Significance
Sympatric speciation (SS) has always been controversial since it was proposed by Darwin. Recently, we showed SS empirically in Spalax by amplified fragment-length polymorphism (AFLP), mitochondrial, and nuclear genomes. Similarly, SS in spiny mice, Acomys, from Evolution Canyon I (EC I), was earlier proposed by mtDNA and AFLP. Here, we show that full transcriptome data substantiates SS under sharp microclimatic and ecological divergence with gene flow, displaying extensive adaptive complexes to slope-specific stresses coupled with habitat choice and signals of reproductive isolation. Remarkably, strong natural selection across a sharply divergent ecological microsite overrules gene flow and advances SS, which is common at EC I. Because microsite ecological, geological, edaphic, and climatic divergences are widespread in nature, we conclude that SS might be a common mode of speciation.
Keywords: adaptive ecological speciation, microclimate, natural selection, RNA-seq
Abstract
Spiny mice, Acomys cahirinus, colonized Israel 30,000 y ago from dry tropical Africa and inhabited rocky habitats across Israel. Earlier, we had shown by mtDNA that A. cahirinus incipiently sympatrically speciates at Evolution Canyon I (EC I) in Mount Carmel, Israel because of microclimatic interslope divergence. The EC I microsite consists of a dry and hot savannoid “African” slope (AS) and an abutting humid and cool-forested “European” slope (ES). Here, we substantiate incipient SS in A. cahirinus at EC I based on the entire transcriptome, showing that multiple slope-specific adaptive complexes across the transcriptome result in two divergent clusters. Tajima’s D distribution of the abutting Acomys interslope populations shows that the ES population is under stronger positive selection, whereas the AS population is under balancing selection, harboring higher genetic polymorphisms. Considerable sites of the two populations were differentiated with a coefficient of FST = 0.25–0.75. Remarkably, 24 and 37 putatively adaptively selected genes were detected in the AS and ES populations, respectively. The AS genes involved DNA repair, growth arrest, neural cell differentiation, and heat-shock proteins adapting to the local AS stresses of high solar radiation, drought, and high temperature. In contrast, the ES genes involved high ATP associated with energetics stress. The sharp ecological interslope divergence led to strong slope-specific selection overruling the interslope gene flow. Earlier tests suggested slope-specific mate choice. Habitat interslope-adaptive selection across the transcriptome and mate choice substantiate sympatric speciation (SS), suggesting its prevalence at EC I and commonality in nature.
The origins of species—the “mystery of mysteries” (1) and “the most important event in Evolution” (2)—has challenged evolutionary biologists for over two centuries and is still controversial as to its mode and tempo (2, 3). Sympatric speciation (SS), the process by which new species arise in the absence of geographical barriers in free breeding populations with ongoing gene flow, was first proposed by Darwin (1) and proved as possible in theory by mathematical models (4–7). Recently, an increasing number of empirical studies described SS in both plants and animals (SI Appendix, Suggested Readings). We have added field evidences supporting SS in blind subterranean mole rats, Spalax galili (8, 9), which until our discovery, were considered to speciate only peripatrically or allopatrically.
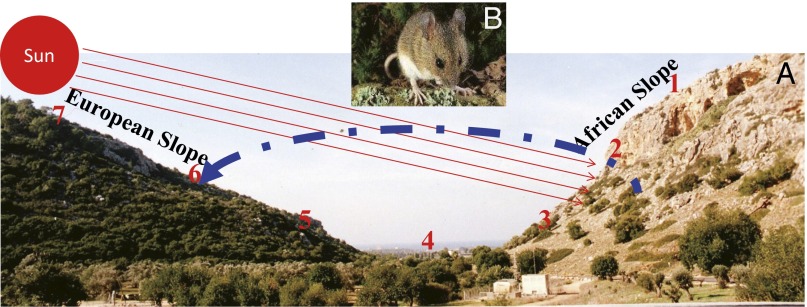
Likewise, we identified SS in five organisms from bacteria to mammals at Evolution Canyon I (EC I), Mount Carmel, Israel (10). These organisms include Bacillus simplex (11), wild barley [Hordeum spontaneum (10, 12)], fruit flies [Drosophila melanogaster (13, 14)], beetles [Oryzaephilus surinamemsis (15)], and spiny mice [Acomys cahirinus (16)]. All five model organisms display evolution in action of microclimatic adaptation and incipient sympatric adaptive ecological speciation on the tropical and temperate abutting slopes of EC I that are, on average, 250 m apart (10). These phylogenetically distant species converge in their microclimatic adaptations to the hot, dry, African, south-facing slope (AS) and the opposite cool, humid, forested, European, north-facing slope (ES) (Fig. 1) (10). The interslope drastically contrasting microclimatic divergence (17) is the driver of sharp ecological divergence between the forested ES and the savannoid AS.
