Significance
The ribosome is a large and complex RNA–protein complex that synthesizes proteins in all kingdoms of life. The sequences of bacterial ribosomes differ, and antibiotics exploit these differences to inhibit the ribosome and stop cell growth. Mutations in the ribosome can interfere with antibiotic binding and render the bacteria resistant to the antibiotic. Using a combination of structural, biophysical, and biochemical experiments, we show how rRNA mutations alter the structure of the ribosome at the antibiotic binding site and result in divergent antibiotic susceptibility profiles.
Keywords: negamycin, tetracycline, tRNA selection, ribosome structure, antibiotic resistance
Abstract
Mutations conferring resistance to translation inhibitors often alter the structure of rRNA. Reduced susceptibility to distinct structural antibiotic classes may, therefore, emerge when a common ribosomal binding site is perturbed, which significantly reduces the clinical utility of these agents. The translation inhibitors negamycin and tetracycline interfere with tRNA binding to the aminoacyl-tRNA site on the small 30S ribosomal subunit. However, two negamycin resistance mutations display unexpected differential antibiotic susceptibility profiles. Mutant U1060A in 16S Escherichia coli rRNA is resistant to both antibiotics, whereas mutant U1052G is simultaneously resistant to negamycin and hypersusceptible to tetracycline. Using a combination of microbiological, biochemical, single-molecule fluorescence transfer experiments, and X-ray crystallography, we define the specific structural defects in the U1052G mutant 70S E. coli ribosome that explain its divergent negamycin and tetracycline susceptibility profiles. Unexpectedly, the U1052G mutant ribosome possesses a second tetracycline binding site that correlates with its hypersusceptibility. The creation of a previously unidentified antibiotic binding site raises the prospect of identifying similar phenomena in antibiotic-resistant pathogens in the future.
The ribosome is an intricate RNA–protein assembly that translates mRNA into protein. Distinct structural classes of translation inhibitors bind to discrete sites on the ribosome, inhibiting protein synthesis at one or more stages of the translation cycle. The mechanisms of action and structural basis of ribosomal inhibition have been defined for multiple classes of antibiotics that target the ribosome (1). Given that the RNA component of the ribosome plays a critical role in all stages of protein synthesis, translation inhibitors typically bind to rRNA, and consequentially, target-based antibiotic resistance mutations are often associated with discrete rRNA nucleotide changes (2).
The simultaneous loss of antibiotic susceptibility after the acquisition of a single rRNA nucleotide change to combinations of structurally discrete antibiotics may occur when the individual agents target a shared or partially overlapping binding site on the ribosome (1). Various genetic tools have been used to isolate antibiotic resistance mutations and characterize functionally important sites on the bacterial ribosome (3–5). A c-omplementary design-focused approach to understanding antibiotic specificity and eukaryotic toxicity can be achieved through the construction of mutant ribosomes with site-directed changes (6, 7). X-ray crystallography can subsequently be applied to visualize structural changes associated with specific rRNA mutations (8, 9). Minor structural changes may suffice to yield resistance when the mutated nucleotide is directly involved in the antibiotic binding site, such as is the case for a guanine to adenine base substitution that alters susceptibility to macrolides without affecting the binding mode of the antibiotic to the ribosome (9). Other localized mutations, including streptomycin resistance mutations, significantly alter the position of the bases and base pairing without changing the conformation of the phosphate backbone (8). Resistance mutations can also occur at sites that are not involved in antibiotic binding but where mutation causes structural perturbations that propagate toward the antibiotic binding site (10). Characterizing the structural and mechanistic impact of acquired antibiotic resistance mutations of the ribosomes in bacterial pathogens is essential to understand their role in clinical efficacy. A link between rRNA sequence and antibiotic-induced perturbations of translation could reveal strategies for leveraging sequence information to guide additional antibiotic discovery as well as optimize treatment options.
Negamycin is a natural bactericidal antibiotic that displays activity against a broad spectrum of Gram-negative pathogens, including Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii (11, 12). Tetracycline is broad spectrum antibiotic with bacteriostatic activity that is used to treat a variety of bacterial infections (1). The mechanism of action for both antibiotics is to interfere with tRNA binding to the aminoacyl-tRNA (aa-tRNA) site of the small 30S ribosomal subunit (11, 12). The clinical utility of tetracycline has been reduced because of the rise of antibiotic resistance, providing incentive for the introduction of tigecycline into the clinic (13).
Although negamycin and tetracycline share an overlapping binding site on the 30S ribosomal subunit, differential antibiotic susceptibility profiles have recently been observed for negamycin-resistant mutants (11, 12). One of these resistance mutants displays negamycin resistance and tetracycline hypersusceptibility that cannot be reconciled with any currently available structural data. In this study, we evaluate a negamycin-resistant strain to understand its unexpected switch to tetracycline hypersusceptibility. Microbiological, biochemical, and single-molecule FRET (smFRET) experiments as well as X-ray crystallography evidence suggest that this switch in susceptibility is caused by a previously unidentified tetracycline binding site in the small ribosomal subunit.
Results and Discussion
Negamycin Resistance Mutations.
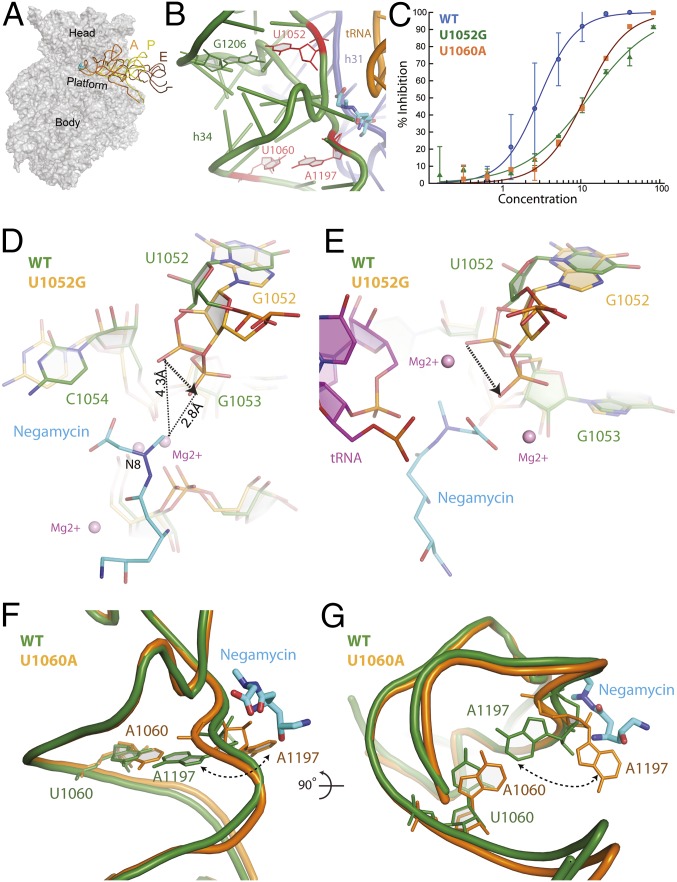
Two previously identified negamycin resistance mutations, U1052G and U1060A, localize to the head domain of the 30S subunit positioned within helix 34 (h34) and are in close proximity to the negamycin binding site (Fig. 1 A and B) (11, 12). These mutations cause high levels of negamycin resistance, with minimum inhibitory concentration (MIC) values that are approximately eight- (U1052G) and fourfold (U1060A) greater than wild-type susceptibility levels (Table 1). In addition, the U1052G or U1060A mutants are resistant to translation inhibition by negamycin in luminescence-based in vitro-coupled transcription–translation systems using purified components in line with MIC values (Fig. 1C and Table 1).
