA | III | + | + | + | − | 02:01 | 02:01 |
| p2 | 49 | IIA | III | NC | + | + | + | 02:01 | 03:01 |
| p3 | 55 | IIB | II | − | + | + | − | 02:01 | 01:01 |
| p4 | 54 | IIIA | II | NC | + | + | − | 02:01 | 02:01 |
| p5 | 66 | IIIA | II | + | + | + | − | 02:01 | 01:01 |
| p6 | 44 | IA | II | NC | + | + | − | 02:01 | 29:01 |
| p7 | 30 | IA | III | NC | + | + | + | 02:01 | 24:03 |
| p8 | 35 | IA | III | − | + | + | − | 02:01 | 31:01 |
| p9 | 45 | IIA | II | NC | + | + | − | 02:01 | 03:01 |
| p10 | 53 | IA | II | + | + | + | − | 02:01 | 03:01 |
| p11 | 47 | IA | II | NC | + | + | − | 02:01 | 03:01 |
| p12 | 53 | IIA | III | NC | + | + | − | 02:01 | 01:01 |
| p13 | 56 | IV | II | NC | − | − | + | 02:01 | 03:01 |
| p14 | 71 | IIA | III | + | + | − | − | 02:01 | 03:01 |
| p15 | 63 | IV | II | NC | − | − | − | 02:01 | 32:02 |
| p16 | 60 | IA | III | − | − | − | − | 02:01 | 02:01 |
| p17 | 57 | IIA | III | − | + | + | − | 02:02 | 30:02 |
| p18 | 29 | IIA | III | − | + | + | − | 02:06 | 33:01 |
| p19 | 42 | IIIA | III | − | + | + | − | 02:06 | 74:01 |
| p20 | 53 | IA | I | + | + | + | − | 02:06 | 74:01 |
| p20 | 53 | IA | II | − | + | + | − | 02:06 | 74:01 |
Samples were collected at The City of Hope Beckman Research Institute and University of Colorado Hospital Breast Center. NC, not collected; −, tumor negative; +, tumor positive.
Tumor expresses ER, PR, or Her2.
Fig. 2.
Pairing efficiency of patient-derived TCRs exhibits predominant pairing with a single partner. (A) Sequence read counts, cell counts, and unique TCR pairs are shown for each tissue repertoire analyzed (n = 21 TIL, n = 16 LN, n = 25 PBL). (B) Frequency of pairing of the dominant TCR pair out of all recovered pairs for either the dominant alpha- or beta-TCR.
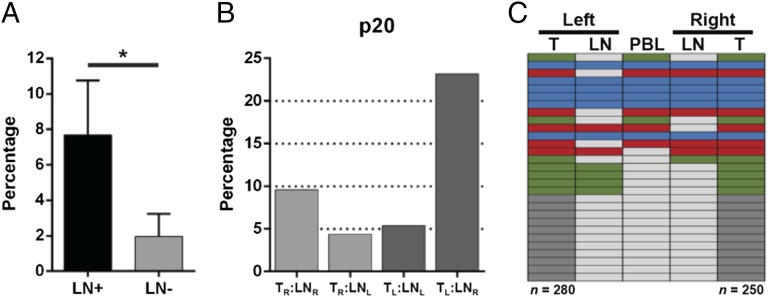
We were interested in comparing the repertoires from TILs with the repertoire of either their corresponding tumor-involved (LN+) or tumor-free (LN−) sentinel LN T cells (Fig. 3A). We found that 7.7% of the TIL TCR repertoire was also found in the corresponding LN+ repertoire, as opposed to only 2% when comparing the TIL repertoire with the LN− repertoires (Fig. 3A). We then determined the T-cell repertoire heterogeneity in tissues from distinct spatial regions of a patient who presented with estrogen receptor (ER)+/progesterone receptor (PR)+ tumors in each breast (p20; Table 1). The right LN was tumor-involved, whereas the left LN was tumor-free. A total of 9.6% of the TIL TCRs from the right tumor were found in the right LN, whereas 5.4% of left TIL TCRs were found in the left LN (Fig. 3B). Comparison of the TIL repertoires revealed 11.2% of the right TIL TCRs were found in the left TIL TCR repertoire and 10% for the reverse analysis, with similar results observed for the LNs (8.3% and 13.2%, respectively). Of the 29 TCRs common to both TIL repertoires, five were found in all tissues collected from p20 (Fig. 3C).
