Abstract
Phylogeography, and its extensions into comparative phylogeography, have their roots in the layering of gene trees across geography, a paradigm that was greatly facilitated by the nonrecombining, fast evolution provided by animal mtDNA. As phylogeography moves into the era of next-generation sequencing, the specter of reticulation at several levels—within loci and genomes in the form of recombination and across populations and species in the form of introgression—has raised its head with a prominence even greater than glimpsed during the nuclear gene PCR era. Here we explore the theme of reticulation in comparative phylogeography, speciation analysis, and phylogenomics, and ask how the centrality of gene trees has fared in the next-generation era. To frame these issues, we first provide a snapshot of multilocus phylogeographic studies across the Carpentarian Barrier, a prominent biogeographic barrier dividing faunas spanning the monsoon tropics in northern Australia. We find that divergence across this barrier is evident in most species, but is heterogeneous in time and demographic history, often reflecting the taxonomic distinctness of lineages spanning it. We then discuss a variety of forces generating reticulate patterns in phylogeography, including introgression, contact zones, and the potential selection-driven outliers on next-generation molecular markers. We emphasize the continued need for demographic models incorporating reticulation at the level of genomes and populations, and conclude that gene trees, whether explicit or implicit, should continue to play a role in the future of phylogeography.
Keywords: monsoon tropics, introgression, comparative phylogeography, species trees, coalescent theory
Phylogeography is being revolutionized by a whole-genome perspective driven by next-generation sequencing (NGS) in combination with development of coalescent-based methods of analysis within and among species. The classical phylogeographic foundation from which genome-scale phylogeography has grown was established in the decades spanning the early 1980s’ emphasis on animal mtDNA (1, 2) to the mid-2000s, just before the first genome-wide surveys of genetic variation in humans (3, 4). By the early 2000s, phylogeographic surveys of nonmodel species typically included a handful of loci, mostly using methods that facilitated a locus-by-locus phylogeographic analysis (5, 6). There are now a growing number of studies realizing a distant goal of phylogeography, geographically informed whole-genome resequencing (7, 8), as well many more sampling subgenomes through varied approaches (9–12). With the expansion to genome-wide analyses afforded by NGS, phylogeographic analysis has necessarily expanded its analytical toolkit.
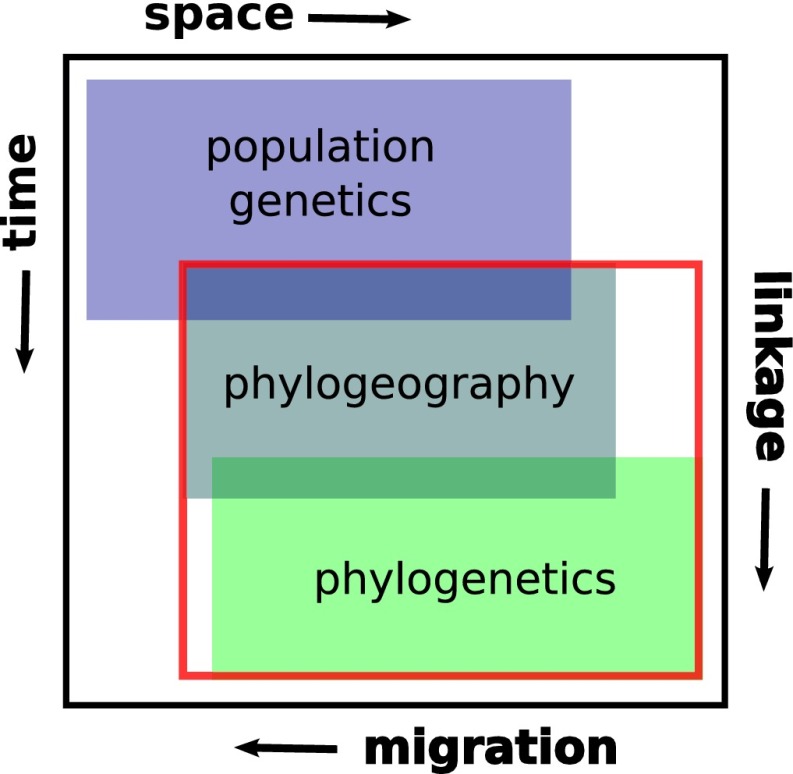
The increasingly routine analysis of genome-scale data has blurred the disciplinary boundaries between phylogeography and its sister discipline, population genetics, and has allowed phylogeography to contribute to endeavors such as scans for selection and association mapping (13). Indeed, with burgeoning data and increasing applications of related analytical tools, such as site-frequency spectra and coalescent simulations, we can ask whether and how phylogeography is now distinct from population genetics (13). We contend that there is still value in the original conception of phylogeography as a bridge between population biology and phylogenetics (1) (Fig. 1). This bridge can be thought of across geography and time, as is often the case with practitioners, or across gradients of migration rates and linkage disequilibrium (14), with the former decreasing and the latter increasing from the population to phylogenetic scale. That phylogeography sits centrally in this process-oriented space emphasizes the importance of understanding interactions between reticulation (gene flow/introgression and recombination), drift, and protracted isolation. This combination of processes sets phylogeography apart from traditional population genetics and phylogenetics.
Fig. 1.
Diagram classifying the disciplines of population genetics, phylogeography, and phylogenetics. Traditionally, we think of these respective disciplines as being concerned with variation among organisms arising over short, intermediate, and long temporal (and often spatial) scales. Increasingly, with large quantities of data, there are opportunities to classify studies according to the way different processes are inferred to have shaped datasets. For example, it is likely that migration among populations is common in “population genetics” datasets and rarer in phylogenetics. Similarly, recombination is likely to reduce the detectable effects of linkage in population genetic datasets, such that the effects of linkage likely lead to larger haplotype blocks in studies at the “phylogeographic” scale. In this way, different studies might form, and next-generation methods might facilitate, a continuum from population genetics to phylogenetics. In this review, we focus on studies spanning the part of this continuum spanning phylogeography and shallow phylogenetics, indicated by the red box.
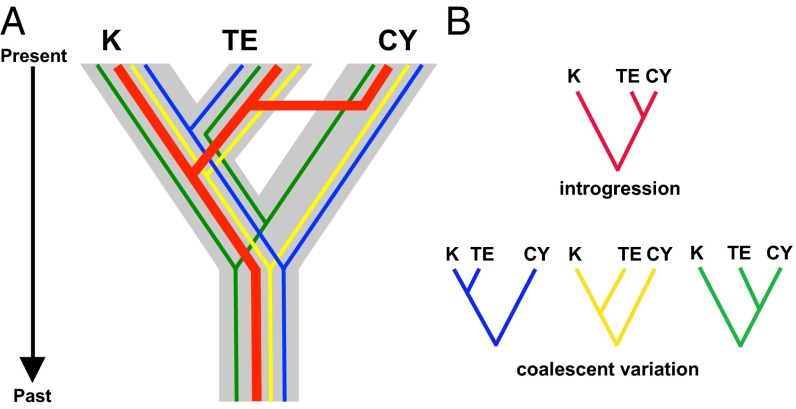
Scanning entire genomes of closely related organisms has unleashed a level of heterogeneity of signals that was largely of theoretical interest in the PCR era. This genomic heterogeneity is profoundly influencing our basic concepts of phylogeography and phylogenetics, and indeed our views of speciation processes. It is now routine to encounter a diversity of gene trees across the genome that is often as large as the number of loci surveyed (15, 16). Aside from variation induced by the coalescent process within and across species, we are only beginning to understand how such gene tree heterogeneity arises (16, 17). Recognition of this heterogeneity has driven the development of phylogenetic methods for accommodating such conditional independence of gene trees, so-called “species tree” methods (18–21). For phylogeographic analyses, at the transition from population structure to phylogenetic divergence, incomplete lineage sorting (ILS) is prevalent where populations have been separated for less than 4Ne generations, where Ne is the effective population size (22, 23). Another increasingly evident source of heterogeneity is introgression among species (16) (Fig. 2), the converse of the deep phylogeographic structure often observed in low-dispersal taxa. Such reticulation has long been recognized in plants, or in microbial systems, where horizontal gene transfer is an established paradigm. Increasingly, zoologists are also finding evidence for extensive movement of genes between phenotypically divergent taxa (24, 25), including nonsister species. Such observations have increased attention to models of “speciation-with-gene-flow” (26). The new genome-scale analyses are causing evolutionary biologists to reevaluate the very nature of species (27, 28), which, in some cases, appear to maintain phenotypic distinctiveness despite extensive gene flow across most of the genome (29–32), and to recognize introgression as an important source of adaptive traits in a variety of study systems (33–35). Analytically, evidence of introgression among species is driving the emergence of network models of diversification (36). Clearly genome-scale biology and the abundant reticulations across the “Tree of Life” are turning much of evolutionary biology, including phylogeography, on its head.