Fig. 1.
Evolution Canyon I model in Israel. (A) The cross-section of EC I. The sharp divergence of savanna African slope (AS) and forested, European slope (ES) habitats is seen in the cross-section of EC I. Samples were collected at station 2 of the tropical, hot, dry, savannoid, south-facing AS and station 6 of the abutting temperate, cool, humid, forested, north-facing ES. (B) The spiny mouse, A. cahirinus.
African tropical spiny mice, A. cahirinus, colonized Israel some 30,000 y ago (18) in the Upper Pleistocene and extended across all rocky habitats in Israel. At EC I, Acomys first inhabited the tropical hot and dry AS and then, dispersed to the temperate cooler and humid ES (16). Although the interslope distance of Acomys populations at EC I is, on average, only 250 m apart, the animals from the two slopes display different body size and pelage color (19) as well as allozymic, DNA (20), mtDNA, and amplified fragment-length polymorphism (AFLP) differences (16).
This evolutionary divergence between the sympatrically evolving species was shown by phenomics (morphology, physiology, and behavior) and genomics (mtDNA and AFLP markers), which supported incipient SS with gene flow (16). Our prime goals in this Acomys transcriptome study were to expand and deepen our earlier study on Acomys SS at EC I based on mtDNA and AFLP markers and explore SS through the insights of the entire transcriptome analysis.
We posed here the following questions. Does SS emerging across sharp microsite ecological divergence also involve adaptive complexes across the transcriptome, which was earlier shown by the mitochondrial genome (16) as well as in our Spalax study (9)? Will it follow the pattern that we identified in the SS of S. galili, in which the entire genome was involved in the intersoil population divergence (9), but under EC I microclimate, might it replace soil as the major evolutionary driver? This study, indeed, revealed that the entire transcriptome is involved in SS, but at EC I, SS is driven by interslope climatic stresses.
Results
Transcriptome Sequencing and Assembly.
The animals used for this study were collected from the ASs and ESs of EC I (Fig. 1A) in July of 2013 (Fig. 1B and SI Appendix, Table S1). After leaving the animals in the laboratory for about 6 h, the whole brain was harvested and then immediately submerged into liquid nitrogen. Ten cDNA libraries were constructed and sequenced. Altogether, 55.5 Gb clean data were left for 10 individuals after stringent quality control and data filtering. After de novo assembly of all of these clean reads, in total, 539,605 unigenes were generated. The N50 for the unigenes is 2,354 bp, whereas the mean length is 1,075 bp.
In total, 69,126 coding sequences were found from all of the unigenes, and 67,979 of them showed homology with known proteins from the Nonredundant (Nr) and Swiss-Prot database; the other 1,147 were newly predicted with software. Of all of the unigenes, 52,494, 169,642, 47,592, 61,690, 62,748, 23,741, and 12,633 (SI Appendix, Fig. S1) were found to show homology with sequences from the Nr database, the National Center for Biotechnology Information databases of nucleotide (Nt), the Swiss-Prot database, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (SI Appendix, Fig. S2), the protein families (Pfam) database, the gene ontology (GO) database (SI Appendix, Fig. S3), and the Cluster of Orthologous Groups (COG) of proteins database (SI Appendix, Fig. S4), respectively.