Fig. 3.
Comparison of TIL repertoires with the corresponding sentinel LN repertoires. (A) Number of TCRs out of the total TIL repertoire that were also present in the tumor-infiltrated (LN+, n = 5) or non–tumor-infiltrated (LN−, n = 5) sentinel LN repertoires is shown as a percentage (*P = 0.01). Patient 20 presented with tumors in each breast. The tumors were resected, along with the sentinel LN from each axilla. (B) Repertoires of the TILs and LNs [either left (L) or right (R)] were compared with each other and percentage of overlap was determined as in A. (C) Twenty-nine TCRs shared between the tumor repertoires of patient 20 are shown, with TCRs found in all five (blue), four (red), three (green), or TIL-only (dark gray) repertoire. TCRs not found in a specific repertoire are represented in light gray. n, total number of unique TCR sequences in the TIL.
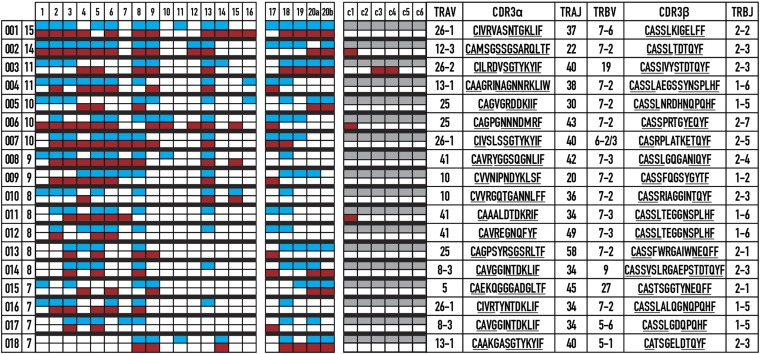
With the goal of ultimately activating endogenous breast cancer-specific immune responses present in vaccine recipients, we next determined which TCRs from the tumor samples were shared across patients, but not tumor-free controls. We hypothesized that tumor-specific T cells are present in both the tumor and the blood of patients with cancer, but exist at a lower frequency in cancer-free controls. Shared TCRs from 21 patient tumors (the majority of which were ER+PR+; Table 1) and corresponding PBLs were identified, and compared with the PBLs from six HLA-A2+ (Fig. 4) and three HLA-A2− (data not shown) healthy controls of a similar age range to the patients. The degree of overlap between the repertoires of different patients either in the PBLs or the tumor was not significantly different. Strikingly, we found 18 TCRs shared across seven or more patient tumor repertoires, many of which were also detected in the patient blood repertoires (Fig. 4 and Fig. S1A). The shared TCRs were encoded by various nucleotide sequences, suggesting antigen-based selection and expansion of these TCRs (Table S5). Additionally, only four shared TCRs were found in PBLs of three HLA-A2+ control donors, suggesting that the shared TCRs were predominantly restricted to patients with cancer (Fig. 4). None of the TCRs were present in the blood from healthy HLA-A2− controls (data not shown). We found no significant differences in the number of unique TCRs found exclusive to a single repertoire across all tissue samples (Fig. S1B). The TCRs encoded a range of V, J, and complementarity determining region 3 (CDR3) sequences on the alpha-chain, and had restricted V-beta use. The CDR3 lengths averaged 14 aa for alpha and 15 aa for beta. Shared TCRs 005 and 010 were notable because they were mostly detected in the tumor repertoires. These specific TCRs may bind to an antigen expressed by the tumor, which will be determined in future experiments and may be useful in the development of immunotherapies.
