Significance
Niemann–Pick C1 protein (NPC1) is a late-endosomal membrane protein required for transport of LDL-derived cholesterol into cells and Ebola virus entry; mutations cause Niemann–Pick type C disease. NPC1 contains a “sterol-sensing domain” (SSD) that also appears in several key regulatory proteins of cholesterol biosynthesis, uptake, and signaling. We present here the crystal structure of a large portion of human NPC1, which reveals the architecture of the SSD, including a cavity that is accessible both vertically to the endosome lumen and laterally to the “luminal” leaflet of the lipid bilayer. We propose that NPC1’s SSD functions in a pocket-relay system for cholesterol transport, the activity of which is regulated by the cholesterol concentration of the adjacent lipid bilayer.
Keywords: endosomal membrane, cholesterol traffic, sterol-sensing domain, crystal structure, allostery
Abstract
Niemann–Pick C1 protein (NPC1) is a late-endosomal membrane protein involved in trafficking of LDL-derived cholesterol, Niemann–Pick disease type C, and Ebola virus infection. NPC1 contains 13 transmembrane segments (TMs), five of which are thought to represent a “sterol-sensing domain” (SSD). Although present also in other key regulatory proteins of cholesterol biosynthesis, uptake, and signaling, the structure and mechanism of action of the SSD are unknown. Here we report a crystal structure of a large fragment of human NPC1 at 3.6 Å resolution, which reveals internal twofold pseudosymmetry along TM 2–13 and two structurally homologous domains that protrude 60 Å into the endosomal lumen. Strikingly, NPC1's SSD forms a cavity that is accessible from both the luminal bilayer leaflet and the endosomal lumen; computational modeling suggests that this cavity is large enough to accommodate one cholesterol molecule. We propose a model for NPC1 function in cholesterol sensing and transport.
Cholesterol is a critical component of cellular membranes, and it is either synthesized de novo or supplied from the diet. Although amphiphilic, cholesterol is only poorly soluble in water. Therefore, cholesterol associates with soluble proteins for transport between compartments (1), either as a single molecule or in the form of large lipoprotein particles (2). Cholesterol also functions as a covalently attached ligand in hedgehog-mediated signal transduction (3). Not surprisingly, many proteins involved in cholesterol biosynthesis, transport, or signaling pathways are polytopic, integral membrane proteins (4). Structures of the critical transmembrane region of these polytopic membrane proteins have thus far not been determined, except for an NADPH-dependent reductase in the cholesterol biosynthetic pathway (5), the structure of which yielded insights into the mechanism of intramembrane catalysis.
A subgroup of the polytopic integral membrane proteins of cholesterol-related pathways shares a highly conserved region comprised of five transmembrane segments (TMs) that are thought to represent a key regulatory element in response to cholesterol in the bilayer (6). This transmembrane region has been termed the “sterol-sensing domain” (SSD) (7, 8). Because crystal structures of these SSDs have not been determined, it has remained unclear precisely how an SSD detects cholesterol in the bilayer and conveys this information to the rest of the protein to influence its activity, stability, or trafficking.
Niemann–Pick C1 protein (NPC1) is an SSD-containing, ubiquitous, cholesterol-trafficking protein in the cholesterol uptake pathway (9). Cholesterol is transported throughout the body as cholesterol esters that are packaged into lipoprotein particles including “low-density lipoprotein” (LDL) (2, 10). LDL is endocytosed and transported to late endosomes and lysosomes, where the ∼25-nm particle is subject to lipolysis by lysosomal acid lipase (11, 12) at the low pH of this compartment. The liberated cholesterol is then bound, one molecule at a time, to a soluble intralysosomal protein, termed Niemann–Pick type C2 (NPC2) (13). A crystal structure of the 15-kD NPC2 revealed a cholesterol-binding pocket with distinct polarity: The iso-octyl moiety is located at the deep end and the hydroxyl group at the opening (14). The NPC2-bound cholesterol is then thought to be transferred to another pocket located in the ∼230-residue, N-terminal domain (NTD) of NPC1 that binds cholesterol in an opposite orientation (15–17) (Fig. 1A). Notably, the NTD of NPC1 is connected via a proline-rich region to TM 1 (17). This flexible region has been suggested to facilitate cholesterol transfer from NPC1’s NTD pocket to another, downstream site within NPC1 (15). Although the exact location and structure of this second cholesterol binding site has not been determined precisely, its existence is strongly implied from multiple, independent lines of evidence (18–20). However, the relationship of these two binding sites, and if they interact directly, is not clear.
Fig. 1.
