Abstract
Remote island archipelagos offer superb opportunities to study the evolution of community assembly because of their relatively young and simple communities where speciation contributes to the origin and evolution of community structure. There is great potential for common phylogeographic patterns among remote archipelagos that originate through hotspot volcanism, particularly when the islands formed are spatially isolated and linearly arranged. The progression rule is characterized by a phylogeographic concordance between island age and lineage age in a species radiation. Progression is most likely to arise when a species radiation begins on an older island before the emergence of younger islands of a hotspot archipelago. In the simplest form of progression, colonization of younger islands as they emerge and offer appropriate habitat, is coincident with cladogenesis. In this paper, we review recent discoveries of the progression rule on seven hotspot archipelagos. We then discuss advantages that progression offers to the study of community assembly, and insights that community dynamics may offer toward understanding the evolution of progression. We describe results from two compelling cases of progression where the mosaic genome may offer insights into contrasting demographic histories that shed light on mechanisms of speciation and progression on remote archipelagos.
Keywords: progression rule, speciation, priority effect, community assembly, radiation zone
Evolutionists are drawn to the study of island biotas for their unique species (1–5), as refuges of extant, “relict” organisms (6, 7), and most famously, for their displays of adaptive radiation (8, 9). Likewise, ecologists have long recognized the value of islands as microcosms of the processes of community assembly (10, 11). In recent years, there has been growing interest in combining these elements to study the evolution of community assembly, with particular focus on islands within (and beyond) the “radiation zone” (10), where in situ speciation can be a major contributor to the origin of ecological communities (12, 13). Under such circumstances, rules for community assembly can be illuminated by comparative phylogeographic approaches, revealing common evolutionary histories of codistributed, endemic taxa both within and between island archipelagos.
Remote island archipelagos offer relatively simple arenas for the evolutionary dynamics of community assembly because they are generally small in size and are often characterized by spatial isolation beyond the probable dispersal range of most organisms (14, 15). As G. G. Simpson (16) hypothesized, the probability of colonization should decline with increasing remoteness of an island from a mainland source pool, which he aptly named “sweepstakes” dispersal. Moreover, remote island archipelagos are veritable specks of land in a wide ocean world, again reducing the probability of colonization. Evidence of these hypothesized filters to colonization shows that remote islands frequently harbor disharmonious biotas (where there is an imbalance of taxonomic representation compared with mainland source pools). Moreover, the degree of disharmony increases with increasing distance from a probable source pool offering additional evidence of these spatial sieves and the potential for a relatively simpler community assembly (17). Together with small size and extreme isolation, the assemblage of biotas may be further reduced in remote oceanic archipelagos because they are generally formed without life due to the nature of their geological origins (18). In contrast, species and community evolution in island systems in close proximity to continental species pools (e.g., continental “fragment” islands) (17) can be extraordinarily complex (19) in particular because they are often formed with a full complement of species and have more frequent connections with source pools.
Along with the effects of small size, spatial isolation, and dispersal origins on community assembly of remote islands, common phylogeographic patterns may further enhance the “laboratory” for study of the evolution of community assembly. The evidence of disharmony, and potential for geographically diverse source pool origins (17–21) might lead us to expect somewhat haphazard phylogeographic patterns in taxa distributed across multiple remote archipelagos (18), for example, as appears to be the case for the spider genus Tetragnatha across Polynesia (22). However, once a lineage has initially established within a remote archipelago, it is largely cut off from its source population (qualified, of course, on the dispersal ability of the taxon). Thereafter, colonization among the constituent islands is considerably more predictable, and geological history at this more restricted geographic scale inspires hypotheses of common phylogeographic patterns among codistributed taxa (23, 24).
Many remote archipelagos are volcanic in origin and sometimes exist in a linear age progression. These “hotspot” archipelagos are formed by molten lava rising from relatively fixed spots on the sea floor. Plate tectonic drift creates a conveyor belt motion that carries newly formed islands away from the hotspot location in a consistent direction at a steady rate, the mechanism that generates the linear age progression (25, 26). These geological origins create a geographical and chronological context for hypotheses of phylogeographic congruence among codistributed taxa that track the ages of the islands, termed the “progression rule” (Figs. 1 and 2).
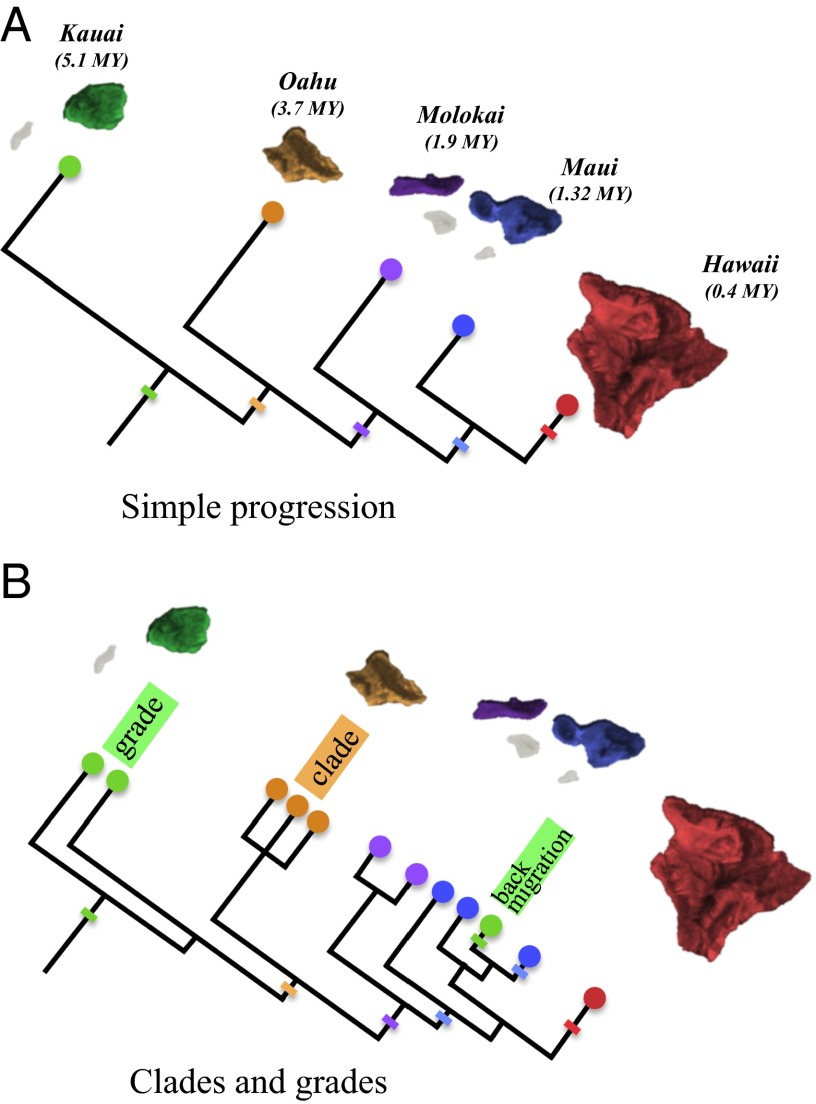
Fig. 1.
Hypothetical progression patterns. (A) A simple progression pattern with cladogenesis coincident with interisland migration. Hatch marks show interisland migrations. (B) A complex progression showing clades, grades, and back migration. Hatch marks show interisland migrations. Colors correspond to islands: green, Kauai; orange, Oahu; purple, Molokai; blue, Maui; and red, Hawaii.
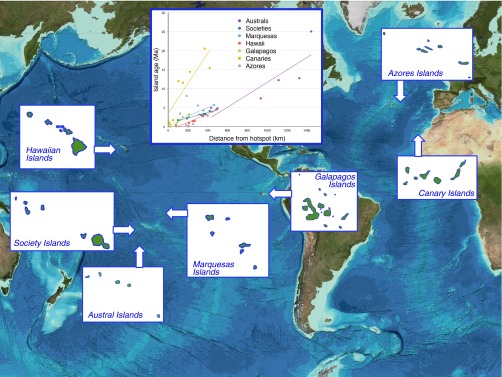
Fig. 2.
Map of seven archipelagoes that show a variable geological age chronology. (Inset) Relationship between island age and distance from the hotspot for each archipelago (estimated in the case of the Azores) (106). This reflects the match of the geographical arrangement of islands to their age, with tighter relationships in archipelagoes such as that of Hawaii, in which islands are produced in a conveyor belt fashion. The figure includes only islands of area >10 km2, as smaller islands tend to be atolls with communities limited to coastal strand. Trend lines are shown (except for the Azores in which there is no clear trend line). Background image reproduced from the GEBCO world map 2014, www.gebco.net.
