Significance
Adipogenesis is vital for animals to maintain energy balance by storing lipid. As a crucial integrator of nutrient, energy, and metabolite signals, mammalian target of rapamycin (mTOR) reportedly regulates adipogenesis. However, conventional cell cultures are limited to mimicking in vivo conditions, and previous reports do not provide the localization information of mTOR during adipogenesis. Here, we developed an automated microfluidic large-scale integration platform for differentiating human adult stem cells into mature adipocytes with standardized nutrient availability over weeks. With an integrated multiplexed immunoassay, we detected interaction, phosphorylation, and abundance changes of mTOR under the defined conditions. High-content analysis of single-cell data revealed that mTOR complex 1 changes its subcellular position in a temporally synchronized process with lysosomes during adipogenesis.
Keywords: microfluidics, stem cell differentiation, adipogenesis, mTORC1 regulation, multiplexed PLA
Abstract
Mammalian target of rapamycin (mTOR) is a central kinase integrating nutrient, energy, and metabolite signals. The kinase forms two distinct complexes: mTORC1 and mTORC2. mTORC1 plays an essential but undefined regulatory function for regeneration of adipose tissue. Analysis of mTOR in general is hampered by the complexity of regulatory mechanisms, including protein interactions and/or phosphorylation, in an ever-changing cellular microenvironment. Here, we developed a microfluidic large-scale integration chip platform for culturing and differentiating human adipose-derived stem cells (hASCs) in 128 separated microchambers under standardized nutrient conditions over 3 wk. The progression of the stem cell differentiation was measured by determining the lipid accumulation rates in hASC cultures. For in situ protein analytics, we developed a multiplex in situ proximity ligation assay (mPLA) that can detect mTOR in its two complexes selectively in single cells and implemented it on the same chip. With this combined technology, it was possible to reveal that the mTORC1 is regulated in its abundance, phosphorylation state, and localization in coordination with lysosomes during adipogenesis. High-content image analysis and parameterization of the in situ PLA signals in over 1 million cells cultured on four individual chips showed that mTORC1 and lysosomes are temporally and spatially coordinated but not in its composition during adipogenesis.
Adipose tissue has evolved to serve as the major energy reserve under glucose-limiting conditions, and in times of high glucose availability, energy is stored in the adipose tissue as lipids (1). Maintenance and regeneration of adipose tissue are organized by adult stem cells of mesenchymal origin (2). The extreme increase of adipose depots in cell size and number under certain age conditions is the hallmark of obesity (3). Concomitantly, the incidence of obesity is considered a major risk factor for type 2 diabetes mellitus (4). Therefore, there is great interest in recapitulating adipose stem cell differentiation in vitro for disease modeling and developmental adipose tissue studies.
In vitro differentiation of pluripotent stem cells into mature adipocytes has been achieved and requires, at a minimum, the growth factor insulin, the antiinflammatory steroid dexamethasone, and the nonselective phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBMX) (5). A large variation in concentration and stimulation time ranges of the three chemicals has been used for in vitro differentiation of adipocytes (6). Additionally, external factors, such as cell heterogeneity of the initial adipose-derived stem cell population, cell densities, nutrient availability, oxygen levels, and/or secreted cell factors, change the differentiation landscape of the stem cells, which leads to low correlation between in vitro differentiation studies (7–9).
Despite the difficulties involved in in vitro adult stem cell culture approaches, the transcriptional program occurring during adipogenesis has been resolved in mouse (10) and to a lesser extent, human adipose-derived stem cells (hASCs) (11). Hormonal and nutrient signals that initiate and regulate the adipose cell maturation process, however, are still elusive but of central importance for therapeutic development.
One key signaling regulator for the induction of adipogenesis is the mammalian target of rapamycin (mTOR) (12). In humans, this conserved serine/threonine kinase forms two distinct functional complexes [i.e., the mammalian target of rapamycin complex 1 (mTORC1) (13) and mTORC2 (14)]. Each of the complexes governs specific cell growth, proliferation, and survival signals during various developmental and homeostatic processes. Activity and substrate specificity of the two mTORCs are tightly regulated by their interaction partners, subcellular localization, and expression levels (15). The mTORCs can be distinguished from one another by their differential sensitivity to rapamycin (16, 17). In an early experiment, it was shown that the addition of rapamycin to the differentiation mixture impairs adipogenesis in mouse and human mesenchymal stem cells (18). In fact, rapamycin inhibits the activity and expression of transcription factor peroxisome proliferator-activated receptor γ (PPARγ) (19), which is the master regulator of adipogenesis. The central importance of mTORC1 in the signaling network of adipocyte stem cell differentiation was fueled by the accumulating knowledge that mTORC1 integrates and transduces signals regulating energy balance (20). Specifically, mTORC1 is involved in the regulation of lipolysis (21) and lipogenesis (22), which are the central functions of adipocytes. One of the limitations that has hindered elucidation of the precise regulatory function of mTORC1 and its signaling network during adipogenesis is the lack of in situ analytical methods that can resolve its protein complex composition, changes in temporal interactions, and cellular localization independent of mTORC2.
In this work, we developed (i) a microfluidic large-scale integration (mLSI) cell culture platform for robust human adult stem cell differentiation over longer timescales and (ii) a chip-integrated multiplex in situ proximity ligation assay (mPLA) to detect the two mTORCs selectively in single cells. With the microfluidic cell culturing chip platform, we automated and standardized hASC differentiation for 128 adipose-derived stem cell cultures in parallel. The mPLA was characterized and benchmarked with the known response of the composition and localization changes of mTORC1 to amino acid deprivation and recovery. Lastly, by combining the chip platform and the in situ protein assay, we resolved mTORC1 dynamics during adipogenesis and revealed lysosome-related position changes of the complex.
Results
Adipogenesis on Chip.
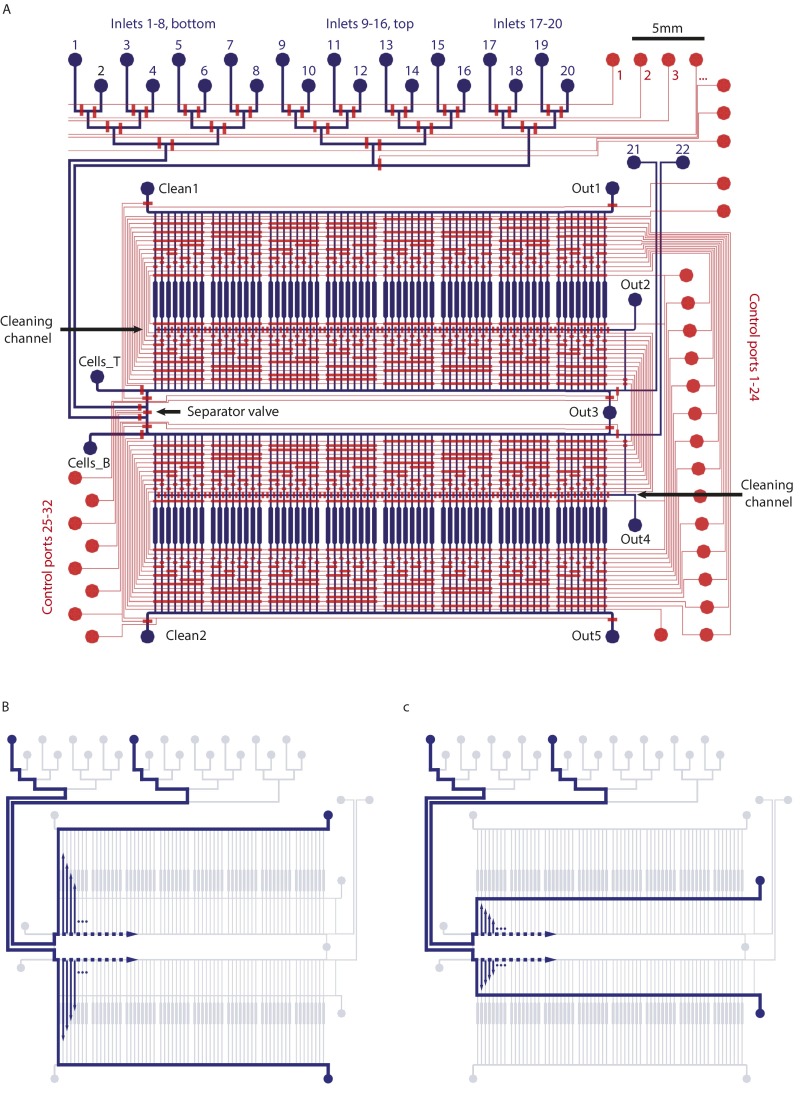
A two-layered polydimethylsiloxane (PDMS) mLSI chip was developed to automate and standardize differentiation and long-term culturing of hASCs (Fig. 1A). The chip platform contains two symmetrically arranged blocks of 64 individually addressable cell culture chambers. The flow channel network was optimized to ensure cross-contamination–free cell feeding. To achieve this task, it was, in particular, required to include medium purge lines before and after the cell culture chambers, synchronizing the pneumatic membrane valves for fluid routing and with external hardware components including fluid reservoirs. A detailed program to establish long-term cell culture on the mLSI chip is given in SI Text, mLSI Chip Operations and Fig. S1. To characterize the differentiation of hASCs on chip, about 220 ± 55 cells were introduced in each cell culture chamber. During the first 4 d, all hASCs were cultured with growth medium on the chip. Medium exchange was performed every 1 h with a feeding pulse of 15 s and a flow rate of 0.46 µL/min, which ensured a complete replacement of the 38-nL volume of the cell culture chamber. From the fifth day, the growth medium was replaced with differentiation medium in two cell culture chambers every 12 h. The two 64-cell culture blocks on the chip were treated equally to provide four replicates for each of 30 time points. The remaining eight cell culture chambers per block were used as no differentiation controls, and their positions were equally spaced within the two cell culture blocks. After 14 d of differentiation (DOD), all cell cultures were chemically fixed. The resulting culture array maintained the time trajectory of adipogenesis and was used for downstream lipid and protein analysis.