Fig. 2.
Sources of gene tree heterogeneity among diverging lineages. The three lineages KIM (K), TE, and CY are representative of northern Australia populations (Fig. 3). (A) These lineages are arranged by true evolutionary relationships depicted in gray: the “species” coalescent. (B) Within the species tree are gene trees colored in blue, yellow, red, and green. These gene trees represent independent coalescent histories and highlight various sources of gene tree discordance. Blue, green, and yellow gene trees highlight variation due to differing mutation rates and stochastic coalescent histories (including ILS), and red depicts effects of introgression among nonsister lineages. The MSC model allows for estimation of species trees, given mutational and coalescent variance. However, introgression, if extensive, can yield an incorrect species tree (e.g., [K, (TE,CY)] in this case) using most available methods. Additionally, distinguishing introgression from ILS in gene trees can be challenging and mostly relies on branch lengths in gene trees (as in IM models).
In this paper, we explore the themes of reticulation and the genomics of speciation as key processes across the phylogeography–phylogenetics continuum. Reticulation presents challenges to many concepts and methods in phylogeography and speciation that were not as evident when we were locus-poor in the PCR era. Reticulation in the form of recombination causes gene trees to depart from a strictly bifurcating pattern, hence posing challenges for some methods of reconstructing evolutionary history (37). Recombination has also long been known to play a central role in speciation (38), and the suppression of recombination, such as occurs in chromosomal inversions, can dramatically reduce the local genomic rate of gene flow (39, 40). Reticulation via gene flow is to be expected among intraspecific lineages that are the classical domain of phylogeography (Fig. 2). Finally, reticulation is becoming increasingly conspicuous at the level of diverging species and adaptive radiations (25, 35, 41–44), and is causing biologists to consider “ephemeral species,” with frequent lineage mergers, not only among sister taxa but between lineages with any geographic co-occurrence (45). The goal, however imperfect it is now realized, is to develop and apply models that integrate phylogeography, demography, and genome evolution in ways that will allow more nuanced interpretation of myriad interacting evolutionary processes from patterns of genomic diversity (46). Acknowledging and modeling reticulation at various levels in the hierarchy of life will be an important part of reaching this goal.
Key Processes of Divergence and Reticulation in Nature
Comparative Phylogeography Across the Australian Monsoonal Tropics.
In essence, comparative phylogeography is about establishing commonalities of spatial patterns of genetic and gene tree diversity across codistributed species (47, 48). Combined with population genetic (coalescent) and spatial modeling (49), this effort has yielded insights into biogeographic history, such as locations of refugia and expansion areas (50) and the varying effects of ecological or physical dispersal barriers. In a comparative setting, such studies can identify how landscape features and regional climatic variation have interacted with the varying ecologies of species to shape current diversity (51) and how these interactions can influence speciation processes (52, 43).
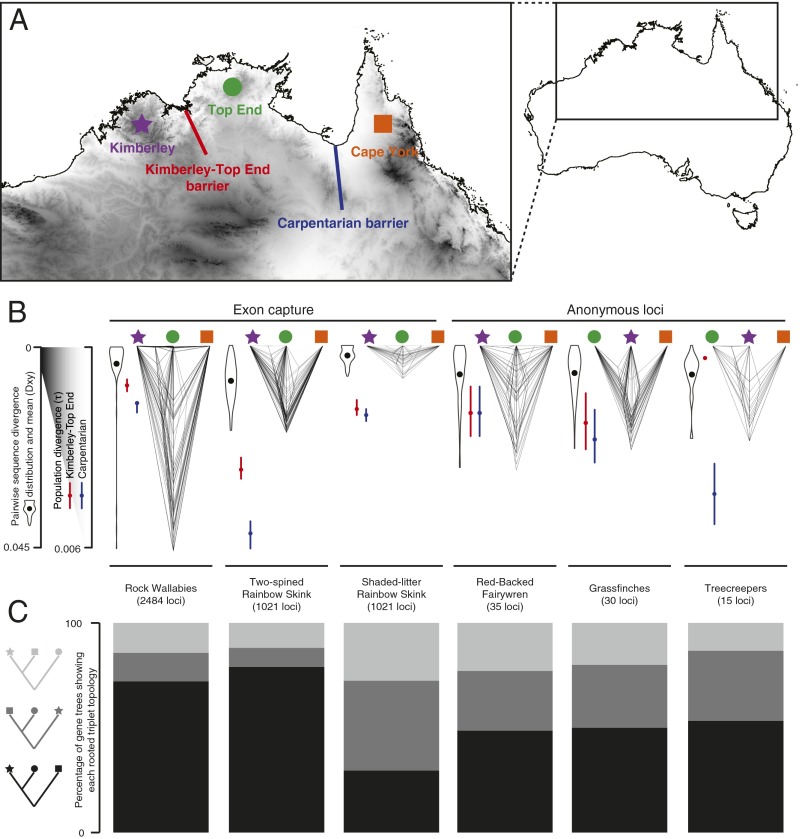
To explore divergence vs. reticulation processes in a comparative context, we focus on phylogeographic data for ecologically diverse species from northern Australia, a vast stretch of monsoonal savanna and woodlands with interspersed ancient sandstone plateaus (54) (Fig. 3). That this rich tropical fauna is biogeographically structured has long been known (55) and formalized using cladistic biogeography by Cracraft (56), who recognized a basal dichotomy across the treeless “Carpentarian Barrier” (CB) separating the Kimberley (KIM) and Top End (TE) faunas from the faunas in Cape York (CY) and the eastern Australian forest (EF), as well as New Guinea. KIM and TE are, in turn, separated by hot, low-relief, and relatively dry regions associated with several smaller barriers (57), collectively referred to here as the Kimberley-Top End Barrier (KTEB). Early sequence-based phylogeographic studies in babblers [Pomatostomus (58)] reported deep divergence across the CB relative to divergence within the eastern and western regions of the continent. Subsequent multilocus analyses of several avian systems revealed mostly Pleistocene divergences across the CB for congeners (59–61), as well as within species (62, 63). Some studies examining divergence across the region have discovered clines (64) or complex reticulate patterns in the form of introgression; for example, in butcherbirds (Cracticus), populations east of the KTEB are introgressed with mtDNA from populations of arid-adapted species to the south that expanded during the last glacial maximum, whereas populations west of the KTEB are not introgressed (65).
Fig. 3.
Gene tree heterogeneity in multilocus phylogeographic datasets of birds (Red-Backed Fairywren, Malurus melanocephalus; Poephila grassfinches; Climacteris treecreepers), skinks (Two-Spined Rainbow Skink, C. amax; Shaded-Litter Rainbow Skink, C. munda), and mammals (Petrogale rock-wallabies) across northern Australia. (A) Map of northern Australia showing the KTEB and CB that separate the KIM, TE, and CY faunas. (B) Cloudograms illustrate topological and branch length variation of gene trees. Violin plots represent the distribution of pairwise sequence divergences across the CB, and black dots indicate mean pairwise sequence divergence, or Dxy. Red dots and lines are estimates and 95% confidence intervals of population divergence across the KTEB, whereas green dots and lines are estimates and 95% confidence intervals of population divergence across the CB. (C) Distribution of rooted triplets shows that gene trees exhibiting deeper divergence times across the CB than the KTEB are the most frequent in all taxa except the Shaded-Litter Rainbow Skink. Additional details are provided in SI Text.
Fewer multilocus phylogeographic studies have been conducted for mammals across the monsoonal tropics, yet some common themes emerge. Rock-wallabies (Petrogale), specialists of rocky habitats as the name implies, are strongly structured across the disjunct sandstone plateaus of the region, with deeper divergences across the CB than across the KTEB (66, 67) (Fig. 3). Other macropod species have different degrees of geographic and genetic discontinuity across the CB, suggesting the species’ ecology has played a key role in their ability to adapt and persist across this region. The antilopine wallaroo (Macropus antilopinus), a savanna-woodland specialist, has a disjunct distribution but shallow divergence on either side of the CB grasslands, suggesting recent gene flow or range expansion. By comparison, a more ecologically generalized and widespread congener, the common wallaroo (Macropus robustus), has a more continuous distribution but substantial divergence across the CB (68). Preliminary analyses of other mammals suggest deep divergences across the CB, but require more expansive molecular study (69, 70).
