Significance
Using a newly developed technology, HaloTag nucleic acid programmable protein array (HaloTag-NAPPA), we increase the capacity of in situ protein microarray technology several-fold, such that proteome-scale screening becomes feasible. Many examples of novel protein–protein interactions (PPIs) among plant signaling pathways were observed. With few exceptions, nearly all of these connections are undocumented in the existing literature. This study has resulted in an important new resource for the plant biology community—a plant transcription factor-anchored protein–protein interaction network map. Such transcription factor- and transcriptional regulator-based PPI networks may help in the identification of novel genes for use in the improvement of agronomic traits such as grain quality, disease resistance, and stress tolerance.
Keywords: protein arrays, interactome, hormone, systems biology, Arabidopsis thaliana
Abstract
Protein microarrays enable investigation of diverse biochemical properties for thousands of proteins in a single experiment, an unparalleled capacity. Using a high-density system called HaloTag nucleic acid programmable protein array (HaloTag-NAPPA), we created high-density protein arrays comprising 12,000 Arabidopsis ORFs. We used these arrays to query protein–protein interactions for a set of 38 transcription factors and transcriptional regulators (TFs) that function in diverse plant hormone regulatory pathways. The resulting transcription factor interactome network, TF-NAPPA, contains thousands of novel interactions. Validation in a benchmarked in vitro pull-down assay revealed that a random subset of TF-NAPPA validated at the same rate of 64% as a positive reference set of literature-curated interactions. Moreover, using a bimolecular fluorescence complementation (BiFC) assay, we confirmed in planta several interactions of biological interest and determined the interaction localizations for seven pairs. The application of HaloTag-NAPPA technology to plant hormone signaling pathways allowed the identification of many novel transcription factor–protein interactions and led to the development of a proteome-wide plant hormone TF interactome network.
A major objective in the postgenomic era is to assign detailed molecular function(s) to the many protein-coding genes that remain uncharacterized even in model organisms such as the reference plant Arabidopsis thaliana (1). It is estimated that Arabidopsis contains more than 2,000 transcription factors and transcriptional regulators (hereafter “TFs”), most of which are uncharacterized (2). TFs function as key players in plant hormone signal transduction pathways and are responsible for directing the widespread changes in gene expression that are essential for the regulation of growth and development in plants (3, 4). The TFs within these transcriptional regulatory networks do not work independently but rather undergo complex interactions with other proteins (2). Understanding how these TFs interact with other proteins will ultimately lead to a greater comprehension of biological systems.
For determination of physical protein–protein interactions (PPIs), protein microarrays (5–10) are complementary to other PPI technologies such as the yeast two-hybrid system (Y2H) (11) and protein complex purification coupled with mass spectrometry (AP-MS) (12, 13). With conventional protein microarrays it is necessary to purify thousands of in vivo expressed proteins and to spot these purified proteins on a solid surface (5–8). In contrast, in situ synthesis protein microarray technologies simplify protein microarray fabrication by circumventing the steps of in vivo protein expression and purification (9, 10, 14, 15). This streamlining facilitates an increase in the number of target genes that can be assayed, allowing for thousands of protein-encoding plasmids to be spotted at lower cost and in less time. Such a large scale has been achieved with the nucleic acid programmable protein array (NAPPA) technology (9, 10), which uses a cell-free expression system and antibody-based protein capture to create protein microarrays from DNA microarrays. Current NAPPA technology, however, has been hampered by the low efficiency of antibody-based protein capture, which requires large spot sizes, resulting in low spot density (∼2,000 spots per array) (10). Here we report the development of an improved high-affinity capture technique for the fabrication of in situ synthesized NAPPA protein microarrays, using a set of 12,000 Arabidopsis ORFs (16, 17). We applied this improved NAPPA to the mapping of a protein–protein interaction network for a set of 38 Arabidopsis hormone response TFs. This study of the TF interactome network with a newly developed technology reveals hitherto hidden cross-talk of plant signaling pathways that regulate multiple TFs, providing a rich dataset complementary to that of previous interactome studies (16).
Results
Development of a HaloTag-Based NAPPA Microarray System.
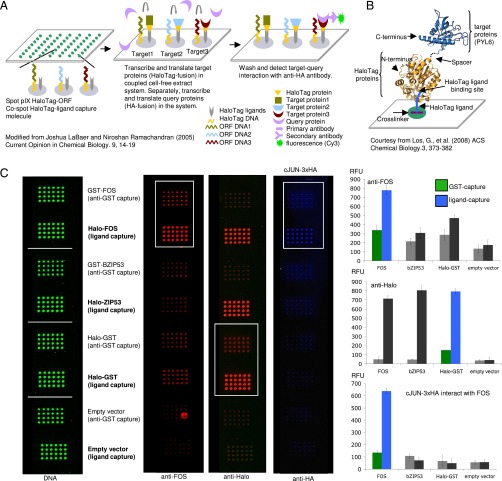
Previous NAPPA technology used an anti-GST antibody to anchor in vitro expressed C-terminal GST fusion proteins to an aminosilanized glass slide (9, 10). We modified this original system by incorporating a high-affinity capture tag, the HaloTag (Promega), to immobilize nascent proteins on the array surface (18). HaloTag fusion proteins irreversibly bind to a small chloroalkane ligand that is cospotted on the array. This approach yielded rapid and high-affinity capture of expressed fusion proteins with minimal lateral protein diffusion (19) (Fig. 1 A and B).
Fig. 1.
(A) The NAPPA assay. Plasmid DNA, cross-linker, and HaloTag ligand are spotted on glass slides. Addition of coupled transcription–translation reagent results in protein expression and localized protein capture. Coexpression of an epitope-tagged query protein enables detection of protein interactions by immunodetection. Modified from ref. 63. (B) Schematic of the HaloTag protein interacting with its chloroalkane ligand. Courtesy of ref. 18. (C) HaloTag gives higher yields of active protein compared with GST-antibody. (C, Left) Amount of deposited plasmid DNA as measured with PicoGreen. (C, Middle) FOS protein as detected by an anti-FOS antibody, Halo-tagged proteins detected by an anti-Halo antibody, and protein interaction between FOS and 3×HA-JUN as detected by an anti-HA antibody (from left to right). (C, Right) Signal quantification of arrays is shown. The y axis represents relative fluorescence units (RFUs). Colored histogram bars indicate signals corresponding to boxed regions on the arrays. Error bars represent SE of the signal intensity.