Common evolutionary histories of taxa, as well as deviations, can have a profound impact on our understanding of community assembly in the radiation zone. In this paper, we examine the progression rule to gain insight into the process of community assembly. In the following sections, we first present a brief overview of the progression rule from exemplar remote oceanic archipelagos. We then consider a simple question: can the progression rule be explained on the basis of island hot spot theory alone? We evaluate recent studies of progression from remote oceanic islands, highlighting two cases where contrasting phylogeographic histories are implicated within the same species radiations. The potential for distinct genetic histories held within the same demographic taxon, a phenomenon now widely recognized (27), illuminates the link between ecological community assembly and evolutionary assembly in isolated archipelagos.
The Progression Rule in Comparative Island Phylogeography
In the context of islands, a progression rule (or pattern) refers to a phenomenon of phylogeographic concordance with island age, whereby older lineages map to older islands within an archipelago, and younger lineages map to progressively younger islands in that system (Fig. 1A) (23). Progression is hypothesized to result from the early colonization by a lineage when older islands existed but younger islands had not yet formed. Subsequent colonization occurs as new islands form. In his seminal work on phylogenetic theory, Hennig (28) proposed the progression rule as a consequence of a general speciation mechanism, wherein species ranges consist of an ancestral, central portion within an older geographic area, and a derived, peripheral portion in a younger geographic area. Progression becomes evident in the phylogeny once speciation occurs; the ancestral, central portion splits from the derived, peripheral portion of the ancestral species. Thus, the ancestral lineage is concordant with older area, whereas the derived lineage is concordant with newer geological areas (Fig. 1A). Hennig’s (28) model would not predict a progression to the extent of present day discussions, unless the next new habitat that forms is spatially closer to the newest of the previous habitats.
Oceanic hotspot archipelagos offer, at a minimum, the starting conditions where progression could begin. Indeed, at least superficially, progression is not difficult to explain, as colonists of younger islands are most likely to come from spatially proximate, older islands. However, there is ample variation among taxonomic lineages in both the degree and form of progression (see below). In general, progression is more likely to evolve with (i) increasing spatial isolation of the archipelago, (ii) increasing spatial linearity of the islands, and (iii) enough dispersal to ensure colonization, such that when new habitat arises after the emergence of a new island, propagules are spatially poised to colonize from the next oldest habitat.
Variation in the starting conditions for progression may arise for a variety of reasons (we treat the persistence of progression below). For example, some archipelagos are less isolated from potential source areas (relative to the dispersal ability of the organisms) than others, resulting in repeated colonizations by the same lineages, producing conditions for either biotic turnover or anagenesis, rather than the within-archipelago cladogenesis associated with the progression pattern (15). Effectively, too much dispersal prevents the development of a progression pattern. In addition, although less likely, some organisms may be dispersal limited, and while habitat is available, they do not disperse to it predictably. In addition, some archipelagos do not show a linear spatial arrangement concordant with age, resulting in potential within archipelago colonizations from islands of mixed age. This nonlinear age arrangement would interfere with the older to younger colonization and ensuing phylogeographic pattern characteristic of progression. Likewise, we would not predict a progression for taxa arriving after the current major islands came into existence.
Progression can be complex; Funk and Wagner (29) discuss a variety of reasons for this complexity, providing a general framework for interpreting more complex patterns. A strict progression pattern is the simplest, where all cladogenic events arise coincident with migration to newly arisen volcanoes along a linear chronological island sequence (Fig. 1A). Funk and Wagner (29) discuss this pattern with reference to Hawaiian endemics, where it is rare for taxa to conform exactly to this simple manifestation of the progression rule, although some taxa do illustrate the progression perfectly (30).
Another more common pattern shows older to younger island colonization followed by some degree of within island speciation (a pattern of progressive clades; Fig. 1B (29). This progression pattern would arise when the interisland colonists from an older island migrate early in the history of the extant, older island clade. Progressive clades can give way to progressive grades (Fig. 1B), where a formerly monophyletic clade within an older island becomes paraphyletic when a subsequent colonization to a younger island occurs by a terminal taxon within that clade. In addition to these basic progression histories, instances of back-migration (migrations from younger to older islands) may also occur (Fig. 1B).
Armed with some understanding of the complexities of the progression pattern, we now examine progression in some well-recognized hotspot archipelagoes, with particular focus on the Pacific Ocean (Fig. 2).
Hawaiian Islands.
Progression has been established most famously as a general pattern in the Hawaiian archipelago (Fig. 2), where the formation of new, colonizable habitats does follow such a chronological sequence (23). Hawaii’s indigenous biota is highly disharmonious, and chance colonization has undoubtedly played a large part in the initial establishment of the native diversity that we see today. However, within the Hawaiian Islands, a taxonomically broad expression of the progression rule is evident, including examples from plants [e.g., Hawaiian silverswords (31); Schiedea (32); Psychotria (33); and lobeliads (34)], insects [e.g., Megalagrion damselflies (35); Laupala crickets (36); Banza katydids (37); Hyposmocoma moths (38); and picture-winged Drosophila (30, 39)], land snails [e.g., succineid species (40)], spiders [e.g., Orsonwelles (41)], and birds (42), among many others. Funk and Wagner (23) documented progression in 18 of 25 endemic lineages. More recent reviews (43, 44) have further summarized this impressive phylogeographic result.
Austral Islands.
The Austral Archipelago (Fig. 2) is considered geologically continuous with the Cook Islands (located to the northwest), which together were formed from repeated episodes of volcanism at several sites (45, 46). Like the other Pacific hotspots, they are sequentially ordered from southeast to northwest by increasing age, although there has been secondary volcanic activity in the older islands, making them older than other Pacific hotspots (26) (Fig. 2, Inset, and Table S1). Among independent lineages of spiders, in particular the crab spider Misumenops rapaensis (47), the orb web spider Tangaroa tahitiensis (Uloboridae) (48), and Rhyncogonus weevils (49), a similar pattern of sequential colonization of islands is emerging, with large genetic distances between island populations. Thus, despite the modest extent and topography of the Austral islands and the widespread and generalist nature of their taxa, all studies to date show strong support for a progression rule in this archipelago.
Table S1.
Remote archipelago life history traits
| Archipelago/islands | Longitude/latitude | Distance from hot spot (km) | Age (range) (My) | Rate plate movement (cm/y) | Area (km2) | Altitude (m) | Nearest continent |
| Marquesas Islands | |||||||
| Eiao | 8.00°S/140.68°W | 465 | 5.75 ± 4.98 | 7 | 40 | 578 | South America |
| Nuku Hiva | 8.86°S/140.10°W | 355 | 4.34 ± 2.99 | 7 | 345 | 1,227 | South America |
| Ua Huka | 8.92°S/139.55°W | 312 | 3.24 ± 1.36 | 7 | 82 | 884 | South America |
| Ua Pou | 9.40°S/140.08°W | 300 | 5.61 ± 1.78 | 7 | 105 | 1,203 | South America |
| Hiva Oa | 9.76°S/139.00°W | 210 | 2.55 ± 1.66 | 7 | 318 | 1,276 | South America |
| Tahuata | 9.92°S/139.08°W | 200 | 2.09 ± 1.82 | 7 | 61 | 1,050 | South America |
| Fatu Hiva | 10.48°S/138.65°W | 123 | 2.55 ± 1.33 | 7 | 85 | 960 | South America |
| Society Islands | |||||||
| Maupiti | 16.45°S/152.25°W | 488 | 4.60 ± 3.94 | 7 | 12 | 380 | Australia |
| Bora Bora | 16.45°S/151.87°W | 420 | 4.15 ± 3.12 | 7 | 29 | 727 | Australia |
| Tahaa | 16.62°S/151.49°W | 380 | 3.30 ± 2.60 | 7 | 91 | 590 | Australia |
| Raiatea | 16.82°S/151.43°W | 360 | 3.4 ± 2.4 | 7 | 173 | 1,017 | Australia |
| Huahine | 16.72°S/151.10°W | 335 | 3.15 ± 1.95 | 7 | 75 | 669 | Australia |
| Moorea | 17.57°S/150.00°W | 190 | 2.25 ± 1.20 | 7 | 142 | 1,207 | Australia |
| Tahiti | 17.35°S/149.30°W | 100 | 1.75 ± 0.75 | 7 | 1,040 | 2,241 | Australia |
| Austral Islands | |||||||
| Rimatara | 22.67°S/153.42°W | 1445 | 25.1 ± 4.9 | 7 | 18 | 95 | Australia |
| Rurutu | 22.47°S/151.33°W | 1325 | 12.8 ± 0.3 | 7 | 24 | 390 | Australia |
| Tubuai | 23.38°S/149.45°W | 1115 | 12.3 ± 8.1 | 7 | 45 | 402 | |
| Raivavae | 23.86°S/147.67°W | 935 | 7.5 ± 5.7 | 7 | 18 | 437 | |
| Rapa | 26.60°S/144.33°W | 435 | 5.0 ± 4.1 | 7 | 40 | 650 | South America |