Low-dispersal species, such as lizards and frogs, have been the subject of a burst of recent multilocus phylogeographic studies across the region, including early applications of NGS in this context. Relative to birds and mammals, these taxa exhibit phylogeographic structure at a finer spatial scale, often with cryptic species and greater phylogenetic depth among regions, possibly reflecting a combination of lower dispersal and higher localized persistence through cycles of harsh climate. Deep structure across the CB, and often also the KTEB, is observed across phylogenetically and ecologically diverse reptiles, including species complexes of agamid lizards (71), rainbow skinks (12, 72), several species complexes of geckos (73, 74), and toadlet frogs (75). In many cases, the divergence across the CB appears at deep phylogenetic scales rather than within species. For example, Carlia rainbow skinks have radiated across the KIM and TE, yet these taxa diverged from the eastern species of Carlia in the mid-Miocene (72). Analyses of ∼2,000 exons for the Two-Spined Rainbow Skink (Carlia amax) inferred recent population expansion from western KIM across the KTEB to the western TE, emphasizing that the KTEB is a more porous filter than the CB (12). Studies of low-dispersal taxa (12, 73, 75) are also revealing congruent patterns at a finer scale than the major barriers envisioned by Cracraft (56). These congruent patterns include deep structuring between offshore islands and mainland populations and an unexpected north-south split from the TE to the northern desert region (12, 73, 75). Closely related northern desert taxa often have ranges that are more widespread than those ranges across the savannas and sandstones to the north, and sometimes with evidence of broad-scale introgression (12, 73, 75). For example, in contrast to strong phylogeographic structuring within C. amax, an arid-adapted congener, Carlia munda (Shaded-Litter Rainbow Skink) includes a single widespread clade from the west coast across the northern desert to the east coast (Fig. 3).
Gene Tree Heterogeneity Across the CB.
The complex landscapes and dynamic climate history across this region have resulted in a combination of often strongly vicariant processes across the CB and a mix of divergence and dispersal or introgression across the KTEB. Given that gene tree heterogeneity arises from both ILS and gene flow between populations (Fig. 2), we can expect to see a more dominant phylogenetic signal across the CB in which the deepest split for a majority of gene trees spans the CB, with fewer loci having their deepest split across the KTEB, or between the CY/EF and TE (Fig. 3). We explored this hypothesis for exemplar avian, mammal, and lizard taxa for which we had multilocus sequence data spanning these geographic regions (Fig. 3 and Tables S1 and S2). As expected, among four-tip gene trees (one allele sampled for the KIM, TE, and CY/EF plus outgroup), we found diverse gene tree distributions across the region, with gene trees exhibiting deeper divergence times across the CB than the KTEB being the most frequent (Fig. 3 and Table S3). An exception to this pattern is C. munda, the more arid-adapted lizard, in which the dominant gene tree is one in which the TE and CY alleles are sisters, implying a more isolation-by-distance than vicariance model (76). Analyzing the larger datasets in which these simple gene trees are embedded with coalescent models (77) uniformly suggests deeper population divergence and speciation across the CB than across the KTEB, although these divergences are quite close temporally in several cases (Fig. 3 and Fig. S1). Although our sample sizes are small, the analysis also suggests that the highest genetic diversity currently segregating within each complex varies among regions; in Fairy Wrens and wallabies, the highest diversity is in the CY/EF, whereas diversity is similar among regions in C. amax. Finally, the analysis does not support a key prediction of the simplest vicariance scenario: that the effective population sizes of descendant lineages are smaller than the sizes of the ancestral populations inhabiting the area before the vicariant event. Our estimates of ancestral Ne for at least four of the six species groups are smaller than for contemporary lineages in the KIM, TE, or CY. A challenge with our brief analysis of comparative phylogeography across the monsoon tropics of Australia is the diversity of markers, which prevents easy comparison across groups due to differences in substitution rate. Use of a common set of markers, such as provided by various forms of target capture (78–80), whether coding or noncoding, will be an important focus of future research.
Reticulation Driven by Ecology and Introgression.
Our comparative phylogeographic analysis for northern Australia highlights the complex mix of divergence and reticulation and diverse spatial and temporal scales of phylogeographic structure that can emerge. Much of this heterogeneity appears to relate to differences among species in their capacity to persist or disperse across the landscape as climates oscillated over the Quaternary. Whereas it is convenient to focus on common patterns of divergence, in a classic vicariance mindset, closer attention to differing outcomes of reticulation, such as we have seen when comparing the results for C. munda with the results for other taxa spanning the CB, will yield more insight into speciation processes (81).
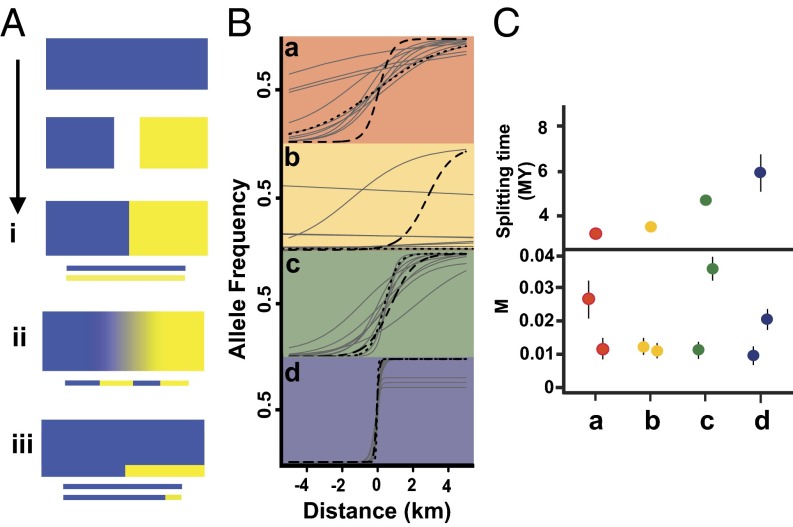
Following secondary contact, genetically distinct populations can form “tension zones,” maintained over time by a balance between dispersal and selection against hybrids (82), progressively merge via introgression [i.e., ephemeral taxa (46)], or overlap while maintaining their integrity (Fig. 4). A special case of introgression occurs when an expanding lineage overrides a static (relictual) one, but is itself invaded by genes from the resident population due to sequential founder events during the spatial expansion (83). Over time, introgressed chromosome segments will recombine between lineages, leading to a mosaic of coalescent histories within and across loci. These reticulation events can manifest at two scales: in genetic clines, for single-nucleotide polymorphisms (SNPs) at the contact zone(s) themselves, and in lineage-scale migration, as estimated from allopatric populations using isolation-migration (IM) models (Fig. 4). Genome-scale data are enabling new approaches (reviewed in 84), including genomic clines (85) and analyses of lengths of introgressed haplotype blocks (86). Given estimates of recombination rate, the length of immigrant haplotypes can, in principle, be used to estimate the timing of recent introgression events at a lineage scale, a parameter that has proved difficult to infer from IM models (87).
Fig. 4.
Contrasting processes and views of introgression. (A) Progression over time from population splitting, divergence in isolation, and secondary contact, with alternate outcomes: (i) tension zone, (ii) merging, and (iii) overriding of expanding population (blue) over the resident population with introgression from yellow→blue for some genes. (B and C) Contrasting perspectives on introgression among cryptic lineages of Australian Wet Tropics lizards at the local scale (B, contact zone) vs. lineage-scale estimates from IM analyses (modified from ref. 53). Note decreasing introgression at contact zones with increasing divergence time of lineage pairs, but no corresponding signal of decreasing migration at the lineage scale.
In the context of comparative phylogeography, insights into reticulation processes can be gleaned by comparing outcomes for taxa with varying ecologies and lineage divergence times across a common geographic and paleoenvironmental setting. Suture zones can be useful for this purpose, where multiple taxa have co-occurring contact zones (88). The fauna endemic to the rainforest of northeastern Australia are a case in point. Climate-driven fluctuations of rainforest-dependent taxa on mountain tops have resulted in spatially concentrated contact zones between morphologically indistinguishable but genetically distinct lineages (52). A comparative analysis of clines and genetic disequilibria across different contact zones (53) revealed less introgression and stronger genetic disequilibrium between more divergent lineage pairs, showing that reproductive isolation between these phenotypically cryptic lineages scales with divergence time (Fig. 4). However, at the lineage scale, levels of gene flow inferred from IM analyses of comparative transcriptomes are generally low and do not scale with divergence time (Fig. 4). These contrasting patterns remind us that estimates of gene flow are often averaged over the entire divergence history.
The Nexus of Comparative Phylogeography and Speciation Genomics.