Fig. 1.
Negamycin resistance mutations. (A) Negamycin binds near the A site on the head domain of the 30S subunit. A, P, and E represent bound tRNA molecules. (B) Negamycin resistance mutations U1052, U1060 (11, 12), and A1197 (12) are positioned on h34 and in close proximity to the negamycin binding site. Previously identified resistance mutations are colored in red. (C) Translation activity of wild-type (blue circles; IC50 value of 2.9 µM), U1052G (green triangles; IC50 value of 13 µM), and U1060A (orange squares; IC50 value of 11 µM) ribosomes in the presence of negamycin in a luminescence-based in vitro-coupled transcription–translation assay with purified components. (D) Mutation U1052G significantly distorts the phosphate backbone of h34. The U1052G mutant (orange) has been superimposed on the wild-type E. coli structure (green) bound to negamycin. (E) Superposition of the U1052G mutant from E. coli (orange) on the mRNA–tRNA–negamycin complex from T. thermophilus (green) (12). (F) Acquisition of the U1060A mutation causes a localized perturbation that drives nucleotide A1197 into a flipped out conformation. The U1060A mutant structure (orange) has been superimposed on the wild-type E. coli structure bound to negamycin (green). (G) This panel is rotated 90° along the horizontal axis, illustrating the steric clash with negamycin.
Table 1.
Biochemical and antimicrobial activity of negamycin, tetracycline, and tigecycline on wild-type and mutant E. coli strain SQ171
| Strain | NMY | TET | TIG |
| Wild-type | 32 (2.9) | 2 (15) | 2 (0.28) |
| U1052G | 256 (13) | 0.5 (2.7) | 2 (0.36) |
| U1060A | 128 (11) | 16 (51) | 8 (2.1) |
MIC measurements are in micrograms per milliliter (n = 3). IC50 values (in parentheses) obtained in luminescence-based in vitro-coupled transcription–translation assays with purified components are shown in micromolar (n = 3). NMY, negamycin; TET, tetracycline; TIG, tigecycline.
U1052G and U1060A Inhibit Negamycin Binding.
To visualize the structural impact that the U1052G and U1060A mutations have on the negamycin binding site, X-ray crystallographic structures of the wild-type strain SQ171 Escherichia coli 70S ribosome and negamycin resistance mutants were determined in their apo conformations (Table S1).
Table S1.
Crystallographic data
| Data and refinement | Wild-type | U1052G Apo | U1060A Apo | Wild-type TET | U1052G TET | Wild-type TIG | U1052G TIG |
| Data reduction | |||||||
| Beam line | 17-ID | 17-ID | 17-ID | 17-ID | 17-ID | 17-ID | 17-ID |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | |||||||
| a, b, c (Å) | 211.81, 434.22, 622.77 | 213.04, 436.85, 628.03 | 211.55, 433.65, 622.03 | 212.31, 434.58, 624.92 | 212.41, 435.69, 625.25 | 212.18, 434.26, 624.26 | 211.72, 435.19, 622.83 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 47.17–3.26 (3.27–3.26) | 48.39–3.03 (3.04–3.03) | 48.13–3.32 (3.33–3.32) | 48.16–2.8 (2.80–2.79) | 48.61–3.00 (3.01–3.00) | 48.51–2.96 (2.97–2.96) | 48.525–3.10 (3.11–3.10) |
| I/σI | 10.9 (2.1) | 10.8 (2.0) | 127.3 (32.2) | 188 (16.2) | 12.9 (2.2) | 207.3 (15.5) | 12.8 (2.0) |
| CC(1/2) | 0.988 (0.587) | 0.988 (0.646) | 0.989 (0.640) | 0.983 (0.502) | 0.987 (0.610) | 0.990 (0.540) | 0.993 (0.654) |
| Completeness (%) | 92.8 (83.2) | 96.8 (67.6) | 91.8 (51.5) | 92.1 (30.3) | 99.2 (67.1) | 97.1 (53.5) | 99.7 (79.5) |
| Rmerge | 0.115 (0.515) | 0.126 (0.555) | 0.119 (0.539) | 0.098 (0.037) | 0.142 (0.724) | 0.117 (0.595) | 0.122 (0.575) |
| No. of crystals | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| Total no. of reflections | 2,434,858 (16,991) | 5,238,635 (19,523) | 3,615,497 (37162) | 5,438,737 (7,371) | 8,618,637 (27,571) | 4,505,450 (9,359) | 6,133,491 (28,625) |
| No. of unique reflections | 818,609 (7,264) | 1,084,668 (7,578) | 767,822 (8,916) | 1,291,300 (4,211) | 1,135,145 (7,603) | 1,150,983 (6,297) | 1,027,314 (8,221) |
| Multiplicity | 3.0 (2.3) | 4.8 (2.6) | 4.7 (2.6) | 4.2 (1.8) | 7.6 (3.6) | 3.9 (1.5) | 6.0 (3.5) |
| Refinement | |||||||
| Resolution (Å) | 3.2 | 3.0 | 3.3 | 2.8 | 3.0 | 3.0 | 3.1 |
| Rwork/Rfree (%) | 0.19/0.22 | 0.17/0.20 | 0.18/0.22 | 0.20/0.22 | 0.17/0.19 | 0.21/0.22 | 0.17/0.19 |
| PDB ID code | 5IT8 | 5JC9 | 5J88 | 5J5B | 5J7L | 5J91 | 5J8A |
Rmerge = Σ|I − <I>|/ΣI, where I is the integrated intensity of a given reflection, and <I> is the average intensity of multiple observations of symmetry-related reflections. Rwork = Σ|Fo − Fc|/ΣFo, where Fo and Fc are observed and calculated structure factors, respectively. Rfree was calculated from a 5% subset of reflections that were excluded from the refinement. Parentheses indicate highest resolution shell. CC(1/2), correlation coefficient; TET, tetracycline; TIG, tigecycline.