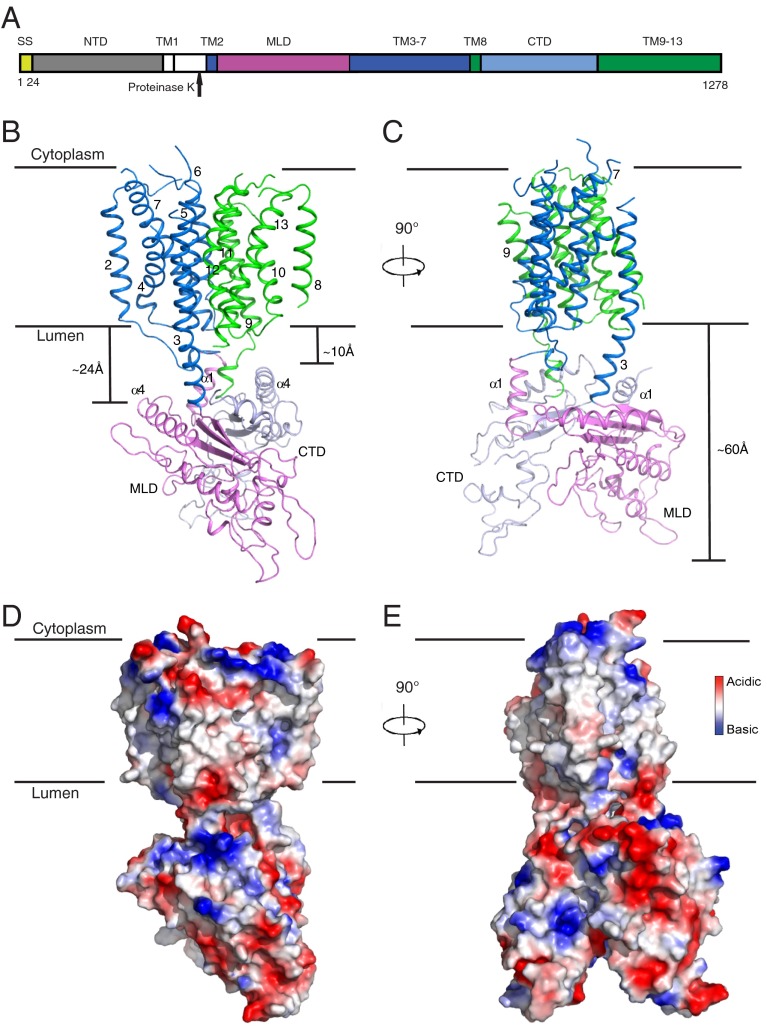
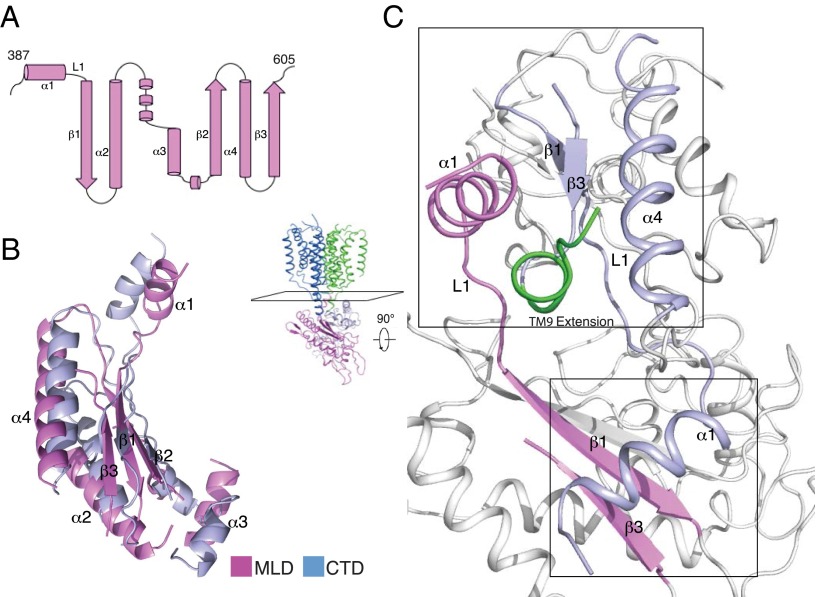
Structural overview of human NPC1. (A) Schematic representation of NPC1 domains with the part after the proteinase K site crystallized here in color. The structures of the NTD and MLD have been previously solved. (B) Overall structure of NPC1*. The structure is shown in perspectives horizontal to the plane of the lipid bilayer and rotated by 90° around a vertical axis; structures are shown either in ribbon presentation (B and C) or in space-filling models using a graded color scheme to indicate the electrostatic surface potential (D and E). The 12 TMs are numbered and divided into an N-terminal (TM 2–7, blue) and a C-terminal half (TM 8–13, green). The two black lines show the approximate location of the lipid bilayer. The MLD is shown in magenta and the CTD in light blue; numbered helices are involved in interdomain interactions. The distinct distances of MLD and CTD from the lysosomal membrane are indicated. (C) View of NPC1 rotated 90° relative to B. The extended helices of TM 3 and TM 9 form the connection between the TMs and the luminal domains. The total distance from the membrane is indicated. (D) Electrostatic surface representation of NPC1 in the same orientation as B. (E) Electrostatic surface representation of NPC1 in the same orientation as C.
Mutations in NPC1 cause excessive accumulation of cholesterol and glycosphingolipids in lysosomes within multiple tissues, including the liver, spleen, and nervous system. This accumulation induces a fatal lysosomal storage disease termed Niemann–Pick type C (NPC), most often diagnosed in early childhood and usually leading to death before age 20 (21, 22). In addition, NPC1 has been shown to be required for Ebola and Marburg virus entry (23, 24) into the cytoplasm, serving as an intraluminal receptor after these viruses have been internalized. Recently a crystal structure of a complex between the middle luminal domain (MLD) of NPC1 and the cleaved glycoprotein (GP) of the Ebola virus has been reported (25, 26). Some NPC1 inhibitors can prevent viral infection (27), and the Ebola virus was completely noncontagious in Npc1−/− mice (28).
In this paper, we report the crystal structure of a large fragment of human NPC1 that contains 12 transmembrane helices, the SSD, and two luminal domains, one of which is the Ebola virus-binding domain. The SSD includes a two-way cavity open to both the endosomal lumen and the luminal leaflet of the lipid bilayer. These features could explain how NPC1 transports cholesterol across the lipid bilayer.
Results and Discussion
Expression and Crystallization of NPC1.