So how will a fully genomic perspective enrich our understanding of the nexus between comparative phylogeography and speciation? A plethora of recent whole-genome comparisons among sister taxa reveal fascinating, but complex, heterogeneity of divergence across the genome (reviewed in ref. 84). The most common outcome among recently diverged taxa is stronger differentiation on X and Z sex chromosomes than autosomes and scattered “islands” of high divergence against a background of low divergence. Islands of divergence were initially taken as suggesting locations of incompatible genes in the context of ongoing gene flow (89). However, it is also possible that they reflect varying levels of background selection in the absence of gene flow (90, 91), leading to reinterpretation of some high-profile examples (92).
A key factor emerging from these studies and earlier scans of intraspecific diversity is the strong effect of recombination rate variation on the spatial patterning of genomic diversity, mediated most strongly by hitchhiking (93). Thus, we expect to see reduced within-lineage diversity in regions of low recombination, with a corresponding increase in divergence using measures that are sensitive to levels of within-lineage diversity [e.g., Wright's fixation index Fst (94)]. Paradoxically, it has also been proposed that low-recombination regions, as might occur within chromosomal inversions or near centromeres, will accumulate locally adapted alleles, thereby contributing to genetic incompatibility between lineages (95). Empirical evidence for this proposal is mixed, but there are some positive examples (96–98). Finally, genome comparisons among closely related taxa have also highlighted introgression of adaptive alleles from one lineage to another (35, 99), an old concept reborn (100). Such alleles can readily flow across contact zones even if there is strong hybrid breakdown. Analytical challenges aside, such cases point to the exciting prospect of understanding how adaptive evolution influences divergence and reticulation among lineages.
One limitation of many of the above analyses of genomic divergence during speciation is that the historical biogeographic and environmental setting of isolation and reconnection of diverging lineages over time is often not well established (84). Understanding these processes is the core business of phylogeography, and closer interaction between analyses of historical biogeography and speciation genomics can be expected to bear fruit. Conversely, in the genomic era, comparative phylogeographers will not just have to master details of environmental history, species’ ecology, and the plethora of methods for NGS and demographic inference but will also have to comprehend effects of selection and recombination rate variation across the genome. This challenge is exciting and will serve to strengthen further the link between population genomics and phylogenetics.
Reconstructing Processes of Divergence and Reticulation
Evolution of Molecular Markers in the Next-Generation Era.
Having outlined some of the key processes of divergence and reticulation observable with genomic data, we now ask: How do we reconstruct phylogeographic history in the era of NGS? What new practical and analytical challenges do the increased detail afforded by NGS bring to phylogeographic reconstruction? The glimpse of comparative phylogeography across the CB in northern Australia makes clear the implications of one of the key components of any effort at reconstructing demographic history, namely, how we select molecular markers and the need for easy comparison across datasets. As a start, we may ask: Has NGS finally liberated phylogeographers from the constraints of marker choice, allowing unfettered access to the most appropriate markers for the questions being asked? Which combinations of markers may promote the further integration of phylogeography and phylogenetics? In these still-early days of next-generation phylogeography, marker choice is still constrained somewhat by technical and resource considerations, and will remain so until whole-genome sequencing of individuals or at least exemplars of the clades being studied becomes routine. The emergence of several widely used NGS platforms and marker suites in the past few years illustrates this point. For example, the flanking regions of ultraconserved elements (UCEs) have been promoted as suitable for phylogeographic questions, with the advantage that they are variable and their presence in can be predicted in uncharacterized genomes (101). Another comparison (102) found similar phylogeographic resolution between exons (drawn randomly from transcriptomes) vs. anchored hybrid enrichment (AHE) loci, which mostly target conserved exons (96, 97). Exon capture has been effectively used to study diverging lineages of both vertebrates and invertebrates (e.g., 12, 98, 103) and is particularly appropriate for retrieving genomic data from museum specimens (80, 104, 105).
Arguably, most UCE, AHE, or exon capture loci that have been used thus far for next-generation phylogeography are under mild or even strong purifying selection. Such selection is not necessarily a problem; after all, much of the animal mitochondrial genome, despite its high variability, is under purifying selection. However, purifying selection will likely reduce variation and bias the site-frequency spectrum toward low-frequency variants in a manner similar to, but less extreme than, selective sweeps, making gene trees compressed toward the tips (106). There is also clear evidence that loci in the vicinity of exons exhibit reduced levels of ILS compared with anonymous genomic regions (107). So long as researchers frame their findings within the context of the diversity of loci found throughout the genome, exons and UCEs are likely to remain a powerful force in phylogeography. The pervasiveness of natural selection, particularly for species with large effective population sizes (108), is, however, a force with which phylogeographers have not yet fully come to grips. One wonders whether any of the loci used in phylogeography in the next-generation era are genuinely neutral.
The approach using restriction site associated DNA sequences, or RAD-seq, is a popular application of NGS to phylogeography, and yields large but sometimes patchy matrices of relatively short and mostly noncoding loci (109), which are often analyzed in the form of SNPs. Such markers can be powerful measures of phylogeographic structure and, in some cases, seem relatively free of strong selection (110). Within-locus recombination is irrelevant to SNPs, whereas recombination may pose challenges for analysis of the longer loci such as are generated by target capture and AHE. RAD-seq loci are less amenable to the type of gene tree building that has characterized phylogeography (78, 111, 112), but few NGS loci of any kind yield highly resolved gene trees when applied on a phylogeographic scale. The power of next-generation methods lies primarily in generating more independent loci, although the phylogenetic informativeness of individual loci also plays a role, especially in species tree reconstruction (18). Going forward, it will be important to compare the behavior and informativeness of different types of markers and genomic compartments explicitly in phylogeographic settings (16, 17).
The ideal phylogeographic marker in the next-generation era presumably depends on the questions being asked and the temporal and taxonomic scales over which comparisons are made. Whereas introns and anonymous loci were popular sequence-based markers in the PCR era (113), and continue to be captured by various NGS approaches (79, 114, 115), targeting of such unconstrained sequence-based markers has made few inroads in the next-generation era, presumably because compared with exons (116), such loci are difficult to predict, and therefore capture, in unknown genomes using probes from other species. Herein lies a conflict between the ease of retrieving markers and their variability within species: Until whole-genome sequencing of phylogeographic exemplars allows us to design probes that are optimal for a given species, and consistent across taxa, the practicalities of easily and cheaply capturing large numbers of loci may tend the field toward conserved loci. Ultimately, phylogeographers should embrace a diversity of marker types even within individual studies, not only to allow the phylogeographic history of different marker types to illuminate each other but also to study genomic diversity and history in an unbiased way that facilitates the discovery of genomic loci underlying adaptation.
Insight into Processes of Reticulation from Gene Tree Outliers.
Gene tree outliers, like Fst outliers, may be important indicators of nonneutral or locus-specific processes in the genome. We used a newly proposed gene tree outlier approach, KDEtrees (117), to explore the behavior of gene tree distributions in empirical phylogeographic and low-level phylogenetic datasets of several marker types (Fig. S2 and Table S4). KDEtrees appears effective at identifying loci that result from horizontal gene transfer or are clear outliers, such as gene trees generated by a species tree different from the majority. However, it is unclear how KDEtrees behaves when confronted with loci influenced largely by demographic processes, or how the number of outliers varies by marker type. Although our sample size is small, our analysis of eight datasets (Fig. S2 and Table S2) suggests that many phylogeographic and transcriptome datasets harbor surprisingly few gene tree outliers, and so conform well to expected distribution based on overall patterns and levels of divergence. For example, given our chosen level of sensitivity (λ = 1.5), for which we expect roughly 5% of gene trees to exhibit outlier behavior simply by chance, none of the datasets we analyzed contains a significant number of outliers. In the future, the KDEtrees approach, and other methods (118), should be a useful tool to explore gene tree heterogeneity within and between datasets.
Methods for Detecting Reticulation: Recombination and Introgression
Reticulation and Phylogenetic Networks.
As we have seen, as phylogeography and speciation studies begin to probe the genomes of diverse species on a large scale, reticulation, in the forms of introgression and recombination, appears much more common than previously supposed. Accordingly, a major challenge going forward is to incorporate reticulation as a standard component of phylogeographic analysis. Many computational methods targeted at the phylogeography–phylogenetics continuum necessarily ignore some kinds of reticulation. Key examples include models to estimate species trees from multiple unlinked loci using the multispecies coalescent (MSC) model (21). MSC methods ignore two fundamental aspects of reticulation: recombination within loci and postspeciation hybridization. Some MSC methods (119) are known via simulations and theoretical arguments to be robust to reticulations, such as introgression, particularly when datasets are large and when introgression is confined to a subset of loci. However, other MSC methods are not robust to such model violations (21). It is not surprising that phylogeographers are among the most comfortable working with MSC methods because of the similarity in assumptions they apply to multilocus datasets. At the same time, due to their familiarity with reticulating lineages, phylogeographers are the most likely to identify shortcomings arising from the inherent simplifications of standard MSC models.