Fig. 1.
Adipogenesis on an mLSI chip. (A) Image of the mLSI chip for automated cell culturing. Blue and red dye-filled channels show the fluidic (bottom layer) and control (upper layer) channels, respectively, within the two-layered PDMS chip. Inlet ports for fluids and the corresponding control lines are omitted (SI Text and Fig. S1). The flow path from an inlet port through a cell culture chamber toward the outlet is indicated with a dashed line. The enlarged image on the right shows the dimensions of a cell culture chamber filled with 287 hASCs. White lines, blue dots, and red areas denote the cell chamber boundaries, cell nuclei, and cell cytoplasm, respectively. (B) Images from left to right show lipid accumulation in hASCs after 0, 2, 8, and 14 DOD. Red, blue, and green colors are the counterstains for the cell cytoplasm (Pallodin-Atto 643), nuclei (Hoechst 3342), and LDs (Bodipy 493), respectively. (Scale bar: 20 µm.) (C) The graphs show (Left) the mean LD size and (Right) number (black lines) accumulating in the hASCs during chemically induced adipogenesis on chip. The enclosing dashed lines show the SD among the hASCs. The red lines denote the lipid accumulation with SD (n = 2,200 cells) for induced hASCs in a 96-well plate.
Fig. S1.
Schematic illustrations of the microfluidic device used for long-term culturing hASCs. (A) Overview of the complete chip design. The two-layer PDMS device consists of flow (blue) and control (red) layers. All fluidic inlet and outlet ports are indicated. (B) Flow operation to feed all cell culture chambers sequentially from an inlet port. The upper and lower culture banks are parallel addressed. (C) Flow operation to clean the multiplexer for addressing the cell culture chamber. Without this functionality, cross-contamination between cell culture chambers is observed.
Lipid accumulation has been used as the primary indicator for the differentiation of mesenchymal stem cells into mature adipocytes (7). Fig. 1B shows representative fluorescence images at different times of hASCs that were chemically induced to undergo adipogenesis on chip. Fig. 1C shows the mean lipid droplet (LD) number and area per cell during 14 DOD. Each 12-h data point is an average value of at least 2,200 cells acquired in three different chip runs. LD accumulation, as measured by absolute area, increases steadily during 14 DOD, whereas the LD number increases only up to day 10 and then, reaches a plateau. Initial formation of multilocular LDs in hASCs during adipogenesis with subsequent merging into larger LDs has been previously reported (23).
LD accumulation within hASCs during adipogenesis is dependent on the time gap between the feeding cycles of the cell cultures on chip (Fig. S2). Longer time gaps between the feeding cycles led to lower LD accumulation rates. For comparison and standardization of hASC adipogenesis on chip, we measured LD accumulation rates of hASCs in 96-well plates; 100 µL growth and differentiation medium in each well was exchanged every 2 d over the same time as on the chip. The red line in Fig. 1C denotes the off-chip LD accumulation results for hASCs differentiated in a 96-well plate. Despite the volume and feeding differences, LD accumulation in the 96-well plate was comparable with the hourly feeding cycle on chip. Therefore, a time gap of 1 h between the feeding cycles was chosen for all following experiments. The correlation coefficient of LD accumulation from different chip experiments was higher than 0.92, which shows the reproducibility of the differentiation process (Fig. S3).
Fig. S2.
Correlation between cell feeding frequency on chip and LD accumulation. (A) Cell culture feeding frequency. Each bar represents a feeding pulse of 15 s at a flow rate of 0.46 µL/min. (B and C) Average LD (B) number and (C) size after 14 DOD in hASCs. Each bar corresponds to the feeding frequency in A, which was performed during the whole differentiation time.
Fig. S3.
Chip to chip variation and comparison of the LD accumulation on and off chip. (A) Representative images of hASCs after 14 DOD on and off chip. Differentiation conditions are described in the text. (Scale bar: 20 μm.) (B) LD accumulation in hASC measured on two different chip platforms after 14 DOD under the same culture program. (C) LD accumulation in hASC after 14 DOD measured in statistical repeats off chip.
To confirm hASCs adipogenesis at the protein level, we analyzed the temporal transcription factor expression levels of PPARγ, CCAAT/enhancer-binding protein β (CEBPβ), and CEBPα in the cell lysates of the off chip–cultured hASCs by Western blotting. As expected, we found the sequential expression of the three transcription factors as described previously during adipogenesis (11, 24) (Fig. S4).
Fig. S4.
Western blot protein quantification. Protein expression levels of (A) C/EBPβ, C/EBPα, PPARγ, mTOR, p-mTOR, and Actin as well as (B) Raptor, pS6, S6, and Actin in hASCs during 14 DOD. Cells were grown in standard six-well plates. mTOR, Raptor, and p-mTOR (Ser-2448) were tested to confirm mPLA results. Note that the increase of the mTOR phosphorylation on 4 DOD is accompanied by the phosphorylation of the S6 protein. MW, molecular mass.
mPLA.
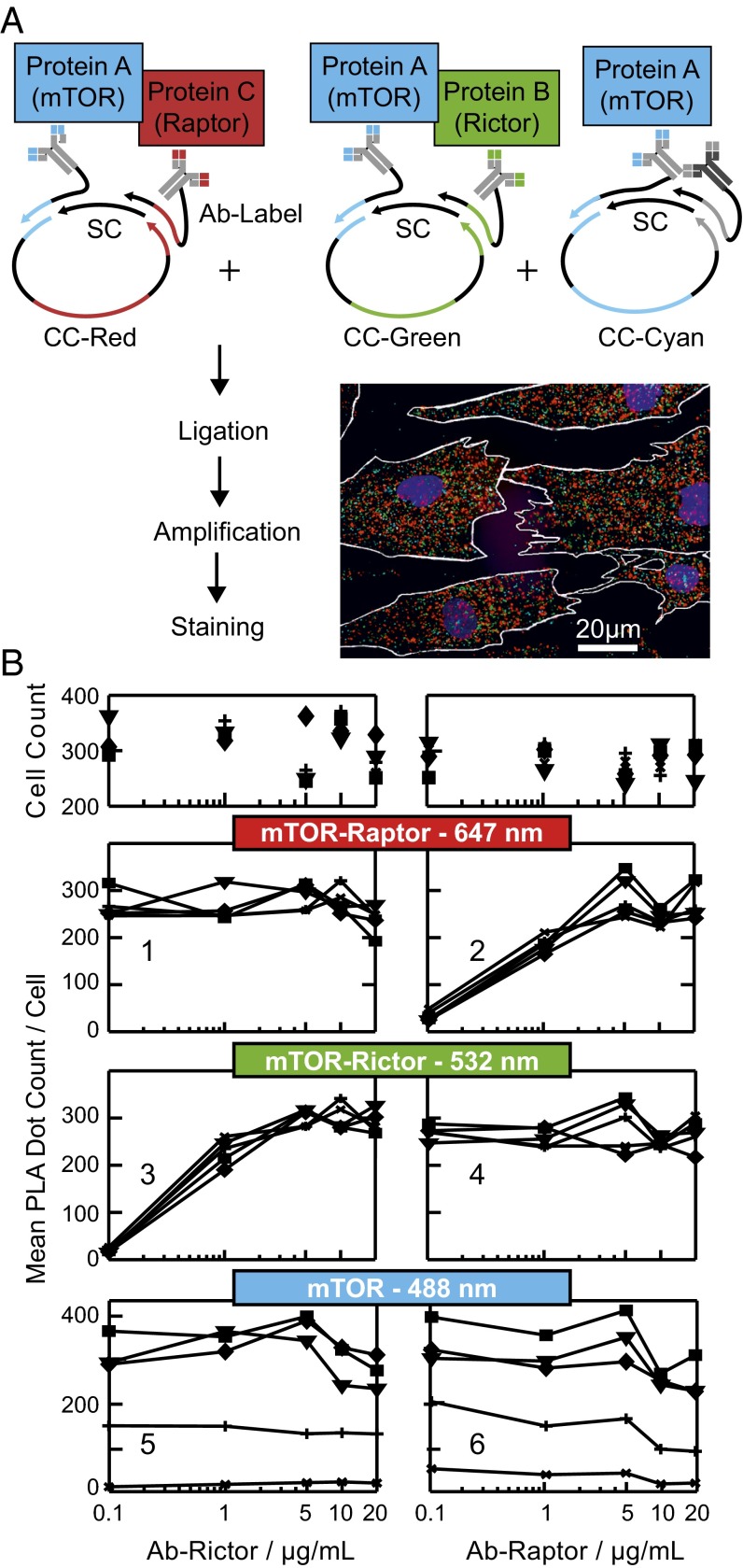
For studying mTORC1 signaling in hASCs during adipogenesis, we developed and applied an mPLA for simultaneous quantitation of the abundance, localization, modifications, and interactions of key proteins (25) in the mTORC1 pathway. The principle of the mPLA is illustrated in Fig. 2A. In short, in an mPLA, primary antibodies are labeled with barcode oligonucleotides. On binding of the antibodies to their targets within a fixed cell, their proximity is tested by addition of oligonucleotides bridging two specific barcode labels. Two bridging oligonucleotides are used per proximity test to form a circular DNA template that can be amplified by rolling circle amplification. The product is an ssDNA polymer that is ∼0.5 µm in diameter and detectable by fluorescence microscopy at low magnification. Decoding the proximity event of an antibody pair is achieved by addition of a unique event sequence in one of two bridging oligonucleotides. Thus, the information of the two barcodes of the antibody pair is transferred onto the DNA template and on amplification onto the proximity ligation assay (PLA) dot, from where the proximity signal can deciphered with fluorescent probe complementary to the event sequence. Color coding of the fluorescent probes allows the parallel detection of PLA events. Details of the PLA template designs and oligonucleotide sequences are given in Fig. S5 and Table S1, respectively.