Fig. 4.
Sharing of TCR pairs across breast cancer tumors reveals a shared response among HLA-A2+ patients. TCR pairs shared between seven or more patient tumors are listed (Right) with the number of patients (Left) where the specific TCR pairs were identified. A color value was assigned corresponding to the presence of the TCR in a repertoire in the tumor (blue), in the blood (red), not found (white), or not done (gray). Amino acids corresponding to the germ-line V and J sequences are underlined. Patients 1–16 are HLA-A2:01+ (HLA-A genotypes of other samples are shown in Table 1).
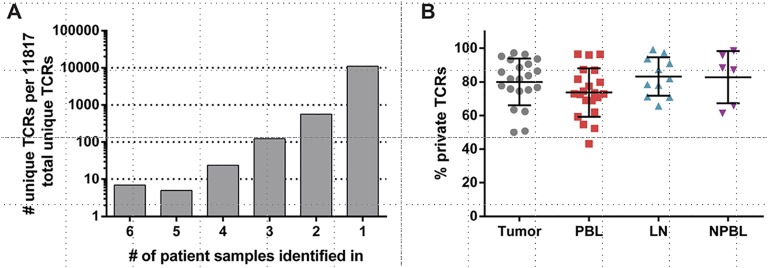
Fig. S1.
TCR repertoire overlap among patients with breast cancer. (A) Number of unique TCRs shared between six or fewer patient tumor repertoires is shown. The TCRs shared between greater than six patient tumor repertoires is shown in Fig. 4. (B) Percentage of TCRs from a given patient repertoire unique to that repertoire (“private” TCRs) is shown. NPBL, control PBL. Error bars represent the SEM (n = 21 tumor, n = 21 PBL, n = 11 LN, and n = 6 NPBL).
Table S5.
Multiple DNA sequences encode the tips of the CDR3 loops of the shared TCRs
| TCR | Tumor alpha | Tumor beta | Tumor alpha/beta | PBL alpha | PBL beta | PBL alpha/beta |
| 001 | 8 | 5 | 12 | 19 | 8 | 26 |
| 002 | 13 | 4 | 16 | 2 | 1 | 2 |
| 003 | 1 | 1 | 1 | 24 | 13 | 36 |
| 004 | 1 | 1 | 1 | 1 | 1 | 1 |
| 005 | 2 | 2 | 3 | 5 | 5 | 9 |
| 006 | 3 | 3 | 5 | 4 | 5 | 8 |
| 007 | 1 | 1 | 1 | 1 | 6 | 6 |
| 008 | 3 | 2 | 4 | 1 | 2 | 2 |
| 009 | 1 | 1 | 1 | 1 | 1 | 1 |
| 010 | 1 | 1 | 1 | 8 | 1 | 8 |
| 011 | 2 | 1 | 2 | 5 | 2 | 6 |
| 012 | 1 | 1 | 1 | 2 | 3 | 4 |
| 013 | 1 | 1 | 1 | 1 | 1 | 1 |
| 014 | 3 | 3 | 5 | 1 | 1 | 1 |
| 015 | 7 | 1 | 7 | 3 | 1 | 3 |
| 016 | 1 | 1 | 1 | 1 | 1 | 1 |
| 017 | 1 | 1 | 1 | 1 | 1 | 1 |
| 018 | 3 | 6 | 8 | 5 | 1 | 5 |
Each TCR alpha and beta chain was analyzed apart and together for the number of nucleotide sequences found across all patients in the tissue types indicated. The DNA sequence-encoding amino acids between the second amino acid C-terminal of the V Cys and 4 aa N-terminal to the J Phe of the CDR3 were analyzed to focus on CDR3 sequences and not allelic differences in the germ line.