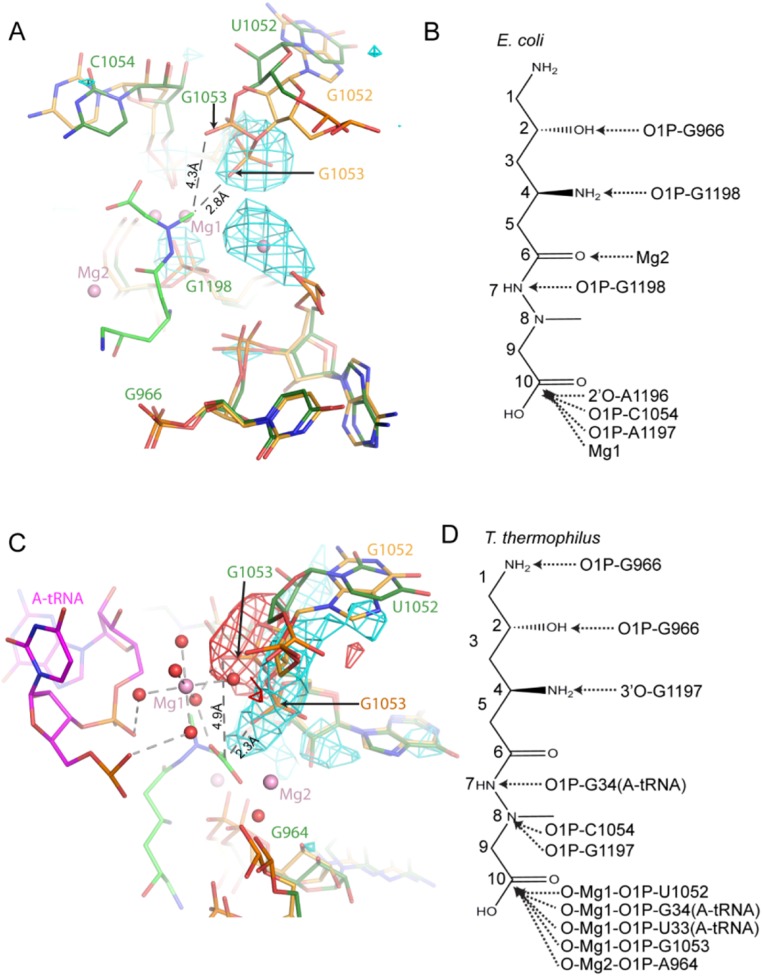
Structural analysis revealed that the wobble base pair that is normally seen between nucleotides U1052 and G1206 is disrupted in the U1052G mutant structure (Fig. 1 B and D). The guanine substitution at this position disrupts the carbonyl–imino hydrogen bond between the O6 oxygen of nucleotide G1206 and the N3 nitrogen of nucleotide U1052. Detailed analysis of the crystallographic data also showed a localized widening of h34 and the displacement of the phosphate group of nucleotide G1053 by ∼2 Å from its position in the apo conformation (Fig. 1D). The negamycin-bound structure of the wild-type 70S E. coli ribosome shows that the phosphate oxygen of nucleotide G1053 is within 4.3 Å of the methyl group on N8 of negamycin (12). Distinctly, in the U1052G mutant, the phosphate oxygen of nucleotide G1053 is positioned 1.5 Å closer to the methyl group on N8 of negamycin, a change that is predicted to increase the degree of steric hindrance between h34 and the bound negamycin molecule, thus providing a structural basis for negamycin resistance (Fig. 1D and Fig. S1).
Fig. S1.
U1052G mutant inhibits negamycin binding. (A) The U1052G mutation displaces the h34 phosphate backbone into the negamycin binding site of the Escherichia coli structure. The U1052G mutant (yellow) is superimposed on the wild-type (green) ribosome. (B) Interactions formed between negamycin and the E. coli ribosome. (C) Superposition of the apo U1052G mutant onto the wild-type negamycin-bound Thermus thermophilus structure with a bound tRNA molecule. The U1052G mutant is shown in yellow, the T. thermophilus ribosome is colored green, and the bound tRNA molecule is presented in magenta. (D) Interactions formed between negamycin and the T. thermophilus ribosome with a bound tRNA molecule. Fo-Fc difference electron densities contoured at 3σ are shown in cyan (positive) and red (negative). Dashed lines indicate distances between atoms. Negamycin is shown as a stick model, with green carbon atoms, blue nitrogen atoms, and red oxygen atoms. Magnesium ions are shown as pink spheres, and coordinating water molecules are shown as red spheres.
Similarly to U1052, nucleotide U1060 is also found in h34 (Fig. 1B), base-pairing with A1197 in the apo conformation of the wild-type ribosome (Fig. 1B). Examination of the X-ray crystallographic data for the U1060A mutant shows that introduction of A1060 drives nucleotide A1197 into a flipped out conformation (Fig. 1 F and G). This localized conformational change is expected to disrupt the hydrogen-bonding pattern between the phosphate group of A1197 and negamycin to, thereby, confer resistance.
A comparison of the apo crystal structures of the U1052G and U1060A mutants with the structures of mRNA–tRNA-bound Thermus thermophilus and E. coli 70S ribosomes in complex with negamycin (11, 12) lends additional support to these observations (Fig. S1). In the T. thermophilus structure with a bound tRNA molecule, the phosphate oxygens of nucleotides U1052 and G1053 indirectly interact with negamycin and the tRNA molecule through a coordinated Mg2+ ion (Fig. 1E and Fig. S1). The phosphate group of A1197 also engages negamycin through a bound Mg2+ ion. Acquisition of the U1052G mutation and localized conformational changes that accompany the distortion of h34 would position the phosphate group of nucleotide G1053 within 2 Å of the C1 carboxyl of negamycin, 3 Å closer than in the wild-type ribosome (Fig. 1E and Fig. S1). This conformational change is expected to disrupt multiple electrostatic interactions between the phosphate backbone of nucleotides G1053 and U1052 with the tRNA molecule, thereby destabilizing tRNA binding. These distortions collectively define the structural basis for altered negamycin susceptibilities for these acquired resistance mutations.
U1052G Is Hypersusceptible to Tetracycline.
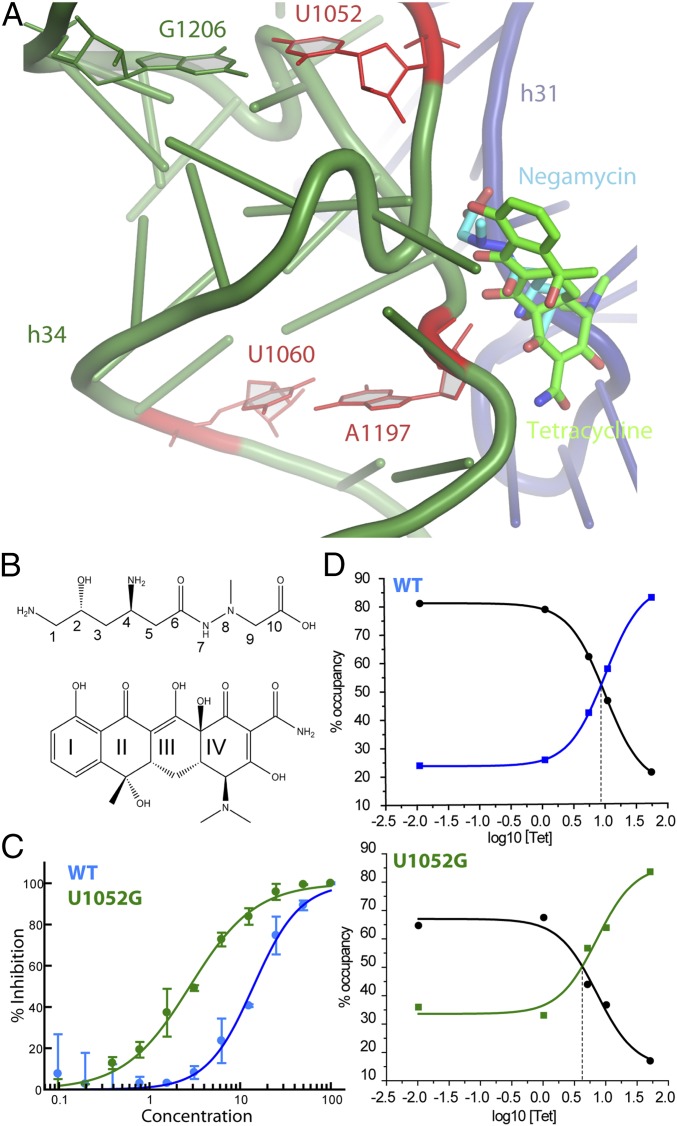
The binding of chemically distinct antibiotics at or near the same region of the ribosome often results in cross-resistance (1). The translation inhibitors negamycin and tetracycline occupy a common binding site and interfere with tRNA binding to the aa-tRNA site of the 30S ribosomal subunit (Fig. 2 A and B). Given this physical overlap, we examined the susceptibility of the U1052G and U1060A mutants to tetracycline. Cell growth assays previously revealed that the U1052G mutant is uniquely resistant to negamycin but hypersusceptible to tetracycline (Table 1). The MIC value of tetracycline for the U1052G mutant was fourfold lower than that of the wild-type ribosome (12) (Table 1). Using a luminescence-based in vitro-coupled transcription–translation system using purified components, we found that the IC50 of tetracycline for the U1052G mutant is ∼5.5-fold lower than that for wild-type ribosomes (Fig. 2C). By contrast, the U1060A mutant was resistant to tetracycline in both MIC measurements and in vitro-coupled transcription–translation assays (Table 1).