Compared with the original NAPPA method, our HaloTag fusion arrays showed a higher signal-to-noise ratio than seen with GST-based arrays (Fig. 1C). The HaloTag method captured up to four times more protein than the anti-GST antibody method, as revealed by direct detection of captured proteins using an anti-HaloTag antibody or a gene-specific antibody targeting the protein product of the human Fos proto-oncogene (FOS) (Fig. 1C). We used the human Jun proto-oncogene fused to a triple hemagglutinin tag (3×HA-JUN) to probe the array for PPIs and found that the signal-to-noise ratio of the well-known JUN–FOS pair was four times greater with the HaloTag capture system than with anti-GST antibody capture (Fig. 1C). The increased signal from the HaloTag spots most likely arises from the small molecular size of the HaloTag capture ligand compared with the more bulky anti-GST antibody. Thus, a higher ratio of capture molecules to DNA can be spotted, resulting in more efficient capture of expressed protein. HaloTag-NAPPAs offer several advantages over conventional protein arrays. The synthesis and capture of proteins in situ eliminate the cumbersome process of expressing and purifying individual proteins and hence avoids compromising protein functional integrity. Additionally, because DNA is spotted instead of proteins, the arrays require no special storage conditions and have an extended shelf life of greater than 12 mo.
Detecting Protein Interactions on High-Density HaloTag-NAPPAs.
We transferred a set of 12,000 Gateway pENTR ORF clones, representing 40% of all Arabidopsis protein-coding genes and including ∼900 TFs (16, 17), into a custom-designed in vitro HaloTag expression vector (pIX-Halo:ccdB; see Materials and Methods for vector details; Dataset S1). Plasmid DNA was prepared from these 12,000 clones and robotically spotted at high density onto three independent sets of custom arrays, AtNAPPA01, AtNAPPA02. and AtNAPPA03. Each array contained ∼4,600 unique Arabidopsis ORFs spotted in duplicate (9,200 spots) at 250-µm spacing, representing a total of ∼27,600 features in duplicate across the three different arrays (Fig. 2A).
Fig. 2.
High-density arrays each containing ∼4,600 unique ORFs spotted in duplicate. (A) AtNAPPA02 high-density array showing DNA staining by PicoGreen. (B) Global protein expression levels generated by the submerge method and detected with an anti-Halo antibody are similar for most ORFs. (C) Protein interaction between VND4 and 3×HA-VND7 as detected by an anti-HA antibody. (D, Top) The sorted histogram of B indicates signal intensity (black), median intensity (red), median intensity 2× (green), and median intensity 0.5× (blue) from all ORF clones on the array. (D, Bottom) Signal quantification of the PPI array (C) is shown. The histogram indicates signal intensity (blue) corresponding to boxed regions on the arrays (C, Right, Top). The signals from candidate interactors are significantly above Halo average signal + 3-SD cutoff (black arrows).
We also developed a “submerged protein expression” technique to produce proteins on the HaloTag-NAPPAs. This “submerge” protocol differed in both the concentration and volume of T7 polymerase and wheat germ expression components relative to our first-generation NAPPA protocol (HybriWell gasket method; Fig. 1C and SI Appendix, Fig. S1). The larger volume of the submerge protocol potentially facilitates local diffusion by alleviating the space constraints on protein synthesis that were imposed by the previously used HybriWell gasket incubation chambers. In turn, this allows noncaptured HaloTag proteins to diffuse away from the site of synthesis more easily, resulting in more focused spots and ultimately enabling higher spot density (see Materials and Methods for details).
After applying the submerge protocol, we assessed global protein expression across the HaloTag-NAPPAs using an anti-HaloTag antibody. Over 80% of the spots produced signals within twofold of the median, indicating that the protein levels produced by the submerge protocol are constant for most ORFs (Fig. 2 B and D). Sixteen percent of the proteins on the arrays fell outside this range, with 8% showing no or lower expression (Dataset S1). The predicted average mass of expressed fusion proteins was 72.6 kDa. We tested the submerge HaloTag-NAPPA protocol with known protein-interaction candidates (Fig. 2C). Protein query VND7 (AT1G71930) heterodimerized with closely related family member VND4 (AT1G12260), in agreement with previous reports of VND7 heterodimerization (20) (Fig. 2 C and D). These results show that immobilized proteins on the array are likely to be properly folded and capable of specific interactions. Among the thousands of proteins on the HaloTag-NAPPAs, we also detected binding of VND7 to novel proteins such as AT1G20960 and AT3G13660 (Fig. 2 C and D).
An Arabidopsis Hormone Response Transcription Factor Interactome Network.
Plant hormones are a group of structurally unrelated small molecules central to the integration of environmental cues and growth and development (21). Discovering novel protein–protein interactions among key players in these pathways is expected to provide insight into the mechanisms by which signals from the different pathways are integrated. Therefore, as a first application of HaloTag-NAPPA for mapping protein–protein interactions, we selected a set of 38 Arabidopsis TFs with known roles in plant hormone signaling pathways [ethylene (ET), brassinosteroid (BR), salicylic acid (SA), jasmonic acid (JA), auxin (indole acetic acid; IAA), cytokinin (CK), gibberellin (GA), or abscisic acid (ABA)] that mediate essentially all developmental decisions and stress responses in plants (Datasets S2 and S3).
Because most TFs work in complexes with other proteins (2), identification of additional proteins that interact with hormone TFs is expected to reveal novel hormone signaling components and novel connections among known signaling pathways. Moreover, for several of the proteins in this subset, few or no interactions were identified in a large-scale Y2H-based interactome mapping experiment (16), partly due to autoactivation (22).
We probed the three HaloTag-NAPPAs with each of the 38 TFs using the submerged protein-expression technique. We scored an interaction as positive when the average signal intensity from duplicate protein spots was more than 3 SDs above the median of the negative control, which consisted of empty vector encoding only the 33-kDa HaloTag. By this criterion, HaloTag-NAPPA screens of the 38 TFs produced a TF-NAPPA dataset containing 3,580 interactions among 2,238 proteins (Dataset S4).
We determined the overlap of interactions between the TF-NAPPA and literature-curated binary interactions (LCIs) in BioGrid (thebiogrid.org) and IntAct (www.ebi.ac.uk/intact), excluding 124 interactions reported only in our previously published Arabidopsis Interactome 1 (AI-1) interactome dataset (16). The HaloTag-NAPPA TF screen recapitulated a statistically significant 8 of 461 binary LCI interactions in the shared clone space (P = 0.0265, χ2 test with Yates’ correction; hereafter “χ2”) (Dataset S4). The moderate albeit significant overlap is probably explained by our previous observation (23) that different interaction assays detect different subsets of “true” interactions, largely as a consequence of different assay biochemistry, fusion tags, and so forth. Thus, the low overlap suggests that the HaloTag-NAPPA is highly complementary to the widely used Y2H assay.