Conceptually, networks subsume trees; networks are trees with reticulation (figure 1 of ref. 120). Genome-scale evidence for introgression is renewing enthusiasm for coalescent phylogenetic models that allow for hybridization between diverging lineages. Several recent phylogenomic datasets, including those datasets analyzing human populations as well as distantly related lineages of birds or mammals (121, 122), have noted signals for reticulation in the form of ancient interlineage hybridization. Phylogenomic network models based on the MultiSpecies Network Coalescent (MSNC) (36, 123, 124) are likely to be an important new tool for phylogeneticists in general and phylogeographers in particular. Early studies suggest that application of the MSNC to genome sequences from diverging species will yield new insights into complex evolutionary histories of divergence and reticulation (125).
Better insight into the presence of reticulation at the level of populations need not involve computationally intensive algorithms. For example, application of simpler SNP-based tests of introgression and admixture [e.g., the “ABBA-BABA” test (126)] will help flag phylogeographic scenarios that may be more complex than originally envisioned. Although they have yet to make inroads into the phylogeography of nonmodel species, a suite of recently developed drift (F) statistics, related to but distinct from Wright’s F-statistics, provide simple and powerful metrics to test various models of population history, such as whether populations are related in a tree-like fashion [127, 128; reviewed by Peter (129)]. Tools for model selection in phylogeography (130–136) will also be critical for determining whether reticulation at the population level is an important part of the demographic history under study.
Capturing Heterogeneity with the Sequentially Markovian Coalescent.
Recombination within loci violates assumptions of most phylogenomic analyses, whether informed by the MSC or not. The departure from the assumptions of the MSC could be particularly acute for datasets consisting of sequences from long loci relative to the distance over which linkage disequilibrium decays, which can be <1 kb in many organisms. The one simulation study exploring effects of intralocus recombination (without introgression) on the performance of species tree methods (37) found little effect, and then only on very short trees, as is typical of phylogeographic datasets. However, Potter et al. (12) observed incongruent and less resolved species trees among lineages of C. amax (as in Fig. 3) when using full-length exons compared with the longest nonrecombining segments of these loci. Still, phylogeography is no stranger to intralocus recombination. Several phylogeographic models have been adapted to incorporate recombination (137, 138), using information from the joint site frequency spectrum among loci and other data. If justified, such models could be adapted to MSC and MSNC methods to allow for intralocus recombination. Additionally, several postgenome phylogeographic models have emerged that incorporate recombination via the sequentially Markovian coalescent, a new approach that models the coalescent site-by-site along the genome, exploiting the variation in site patterns among linked SNPs (139–141). Sequentially Markovian coalescent models have obvious applications in traditional species tree methods and may alleviate lingering concerns about recombination. A final means of addressing the issue of intralocus recombination in phylogeography is by using SNP data, which obviates intralocus recombination. Phylogeographic models using SNP data have been available for a number of years (e.g., 142, 143), and several MSC methods (144, 145) now use linked or unlinked SNP data to estimate phylogenetic trees without explicitly estimating constituent gene trees. It remains to be seen whether the limited genealogical information in SNPs is compensated for by the large number of SNPs that can be collected in typical phylogenomic datasets.
Conclusion
Phylogeography has come a long way from its origins of analyzing single gene trees across geography (1). Sophisticated statistical inference, integration with spatial modeling, model choice, parameter estimation, and now access to sequence or SNP data for thousands of loci have all enriched the field tremendously. It will be interesting to see how closely future phylogeographers adhere to its conceptual roots, the “mitochondrial DNA bridge,” as mirrored in gene trees empirically derived from nuclear sequence data. On the one hand, extensive reticulation in the form of recombination and the convenience of analyzing large numbers of unlinked SNPs with rapid parametric tests may be ushering in an era of phylogeography beyond gene trees, or, at the very least, an era that acknowledges them only implicitly, via connections with coalescent theory (146, 147). On the other hand, some of the currently popular methods of locus capture are showing promise for capturing genetic diversity in the form of gene trees, even if weakly resolved at lower taxonomic levels. What seems clear is that next-generation approaches are pushing phylogeography toward a future dominated by SNPs or sequence data for thousands of loci, a positive development that we believe will help bridge the phylogeography–phylogenetics continuum as envisaged by Avise et al. (1).
In the future, we can expect integration of phylogeography with increased understanding of genome organization and parameters, such as variation in recombination rate across the genome (46). We can also expect continuing integration of phylogeography with speciation biology and with analyses of adaptive variation, including phenotype-genotype associations (13). Almost by definition, phylogeography will retain its distinctions from sister disciplines like population genetics by its emphasis on broad geographic sampling and the natural history origin of the questions it seeks to answer. One of the thrills of phylogeographic research is the ability of researchers to absorb cutting-edge technologies that now put whole-genome variation within our grasp, yet also retain the exploratory field spirit that has motivated the discipline since its inception. With such a detailed view of genomic variation across geography, reticulation is likely to be omnipresent, pushing phylogeography to reinvent itself, question its foundations, and strive for new syntheses.
Supplementary Material
Acknowledgments
We thank Mark Eldridge (Australian Museum Research Institute) for access to tissues and data of rock-wallabies; Ke Bi and Sonal Singhal for assistance with Carlia data collection and preparation of Fig. 4; Brant Faircloth, John McCormack, and Robb Brumfield for assistance in acquisition of online datasets; and John Wakeley, Rudy Yoshida, and Alan Lemmon for helpful discussion. S.V.E.’s research is supported by the US National Science Foundation and Harvard University. S.P., J.G.B., and C.M. are supported by grants from the Australian Research Council.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution X: Comparative Phylogeography,” held January 8–9, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_X_Comparative_Phylogeography.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601066113/-/DCSupplemental.