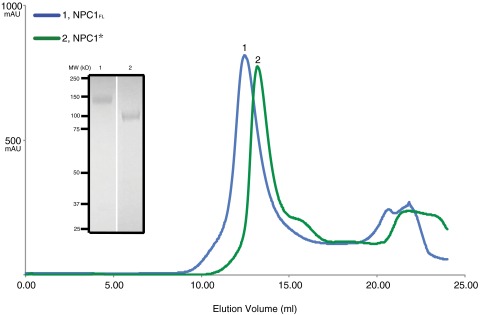
We subcloned cDNA of human NPC1 into different vectors for eukaryotic expression and eventually chose the baculovirus-mediated gene transduction of mammalian cells (BacMam) system (29). Infected cells yield ∼0.5–1 mg/L of full-length protein. After screening detergents in the purification process, the full-length protein (NPC1FL) was purified and eluted as a monodisperse peak during gel filtration (Fig. S1). NPC1FL failed to crystallize, which led us to search for a crystallizable large fragment. Digestion with various proteases produced suitably large and stable fragments of NPC1 as assessed by size exclusion chromatography (Fig. S1). Mass spectrometric analyses of a proteinase K fragment, designated NPC1*, showed that it is comprised of residues 314 to the carboxyl-terminus (1278), including 12 of the 13 TMs with two soluble domains facing the lysosomal lumen; it lacks only the NTD (for which a crystal structure has been reported) (15) and its adjacent downstream TM 1 (Fig. 1A). Notably, NPC1 without the NTD has been reported to bind sterols (20). Moreover, mutations in the SSD (P691S) abolish sterol binding (18, 19). Therefore, NPC1 harbors another sterol-binding site besides that in the NTD, most likely within the SSD.
Fig. S1.
Expression and purification of human NPC1 protein. Representative Superdex 200 10/30 gel-filtration chromatogram of human full-length NPC1 (NPC1FL, residues 1–1278, blue curve) and digested protein for crystallization (NPC1*, residues 314–1278, green curve). (Inset) Peak fractions of NPC1FL (1) and NPC1* (2).
Overall Structure of NPC1.
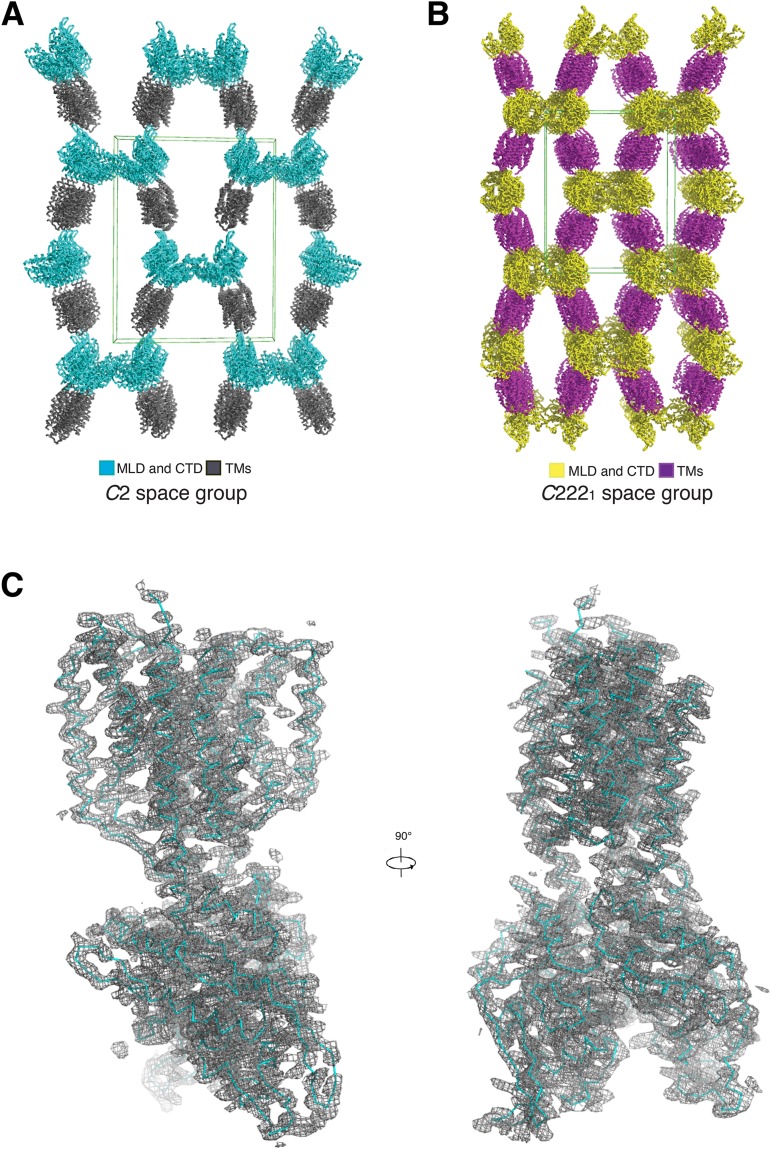
After screening more than 10,000 conditions, we eventually obtained suitable crystals of NPC1* for X-ray data collection. NPC1* was crystallized in two space groups. The initial phases in the C2221 space group were determined by Ta6Br12-based single-wavelength anomalous dispersion (SAD). We then performed cross-crystal electron density averaging with the C2 space group and extended this density to the native data of 3.6 Å resolution for building the initial model (Fig. S2 and Table S1). Using the coordinates of the MLD (26) improved the electron density of the luminal domain and confirmed the assignment of residues. Due to its higher resolution, we analyzed the structure of the C2 space group in this manuscript (Fig. S2).
Fig. S2.
Crystal packing and representative 2Fo-Fc electron density map. (A) Crystal packing of the C2 space group. The transmembrane domains are shown in gray, and the luminal domains are shown in cyan. The green box shows the three dimensions of the C2 unit cell. (B) Crystal packing of the C2221 space group. The transmembrane domains are shown in purple, and the luminal domains are shown in yellow. The green box shows the three dimensions of the C2221 unit cell. (C) Overall view of the 2Fo-Fc electron density, contoured at 1.5 σ, in one asymmetric unit.
Table S1.
