Significance
Cytosine deamination appears to be largely responsible for spontaneous mutations in the modern world. Because of its sensitivity to temperature (Q10 = 4), that reaction would have furnished a mechanism for rapid evolution on a warm earth. As the temperature fell from 100° to 25 °C, the rate of cytosine-based mutation would have fallen by a factor of more than 4,000, with a corresponding increase in the stability of genetic information. Other potentially mutagenic events are known to be even more sensitive to temperature, and would presumably have led to an even steeper decline in the rate of spontaneous mutation as the earth cooled.
Keywords: spontaneous mutation, heat mutagenesis, cytosine deamination, HIV-1 protease
Abstract
The hydrolytic deamination of cytosine and 5-methylcytosine residues in DNA appears to contribute significantly to the appearance of spontaneous mutations in microorganisms and in human disease. In the present work, we examined the mechanism of cytosine deamination and the response of the uncatalyzed reaction to changing temperature. The positively charged 1,3-dimethylcytosinium ion was hydrolyzed at a rate similar to the rate of acid-catalyzed hydrolysis of 1-methylcytosine, for which it furnishes a satisfactory kinetic model and a probable mechanism. In agreement with earlier reports, uncatalyzed deamination was found to proceed at very similar rates for cytosine, 1-methylcytosine, cytidine, and cytidine 5′-phosphate, and also for cytosine residues in single-stranded DNA generated from a phagemid, in which we sequenced an insert representing the gene of the HIV-1 protease. Arrhenius plots for the uncatalyzed deamination of cytosine were linear over the temperature range from 90 °C to 200 °C and indicated a heat of activation (ΔH‡) of 23.4 ± 0.5 kcal/mol at pH 7. Recent evidence indicates that the surface of the earth has been cool enough to support life for more than 4 billion years and that life has been present for almost as long. If the temperature at Earth's surface is assumed to have followed Newton's law of cooling, declining exponentially from 100 °C to 25 °C during that period, then half of the cytosine-deaminating events per unit biomass would have taken place during the first 0.2 billion years, and <99.4% would have occurred during the first 2 billion years.
In the absence of enzymes, most biological reactions take place so slowly that they can be followed conveniently only at elevated temperatures (1). Among those reactions are several that may result in spontaneous mutation, notably the hydrolytic deamination of cytosine and 5-methylcytosine, which generate uracil and thymine, respectively. These deamination reactions have been shown to account for many of the single-site mutations that lead to inherited diseases in humans (2) and for the marked bias of spontaneous mutation toward increased AT content in microorganisms (3, 4). Rates of mutation have long been known to increase with increasing temperature, a phenomenon that Drake has termed “heat mutagenesis” (5).
There is widespread (6–8), if not universal (9), agreement that life originated when the earth was warmer—perhaps much warmer—than it is today. A recent isotopic analysis of carbon inclusions in zircons from the Jack Hills in Western Australia indicates that life may have emerged as early as 4.1 billion years ago (10), shortly after water first appeared at the surface in liquid form (11). Several present-day organisms thrive at temperatures near the boiling point of water. Thus, Ignisphera aggregans (12) and Pyrococcus horikoshii (13) exhibit optimal growth temperature of 92° and 98 °C, respectively, whereas another organism isolated from a hydrothermal vent has been reported to grow at 121 °C (14). Moreover, reconstructions of ancestral proteins, with amino acid sequences inferred from the sequences of their modern descendants, have been shown to be remarkably thermostable, with melting temperatures ∼30 °C higher than those of proteins from their modern descendants (15, 16).
If, as those findings suggest, life arose while the surface of the earth was still warm, and if early mechanisms for replication involved molecules bearing some resemblance to the components of modern DNA, then the thermal stabilities of those components are of special interest. To appreciate the burden borne by systems for nucleic acid repair in thermophiles, and the problems that may have been faced by primordial organisms before the advent of sophisticated systems for DNA repair, it would be desirable to have accurate information about the rate at which cytosine deamination and related reactions proceed at elevated temperatures. Several values have been reported for the enthalpy of activation (ΔH‡) for the neutral hydrolysis of cytosine derivatives, lying in the range between 12.2 and 29.0 kcal/mol (Table 1). The compounding effect of temperature on reaction rates, first identified by Harcourt and Essen (17), is such that Q75, here defined as the ratio of the reaction rate at 100 °C to the rate at 25 °C, is 70-fold if ΔH‡ = 12 kcal/mol, but 20,000-fold if ΔH‡ = 29 kcal/mol. In the present work, we set out to resolve these discrepancies by extensive Arrhenius analysis of rate constants gathered over a wide range of temperatures.