| Hawaiian Islands | |||||||
| Kauai | 22.50° N/159.50° W | 500 | 4.7–5.2 | 7 | 1,435 | 1,598 | North America |
| Oahu | 21.50° N/157.95° W | 353 | 3.4–2.2 | 7 | 1,583 | 1,227 | North America |
| Molokai | 21.15° N/157.00° W | 268 | 1.8–1.3 | 7 | 678 | 1,515 | North America |
| Lanai | 0.85° N/156.95° W | 250 | 1.50 | 7 | 358 | 1,027 | North America |
| Maui | 20.75° N/156.25° W | 210 | 1.3–0.8 | 7 | 1,903 | 3,052 | North America |
| Hawaii | 19.50° N/155.50° W | 79 | 0.2–0.6 | 7 | 10,434 | 4,169 | North America |
| Galapagos Islands | |||||||
| Espanola | 1.38° S/89.67° W | 205 | 3.0–3.5 | 7 | 60 | 206 | South America |
| San Cristobal | 0.87° S/89.44° W | 215 | 2.4–4.0 | 7 | 557 | 730 | South America |
| Santa Cruz | 0.62°S/ 90.37°W | 110 | 1.1–2.3 | 7 | 986 | 864 | South America |
| Floreana | 1.31° S/90.43° W | 125 | 1.5–2.3 | 7 | 173 | 640 | South America |
| Santiago | 0.26° S/90.72° W | 90 | 0.8–1.4 | 7 | 572 | 905 | South America |
| Isabela | 0.83° S/91.14° W | 20 | 0.5–0.8 | 7 | 4,670 | 1,707 | South America |
| Fernandina | 0.41° S/91.48° W | 20 | 0.03–0.07 | 7 | 642 | 1,476 | South America |
| Canary Islands | |||||||
| Lanzarote | 29.05° N/13.59° W | 420 | 15.50 | 1.2 | 846 | 670 | Africa |
| Fuerteventura | 28.40° N/14.01° W | 370 | 20.00 | 1.2 | 1,660 | 812 | Africa |
| Gran Canaria | 27.92° N/ 15.55° W | 220 | 14–16 | 1.2 | 1,560 | 1,949 | Africa |
| Tenerife | 28.29° N/ 16.63° W | 150 | 11.6–7.5 | 1.2 | 2,034 | 3,718 | Africa |
| La Gomera | 28.10° N/ 17.22° W | 105 | 11 ± 9 | 1.2 | 370 | 1,487 | Africa |
| La Palma | 28.71° N/ 17.91° W | 50 | 2.00 | 1.2 | 708 | 2,426 | Africa |
| El Hierro | 27.73° N/ 18.02° W | 10 | 1.10 | 1.2 | 269 | 1,501 | Africa |
| Azores Islands | |||||||
| São Miguel | 37.78° N/ 25.50° W | 156 | 4.01 | 759 | 1,105 | Europe | |
| Pico | 38.46° N/ 28.32° W | 0 | 0.25 | 446 | 2,351 | Europe | |
| Terceira | 38.72° N/ 27.22° W | 100 | 3.52 | 403 | 1,021 | Europe | |
| São Jorge | 38.67°N/ 28.12°W | 17 | 0.55 | 246 | 1,053 | Europe | |
| Faial | 38.59° N/ 28.70° W | 7 | 0.73 | 173 | 1,043 | Europe | |
| Flores | 39.45° N/ 31.19° W | 160 | 2.16 | 143 | 886 | Europe | |
| Santa Maria | 36.98° N/ 25.11° W | 240 | 8.12 | 97 | 587 | Europe | |
| Graciosa | 39.05° N/ 28.01° W | 30 | 2.5 | 62 | 402 | Europe | |
| Corvo | 39.70° N/ 31.11° W | 168 | 0.71 | 17 | 718 | Europe |
Society Islands.
Age progression within the Society Islands is in good agreement with the fixed hotspot hypothesis (26), although the islands are considerably smaller in size than those of the Hawaiian chain (Fig. 2 and Table S1). However, there is considerable topographic diversity. Among insects, blackflies in the genus Simulium (50, 51) show no evidence of a progression rule, which may be because of extinctions of habitat-specialized species on the older islands, due to loss of habitat. Likewise, among weevils in the genus Rhyncogonus, there appear to have been multiple independent colonizations of the island of Tahiti from neighboring island chains and no evidence of progression from older to younger islands (49, 52). A similar conclusion results from phylogenetic analyses of Polynesian reed-warblers (genus Acrocephalus; 53). One reason that has been suggested is that there are ancient islands in close proximity to the Societies that may have served as a source of propagules (48).
Marquesas Islands.
The chronological arrangement of the Marquesas Islands is not strictly regular (26). Nevertheless, among birds, an approximate progression from older to younger islands is found in the genus Pomarea (54). Among spiders, the endemic taxa fall into a northern and southern lineage (55), consistent with the progression, although without strong phylogenetic support. A similar pattern of northern and southern lineages is found in Rhyncogonus weevils (52) and partulid land snails of the genus Samoana (56).
Galapagos.
The Galapagos Islands, although more geographically clustered than other Pacific hotspots (57), are still arranged chronologically (Fig. 2). This spatial arrangement implies a potential diversification sequence from southeast to northwest, paralleling the geological formation of the archipelago’s islands (58). Some lineages in the Galapagos show a very clear progression from older to younger islands (59), well illustrated by the Galapagos giant tortoise (Geochelone nigra), in which both the species-level phylogeographic pattern based on mtDNA data and the pattern of lineage sorting suggest diversification in parallel with the island geological formation (60, 61). One of the two lineages of Galapagos lava lizards (Microlophus) (62, 63) has also diversified in concert with the geological formation of island clusters of similar age (64). Among marine iguanas (Amblyrhynchus cristatus), mtDNA shows population differentiation concordant with geographical isolation of populations across the archipelago, a result largely in agreement with nuclear microsatellite data (65). Among birds, Galapagos mocking birds also appear to follow the progression rule (66), although Darwin’s finches show limited evidence of diversification closely associated with the geological formation of the islands (2). A progression pattern has been inferred for Galapagos bulimulid land snails (67). In contrast, insects including Galapagos flightless weevils (Galapaganus) (68) and the microlepidopteran genus Galagete (69) do not follow the progression rule. Evidently, many Galapagos terrestrial faunal groups follow the progression rule, the major exceptions coming from more vagile lineages (Galapagos finches, Galagete lepidopterans, and Galapaganus weevils) (59). Interestingly, in no situation is adaptive radiation associated with progression.
Canary Islands.
Phylogeographically, the Canary Islands are the most thoroughly studied of all of the Atlantic island groups (70). In this archipelago, the islands farther from the mainland are younger, and those closer to the mainland (Fuerteventura and Lanzarote) are older (Fig. 2, Inset). Compared with the Pacific volcanoes, the archipelago is much older and moves much more slowly. Older islands closer to the continent are drier, as well as lower in elevation; thus, Fuerteventura and Lanzarote do not contain any wet forest habitats. The progression rule appears to be a common pattern of colonization shown by several groups of organisms (70–72). However, species may, at least sometimes, also disperse from younger to older islands, even when these islands are occupied by close relatives (70). Among spiders, a progression rule has been documented for Loxosceles (73) and Dysdera (74, 75), whereas Pholcus and Spermophorides probably colonized the older Fuerteventura and Lanzarote from the younger Gran Canaria (76, 77). The mixed support for a progression across the Canary Islands presents the possibility of an alternative explanation for the origin of the endemics of the oldest islands, which could be the result of secondary replacement of its original fauna by new colonists better adapted to increasingly arid conditions (78). Older islands in the Canaries may show loss of old resident species due to aridification and orographic simplification; in some lineages, this may have been compensated by colonization and subsequent diversification of new, better-adapted organisms that could take advantage of empty niches and new opportunities.
Azores Islands.
The Azores have also been used for examination of the role of island progression in dictating biogeographic patterns (79). However, the islands are located over a complex microplate rather than a single hotspot. Nevertheless, there is some evidence for a directional mode of dispersal from older to younger islands [plants (80, 81); arthropods (82, 83)], although a simple progression rule is not common [yet Van Riel (84) presents intriguing evidence from gastropods]. Much of the current evidence consists of age estimates of haplotypes/alleles rather than robustly structured cladogenetic branching between islands. This pattern may be in part due to young radiations, since much of the landmass in the Azores being younger than 1 Ma. Moreover, the islands have suffered extensive habitat modification due to more recent volcanic activity and deforestation of native forests over many centuries of occupation (79).
Persistence of the Progression Pattern and the Evolution of Community Assembly
As with most evolutionary processes, community assembly in the radiation zone happens more slowly than can be observed. Progression patterns on remote oceanic archipelagos are valuable in that they suggest a temporal frame of reference among extant communities as well as comparative insights into past events. For example, a progression pattern is typically interpreted as evidence of an historical path of colonization. A phylogenetic estimate that allies early branches with older islands and more distal branches with younger islands suggests that a lineage has been established within a given archipelago because the earliest islands were habitable by that taxon. Subsequent colonizations occur coincident with the emergence of younger islands, initiating new communities at progressively younger times. In archipelagos that meet conditions for progression, codistributed taxa with common phylogeographic histories therefore can be studied as part of their communities on specific islands in the sequence, representing slices of time in the history of the archipelago. Thus, a progression pattern provides snapshots of communities through time, for as long as the oldest islands have been supporting such communities.