We considered the possibility that the query TFs may bind to the spotted plasmid DNA on the arrays instead of the displayed protein. To systematically address this potential artifact, we took advantage of a recently published protein binding microarray (24) dataset describing DNA-recognition motifs for our query TFs. We queried all ORFs (including the junction between universal linkers and ATG and STOP codons; hereafter “ORF region”) for the presence of binding motifs for our bait TFs. Any signal due to motifs in the vector backbone was subtracted as background in our scoring scheme and therefore not included in this analysis. For short binding motifs (on average eight bases), we observed that ∼55% of the 11,630 ORFs spotted on the HaloTag-NAPPA had predicted TF binding motifs (CIS-BP; cisbp.ccbr.utoronto.ca; Dataset S5). For each TF, we assessed whether there was a statistical association between the TF-NAPPA–positive spots (NAPPA+) and the presence of a binding motif for the respective TF in the corresponding ORF. According to this analysis the TFs fell into two groups. For nine TFs, the NAPPA+ spots were not significantly associated with the presence of the binding motif for the query TF in the encoding ORFs (Dataset S5; P > 0.1, χ2). This suggests that for these TFs, binding is unlikely to be mediated by protein–DNA interactions, especially in light of the large number of NAPPA-negative (NAPPA−) spots containing the motif. In contrast, 13 of the 22 TFs in CIS-BP showed a significant association (P < 0.05, χ2) between putative protein–protein interactions detected by HaloTag-NAPPA and the presence of the binding motif. For KNAT3, ORFs of all identified protein-interaction partners contained the KNAT3 binding motif (Dataset S5; P = 0.036, χ2), thus raising the possibility that NAPPA+ signal for these TFs may be caused by protein–DNA instead of protein–protein interactions. At the same time, for each TF, a much larger proportion of ORFs containing the binding motif was NAPPA−, indicating that the presence of a motif per se is insufficient for TF binding. Therefore, we tested whether the number of TF binding motifs per ORF could play a role by comparing the number of binding motifs in the ORFs of putative protein interactors with the background distribution for all ORFs on the array containing the motif (SI Appendix, Fig. S2 and Dataset S5). Again, the result was not uniform; for some query TFs the distribution of binding-motif counts in the NAPPA+ spots was statistically indistinguishable from the background distribution, indicating that the number of TF-binding sites in these ORFs did not affect its detection in the PPI assay. This group of TFs included ARF7, EIN3, VND7, and EDF1 (Dataset S5; P > 0.1, Wilcoxon rank-sum test). In contrast, for a different subset, for example ABI5, TGA1, or MYC2, the motif-count distribution of the NAPPA+ spots was significantly shifted toward higher counts compared with the background distribution (Dataset S5; P < 0.0001, Wilcoxon rank-sum test). Thus, even though the large subset of high–motif-count ORFs that were NAPPA− precludes a simple causality of the NAPPA signal being simply due to DNA binding, we cannot exclude the possibility that DNA binding may play a role for a subset of query TFs. We therefore separated those interactions that could be due to DNA binding from all others. As a “core” dataset, we classified 750 TF-NAPPA+ pairs that did not contain a TF binding motif in the ORF region or that involved queries without a DNA binding domain (e.g., RGA); as “noncore,” we classified 2,830 putative interaction pairs for which the ORF region contained at least one motif for the query TF or where the motif was unknown (Datasets S2 and S4).
Evaluation of the Quality of the TF-NAPPA Dataset by in Vitro Pull-Down Assay.
A critical question for any new technology regards the quality of the obtained data. To address this, we followed an approach we previously described in several publications (16, 23, 25, 26) in which a subset of interactions from a new dataset is systematically validated in a second interaction assay, in our case a pull-down assay. The challenge of this approach is that no assay can detect all interactions and each assay has a different interaction-detection profile (25, 27). To estimate the false-discovery rate of a new dataset, it is therefore important to measure the sensitivity and background of the validation assay by benchmarking this against sets of control pairs: a positive reference set (PRS) of well-documented interactions and a random reference set (RRS) composed of random protein pairs (27). Subsequently, the retest rate of the new dataset in the validation assay can be interpreted in light of these benchmarking data. To measure the sensitivity and background of our pull-down assay, we benchmarked it against (i) a set of 49 well-documented interactions from the literature (PRS; AtPRS_v1s) and (ii) a set of 69 random protein pairs for which there was no evidence of interaction in the literature (RRS) (25). The 49 AtPRS_v1s pairs are a random subset of our original set of 118 PRSs (AtPRS_v1) (16). All interactions in AtPRS_v1 were collected from the IntAct database and are a manually recurated subset of interactions that are supported by at least two independent peer-reviewed publications (28).
In the benchmarking experiments using these reference sets, the pull-down assay detected 31 of the 49 AtPRS_v1s pairs (63%) and only 2 of the 69 RRS pairs (3%), thus defining the sensitivity and background of the assay (SI Appendix, Fig. 3 and Dataset S6). To systematically validate our dataset, we randomly picked 125 TF-NAPPA interactions from AtNAPPA01 (corresponding to ∼10% of interactions from this dataset) and tested these in the benchmarked validation assay (Dataset S6). Of the 125 random pairs, 81 (64%) scored positive, which is statistically indistinguishable from the PRS (P = 0.8618, Fisher’s exact test; Fig. 3 and SI Appendix, Fig. S3A). Among the core and noncore subsets, 20 of the 31 core interactions (65%) and 61 of the 94 noncore pairs (65%) scored positive (Fig. 3, SI Appendix, Figs. S3C and S4, and Dataset S6). The observation that the validation rates for the full set of HaloTag-NAPPA positives and for the individual NAPPA core and noncore subsets were indistinguishable from the PRS indicates that TF-NAPPA interactions are equal in quality to well-documented interactions reported in the literature.
Fig. 3.
TF-NAPPA protein–protein interactions replicated by pull-down assay. The histogram shows the proportion of positive scoring pairs from the PRS dataset, the set of randomized sample pairs in NAPPA, and the set of sample pairs positive in NAPPA. Error bars represent SE of the proportion. PRS pairs (yellow bar) were significantly higher than RRS pairs (blue bar) in NAPPA (P < 0.0001, Fisher’s exact test). The data from sample pair positives (green bar) in NAPPA are indistinguishable from PRS (P = 0.8618, Fisher’s exact test).
The high validation rate of the noncore subset was slightly surprising. However, upon closer inspection, we noticed that this is likely due to the large size and heterogeneity of the noncore data, which include true protein–protein interactions and a small subset of putative protein–DNA interactions. For example, all noncore interactions for ABI5 (5), ARR14 (2), and MYC2 (10) could be confirmed by the secondary assay, whereas for ZIP63 a clear difference emerged between the core interactions, where 10 of 13 (77%) were validated, and the noncore interactions, where only 5 of 21 (24%) were validated (Dataset S6). Thus, in the absence of additional evidence, we maintain our distinction of core and noncore data, but consider most of the interactions even in the noncore dataset as likely protein–protein interactions and will include these in our biological analysis. Taken together, the validation results using a benchmarked validation assay indicate that the HaloTag-NAPPA system produces reliable and high-quality protein–protein interaction data that are statistically indistinguishable from literature interactions.
HaloTag-NAPPA Reveals a Potential Role of TFs in Plant Hormone Cross-Regulation.