Data collection and refinement statistics
| NPC1* | Native C2 | Ta-SAD C2221 | Native C2221 |
| Data collection | |||
| Wavelength, Å | 1.0000 | 1.2546 | 1.0000 |
| Space group | C2 | C2221 | C2221 |
| Unit cell | |||
| a, b, c, Å | 181.75, 222.97, 63.17 | 172.85, 221.33, 108.24 | 176.63, 222.78, 109.25 |
| α, β, γ, ° | 90, 105.31, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution, Å | 50∼3.60 (3.67∼3.60)† | 50∼5.0 (5.18∼5.0) | 50∼3.8 (3.94∼3.8) |
| Rmerge, % | 12.5 (80.6) | 16.3 (63.6) | 17.3 (61.9) |
| I/σI | 9.5 (1.01) | 12.8 (1.30) | 5.66 (1.12) |
| Completeness, % | 96.5 (85.3) | 86.3 (57.2) | 83.1 (64.2) |
| Redundancy | 3.0 (2.2) | 6.2 (3.9) | 3.3 (2.7) |
| Refinement | |||
| Resolution, Å | 50∼3.6 | ||
| No. reflections | 22,222 | ||
| Rwork/Rfree, %‡ | 32.60/34.67 | ||
| Molecules in asymmetric unit | 1 | ||
| No. atoms of protein | 5,883 | ||
| B factors of protein | 113.59 | ||
| Ramachandran plot, % | |||
| Favored/allowed/disallowed | 86.8/12.7/0.54 | ||
| rmsd in: | |||
| Bond distances, Å | 0.0072 | ||
| Bond angles, ° | 1.139 |
Values in parentheses are for the highest resolution shell.
Rfree was calculated with 5% of the reflections selected in the thin shell.
The overall dimensions of NPC1* are 105 × 55 × 45 Å (Fig. 1 B and C). Two regions can be distinguished in the structure: (i) a large and compact membrane-spanning domain formed by 12 TMs showing internal pseudotwofold symmetry and (ii) a large extension formed by two lumen-exposed domains, MLD and C-terminal luminal domain (CTD), each folding into a structurally similar, elongated curved rod, with the two rods intertwining with each other at their concave surfaces and forming a 60 Å-long protrusion into the lysosomal lumen (Fig. 1 B–E, Fig. S3, and Movie S1).
Fig. S3.
NPC1 secondary structure schematic. (A) The secondary structure of NPC1 with the crystallized domains in color. (B) NPC1 secondary structure overlay on amino acid sequence; residues unresolved in the final structure are represented by dots. The major structural elements, colored according to A, are indicated.
The Transmembrane Domain.
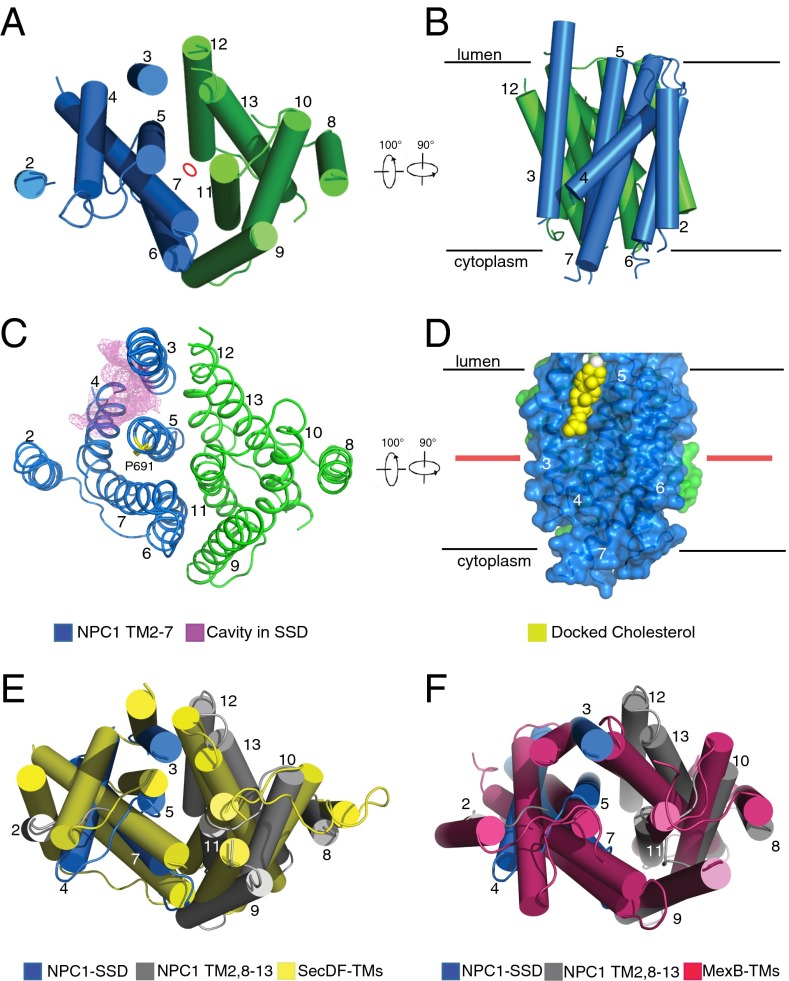
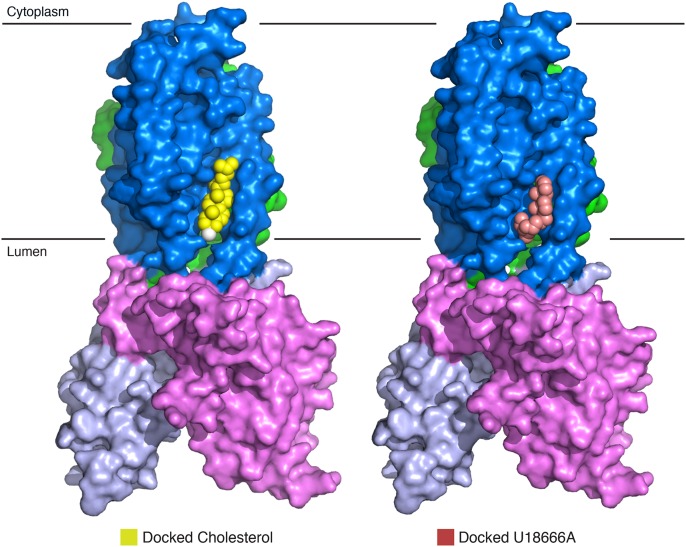
The TMs of NPC1* display internal, pseudotwofold symmetry around a central axis parallel to the TMs (Fig. 2A). Strikingly, TM 3–5 of NPC1’s SSD delineate a large cavity that is accessible both from the lipid bilayer and the lysosome lumen (Fig. 2 B and C). This largely hydrophobic cavity measures ∼24 × 8 × 8 Å (Movie S1). AutoDock (30) analysis showed that this SSD cavity is large enough to accommodate one molecule of cholesterol (Fig. 2D). This cavity also can be fitted with U18666A, a cholesterol-trafficking inhibitor (Fig. S4).