The history of community assembly over evolutionary time also may be informed by the geographical polarity evident in the progression pattern of a lineage. In an archipelago with a linear spatial and chronological sequence, like those often present in hotspot regions, a simple progression pattern is coincident with a unidirectional path of colonization from older to younger islands. A possible explanation for this is that a regular supply of propagules on older islands maintains the constant potential for colonization as new habitat emerges with the origin of a new island. This dynamic suggests that once new habitat is available (on progressively younger islands), colonists from spatially proximate, older habitat have the potential to arrive, establish, and for many such radiations, diverge.
Biotic Resistance on an Evolutionary Timescale.
Although a progression rule may be a valuable tool to understand the evolution of community assembly, there is also the possibility that community assembly may help us understand the development of the progression rule. One of the more prominent features of the progression pattern, and oceanic colonization pathways in general, is that they are largely unidirectional (85). In hotspot regions, the chronological sequence of the archipelago landscape promotes a progression pattern because a probable source of colonists is available to establish the next stepping stone in the series once the next island habitat emerges. Thus, the initial progression arising from older to younger island colonizations seems relatively easy to explain.
On the other hand, dispersal vectors (wind, ocean currents or biological vehicles) (18) are not expected to promote a unidirectional migration route (e.g., in Hawaii, complex wind currents have the potential to bring propagules in a variety of directions, and tropical storm tracks in particular, at times flow in a reverse direction to island age). Indeed, radiations that evidently postdate the Hawaiian archipelago formation [e.g., the spider genus Havaika (86); the plant genus Tetramolopium; (87); some Hawaiian birds, (88)] show colonization routes at odds with the chronosequence, which suggests that there is no physical feature dictating an older to younger colonization route after the islands exist.
Although an early progression pattern arising from the initial colonization opportunities in hot spot archipelagos might often be expected, the unpredictable nature of dispersal might be expected to erase the pattern over evolutionary time. Drawing on ideas dating back to MacArthur and Wilson (10), it has been argued that an apparent unidirectional pathway of colonization toward younger islands, and by extension the progression rule, may be bolstered by niche preemption, a type of priority effect (14, 17, 85, 89, 90).
Priority effects, which arise from the impact that species have on one another within a community, depend on the order of arrival in the community (90). By this process, once the first colonist (a founder) establishes in a newly available habitat, it soon monopolizes resources in the critical dimensions of the niche and blocks subsequent propagules from establishing. Key to the concept of priority effect is order of arrival. Although priority effects can be positive or negative (90), this manifestation of the priority effect is negative, yielding advantage to the first colonist to arrive: once a niche has been filled, it is more difficult for ecologically similar individuals to enter (10).
Priority effects would have an impact on phylogeographic patterns potentially in two ways. First, and most dramatically, patterns that suggest older to younger island colonization would unfold over evolutionary time as a hotspot archipelago forms in a spatiotemporal sequence. On each successively younger island, initial (early) colonists would arrive from the most proximate older island, establish in an empty habitat, and increase in population size and density, i.e., preempting the relevant niche. At any point in the future, ecologically similar propagules from younger islands back-colonizing to older islands would be second to arrive (late invaders), suffer a disadvantage and fail to establish due to arriving in the pre-empted niche (17). Thus, the ecological consequences of niche preemption by the early colonists is that back-migrations are discouraged from establishing. Second, niche-preemption may further accentuate the initial older to younger island progression pattern by bolstering the genetic legacy of the early colonists via a “founder takes all” dynamic, whereby repeated older to younger island colonizations are likewise thwarted, again due to niche preemption (85). Both of these scenarios suggest that ecological dynamics and species interactions are important in giving rise to progression. In other words, to explain features of a progression rule pattern, particularly for a species radiation where all available habitat has been colonized (suggesting propagules are not rare), but speciation has occurred in concert with interisland colonization (suggesting that gene flow has terminated), priority effects may be critical.
Moreover, the ability of new colonists to enter a community is frequently thought to decline as the diversity of species already in the community increases; the established species are considered to dictate biotic resistance against further colonization or invasion (91–93). Thus, biotic resistance results from a complementarity effect; the larger the number of species in a community, the more resource used, in turn creating more community resistance to new invaders. Diversity can also generate biotic resistance through a selection effect under which the most effective competitors, such as those most similar to focal invaders, are more likely to be present in more diverse mixes (91, 94). Priority effects can play a synergistic role in each of these dynamics. Curiously, although the consequences of priority effects are well understood over evolutionary time scales, the opposite appears to be true at ecological time scales.
Biotic Turnover on an Ecological Timescale.
MacArthur and Wilson (10) proposed that species richness on islands is the result of a dynamic balance between stochastic immigrations and extinctions. Their thinking was a radical shift from most community assembly theory with rejection of a deterministic equilibrium driven by species interactions. Today, the proximity to the mainland, the size of the mainland species pool, and the area of the island are hypothesized to be the three main factors explaining species richness on islands. The Equilibrium Theory of Island biogeography assumes a dynamic equilibrium with continual species turnover, because of colonization and extinction events that constantly modify community composition, a proposition subsequently developed in metapopulation ecology (95). Thus, biotic turnover prevents genetic differentiation from accumulating, and thereby continually erases the development of a progression pattern.
How Do We Reconcile the Concepts of Biotic Turnover with Biotic Resistance?
Based on many (perhaps most) models of ecological community assembly, biotic resistance seems unlikely. So why, then, is biotic resistance so apparent on an evolutionary timescale (96)? Put another way, how does ecological turnover (and associated ecological processes) give way to the biotic resistance and community “lock up” dynamics that might sustain a progression pattern over evolutionary timescales?
Possible hypotheses to explain community lock up dynamics that maintain progression patterns might be as follows. (i) Species and/or communities might be impervious to subsequent invasion due to “high density blocking” (85). This hypothesis posits that, although additional colonizations occur (e.g., through back migration), numerical dominance of the early, resident species deters establishment or dilutes the genetic contributions of late colonists with similar niche requirements. A prediction of this hypothesis is that the more similar late colonists are to resident species, the less likely they are to successfully invade. A genetic manifestation of this idea is that the distinct alleles of late invaders that manage to hybridize (admix) with residents have a high likelihood of extinction, thereby obscuring repeated- and back-colonization events. (ii) Late invaders suffer a mating disadvantage, or mate with residents but have no genetic impact, because they either fail to reproduce or their offspring have low fitness. (iiia) Early invaders have adapted to conditions specific to each island, so that late invaders are always inferior competitors, or (iiib) early colonizers undergo ecological release, resulting in extensive niche overlap among members of the early community; over time, and as the environment becomes more stable, they may specialize, placing subsequent colonizers at a disadvantage. Alternatively, (iv) after some initial turnover, the combined island communities lock in at a state where “by chance” no island can be invaded. Although it is difficult to separate these possible explanations (and they may not be mutually exclusive), genomic data promises an opportunity at least to test whether priority effects are real, and if so, what consequences would arise for genetic admixture and introgression.
The Progression Rule and the Mosaic Genome
To address the lock up hypothesis will require a multidisciplinary approach. However, the first explanation suggested above, that late invaders hybridize with species in residence and their genes admix into the resident population, falls in the domain of comparative phylogeography. This hypothesis is predicated on the idea of a mosaic genome where specific regions can have separate historical identities (27, 97–100), some reflecting a history of progression, and others reflecting additional demographic complexities (such as back migrations). Teasing apart complex histories within the same species lineage requires separate analyses of distinct hereditary units. Whether sufficient data exist to mark these separate histories is an empirical issue, but to be sure, if we combine data across data partitions that represent distinctive histories, we will lose insights into these complexities.
In a recent phylogeographic context, it is possible for different gene partitions to reveal conflicting but true histories that manifest more than one of these progression patterns within the same lineage. Such situations are extremely valuable because they suggest a multidimensional insight into past demography of the lineage.