After confirming the biochemical validity of our interactions, we aimed to gather evidence for their biological validity and the insights they may shed on phytohormone signal transduction. The TF-NAPPA dataset includes protein interactions among TFs from several key Arabidopsis hormone signaling pathways (Fig. 4 and Dataset S7). In addition to the previous systematic tests, we also validated several of these biologically interesting novel interactions by in vitro pull-down assays (SI Appendix, Fig. S5). Of 46 interactions selected based on their role in hormone regulatory processes, 34 (73.9%) could be validated using the in vitro pull-down assay (Fig. 4, SI Appendix, Fig. S5, and Dataset S7). Consistently, 9 of 11 positive control interactions from the literature, but only 3 of 17 stringent negative controls, scored positive in this assay (Fig. 4, SI Appendix, Fig. S5, and Dataset S7). This “negative set” was composed of family members of protein–protein interaction pairs from the TF-NAPPA dataset for which there was no evidence of interaction in the HaloTag-NAPPA screen or the literature. Such positive interactions may have been missed in our original screen, or may be due to structural similarities between family members.
Fig. 4.
Three case studies from the TF-NAPPA dataset that highlight how Arabidopsis TFs may control and contribute to plant signaling pathways. Arrows indicate the direction in which the protein–protein interactions were discovered. Bidirectional arrows indicate reciprocal detection. (A) Protein–protein interactions in TF-NAPPA suggest that DELLA proteins interact with MYC2, a key transcriptional activator of JA responses. (B) A subnetwork of ABA, other hormones, and nitrate cross-talk extracted from TF-NAPPA interactions, literature-curated interactions, and the AI-1 binary experimental PPI dataset (22). PYL6 directly interacts with TGA1, which acts as a connector to CIPK23, the master regulator of the nitrate signaling pathway. (C) Network communities in the ethylene pathway. TF-NAPPA suggests interplay between the ethylene and gibberellin and light signaling pathways.
Gibberellin and Jasmonate.
Considerable cross-talk between the GA and JA signaling pathways has been documented (29). Up-regulation of JA-responsive genes is delayed in loss-of-function quadruple della mutants (30). DELLA proteins were previously identified as key negative regulators in the GA response pathway (31). In our HaloTag-NAPPA dataset, MYC2, a key TF of the JA signaling pathway, interacts with the DELLA proteins RGL1 and GAI (Fig. 4A, SI Appendix, Fig. S5A, and Dataset S7). The interaction between MYC2 and DELLAs (RGA1, RGL1, and GAI1) was confirmed by an in vitro pull-down assay (Fig. 4A, SI Appendix, Fig. S5A, and Dataset S7). These interactions were independently detected by the Y2H assay (32). GA-triggered degradation of DELLA proteins has previously been proposed to release JAZ proteins, allowing them to bind MYC2 and thus inhibiting the JA response (29). Our findings suggest a new role for DELLA proteins as direct regulators of MYC2, permitting cross-talk between the GA and JA signaling pathways. Interaction with DELLA proteins might prevent MYC2 from being accessed and inhibited by JAZ proteins. Alternatively, this interaction may provide a more efficient route for GA to interfere with JA signaling by directly targeting MYC2 and perhaps its closely related homologs MYC3 and MYC4 (33) rather than by targeting all 12 JAZ proteins. Our finding could support an alternative scenario where DELLAs interacting with MYC2 regulate JA signaling negatively via DELLA–JAZ interactions.
Abscisic Acid, Nitrogen, and Other Hormones.
In our TF-NAPPA network, we identified a subnetwork of proteins involved in the ABA signaling pathway. ABA regulates a broad range of plant traits, particularly those involved in seed development (34) and the adaptation to environmental stress (35). The HaloTag-NAPPA interactions expanded the known ABA network (Fig. 4B), relative to interactions previously reported in the literature (36), by at least 10 protein interactions, including ABI5, bHLH064, MYC2, BZR1, bZIP63, ARR14, bZIP53, and TGA1 (Fig. 4B, SI Appendix, Fig. S5B, and Dataset S7). These eight TFs interacted with PYL6, a member of the recently identified PYR/PYL/RCAR ABA receptor family (37–40). Consistent with previous observations (41, 42), the TF-NAPPA network also revealed molecular links between the ABA and nitrate signaling pathways via ARR14 and TGA1, which have been found to regulate nitrate response (43), although the molecular mechanism of this connection remains unclear. Our TF-NAPPA network provides a hypothesis that ABA and nitrate signaling cross-regulation occurs through protein–protein interactions between PYL6 and TGA1, and also suggests an interaction of the ABA signaling pathway with other plant hormones such as brassinosteroid (BZR1) or jasmonic acid (MYC2). To support these interactions, we tested each of these HaloTag-NAPPA–positive protein pairs in the Y2H assay (44). One of these interactions (MYC2–PYL6) was confirmed by Y2H (SI Appendix, Fig. S6) and pull-down assays (Fig. 4B, SI Appendix, Fig. S5B, and Dataset S7). These results point to a possible connection between ABA and JA signaling pathways via an ABA receptor (45). However, the other pairs tested by Y2H assay, such as TGA1–PYL6 and ABI5–PYL6, showed no interaction (SI Appendix, Fig. S6). The limited overlap between pull-down and Y2H assays has been described before, and is likely due to the different assay principles and resulting biases inherent to these assays (27).
Ethylene, Gibberellin, and Light.
Interconnections between the gaseous plant hormone ET and GA and light-responsive pathways have been reported (46–48). ET is responsible for maintaining the apical hook as a seedling emerges from the soil to protect the developing shoot meristem. Exogenous application of ET inhibits hypocotyl elongation in the dark (49). A previous study reported that the master regulatory TF of the ET response, EIN3, targeted genes encoding the chlorophyll biosynthesis enzymes PORA and PORB, and that EIN3 cooperates with PIF1 to optimize the deetiolation of the Arabidopsis seedling (50). ET and GA likely act in an opposing manner to regulate seedling growth in the light (51). We found that EIN3 interacted with PIF4, another TF involved in light signaling (Fig. 4C, SI Appendix, Fig. S5C, and Dataset S7). PIF4 negatively interacts with phytochrome B (52) and acts as an activator of the GA transcriptional response (53). Further interplay between the ET and GA and light signaling pathways may occur through interaction of EIN3 and the DELLA protein RGA1 (Fig. 4C, SI Appendix, Fig. S5C, and Dataset S7), upstream of PIF4. The HaloTag-NAPPA interactome revealed novel potential interactions between the ET, GA, and light signaling pathways.
The interactions described in these examples reveal the wide range of hypotheses about the mutual modulation of hormone and nutrient signaling pathways that can be derived from our dataset, and demonstrate the power of our HaloTag-NAPPA platform to uncover unexpected network complexity even in well-characterized signaling systems.
In Planta Visualization of Protein–Protein Interactions by Bimolecular Fluorescence Complementation.