Fig. 2.
Structure of NPC1 transmembrane domains. (A) View from the lumen of the transmembrane helices perpendicular to the view in Fig. 1B. The pseudosymmetry center is labeled by a red oval. (B) Side (membrane) view of the transmembrane helices after two 90° rotations relative to A. (C) The cavity in the SSD in the same orientation as A. The transmembrane cavity (purple mesh) opens to the lumen and is continuous with the membrane inner leaflet. (D) Modeling of cholesterol (yellow) in the SSD binding pocket after two 90° rotations relative to C. The red line approximates the middle of the lipid bilayer. Docking of cholesterol was performed by AutoDock (30). (E) Structural comparison between NPC1 and SecDF. (F) Structural comparison between NPC1 and MexB in the same orientation as A. The SSD of NPC1 is shown in blue, and the other TMs of NPC1 are shown in gray. TM 4 of NPC1 undergoes an obvious movement to open the cavity in the SSD.
Fig. S4.
Surface representation of NPC1* with modeled cholesterol and U18666A. Cholesterol and U18666A were docked using AutoDock.
A Dali server search (31) for homologous structures showed that the transmembrane domain of NPC1 shares a similar transmembrane helix arrangement with prokaryotic Resistance–Nodulation–Cell Division (RND)-type transporters [AcrB (32), MexB (33), CusA (34), and SecDF (35)] (Fig. 2 E and F). The root-mean-square deviation (rmsd) of the superimposition of Cα traces of transmembrane domains of NPC1 and RND transporters is about 3 Å (z score above 10), which is consistent with previously noted sequence similarities and predicted topologies (36, 37). For the evolutionary history of NPC1 (and of the similar, intestine-localized, NPC1-like1), we suggest that several regions of DNA were recombined to code for NPC1’s distinct domains: (i) a novel cholesterol-engulfing domain (NTD) that became membrane-anchored via its downstream TM 1 and (ii) two RND-derived domains but arranged in reverse order—the primary structure of RND transporter TM 7–12 resembles more closely TM 2–7 of NPC1 and vice versa, and that of RND transporter TM 1–6 is more similar to TM 8–13 of NPC1. This structural similarity is reflected by the sequence alignment of human NPC1 with proteins of known structure; TM 8–13 in NPC1 show ∼40% similarity with TM 1–6 in AcrB or MexB.
Luminal Domains.
The MLD and CTD are attached to the membrane region of NPC1 via “stalks” that consist primarily of extended regions of TM 2 and 3 linking the MLD and TM 8 and 9 connecting the CTD (Fig. 1 B and C). The MLD and CTD fold into similar, elongated, curved structures (Fig. 3 A and B). They intertwine with each other through their concave surfaces, especially at their membrane-proximal ends, and protrude 60 Å into the endosomal lumen (Fig. 1 B and C and Movie S1). The electrostatic surface potential of the luminally exposed domains is primarily negative. Notably, the longer stalk region of the MLD (∼24 Å) leaves a larger gap between the MLD and the luminal aspect of the membrane region (i.e., the SSD) than the shorter stalk (∼10 Å) connecting the CTD to the luminal membrane (Fig. 1B).
Fig. 3.
Structure of NPC1 luminal domains. (A) Secondary structure schematic of the MLD. The major secondary elements are labeled. (B) Alignment of the MLD and CTD major secondary structures. The loops of both domains are omitted for clarity. The colors of both domains are specified. (C) The main interactions between the MLD and CTD after a 90° rotation relative to Fig. 1B from the point of view of the TMs, along a plane indicated in the whole structure. Upper box shows the interface of the extended helix of TM 9 (green) with the MLD and CTD. Lower box shows the interface of β1 and β3 of the MLD with α1 of the CTD.
There are two interfaces between the MLD and CTD: (i) the N-terminal end of the extended TM 9 helix mediates the interaction with α1 of MLD and α4 of CTD, and the loops between α1 and β1 of the MLD and CTD (referred to as L1) make direct contact (Fig. 3C, upper box), and (ii) α1 of the CTD binds to β1 and β3 of the MLD (Fig. 3C, lower box), and this interaction directly contributes to keeping the MLD farther away from the membrane.
Structural Insights into Previous Studies.
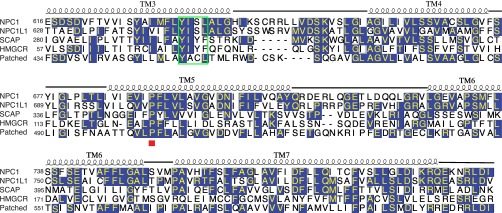
Our work sheds light on other SSD-containing proteins (Fig. S5). For example, Y298 in SCAP is part of a YIYF tetrapeptide motif that is necessary for SCAP (38) and HMGCR (39) binding to the protein Insig. Y298/I299 is conserved in the SSD of NPC1 (Y634/I635 in TM 3), exposed to the membrane at the edge of the transmembrane domain, suggesting that the corresponding helix of SCAP or HMGCR could potentially have enough room to bind Insig directly (Fig. 4 A and B). P691, in the center of TM 5, could potentially influence the rigidity of the SSD cavity (Fig. 4B).
Fig. S5.
Sequence alignment of NPC1-SSD with the SSDs of human NPCL1L, SCAP, HMGCR, and Patched. Secondary structural elements of NPC1-SSD are indicated above the sequences. Invariant amino acids are highlighted in blue. A red box under the sequences highlights location of NPC1 proline 691. Sequence alignment was done with ClustalW.