References
- 1.Avise JC, et al. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 1987;18:489–522. [Google Scholar]
- 2.Moritz C, Dowling TE, Brown WM. Evolution of animal mitochondrial-DNA–relevance for population biology and systematics. Annu Rev Ecol Syst. 1987;18:269–292. [Google Scholar]
- 3.Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat Rev Genet. 2009;10(11):745–755. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolman G, Moritz C. A multilocus perspective on refugial isolation and divergence in rainforest skinks (Carlia) Evolution. 2006;60(3):573–582. [PubMed] [Google Scholar]
- 6.Brito PH, Edwards SV. Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica. 2009;135(3):439–455. doi: 10.1007/s10709-008-9293-3. [DOI] [PubMed] [Google Scholar]
- 7.Wallberg A, et al. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat Genet. 2014;46(10):1081–1088. doi: 10.1038/ng.3077. [DOI] [PubMed] [Google Scholar]
- 8.Jones FC, et al. A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr Biol. 2012;22(1):83–90. doi: 10.1016/j.cub.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey MG, Brumfield RT. Genomic variation in a widespread Neotropical bird (Xenops minutus) reveals divergence, population expansion, and gene flow. Mol Phylogenet Evol. 2015;83:305–316. doi: 10.1016/j.ympev.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Smith BT, Harvey MG, Faircloth BC, Glenn TC, Brumfield RT. Target capture and massively parallel sequencing of ultraconserved elements for comparative studies at shallow evolutionary time scales. Syst Biol. 2014;63(1):83–95. doi: 10.1093/sysbio/syt061. [DOI] [PubMed] [Google Scholar]
- 11.McCormack JE, et al. Next-generation sequencing reveals phylogeographic structure and a species tree for recent bird divergences. Mol Phylogenet Evol. 2012;62(1):397–406. doi: 10.1016/j.ympev.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Potter S, Bragg JG, Peter BM, Bi K, Moritz C. Phylogenomics at the tips: Inferring lineages and their demographic history in a tropical lizard, Carlia amax. Mol Ecol. 2016;25(6):1367–1380. doi: 10.1111/mec.13546. [DOI] [PubMed] [Google Scholar]
- 13.Edwards SV, Shultz AJ, Campbell-Staton SC. Next-generation sequencing and the expanding domain of phylogeography. Folia Zool (Brno) 2015;64(3):187–206. [Google Scholar]
- 14.Slatkin M. Linkage disequilibrium--understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9(6):477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song S, Liu L, Edwards SV, Wu S. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc Natl Acad Sci USA. 2012;109(37):14942–14947. doi: 10.1073/pnas.1211733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontaine MC, et al. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347(6217):1258524. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nater A, Burri R, Kawakami T, Smeds L, Ellegren H. Resolving evolutionary relationships in closely related species with whole-genome sequencing data. Syst Biol. 2015;64(6):1000–1017. doi: 10.1093/sysbio/syv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Xi Z, Wu S, Davis CC, Edwards SV. Estimating phylogenetic trees from genome-scale data. Ann N Y Acad Sci. 2015;1360:36–53. doi: 10.1111/nyas.12747. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Yu L, Kubatko L, Pearl DK, Edwards SV. Coalescent methods for estimating phylogenetic trees. Mol Phylogenet Evol. 2009;53(1):320–328. doi: 10.1016/j.ympev.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 20.Knowles LL, Kubatko LS. Estimating species trees: An introduction to concepts and models. In: Knowles LL, Kubatko LS, editors. Estimating Species Trees: Practical and Theoretical Aspects. Wiley–Blackwell; New York: 2010. pp. 1–14. [Google Scholar]
- 21.Edwards SV. Inferring species trees. In: Kliman R, editor. Encyclopedia of Evolutionary Biology. Elsevier; New York: 2016. [Google Scholar]
- 22.Neigel JE, Avise JC. Phylogenetic relationships of mitochondrial DNA under various demographic models of speciation. In: Karlin S, Nevo E, editors. Evolutionary Processes and Theory. Academic; New York: 1986. pp. 515–534. [Google Scholar]
- 23.Rosenberg NA. The probability of topological concordance of gene trees and species trees. Theor Popul Biol. 2002;61(2):225–247. doi: 10.1006/tpbi.2001.1568. [DOI] [PubMed] [Google Scholar]
- 24.Novick PA, Basta H, Floumanhaft M, McClure MA, Boissinot S. The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals. Mol Biol Evol. 2009;26(8):1811–1822. doi: 10.1093/molbev/msp090. [DOI] [PubMed] [Google Scholar]
- 25.Lamichhaney S, et al. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518(7539):371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 26.Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends Genet. 2012;28(7):342–350. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Mallet J. Hybridization, ecological races and the nature of species: Empirical evidence for the ease of speciation. Philos Trans R Soc Lond B Biol Sci. 2008;363(1506):2971–2986. doi: 10.1098/rstb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallet J, Besansky N, Hahn MW. How reticulated are species? BioEssays. 2016;38(2):140–149. doi: 10.1002/bies.201500149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malinsky M, et al. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350(6267):1493–1498. doi: 10.1126/science.aac9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mavárez J, et al. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441(7095):868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 31.Dasmahapatra KK, et al. Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487(7405):94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soria-Carrasco V, et al. Stick insect genomes reveal natural selection’s role in parallel speciation. Science. 2014;344(6185):738–742. doi: 10.1126/science.1252136. [DOI] [PubMed] [Google Scholar]
- 33.Nosil P, Feder JL. Genomic divergence during speciation: Causes and consequences. Philos Trans R Soc Lond B Biol Sci. 2012;367(1587):332–342. doi: 10.1098/rstb.2011.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nosil P, Crespi BJ. Does gene flow constrain adaptive divergence or vice versa? A test using ecomorphology and sexual isolation in Timema cristinae walking-sticks. Evolution. 2004;58(1):102–112. doi: 10.1111/j.0014-3820.2004.tb01577.x. [DOI] [PubMed] [Google Scholar]
- 35.Rheindt FE, Edwards SV. Genetic introgression: An integral but neglected component of speciation in birds. Auk. 2011;128(4):620–632. [Google Scholar]
- 36.Nakhleh L. Computational approaches to species phylogeny inference and gene tree reconciliation. Trends Ecol Evol. 2013;28(12):719–728. doi: 10.1016/j.tree.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanier HC, Knowles LL. Is recombination a problem for species-tree analyses? Syst Biol. 2012;61(4):691–701. doi: 10.1093/sysbio/syr128. [DOI] [PubMed] [Google Scholar]
- 38.Butlin RK. Recombination and speciation. Mol Ecol. 2005;14(9):2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 39.Nishikawa H, et al. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet. 2015;47(4):405–409. doi: 10.1038/ng.3241. [DOI] [PubMed] [Google Scholar]
- 40.Joron M, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477(7363):203–206. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermansen JS, et al. Hybrid speciation through sorting of parental incompatibilities in Italian sparrows. Mol Ecol. 2014;23(23):5831–5842. doi: 10.1111/mec.12910. [DOI] [PubMed] [Google Scholar]
- 42.Delmore KE, et al. Genomic analysis of a migratory divide reveals candidate genes for migration and implicates selective sweeps in generating islands of differentiation. Mol Ecol. 2015;24(8):1873–1888. doi: 10.1111/mec.13150. [DOI] [PubMed] [Google Scholar]
- 43.Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491(7426):756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 44.Yeaman S. Hybridization and the porous genome: Patterns of isolation and introgression in manakins. Mol Ecol. 2013;22(12):3195–3197. doi: 10.1111/mec.12314. [DOI] [PubMed] [Google Scholar]
- 45.Rosenblum EB, et al. Goldilocks Meets Santa Rosalia: An Ephemeral Speciation Model Explains Patterns of Diversification Across Time Scales. Evol Biol. 2012;39(2):255–261. doi: 10.1007/s11692-012-9171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cutter AD. Integrating phylogenetics, phylogeography and population genetics through genomes and evolutionary theory. Mol Phylogenet Evol. 2013;69(3):1172–1185. doi: 10.1016/j.ympev.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Bermingham E, Moritz C. Comparative phylogeography: Concepts and applications. Mol Ecol. 1998;7(4):367–369. [Google Scholar]
- 48.Avise JC. Phylogeography: The History and Formation of Species. Harvard Univ Press; Cambridge, MA: 2000. [Google Scholar]
- 49.Knowles LL. Statistical phylogeography. Annu Rev Ecol Evol Syst. 2009;40:593–612. [Google Scholar]
- 50.Hewitt GM. Quaternary phylogeography: The roots of hybrid zones. Genetica. 2011;139(5):617–638. doi: 10.1007/s10709-011-9547-3. [DOI] [PubMed] [Google Scholar]
- 51.Carnaval AC, et al. Prediction of phylogeographic endemism in an environmentally complex biome. Proc Biol Sci. 2014;281(1972):20141461. doi: 10.1098/rspb.2014.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moritz C, et al. Identification and dynamics of a cryptic suture zone in tropical rainforest. Proc Biol Sci. 2009;276(1660):1235–1244. doi: 10.1098/rspb.2008.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singhal S, Moritz C. Reproductive isolation between phylogeographic lineages scales with divergence. Proc Biol Sci. 2013;280(1772):20132246. doi: 10.1098/rspb.2013.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowman DM, et al. Biogeography of the Australian monsoon tropics. J Biogeogr. 2010;37(2):201–216. [Google Scholar]
- 55.Keast A. Bird speciation on the Australian continent. Bull Mus Comp Zool. 1961;123:303–495. [Google Scholar]
- 56.Cracraft J. Origin and evolution of continental biotas: Speciation and historical congruence within the Australian avifauna. Evolution. 1986;40(5):977–996. doi: 10.1111/j.1558-5646.1986.tb00566.x. [DOI] [PubMed] [Google Scholar]
- 57.Eldridge MD, Potter S, Cooper SJ. Biogeographic barriers in north-western Australia: An overview and standardisation of nomenclature. Aust J Zool. 2011;59(4):270–272. [Google Scholar]
- 58.Edwards SV. Long-distance gene flow in a cooperative breeder detected in genealogies of mitochondrial DNA sequences. Proc Biol Sci. 1993;252(1335):177–185. doi: 10.1098/rspb.1993.0063. [DOI] [PubMed] [Google Scholar]
- 59.Jennings WB, Edwards SV. Speciational history of Australian grass finches (Poephila) inferred from thirty gene trees. Evolution. 2005;59(9):2033–2047. [PubMed] [Google Scholar]
- 60. Balakrishnan CN Lee JY, Edwards SV (2010) Phylogeography and phylogenetics in the nuclear age. Searching for the Causes of Evolution: From Field Observations to Mechanisms, eds Grant P, Grant R (Princeton Univ Press, Princeton), pp 65–88.