Although we are able to validate many of the novel HaloTag-NAPPA protein–protein interactions by pull-down assay, because HaloTag-NAPPA is an in vitro assay the question arises as to whether these interactions are functionally relevant in vivo. We therefore used a bimolecular fluorescence complementation (BiFC) assay in tobacco leaves (54) to validate 15 novel interactions in planta, of which 7 (46%) scored positive. Similarly, only one of our two positive controls scored positive, even though both control pairs were tested in two assay orientations, whereas 8 of 15 test pairs were tested in only a single orientation. Specifically, we assessed the in planta relevance of five interactors of the ABA coreceptor PYL6, which we found to interact with ABI5, BZR1, MYC2, bHLH064, and TGA1 (Dataset S8). To ensure specificity, we included as a negative control a close family member of PYL6, PYR1, which did not interact with any of these five proteins by HaloTag-NAPPA. Consistent with results of the HaloTag-NAPPA assays, all PYL6-interaction partners identified by NAPPA were able to reconstitute YFP signals in planta (BiFC-positive) and specifically localized to the nucleus (Fig. 5A, Top). These same proteins were unable to interact with PYR1 (BiFC-negative) (Fig. 5A, Middle). Interestingly, PYL6-YFP alone localized not only to the nucleus but also to the cytosol in tobacco leaves (Fig. 5B, Top). The nuclear-specific fluorescence patterns of BiFC suggest that only the nuclear-localized PYL6 interacts with ABI5 (ABA), BZR1 (brassinosteroid), bHLH064 (cytokinin), MYC2 (jasmonic acid), and TGA1 (SA) proteins.
Fig. 5.
BiFC visualization of protein–protein interactions in tobacco leaves (Insets show a magnification of a representative nucleus). (A, Top) PYL6 interactions with the TFs BZR1, ABI5, MYC2, TGA1, and bHLH064 (AT2G18300) take place in the nucleus. (A, Middle) No reconstituted split-YFP fluorescence was observed when these five TFs were tested with the PYL6− family member PYR1, thus supporting the specificity of the PYL6 interactions. (A, Bottom) No reconstituted split-YFP fluorescence was observed when these five TFs were tested with CYFP-empty vector. (B) YFP-tagged protein localization in tobacco leaves. (B, Top) Five TFs, BZR1, ABI5, MYC2, TGA1, and bHLH064 (AT2G18300), are localized to the nucleus. (B, Bottom) Both PYL6-YFP and PYR1-YFP localized to both the cytosol and nucleus. (C, Top) Two additional interactions from the TF-NAPPA dataset, TGA1–CIPK23 and GAI–ZML1, showed reconstituted split-YFP fluorescence in the nucleus, whereas the positive control pair, CIPK23–CHL1, showed reconstituted split-YFP fluorescence in the plasma membrane, as previously reported (52). (C, Bottom) No reconstituted split-YFP fluorescence was observed when NYFP-CIPK23, -TGA1, and -GAI were tested against CYFP-empty vector. (D) Fluorescence of CIPK23-YFP localized to the cytosol, plasma membrane, and nucleus. CHL1-YFP localized to the plasma membrane.
We also tested by BiFC three additional interaction pairs, GAI–ZML1, CIPK23–TGA1, and a positive control, CIPK23–CHL1 (Fig. 5C, Top). GAI–ZML1 and CIPK23–TGA1 reconstituted YFP signals that specifically localized to the nucleus. CIPK23–CHL1 reconstituted YFP signal at the cell membrane, as expected. CHL1 is a known interactor of CIPK23 at the cell membrane (55). We also observed the localization of CIPK23-YFP and CHL-YFP alone (Fig. 5D, Right). CHL1-YFP localized to the cell membrane, whereas CIPK23-YFP was detected in both the cytosol and nucleus. Taken together, these observations indicate that nuclear CIPK23 interacts with the transcription factor TGA1 and that cytosolic CIPK23 interacts with membrane-localized CHL1.
Altogether, we confirmed in planta several HaloTag-NAPPA PPIs that are of specific biological interest (Figs. 4 B and C and 5 and Dataset S8). The localization of individual proteins in comparison with the localization of the reconstituted proteins pair suggests that many protein–protein interactions depend on the cellular compartmentalization of different signaling processes. This compartmentalization of protein–protein interactions stresses the importance of subcellular location in the regulation of hormone response pathways.
Discussion
We developed an improved NAPPA methodology, HaloTag-NAPPA, and used it to map interactions for Arabidopsis TFs involved in hormone signaling. Using systematic validation in a benchmarked interaction assay, we demonstrated that the TF-NAPPA dataset has a quality that is comparable to interactions reported in the literature. The high quality of the data was further confirmed by the validation of interactions of biological interest by BiFC. Our conclusion that most NAPPA+ spots are due to protein–protein as opposed to protein–DNA interactions is supported by DNase experiments, in which NAPPA+ spots were unaffected by DNase digestion of the spotted DNA (56). Despite the significant overlap with previously described interactions the overlap was moderate, indicating that HaloTag-NAPPA is complementary to Y2H assays or AP-MS (16, 57). This is supported by the results of the validation assay, which equally reproduced TF-NAPPA and literature interactions. Somewhat surprising was the large number of interaction partners that we identified for the 38 bait TFs. We cannot completely rule out that some pairs in the noncore subset might be due to protein–DNA interactions; therefore, these interactions should be investigated with the appropriate caution. However, a more important reason for the large number of interactions appears to be our selection of bait proteins that are critical components of some of the most vital plant signaling pathways. By selecting TFs of central developmental and defense signaling pathways, it is likely that we inadvertently picked more highly connected proteins and family members. This is not per se unusual: in other systems also, central proteins have a very large number of interaction partners; for example, for the human tumor suppressor protein TP53 >2,500 interaction partners have been described (IntAct). Similarly, a high connectivity is not limited to TFs, as the Arabidopsis LOW SULFUR UP-REGULATED proteins are similarly characterized by a high connectivity. The existence of such highly connected proteins is typical for scale-free networks; however, the high connectivity of some hubs is not representative of the connectivity of all proteins in the network. Finally, the results obtained for individual family members cannot be taken as representative of the entire protein family. In our previously published Arabidopsis interactome network (16) a very high connectivity was observed for the TFs ANAC089 and TCP14 (222 and 102 interactors, respectively), whereas for the family members ANAC019 (10) and ANAC072 (1) or TCP15 (40) and TCP1 (2), respectively, many fewer interaction partners were identified in the same experiment. Thus, within one protein family, there are highly connected proteins and much less connected family members.
It is likely that assay biochemistry plays an important role in this: HaloTag-NAPPA functions by anchoring target proteins to a solid support, which may limit the accessibility of the target proteins, whereas the low-throughput pull-down validation assay is not subject to this limitation. Thus, once interactions are identified despite the anchoring, these can be validated by the second assay. Importantly, in addition, a microtiter plate-based version of the array technology, wNAPPA, showed a limited overlap with Y2H-positive interactions in a systematic comparison using human interaction pairs (25), thus reinforcing the conclusion that NAPPA technology is complementary to other interaction assays. Biologically, the TF-NAPPA dataset confirms known interactions, provides biochemical support for genetic and molecular biological observations, and illuminates previously unknown cross-regulation of plant hormone signaling pathways.