Table 1.
Enthalpies of activation for cytosine deamination
| Cytosine derivative (ref) | ΔH‡ (kcal/mol) | pH | Method | Temperature (°C) | No. points |
| Cytarabine (38) | 16.4 | 6.9 | UV | 24–47 | 4 |
| Cytosine (21) | 11.6 | 7.0 | UV | 70–90 | 4 |
| Cytidine (21) | 12.4 | 7.0 | UV | 70–90 | 4 |
| dCMP (21) | 28.4 | 7.4 | UV | 70–95 | 3 |
| ssDNA (21) | 28.4 | 7.4 | 14C chromat. | 70–95 | 3 |
| ssDNA (22) | 27.4 | 7.4 | Reversion | 30–90 | 13 |
| Cytidine (39) | 22.1 | 6.8 | UV | 80–157 | 9 |
| 2'-Deoxycytidine (37) | 24.8 | 6.8 | 1H-NMR | 90–137 | 6 |
| 1-Methylcytosine (37) | 24.6 | 6.8 | 1H-NMR | 90–137 | 6 |
| Cytosine (37) | 22.9 | 6.8 | 1H-NMR | 90–137 | 6 |
| Cytosine (this work) | 23.4 | 7.0 | 1H-NMR | 70–180 | 12 |
| 1-Methylcytosine-H+ (this work) | 23.0 | 2.4 | 1H-NMR | 90–200 | 12 |
| Average by 1H-NMR | 23.7 | 1H-NMR | |||
| Other C-N cleavages | |||||
| Acetamide (40) | 23.8 | 6.8 | 1H-NMR | ||
| Ac-gly—gly-NHMe (41) | 22.9 | 6.8 | 1H-NMR |
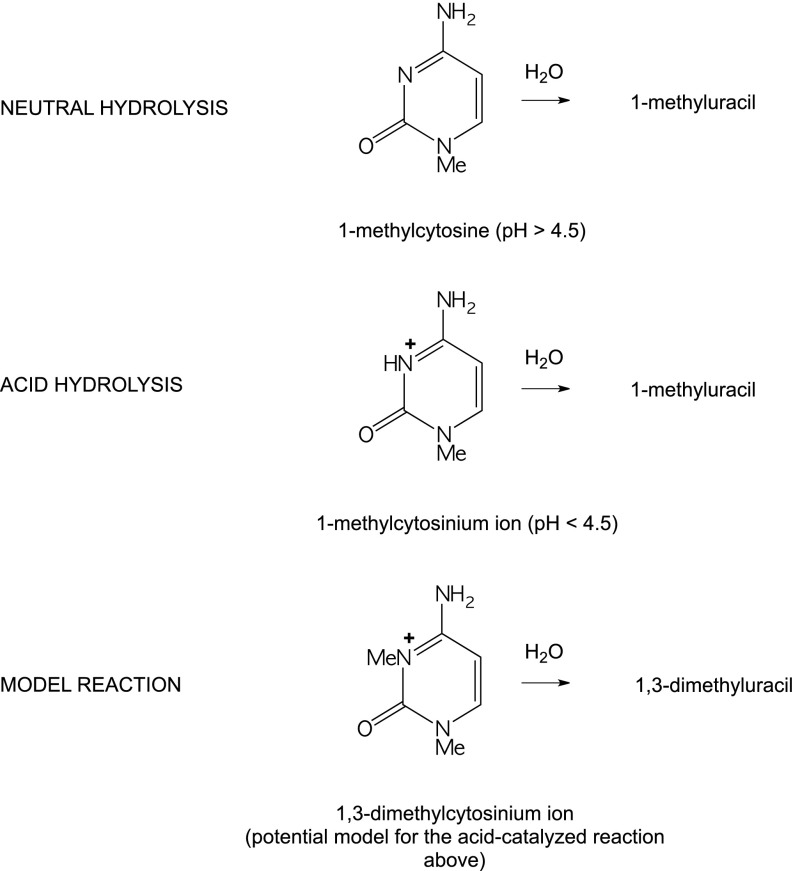
Earlier work has shown that both the enzymatic (18) and the nonenzymatic (19) deamination of cytidine proceed by 3,4-addition of water to form a tetrahedral intermediate, followed by elimination of ammonia. The nonenzymatic deamination of cytosine derivatives is subject to acid catalysis (20), but approaches a constant rate at pH values between 5 and 9 (21). At higher pH values, a base-catalyzed reaction sets in (21). In the present experiments, we tested the postulate that the acid-catalyzed reaction involves attack by water on the 3-protonated cytosinium ion, by comparing the behavior of 1-methylcytosine with that of 1,3-dimethylcytosinium ion (Me2C+) at various pH values (Fig. 1). To determine the effects of substituents, we also examined the kinetics of neutral hydrolysis of cytosine, 1-methylcytosine, 5-methylcytosine, cytidine, and cytidine 5′-phosphate. In experiments comparable with those reported by earlier investigators (22, 23), we extended those measurements to include cytosine residues in single-stranded DNA, using a phagemid in which we sequenced an insert representing the gene of the HIV-1 protease to compare the rates of cytosine-deaminating events with those of other potentially mutagenic reactions in single-stranded DNA (ssDNA).