Fig. 2.
Effect of U1052G mutation on tRNA selection in the presence of negamycin and tetracycline. (A) The overlapping binding sites of negamycin and tetracycline on the small subunit A site. Negamycin and tetracycline are shown in blue and green, respectively. (B) Chemical structures of (Upper) negamycin and (Lower) tetracycline. The ring nomenclature is indicated. (C) In vitro-coupled transcription–translation activity of wild-type (blue; IC50 value of 15 µM) and U1052G (green; IC50 value of 2.7 µM) ribosomes in the presence of tetracycline. (D) smFRET imaging of aa-tRNA selection. Real-time delivery of ternary complex to wild-type and U1052G ribosomes in the presence of increasing concentrations of tetracycline. Fraction occupancy refers to the GA (intermediate FRET; blue and green squares) and the accommodated (high-FRET; black circles) states for all molecules.
Mechanistic Impact of Tetracycline on U1052G Ribosomes.
To explore the mechanism of the tetracycline hypersusceptibility of the U1052G ribosome, single-turnover, presteady-state, smFRET experiments were performed to follow the process of aa-tRNA selection. Single-molecule imaging methods enable this process to be tracked by time-dependent changes in FRET as acceptor-labeled aa-tRNA enters the A site of donor-labeled ribosome complexes (14). As previously described (12), we monitored the binding of acceptor (LD650)-labeled Phe-tRNAPhe in ternary complex with Elongation Factor Tu (EF-Tu) and GTP to surface-immobilized 70S in-itiation complexes bearing a donor (Cy3)-labeled initiator tRNAfMet within the P site. In this system, time-dependent changes in FRET reflect initial selection events, in which the ternary complex reversibly binds at the A site and proceeds through codon recognition (CR; low-FRET) and GTPase-activated (GA; intermediate FRET) states followed by proofreading events after GTP hydrolysis, which ultimately drive the incoming aa-tRNA from the GA state to its fully accommodated (high-FRET) position within the pretranslocation ribosome complex. Previous investigations have shown that negamycin has no detectable impact on the selection of cognate aa-tRNA but instead, compromises the ribosome’s ability to reject those that are near-cognate (12). Negamycin’s impact on miscoding is, however, significantly suppressed by the U1052G resistance mutation (12). By contrast, tetracycline sterically blocks aa-tRNA entry, resulting in cognate and near-cognate inhibition of initial selection between CR and GA states (15, 16).
The mechanistic impact of the U1052G mutation was assessed in this context in the absence and presence of increasing concentrations of tetracycline. As anticipated, tetracycline inhibited tRNA selection in a concentration-dependent manner, preventing aa-tRNA accommodation and the accumulation of high-FRET precomplex configurations (Fig. 2D). In the absence of tetracycline, the U1052G ribosome exhibited a somewhat slower rate of initial selection as evidenced by the extended period needed for GA-state passage (approximately three- to fourfold longer than that of the wild-type; ∼150–200 ms). This defect may be attributed to the displacement of the G1053 phosphate group from the tRNA molecule in the A site (Fig. 1E). The defect was enhanced in the presence of tetracycline, giving rise to an apparent hypersusceptibility to drug inhibition (approximately threefold) compared with wild-type ribosomes. These data are in agreement with those obtained in the aforementioned cell growth and in vitro translation reactions, suggesting that the observed hypersusceptibility specifically arises from defects in tRNA selection.
Structurally Defining U1052G Hypersusceptibility.
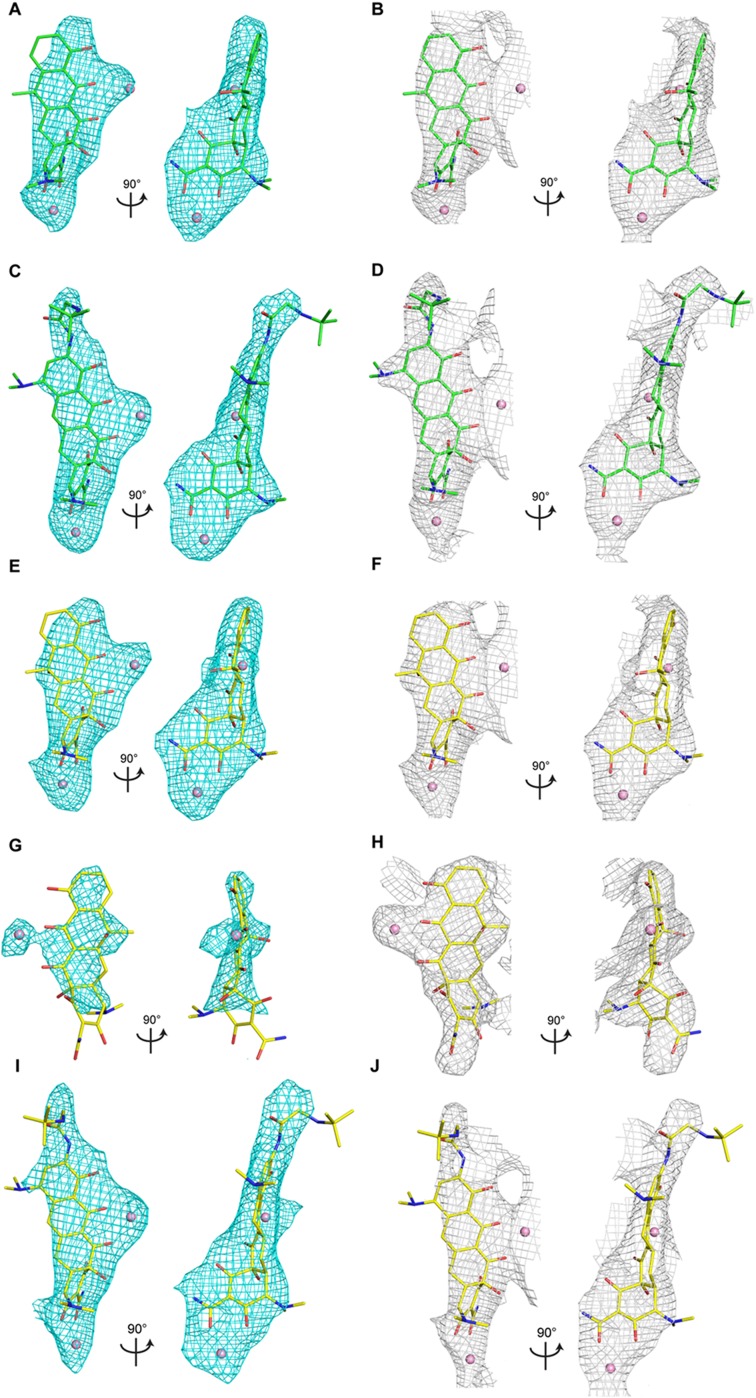
Despite the overlapping binding sites, negamycin and tetracycline possess chemically distinct features (Fig. 2B) that form different sets of electrostatic and van der Waals interactions, specifically with h31 and h34 (11, 12, 17). To visualize the structural basis for the confounding tetracycline hypersusceptibility of negamycin resistance mutant U1052G, the crystallographic structures of the wild-type E. coli ribosome and the U1052G mutant in complex with tetracycline were determined at 2.89- and 3.00-Å resolution, respectively (Fig. S2 and Table S1).