Simple sequence repeats (SSRs) are important molecular markers in breeding, mapping, and population genetics. Here, we found 112,355 SSRs from 94,535 contigs across the transcriptome, among which 3,774 compound SSRs were found, and 44,287 primers were designed and could be used as molecular markers. The repeat type frequency and SSR classification are also listed in Dataset S1. The distribution of the SSR motif is shown in SI Appendix, Fig. S5.
Variation Calling and Population Structure.
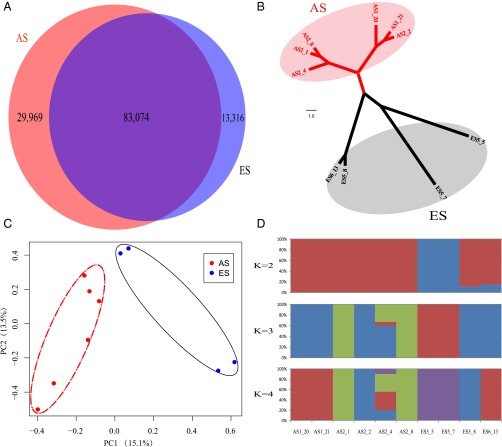
After variation calling, 113,043 and 96,390 SNPs were detected from the AS and ES populations, respectively. There are 29,969 and 13,316 SNPs unique to AS and ES populations, respectively (Fig. 2A). Kinship within each group was tested, and no siblings were found in any population (SI Appendix, Fig. S6). A neighbor-joining (NJ) tree was constructed based on the SNPs detected above. Six individuals from the AS formed one clade, four individuals from the ES formed the other clade, and they were separated into AS and ES clusters, respectively (Fig. 2B), which agrees with their environmental origin (Fig. 1A). Principle component analysis (PCA) was also conducted, and all of the animals could be divided into AS and ES populations according to their origin by the first eigenvector. The first and second components explain 15.1% and 13.5% genetic difference of the opposite slope populations (Fig. 2C), respectively. Both NJ tree and PCA showed two clear-cut AS and ES populations, which are congruent with the ecological sharp interslope divergence. To estimate the individual ancestry and admixture proportions, we also performed a population genetic structure analysis with a K value from one to four. Cross-validation (CV) for K values was from one to four (SI Appendix, Fig. S7). When the K value was one, it displayed the lowest CV. When K was set to two, the animals from the AS were clustered into one group, and the animals from the ES were almost in the other group; however, two of them were recombinants. The genetic proportion of AS is larger than that of the ES proportion in the two recombinants (Fig. 2D). When the K was increased from two to three and then to four, both the AS and ES populations were separated into subpopulations (Fig. 2D).
Fig. 2.
Population divergence of spiny mouse, A. cahirinus, at EC I. (A) Venn diagram of SNPs unique to the AS and ES populations. (B) NJ tree of the AS (red) and ES (gray) populations. (C) PCA of the AS and ES populations. (D) Population genetic structure of the AS and ES populations when K was set to two, three, or four. Note that AS is the south-facing slope, whereas ES is the north-facing slope. The ecological divergence between the slopes is like between two continents, despite the very short distance separating them (on average 250 m).
The genetic diversity of the two opposite populations was measured by calculating the π-value. It was significantly (P = 4.4 × 10−15) higher in the AS population, with a mean value of 10.4 × 10−5, than in the ES population, with a mean value of 9.98 × 10−5.
Natural Selection and Genetic Divergence.
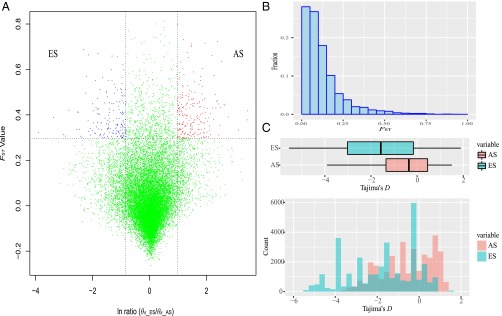
Genes subjected to selection were also inferred from ln(θπ_ES/θπ_AS) and FST. Transcripts under putative selection for the AS and ES populations are marked in red and blue dots, respectively, in Fig. 3A. There were 24 genes that were subjected to natural selection in the AS population, whereas 47 putative selected genes were found in the ES population. We also calculated the SNPs that were unique to each population. The SNPs unique to the AS population were located in 134 genes, and the SNPs unique to the ES population were located in 150 genes.