Many of the connections we identified are undocumented, and hence provide useful hypotheses for plant biologists. The resulting PPI network map provides a source of candidate TFs for future studies and possible targets of genetic manipulation of plant hormone response pathways. Besides plants, HaloTag-NAPPA can readily be applied to other organisms, particularly mammalian model systems for which large ORF clone collections already exist (58). We envision that proteome-scale HaloTag-NAPPA could be adapted for other applications, including identification of small molecule–protein interactions (59) and discovery of posttranslational modifications (59).
Materials and Methods
HaloTag-NAPPA Implementation and Optimization.
Initial testing and comparison of the HaloTag- and anti-GST NAPPA systems were performed using a first-generation set of custom, in vitro expression vectors: pEU-T7/SP6-GW-Halo:ccdB and pEU-T7/SP6-GW-GST:ccdB (see below for vector details). pENTR-ORFs (pENTR-223-Sfi) were transferred by Gateway LR recombination into the pEU-T7/SP6-GW-GST/Halo:ccdB destination vectors. Plasmid DNA was prepared from 100 mL overnight culture using NucleoBond Xtra Midiprep columns (Macherey-Nagel Midi-XL) according to the manufacturer’s recommendations. For array fabrication, a spotting mix containing a final concentration of 1.5 μg/μL plasmid DNA, 2 mM amine-O4 chloroalkane ligand (Promega) or 1:100 anti-GST polyclonal antibody (GE Healthcare), 3.6 mg/mL BSA (Sigma), and 1.25 mg/mL BS3 (Pierce) was spotted onto aminosilane-coated glass slides (CMT-GAPS II; Corning) using a Molecular Dynamics Gen III microarray spotter. Spotted DNA was detected as follows. Slides were blocked for 1 h at room temperature (RT) in SuperBlock (TBS) (Pierce) and then incubated in 20 mL 1:600 PicoGreen (Invitrogen) for 5 min. Slides were washed in PBS, quickly rinsed in dH2O, and dried with filtered compressed air. Signal was measured using Cy3 excitation/emission parameters on a Molecular Dynamics Gen III scanner. Protein detection conditions are described in HybriWell Gasket Assay below.
Vector Construction.
The vectors pEU-T7/SP6-GW-Halo:ccdB and pEU-T7/SP6-GW-GST:ccdB were constructed by inserting the SP6 promoter sequence downstream of the T7 promoter in pEU3-N11 (Proteios), followed by the ligation of the ccdB-CAMR Gateway cassette into the blunt-ended Acc65I and BamHI sites of pEU-T7/SP6 to form pEU-T7/SP6-GW. Nucleotides 1–961 of the HaloTag v7 sequence (Promega) were PCR-amplified from pFN19A (Promega) and inserted into the EcoRV and Xho sites of pEU-T7/SP6-GW to form pEU-T7/SP6-GW-Halo:ccdB. Note that the three terminal amino acids (ISG) of the HaloTag were modified to LGT. To make pEU-T7/SP6-GW-GST:ccdB, EcoRV and Xho sites were added to the GST-coding sequence by PCR amplification and cloned into the EcoRV and Xho sites of pEU-T7/SP6-GW. To make pIX-Halo:ccdB, the vector backbone of pEU-T7/SP6-GW-Halo, -GST, and -3×HA was replaced with the backbone of pCITE4b using EcoRI sites, resulting in the in-frame addition of a 6×His tag and T7 terminator sequence following the attR2 Gateway recombination sites. The pIX-Halo-His6× control plasmid (HaloTag-only control plasmid) was created by digesting pIX-Halo:ccdB with Xho to remove the Gateway recombination cassette followed by self-ligation. Vectors and complete vector sequences of pIX-Halo:ccdB can be found at the Arabidopsis Biological Resource Center (https://abrc.osu.edu).
HybriWell Gasket Assay.
Capture comparison arrays containing Halo-ORF and GST-ORF plasmids were expressed as follows. Slides were blocked for 1 h in SuperBlock (TBS). A HybriWell gasket (Grace Bio-Labs) was applied to the preblocked array and filled with 145 μL per slide of T7 coupled Transcription/Translation System (TNT) (Promega)-coupled wheat germ expression mix (Promega) according to the manufacturer’s recommended concentrations (9, 10). Slides were then incubated for 1.5 h at 30 °C. For protein–protein interaction assays, 200–500 ng of query plasmid DNA (pEU-T7/SP6-GW-3×HA-ORF) was added to the TNT expression mix in the HybriWell gasket chamber, incubated for 1.5 h at 30 °C, and subsequently incubated at 15 °C for an additional 2 h. Slides were washed three times for 5 min each in blocking buffer [5% (wt/vol) nonfat milk + 1× PBS + 0.1% Tween-20 (PBST)] and then blocked for 1 h at RT. Primary antibodies [anti-HA, mouse monoclonal, 1:500, Covance; anti-GST, mouse monoclonal, 1:200, Cell Signaling; anti-HaloTag, rabbit polyclonal, 1:200 (vol/vol), Promega; anti-FOS, rabbit polyclonal, 1:200 (vol/vol), Calbiochem] were diluted and 300 μL was applied to each slide, covered with a LifterSlip (Erie Scientific), and incubated at RT for 1 h. Slides were washed three times for 5 min each in blocking buffer, and then 300 μL secondary antibody [anti-mouse HRP-coupled, 1:200 (vol/vol), GE Healthcare; anti-rabbit HRP-coupled, 1:1,500 (vol/vol), Jackson] was applied, covered with a LifterSlip, and incubated at RT for 1 h in a humidity box. Slides were washed two times for 5 min each in PBST and two times for 5 min each in PBS. Slides were briefly rinsed in water, and 500 μL of a 1:100 (vol/vol) dilution of tyramide signal amplification (TSA)-Cy3 (PerkinElmer) in amplification buffer was applied and incubated for 10 min at RT. Slides were rinsed in dH2O and immediately dried using filtered compressed air.
Construction of a HaloTag-Fused Arabidopsis ORF Collection and Microarray Development.
SI Appendix, Fig. S7 summarizes the pipeline for the generation of the pIX-Halo expression clones and the subsequent construction of the high-density HaloTag-NAPPA microarrays. Plasmid DNA from AtORFeome2.0 clones in the Gateway-compatible entry vector was recombined using Gateway LR clonase II (Life Technologies) into the pIX-Halo:ccdB destination vector. The prepared plasmid DNA (Qiagen) was combined with the microarray spotting mixture components 1.25 mg/mL BS3 cross-linker (Pierce), 3.6 mg/mL BSA (Sigma), 25% DMSO (Sigma), and 2 mM chloroalkane ligand (Promega) and spotted onto an aminosilane-coated glass slide (CMT-GAPS II; Corning) using a Molecular Dynamics Gen III microarray spotter (60, 61).