Fig. 2.
mPLA for on-chip analysis of protein–protein interaction and protein expression in hASCs. (A) PLA workflow to target three protein events in parallel within a single cell. The multicolor fluorescence image shows representative PLA results for hASCs after in situ staining. The red, cyan, and green PLA dots report on the mTOR–Raptor and mTOR–Rictor protein interactions and mTOR expression levels, respectively. CC and SC denote for the shared and coding connector strand, respectively. (B) Orthogonality of the in situ mPLA system. Rows 2–4 show the PLA dot counts per cell for three individual test systems on increasing (Left) Rictor or (Right) Raptor Ab concentrations. The Ab concentration for mTOR was constant (10 µg/mL) in all experiments, whereas the secondary Ab targeting the mTOR Ab changed from 0.1 (*), 1 (+), 5 (▼), and 10 (■) to 20 µg/mL (♦). Row 1 shows the cell count number used to extract the corresponding PLA result in rows 2–4.
Fig. S5.
DNA design for the mPLA. One PLA template consists of four ssDNAs: a coding connector (CC), a shared connector (SC), and two antibody labels named adaptor strands (ASs). The CC specifically hybridizes to adaptors 1 and 2 on the indicated AS1 and AS4 sequences, respectively. Furthermore, the CC contains a probe sequence in its midsegment. The SC hybridizes to the AS2 and AS3 on adaptors 1 and 2, respectively. The sequences of the three CCs, six adaptors, and SC DNA strands used within this study are given in Table S1.
Table S1.
Sequences of the used oligonucleotides
| Oligos and names | Sequences (5′–3′) |
| Adapters | |
| Shared adapter | [ThiC6]AAAAAAAAAAATAGTTCGGTCGAAGTTAGTCC[Inv-dA] |
| Atto 647 adapter | [ThiC6]AAAAAAAAAAGAATGGAACCTCGCTAGAACGT |
| Atto 488 adapter | [ThiC6]AAAAAAAAAAGAATGGAACCTATCAATGCCTG |
| Cy3 adapter | [ThiC6]AAAAAAAAAAGAATGGAACCTGCACAGCTAAT |
| Shared connector | |
| Short connector | [Phos]AGGTTCCATTCAAAGGACTAACTTC |
| Coding connectors | |
| Atto 647 long connector | [Phos]GACCGAACTATCTAGTGCTGGATGATCGTCCCCCCTGCACCTCAAAACACCCTAACGTTCTAGCG |
| Atto 488 long connector | [Phos]GACCGAACTATGTATCTGCTTATGTCGCCCGGGACAATCATCTAAGTATTAATCCAGGCATTGAT |
| Cy3 long connector | [Phos]GACCGAACTATAGCGATCTGCGAGACCGTATTATTGAAAAAGAACAAATTAAGAATTAGCTGTGC |
| Detection probes | |
| Atto 647 detection probe | [Atto 647]CTAGTGCTGGATGATCGTCC[Atto 647] |
| Atto 488 detection probe | [Atto 488]GTATCTGCTTATGTCGCCCG[Atto 488] |
| Cy3 detection probe | [Cy3]AGCGATCTGCGAGACCGTAT[Cy3] |
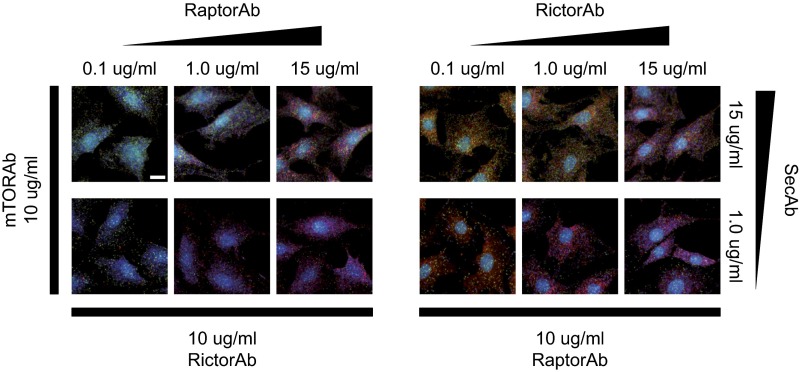
We first sought to test the orthogonality of the mPLA and concomitantly, establish an in situ assay for specifically detecting the intracellular mTORC1 (see SI Text and Fig. S6). For this bioengineering step, the protein interaction between mTOR and regulatory-associated protein of mTOR (Raptor) was targeted in undifferentiated hASCs. Additionally, the mTORC2 complexes, which are represented by the mTOR interaction with rapamycin-insensitive companion of mammalian target of rapamycin (Rictor), and total mTOR abundance were quantified. Fig. 2A, Inset shows a representative multicolor fluorescence image for interactions between mTOR and Raptor, mTOR and Rictor, and total mTOR represented by red, green, and blue PLA dots, respectively. Fig. 2B shows the PLA dot counts per cell for the mTOR–Raptor (Fig. 2B, row 2), the mTOR–Rictor (Fig. 2B, row 3), and the mTOR (Fig. 2B, row 4) proximity tests as a function of the anti-Raptor antibody (Ab-Raptor), the anti-Rictor antibody (Ab-Rictor), and secondary antibody concentrations, respectively. The concentration of the shared mTOR antibody was constant in all experiments. The PLA dot count per cell for the mTOR–Raptor proximity test increased with the anti-Raptor concentration (Fig. 2B, row 2). The titration curve was linear in the semilog plot between antibody concentrations of 0.1 and 10 µg/mL and gets saturated at higher concentrations, where PLA signals start to merge and can no longer be optically resolved as single dots. The same result was observed for the mTOR–Rictor and mTOR PLAs. Most importantly, the antibody titration curves show that the three PLA signals for the individual proximity tests are independent of each other. Representative images for all conditions are given in Fig. S7.
Fig. S6.
Cell culture chip used for the orthogonality test of the single PLAs. (A) Layout of the chip. Cell culture chambers are arranged in matrix configuration, which enables 2D antibody screening. (B) Implementation of the antibody titration experiment on the matrix cell culture chip. hASCs were grown for 24 h before fixation. Next, antibodies with various concentrations were applied. Each row and column had one defined antibody concentration, which is indicated within the plot.
Fig. S7.
Representative PLA images obtained within the orthogonal test experiment. The red, blue, and green PLA dots denote the mTOR–Raptor and mTOR–Rictor proximities and the mTOR abundances, respectively. The number of PLA dots per cell changed according to the varied antibody concentration. (Scale bar: 20 μm.)
In the next step, we aimed to decipher if the mPLA system can detect changes in protein interactions and subcellular localization of mTORC1 in hASCs. To answer this question, the well-documented mTORC1 regulation by amino acids was exploited (26, 27). In culture, withdraw of amino acids from HeLa and Hek293 cells leads to a dissociation of the mTORC1 from the GTPase-activating protein named Rag (28). The Rag proteins are a family of four heterodimeric GTPases (A–D) that reside at the late endosomal/lysosomal compartment and can interact directly with Raptor within the mTORC1 (26). Accompanied by the loss of the Raptor–Rag interaction, the localization of mTOR and Raptor changes from the perinuclear region to a diffuse distribution in the cytoplasm (28). Both protein interaction and localization changes of mTORC1 proteins can be reinstated with a short amino acid stimulation pulse (26, 27). In contrast, the Rag GTPases do not change their subcellular positions during poststarvation amino acid recovery.