Fig. S2.
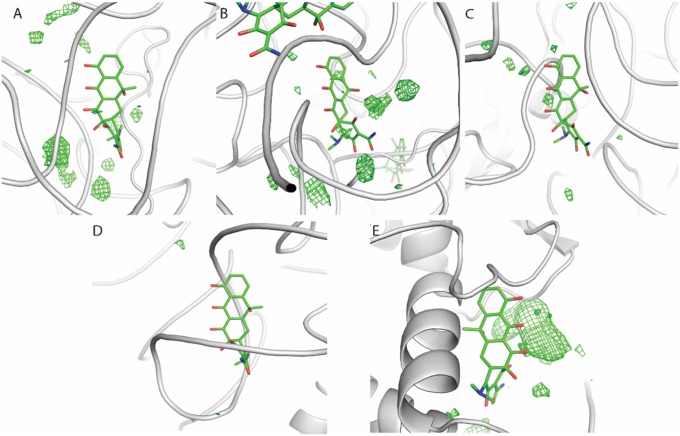
Electron density maps. Positive Fo-Fc difference density (cyan; contoured at 3σ) and 2Fo-Fc density (gray; contoured at 1σ) showing bound tetracycline or tigecycline to the wild-type and mutant ribosome. (A and B) Tetracycline bound to the wild-type ribosome. (C and D) Tigecycline bound to the wild-type ribosome. (E and F) Tetracycline bound to the U1052G mutant. (G and H) Tetracycline bound to the novel tetracycline site in the U1052G mutant. The electron density map shown in H is feature-enhanced (3). (I and J) Tigecycline bound to the U1052G mutant.
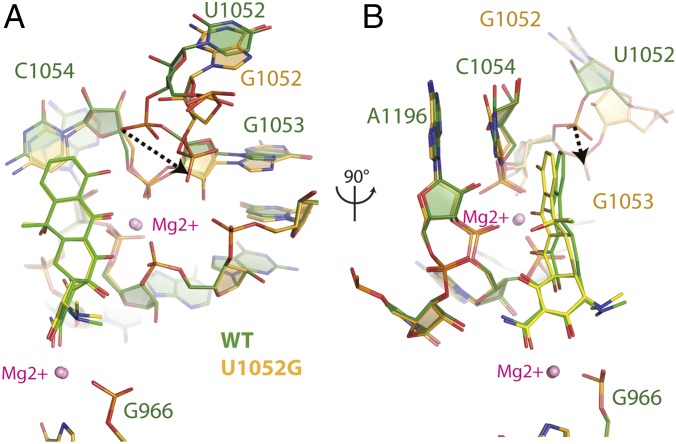
Clear, unambiguous difference electron density, consistent with the chemical structure of tetracycline, was observed near h34 of the small subunit (Fig. S2). Although the conformation of the phosphate backbone of h34 is significantly perturbed in the U1052G mutant, the majority of interactions between tetracycline and the ribosome are retained. The bound tetracycline molecule in the U1052G mutant is slightly rotated relative to the wild-type structure (Fig. 3). This subtle change in orientation has little impact on the van der Waals stacking arrangement that Ring I makes with nucleotide C1054. The indirect Mg2+-mediated interaction with nucleotide G966 from h31 is also retained (Fig. 3). However, acquisition of the U1052G mutation positions the hydroxyl group of Ring I farther away from the phosphate group of G1053, resulting in the loss of a hydrogen bond with h34 (Fig. 3A).
Fig. 3.
Structural perturbations to tetracycline binding to U1052G mutant ribosomes. (A) Superposition of the tetracycline-bound crystal structures of the wild-type and U1052G mutant ribosome. The dotted arrow represents the displacement of the phosphate group of nucleotide G1053 in the U1052G mutant. (B) This panel is rotated 90° along the vertical axis. wild-type is green, and the U1052G mutant is yellow. Magnesium ions are shown as pink spheres.
Previously Unidentified Tetracycline Binding Site.
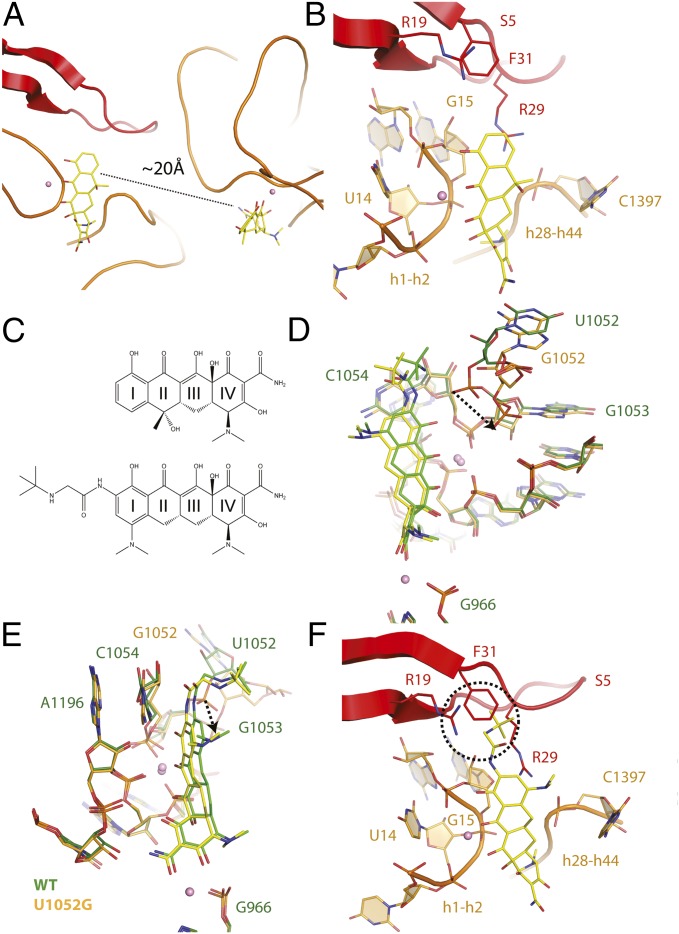
Analysis of the tetracycline-bound crystal structure of the U1052G mutant unexpectedly revealed a second tetracycline molecule bound ∼20 Å from its established binding site (Fig. 4A and Fig. S2). By contrast, no evidence of a second tetracycline molecule was found in either wild-type or U1060A mutant ribosomes (Fig. S3). Tetracycline binding at this position has not been previously described (17–20).