Fig. 3.
Natural selection on the two A. cahirinus opposite slope populations. (A) Distribution of ln ratio (π_ES/π_AS) and FST of each transcript. Red and blue dots (AS and ES, respectively) represent transcript under putative selection (corresponding to P < 0.05, where FST > 0.295 and ln ratio > 1). (B) FST distribution of the AS and ES populations. (C) Tajima’s D distribution of the AS and ES populations. Tajima’s D for the ES population is smaller than that for the AS population. The AS and opposite ES populations were marked in red and blue, respectively.
Interestingly, of 24 putatively selected genes from the AS population, RD23B, XRCC3, HSP105, and LMTK1 were found to be related to DNA repair, heat shock protein, growth arrest, and neural differentiation, all of which are associated with the local stresses affecting the AS population: high solar radiation, temperature, and drought. ATPF2 was putatively selected in the ES population related to ATP synthesis (i.e., energetics).
The population differentiation statistic was also estimated by calculating the FST between the two opposite populations. A large part of the FST was below 0.25; however, there are considerable sites with differentiation coefficients of ∼0.25–0.75 (Fig. 3B). The FST distribution of the two opposite populations displayed a clear L shape, which showed that some of the loci already diverged deeply. Tajima’s D test was performed on both of the AS and ES populations to assess the selection. The distribution of Tajima’s D for both ES and AS populations is shown in Fig. 3C. Most of the D values for both of the two populations were negative. Notably, some of the intervals of ES population are <−4, but there is no such area in the AS population. Some of the intervals of the AS population display Tajima’s D > 1, which does not appear in the ES population (Fig. 3C).
Discussion
SS and EC I in the Israeli Galapagos.
EC I in Nahal Oren has been dubbed the “Israeli Galapagos,” because several organisms, including soil bacterium, wild barley, fruit flies, beetles, and spiny mice, have been described as species undergoing adaptive incipient ecological SS (review and reanalysis are in ref. 10; see also refs. 13–16). The first analysis of incipient SS of spiny mice, A. cahirinus (16), was based on mtDNA and AFLP. Both showed significantly higher genetic diversity on the AS, the more stressful slope, which was also true regionally in Israel both genotypically (20) and phenotypically (21). A. cahirinus in EC I was smaller in size, longer in extremities (20), and lighter in color on the AS than on the ES and in the Israeli Negev desert (21). In complete mtDNA, 25% of the haplotypes at EC I were slope-biased. Phenotypically, the opposite slope’s populations also showed adaptive morphology, physiology, and behavior divergence paralleling regional populations across Israel (16, 20). Preliminary tests indicated slope-specific mate choice. The conclusion of that earlier study was that Acomys undergo incipient SS at EC I based on both habitat and mate choice slope selections caused by interslope microclimatic natural selection. Transcriptome sequencing has been an effective method of developing new SSR markers. The expressed sequencing tag SSR could be used for not only the source species but also, related species (22). The molecular markers identified from the transcriptome and primers designed could be used in the future for population genetics study (23), such as population diversity study, genome mapping, gene tagging, and quantitative trait loci analyses (24).
Population Genetic Divergence.