Discussion
To identify alpha–beta TCR pairs from tumors of patients with breast cancer as tools for immunotherapies, we modified and expanded a previously published method of single-cell emulsion RT-PCR (19). With this enhanced method, we analyzed the T-cell repertoire ex vivo, unaffected by skewing or cell death during culture (14, 21). Single-cell emulsion RT-PCR provides information about the identity of individual T cells by analyzing the TCR chains as a pair, eliminating the need for computer-based algorithmic pairing or mix-and-match of separate alpha- and beta-chains. This high-throughput technique can be applied to large diverse populations, limited numbers of cells, and/or low-diversity populations.
The pairing efficiency of TCRs from T cells from primary tissue is ∼90%, although the calculation to obtain this number requires the assumption that a given alpha or beta only pairs with one other chain in the repertoire. Emulsion RT-PCR provides a qualitative profile of the TCR repertoire, but due to minute primer-binding differences, TCR transcript abundance and/or stability, and/or cDNA conversion efficiency, it is not quantitative. Therefore, to determine the population dynamics of a given emulsion RT-PCR–derived pair precisely, this technique must be paired with more established TCR profiling methods, such as single-cell RT-PCR from sorted cells, cloning of tumor T cells, and/or RNA- or DNA-based bulk sequencing (11, 22–25). Additionally, our observation that differences in TCR transcript abundance in laboratory-generated T-cell hybridomas results in differential recovery of TCR sequences has implications in profiling diverse T-cell pools. Naive and tolerant T cells express less TCR transcript than activated functional counterparts. Directly sequencing these diverse populations may lead to enrichment for activated T cells and loss of T cells that are naive or hyporeactive. Sorting populations of interest will likely enhance recovery of target populations by normalizing TCR transcript levels.
We obtained an average of 10 reads per input cell, with an average of ∼500,000 sequences per sample; 1–3% of the TCR sequences appeared once and were dropped from the analysis as potential sequencing errors and because we were ultimately interested in T cells that expanded in response to the tumor. It has been estimated that the size of the TCR-beta repertoire in an individual is ∼3 × 106 unique TCRs, drawing from an estimated 5 × 1011 possible sequences (26). Because the PBL is so heterogeneous with numerous infrequent clones, discarding the singly sequenced TCRs may have removed bona fide antitumor TCRs. Additionally, we assume that the tumor samples we analyzed are representative of the entire tumor. However, spatial differences in immune cell infiltrates (27) render it likely that some antitumor TCRs may not have been sampled.
Isolation and cloning of T cells from primary tumors has difficulties stemming from issues such as T-cell exhaustion and anergy, as well as the deleterious environment of the tumor (28). We expected that isolation of T cells from the sentinel LN would provide a potential source of tumor-specific T cells, less affected by the tumor environment. We showed that tumor-infiltrated LNs are nearly fourfold more likely to contain TCRs that are also present in the TIL from the primary tumor than tumor-free LNs (Fig. 3A). Therefore, future studies aimed at identifying tumor-specific TCRs could use tumor-involved LNs as an alternative tissue source.
The analysis of the alpha–beta pairs from breast cancer TILs and blood shows a predominance of TCR variable region beta (TRBV) 7 family-member TCRs, suggesting that this V-beta family is responsible for tumor recognition. Alternatively, the TRBV7 primers may confer bias due to mispriming, because there are eight primers for the TRVB7 family in these assays (Fig. 4 and Table S2). However, there are also eight primers for the TRVB6 family, and we only identified one shared TCR using this V-beta gene. Comprehensive analysis of V-beta use in healthy individuals does not show dominance in TRBV7 use (26, 29). Furthermore, it has been described previously that specific V-beta subsets are enriched within the tumor (29, 30), and analysis of colorectal tumors has shown that only one-third of shared V-beta TCRs are from the TRBV7 family (13). However, use of V-region primers designed to bind closer to the 5′ end of the variable genes, coupled with the rapid development of sequencers delivering greater read length and depth, will be instrumental in expanding the limits and sensitivity of emulsion RT-PCR.