Protein–Protein Interaction Assay Using the Microarray Submerge Assay.
The functional proteins were synthesized in situ from spotted HaloTag-ORF plasmid DNA. The HaloTag-ORF fusion proteins were translated using the TNT wheat germ system (Promega) by submerging the entire glass slide into a mixture containing the following components: 1.2 mL diethylpyrocarbonate (DEPC) water, 15% wheat germ extract, 30 μL 10× buffer, 7.5 μL amino acid mix −Met, 7.5 μL amino acid mix −Leu/−Cys, and 15 μL each of T7 polymerase and RNase inhibitor (Promega). 3×HA-fused TF proteins were independently expressed as a query using the TNT system (Promega) according to the recommendations of the manufacturer. Expressed query proteins were applied to HaloTag-NAPPA microarrays and incubated at 4 °C for 2 h. Subsequently, HaloTag-NAPPAs were washed three times with 1× PBST containing 5% nonfat dry milk and blocked overnight at 4 °C, and subsequently incubated with mouse anti-HA monoclonal antibody [HA.11 clone16B12, 1:500 (vol/vol), Covance] for 1 h at RT. After further washes, arrays were incubated with anti-mouse HRP-coupled secondary antibody [1:500 (vol/vol), GE Healthcare] for 1 h at RT. Arrays were then washed before adding Cy3-conjugated TSA substrate (Molecular Probes) according to the recommendations of the manufacturer. Fluorescence was detected using a PowerScanner (Tecan). Because the signal-to-noise ratios were very uniform for all features across the glass slide with the submerged assay conditions, we performed all protein–protein interaction assays for the 38 TFs with this method (SI Appendix, Fig. S1).
Data Scanning, Quantification, and Data Processing.
Feature extraction and image analysis software (Array-Pro Analyzer; Tecan) was used to locate and delineate every spot in the array and to integrate each spot’s intensity. Signal intensity was normalized by subtracting the background intensity from the intensity of each microarray spot using Array-Pro Analyzer. From the two duplicate spots on each array, we calculated the normalized average signal intensities. Those data points whose intensity was greater than 3 SDs above the negative control (pIX-Halo vector backbone) median were selected as putative TF interactors (62). Protein levels of background clones measured with the use of an anti-HA antibody show that ∼89%, ∼91%, and ∼86% of signal levels were within threefold of the median on each array, AtNAPPA01, 02, and 03, respectively. Single experiments using duplicated spots on one glass slide were performed for each TF with the NAPPA protein–protein interaction method. Duplicated spots yielded very consistent results (SI Appendix, Fig. S8).
Pull-Down Assay.
The pull-down assay was generally performed using HaloLink magnetic beads according to the recommendations of the manufacturer (Promega). The corresponding Arabidopsis ORFs were transferred by Gateway LR recombination into both pIX-Halo:ccdB and pIX-3×HA:ccdB destination vectors. Competent bacteria (Escherichia coli, strain DH5α-T1R) were transformed with the resulting LR recombination products. After selection of transformants in liquid terrific broth medium containing 50 µg/mL carbenicillin, plasmid DNA was extracted and purified using DNA Miniprep Kits (Qiagen). Bait proteins in pIX-Halo were expressed using the TNT T7 Coupled Wheat Germ Extract System (Promega) according to the recommendations of the manufacturer. Twenty-five microliters of bait proteins (pIX-Halo-ORFs) was mixed and rotated with 25 μL Halo magnetic beads in a total volume of 150 μL buffer (PBS + Nonidet P-40) at RT for 1 h. Subsequently, beads with HaloTag fusion protein baits were washed and added to 25 μL prey protein fused with 3×HA and subsequently rotated at RT for 1 h. The mixture was subsequently washed three times with 500 μL washing buffer, and washed beads were boiled in 15 μL SDS sampling buffer at 80 °C for 10 min. Bait and prey proteins (2.5 μL; 10% input) were loaded as a control to indicate the original protein amount.
Selecting the Positive Reference Set.
Our PRS (named “AtPRS_v1s”) consisted of a randomly picked subset of the original PRS (hereafter “AtPRS_v1”) described previously (16). All interactions in the PRS were collected from the IntAct database and are part of the filtered subset that is supported by at least two independent peer-reviewed publications. Moreover, the more than 200 publications supporting these interactions were manually recurated to ensure a high quality of the supporting experiments, namely completeness of controls (28). However, because AtPRS_v1 was initially assembled to assess the (AI-1 dataset) (16), it contains no pairs that were identified for the first time in this large-scale Y2H experiment (although some interactions were “rediscovered”). Among the 49 interactions in the AtPRS_v1s subset, 7 scored positive when tested in our Y2H system. Due to the labor intensity of the pull-down experiments, only a subset of the original AtPRS_v1 (16) was used for its benchmarking and, to avoid introducing any bias, the 49 pairs of AtPRS_v1s were picked randomly from the 118 interactions making up AtPRS_v1.
Yeast Two-Hybrid Assay.
PYL6 was cloned in-frame by USERTM (https://www.neb.com/applications/cloning-and-synthetic-biology/user-cloning) (New England BioLabs) cloning in pGBT9.BS, and the TFs MYC2, ABI5, BZR1, TGA1, and bHLH064 were cloned in-frame with pGAD.GH. Plasmids were cotransformed into the yeast strain PJ69-4A, and transformants were selected on SD media without leucine and tryptophan. Drop tests were done with 5 μL 1:10 dilutions starting at an OD600 of 1 until 0.001 in media without leucine, tryptophan, and histidine with the addition of 2.5 mM 3-amino-1,2,4-triazole. Plates were grown at 30 °C for 13 d.
Bimolecular Fluorescence Complementation Assay.
ORFs were cloned into either pNY0 or pCY0 vector to produce N-terminal half YFP (nYFP) or C-terminal half YFP (cYFP), and were transformed into Agrobacterium strain GV3101 by electroporation. The agrobacteria harboring either pNY0 or pCY0 was grown at 28 °C overnight and centrifuged at 4,000 × g for 15 min. Cell pellets were resuspended with infiltration buffer (10 mM Mes, 10 mM MgCl2, 0.1 mM acetosyringone) and kept at RT for 2 h. Appropriate pairs of the cell suspensions were mixed with agrobacteria expressing p19 and were adjusted to OD600 0.5–0.6. The cell suspensions were infiltrated into Nicotiana benthamiana leaves and incubated for 3 d. The leaves were observed by confocal microscopy (Leica). All fluorescent images were taken as a single optical section.