Fig. 1.
1,3-Dimethylcytosine as a potential model for the acid-catalyzed hydrolysis of 1-methylcytosine.
Experimental Procedures
Rates of Deamination of Cytosine Derivatives.
1,3-Dimethylcytosine was prepared by the method of Yamauchi (24). Other cytosine derivatives were obtained from Sigma Chemical. Rates of hydrolysis of cytosine derivatives (0.02 M) were measured in potassium acetate, phosphate, borate, and carbonate buffers (0.1 M). Solutions were flushed with argon, sealed under vacuum in quartz tubes, and heated for various time intervals in Thermolyne 47099 furnaces. After cooling with ice, samples of the product mixtures (0.025 mL) were diluted with D2O-containing pyrazine as an internal reference for calibrating integrated proton signal intensities. 1H NMR spectra were acquired with a Bruker AVANCE 500 MHz spectrometer equipped with a cryoprobe, over one to four transients. Samples were heated in triplicate, over a time period long enough for the reaction to proceed to between 10% and 90% completion. The integrated intensities of reactants and products were used to follow the progress of reaction, and the disappearance of starting material followed first-order kinetics to at least 90% completion. Logarithms of the resulting rate constants were plotted as a function of the reciprocal of absolute temperature. Linear regression was used to obtain the slope and intercept of this Arrhenius plot.
Isolation of ssDNA Containing the Antisense Strand of the HIV-1 Protease Gene (pro).
Primers were constructed so that the 5′ ends contained portions of overlap with the plasmid pBlueScript+ that contains an f1 phage origin of replication (phagemid). The 3′ primer ends were designed to amplify a region of the infectious molecular clone pNL4-3 (25) from nucleotides 2006–2599 HIV-1 strain [HXB2 (GenBank accession no. K03455) numbering], which contains the entirety of the gene encoding for the viral protease, as well as some surrounding coding region. This region was PCR amplified from a convenient plasmid subclone of pNL4-3 and cloned into pBlueScript+ spanning from the EcoRI site to the HindIII site using the Gibson Assembly method (New England BioLabs). This orientation of pro was chosen to allow for expression of ssDNA corresponding to the antisense strand of pro. The construct was confirmed by Sanger sequencing.
The construct was transformed as a plasmid into 5-α F′ Iq Chemically Competent Escherichia coli (New England BioLabs) after which the cells were diluted to obtain isolated colonies on an agar plate containing ampicillin. LB medium was inoculated with one colony and incubated at 37 °C for 6 h. The culture was then inoculated with M13KO7 Helper Phage (New England BioLabs) and incubated at 37 °C for an additional hour to allow rescue of ssDNA from the pBlueScript+ in phage particles. The ssDNA was purified from the culture supernatant using a QIAprep Spin M13 Kit (Qiagen). The elution buffer was then exchanged for 1× phosphate buffered solution modified by addition of EDTA to 1 mM and titrated to a final pH of 8.0 using diafiltration spin columns Vivaspin 6 30-kDa molecular weight cut-off (GE Healthcare Bio-Sciences). The buffered ssDNA was stored at −20 °C. For experimental analysis, the ssDNA was divided into six aliquots and incubated at 90 °C for 0, 1, 2, 4, 8, and 16 h in a thermal cycler.