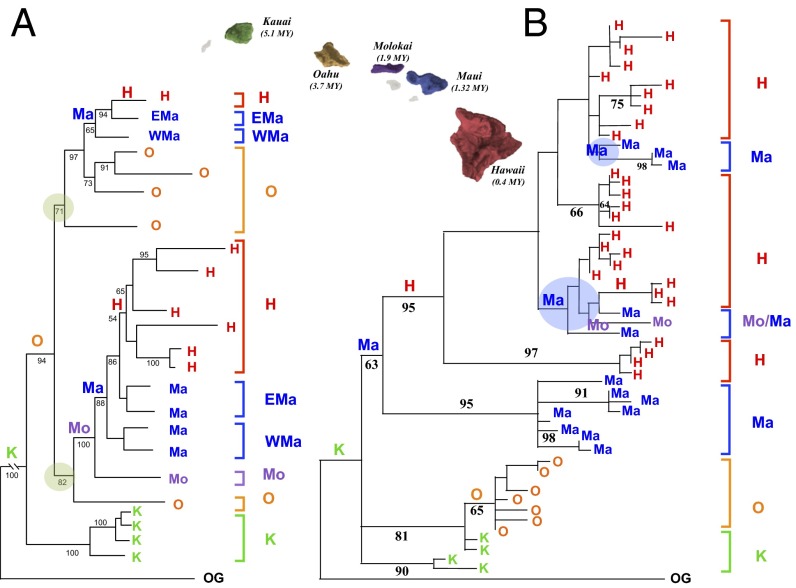
A potential example comes from the Hawaiian cricket genus Laupala, a morphologically cryptic group of 38 flightless, single island endemic, species (1). Multiple datasets reveal a compelling case for progressive clades and grades in Laupala, the most resolved of which emerges from a nuclear (presumably) phylogeny based on amplified fragment length polymorphisms (36) (Fig. 3A). The phylogeny is well supported, is concordant with the species taxonomy, and is consistent with a less resolved nuclear sequence phylogeny (101). The area cladogram shows that the group began on Kauai (or an older island) and split into two species groups once on the younger Oahu. Subsequent diversification occurred via fine-scale progression to increasingly younger islands, colonizing first either Molokai (absent in one group) or West Maui, then East Maui, and, last, Hawaii Island. Thus, the nuclear phylogeny suggests a unidirectional pattern with rare interisland migrations.
Fig. 3.
Progression and the mosaic genome. (A) Area cladogram derived from the nuclear AFLP phylogeny of Laupala (36), a genus of flightless crickets endemic to Hawaii. Two species groups are shown (green discs overlaying resolved nodes). An overall progression is evident, with two progression subpatterns in each species group. Minimum interisland migrations are marked by K (Kauai), O (Oahu), Mo (Molokai), Ma (Mauai), and H (Hawaii). Additional abbreviations include OG (outgroup), EMa (East Maui), and WMa (West Maui). A neighbor joining tree based on Nei-Li distances is shown. Terminal taxa are species represented by 2–10 individuals color coded to the island to which they are endemic. Bootstrap values are shown for 1,000 neighbor joining replicates below branches. (B) An area cladogram derived from the mtDNA phylogeny of Laupala (101). Shown is the maximum parsimony tree with the highest likelihood score; terminal taxa are unique sequences from concatenated 12s, 16s, and tRNAval regions. The mtDNA topology is not congruent with the AFLP tree, and species groups are not evident in the mtDNA tree. An overall progression is evident, with two back migrations marked by blue discs. One possible minimum interisland migration scenario is shown, marked by K (Kauai), O (Oahu), Mo (Molokai), Ma (Maui), and H (Hawaii). Bootstrap values are shown for 1,000 parsimony replicates below branches.
However, the unidirectional pattern of colonization is contradicted by the mtDNA phylogeny (Fig. 3B). Although there is a general pattern of progression in the mtDNA area cladogram as well, there is extensive conflict between a mtDNA tree and both the nuclear DNA trees (101). Importantly, the phylogeographic patterns in the mtDNA include two potential back migrations between Hawaii and Maui and one additional back migration between Maui and Molokai. These back-migrations are between neighboring islands, a trend shared by other taxa where back migrations occur over a pattern of progression (29). In short, the mtDNA evidence suggests that there has been considerably more interisland movement than is revealed by the nuclear partitions.
A recent study of the flightless, species-rich weevil Laparocerus (71) from the Canary and Madeira Islands comes to a similar conclusion. The mtDNA and nuclear gene trees sampled from Laparocerus show conflicting topologies, similar to Laupala, along with a progression from older to younger islands evident in both data partitions. Like Laupala, the nuclear phylogeographic history is simpler, whereas the mtDNA phylogeographic history captures considerably more interisland movement. Although it is unclear in the case of Laupala, the nuclear data show evidence of subsequent admixture in cases of multiple colonization in Laparocerus. Importantly, in both systems, the patterns of mtDNA variation do not conform to expectations of incomplete lineage sorting, and instead appear to be informative about interisland colonization patterns.
Recent phylogeographic studies of Hawaiian planthoppers and spiders using next-generation sequencing approaches are starting to reveal similar patterns of higher than expected movement, at least between the younger islands. Together, the data suggest that there is some movement between islands, at least among younger islands (102). The promise of these new genetic data technologies is that the heterogeneity of progression patterns can be fully investigated, and potentially lead to an understanding of the causes of such heterogeneous genetic signatures.
Discussion
The progression rule is one of the most pervasive phylogeograpic patterns yet documented, at least for the well-studied remote archipelago of Hawaii. There is considerable evidence accumulating from two other well-studied archipelagos, the Canary Islands (70) and the Galapagos (59, 71), despite the complicating factors of spatial nonlinearity and greater proximity to continents. Additional evidence is accumulating for progression in other remote archipelagos as well (summarized here for the Australs and the Marquesas) (53). Evidence for progression, thus far, is mixed in the Azores, presumably due to the geological complexity of the hotspot (79), and the Society Islands, perhaps due to the close proximity to much older islands, meaning that there were other islands that served as a source of colonists, or a “bridge” between other archipelagoes to the younger islands of the Society chain.
Progression is a feature that emerges over evolutionary time and can only develop once a community transitions from biotic turnover (manifest on an ecological timescale) to biotic resistance (on an evolutionary timescale). To a large degree, the transition from biotic turnover to biotic resistance mirrors the transition from population genetic cohesion to the genetic differentiation that characterizes speciation. In species formation, eventually, a nascent species closes genetic connections with its past (represented by its sister species) and persists as an independent evolutionary entity. Likewise, in the transition from ecological turnover to biotic resistance, a new community forms. Whereas formerly, the community was in a dynamic state of revision due to immigration and extinction (under some theories), eventually, the community resists repeated immigration of colonists. The speciation process occurs within the context of this community transition, and embodies the zone of radiation.
The geographic and chronological settings of remote, hotspot archipelagos offer conditions under which progression patterns can arise. However, the ecological features of the organisms involved play an important role in the subsequent patterns of diversification, and the rate at which they evolve. Key are the rates of dispersal and likelihood of colonization and establishment (103). Obviously, if little to no dispersal occurred, a progression pattern would be unlikely to arise as the species would likely remain in the ancestral habitat and not radiate (e.g., see potential examples reviewed in ref. 21). Likewise, if dispersal and subsequent gene flow facilitated panmixia, neither speciation nor progression would be expected to arise because biotic turnover would prevail. However, if dispersal were infrequent but predictable (the likely condition for the majority of taxa), colonization would occur as new island habitats become available. Gene flow would be reduced, and depending on its magnitude, would not continue to homogenize gene frequencies between founder and source.
Once the founder population, now the resident species, established and has filled the available niche, priority effects could come into play by a number of possible mechanisms. (i) The community might be resistant to subsequent invasion due to the monopolization of resources by the numerically dominant resident species; the more similar late colonists are to resident species, the less likely they are to successfully invade. (ii) Late colonists may suffer incompatible mating encounters with the numerically dominant resident species, leading to a failure to reproduce. (iii) The resident species may have adapted to local conditions, giving it a competitive edge over later colonists. The last mechanism involves natural selection and competitive exclusion, whereas numerical dominance drives the first, and may contribute to the second interaction. These possible mechanisms could, in theory, be tested with appropriate experiments.
Depending on the degree of differentiation, the late colonist may nonetheless hybridize with the resident species and its genes may diffuse into the resident species gene pool, leaving some trace of its attempt at establishment. As discussed in the two examples described earlier, mosaic histories contained within the genome of a given lineage may harbor evidence regarding how a progression pattern evolves in this intermediate dispersal domain. It seems unlikely that species radiations manifesting a progression have occurred in the clean manner that a phylogeny might suggest. Even in the most straightforward cases, such as the patterns of progressive clades and grades in the amplified fragment length polymorphism (AFLP) phylogeographic pattern in Laupala crickets, additional data cautions us from concluding that interisland migrations do not occur. More efforts are needed to examine contrasting evidence of progression patterns as they may provide insights into how the pattern develops. If we can understand the development of progression, we may begin to understand how ecological turnover (where progression would not arise) gives way to biotic resistance (where progression could arise).
As elucidated by Fukami (90), priority effects by niche preemption are most likely to occur when two species show highly overlapping resource use, the first colonist has a high impact on the overlapping niche dimensions, and when the growth rate of the late colonist is heavily dependent on the environment. Thus, we might expect that ecological shifts create opportunities for late colonists to escape the impact of priority effects as a result of exploring new niche dimensions. Highly dynamic adaptive radiations that appear to violate the progression rule via back migration, such as some branches of the Hawaiian picture-winged Drosophila radiation (30), provide an opportunity to test this idea. Perhaps herein lies the explanation for the finding that adaptive radiations in the Galapagos were not found to adhere to the progression rule (59). Likewise, species that have more plastic attributes to their resource use might have a heightened immunity to priority effects, which would act to discourage the development of a progression pattern in the lineage. Better characterization of ecological traits, reproductive behaviors, divergence times and genetic admixture among lineages of island radiations should allow for more rigorous evaluation of priority effects on the development of progression within a phylogenetic context.