Colonization of a few individuals to a new environment may lead to extensive genetic changes, thus triggering reproductive isolation (25), which is the hallmark of speciation. This study, based on analysis of the entire transcriptome, substantiated and expanded the preliminary analysis in the work by Hadid et al. (16). To avoid comparing the differences originating from different families, kinships between animals were estimated (SI Appendix, Fig. S6), and the results showed that no animals from the same population are from the same biological family. Thus, it seems justified to assume that the samples were collected randomly and that the differences between them could display the population difference. The environmental stresses of the two slopes are different in temperature, humidity, and UV solar radiation, which are all substantially higher on the AS than on the ES (17, 26), although they are, on average, only 250 m apart. The interslope divergent stresses of the two slopes (Fig. 1A) are driving the slope-specific population adaptations to resist local stresses across the transcriptome (Fig. 3 A and C) and drive incipient ecological SS of the two opposite populations, each adapted to its unique ecology: tropical, dry, and hot on the AS and temperate, humid, and cool on the ES. It seems that large proportions of the entire transcriptome are involved in this interslope divergence, dramatically deciphering opposite slope adaptations of local populations. Natural selection overrules the gene flow (27); thus, transcriptome results fully substantiate, on a grand scale, the involvement of many adaptive complexes in resisting slope-specific environmental stresses. In the NJ tree, the individuals from the AS are clustered into the AS population and so are the individuals from the ES (Fig. 2B). In PCA, the first eigenvector separates all of the individuals into two clusters, AS group and ES group (Fig. 2C), which meet the divergent ecologies’ slope-specific stresses. These results show clearly that the two interslope populations are genomically divergent in the face of gene flow. In ancestry estimation analysis, the CV was the smallest when K was set to one (SI Appendix, Fig. S7); this suggests that all of the animals are from the same ancestry: animals first colonized the savannoid south-facing AS and then, dispersed to the forested, north-facing slope ES. However, other than K = 1, the CV was the second smallest when K was set to two, which means that all of the individuals are clustered into two populations. All of the animals from the AS population are in one cluster; however, the animals in the ES population are in two colors, suggesting the ancestral genetic background of the AS (Fig. 2D).
Ongoing Gene Flow Between the Two Slopes.
Rodent species diversity and microhabitat distribution across the opposing slopes were assessed by the capture, mark, and recapture method at EC I for 1 y (28). Most trapped animals on the AS were the African originated A. cahirinus. By contrast, on the ES, the vast majority of rodents were the European-originated Apodemus mystacinus and Apodemus flavicolis, with only 1/12 being A. cahirinus. The latter, the incipient new species of Acomys, confronted an ecologically novel cool and humid habitat on the ES and had to adapt to it, evolving the new incipient sympatric species described here based on the entire transcriptome. Incidentally, on the ES, Acomys densities decreased from the rocky top to the densely forested bottom. An advanced study of 460 recorded Acomys marked by chips conducted for 3 y (1996–1999) (table S8 in ref. 16) found five long-term migrations of Acomys from extreme stations between the slopes. This result indicates that a very low and limited gene flow occurs by interslope migrants, both males and females, from the AS to the ES and from the ES to the AS, permitting free interbreeding (i.e., confirming SS). The evidence of a low interslope gene flow suggests an indirect high level of habitat selection, an important hallmark of speciation, coupled with the preliminary evidence of slope-specific mate choice (16). Data in the work by Hadid et al. (16) indicate that the colonization of EC I by Acomys started on the AS, and it was only later that Acomys from the AS moved to the ES, speciating sympatrically on that forested ES. Moreover, the sampled Acomys have a common origin (SI Appendix, Fig. S7) (figure 3 in ref. 16), suggesting that the ES Acomys derived from the AS and not from a different outside region; this substantiates SS and rejects the secondary contact hypothesis from other Acomys populations, which all differ in Cytochrome b, whereas all EC I Acomys clusters had a similar Cytochrome b marker. It is tempting to speculate that Acomys carmelensis (akin to A. cahirinus), which were described by George Haas in the Natufian–Neolithic site (12,000–13,000 y ago) of the Abu-Usba cave on the upper ES slope of EC I at lower Nahal Oren, Mount Carmel (18), may represent the fossil of A. cahirinus, a sympatric species that migrated from the AS to the ES, representing the new sibling species of A. cahirinus on the ES.
Genetic diversity was significantly higher in the AS population than in the ES population, which was shown by both π and θ, estimates displaying a positive association between environmental stresses and genetic variation: the higher the stress, the higher the genetic diversity or polymorphism, which was shown earlier at global, regional, and local scales, in which the polymorphism was shown to be positively associated with environmental stress (29).