- 61.Toon A, Hughes JM, Joseph L. Multilocus analysis of honeyeaters (Aves: Meliphagidae) highlights spatio-temporal heterogeneity in the influence of biogeographic barriers in the Australian monsoonal zone. Mol Ecol. 2010;19(14):2980–2994. doi: 10.1111/j.1365-294X.2010.04730.x. [DOI] [PubMed] [Google Scholar]
- 62.Lee JY, Edwards SV. Divergence across Australia’s Carpentarian barrier: Statistical phylogeography of the red-backed fairy wren (Malurus melanocephalus) Evolution. 2008;62(12):3117–3134. doi: 10.1111/j.1558-5646.2008.00543.x. [DOI] [PubMed] [Google Scholar]
- 63.Kearns AM, Joseph L, Omland KE, Cook LG. Testing the effect of transient Plio-Pleistocene barriers in monsoonal Australo-Papua: Did mangrove habitats maintain genetic connectivity in the Black Butcherbird? Mol Ecol. 2011;20(23):5042–5059. doi: 10.1111/j.1365-294X.2011.05330.x. [DOI] [PubMed] [Google Scholar]
- 64.Rollins LA, Svedin N, Pryke SR, Griffith SC. The role of the Ord Arid Intrusion in the historical and contemporary genetic division of long-tailed finch subspecies in northern Australia. Ecol Evol. 2012;2(6):1208–1219. doi: 10.1002/ece3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kearns AM, Joseph L, Toon A, Cook LG. Australia’s arid-adapted butcherbirds experienced range expansions during Pleistocene glacial maxima. Nat Commun. 2014;5:3994. doi: 10.1038/ncomms4994. [DOI] [PubMed] [Google Scholar]
- 66.Potter S, Eldridge MD, Taggart DA, Cooper SJ. Multiple biogeographical barriers identified across the monsoon tropics of northern Australia: Phylogeographic analysis of the brachyotis group of rock-wallabies. Mol Ecol. 2012;21(9):2254–2269. doi: 10.1111/j.1365-294X.2012.05523.x. [DOI] [PubMed] [Google Scholar]
- 67.Potter S, Cooper SJ, Metcalfe CJ, Taggart DA, Eldridge MD. Phylogenetic relationships of rock-wallabies, Petrogale (Marsupialia: Macropodidae) and their biogeographic history within Australia. Mol Phylogenet Evol. 2012;62(2):640–652. doi: 10.1016/j.ympev.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 68.Eldridge MD, Potter S, Johnson CN, Ritchie EG. Differing impact of a major biogeographic barrier on genetic structure in two large kangaroos from the monsoon tropics of Northern Australia. Ecol Evol. 2014;4(5):554–567. doi: 10.1002/ece3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malekian M, Cooper SJB, Carthew SM. Phylogeography of the Australian sugar glider (Petaurus breviceps): Evidence for a new divergent lineage in eastern Australia. Aust J Zool. 2010;58(3):165–181. [Google Scholar]
- 70.Aplin KP, Rhind SG, Have JT, Chesser RT. Taxonomic revision of Phascogale tapoatafa (Meyer, 1793) (Dasyuridae; Marsupialia), including descriptions of two new subspecies and confirmation of P. pirata Thomas, 1904 as a ‘Top End’ endemic. Zootaxa. 2015;4055(1):1–73. doi: 10.11646/zootaxa.4055.1.1. [DOI] [PubMed] [Google Scholar]
- 71.Melville J, Ritchie EG, Chapple SN, Glor RE, Schulte JA., 2nd Evolutionary origins and diversification of dragon lizards in Australia’s tropical savannas. Mol Phylogenet Evol. 2011;58(2):257–270. doi: 10.1016/j.ympev.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 72.Dolman G, Hugall AF. Combined mitochondrial and nuclear data enhance resolution of a rapid radiation of Australian rainbow skinks (Scincidae: Carlia) Mol Phylogenet Evol. 2008;49(3):782–794. doi: 10.1016/j.ympev.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 73.Moritz C, et al. Multilocus phylogeography reveals nested endemism in a gecko across the monsoonal tropics of Australia. Mol Ecol. 2016;25(6):1354–1366. doi: 10.1111/mec.13511. [DOI] [PubMed] [Google Scholar]
- 74.Oliver PM, Couper PJ, Pepper M. Independent transitions between monsoonal and arid biomes revealed by systematic revison of a complex of Australian geckos (Diplodactylus; Diplodactylidae) PLoS One. 2014;9(12):e111895. doi: 10.1371/journal.pone.0111895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Catullo RA, Lanfear R, Doughty P, Keogh JS. The biogeographical boundaries of northern Australia: Evidence from ecological niche models and a multi‐locus phylogeny of Uperoleia toadlets (Anura: Myobatrachidae) J Biogeogr. 2014;41(4):659–672. [Google Scholar]
- 76.Cracraft J. Origin and evolution of continental biotas: Speciation and historical congruence within the Australian avifauna. Evolution. 1986;40:977–996. doi: 10.1111/j.1558-5646.1986.tb00566.x. [DOI] [PubMed] [Google Scholar]
- 77.Rannala B, Yang Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164(4):1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brumfield R, Nickerson DA, Beerli P, Edwards SV. The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol Evol. 2003;18:249–256. [Google Scholar]
- 79.Lemmon AR, Lemmon EM. High-throughput identification of informative nuclear loci for shallow-scale phylogenetics and phylogeography. Syst Biol. 2012;61(5):745–761. doi: 10.1093/sysbio/sys051. [DOI] [PubMed] [Google Scholar]
- 80.Jones MR, Good JM. Targeted capture in evolutionary and ecological genomics. Mol Ecol. 2016;25(1):185–202. doi: 10.1111/mec.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harrison RG, Larson EL. Hybridization, introgression, and the nature of species boundaries. J Hered. 2014;105(Suppl 1):795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- 82.Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- 83.Ray N, Excoffier L. Inferring past demography using spatially explicit population genetic models. Hum Biol. 2009;81(2-3):141–157. doi: 10.3378/027.081.0303. [DOI] [PubMed] [Google Scholar]
- 84.Payseur BA, Rieseberg LH. A genomic perspective on hybridization and speciation. Mol Ecol. March 9, 2016 doi: 10.1111/mec.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gompert Z, Buerkle CA. A powerful regression-based method for admixture mapping of isolation across the genome of hybrids. Mol Ecol. 2009;18(6):1207–1224. doi: 10.1111/j.1365-294X.2009.04098.x. [DOI] [PubMed] [Google Scholar]
- 86.Pool JE, Nielsen R. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics. 2009;181(2):711–719. doi: 10.1534/genetics.108.098095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sousa VC, Grelaud A, Hey J. On the nonidentifiability of migration time estimates in isolation with migration models. Mol Ecol. 2011;20(19):3956–3962. doi: 10.1111/j.1365-294x.2011.05247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dasmahapatra KK, Lamas G, Simpson F, Mallet J. The anatomy of a ‘suture zone’ in Amazonian butterflies: A coalescent-based test for vicariant geographic divergence and speciation. Mol Ecol. 2010;19(19):4283–4301. doi: 10.1111/j.1365-294X.2010.04802.x. [DOI] [PubMed] [Google Scholar]
- 89.Wu CI. The genic view of the process of speciation. J Evol Biol. 2001;14(6):851–865. [Google Scholar]
- 90.Payseur BA, Nachman MW. The genomics of speciation: Investigating the molecular correlates of X chromosome introgression across the hybrid zone between Mus domesticus and Mus musculus. Biol J Linn Soc Lond. 2005;84(3):523–534. [Google Scholar]
- 91.Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23(13):3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- 92.Burri R, et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 2015;25(11):1656–1665. doi: 10.1101/gr.196485.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134(4):1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nachman MW, Payseur BA. Recombination rate variation and speciation: Theoretical predictions and empirical results from rabbits and mice. Philos Trans R Soc Lond B Biol Sci. 2012;367(1587):409–421. doi: 10.1098/rstb.2011.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biol. 2010;8(9):e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lemmon EM, Lemmon AR. High-throughput genomic data in systematics and phylogenetics. Annu Rev Ecol Evol Syst. 2013;44:99–121. [Google Scholar]
- 97.Lemmon AR, Emme SA, Lemmon EM. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst Biol. 2012;61(5):727–744. doi: 10.1093/sysbio/sys049. [DOI] [PubMed] [Google Scholar]
- 98.Zhou L, Bawa R, Holliday JA. Exome resequencing reveals signatures of demographic and adaptive processes across the genome and range of black cottonwood (Populus trichocarpa) Mol Ecol. 2014;23(10):2486–2499. doi: 10.1111/mec.12752. [DOI] [PubMed] [Google Scholar]
- 99.Hedrick PW. Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol. 2013;22(18):4606–4618. doi: 10.1111/mec.12415. [DOI] [PubMed] [Google Scholar]
- 100.Lewontin R, Birch L. Hybridization as a source of variation for adaptation to new environments. Evolution. 1966;3:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. [DOI] [PubMed] [Google Scholar]