The study of community assembly in and beyond the radiation zone is exciting in part because it brings together two fairly disparate disciplines: the ecological study of community structure and the evolutionary study of the origin of species. The presence of progression, and its more nuanced manifestation revealed by the mosaic genome, is fortunate for the study of community assembly because the pattern provides a temporal framework for both ecological and evolutionary studies of communities and their interrelationship. The key importance of the progression pattern is that multiple lineages are establishing and assembling, interacting and adapting, over a similar timeframe that plays out over extended evolutionary time. Thus, we can measure ecological metrics at different time slices of the community assembly process to find how properties (species diversity, abundance, body size distributions, trophic interactions) change over time (104, 105), and how the origin of new species affects these properties. Moreover, as genomic data become available across multiple lineages that appear to follow a progression in a given system (9), we are gaining insight into how taxa differ in mode, rates, and patterns of establishment and diversification (13). Integrating multidimensional datasets across stages of the progression will allow us to understand how interactions develop and evolve and the importance of such interactions in dictating properties of stability, turnover, and the evolution of biotic resistance in a community. Effectively, with multiple lineages being formed over the same timeframe, we can examine the feedback between ecology and evolution and hence generate insights into the processes involved in the formation and loss of biodiversity.
Acknowledgments
We thank Francisco Ayala, John Avise, and Brian Bowen for the invitation to participate in the symposium and special issue. We thank two reviewers for comments that helped improve the manuscript. We also thank our students and colleagues of the Dimensions of Hawaiian Biodiversity team for many stimulating discussions. This work was supported by National Science Foundation (NSF) Division of Environmental Biology Grants 1241060 (to K.L.S.) and 1241253 (to R.G.G.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution X: Comparative Phylogeography, ” held January 8–9, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_X_Comparative_Phylogeography.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601078113/-/DCSupplemental.
References
- 1.Otte D. The Crickets of Hawaii: Origin, Systematics and Evolution. The Orthopterists' Society; Philadelphia: 1994. p. 338. [Google Scholar]
- 2.Grant P-R, Grant B-R. How and why species multiply. In: Grant P-R, Grant B-R, editors. The Radiation of Darwin’s Finches. Princeton Univ Press; Princeton: 2008. [Google Scholar]
- 3.Rubinoff D, Haines W-P. Web-spinning caterpillar stalks snails. Science. 2005;309(5734):575. doi: 10.1126/science.1110397. [DOI] [PubMed] [Google Scholar]
- 4.Yoder A, et al. Geogenetic patterns in mouse lemurs (genus Microcebus) reveal the ghosts of Madagascar's forests past. Proc Natl Acad Sci USA. 113:8049–8056. doi: 10.1073/pnas.1601081113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen BW, et al. Comparative phylogeography of the ocean planet. Proc Natl Acad Sci USA. 113:7962–7969. doi: 10.1073/pnas.1602404113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood HM, Gillespie R-G, Griswold C-E, Wainwright P-C. Why is Madagascar special? The extraordinarily slow evolution of pelican spiders (Araneae, Archaeidae) Evolution. 2015;69(2):462–481. doi: 10.1111/evo.12578. [DOI] [PubMed] [Google Scholar]
- 7.Buckley T-R, Attanayake D, Bradler S. Extreme convergence in stick insect evolution: Phylogenetic placement of the Lord Howe Island tree lobster. Proc Biol Sci. 2009;276(1659):1055–1062. doi: 10.1098/rspb.2008.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Losos J-B, Ricklefs R-E. Adaptation and diversification on islands. Nature. 2009;457(7231):830–836. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie R-G. Island time and the interplay between ecology and evolution in species diversification. Evol Appl. 2015;9(1):53–73. doi: 10.1111/eva.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacArthur R-H, Wilson E-O. The Theory of Island Biogeography. Princeton Univ Press; Princeton: 1967. [Google Scholar]
- 11.Warren B-H, et al. Islands as model systems in ecology and evolution: Prospects fifty years after MacArthur-Wilson. Ecol Lett. 2015;18(2):200–217. doi: 10.1111/ele.12398. [DOI] [PubMed] [Google Scholar]
- 12.Heaney L-R. Is a new paradigm emerging for oceanic island biogeography? J Biogeogr. 2007;34(5):753–757. [Google Scholar]
- 13.Rominger A-J, et al. Community assembly on isolated islands: Macroecology meets evolution. Glob Ecol Biogeogr. July 14, 2015 doi: 10.1111/geb.12341. [DOI] [Google Scholar]
- 14.Gillespie R-G, Baldwin B-G. Island biogeography of remote archipelagos: Interplay between ecological and evolutionary processes. In: Losos J, Ricklefs R, editors. The Theory of Island Biogeography Revisited. Princeton Univ Press; Princeton: 2009. pp. 358–387. [Google Scholar]
- 15.Rosindell J, Phillimore A-B. A unified model of island biogeography sheds light on the zone of radiation. Ecol Lett. 2011;14(6):552–560. doi: 10.1111/j.1461-0248.2011.01617.x. [DOI] [PubMed] [Google Scholar]
- 16.Simpson G-G. Mammals and land bridges. J Wash Acad Sci. 1940;30:137–163. [Google Scholar]
- 17.Gillespie R-G, Roderick G-K. Arthropods on islands: Colonization, speciation, and conservation. Annu Rev Entomol. 2002;47:595–632. doi: 10.1146/annurev.ento.47.091201.145244. [DOI] [PubMed] [Google Scholar]
- 18.Gillespie R-G, et al. Long-distance dispersal: A framework for hypothesis testing. Trends Ecol Evol. 2012;27(1):47–56. doi: 10.1016/j.tree.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Steppan S-J, Zawadzki C, Heaney L. Molecular phylogeny of the endemic Philippine rodent Apomys (Muridae) and the dynamics of diversification in an oceanic archipelago. Biol J Linn Soc Lond. 2003;80(4):699–715. [Google Scholar]
- 20.de Queiroz A. The resurrection of oceanic dispersal in historical biogeography. Trends Ecol Evol. 2005;20(2):68–73. doi: 10.1016/j.tree.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Keeley S-C, Funk V-A. In: The Biology of Island Floras. Bramwell D, Caujapé-Castells J, editors. Cambridge Univ Press; Cambridge, UK: 2011. pp. 57–88. [Google Scholar]
- 22.Gillespie R-G. Colonization of remote oceanic islands of the Pacific: Archipelagos as stepping stones? J Biogeogr. 2002;29(5-6):655–662. [Google Scholar]
- 23.Funk V-A, Wagner W-L. In: Hawaiian Biogeography Evolution on a Hot Spot Archipelago. Wagner WL, Funk VA, editors. Smithsonian Institution Press; Washington, DC: 1995. [Google Scholar]
- 24.Avise J. Phylogeography: The History and Formation of Species. Harvard Univ Press; Cambridge, MA: 2000. [Google Scholar]
- 25.Wilson J-T. A possible origin of the Hawaiian Islands. Can J Phys. 1963;41(6):863–870. [Google Scholar]
- 26.Clouard V, Bonneville A. Ages of seamounts, islands, and plateaus on the Pacific plate. In: Foulger G-R, Natland J-H, Presnall D-C, Anderson D-L, editors. Plates, Plumes, and Paradigms. Geological Society of America; Boulder, CO: 2005. pp. 71–90. [Google Scholar]
- 27.Seehausen O, et al. Genomics and the origin of species. Nat Rev Genet. 2014;15(3):176–192. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- 28.Hennig W. Phylogenetic Systematics. Univ of Illinois Press; Urbana, IL: 1966. [Google Scholar]