Tajima’s D Suggests the Operation of Natural Selection on the Contrasting Slopes.
Some intervals of Tajima’s D for the AS population are >1, which may be caused by the balancing selection for diverse allele content; this is congruent with the genetic diversity comparison, which shows significantly higher genetic diversity in the AS population (30). The fact that Tajima’s D value < −4 was only in the ES population indicates that this population is under stronger positive selection or that the population is expanding (30). The spiny mouse, A. cahirinus, displays higher population density in the AS than in the ES (16). They migrated to the less comfortable ES environment (on average, 250 m apart), which represents a totally different biome. A. cahirinus originated from tropical Africa and prefers hot and dry environments, like the AS in EC I. They colonized the AS first and then, migrated to the cooler, more humid ES environment, where different temperate stresses operated. The L-shaped FST distribution shows that the two slope populations diverged, with limited ongoing gene flow (31).
Putatively Selected Genes and Adaptation.
Genes play pivotal roles in organism adaptation to local environments. In this study, several genes were identified as putatively selected genes, which respond to the slope-specific stresses. RD23B (Gene ID 100306736) is the UV excision repair protein RAD23 (Gene ID 856674) homolog B, and it participates in DNA repair. This gene was under selection, because it might have been involved in the nucleotide excision repair and advances DNA repair (32, 33) damaged by high UV, typical of the AS, up to eightfold higher than UV of the ES in EC I (17). The gene XRCC3 (Gene ID 7517) encodes an RecA/Rad51-related protein, and it promotes homology-directed DNA damage repair in mammalian cells (34) and correct chromosome segregation (35). HSP105 (Gene ID 15505) is a major heat shock protein, and it can protect against stress-induced apoptosis (36) and reduce the aggregation of denatured proteins and cytotoxicity (37). LMTK1 (Gene ID 9625) regulates neuronal cell differentiation (38, 39) and is also selected in the AS. Because the AS stresses are characterized by high UV, high temperature, and drought, these genes were positively selected to adapt spiny mice of the AS to its slope-specific stresses.
Conclusions and Prospects
SS was proposed by Darwin (1) in 1859 and has been debated until now. We have shown in several empirical studies that SS exists in nature (8, 10, 16), and like SS in Spalax, it affects the genome (9), transcriptomes, genetic editing, and microRNA. The spiny mouse, A. cahirinus (16), showed SS by using mtDNA and AFLP molecular markers. Here, we substantiated these findings based on the entire transcriptome analysis. In the future, whole-genome sequencing and whole-genome methylation study will be explored to show how natural selection shaped SS at different levels caused by sharp abutting but divergent ecologies, highlighting the incipient SS model. SS might be common in nature because of the commonality of sharply divergent ecologies, geologies, soil, climatic, and biotic abutting habitats, where SS could proceed with gene flow.
Methods
The experiments on animals in this study have been performed following the rules and guidelines of the Ethics Committee of the Institute of Apicultural Research, Chinese Academy of Agricultural Sciences. All of the animals were collected from EC I in 2013. Total RNA was isolated from the whole brain. Libraries were prepared using TruSeq RNA Library Preparation Kit v2 following the manual, and each one was prepared with a unique barcode. Pair end sequencing was performed on HiSeq 2000. After data quality control, all of the reads were assembled by Trinity (40). Reads were mapped to the reference transcriptome, and variations were detected by GATK. A population genetic structure study was conducted by NJ tree analyses, PCA, and ancestry and admixture estimation analyses. Genetic diversity was investigated by calculating and comparing π. Natural selection was detected by Tajima’s D, FST, and θπ.