Fig. 4.
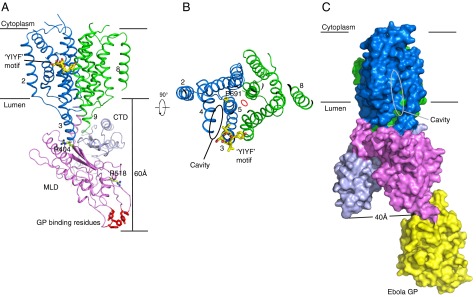
Residues of NPC1 involved in binding other proteins. (A) Two residues, R404 and R518, previously shown to affect NPC2 binding are shown in stick form in green. The Ebola GP binding residues (from PDB ID code 5F1B) are colored in red. The YIYF motif is in TM 3 and shown in stick form in yellow. P691 lies in TM 5 and is colored in yellow. (B) Top view of NPC1. The cavity of the SSD is indicated by an oval. (C) Surface representation of NPC1 with domains colored as in A rotated by 90°, shown in complex with Ebola GP in yellow. The SSD cavity is highlighted by a gray oval.
Previous studies have shown that the MLD of NPC1 binds NPC2 in vitro and in vivo and that two disease-causing mutations (R404Q and especially R518Q) in NPC1 disrupt the NPC2–MLD interaction (40). In our structure, R518 is exposed at the end of the short helix α3 on top of the MLD, clearly free to interact with NPC2 (Fig. 4A). R404 is buried in the MLD, interacting with E606 in the part of the stalk that is the amino terminus of TM 3 (Fig. 4A). Mutation of this residue to glutamine would likely destabilize the conformation of the protein.
A recently reported structure of the MLD bound to a GP of the Ebola virus (26) is essentially identical to our MLD structure; the viral GP binding sites consist of loops located at the most membrane-distant region of the MLD (Fig. 4A). By docking the crystal structure of GP to our NPC1* (Fig. 4C), we could visualize this interaction as separate from the CTD (about 40 Å away) and confirm the MLD alone is involved in viral entry (25). Our data do not provide information regarding whether or not the Ebola virus GP also interacts with the NPC1 NTD.
Model for Cholesterol Sensing.
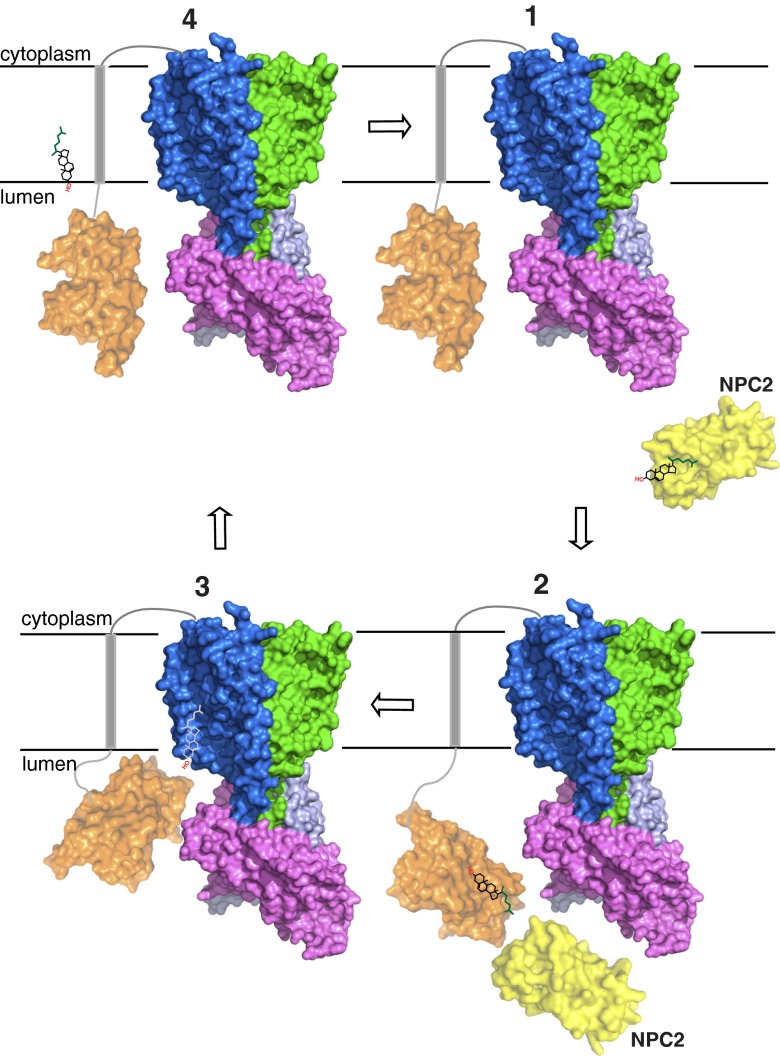
Based on our findings, we propose a working model for cholesterol transport across the lysosome membrane (Fig. 5). Unesterified cholesterol (state 1, top right) binds first to NPC2, which then docks to the NTD of NPC1, permitting cholesterol transfer between their sterol binding pockets (state 2). The NTD is linked to TM 1 and the rest of NPC1 by a loop consisting of residues 247–266 that includes eight conserved prolines (17). The height of the protein may help NPC1 reach through the glycocalyx that lines the inner surface of the lysosome membrane (15, 41). This would permit the NTD to reorient in such a way as to allow cholesterol transfer to the transmembrane domain (state 3). We suggest that luminal entry is used for transfer of cholesterol from NPC1’s NTD binding site. The newly deposited cholesterol could then exit via the membrane pocket’s lateral opening and join the pool of cholesterol in the luminal leaflet of the lysosomal lipid bilayer. Conversely, the lateral opening of the membrane pocket would provide access for free cholesterol from the lipid bilayer (state 4).