- 29.Funk V-A, Wagner W-L. Biogeographic patterns in the Hawaiian Islands. In: Wagner WL, Funk VA, editors. Hawaiian Biogeography: Evolution on a Hot Spot Archipelago. Smithsonian Institution Press; Washington, DC: 1995. pp. 379–419. [Google Scholar]
- 30.Magnacca K-N, Price D-K. Rapid adaptive radiation and host plant conservation in the Hawaiian picture wing Drosophila (Diptera: Drosophilidae) Mol Phylogenet Evol. 2015;92:226–242. doi: 10.1016/j.ympev.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Baldwin B-G, Robichaux R-H. Historical biogeography and ecology of the Hawaiian silversword alliance (Asteraceae): New molecular phylogenetic perspectives. In: Wagner WL, Funk VA, editors. Hawaiian Biogeography: Evolution on a Hot Spot Archipelago. Smithsonian Institution Press; Washington, DC: 1995. pp. 259–287. [Google Scholar]
- 32.Nepokroeff M, Sytsma K-J, Wagner W-L, Zimmer E-A. Reconstructing ancestral patterns of colonization and dispersal in the Hawaiian understory tree genus Psychotria (Rubiaceae): A comparison of parsimony and likelihood approaches. Syst Biol. 2003;52(6):820–838. [PubMed] [Google Scholar]
- 33.Ree R-H, Smith S-A. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57(1):4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 34.Givnish T-J, et al. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae) Proc Biol Sci. 2009;276(1656):407–416. doi: 10.1098/rspb.2008.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan S, Simon C, Polhemus D. Molecular systematics and adaptive radiation of Hawaii’s endemic Damselfly genus Megalagrion (Odonata: Coenagrionidae) Syst Biol. 2003;52(1):89–109. doi: 10.1080/10635150390132803. [DOI] [PubMed] [Google Scholar]
- 36.Mendelson T-C, Shaw K-L. Sexual behaviour: Rapid speciation in an arthropod. Nature. 2005;433(7024):375–376. doi: 10.1038/433375a. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro L-H, Strazanac J-S, Roderick G-K. Molecular phylogeny of Banza (Orthoptera: Tettigoniidae), the endemic katydids of the Hawaiian Archipelago. Mol Phylogenet Evol. 2006;41(1):53–63. doi: 10.1016/j.ympev.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Haines W-P, Schmitz P, Rubinoff D. Ancient diversification of Hyposmocoma moths in Hawaii. Nat Commun. 2014;5:3502. doi: 10.1038/ncomms4502. [DOI] [PubMed] [Google Scholar]
- 39.Bonacum J, O’Grady P-M, Kambysellis M, Desalle R. Phylogeny and age of diversification of the planitibia species group of the Hawaiian Drosophila. Mol Phylogenet Evol. 2005;37(1):73–82. doi: 10.1016/j.ympev.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Rundell R-J, Holland B-S, Cowie R-H. Molecular phylogeny and biogeography of the endemic Hawaiian Succineidae (Gastropoda: Pulmonata) Mol Phylogenet Evol. 2004;31(1):246–255. doi: 10.1016/j.ympev.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 41.Hormiga G, Arnedo M, Gillespie R-G. Speciation on a conveyor belt: Sequential colonization of the hawaiian islands by Orsonwelles spiders (Araneae, Linyphiidae) Syst Biol. 2003;52(1):70–88. doi: 10.1080/10635150390132786. [DOI] [PubMed] [Google Scholar]
- 42.VanderWerf E-A, Young L-C, Yeung N-W, Carlon D-B. Stepping stone speciation in Hawaii’s flycatchers: Molecular divergence supports new island endemics within the elepaio. Conserv Genet. 2010;11(4):1283–1298. [Google Scholar]
- 43.Roderick G-K, Gillespie R-G. Speciation and phylogeography of Hawaiian terrestrial arthropods. Mol Ecol. 1998;7(4):519–531. doi: 10.1046/j.1365-294x.1998.00309.x. [DOI] [PubMed] [Google Scholar]
- 44.Cowie R-H, Holland B-S. Molecular biogeography and diversification of the endemic terrestrial fauna of the Hawaiian Islands. Philos Trans R Soc Lond B Biol Sci. 2008;363(1508):3363–3376. doi: 10.1098/rstb.2008.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dickinson W-R. Geomorphology and geodynamics of the Cook-Austral Island Seamount chain in the South Pacific Ocean: Implications for hotspots and plumes. Int Geol Rev. 1998;40(12):1039–1075. [Google Scholar]
- 46.Bonneville A, et al. Arago Seamount: The missing hotspot found in the Austral Islands. Geology. 2002;30(11):1023–1026. [Google Scholar]
- 47.Garb J-E, Gillespie R-G. Island hopping across the central Pacific: Mitochondrial DNA detects sequential colonization of the Austral Islands by crab spiders (Araneae: Thomisidae) J Biogeogr. 2006;33(2):201–220. [Google Scholar]
- 48.Gillespie R-G, Claridge E-M, Goodacre SL. Biogeography of the fauna of French Polynesia: diversification within and between a series of hot spot archipelagos. Philos Trans R Soc Lond B Biol Sci. 2008;363(1508):3335–3346. doi: 10.1098/rstb.2008.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Claridge E-M, Gillespie R-G, Brewer M-S, Roderick G-K. Stepping-stones across space and time: Repeated radiation of Pacific flightless broad-nosed weevils (Coleoptera: Curculionidae: Entiminae: Rhyncogonus) J Biogeogr. 2016 in press. [Google Scholar]
- 50.Joy D-A, Conn J-E. Molecular and morphological phylogenetic analysis of an insular radiation in Pacific black flies (Simulium) Syst Biol. 2001;50(1):18–38. [PubMed] [Google Scholar]
- 51.Craig D-A. Geomorphology, development of running water habitats, and evolution of black flies on Polynesian islands. Bioscience. 2003;53(11):1079–1093. [Google Scholar]
- 52.Claridge E-M. 2006. The systematics and diversification of Rhyncogonus (Entiminae: Curculionidae: Coleoptera) in the central Pacific. PhD thesis, University of California, Berkeley.
- 53.Cibois A, et al. Charting the course of reed-warblers across the Pacific islands. J Biogeogr. 2011;38(10):1963–1975. [Google Scholar]
- 54.Cibois A, Thibault J-C, Pasquet E. Biogeography of eastern Polynesian monarchs (Pomarea): An endemic genus close to extinction. Condor. 2004;106(54):837–851. [Google Scholar]
- 55.Gillespie R-G. Marquesan spiders of the genus Tetragnatha. J Arachnol. 2003;31(1):62–77. [Google Scholar]
- 56.Johnson M-S, Murray J, Clarke B. Parallel evolution in Marquesan partulid land snails. Biol J Linn Soc Lond. 2000;69(4):577–598. [Google Scholar]
- 57.White W-M, McBirney A-R, Duncan R-A. Petrology and geochemistry of the Galapagos Islands: Portrait of a pathological mantle plume. J Geophys Res Solid Earth. 1993;98(B11):19533–19563. [Google Scholar]
- 58.Poulakakis N, et al. Historical DNA analysis reveals living descendants of an extinct species of Galápagos tortoise. Proc Natl Acad Sci USA. 2008;105(40):15464–15469. doi: 10.1073/pnas.0805340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parent C-E, Caccone A, Petren K. Colonization and diversification of Galápagos terrestrial fauna: A phylogenetic and biogeographical synthesis. Philos Trans R Soc Lond B Biol Sci. 2008;363(1508):3347–3361. doi: 10.1098/rstb.2008.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caccone A, et al. Phylogeography and history of giant Galápagos tortoises. Evolution. 2002;56(10):2052–2066. doi: 10.1111/j.0014-3820.2002.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 61.Beheregaray L-B, et al. Giant tortoises are not so slow: Rapid diversification and biogeographic consensus in the Galápagos. Proc Natl Acad Sci USA. 2004;101(17):6514–6519. doi: 10.1073/pnas.0400393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wright J-W. 1983. The evolution and biogeography of the lizards of the Galapagos Archipelago: Evolutionary genetics of Phyllodactylus and Tropidurus populations. Patterns of Evolution in Galapagos Organisms, eds Bowman R-I, et al. (AAAS Symposium Volume, San Francisco), pp. 123–155.