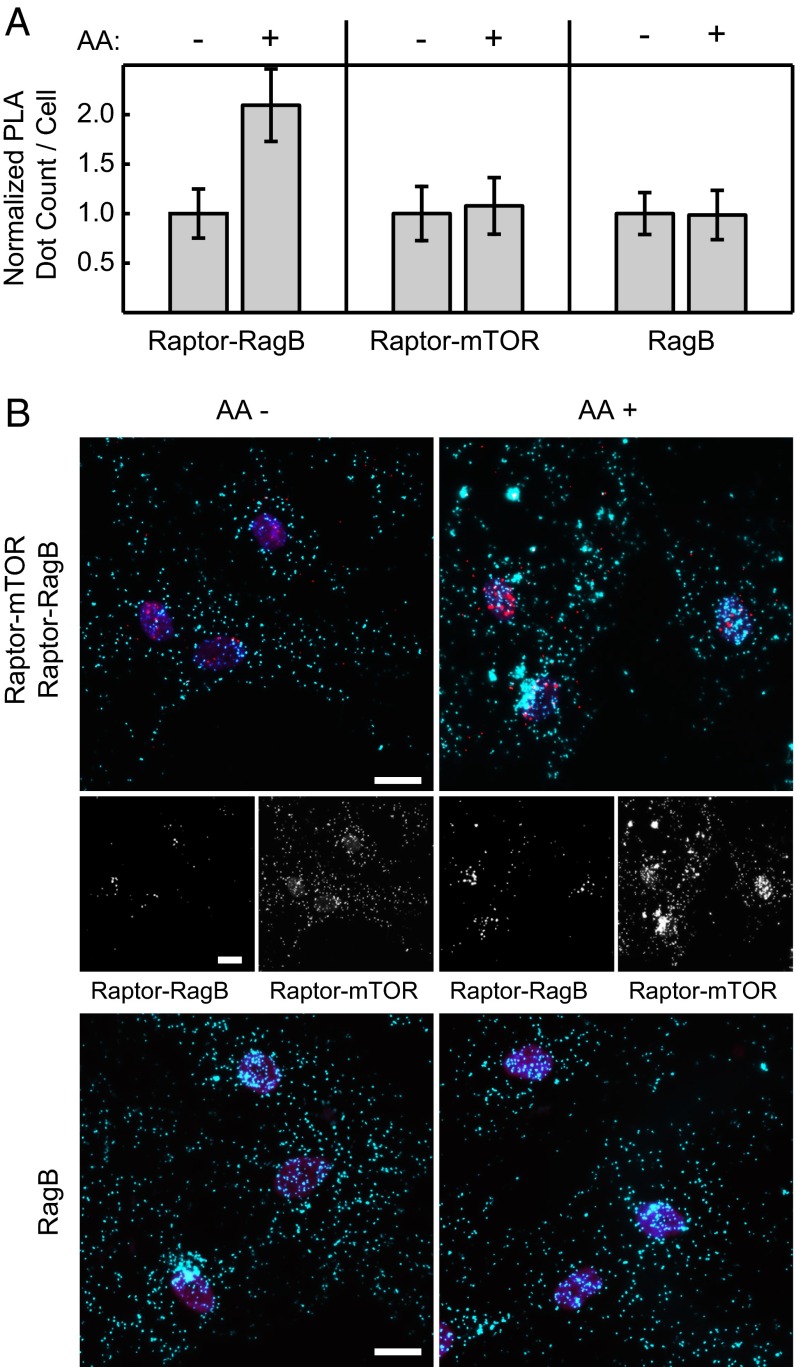
Fig. 3A shows results from the PLA tests for the Raptor–RagB and Raptor–mTOR and the abundance of Raptor and RagB. Of note, the RagB abundance was measured as a subcellular location control in a separate PLA experiment. The PLA dot count per cell for the Raptor–RagB interaction doubled on amino acid stimulation of starved hACSs, whereas the PLA dot counts for mTOR–Raptor, Raptor (omitted), and RagB stayed constant. These results are concordant with previous findings (26, 28). Representative PLA images for the two interactions and RagB under the two conditions are given in Fig. 3B.
Fig. 3.
mTORC1 protein interaction changes on amino acid starvation and recovery in hASCs. (A) Normalized PLA dot count per cell for (Left) the Raptor–RagB , (Center) Raptor–mTOR interactions, and (Right) RagB protein abundance. All hASCs were amino acid (AA)-starved for 50 min, a portion of the cells was replenished with amino acids for 10 min, and all cells were fixed at the same time. (B) Representative PLA images of the hASCs corresponding to the conditions in A. In Top, red and cyan dots denote Raptor–RagB mPLA signal and Raptor–mTOR mPLA signal, respectively. Middle represents mPLA signals in each single channel. In Bottom, cyan dots denote RagB PLA signal. Nuclei (magenta) are stained with DAPI. (Scale bars: 20 μm.)
To characterize changes of PLA dot distributions in single cells, we parameterized the subcellular PLA dot positions. Similar to a previous parameterization approach for FISH probe signals (29), the mean PLA dot distance to the nuclear centroid and the mean PLA dot to dot distance were calculated and normalized to the size of the cell. To investigate the heterogeneity of the amino acid response in the cell population, the distributions of both normalized distance vectors among cells were evaluated with the mean and SD. All six parameters were z-scored against randomized PLA dot distributions (Materials and Methods). Fig. 4A illustrates the calculated PLA dot parameters, and Fig. 4B shows the parameterization results for the Raptor–mTOR, Raptor–RagB, and RagB PLA events (dots) within hASCs under the amino acid-starved and -reinstated conditions. Although the mean PLA dot distance to the nucleus centroid and the PLA dot to dot distance decreased by 1–1.5 SDs for the two interactions, they stayed constant for RagB under the two conditions. The mean SD of the two distance parameters that report on the heterogeneity of the response in the cell population decreased for all three PLA events. Therefore, a homogeneous cell response was observed in the hASCs to amino acid stimulation. The statistical evaluation of the PLA dot distributions confirmed the translocation of the mTORC1 from a spread/diffuse cytoplasmic to an accumulated/perinuclear distribution and the constant position of RagB at late endosomes/lysosomes during amino acid starvation and stimulation. These results show that it is possible to statistically evaluate subcellular localization changes of protein interactions by using mPLA.
Fig. 4.
Analysis of PLA dot distributions. (A) Within a single cell, the mean PLA dot distance to the nuclear centroid and PLA dot to dot distance were calculated. Both values were normalized to the largest distance between the nuclear centroid and cell outline of a given cell. (B) For the analysis of PLA events over a population of cells under starved or replenished with amino acid (AA) condition, the means, standard deviations (STD), and variances of the STDs for both PLA distance vectors were calculated and z-scored against random PLA dot distributions (Materials and Methods).
mTORC1 Dynamics During hASC Adipogenesis.
In this section, we used the validated microfluidic chip-based mPLA technology to investigate interaction changes of the mTORC1 during adipogenesis. Fig. 5A and Fig. S8 shows the PLA dot count changes relative to undifferentiated hASCs for interactions of the mTORC1 with known regulators during adipogenesis. To track mTORC1, we monitored the mTOR–Raptor interaction. The lysosomal/endosomal localization of the mTORC1 was traced with the interactions of Raptor with RagB, the mTOR-activating GTPase Ras homolog enriched in brain (Rheb) (30, 31), and inhibitor protein proline-rich Akt substrate 40 (PRAS40) (32). Furthermore, the protein abundance of mTOR, Raptor, and LAMP1 and the protein interaction between mTOR and Rictor were measured as references. All PLA data were measured with the mPLA except for LAMP1, which was measured in a separate experiment.
Fig. 5.
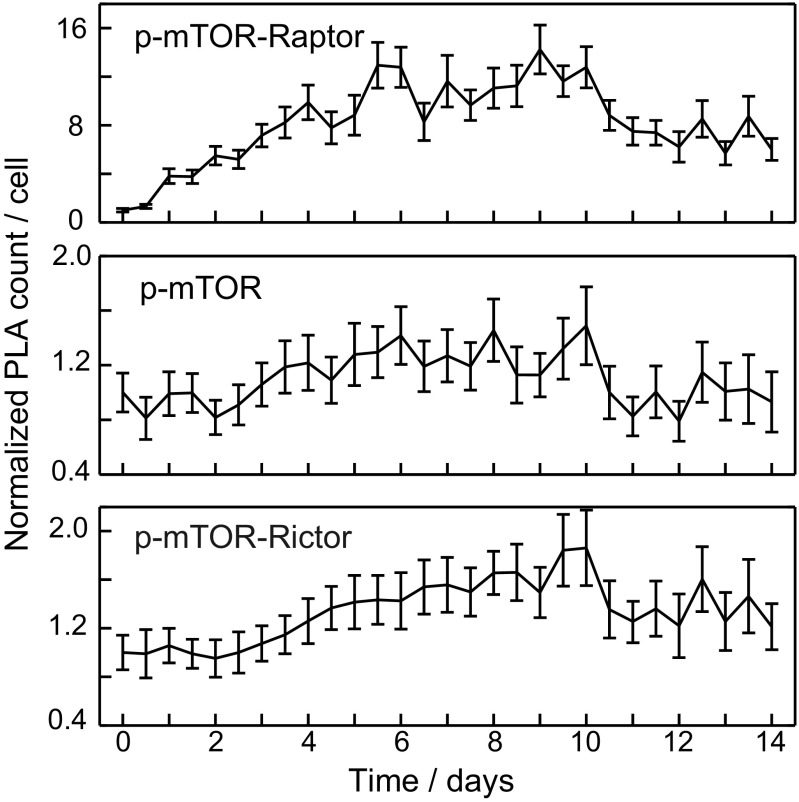
Temporal changes of PLA dot count during hASC adipogenesis for (A) protein–protein interactions within the mTORC1 and (B) expression levels of mTOR, Raptor, and LAMP1 phosphorylated mTOR interacting with Raptor. All PLA dot counts per cell were normalized to the PLA dot count in hASCs before chemical induction. The colored scale highlights fold changes of the PLA dot count per cell. Horizontal lines on the right indicate separate chip experiments.
Fig. S8.
Temporal changes of PLA dot count during hASC adipogenesis for all measured protein–protein proximities within the mTORC1 and expression levels of mTOR, Raptor, and LAMP1. All PLA dot counts per cell were normalized to the PLA dot count in hASCs before chemical induction. Horizontal lines on the right indicate separate chip experiments. Unlike in Fig. 5, we plotted here the normalized PLA dot values with SDs obtained from cell populations with at least 400 cells.
During hASC adipogenesis, the PLA dot count for mTOR remained constant, whereas the signal for Raptor decreased from day 3 by threefold. This finding was confirmed by Western blot analysis (Fig. S4B). Consistently, all tested interactions formed by Raptor decreased on a similar timescale and with similar amplitude during adipogenesis of the hASCs, although the decrease in the Raptor–mTOR interaction was somewhat slower. Normalizing the interaction data with the decrease of Raptor abundance reveals that none of the interactions formed by Raptor changed additionally. These data indicate that mTORC1 abundance changed only in response to a decrease in the scaffold protein Raptor.