Fig. 5.
Model of NPC1 for lipoprotein-derived cholesterol transport in endosomes. States 1 and 2 show cholesterol transfer of NPC2 to the NTD of NPC1 based on previous observations. States 3 and 4 show cholesterol transfer of NPC2 to the NTD of NPC1 based on the data presented here.
The access properties of NPC1’s SSD membrane pocket might enable it to sample, and therefore monitor (i.e., sense), the cholesterol concentration of the lysosome lipid bilayer. Cholesterol occupancy in the SSD pocket might reflect free cholesterol concentration in the lipid bilayer; increasing SSD occupancy might decrease the rate of interpocket transfer from the NPC1 NTD pocket onto the SSD. Differential occupancy of the SSD pocket might allosterically influence the conformation of other parts of NPC1. For example, cholesterol occupancy of the SSD pocket might affect NPC2’s transient association with the MLD of NPC1 (40) in the pathway of interpocket transfer of cholesterol between NPC2 and NPC1 NTD. The working model of NPC1 and the functions of the SSD, which we proposed here, will be tested by future experiments.
The small cytoplasmic region of NPC1 is comprised of the C-terminal tail and six loops that interconnect the TMs. These loops could serve as docking sites for egress of cholesterol using other transporters, like the START (STeroidogenic Acute Regulatory protein-related lipid Transfer) family of proteins, which are involved in cholesterol transfer from endosomes to the cytoplasmic leaflets of the endoplasmic reticulum or other intracellular membranes (42). Our proposed pathway of cholesterol transport in the late endosomal compartment, shown in Fig. 5, is essentially analogous to a “bucket brigade.” Transfer between pockets (a “pocket brigade”) of NPC2, NPC1 NTD, and the membrane-embedded NPC1 SSD must be followed by flipping cholesterol to the cytoplasmic leaflet of the endosomal membrane. How flipping occurs is presently unknown; it might be facilitated by NPC1 or occur spontaneously with subsecond kinetics. Once in the cytoplasmic leaflet of the endosomal membrane, further transport to cytoplasmically exposed membranes might be continued in a pocket brigade manner mediated by various cytoplasmic cholesterol transport proteins (e.g., START). The degree of occupancy of NPC1’s SSD pocket might also affect the rate of cholesterol export from endosome membranes, if other cholesterol-binding protein docking to NPC1 is part of the pocket brigade for distribution of cholesterol to other compartments.
Hence, by serving in this pocket brigade of cholesterol transport and by sensing cholesterol concentration in the endosomal bilayer, NPC1 could represent a critical and highly regulated hub between external cholesterol uptake and internal, biosynthetic pathways. The SSDs of the other integral membrane proteins of cholesterol-related pathways (Fig. S5) may likewise be allosteric sites, gauging the concentration of free cholesterol in the surrounding lipid bilayer to affect each membrane protein's conformation and behavior.
Experimental Procedures
Protein Expression and Purification.
The cDNA of human NPC1 (GI: 83305902) was cloned into pEG BacMam with a C-terminal His8-tag. The protein was expressed using baculovirus-mediated transduction of mammalian HEK-293S GnTI− cells. At 50 h postinfection, the cells were disrupted by sonication in buffer A, containing 20 mM Hepes, pH 7.0, 150 mM NaCl, 1 mM PMSF, and 5 μg/mL each of leupeptin and aprotinin. After low-speed centrifugation, the resulting supernatant was incubated in buffer A with 1% (wt/vol) n-Dodecyl-β-d-maltopyranoside (Anatrace) and 0.2% cholesteryl hemisuccinate (Sigma-Aldrich) for 2 h at 4 °C. The lysate was centrifuged again, and the supernatant was loaded onto a Ni2+-NTA affinity column (Qiagen). After washing twice, the protein was eluted in 20 mM Hepes, pH 7.0, 300 mM NaCl, 300 mM imidazole, 0.02% n-Dodecyl-β-d-maltopyranoside, and 0.004% cholesteryl hemisuccinate and concentrated. The concentrated protein was incubated with 10 μg/mL proteinase K (Sigma-Aldrich) at room temperature for 15 min and purified by Superdex-200 size-exclusion chromatography (GE Healthcare) in a buffer containing 20 mM Mes pH 5.5, 150 mM NaCl, and 0.06% (wt/vol) Cymal-6 (Anatrace). The peak fractions were collected and concentrated to 5∼8 mg/mL for crystallization. DNA constructs were generated using Gibson Assembly (New England Biolabs).
Crystallization.
Crystals were grown at 20 °C by the hanging-drop vapor-diffusion method. Initially, the diffraction and quality of all of the crystals was poor. After optimization, crystals in the C2 or C2221 space group appeared in 2 d in well buffer containing 0.1 M sodium citrate pH 5.5 or sodium acetate pH 5.5, 0.2 M NaCl, 12% (vol/vol) PEG4000, 5% (vol/vol) Jeffamine M-600 pH 7.0, and 5% (vol/vol) PEG P400. The crystals in space group C2 have unit cell dimensions of a = 181.75 Å, b = 222.97 Å, c = 63.17 Å, and β = 105.31°. The crystals in space group C2221 have unit cell dimensions of a = 176.63 Å, b = 222.78 Å, and c = 109.25 Å. Each asymmetric unit contains one molecule either in C2221 or C2 (75–78% solvent content). Tantalum derivatives were obtained by soaking native crystals for 72 h in mother liquor with Ta6Br12 (Jena Bioscience) powder. All crystals were flash-frozen in liquid nitrogen stream with well buffer plus 25% Jeffamine M-600 pH 7.0 for cryoprotection.
Data Collection and Structure Determination.