We discovered 18 unique TCR alpha–beta pairs in tumors and/or blood shared in seven to 15 of 20 patients. The shared TCRs were found sparingly in control blood from female HLA-A2+ donors without breast cancer (Fig. 4), and were absent in HLA-A2− donors, suggesting that the shared TCRs may be tumor-specific. The shared TCRs displayed variation in the encoding CDR3 nucleotide sequences, indicating selection of the TCR is driven by the tumor (Table S5). Although it is possible that we identified TCRs reactive against a common viral antigen(s), the limited recovery of the shared TCRs from control blood makes this conclusion unlikely. Additionally, because 16 of the 20 patients shared the HLA-A2:01 allele and the other four expressed alleles of the HLA-A*02 supertype, it is likely that the TCRs we have identified are HLA-A2:01–restricted and present the same peptide (31). The overall mutational burden of breast cancer is lower than most human cancers (32), and the frequency of more common mutations is less than 10% (17). These observations, coupled with the majority of the patients in this study being ER+/PR+, means it is likely that the recovered shared TCRs are to a nonmutated antigen(s) found in each patient. Additionally, the patients who were not ER+/PR+ contained fewer shared TCRs in their repertoires. Future experiments will determine the antigens responsible for the expansion of these T cells and will contribute to antigen-specific therapies.
Materials and Methods
Cell Lines.
All 5KC T-cell hybridomas were propagated in Spinner modification minimum essential medium (S-MEM) supplemented with essential and nonessential amino acids, penicillin/streptomycin, 7 mM NaHCO3, 75 μM gentamicin, 750 μM sodium pyruvate, 3 μM dextrose, 1.7 μM l-glutamine, 36 nM beta-mercaptoethanol, and 10% FCS. The 5KC cell lines used are listed in Table S4.
Real-Time Quantitative PCR.
RNA from equal numbers of 5KC hybridoma cells (Table S4) was isolated using TRIzol. Real-time quantitative PCR was performed using the following primer sets for the mouse alpha- and beta-TCR constant regions: alpha, GGACATGAAAGCTATGGATTCCAAGAGCAATGGG and CAGGAGGATTCGGAGTCCCATAACTG; beta, GTGGGTGAATGGCAAGGAGGTCCAC and GGTTTGGGTGAGCCCTCTGGC. Samples were amplified in triplicate using Power SYBR Green RNA to Ct 1-Step Mix (Applied Biosystems) and analyzed on a Roche Lightcycler 450.
Tissue Procurement and Processing.
Fresh tumor, LNs, and PBLs were obtained from patients who gave institutional review board (IRB)-approved informed consent [City of Hope (COH) IRB 05091] before tissue collection at the City of Hope Beckman Research Institute (COH IRB 11273) and University of Colorado Hospital Breast Center [Colorado Multiple (COM) IRB #09-0583). Tumor tissue and tumor-infiltrated LNs were separated from fat tissue and minced into pieces up to 2 mm in diameter with scalpel blades. Fragments were then treated with 0.2 Wunsch U/mL Liberase (Roche) and 10 U/mL DNase (Sigma) in RPMI (Life Technologies) for up to 1 h as needed until the tissue dissociated. Enzymatic dissociation was stopped by adding 5 mL of RPMI +10% (vol/vol) FBS. The digested tissue suspension was then filtered through a 70-μm filter followed by a 40-μm filter. Red blood cell (RBC) lysis was performed, if necessary, using RBC Lysis Buffer (Biolegend). LNs without tumor infiltration were similarly treated, but without enzymatic digestion. PBL samples were Ficoll-separated (GE Healthcare) to remove cellular debris and RBCs. T-cell populations were isolated from single-cell suspensions using a CD8 Positive Selection Kit (Stem Cell Technologies or Life Technologies) following the manufacturer’s recommendations and incubated in 10 IU/mL IL-2 (Peprotech) overnight before emulsion RT-PCR.