To obtain complex-specific activity information of the mTOR kinase during the adipogenesis of hASCs, we measured the proximity of an Ser-2448 phosphosite mTOR antibody to anti-Raptor and anti-Rictor antibodies in an mPLA experiment on chip. Fig. 5B shows the result for the p-mTOR–Raptor PLA. From the first DOD, the phosphorylation of the mTOR kinase increased, reached a 16-fold increase at the sixth DOD, and remained high until the last measured DOD. The phosphorylation of the S6 protein, which is a downward signaling indicator for the kinase activity of mTOR, accompanied the p-mTOR phosphorylation (Fig. S4B). Only a minor change of the mTOR Ser-2488 phosphorylation site in complex 2 was detected (Fig. S9).
Fig. S9.
Temporal changes of PLA dot count during hASC adipogenesis for the protein–protein proximities of Raptor and Rictor within Ser-2488–phosphorylated mTOR. Unlike in Fig. 5, we plotted here the normalized PLA dot values with SDs obtained from cell populations with at least 400 cells.
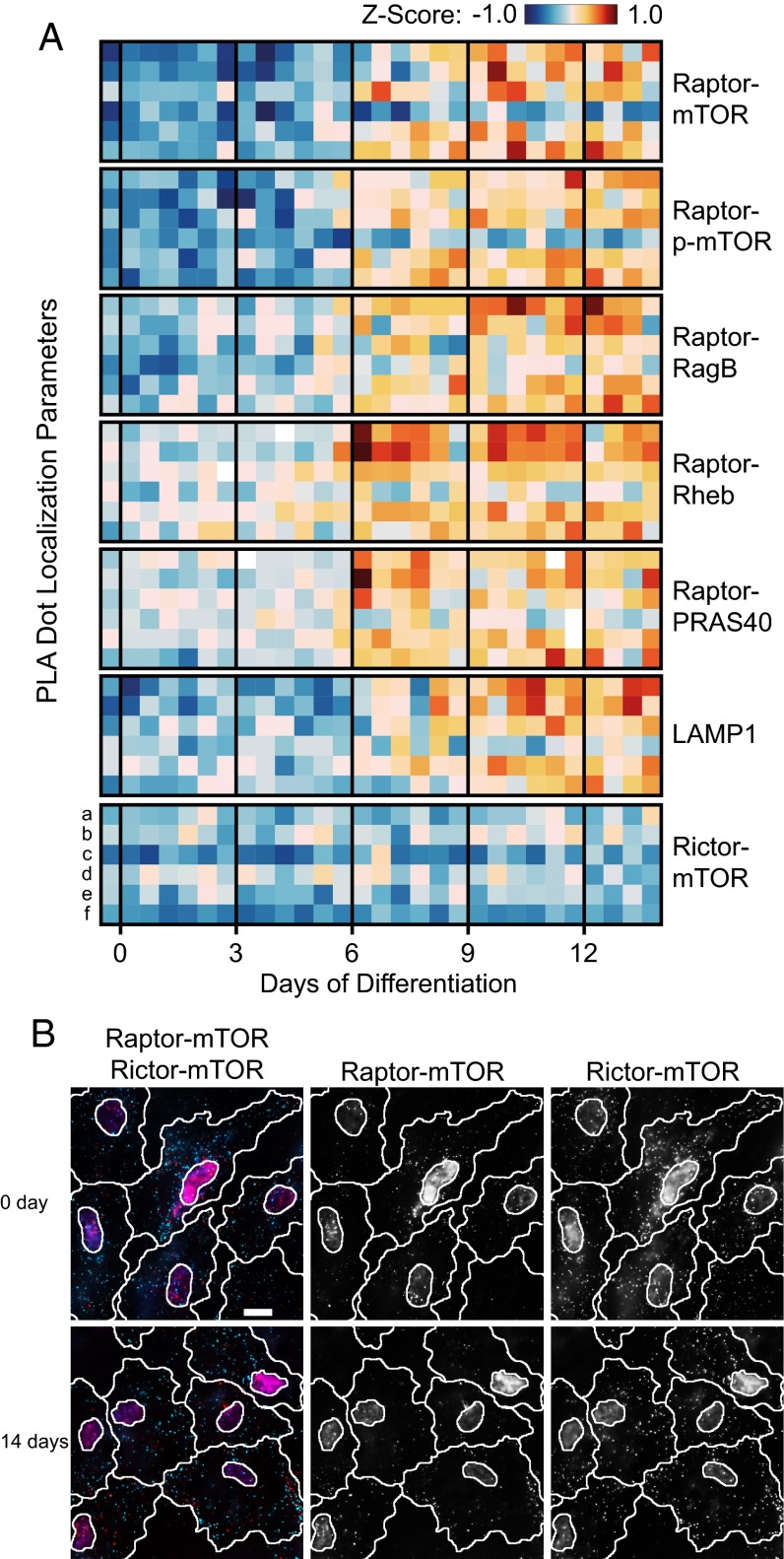
In addition to measuring the abundance and phosphorylation of the mTOR kinase in complex 1, we evaluated the localizations of these interactions within the hASCs during adipogenesis. Fig. 6A shows the mean values and mean SDs of the PLA dot distances to the nuclear centroid and each other. Representative fluorescence images of mTOR–Raptor and mTOR–Rictor interactions during DOD 0 and 14 are shown in Fig. 6B. The parameters were derived in a similar manner to the procedures explained above. Interaction signals for the mTORC1 increased in distance from the nucleus and each other from 6 to 7 DOD. Additionally, the distribution parameters for the SDs of the two PLA distance vectors increased, except those of the PLA dot distances to each other. This increase indicates heterogeneity among the cells for the localization change of mTOR–Raptor interactions. In contrast, PLA dots for the mTOR–Rictor interaction did not change their position parameters over the same time course. Despite the lipid accumulation in adipocytes, PLA distance parameters indicated that the mTORC2 maintains a diffuse cytoplasmic localization in hASC cells during 14 DOD. It should be noted that this signal was measured in the same cells in which the mTOR–Raptor signal was recorded. Single-cell data is of importance, because the adipose-derived stem cells changed, in addition to the lipid content, their morphology from fibroblast-like to spherical during the differentiation to mature adipocytes (33). The visible differences in cell morphology can be observed in Fig. 6B. Thus, measuring mTOR interactions with both Raptor and Rictor in the same cell excludes evaluation differences caused by changes in cell shape and lipid content.
Fig. 6.
mTORC1 localization analysis during the adipogenesis of hASCs. (A) Heat map of the z scores obtained for the PLA distance vectors, where a–f denote the mean distance to the nuclear centroid, mean PLA dot to dot distance, their two SDs, and their two variances over a cell populations under the same condition, respectively. (B) Representative PLA images of the mTOR–Raptor and mTOR–Rictor interactions during 0 and 14 DOD of hASCs. Red and cyan dots denote Raptor–mTOR mPLA signal and Rictor–mTOR mPLA signal, respectively. Nuclei (magenta) are stained with DAPI. (Scale bar: 20 µm.)
The results above (in particular, those for Raptor–RagB interactions) argue that the majority of the mTORC1 stays bound to the lysosomes. Therefore, an increase in the perinuclear–nuclear distance of mTORC1 could be explained by either relocalization of the lysosome together with the mTORC1 or dissociation of the mTORC1 with RagB from the lysosomes. To investigate these two possibilities, we measured the PLA localization profile of the lysosome marker lysosomal-associated membrane protein 1 (LAMP1) during adipogenesis. The mean distance of the LAMP1 PLA dots showed the same quantitative and temporal localization changes as those traced for mTORC1. This result shows that the lysosome changes in accordance to the mTORC1 complex and suggests that the majority of mTORC1 stays bound to the lysosomes during the subcellular position reorganization of the organelle during adipogenesis.
Discussion
Long experimental times are a major limitation to in vitro stem cell differentiation studies. From early on, the intrinsic properties of microfluidic chip technology for automated and precise control of the cellular microenvironment have been suggested to improve in vitro stem cell models and help establish robust disease models (34). In fact, differentiation of murine mesenchymal stem cells to adipocyte-like cells was one of the first differentiation processes performed on a microfluidic chip (35). Despite this achievement, the integration levels by means of a number of cell cultures and analytical capabilities of microfluidic platforms are still limited. In this work, we developed an mLSI chip to optimize long-term cell culture and integrated the mPLA on chip, thus overcoming the current limitations.
The major difference of the programmable miniaturized cell culture chip compared with classical cell culture is the ease of repetitive feeding cycles for cell maintenance. Periodic feeding leads to modulation of consumed and produced substances in the culture media. It is clear that higher frequencies of the feeding pulse lead to a sustained supply of nutrients, oxygen, and external factors and lower concentrations of secreted factors in the cell media. Recently, the influence of periodic medium delivery on human pluripotent stem cell cultures has been systematically investigated (36), and the findings show that, when feeding frequency is optimized, the stem stability and differentiation on chip are more homogeneous than in cell culture flasks. In our work, we observed that the rate of lipid accumulation during hASC differentiation on chip was also dependent on the feeding pulse frequency. This dependency was expected, because lipid accumulation rates decrease under hypoxia (37) and deprivation conditions (38), which can explain the lower lipid accumulation rate in hASCs at lower feed frequencies on chip (Fig. S2). In this multiparameter space, we adjusted the lipid accumulation rate on chip to standard off-chip protocols (Fig. 1C) to make our results comparable. The distribution of LD content per cell in the cell population on and off chip after 14 DOD was comparable, with the exception that about 8% of all cells in the 96-well plates were not accumulating lipids (Fig. S10). Thus, a small gain in homogeneity is obtained on chip. Reproducibility of the lipid accumulation rate during adipogenesis was high on and off chip (r > 0.92) (Fig. S3).