Fig. 4.
Two tetracycline binding sites are present in mutant U1052G. (A) Presence of two tetracycline binding sites in the U1052G mutant. (B) The novel tetracycline binding site is located near the highly conserved 16S central pseudoknot and adjacent to the previously established streptomycin binding site. (C) Chemical structures of (Upper) tetracycline and (Lower) tigecycline. (D) Superposition of the tetracycline-bound crystal structures of the wild-type and U1052G mutant ribosome. The dotted arrow represents the displacement of the phosphate group of nucleotide G1053 in the U1052G mutant. (E) This panel is rotated 90° along the vertical axis. (F) Tigecycline modeled into the second tetracycline binding site of U1052G. The dotted circle illustrates the expected steric clash between the butylglycylamido substituent attached to Ring I of tigecycline with residues found in the S5 protein. In these panels, wild-type is green, and the U1052G mutant is yellow. Magnesium ions are shown as pink spheres.
Fig. S3.
Absence of positive difference density at previously described tetracycline binding sites. The crystal structure of the tetracycline-bound Thermus thermophilus small subunit (PDB ID code 1I97) was superimposed on the crystal structure of the tetracycline-bound U1052G Escherichia coli ribosome. Previously described secondary tetracycline binding sites are shown: (A) h43, (B) h28, (C) h27, (D) h40, and (E) protein S4. Fo-Fc positive difference electron density contoured at 2α is shown in green.
The tetracycline binding site observed in the U1052G mutant structure is located near the highly conserved 16S central pseudoknot and adjacent to the previously established streptomycin binding site (18). This site does not change the conformation of the adjacent rRNA compared with the wild-type ribosome. The fused aryl core of tetracycline forms multiple Mg2+-coordinated interactions with the phosphate backbone of the loop that connects rRNA h1 and h2 to ribosomal protein S5 (Fig. 4B). Ring I of tetracycline also forms van der Waals stacking interactions with the guanidinium group of residue R29 from ribosomal protein S5 (21). Ring IV is positioned ∼5 Å from the functionally important nucleotide, C1397, which is located in the loop that connects h28 and h44 of the small 30S ribosomal subunit (Fig. 4B).
We attempted to determine the function of the novel tetracycline binding with site-directed mutagenesis. Our strategy was to abolish tetracycline binding at the primary binding site while maintaining the structural integrity of the novel binding site to evaluate the hypersusceptibility profile of the negamycin resistance mutant U1052G.
Previous studies have shown that mutations G966U and G1058C cause high levels of resistance to tetracycline (22–24). However, in our hands, all attempts to introduce these mutations combined with the U1052G negamycin resistance mutation failed to produce viable clones. Ribosomal mutations or combinations thereof can result in defective ribosomes with a dominant lethal phenotype (5). To relax the selective pressure on ribosome function in the presence of the U1052G mutation, a randomized plasmid library of positions 965–967 was subsequently constructed. Equivalent libraries have been used to isolate mutants with tetracycline resistance (24). Using this approach, multiple tetracycline-resistant mutants with sequence variations at positions 965–967 were identified. However, all isolates contained the wild-type U1052 nucleotide and not the U1052G mutation. Taken together, we conclude that tetracycline resistance rRNA mutations in this region are difficult to obtain in the presence of the U1052G negamycin resistance mutation, because viable levels of translation could not be achieved in these experiments.
Mutant U1052G Shows No Hypersusceptibilty to Tigecycline.
Tigecycline is a tetracycline derivative with a butylglycylamido substituent that is linked with Ring I (Fig. 4D). This chemical modification results in increased biochemical potency relative to tetracycline (IC50 values of 0.28 µM for tigecycline and 14.5 µM for tetracycline). However, MIC values and in vitro-coupled transcription–translation assay data indicate that mutant U1052G shows no indication of hypersusceptibility toward tigecycline (Table 1).
To understand the structural basis of the lack of hypersusceptibility to tigecycline for the U1052G mutant, we determined the crystallographic structures of the wild-type ribosome and the U1052G mutant in complex with tigecycline at 2.96- and 3.10-Å resolution, respectively (Fig. S2 and Table S1).
The crystal structures of tigecycline bound to the wild-type ribosome and the U1052G mutant revealed that the butylglycylamido substituent is stacked against nucleotide C1054 in its established binding site (Fig. 4 C and E). As expected, the backbone phosphate of nucleotide G1053 from h34 is displaced by ∼4 Å relative to the wild-type ribosome. The plane of the fused aryl ring structure is rotated by ∼8°, bringing Ring I and the butylglycylamido substituent close to C1054 (Fig. 4E). Moreover, this movement positions the hydroxyl group of Ring I in hydrogen-bonding distance with C1054 (Fig. 4E).
Using the U1052G mutant with a second tetracycline molecule bound, we modeled tigecycline into the novel tetracycline binding site (Fig. 4F). This model indicates that the butylglycylamido addition to Ring I of tigecycline would sterically clash with residues found in ribosomal protein S5, thereby precluding binding at this location. Taken together, these results define the structural basis for the lack of hypersusceptibility toward tigecycline for the U1052G mutant.
Conclusions
The data presented here reveal that mutations in h34 in the head domain of the small subunit alter the susceptibility of the ribosome to negamycin, tetracycline, and tigecycline. Ribosomes with the U1052G mutation, although resistant to negamycin inhibition, are hypersusceptible to tetracycline but not tigecycline. Crystallographic analysis of antibiotic-bound ribosomes revealed the presence of a novel tetracycline binding site in the U1052G mutant ribosome near the central pseudoknot within the head domain away from the established tetracycline binding site. smFRET measurements of tRNA selection showed that this mutant displayed a defect in tRNA selection in the presence of tetracycline that correlated with results from in vitro transcription–translation experiments. These findings provide additional evidence that conformational rearrangements between the head and body regions of the small subunit play a role in the tRNA selection process (12).
The persistent emergence of multidrug-resistant bacteria continues to erode the clinical utility of antibiotics. Focused efforts to understand the binding mode and mechanism of action for established classes of antibiotics and discover and develop novel antibiotics are essential (25). The bacterial ribosome remains an important target for antibiotic discovery given the presence of multiple binding sites that interfere with the translation cycle, which is essential for bacterial growth. The integration of single-molecule biophysical techniques and X-ray crystallography to delineate new features of the translation mechanism has enabled structure-guided design strategies that can be combined to modify existing chemical scaffolds to restore clinical utility of existing agents and decipher unpredicted antibiotic susceptibility profiles, including the emergence of new antibiotic binding sites in mutant ribosomes. It will be important in the future to decipher whether other antibiotic resistance mutations that arise in pathogens create novel antibiotic sites that can be exploited for therapeutic benefit.