Supplementary Material
Acknowledgments
We thank Shoji Mano for the generous gifts of BiFC vectors. We also thank Mattia Pelizzola for assistance with data analysis and Mark Zander, Mathew G. Lewsey, Matija Dreze, Samuel J. Pevzner, and Michael E. Cusick for critical reading of the manuscript. We thank Kenichi Hitomi for creation of 3D modeling of the Halo and PYL6 proteins and Rosa Castanon and Anna Bartlett for technical help. This work was supported by grants from the NSF Plant Genome Research Program (DBI-0703905; to M.V. and J.R.E.), Gordon and Betty Moore Foundation (GBMF 3034; to J.R.E.), and NIH (R01GM060396-ES010337; to J.I.S.). J.Y. is supported by the Sumitomo Foundation, Grant for Basic Science Research Project (140384). P.B. is supported by the Deutsche Forschungs Germeinschaft (SFB924) and has received funding from the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (Grant Agreement 648420). F.A. was supported by a Caja Madrid Foundation Fellowship. J.C. and J.R.E. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603229113/-/DCSupplemental.
References
- 1.Meyerowitz EM. 1994. Structure and organization of the Arabidopsis thaliana nuclear genome. Arabidopsis, eds Meyerowitz EM, Somerville CR (Cold Spring Harb Lab Press, Cold Spring Harbor, NY), Vol 27, pp 21–36.
- 2.Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009;50(7):1232–1248. doi: 10.1093/pcp/pcp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long TA, Benfey PN. Transcription factors and hormones: New insights into plant cell differentiation. Curr Opin Cell Biol. 2006;18(6):710–714. doi: 10.1016/j.ceb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126(3):467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 5.Gong W, et al. The development of protein microarrays and their applications in DNA-protein and protein-protein interaction analyses of Arabidopsis transcription factors. Mol Plant. 2008;1(1):27–41. doi: 10.1093/mp/ssm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin YY, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136(6):1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popescu SC, et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA. 2007;104(11):4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293(5537):2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 9.Ramachandran N, et al. Self-assembling protein microarrays. Science. 2004;305(5680):86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 10.Ramachandran N, et al. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008;5(6):535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seebacher J, Gavin AC. SnapShot: Protein-protein interaction networks. Cell. 2011;144(6):1000–1000.e1. doi: 10.1016/j.cell.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 12.Collins SR, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6(3):439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Kühner S, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326(5957):1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 14.He M, Taussig MJ. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method) Nucleic Acids Res. 2001;29(15):e73. doi: 10.1093/nar/29.15.e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He M, et al. Printing protein arrays from DNA arrays. Nat Methods. 2008;5(2):175–177. doi: 10.1038/nmeth.1178. [DOI] [PubMed] [Google Scholar]
- 16.Arabidopsis Interactome Mapping Consortium Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333(6042):601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302(5646):842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 18.Los GV, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3(6):373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, et al. A versatile protein microarray platform enabling antibody profiling against denatured proteins. Proteomics Clin Appl. 2013;7(5–6):378–383. doi: 10.1002/prca.201200062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi M, Kubo M, Fukuda H, Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008;55(4):652–664. doi: 10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- 21.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459(7250):1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 22.Dreze M, et al. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- 23.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158(6):1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun P, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6(1):91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6(1):83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun P. Interactome mapping for analysis of complex phenotypes: Insights from benchmarking binary interaction assays. Proteomics. 2012;12(10):1499–1518. doi: 10.1002/pmic.201100598. [DOI] [PubMed] [Google Scholar]
- 28.Cusick ME, et al. Literature-curated protein interaction datasets. Nat Methods. 2009;6(1):39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou X, Lee LYC, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19(6):884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Navarro L, et al. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18(9):650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 31.Zentella R, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19(10):3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 2012;24(6):2635–2648. doi: 10.1105/tpc.112.098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernández-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 36.Hauser F, Waadt R, Schroeder JI. Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol. 2011;21(9):R346–R355. doi: 10.1016/j.cub.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura N, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61(2):290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santiago J, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60(4):575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 41.Signora L, De Smet I, Foyer CH, Zhang H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001;28(6):655–662. doi: 10.1046/j.1365-313x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- 42.Kanno Y, et al. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA. 2012;109(24):9653–9658. doi: 10.1073/pnas.1203567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez JM, et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014;80(1):1–13. doi: 10.1111/tpj.12618. [DOI] [PubMed] [Google Scholar]
- 44.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 45.Aleman F, et al. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling. Sci Rep. doi: 10.1038/srep28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallego-Bartolomé J, et al. Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 2011;67(4):622–634. doi: 10.1111/j.1365-313X.2011.04621.x. [DOI] [PubMed] [Google Scholar]
- 47.Leivar P, Quail PH. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16(1):19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An F, et al. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012;22(5):915–927. doi: 10.1038/cr.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kendrick MD, Chang C. Ethylene signaling: New levels of complexity and regulation. Curr Opin Plant Biol. 2008;11(5):479–485. doi: 10.1016/j.pbi.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong S, et al. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA. 2009;106(50):21431–21436. doi: 10.1073/pnas.0907670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurepin LV, Pharis RP. Light signaling and the phytohormonal regulation of shoot growth. Plant Sci. 2014;229:280–289. doi: 10.1016/j.plantsci.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21(10):2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451(7177):480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka Y, et al. 2012. Gateway vectors for plant genetic engineering: Overview of plant vectors, application for bimolecular fluorescence complementation (BiFC) and multigene construction. Genetic Engineering—Basics, New Applications and Responsibilities, ed Barrera-Saldaña HA (InTech Europe, Rijeka, Croatia), pp 35–58.
- 55.Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138(6):1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 56.Yu X, et al. Copper-catalyzed azide-alkyne cycloaddition (click chemistry)-based detection of global pathogen-host AMPylation on self-assembled human protein microarrays. Mol Cell Proteomics. 2014;13(11):3164–3176. doi: 10.1074/mcp.M114.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vidal M, Cusick ME, Barabási AL. Interactome networks and human disease. Cell. 2011;144(6):986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.ORFeome Collaboration The ORFeome Collaboration: A genome-scale human ORF-clone resource. Nat Methods. 2016;13(3):191–192. doi: 10.1038/nmeth.3776. [DOI] [PubMed] [Google Scholar]
- 59.Hu S, Xie Z, Qian J, Blackshaw S, Zhu H. Functional protein microarray technology. Wiley Interdiscip Rev Syst Biol Med. 2011;3(3):255–268. doi: 10.1002/wsbm.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yazaki J, et al. Genomics approach to abscisic acid- and gibberellin-responsive genes in rice. DNA Res. 2003;10(6):249–261. doi: 10.1093/dnares/10.6.249. [DOI] [PubMed] [Google Scholar]
- 61.Yazaki J, et al. Embarking on rice functional genomics via cDNA microarray: Use of 3′ UTR probes for specific gene expression analysis. DNA Res. 2000;7(6):367–370. doi: 10.1093/dnares/7.6.367. [DOI] [PubMed] [Google Scholar]
- 62.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438(7068):679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 63. LaBaer J, Ramachandran N (2005) Protein microarrays as tools for functional proteomics. Curr Opin Chem Biol 9(1):14–19. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.