HLA Typing.
We collected PBLs from patients at the time of consent, 1–2 wk before surgery. After Ficoll separation, we determined the HLA serotype by flow cytometry with an HLA-A2 antibody (clone BB7.2; Biolegend). We isolated genomic DNA by TRIzol extraction from 1 × 106 mononuclear cells for high-resolution genotyping from the HLA-A2+ samples (HLA Laboratory, City of Hope).
Emulsion Reverse Transcription-PCR.
An emulsion phase was generated using a Micellula Emulsion and Purification Kit, per the manufacturer’s recommendations (EURx). Three hundred microliters of emulsion phase was added to 50 μL of RT-PCR master mix [1× Qiagen OneStep RT-PCR buffer, 1× Qiagen Q-Solution, 0.4 μM dNTPs, 1 μL Qiagen OneStep RT-PCR enzyme mix, 10 U of RNAseOUT (Life Technologies), 600 nM C-region primers, 600 nM Stepout primer, 60 nM V region primers (Tables S1–S3)] containing 5,000–250,000 T cells and vortexed at high speed for 5 min at 4 °C. The resulting 350-μL reaction was separated into three PCR tubes and subjected to the following RT-PCR regimen: one cycle of 65 °C for 2 min; one cycle of 50 °C for 40 min; one cycle of 95 °C for 15 min; 40 cycles of 94 °C for 30 s, 60 °C for 1 min, and 72 °C for 2 min; and one cycle of 72 °C for 5 min. The three PCR reactions per original emulsion phase were then pooled and purified using the Micellula Emulsion and Purification Kit.
Nested PCR.
Two nested PCR reactions were performed following reverse transcription-PCR. The first nested PCR reaction amplified the entire eluted RT-PCR product in a 100-μL total volume using Taq polymerase (New England Biolabs), 0.2 μM C-region primers, and 1.55 μM blocking oligos (Table S3), and the following cycling conditions were used: 95 °C for 30 s; 30 cycles of 95 °C for 30 s, 52 °C for 30 s, and 68 °C for 1 min; and one cycle of 68 °C for 5 min. The second nested PCR reaction was performed using LA Taq (New England Biolabs) and 2 μL of the first nested PCR and 0.4 μM primers (Table S3) in a 50-μL total volume with the following cycling conditions: 94 °C for 30 s; 20 cycles of 94 °C for 30 s, 53 °C for 30 s, and 65 °C for 1 min; and one cycle of 65 °C for 5 min. The resulting products were gel-purified and quantified using a QuBit Fluorometer (Life Technologies).
Illumina High-Throughput Sequencing.
Separate samples were pooled in equal molar amounts and adjusted to a final concentration of 3.4 μM. Six microliters of the pooled sample was diluted 1:1 with 0.2 M NaOH and incubated at room temperature for 5 min before addition of 1 mL of Hybridization Buffer (Illumina). A 6:10 dilution of the sample was then made in Hybridization Buffer. Six hundred microliters of the diluted sample was loaded on an Illumina MiSeq flow cell, and paired-end 2 × 250 sequencing was performed per the manufacturer’s instructions.
Bioinformatic Processing.