Supplementary Material
Acknowledgments
We thank Robin Permut and Avigdor Beiles (University of Haifa) for commenting on the manuscript and Xiaoying Song for her technical assistance. The following foundations supported this work financially: Agricultural Science and Technology Innovation Program Grant CAAS-ASTIP-2016-IAR, the Ancell–Teicher Research Foundation for Genetics and Molecular Evolution, National Natural Science Foundation of China Grant 31372193, Foundation of Henan Educational Committee Grant 13A180717, and Czech Science Foundation Project 14-31670P.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. SRP076070).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608743113/-/DCSupplemental.
References
- 1.Darwin C. On the Origins of Species by Means of Natural Selection. John Murray; London: 1859. [Google Scholar]
- 2.Mayr E. Animal Species and Evolution. Animal Species and Their Evolution. Belknap; Cambridge, MA: 1963. [Google Scholar]
- 3.Coyne JA, Orr HA. Speciation. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- 4.Gavrilets S. Fitness Landscapes and the Origin of Species (MPB-41) Princeton Univ Press; Princeton: 2004. [Google Scholar]
- 5.Dieckmann U, Doebeli M. On the origin of species by sympatric speciation. Nature. 1999;400(6742):354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- 6.Higashi M, Takimoto G, Yamamura N. Sympatric speciation by sexual selection. Nature. 1999;402(6761):523–526. doi: 10.1038/990087. [DOI] [PubMed] [Google Scholar]
- 7.Kondrashov AS, Kondrashov FA. Interactions among quantitative traits in the course of sympatric speciation. Nature. 1999;400(6742):351–354. doi: 10.1038/22514. [DOI] [PubMed] [Google Scholar]
- 8.Hadid Y, et al. Possible incipient sympatric ecological speciation in blind mole rats (Spalax) Proc Natl Acad Sci USA. 2013;110(7):2587–2592. doi: 10.1073/pnas.1222588110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K, et al. Sympatric speciation revealed by genome-wide divergence in the blind mole rat Spalax. Proc Natl Acad Sci USA. 2015;112(38):11905–11910. doi: 10.1073/pnas.1514896112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nevo E. Evolution in action: Adaptation and incipient sympatric speciation with gene flow across life at “Evolution Canyon”, Israel. Isr J Ecol Evol. 2014;60(2-4):85–98. [Google Scholar]
- 11.Sikorski J, Nevo E. Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at “Evolution Canyons” I and II, Israel. Proc Natl Acad Sci USA. 2005;102(44):15924–15929. doi: 10.1073/pnas.0507944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nevo E, et al. Genomic microsatellite adaptive divergence of wild barley by microclimatic stress in ‘Evolution Canyon’, Israel. Biol J Linn Soc Lond. 2005;84(2):205–224. [Google Scholar]
- 13.Hübner S, et al. Genome differentiation of Drosophila melanogaster from a microclimate contrast in Evolution Canyon, Israel. Proc Natl Acad Sci USA. 2013;110(52):21059–21064. doi: 10.1073/pnas.1321533111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YB, et al. Divergence of Drosophila melanogaster repeatomes in response to a sharp microclimate contrast in Evolution Canyon, Israel. Proc Natl Acad Sci USA. 2014;111(29):10630–10635. doi: 10.1073/pnas.1410372111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharaf K, Hadid Y, Pavlíček T, Eviatar N. Local genetic population divergence in a saw-toothed grain beetle, Oryzaephilus surinamensis (L.) (Coleoptera, Cucujidae) J Stored Prod Res. 2013;53:72–76. [Google Scholar]
- 16.Hadid Y, et al. Sympatric incipient speciation of spiny mice Acomys at “Evolution Canyon,” Israel. Proc Natl Acad Sci USA. 2014;111(3):1043–1048. doi: 10.1073/pnas.1322301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavlícek T, Sharon D, Kravchenko V, Saaroni H, Nevo E. Microclimatic interslope differences underlying biodiversity contrasts in “Evolution Canyon”, Mt. Carmel, Israel. Isr J Earth Sci. 2003;52(1):1–9. [Google Scholar]