Fig. S10.
Distribution of the lipid accumulation in hASCs differentiation in a well plate and on the microfluidic chip. The lipid content is the average total lipid area found in the hASCs after 14 DOD. Note that only the well plate approach shows a fraction of 7% undifferentiated hASCs.
The cell culture chip platform, with its control function over the cellular microenvironment, enabled the precise investigation of the mTOR kinase during adipogenesis. The second required tool for the specific detection of the mTORC was the mPLA. The principle of detecting multiple proximity events in a single cell has been achieved before but with a more complex PLA template design and without proof of orthogonality and dynamic and subcellular position analyses (39). In our benchmark experiments for the mPLA, we confirmed the accumulated knowledge that, under amino acid deprivation conditions, the mTORC1 dissociates from lysosome-localized RagB (26, 28) and changes its cellular distribution from perinuclear to diffuse cytoplasmic (Fig. 4B). Consistent with previous observations (40), the single-cell PLA signal and distance vector analyses showed that the mTOR–Raptor interaction is not affected by amino acid deprivation (Fig. 3A). This result shows that the developed quantitative PLA image analysis is a functional method for determining subcellular protein localization but within defined subcellular resolution boundaries.
The mPLA on chip increases throughput and allows parallel monitoring of different targets within the same cells compared with geometric multiplexing of the PLA in separate microchambers for cell culturing (41, 42). The advantages of the mPLA were exploited for the characterization of the mTORC1 during adipogenesis. Without cellular lysis and related dissociation problems of the mTORCs during immunoprecipitation assays (14), we observed that the mTORC1 changes in abundance and posttranslational modifications but does not change in its composition during adipogenesis. On chemical induction of adipogenesis with insulin, dexamethasone, and IBMX, the phosphorylation of the mTOR kinase in mTORC1 starts within the first 12 h at the residue Ser-2448 (Fig. 5B). The p-mTORC1 level increased up to day 5. At this time point of differentiation, the S6 protein becomes phosphorylated. Both phosphorylation events argue for an increase of the mTOR kinase activity during the adipogenesis (43), which is in agreement with reports in 3T3-L1 cells (44). Down-regulation of the mTORC1 occurs on day 6 through expression changes of the adaptor protein Raptor. Consistently, when correcting the abundance change of Raptor from all PLA signals indicative for the mTORC1, no additional changes were observed. This finding argues that, although the number of mTORC1 is reduced, the composition, including Raptor, Pras40, Rheb, and RagB/D proteins of mTORC1, during adipogenesis is maintained. Of note, the decrease of bound mTOR kinase in complex 1 did not lead to an increase of the mTOR–Rictor interaction. Assuming that the mPLA is equally sensitive for the mTORC1 as for the mTORC2, our data reveal that no free thermodynamic equilibrium exists between the two complexes.
Next to the quantitative mTORC1, information parameterization of the PLA signal revealed that, after 6 DOD, the mTORC1 moves together with the lysosomes, and its localization changes from a perinuclear position to scattered positions distal from the nucleus. mTOR scaffolding and cellular position are two central regulatory mechanisms for protein kinase-defining substrate specificity, accessibility, and/or kinase activity (15). Intracellular lysosomal positioning in response to nutrient availability is a reported regulatory mechanism for the mTORC1 (27). During nutrient starvation, lysosomes cluster near the microtubule organization center. At this subcellular position close to the nucleus, the fusion of lysosomes with autophagosomes is facilitated on mTORC1 activity repression. Conversely, nutrient replenishment after starvation induces lysosomal scattering in the cell, activation of the mTOR, and reduction of autophagosome synthesis (27). The observation that lysosomal positioning also occurs during adipogenesis (Fig. 6A) suggests that this is a general regulatory mechanism, which determines mTORC1 signaling. Autophagy is required for 3T3-L1 cells to differentiate into white adipocytes, presumably by influencing LD formation (45). Therefore, it matches with the previous observation that lysosomal-associated mTOR in the perinuclear region is not suppressing autophagy. Interestingly, repositioning of mTORC1 associated to the lysosomes occurs at maximum phosphorylation signal of the mTOR kinase in complex 1 on DOD 6.
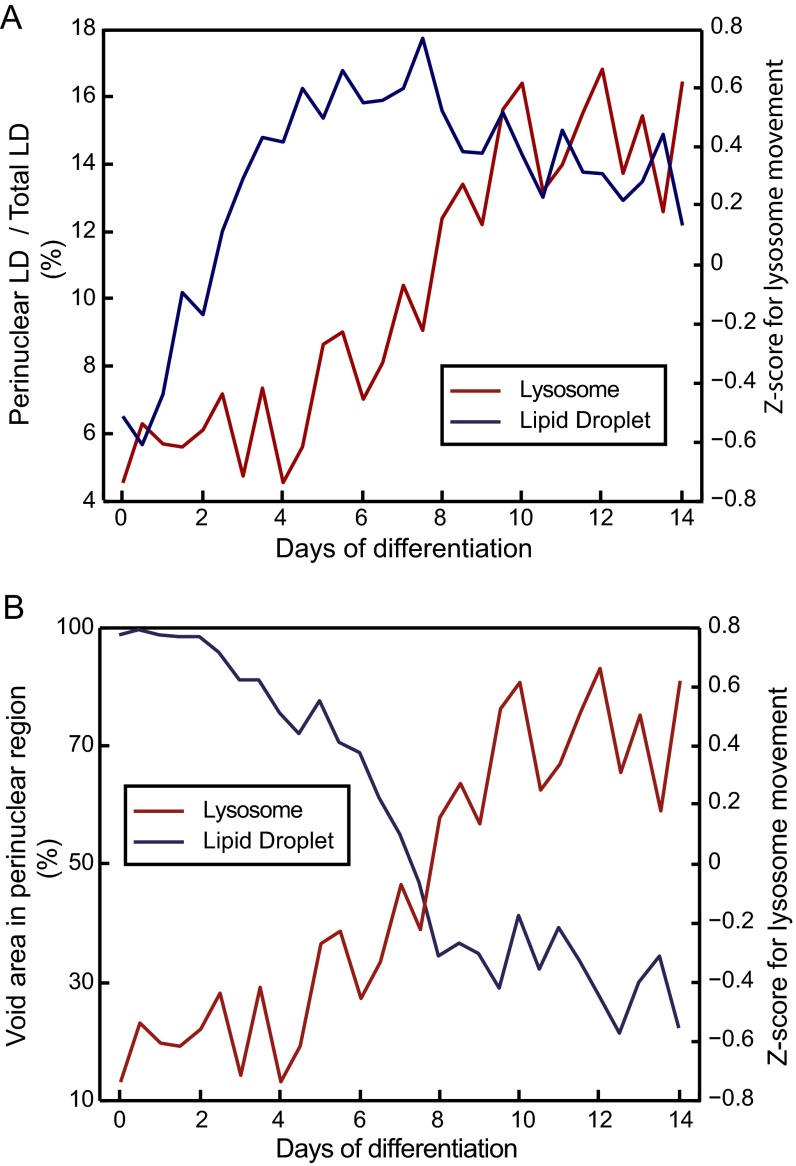
Notably, it cannot be ruled out that mTORC1 repositioning is a secondary effect caused by the repositioning of the lysosomes because of LD accumulation. LD formation starts in the perinuclear region (Fig. S11A), and the movement of lysosomal-associated mTORC1 to a more distal region in the cytoplasm correlates with the decrease of the cytoplasmic space in the perinuclear region (Fig. S11B). Next to the transcriptional program of the adipogenesis, mTORC1 controls the balance between lipogenesis and lipolysis in adipocytes (46). Essential proteins for the LD accumulation and thus, the initiation of lipid storage and lipogenesis are substrates of the mTORC1 as, for example, lipin-1 (47). Therefore, perinuclear accumulation of functional associated events can be expected. On moving outward in more distal and scattered positions in the cytoplasm during the progression of the adipogenesis, mTORC1 can effectively inhibit lipolysis by inactivating lipid-degrading enzymes (48) and lipophagy (49). It is unlikely that lysosomal positioning is only determined by the energy state of the cell, because the reorganization in hASCs during adipogenesis takes place after day 6 of induction. At that time point, cells already accumulate substantial amounts of LDs. In summary, the accumulating knowledge of the mTORC1 functional changes by subcellular positioning argues that movement of mTORC1 belongs to the differentiation program of adipocytes. In future work, it will be interesting to investigate if the lysosomal and mTORC1 positioning determines the cell fate decisions of hASCs by systematic reversal of the chemical induction pulse on chip.
Fig. S11.
Accumulation of LDs in the perinuclear position and lysosmal repositioning. (A) Fraction of LDs in the perinuclear region to the total amount of detected lipid in the cell. The perinuclear region was defined as the circular area surrounding the nucleus with diameter of the nucleus. The plot shows that LDs accumulate first in the perinuclear region. (B) Void area in the perinuclear region is the cellular area without fluorescence signal of LDs. From 4 DOD, the void area is quickly decreasing, and the retrograde lysosomal movement starts accordingly.
Materials and Methods
Microfluidics.