Raw sequence reads were processed using the open-source Galaxy Platform (33–35). Reads were paired by joining the 3′ ends to create a 500-nt-long read with small portions of the C regions on either end. The reads were then clipped from either end using the engineered overlap sequence as the identifier, creating two separate files containing only TCR information from the alpha- or beta-TCR. Nonclipped reads were discarded. The clipped files, which contained identical cluster identifications (IDs) for paired alpha- and beta-TCRs, were then analyzed for their TCR identities using CompleteTCR (https://github.com/kamichiotti/CompleteTCR.git). The CompleteTCR pipeline was built from MiTCR (20), an efficient tool for CDR3 extraction, clonotype assembly, and repertoire diversity estimation, but limited to independent analysis of the alpha- or beta-TCR chain. CompleteTCR allows determination of alpha–beta pairs by manipulating raw MiTCR outputs using an R script. Two modest changes to the MiTCR source code were made because MiTCR assigns each input read a numeric identifier. First, the standard MiTCR results file was written to include a list of the numeric IDs for all reads belonging to each pair. Second, a temporary output file was created to map the sequence identifier for each read in the input FASTQ file to its MiTCR-assigned numeric identifier. No changes were made to the algorithms MiTCR uses for CDR3 extraction, clonotype assembly, or error correction. The R script first annotated the reads of each alpha-clonotype with the appropriate sequence identifiers, repeating the process for the reads of each beta-clonotype. The alpha- and beta-reads were next paired by their sequence identifier, and any read lacking a mate was removed from the dataset. Finally, the frequencies of alpha–beta pairs were calculated. CompleteTCR required Java version 1.7.0 or higher and R version 3.1.0 or higher with the plyr package version 1.8.3 (36) or higher. It was run from the command line via a shell wrapper script that required an input manifest detailing locations of the alpha and beta FASTQ files and their corresponding sample names. These results were further analyzed using standard procedures in Excel.
Statistical Analysis.
The error bars in Figs. 1–3 and Fig. S1B represent the SD of the mean. The LN+ and LN− comparison in Fig. 3 used an unpaired t test with Welch’s correction to determine significance. N refers to the number of samples in each experiment or the number of unique TCR sequences in a sample.
Acknowledgments
We thank Carolyn Charkey, Joan Venticinque, and Susie Brain, the consumer advocates who assisted with this project. This project was supported by Department of Defense (DOD) Congressionally Directed Medical Research Programs Multi-Team Awards BC100597, BC100597P1, and 100597P2 and by University of Colorado Cancer Center Flow Cytometry Shared Resource Cancer Center Support Grant CA046934.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE81313).
See Commentary on page 7944.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606994113/-/DCSupplemental.
References
- 1.Adams S, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32(27):2959–2966. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia Q, Yang Y, Wan Y. Tumor-infiltrating memory T-lymphocytes for prognostic prediction in cancer patients: A meta-analysis. Int J Clin Exp Med. 2015;8(2):1803–1813. [PMC free article] [PubMed] [Google Scholar]
- 3.Loi S, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31(7):860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 4.Ali HR, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–1543. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 5.Lee AH, et al. Different patterns of inflammation and prognosis in invasive carcinoma of the breast. Histopathology. 2006;48(6):692–701. doi: 10.1111/j.1365-2559.2006.02410.x. [DOI] [PubMed] [Google Scholar]
- 6.Peres LdeP, et al. Peptide vaccines in breast cancer: The immunological basis for clinical response. Biotechnol Adv. 2015;33(8):1868–1877. doi: 10.1016/j.biotechadv.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Ernst B, Anderson KS. Immunotherapy for the treatment of breast cancer. Curr Oncol Rep. 2015;17(2):5. doi: 10.1007/s11912-014-0426-9. [DOI] [PubMed] [Google Scholar]
- 8.Mccoy JL, Rucker R, Petros JA. Cell-mediated immunity to tumor-associated antigens is a better predictor of survival in early stage breast cancer than stage, grade or lymph node status. Breast Cancer Res Treat. 2000;60(3):227–234. doi: 10.1023/a:1006405504158. [DOI] [PubMed] [Google Scholar]