Primer ID Library Construction and Sequencing.
We used the previously described protocol (25, 26) to generate the amplicon for the viral protease coding domain as Primer ID sequencing libraries to sequence the antisense strand of HIV-1 protease gene from the incubated ssDNA described above, with the following modifications. We used Platinum Taq HiFi DNA polymerase (Life Technology) for first-strand synthesis of the ssDNA template using the Primer ID primer instead of using reverse transcriptase. The Primer ID primer sequence was GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNNNNCAGTGTAAAACGACGGCCAGT, containing 11 random nucleotides (a 10-nucleotide long Primer ID plus 1 random nucleotide as a buffer for sequencing). We set up separate first-strand DNA synthesis reactions at 400,000, 40,000, and 4,000 copies of the ssDNA template and estimated the value of OD260 for each temperature tested. The thermal cycler conditions for the first-strand synthesis were 95 °C for 5 min, 59 °C for 30 s, and 68 °C for 10 min. We separated the first-strand product from unused primer using Ampure XP PCR purification beads (Beckman Coulter). The first-round PCR amplification forward primer sequence was GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNCAGGAAACAGCTATGAC. After gel purification, libraries were pooled and sequenced using MiSeq 300-base paired-end platform (Illumina, CA). The raw sequence files are available at NIH Sequence Read Archive (www.ncbi.nlm.nih.gov/sra/SRP068325).
Bioinformatics Analysis.
We used the Illumina pipeline to deconstruct the multiplexed pooled libraries to create subsets of sequences representing each of the temperature conditions and with each template dilution. A template consensus sequence was created for each individual ssDNA template in each reaction by creating a consensus sequence for all sequences sharing the same Primer ID tag, as previously described (available at https://github.com/SwanstromLab/PID). Each template consensus sequence was aligned with the reference sequence using MUSCLE (v3.8.1) (27, 28) to remove sequences with insertions/deletions, and the mutation rate and types of substitutions at each position were calculated. We used R (v3.0.0) to plot mutation rates of all cytosine positions and to perform the Shapiro-Wilks normality test to show that the distribution of mutation rates fit a normal distribution.
Results
Influence of pH on the Hydrolysis of 1-Methylcytosine and 1,3-Dimethylcytosine.
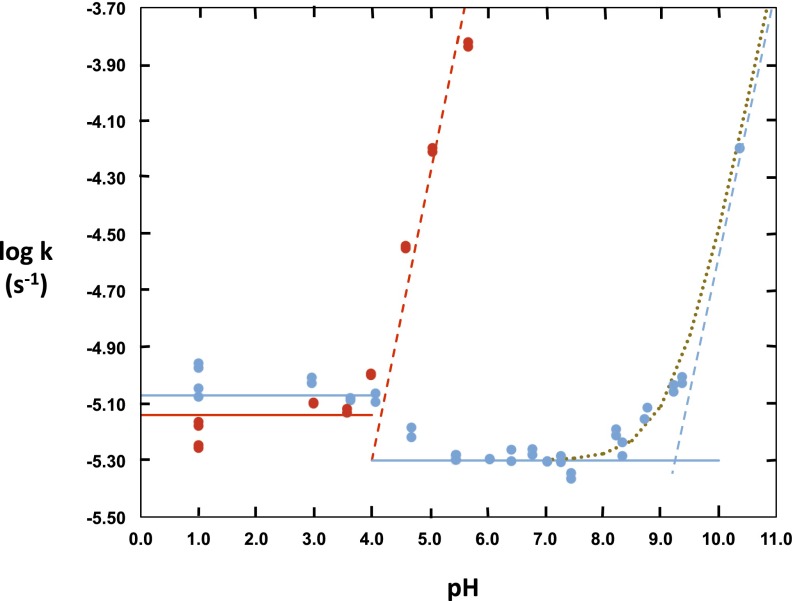
Rate constants for the hydrolysis of 1-methylcytosine and 1,3-dimethylcytosine, measured in buffered solutions at various pH values at 130 °C, are shown in Fig. 2. The pH values of reaction mixtures, measured before and after reaction at 25 °C, were found not to have changed significantly and were corrected to 130 °C using the established heats of proton release from the conjugate acid of each buffer (29). The hydrolysis of 1-methylcytosine (blue line) proceeded at a constant rate in acid solution at pH values between 1 and 4 and at an approximately threefold slower rate at pH values between 5 and 8.5. At pH values above 9.5, the rate of deamination was found to increase rapidly in proportion to the concentration of hydroxide ion. The red line shows the behavior of 1,3-dimethylcytosine, whose hydrolysis proceeds at a constant rate between pH 1 and 4. Although there has been controversy about the structure of the conjugate acid of cytosine derivatives, the scissile proton is now generally believed to combine with the 3-nitrogen atom of 1-substituted cytosine derivatives (Fig. 1) (30). The reactivities of 1,3-dimethylcytosine and 1-methylcytosine were found to be very similar at low pH, confirming that the scissile proton is indeed bound at N3 and indicating that the 1,3-dimethylcytosinium ion furnishes a satisfactory kinetic model and a probable mechanism for hydrolysis of the conjugate acid of 1-methylcytosine. At pH values above 4, where 1-methylcytosine loses its positive charge (Fig. 1), 1,3-dimethylcytosine retains a positive charge, and its rate of hydrolysis was found to increase with increasing pH in a manner consistent with hydroxide attack on the 1,3-dimethycytosinium cation.