Materials and Methods
In Vitro-Coupled Transcription–Translation Assay.
Ribosomes were purified as described previously with minor modifications (26). Briefly, E. coli strain SQ171 cells from an overnight culture were transferred to 1 L LB broth in a 4-L flask and incubated in a 37 °C shaker. Cultures were harvested at the transition from exponential to stationary phase at an OD600 of ∼0.4 and incubated on ice for at least 45 min. Cells were collected by centrifugation at 6,000 × g for 15 min and lysed in a bead beater in buffer AS (20 mM Tris⋅HCl, pH 7.5, 100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 0.15 M sucrose). Cell extracts were centrifuged at 10,000 × g for 100 min in Beckman 45 Ti Tubes. The supernatant was centrifuged through a sucrose cushion as previously described (26). Ribosomal pellets were resuspended in reassociation buffer (20 mM Tris⋅HCl, pH 7.5, 60 mM NH4Cl, 14 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 2 mM TCEP) and centrifuged through a 15–40% (mass/vol) sucrose gradient in reassociation buffer. Fractions corresponding to the 70S ribosome were collected and exchanged into a final buffer containing 20 mM Tris⋅HCl (pH 7.5), 60 mM NH4Cl, 6 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, and 2 mM TCEP. Ribosomes were concentrated to ∼5 mg/mL. In vitro-coupled transcription–translation assays were carried out as described previously using purified components (27).
Purification, Crystallization, and Structure Determination.
An E. coli strain SQ171 has been developed that can be used to express rRNA from either a single genomic rRNA operon or a plasmid-encoded rRNA operon (28). Although this strain expresses RNases that, in principle, may degrade ribosomal preparations, high-quality ribosomes were purified from E. coli strain SQ171 using previously established methods (12). Mutant ribosomes were purified from SQ171/pHKrrnC cells as described previously (26). To confirm the presence of homogeneous populations of rRNA mutants, rRNA was extracted from purified ribosomes and reverse-transcribed. The resulting cDNAs were sequenced to verify the absence of wild-type ribosome sequences.
E. coli 70S ribosomes purified from strains MRE600 and SQ171 crystallize under similar conditions, leading to high-quality crystals that are suitable for structure determination. Crystallization was carried out as previously described (29). Crystals were grown at 20 °C and prepared for vitrification in liquid nitrogen at 4 °C using sequential increasing concentrations of PEG 400 (6–24%). Antibiotics were introduced into the crystals by 2 mM stock solutions into the cryoprotection solution for 16 h. All X-ray diffraction data were collected at cryogenic temperature using synchrotron radiation at beam line ID-17 at the Advanced Photon Source. Diffraction data collected at Industrial Macromolecular Crystallography Association - Collaborative Access Team were processed with XDS (30) and scaled using SCALA (31) as implemented in the autoPROC routines from Global Phasing (32). Oscillations of 0.1° and 40% beam attenuation were used for all data collection experiments. All diffraction data were collected from either one or two crystals.
All liganded structures were solved by molecular replacement using the program MOLREP (33) using the apo 70S E. coli ribosome structure as the search model (PDB ID code 4YBB). Examination of the resulting electron density maps for these liganded complexes showed clear unambiguous difference density with the expected molecular features for these bound compounds (Fig. S2). Sequential rounds of manual rebuilding using Coot (34) and refinement using autoBUSTER (35) with TLS and NCS produced the final models (36). All refinement dictionaries were generated using GRADE (37). The final refinement statistics are shown in Table S1. Sequence alignment of the rrnC and rrnB (which sequence matches the structure of the published E. coli ribosome) rRNAs shows minor differences in sequence between the two operons that are mostly restricted to unstructured parts of the ribosome distant from the A site. Structural alignments were performed in Coot using least square superposition of h34 comprising residues 1,045–1,065. All figures were prepared using PyMOL (38).
Aminoacylation and Fluorescent Labeling of tRNA for Single-Molecule Fluorescence.
tRNAfMet and tRNAPhe from E. coli strain MRE600 were purified as previously described (9) as were aminoacylation, formylation, and fluorescent labeling of tRNA (10). With this approach, Cy3 and photostabilized Cy5 (LD650) dyes (11) were site-specifically attached through either maleimide or N-hydroxysuccinimide chemistry to tRNAfMet(s4 U8) and tRNAPhe(acp3 U47) at naturally occurring modified base residues located near the elbow region of the tRNA body. Charging of tRNAPhe was achieved using recombinant phenylalanyl tRNA synthetase (PheRS) prepared as previously described (10, 12, 13). Dye-labeled tRNAs prepared in this manner are fully competent in tRNA selection, translocation, and peptide bond formation (10).
Preparation of Ribosome Complexes for Single-Molecule Imaging.
Initiation complexes were prepared from 30S and 50S ribosomal subunits (1 μM each) isolated from E. coli as described for crystallographic investigations. Complexes were initiated in vitro on cognate (UUC) or near-cognate (UCU) mRNAs derived from the gp32 gene product bearing a 5′-biotin moiety [5′-biotin: CAA CCU AAA ACU UAC ACA CCC UUA GAG GGA CAA UCG AUG U(UC/CU) AAA GUC UUC AAA GUC AUC] in the presence of IF-1 (2 μM), IF-2 (2 μM), IF-3 (2 μM), 2 mM GTP, and fMet-tRNAfMet(Cy3-s4 U8) in Tris⋅polymix buffer containing 50 mM Tris⋅acetate (pH 7.5), 5 mM Mg(OAc)2, 100 mM KCl, 5 mM NH4OAc, 0.5 mM CaCl2, 0.1 mM EDTA, 5 mM putrescine, and 1 mM spermidine as previously described (10).
Single-Molecule Fluorescence Experiments and Data Processing.
All experiments were performed at 25 °C in Tris⋅polymix buffer containing 1.5 mM β-mercaptoethanol and 100 μM GTP in the presence of an oxygen scavenging environment [2 mM protocatechuic acid, 50 nM protocatechuate 3,4-dioxygenase containing a mixture of triplet-state quenching compounds (1 mM Trolox, 1 mM cyclooctatetraene, 1 mM nitrobenzyl alcohol) (14)]. The ternary complex of EF-Tu•GTP•Phe-tRNAPhe(LD650)-acp3 U47 was prepared following established procedures (15, 16). Ribosome complexes (0.5 nM) programmed with biotinylated mRNA were surface immobilized following brief incubation within PEG-passivated, streptavidin-coated quartz microfluidic devices (13). To avoid contributions of hybrid states formation following accommodation, the amino acid on P-site tRNA was released by incubating immobilized ribosomes with 2 mM puromycin (Sigma) for 10 min before ternary complex delivery. smFRET data were acquired by using a prism-based total internal reflection microscope as previously described (13). The Cy3 fluorophore linked to tRNAfMet was excited by the evanescent wave generated by total internal reflection of a single-frequency light source (Opus 532 nm; Laser Quantum). Photons emitted from both Cy3 and LD650 were collected by using a 1.27 N.A. 60× water immersion objective (Nikon), in which optical treatments were used to spatially separate Cy3 and LD650 frequencies onto two cooled, back-thinned electron multiplying charge coupled device cameras (Evolve 512; Photometrics). Fluorescence data were acquired using MetaMorph acquisition software (Universal Imaging), with an integration time of 100 ms. FRET trajectories were calculated from fluorescence traces by using the formula FRET = ILD650/(ICy3 + ILD650), where ICy3 and ILD650 represent the Cy3 and LD650 fluorescence intensities, respectively. Fluorescence and FRET traces were selected for analysis by using semiautomated smFRET automated analysis software implemented in MATLAB (MathWorks) as previously described (17).