The layout of the flow and control layers of the microfluidic cell culture chip was designed with AutoCAD (Autodesk) and printed onto emulsion photomasks (BVM Maskshop). The mold for the flow circuit was manufactured in a two-layer process described previously (50). Briefly, AZ40-XT Photoresist (DuPont) was used to construct fluidic channels with a rounded profile and a height of 22 µm. The cell chambers were manufactured on top of the AZ40-XT layer with SU-8 3050 Photoresist (MicroChem), and they had a height of 50 µm. The mold for the control circuit was manufactured with SU-8 3025 Photoresist, and all channels had a rectangular profile and a height of 20 µm. Molds were coated with C4F8 following published protocols (51). Sylgard 184 (Dow Corning) was used to replicate PDMS chips from the molds with a process described before (52). Flush protocols for automation of the cell culture process, temporal and chemical induction of the adipogenesis, and the subsequent PLA were controlled through custom scripts that were written in MATLAB (Mathworks) and are described in detail in SI Text.
Cell Culturing.
Primary hASCs were purchased from ZenBio Inc. (lot ASC031903ABD). Adult stem cells were isolated from human s.c. adipose tissue by flow cytometry using CD105 (93.1%) and CD44 (91.7%) as positive surface markers and CD45 (0.2%) and CD31 (undetectable) as negative surface markers. Cells were cultured in EasYFlasks Nunclon Surface (Thermo Scientific) 75-cm2 flasks at 5% (vol/vol) CO2 and 37 °C. As growth medium, we used DMEM/F-12 media substituted with 10% (vol/vol) FCS and 50 U/mL penicillin/streptomycin. Cells were obtained at cell passage 2 and used until passage 5. Adipogenesis of the hASCs was chemically induced with a differentiation medium containing 100 ng/mL insulin, 1 µM dexamethasone, 17 µM d-pantothenic acid, 33 µM biotin, and 15 mM Hepes (pH 7.4) solved in growth medium.
mPLA.
The design of the mPLA is given in detail in Fig. S5, and all of the used oligonucleotide sequences are given in Table S2. The on-chip PLA is described elsewhere (41). In short, hASCs were fixed after 14 d of culturing with 4% (wt/vol) paraformaldehyde in PBS for 12 min. After permeabilizing with 0.1% Triton X-100 in PBS and blocking with blocking solution (250 μg/mL BSA, 2.5 μg/mL sonicated salmon sperm DNA, 5 mM EDTA, 50 mM Tris-buffered saline, 0.05% Tween 20), labeled antibodies (5 μg/mL) were introduced in the cell culture chamber of the mLSI chip and incubated for 4 h. Antibody labeling was done following published protocols (53). Then, the bridging connectors (125 nM) and T4 ligase (0.05 U/μL) were perfused over the cells and incubated for 30 min at 40 °C. In situ-assembled PLA templates were amplified with phi29 polymerase (0.125 U/μL) for 1.5 h at 37 °C.
Table S2.
Transcription factor antibodies used in the Western blot along with supplier information
| Targets | Company | Catalog no. |
| C/EBPβ | Santa Cruz Biotechnology | sc-150 |
| C/EBPα | Santa Cruz Biotechnology | sc-166258 |
| PPARγ | Santa Cruz Biotechnology | sc-7196 |
| β-Actin | Cell Signaling Technology | 8H10D10 |
Western Blot.
hASCs cultured in six-well plates in growth medium were chemically induced for adipogenesis at defined time points with differentiation medium. Medium exchange was performed every 48 h. After 14 d, cells were twice washed with ice-cold PBS and lysed in RIPA Lysis Buffer (Bio-Rad) on ice. Lysates were centrifuged at 16,000 × g for 20 min at 4 °C. The protein concentration of the supernatant was determined with a BCA Assay (Novagen). Gel electrophoresis was performed with cell lysates containing an equal amount of protein (7.5 µg) and Mini-Protean TGX Precasted Gels (Bio-Rad) according to the description of the vendor. Thereafter, the proteins were transferred from the gel to a nitrocellulose membrane using the Bio-Rad Trans-Blot Turbo Transfer System (Bio-Rad). The membranes were blocked in 5% (wt/vol) milk blocking solution and incubated overnight at 4 °C with the primary antibodies. Blotted proteins were detected with an HRP System (Pierce), and secondary antibodies were coupled to HRP (Thermo Fisher Scientific) following the protocol of the vendor.
Antibodies.
For Western blot analysis and PLA, we purchased the antibodies Raptor (PA5-20126), mTOR (PA5-34663), and PRAS40 (PA5-34565) from Thermo Fisher Scientific; p(Ser-2448)-mTOR (5536) from Cell Signaling; Rictor (H00253260-M01) from Abnova; RagB (169101), RagD (169104), and Rheb (6341) from Santa Cruz Biotechnology; and secondary anti-rabbit IgG (Ab7056) from Abcam.
Data Acquisition and Evaluation.
Fluorescence signals from the cell samples were acquired in two iterative rounds on an AxioObserver (Zeiss). In the first round, the three fluorescence signals of the PLA probes (Atto488, Cy3, and Atto647) and the counterstained nucleus (Hoechst 33342) were recorded. In the following step, the cytoplasm and LD were counterstained with Phalloidin-Atto647 (Santa Cruz Biotechnology) and BODIPY493 (Sigma Aldrich), respectively. We then recorded the fluorescence images for the cytoplasm and LD in addition to the nucleus signal. The nucleus signal was used for image registration of the first and second image datasets. Counterstains for the cytoplasm and LD are around 100–1,000 times brighter than the PLA signals, and thus, PLA signals did not influence the LD analysis. All images were processed (i.e., cell and LD segmentation, PLA dot finding, counting, and location of PLA dots in single cells) (SI Text) by custom MATLAB scripts.
SI Text
mLSI Chip Operations.
Cell differentiation and PLAs were carried out on a microfluidic device with 128 separately addressable cell culture chambers as shown in Fig. 1A and Fig. S1A. The chip consists of two PDMS layers, where the upper contained the control channel network (red in Fig. S1A) and the lower contained the fluidic channel network (blue in Fig. S1A). Cross-sections of control and flow channels are separated by thin membranes of PDMS. The 32 control channels can be pressurized individually by a pressure control system with solenoid valves, which deflects the membranes into the flow channels. The pneumatic valves in push-down configuration can, therefore, open and close the flow channels and direct the flow of reagents in the bottom layer. The cell chambers are arranged in two blocks of 64 cell chambers, each with a length of 2.6 mm and width of 0.32 mm (Fig. 1A). Reagents can be plugged into 20 different inlet ports (Fig. S1A) and flushed through any of 128 cell chambers. However, it is also possible to run two mirrored assays on the same chip without cross-contamination. In this case, inlets 1–8 can be used for the bottom row and inlets 9–16 can be used for the top row of cell chambers exclusively. Mixing of reagents is prevented by a closed separator valve. Cells get transferred on the chip through inlet ports Cell_T and Cells_B (Fig. S1A). To clean the inlet multiplexer and flow channels, liquid can be pressed out through waste port Out3. Reagents routed through cell chambers leave the chip by outlets Out1 and Out5 for top and bottom, respectively (Fig. S1B). These outlets can additionally be cleaned by introducing Trypsin through Clean1 and Clean2. Furthermore, the cell chamber multiplexer can be cleaned through two cleaning channels by closing the cell chambers and opening outlets Out2 and Out4 (Fig. S1C).
Before starting the cell culture process, microfluidic chips were placed in an incubation chamber at 37 °C and 5% (vol/vol) CO2. Of note, all introduced fluids were compressed with pressure-adjusted CO2 [5% (vol/vol)] and oxygen level [21% (vol/vol)] at given flow rates with a gas mixer; 10 μg/mL fibronectin solution was loaded into Tygon tubing and plugged into inlet 17. Each cell chamber was automatically flushed for 20 s and incubated for 1 h. The chambers were then preconditioned with growth medium before cells were added. For that purpose, medium was connected to inlet ports 1 and 9, and all top and bottom chambers were flushed in parallel from left (chamber 1) to right (chamber 64) for 20 s. Cells were harvested, resuspended to a concentration of 5 × 106 cells per 1 mL, and manually flushed through cell inlets (Cells_T and Cells_B) into the cell culture chambers until they reached confluency at 220 ± 55 cells per chamber. To ensure equal flow through all chambers, adhered cells in the flow channels were removed by trypsinization by use of the cleaning channels (Fig. S1C). hASCs were cultured for 4 d within growth medium. The medium was replaced every 60 min for 15 s at 0.46 µL/min. Differentiation medium was then plugged in at inlets 2 and 10. Starting with chambers 1 and 2 (top and bottom), every 12 h, two additional cell culture chambers were induced with differentiation medium. At this time point, growth and differentiation media are used sequentially for feeding. To prevent cross-contamination of the two media, the entire multiplexer controlling the addressability of the cell culture chambers was cleaned with the corresponding medium before sending into the cell culture chambers (Fig. S1C). After 14 DOD, 4% (vol/vol) paraformaldehyde (PFA) was connected to inlets 21 and 22, and cell culture chambers were flushed with PFA at high pressure of 1.10 µL/min for 10 s. Fixation was finished by subsequent removal of PFA with PBS (inlets 3 and 11) after 12 min. Cells were permeabilized with 0.1% Triton X-100 (inlets 4 and 12) for 12 min and blocked with blocking solution for 30 min (inlets 5 and 13). Primary antibodies with conjugated adaptors for PLA were diluted to a concentration of 5 μg/mL each in antibody stabilizer (Sigma) and flushed from inlets 6 and 14 into the cell culture chambers. Every 4 h of the total 12-h antibody incubation time, the solution was refreshed. Next, all cell chambers were cleaned with Tris-buffered saline (TBS)-Tween twice and incubated with conjugated secondary antibody for 1 h at 37 °C (if necessary) followed by ligation of connectors (inlet 18) for 1 h at 40 °C. After additional washing with TBS-Tween, the phi29 polymerase mix was applied for 1.5 h at 37 °C (inlet 19) followed by washes with TBS (inlets 8 and 16). In the final steps of the assay, cells were stained with detection probe, DAPI, and phalloidin for 30 min (inlet 20) and washed again with TBS to remove residual fluorescent probes.
mPLA Orthogonality Test on Matrix Chip.