Fig. 2.
Influence of pH on the deamination of 1-methylcytosine (blue) and 1,3-dimethylcytosine (red) at 130 °C. Heats of ionization of acetate (pH 3–5), phosphate (pH 5.5–8.5), and bicarbonate (pH 8.7–10) buffers were used to correct pH values of the samples—actually measured at 25 °C—to the corresponding values at 130 °C. Broken lines with unit slopes have been added as a visual reference.
Influence of Substituents on the Rate of Deamination of Cytosine Derivatives.
When the deamination of cytosine at 130 °C was compared with the deamination of several cytosine derivatives, in 0.1 M potassium phosphate buffer (pH 7.0), these reactions were found to proceed with very similar first-order rate constants: cytosine 6.5 (±0.75) × 10−6 s−1; 1-methylcytosine 5.4 (±0.3) × 10−6 s−1; cytidine 7.0 (±0.1) × 10−6 s−1; cytidine 5′-phosphate 6.8 (±0.2) × 10−6 s−1. The absence of a significant effect of these substituents is noteworthy.
Under the same conditions, 5-methylcytosine was deaminated twice as rapidly as cytosine, with a first-order rate constant of 1.34 (±0.07) × 10−5 s−1. Earlier, Lindahl and Nyberg reported a fourfold enhancement of the rate of deamination of cytidine 5′-phosphate by a 5-methyl substituent at 95 °C (22). It seems reasonable to attribute the modest activating effect of a 5-methyl group to an increase in the polarizability of the substrate in the neighborhood of the scissile C-N bond of the substrate, rendering it more susceptible to nucleophilic attack.
Influence of Temperature on the Rate of Cytosine Deamination.
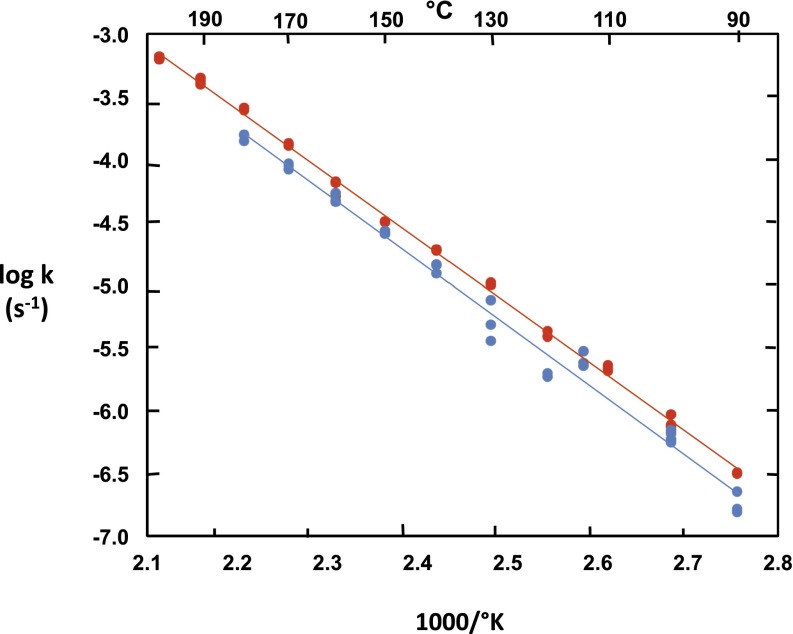
In Fig. 3, first-order rate constants (seconds−1) observed for the deamination of cytosine and 1-methylcytosine at pH 7.0 are plotted as a logarithmic function of the reciprocal of absolute temperature, in the range between 90 °C and 200 °C. The linearity of these Arrhenius plots over a wide range of temperatures suggests the absence of significant heat capacity effects and indicates that the enthalpies of activation are 23.4 kcal/mol for cytosine and 23.0 kcal/mol for 1-methylcytosine. For cytosine
| [1] |
where log k = 7.7 and [d(log k)/d(1,000/Tkelvin)] = −5.23. Table 1 compares these values of ΔH‡ with values reported in the literature for cytosine and several derivatives. The substantial discrepancies between the present ΔH‡ values (averaging 23.7 kcal/mol) and some of those reported in the earlier literature, which vary over the range between 12.2 and 29.0 kcal/mol, may be explained by the reliance of some earlier values on relatively few data points distributed over a relatively limited temperature range and to the fact that they were based on changes in UV spectrum that incorporated the ratios of molar absorbancies at two wavelengths, each of which constitutes a potential source of experimental error. In the present 1H-NMR experiments, the integrated intensities of proton NMR signals allowed the disappearance of reactants and the appearance of products to be monitored independently, with results that were in close accord.