Mutagenesis.
To select for ribosomes with mutations at the secondary tetracycline binding site in the presence of the U1052G mutation, E. coli strain SQ171 cells harboring the U1052G mutation were grown overnight in cation-adjusted Mueller Hinton broth in increasing concentrations of tetracycline (0.5–2 µg/mL) and in the presence of 128 µg/mL negamycin. Overnight cultures were inoculated on Mueller Hinton Media plates supplemented with tetracycline. To construct mutant ribosomes with U1052G and G966U mutations simultaneously, we used site-directed mutagenesis to introduce the G966U mutation into the pHKrrnC or pKK3535 plasmids containing the rRNA operon with the U1052G mutation. No viable mutants could be isolated. To obtain a mutant library with the nucleotides at positions 965–967 fully randomized, we constructed a library of pHKrrnC plasmids with the three positions fully randomized using the GeneArt plasmid construction service. Mutant plasmids were then exchanged with wild-type plasmids in E. coli strain KT101 cells by growing cells in the presence of 10% sucrose and kanamycin (39).
Acknowledgments
We thank C. Vonrhein, G. Bricogne, and A. Scharff for making custom modifications to Global Phasing software and refinement; and E. Code, S. Livchak, and J. Noeske for their guidance with ribosome purification and crystallization. This work was funded by AstraZeneca Pharmaceuticals.
Footnotes
Conflict of interest statement: A.I.C., J.H., E.T.B., S.L.K., P.D., D.B.P., and A.D.F. are (or were) employees of AstraZeneca Pharmaceuticals and may, as a consequence, be shareholders. R.B.A. and S.C.B. have equity interest in Lumidyne Technologies.
This article is a PNAS Direct Submission.
Data deposition: The crystallographic structures and structure factor amplitudes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5IT8, 5J5B, 5J91, 5JC9, 5J7L, 5J8A, and 5J88).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605127113/-/DCSupplemental.
References
- 1.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12(1):35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44(6):393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 3.Cochella L, Green R. Isolation of antibiotic resistance mutations in the rRNA by using an in vitro selection system. Proc Natl Acad Sci USA. 2004;101(11):3786–3791. doi: 10.1073/pnas.0307596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin D, Abdi NM, Fredrick K. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA. 2007;13(12):2348–2355. doi: 10.1261/rna.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yassin A, Fredrick K, Mankin AS. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc Natl Acad Sci USA. 2005;102(46):16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matt T, et al. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc Natl Acad Sci USA. 2012;109(27):10984–10989. doi: 10.1073/pnas.1204073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Fernandez D, et al. 4′-O-substitutions determine selectivity of aminoglycoside antibiotics. Nat Commun. 2014;5:3112. doi: 10.1038/ncomms4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demirci H, et al. Structural analysis of base substitutions in Thermus thermophilus 16S rRNA conferring streptomycin resistance. Antimicrob Agents Chemother. 2014;58(8):4308–4317. doi: 10.1128/AAC.02857-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121(2):257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Blaha G, Gürel G, Schroeder SJ, Moore PB, Steitz TA. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J Mol Biol. 2008;379(3):505–519. doi: 10.1016/j.jmb.2008.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polikanov YS, et al. Negamycin interferes with decoding and translocation by simultaneous interaction with rRNA and tRNA. Mol Cell. 2014;56(4):541–550. doi: 10.1016/j.molcel.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olivier NB, et al. Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome. Proc Natl Acad Sci USA. 2014;111(46):16274–16279. doi: 10.1073/pnas.1414401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhanel GG, et al. The glycylcyclines: A comparative review with the tetracyclines. Drugs. 2004;64(1):63–88. doi: 10.2165/00003495-200464010-00005. [DOI] [PubMed] [Google Scholar]
- 14.Geggier P, et al. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;399(4):576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11(10):1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 16.Jenner L, et al. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci USA. 2013;110(10):3812–3816. doi: 10.1073/pnas.1216691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pioletti M, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20(8):1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407(6802):340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 19.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17(5):555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340(6140):1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirthi N, Roy-Chaudhuri B, Kelley T, Culver GM. A novel single amino acid change in small subunit ribosomal protein S5 has profound effects on translational fidelity. RNA. 2006;12(12):2080–2091. doi: 10.1261/rna.302006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dailidiene D, et al. Emergence of tetracycline resistance in Helicobacter pylori: Multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob Agents Chemother. 2002;46(12):3940–3946. doi: 10.1128/AAC.46.12.3940-3946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerrits MM, Berning M, Van Vliet AH, Kuipers EJ, Kusters JG. Effects of 16S rRNA gene mutations on tetracycline resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2003;47(9):2984–2986. doi: 10.1128/AAC.47.9.2984-2986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonaka L, Connell SR, Taylor DE. 16S rRNA mutations that confer tetracycline resistance in Helicobacter pylori decrease drug binding in Escherichia coli ribosomes. J Bacteriol. 2005;187(11):3708–3712. doi: 10.1128/JB.187.11.3708-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butler MS, Blaskovich MA, Cooper MA. Antibiotics in the clinical pipeline in 2013. J Antibiot (Tokyo) 2013;66(10):571–591. doi: 10.1038/ja.2013.86. [DOI] [PubMed] [Google Scholar]
- 26.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 27.Polikanov YS, et al. Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol Cell. 2015;58(5):832–844. doi: 10.1016/j.molcel.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quan S, Skovgaard O, McLaughlin RE, Buurman ET, Squires CL. Markerless Escherichia coli rrn deletion strains for genetic determination of ribosomal binding sites. G3 (Bethesda) 2015;5(12):2555–2557. doi: 10.1534/g3.115.022301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325(5943):1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Bricogne G, et al. Buster Version V2.11.6. Global Phasing Ltd.; Cambridge, UK: 2011. [Google Scholar]
- 36.Smart OS, et al. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smart OS, et al. Grade V. 1.2.9. Global Phasing Ltd.; Cambridge, UK: 2011. [Google Scholar]
- 38. Schrödinger LLC (2015) The PyMOL Molecular Graphics System, Version 1.8 (Schrödinger, LLC, New York)
- 39.Kitahara K, Yasutake Y, Miyazaki K. Mutational robustness of 16S ribosomal RNA, shown by experimental horizontal gene transfer in Escherichia coli. Proc Natl Acad Sci USA. 2012;109(47):19220–19225. doi: 10.1073/pnas.1213609109. [DOI] [PMC free article] [PubMed] [Google Scholar]