For screening the orthogonality of the mPLA (Fig. 2), we used a successor mLSI chip published and explained elsewhere (41). The chip was refined with optimized layout regarding fluid resistance, enlarged cell culture chambers (1-mm diameter), and cross-contamination–free flushing (Fig. S6A). After cell seeding on chip, the hASCs were cultured with growth medium overnight. Next, the mPLA assay was performed following the same protocol described in SI Text, mLSI Chip Operations. Fig. S6B illustrates the flush direction used to implement the antibody titration experiments within an mPLA on chip.
Features Investigated for Parameterizing PLA Signal Distribution.
For the calculation of the following PLA dot parameters, the cell nuclei outlines and cell outlines were segmented within all PLA images. Additionally, all nuclei centroids were determined. In general, all distance measurements were normalized to the largest distance between nuclear centroid and the outline of the given cell.
(i) The mean distance of PLA signals to the nuclear centroid, Dc, in cell population under the same condition was calculated accordingly:
| [S1] |
where N denotes the number of cells, p is the number of PLA dots in a given cell, and dk is the normalized distance of each PLA dot to the nucleus centroid.
(ii) The mean PLA dot to dot distance, Dd, within a cell population under the same condition was calculated according to
| [S2] |
where da is the normalized distance to every other PLA dot in a given cell.
(iii) The mean of the SD of the PLA dot distance to nuclear centroid in a cell population under the same condition, STDc, was calculated according to
| [S3] |
where STDn is the SD of the PLA dot distances to the nuclear centroid in a given cell. It follows
| [S4] |
where is the mean normalized distance of the PLA dots to the nuclear centroid in a single cell, which is calculated by
| [S5] |
(iv) The mean of the SD of the PLA dot to dot distances in a cell population under the same conditions, STDd, was calculated according to
| [S6] |
where STDi is the SD of the PLA dot to dot distances in a given cell. It follows
| [S7] |
where and denote the mean PLA dot to dot pixel distances for each single PLA dot and the mean PLA dot to dot pixel distances of all PLA dots in a single cell, respectively, which are calculated by Eqs. S8 and S9, respectively:
| [S8] |
and
| [S9] |
(v) The SD of the SD of the PLA dot distance to nuclear centroid in a cell population under the same condition, STDss, was calculated according to
| [S10] |
(vi) The SD of the SD of the PLA dot to dot distances in a cell population under the same conditions, STDdd, was calculated according to
| [S11] |
All of the features derived for each condition were normalized by z scoring against features calculated based on randomized pixels in the same cells. For this ■■■, the same number of pixels as found PLA dots in the given cell was arbitrarily placed into the cell and iterated for 100 times without replacement. For example, 10 PLA dots were identified in the given cell, and we placed 100 × 10 random pixels into the same cell without replacement and each time, calculated the parameters i–vi.
Then, the mean value () and SD (σ) of the features for 100 iterations were calculated. All of the features derived from experiment (x) were z-scored following the equation:
| [S12] |
Acknowledgments
This study was supported by Excellence Initiative of the German Federal and State Governments EXC-294 and German Research Foundation Emmy-Noether Grant ME3823/1-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601207113/-/DCSupplemental.
References
- 1.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444(7121):847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pittenger MF, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Haslam DW, James WPT. Obesity. Lancet. 2005;366(9492):1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 4.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444(7121):840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 5.Hauner H, et al. Promoting effect of glucocorticoids on the differentiation of human adipocyte precursor cells cultured in a chemically defined medium. J Clin Invest. 1989;84(5):1663–1670. doi: 10.1172/JCI114345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott MA, Nguyen VT, Levi B, James AW. Current methods of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 2011;20(10):1793–1804. doi: 10.1089/scd.2011.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 8.Csete M, et al. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. J Cell Physiol. 2001;189(2):189–196. doi: 10.1002/jcp.10016. [DOI] [PubMed] [Google Scholar]
- 9.Wiederer O, Löffler G. Hormonal regulation of the differentiation of rat adipocyte precursor cells in primary culture. J Lipid Res. 1987;28(6):649–658. [PubMed] [Google Scholar]
- 10.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 11.Zuk PA, et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13(12):4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 14.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 15.Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203(4):563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10(3):457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 17.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6(11):1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 18.Bell A, Grunder L, Sorisky A. Rapamycin inhibits human adipocyte differentiation in primary culture. Obes Res. 2000;8(3):249–254. doi: 10.1038/oby.2000.29. [DOI] [PubMed] [Google Scholar]
- 19.El-Chaâr D, Gagnon A, Sorisky A. Inhibition of insulin signaling and adipogenesis by rapamycin: Effect on phosphorylation of p70 S6 kinase vs eIF4E-BP1. Int J Obes Relat Metab Disord. 2004;28(2):191–198. doi: 10.1038/sj.ijo.0802554. [DOI] [PubMed] [Google Scholar]
- 20.Catania C, Binder E, Cota D. mTORC1 signaling in energy balance and metabolic disease. Int J Obes. 2011;35(6):751–761. doi: 10.1038/ijo.2010.208. [DOI] [PubMed] [Google Scholar]
- 21.Chakrabarti P, English T, Shi J, Smas CM, Kandror KV. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59(4):775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci USA. 2010;107(8):3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugihara H, Yonemitsu N, Miyabara S, Toda S. Proliferation of unilocular fat cells in the primary culture. J Lipid Res. 1987;28(9):1038–1045. [PubMed] [Google Scholar]
- 24.Darlington GJ, Ross SE, MacDougald OA. The role of C/EBP genes in adipocyte differentiation. J Biol Chem. 1998;273(46):30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 25.Fredriksson S, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20(5):473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 26.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korolchuk VI, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13(4):453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Battich N, Stoeger T, Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Methods. 2013;10(11):1127–1133. doi: 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
- 30.Inoki K, Li Y, Xu T, Guan K-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17(15):1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280(25):23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- 32.Vander Haar E, Lee S-I, Bandhakavi S, Griffin TJ, Kim D-H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9(3):316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 33.Kubo Y, Kaidzu S, Nakajima I, Takenouchi K, Nakamura F. Organization of extracellular matrix components during differentiation of adipocytes in long-term culture. In Vitro Cell Dev Biol Anim. 2000;36(1):38–44. doi: 10.1290/1071-2690(2000)036<0038:OOEMCD>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 34.Melin J, Quake SR. Microfluidic large-scale integration: The evolution of design rules for biological automation. Annu Rev Biophys Biomol Struct. 2007;36(1):213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 35.Tenstad E, Tourovskaia A, Folch A, Myklebost O, Rian E. Extensive adipogenic and osteogenic differentiation of patterned human mesenchymal stem cells in a microfluidic device. Lab Chip. 2010;10(11):1401. doi: 10.1039/b926738g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giobbe GG, et al. Functional differentiation of human pluripotent stem cells on a chip. Nat Methods. 2015;12(7):637–640. doi: 10.1038/nmeth.3411. [DOI] [PubMed] [Google Scholar]
- 37.Fink T, et al. Induction of adipocyte-like phenotype in human mesenchymal stem cells by hypoxia. Stem Cells. 2004;22(7):1346–1355. doi: 10.1634/stemcells.2004-0038. [DOI] [PubMed] [Google Scholar]
- 38.Cheong HH, Masilamani J, Phan TT, Chan SY. Cord lining progenitor cells: Potential in vitro adipogenesis model. Int J Obes. 2010;34(11):1625–1633. doi: 10.1038/ijo.2010.86. [DOI] [PubMed] [Google Scholar]
- 39.Leuchowius K-J, et al. Parallel visualization of multiple protein complexes in individual cells in tumor tissue. Mol Cell Proteomics. 2013;12(6):1563–1571. doi: 10.1074/mcp.O112.023374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshiro N, et al. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells. 2004;9(4):359–366. doi: 10.1111/j.1356-9597.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- 41.Blazek M, et al. Proximity ligation assay for high-content profiling of cell signaling pathways on a microfluidic chip. Mol Cell Proteomics. 2013;12(12):3898–3907. doi: 10.1074/mcp.M113.032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blazek M, Santisteban TS, Zengerle R, Meier M. Analysis of fast protein phosphorylation kinetics in single cells on a microfluidic chip. Lab Chip. 2015;15(3):726–734. doi: 10.1039/c4lc00797b. [DOI] [PubMed] [Google Scholar]
- 43.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280(27):25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 44.Kim JE, Chen J. regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53(11):2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 45.Singh R, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119(11):3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ricoult SJH, Manning BD. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013;14(3):242–251. doi: 10.1038/embor.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sembongi H, et al. Distinct roles of the phosphatidate phosphatases lipin 1 and 2 during adipogenesis and lipid droplet biogenesis in 3T3-L1 cells. J Biol Chem. 2013;288(48):34502–34513. doi: 10.1074/jbc.M113.488445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zechner R, et al. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15(3):279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Settembre C, Ballabio A. Lysosome: Regulator of lipid degradation pathways. Trends Cell Biol. 2014;24(12):743–750. doi: 10.1016/j.tcb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288(5463):113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 51.Silva Santisteban T, Zengerle R, Meier M. Through-holes, cavities and perforations in polydimethylsiloxane (PDMS) chips. RSC Advances. 2014;4(89):48012–48016. [Google Scholar]
- 52.McDonald JC, Whitesides GM. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc Chem Res. 2002;35(7):491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 53.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]