- 18.Tchernov E. 1968. Succession of rodent faunas during the Upper Pleistocene of Israel. (Paul Parey, Hamburg)
- 19.Singaravelan N, et al. Spiny mice modulate eumelanin to pheomelanin ratio to achieve cryptic coloration in “Evolution Canyon,” Israel. PLoS One. 2010;5(1):e8708. doi: 10.1371/journal.pone.0008708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nevo E, et al. Genotypic and phenotypic divergence of rodents (Acomys cahirinus and Apodemus mystacinus) at “Evolution Canyon”: Micro-and macroscale parallelism. Acta Theriol (Warsz) 1998;43(Suppl 5):9–34. [Google Scholar]
- 21.Singaravelan N, et al. Adaptation of pelage color and pigment variations in Israeli subterranean blind mole rats, Spalax ehrenbergi. PLoS One. 2013;8(7):e69346. doi: 10.1371/journal.pone.0069346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis JR, Burke JM. EST-SSRs as a resource for population genetic analyses. Heredity (Edinb) 2007;99(2):125–132. doi: 10.1038/sj.hdy.6801001. [DOI] [PubMed] [Google Scholar]
- 23.Vukosavljev M, et al. Efficient development of highly polymorphic microsatellite markers based on polymorphic repeats in transcriptome sequences of multiple individuals. Mol Ecol Resour. 2015;15(1):17–27. doi: 10.1111/1755-0998.12289. [DOI] [PubMed] [Google Scholar]
- 24.Varshney RK, Graner A, Sorrells ME. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005;23(1):48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Carson HL. 1971. Speciation and the founder principle. Stadler Genet Symp 3:51–70.
- 26.Nevo E, et al. Drought and light anatomical adaptive leaf strategies in three woody species caused by microclimatic selection at “Evolution Canyon”, Israel. Isr J Plant Sci. 2000;48(1):33–46. [Google Scholar]
- 27.Nevo E. Selection overrules gene flow at “Evolution Canyons,” Israel. Adv Genet Res. 2011;5(3):67–89. [Google Scholar]
- 28.Blaustein L, Kotler BP, Nevo E. Rodent species diversity and microhabitat use along opposing slopes of Lower Nahal Oren, Mount Carmel, Israel. Isr J Zool. 1996;42(4):327–333. [Google Scholar]
- 29.Nevo E. Molecular evolution and ecological stress at global, regional and local scales: The Israeli perspective. J Exp Zool. 1998;282(1‐2):95–119. [Google Scholar]
- 30.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22(8):437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends Genet. 2012;28(7):342–350. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Subba Rao K. Mechanisms of disease: DNA repair defects and neurological disease. Nat Clin Pract Neurol. 2007;3(3):162–172. doi: 10.1038/ncpneuro0448. [DOI] [PubMed] [Google Scholar]
- 33.Weiss E, et al. Cellular prion protein overexpression disturbs cellular homeostasis in SH-SY5Y neuroblastoma cells but does not alter p53 expression: A proteomic study. Neuroscience. 2010;169(4):1640–1650. doi: 10.1016/j.neuroscience.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat Cell Biol. 2000;2(10):757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 36.Hatayama T, Yamagishi N, Minobe E, Sakai K. Role of hsp105 in protection against stress-induced apoptosis in neuronal PC12 cells. Biochem Biophys Res Commun. 2001;288(3):528–534. doi: 10.1006/bbrc.2001.5802. [DOI] [PubMed] [Google Scholar]
- 37.Yamagishi N, Goto K, Nakagawa S, Saito Y, Hatayama T. Hsp105 reduces the protein aggregation and cytotoxicity by expanded-polyglutamine proteins through the induction of Hsp70. Exp Cell Res. 2010;316(15):2424–2433. doi: 10.1016/j.yexcr.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Takano T, et al. LMTK1 regulates dendritic formation by regulating movement of Rab11A-positive endosomes. Mol Biol Cell. 2014;25(11):1755–1768. doi: 10.1091/mbc.E14-01-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takano T, et al. LMTK1/AATYK1 is a novel regulator of axonal outgrowth that acts via Rab11 in a Cdk5-dependent manner. J Neurosci. 2012;32(19):6587–6599. doi: 10.1523/JNEUROSCI.5317-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.