- 9.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11(4):251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 10.Jang M, et al. Characterization of T cell repertoire of blood, tumor, and ascites in ovarian cancer patients using next generation sequencing. Oncoimmunology. 2015;4(11):e1030561. doi: 10.1080/2162402X.2015.1030561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linnemann C, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat Med. 2013;19(11):1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 12.Linnemann C, Mezzadra R, Schumacher TN. TCR repertoires of intratumoral T-cell subsets. Immunol Rev. 2014;257(1):72–82. doi: 10.1111/imr.12140. [DOI] [PubMed] [Google Scholar]
- 13.Sherwood AM, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol Immunother. 2013;62(9):1453–1461. doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich PY, et al. TCR analysis reveals significant repertoire selection during in vitro lymphocyte culture. Int Immunol. 1997;9(8):1073–1083. doi: 10.1093/intimm/9.8.1073. [DOI] [PubMed] [Google Scholar]
- 15.Buhrman JD, et al. Improving antigenic peptide vaccines for cancer immunotherapy using a dominant tumor-specific T cell receptor. J Biol Chem. 2013;288(46):33213–33225. doi: 10.1074/jbc.M113.509554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie B, et al. High-throughput pairing of T cell receptor α and β sequences. Sci Transl Med. 2015;7(301):301ra131. doi: 10.1126/scitranslmed.aac5624. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams R, et al. Amplification of complex gene libraries by emulsion PCR. Nat Methods. 2006;3(7):545–550. doi: 10.1038/nmeth896. [DOI] [PubMed] [Google Scholar]
- 19.Turchaninova MA, et al. Pairing of T-cell receptor chains via emulsion PCR. Eur J Immunol. 2013;43(9):2507–2515. doi: 10.1002/eji.201343453. [DOI] [PubMed] [Google Scholar]
- 20.Bolotin DA, et al. MiTCR: Software for T-cell receptor sequencing data analysis. Nat Methods. 2013;10(9):813–814. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 21.Andersen RS, et al. Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 2012;72(7):1642–1650. doi: 10.1158/0008-5472.CAN-11-2614. [DOI] [PubMed] [Google Scholar]
- 22.Maryanski JL, Attuil V, Bucher P, Walker PR. A quantitative, single-cell PCR analysis of an antigen-specific TCR repertoire selected during an in vivo CD8 response: direct evidence for a wide range of clone sizes with uniform tissue distribution. Mol Immunol. 1999;36(11-12):745–753. doi: 10.1016/s0161-5890(99)00088-7. [DOI] [PubMed] [Google Scholar]
- 23.Ozawa T, Kishi H, Muraguchi A. Amplification and analysis of cDNA generated from a single cell by 5′-RACE: Application to isolation of antibody heavy and light chain variable gene sequences from single B cells. Biotechniques. 2006;40(4):469–470, 472, 474 passim. doi: 10.2144/000112123. [DOI] [PubMed] [Google Scholar]
- 24.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107(4):1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robins HS, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2(47):47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sautès-Fridman C, et al. Tumor microenvironment is multifaceted. Cancer Metastasis Rev. 2011;30(1):13–25. doi: 10.1007/s10555-011-9279-y. [DOI] [PubMed] [Google Scholar]
- 28.Waugh KA, Leach SM, Slansky JE. Targeting Transcriptional Regulators of CD8+ T Cell Dysfunction to Boost Anti-Tumor Immunity. Vaccines (Basel) 2015;3(3):771–802. doi: 10.3390/vaccines3030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madi A, et al. T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity. Genome Res. 2014;24(10):1603–1612. doi: 10.1101/gr.170753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandruzzato S, et al. Large and dissimilar repertoire of Melan-A/MART-1-specific CTL in metastatic lesions and blood of a melanoma patient. J Immunol. 2002;169(7):4017–4024. doi: 10.4049/jimmunol.169.7.4017. [DOI] [PubMed] [Google Scholar]
- 31.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: A revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative ICGC Breast Cancer Consortium ICGC MMML-Seq Consortium ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blankenberg D, et al. Galaxy: A web-based genome analysis tool for experimentalists. Curr Protocol Mol Biol. 2010;Chapter 19:Unit 19.10.1–10.21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giardine B, et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goecks J, Nekrutenko A, Taylor J. Galaxy Team Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickham H. The split-apply-combine strategy for data analysis. J Stat Softw. 2011;40(1):1–29. [Google Scholar]