- 63.Lopez T-J, Hauselman E-D, Maxson L-R, Wright J-W. Preliminary analysis of phylogenetic relationships among Galapagos Island lizards of the genus Tropidurus. Amphib-reptil. 1992;13(4):327–339. [Google Scholar]
- 64.Kizirian D, Trager A, Donnelly M-A, Wright J-W. Evolution of Galapagos Island Lava Lizards (Iguania: Tropiduridae: Microlophus) Mol Phylogenet Evol. 2004;32(3):761–769. doi: 10.1016/j.ympev.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Steinfartz S, et al. Genetic impact of a severe El Niño event on Galápagos marine iguanas (Amblyrhynchus cristatus) PLoS One. 2007;2(12):e1285. doi: 10.1371/journal.pone.0001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arbogast B-S, et al. The origin and diversification of Galapagos mockingbirds. Evolution. 2006;60(2):370–382. [PubMed] [Google Scholar]
- 67.Parent C-E, Crespi B-J. Ecological opportunity in adaptive radiation of Galápagos endemic land snails. Am Nat. 2009;174(6):898–905. doi: 10.1086/646604. [DOI] [PubMed] [Google Scholar]
- 68.Sequeira A-S, Lanteri A-A, Albelo LR, Bhattacharya S, Sijapati M. Colonization history, ecological shifts and diversification in the evolution of endemic Galápagos weevils. Mol Ecol. 2008;17(4):1089–1107. doi: 10.1111/j.1365-294X.2007.03642.x. [DOI] [PubMed] [Google Scholar]
- 69.Schmitz P, Cibois A, Landry B. Molecular phylogeny and dating of an insular endemic moth radiation inferred from mitochondrial and nuclear genes: The genus Galagete (Lepidoptera: Autostichidae) of the Galapagos Islands. Mol Phylogenet Evol. 2007;45(1):180–192. doi: 10.1016/j.ympev.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 70.Juan I, Emerson BC, Orom I, Hewitt G-M. Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends Ecol Evol. 2000;15(3):104–109. doi: 10.1016/s0169-5347(99)01776-0. [DOI] [PubMed] [Google Scholar]
- 71.Faria C-M-A, et al. Evidence for multiple founding lineages and genetic admixture in the evolution of species within an oceanic island weevil (Coleoptera, Curculiondae) super-radiation. J Biogeogr. 2016;43(1):178–191. [Google Scholar]
- 72.Sanmartín I, Van Der Mark P, Ronquist F. Inferring dispersal: A Bayesian approach to phylogeny‐based island biogeography, with special reference to the Canary Islands. J Biogeogr. 2008;35(3):428–449. [Google Scholar]
- 73.Planas E, Ribera C. Uncovering overlooked island diversity: Colonization and diversification of the medically important spider genus Loxosceles (Arachnida: Sicariidae) on the Canary Islands. J Biogeogr. 2014;41(7):1255–1266. [Google Scholar]
- 74.Arnedo M-A, Oromí P, Ribera C. Radiation of the spider genus Dysdera (Araneae, Dysderidae) in the Canary Islands: Cladistic assessment based on multiple data sets. Cladistics. 2001;17(4):313–353. [Google Scholar]
- 75.Macías-Hernández N, Oromi P, Arnedo M-A. Patterns of diversification on old volcanic islands as revealed by the woodlouse‐hunter spider genus Dysdera (Araneae, Dysderidae) in the eastern Canary Islands. Biol J Linn Soc Lond. 2008;94(3):589–615. [Google Scholar]
- 76.López-Mercader N. 2005. Evolutionary processes of the genus Spermophorides (Araneae, Pholcidae) in the Canary Islands. PhD Thesis, Universitat de Barcelona, Barcelona, Spain.
- 77.Dimitrov D, Arnedo M-A, Ribera C. Colonization and diversification of the spider genus Pholcus Walckenaer, 1805 (Araneae, Pholcidae) in the Macaronesian archipelagos: Evidence for long-term occupancy yet rapid recent speciation. Mol Phylogenet Evol. 2008;48(2):596–614. doi: 10.1016/j.ympev.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 78.Cardoso P, Arnedo M-A, Triantis K-A, Borges P-A. Drivers of diversity in Macaronesian spiders and the role of species extinctions. J Biogeogr. 2010;37(6):1034–1046. [Google Scholar]
- 79.Carvalho J-C, Cardoso P, Rigal F, Triantis K-A, Borges P-A. Modeling directional spatio-temporal processes in island biogeography. Ecol Evol. 2015;5(20):4671–4682. doi: 10.1002/ece3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rumeu B, et al. The colonization history of Juniperus brevifolia (Cupressaceae) in the Azores Islands. PLoS One. 2011;6(11):e27697. doi: 10.1371/journal.pone.0027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Díaz-Pérez A, Sequeira M, Santos-Guerra A, Catalán P. Multiple colonizations, in situ speciation, and volcanism-associated stepping-stone dispersals shaped the phylogeography of the Macaronesian red fescues (Festuca L., Gramineae) Syst Biol. 2008;57(5):732–749. doi: 10.1080/10635150802302450. [DOI] [PubMed] [Google Scholar]
- 82.Parmakelis A, et al. Comparative phylogeography of endemic Azorean arthropods. BMC Evol Biol. 2015;15:250. doi: 10.1186/s12862-015-0523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amorim I-R, Emerson B-C, Borges P-A-V, Wayne R-K. Phylogeography and molecular phylogeny of Macaronesian island Tarphius (Coleoptera: Zopheridae): Why are there so few species in the Azores? J Biogeogr. 2012;39(9):1583–1595. [Google Scholar]
- 84.Van Riel P, et al. Molecular systematics of the endemic Leptaxini (Gastropoda: Pulmonata) on the Azores islands. Mol Phylogenet Evol. 2005;37(1):132–143. doi: 10.1016/j.ympev.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 85.Waters J-M, Fraser C-I, Hewitt G-M. Founder takes all: Density-dependent processes structure biodiversity. Trends Ecol Evol. 2013;28(2):78–85. doi: 10.1016/j.tree.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 86.Arnedo M-A, Gillespie R-G. Species diversification patterns in the Polynesian jumping spider genus Havaika Prószyński, 2001 (Araneae, Salticidae) Mol Phylogenet Evol. 2006;41(2):472–495. doi: 10.1016/j.ympev.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 87.Lowrey T-K. Phylogeny, adaptive radiation, and biogeography of Hawaiian Tetramolopium (Asteraceae: Astereae) In: Wagner W-L, Funk V-A, editors. Hawaiian Biogeography. Smithsonian Institution Press; Washington, DC: 1995. pp. 195–220. [Google Scholar]
- 88.Fleischer R-C, McIntosh C-E. Molecular systematics and biogeography of the Hawaiian avifauna. Stud Avian Biol. 2001;22:51–60. [Google Scholar]
- 89.Fukami T, Beaumont H-J-E, Zhang X-X, Rainey P-B. Immigration history controls diversification in experimental adaptive radiation. Nature. 2007;446(7134):436–439. doi: 10.1038/nature05629. [DOI] [PubMed] [Google Scholar]
- 90.Fukami T. Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst. 2015;46:1–23. [Google Scholar]
- 91.Chapin F-S, 3rd, et al. Consequences of changing biodiversity. Nature. 2000;405(6783):234–242. doi: 10.1038/35012241. [DOI] [PubMed] [Google Scholar]
- 92.Kennedy T-A, et al. Biodiversity as a barrier to ecological invasion. Nature. 2002;417(6889):636–638. doi: 10.1038/nature00776. [DOI] [PubMed] [Google Scholar]
- 93.Hooper D-U, et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol Monogr. 2005;75(1):3–35. [Google Scholar]
- 94.Loreau M, Hector A. Partitioning selection and complementarity in biodiversity experiments. Nature. 2001;412(6842):72–76. doi: 10.1038/35083573. [DOI] [PubMed] [Google Scholar]
- 95.Hanski I. Metapopulation Ecology. Oxford Series in Ecology and Evolution. Oxford Univ Press; Oxford, UK: 1999. [Google Scholar]
- 96.Strauss S-Y, Webb C-O, Salamin N. Exotic taxa less related to native species are more invasive. Proc Natl Acad Sci USA. 2006;103(15):5841–5845. doi: 10.1073/pnas.0508073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maddison W-P. Phylogenetic histories within and among species. Experimental and molecular approaches to plant biosystematics. In: Hoch P-C, Stevenson A-G, editors. Monographs in Systematics. Vol 53. Missouri Botanical Garden; St. Louis: 1995. pp. 273–287. [Google Scholar]
- 98.Baum D-A, Shaw K-L. Genealogical perspectives on the species problem. In: Hoch P-C, Stephenson A-G, editors. Experimental and Molecular Approaches to Plant Biosystematics. Missouri Botanical Garden; St. Louis: 1995. pp. 289–303. [Google Scholar]
- 99.Wu C-I. The genic view of the process of speciation. J Evol Biol. 2001;14(6):851–865. [Google Scholar]
- 100.Shaw K-L. The genealogical view of speciation. Commentary on C-I. Wu, The genic view of the process of speciation. J Evol Biol. 2001;14(6):880–882. [Google Scholar]
- 101.Shaw K-L. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: What mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc Natl Acad Sci USA. 2002;99(25):16122–16127. doi: 10.1073/pnas.242585899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roderick G-K, Croucher P-J-P, Vandergast AG, Gillespie R-G. Species differentiation on a dynamic landscape: Shifts in metapopulation and genetic structure using the chronology of the Hawaiian archipelago. Evol Biol. 2012;39(2):192–206. doi: 10.1007/s11692-012-9184-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buckley H-L, Paterson A-M, Cruickshank R-H. The founder space race: A response to Waters et al. Trends Ecol Evol. 2013;28(4):189–190. doi: 10.1016/j.tree.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 104.Loreau M, Naeem S, Inchausti P. Biodiversity and Ecosystem Functioning: Synthesis and Perspectives. Oxford Univ Press; Oxford, UK: 2002. [Google Scholar]
- 105.Harmon L-J, et al. Evolutionary diversification in stickleback affects ecosystem functioning. Nature. 2009;458(7242):1167–1170. doi: 10.1038/nature07974. [DOI] [PubMed] [Google Scholar]
- 106.Cannat M, et al. Mid-Atlantic Ridge–Azores hotspot interactions: Along-axis migration of a hotspot-derived event of enhanced magmatism 10 to 4 Ma ago. Earth Planet Sci Lett. 1999;173(3):257–269. [Google Scholar]