- 101.McCormack JE, Hird SM, Zellmer AJ, Carstens BC, Brumfield RT. Applications of next-generation sequencing to phylogeography and phylogenetics. Mol Phylogenet Evol. 2013;66(2):526–538. doi: 10.1016/j.ympev.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 102.Brandley MC, et al. Evaluating the performance of anchored hybrid enrichment at the tips of the tree of life: A phylogenetic analysis of Australian Eugongylus group scincid lizards. BMC Evol Biol. 2015;15(1):62. doi: 10.1186/s12862-015-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hugall AF, O’Hara TD, Hunjan S, Nilsen R, Moussalli A. An exon-capture system for the entire class Ophiuroidea. Mol Biol Evol. 2016;33(1):281–294. doi: 10.1093/molbev/msv216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rowe KC, et al. Museum genomics: Low-cost and high-accuracy genetic data from historical specimens. Mol Ecol Resour. 2011;11(6):1082–1092. doi: 10.1111/j.1755-0998.2011.03052.x. [DOI] [PubMed] [Google Scholar]
- 105.Bi K, et al. Unlocking the vault: Next-generation museum population genomics. Mol Ecol. 2013;22(24):6018–6032. doi: 10.1111/mec.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 107.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483(7388):169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Corbett-Detig RB, Hartl DL, Sackton TB. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 2015;13(4):e1002112. doi: 10.1371/journal.pbio.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Andrews KR, et al. Trade-offs and utility of alternative RADseq methods: Reply to Puritz et al. Mol Ecol. 2014;23(24):5943–5946. doi: 10.1111/mec.12964. [DOI] [PubMed] [Google Scholar]
- 110.Dierickx EG, Shultz AJ, Sato F, Hiraoka T, Edwards SV. Morphological and genomic comparisons of Hawaiian and Japanese Black-footed Albatrosses (Phoebastria nigripes) using double digest RADseq: Implications for conservation. Evol Appl. 2015;8(7):662–678. doi: 10.1111/eva.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Knowles LL. Estimating species trees: Methods of phylogenetic analysis when there is incongruence across genes. Syst Biol. 2009;58(5):463–467. doi: 10.1093/sysbio/syp061. [DOI] [PubMed] [Google Scholar]
- 112.Hare M. Prospects for nuclear gene phylogeography. Trends Ecol Evol. 2001;16:700–706. [Google Scholar]
- 113.Hare MP, Karl SA, Avise JC. Anonymous nuclear DNA markers in the American oyster and their implications for the heterozygote deficiency phenomenon in marine bivalves. Mol Biol Evol. 1996;13(2):334–345. doi: 10.1093/oxfordjournals.molbev.a025593. [DOI] [PubMed] [Google Scholar]
- 114.Barrow LN, Bigelow AT, Phillips CA, Lemmon EM. Phylogeographic inference using Bayesian model comparison across a fragmented chorus frog species complex. Mol Ecol. 2015;24(18):4739–4758. doi: 10.1111/mec.13343. [DOI] [PubMed] [Google Scholar]
- 115.Perez MF, Carstens BC, Rodrigues GL, Moraes EM. Anonymous nuclear markers reveal taxonomic incongruence and long-term disjunction in a cactus species complex with continental-island distribution in South America. Mol Phylogenet Evol. 2016;95:11–19. doi: 10.1016/j.ympev.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 116.Bragg JG, Potter S, Bi K, Moritz C. Exon capture phylogenomics: Efficacy across scales of divergence. Mol Ecol Resour. August 20, 2015 doi: 10.1111/1755-0998.12449. [DOI] [PubMed] [Google Scholar]
- 117.Weyenberg G, Huggins PM, Schardl CL, Howe DK, Yoshida R. kdetrees: Non-parametric estimation of phylogenetic tree distributions. Bioinformatics. 2014;30(16):2280–2287. doi: 10.1093/bioinformatics/btu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gruenstaeudl M, Reid NM, Wheeler GL, Carstens BC. Posterior predictive checks of coalescent models: P2C2M, an R package. Mol Ecol Resour. 2015;16(1):193–205. doi: 10.1111/1755-0998.12435. [DOI] [PubMed] [Google Scholar]
- 119.Liu L, Yu L, Pearl DK, Edwards SV. Estimating species phylogenies using coalescence times among sequences. Syst Biol. 2009;58(5):468–477. doi: 10.1093/sysbio/syp031. [DOI] [PubMed] [Google Scholar]
- 120.Edwards SV, et al. Implementing and testing the multispecies coalescent model: A valuable paradigm for phylogenomics. Mol Phylogenet Evol. 2016;94(Pt A):447–462. doi: 10.1016/j.ympev.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 121.Hallström BM, Janke A. Mammalian evolution may not be strictly bifurcating. Mol Biol Evol. 2010;27(12):2804–2816. doi: 10.1093/molbev/msq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stenz NWM, Larget B, Baum DA, Ané C. Exploring Tree-Like and Non-Tree-Like Patterns Using Genome Sequences: An Example Using the Inbreeding Plant Species Arabidopsis thaliana (L.) Heynh. Syst Biol. 2015;64(5):809–823. doi: 10.1093/sysbio/syv039. [DOI] [PubMed] [Google Scholar]
- 124.Park HJ, Nakhleh L. Inference of reticulate evolutionary histories by maximum likelihood: The performance of information criteria. BMC Bioinformatics. 2012;13(Suppl 19):S12. doi: 10.1186/1471-2105-13-S19-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wen D, Yu Y, Hahn MW, Nakhleh L. Reticulate evolutionary history and extensive introgression in mosquito species revealed by phylogenetic network analysis. Mol Ecol. January 25, 2016 doi: 10.1111/mec.13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28(8):2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192(3):1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Peter BM. Admixture, population structure, and F-statistics. Genetics. 2016;202(4):1485–1501. doi: 10.1534/genetics.115.183913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hickerson MJ, et al. Phylogeography’s past, present, and future: 10 years after Avise, 2000. Mol Phylogenet Evol. 2010;54(1):291–301. doi: 10.1016/j.ympev.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 131.Tsai YHE, Carstens BC. Assessing model fit in phylogeographical investigations: An example from the North American sandbar willow Salix melanopsis. J Biogeogr. 2013;40(1):131–141. [Google Scholar]
- 132.Carstens BC, et al. Model selection as a tool for phylogeographic inference: An example from the willow Salix melanopsis. Mol Ecol. 2013;22(15):4014–4028. doi: 10.1111/mec.12347. [DOI] [PubMed] [Google Scholar]
- 133.Ray N, Currat M, Foll M, Excoffier L. SPLATCHE2: A spatially explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics. 2010;26(23):2993–2994. doi: 10.1093/bioinformatics/btq579. [DOI] [PubMed] [Google Scholar]
- 134.Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9(10):e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chan YL, Schanzenbach D, Hickerson MJ. Detecting concerted demographic response across community assemblages using hierarchical approximate Bayesian computation. Mol Biol Evol. 2014;31(9):2501–2515. doi: 10.1093/molbev/msu187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xue AT, Hickerson MJ. The aggregate site frequency spectrum for comparative population genomic inference. Mol Ecol. 2015;24(24):6223–6240. doi: 10.1111/mec.13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Becquet C, Przeworski M. A new approach to estimate parameters of speciation models with application to apes. Genome Res. 2007;17(10):1505–1519. doi: 10.1101/gr.6409707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Naduvilezhath L, Rose LE, Metzler D. Jaatha: A fast composite-likelihood approach to estimate demographic parameters. Mol Ecol. 2011;20(13):2709–2723. doi: 10.1111/j.1365-294X.2011.05131.x. [DOI] [PubMed] [Google Scholar]
- 139.Marjoram P, Wall JD. Fast “coalescent” simulation. BMC Genet. 2006;7:16. doi: 10.1186/1471-2156-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang Y, et al. A new method for modeling coalescent processes with recombination. BMC Bioinformatics. 2014;15:273. doi: 10.1186/1471-2105-15-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Beerli P, Palczewski M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010;185(1):313–326. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152(2):763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. Inferring species trees directly from biallelic genetic markers: Bypassing gene trees in a full coalescent analysis. Mol Biol Evol. 2012;29(8):1917–1932. doi: 10.1093/molbev/mss086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chifman J, Kubatko L. Quartet inference from SNP data under the coalescent model. Bioinformatics. 2014;30(23):3317–3324. doi: 10.1093/bioinformatics/btu530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5(10):e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 2009;24(6):332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.