The data were collected at Advanced Light Source beamline 8.2.1 and the Advanced Photon Source beamline ID24-C at 100 K. All datasets were processed using HKL2000 (43). The anomalous signal in the Ta6Br12-derivative data was further magnified with the local-scaling algorithm using the program SOLVE (44). Then, the Ta sites were determined using the program SHELXD (45). The identified sites were refined and the initial phases were generated in the program PHASER (46) with the single-wavelength anomalous dispersion (SAD) experimental phasing module. Cross-crystal density averaging along with solvent flattening and histogram matching were performed using DMMulti (47). The initial model was built in Crystallographic Object-Oriented Toolkit (COOT) (48) manually. Initially, large aromatic/hydrophobic residues were assigned to facilitate the register of the transmembrane helices. Because the membrane domain of NPC1 has a topology similar to RND transporters, the sequence homology between NPC1 and structurally known RND transporters was used to facilitate model building of the transmembrane domain. The coordinates of the MLD (26) improved the electron density of the luminal domain and confirmed the assignment of residues. The structure was refined with PHENIX.REFINE (Python-Based Hierarchical Environment for Integrated Xtallography) (49) at a 3.6 Å resolution. Due to flexibility and limited resolution, the six linker regions and three loops of the CTD were not visible in the electron density map and were not included in the final structure (Fig. S3). Model validation was performed with MolProbity (50). All figures were generated with PyMOL.
Supplementary Material
Acknowledgments
We thank George F. Gao for sharing the coordinates of the free NPC1-C protein (MLD). We also thank Erik Debler and Yi Ren for constructive comments in structure refinement and manuscript preparation. We acknowledge the staff of the Berkeley Center for Structural Biology at the Advanced Light Source and the Northeastern Collaborative Access Team at the Advanced Photon Source for assistance with data collection. This work was supported by funds from the Rockefeller University and the Howard Hughes Medical Institute (G.B., Investigator). X.L. is a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5I31).
See Commentary on page 7941.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607795113/-/DCSupplemental.
References
- 1.Maxfield FR, van Meer G. Cholesterol, the central lipid of mammalian cells. Curr Opin Cell Biol. 2010;22(4):422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 3.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274(5285):255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 4.Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–157. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Roberti R, Blobel G. Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum. Nature. 2015;517(7532):104–107. doi: 10.1038/nature13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87(3):415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 7.Kuwabara PE, Labouesse M. The sterol-sensing domain: Multiple families, a unique role? Trends Genet. 2002;18(4):193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Carstea ED, et al. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science. 1997;277(5323):228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 10.Roth MG. Clathrin-mediated endocytosis before fluorescent proteins. Nat Rev Mol Cell Biol. 2006;7(1):63–68. doi: 10.1038/nrm1783. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J Biol Chem. 1975;250(21):8487–8495. [PubMed] [Google Scholar]
- 12.Du H, Grabowski GA. Lysosomal acid lipase and atherosclerosis. Curr Opin Lipidol. 2004;15(5):539–544. doi: 10.1097/00041433-200410000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Naureckiene S, et al. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290(5500):2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 14.Xu S, Benoff B, Liou HL, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J Biol Chem. 2007;282(32):23525–23531. doi: 10.1074/jbc.M703848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon HJ, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137(7):1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Infante RE, et al. Purified NPC1 protein. I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J Biol Chem. 2008;283(2):1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- 17.Infante RE, et al. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem. 2008;283(2):1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- 18.Ohgami N, et al. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc Natl Acad Sci USA. 2004;101(34):12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu F, et al. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife. 2015;4:4. doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohgane K, Karaki F, Dodo K, Hashimoto Y. Discovery of oxysterol-derived pharmacological chaperones for NPC1: Implication for the existence of second sterol-binding site. Chem Biol. 2013;20(3):391–402. doi: 10.1016/j.chembiol.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Yanjanin NM, et al. Linear clinical progression, independent of age of onset, in Niemann-Pick disease, type C. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(1):132–140. doi: 10.1002/ajmg.b.30969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pentchev PG. Niemann-Pick C research from mouse to gene. Biochim Biophys Acta. 2004;1685(1-3):3–7. doi: 10.1016/j.bbalip.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477(7364):340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Côté M, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477(7364):344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller EH, et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31(8):1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, et al. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell. 2016;164(1-2):258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhein BA, Maury WJ. Ebola virus entry into host cells: Identifying therapeutic strategies. Curr Clin Microbiol Rep. 2015;2(3):115–124. doi: 10.1007/s40588-015-0021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbert AS, et al. Niemann-pick C1 is essential for ebolavirus replication and pathogenesis in vivo. MBio. 2015;6(3):e00565-15. doi: 10.1128/mBio.00565-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr Purif. 2008;62(2):160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419(6907):587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 33.Nakashima R, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500(7460):102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 34.Long F, et al. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467(7314):484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukazaki T, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474(7350):235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng TT, et al. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1(1):107–125. [PubMed] [Google Scholar]
- 37.Davies JP, Chen FW, Ioannou YA. Transmembrane molecular pump activity of Niemann-Pick C1 protein. Science. 2000;290(5500):2295–2298. doi: 10.1126/science.290.5500.2295. [DOI] [PubMed] [Google Scholar]
- 38.Adams CM, et al. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem. 2004;279(50):52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 39.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell. 2003;11(1):25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 40.Deffieu MS, Pfeffer SR. Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc Natl Acad Sci USA. 2011;108(47):18932–18936. doi: 10.1073/pnas.1110439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Deffieu MS, Lee PL, Saha P, Pfeffer SR. Glycosylation inhibition reduces cholesterol accumulation in NPC1 protein-deficient cells. Proc Natl Acad Sci USA. 2015;112(48):14876–14881. doi: 10.1073/pnas.1520490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soccio RE, Breslow JL. StAR-related lipid transfer (START) proteins: Mediators of intracellular lipid metabolism. J Biol Chem. 2003;278(25):22183–22186. doi: 10.1074/jbc.R300003200. [DOI] [PubMed] [Google Scholar]
- 43.Otwinowski ZMW. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol. 2002;58(Pt 10 Pt 2):1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowtan K. ‘dm’: An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett J Prot Crystallogr. 1994;31:34–38. [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.