Fig. 3.
Temperature dependence of rate constants for the deamination of 1-methylcytosine at pH 2.45 (red) and cytosine at pH 7.0 in phosphate buffer (blue), in the range between 90° and 200 °C. For 1-methylcytosine, k25 = 2.8 × 10−10 s−1, ΔH‡ = 23.0 kcal/mol. For cytosine, k25 = 1.6 × 10−10 s−1, ΔH‡ = 23.4 kcal/mol.
For comparison with other molecules equipped with sp2-hybridized amino groups similar to those in cytidine, Table 1 also includes ΔH‡ values that have been reported for the neutral hydrolysis of two carboxylic acid amides (31). Acetamide and the glycylglycine peptide bond of acetylglycylglycine N-methylamide have been found to undergo hydrolysis with ΔH‡ values very similar to those observed for cytosine and its derivatives in the present experiments.
Cytosine Deamination in ssDNA.
We monitored the spontaneous deamination of cytosine in ssDNA by measuring the change in base-pairing capacity using deep sequencing. In our deep sequencing protocol, each starting template was marked with a unique sequence tag (Primer ID) to allow the number of templates sequenced to be determined and to allow the repetitive sequences of these starting templates to be pooled to make a very accurate consensus sequence obtained for each template. Template sequence recovery of more than 90% was achieved by producing an average of 30-fold sequencing coverage per template. To calculate the mutation rate, we chose template titrations that yielded at least 1,000 but less than 10,000 template consensus sequences. The paired-end sequencing protocol gives an R1 sequence from one end and an R2 sequence from the other end. We did not include the last 50 nucleotides from the R2 end because the sequencing quality dropped dramatically toward the end. Overall, the sequenced region was 495 bp (278 for R1 and 217 for R2 after trimming of last 50 nucleotides) and contained 128 deoxycytidine positions, allowing us to sample between 122,368 and 676,543 deoxycytidines, spread across the templates, at each temperature. Less than 1% of the template consensus sequences had deletions, and those were excluded from mutation rate analysis. Cytosine-to-thymine transition mutations increased with increasing time of incubation at 90 °C (Fig. 4), whereas other types of substitution (A > C, A > G, A > T, C > A, C > G, G > A, G > C, G > T, T > A, T > C, T > C) showed no detectable increase and would have been detected if they had occurred at an estimated 20% of the rate of C > T substitution. The baseline mutation rate, at zero time, was 0.184 per 1,000 cytosines sequenced, reflecting the sequencing error and the presence of any preexisting C > T deamination events. After a 16-h incubation at 90 °C, the C-to-T mutations increased to 5.8 per 1,000 deoxycytidines sequenced. In Fig. 4, the C-to-T mutation rate is plotted as a function of incubation time and shows a strong linear correlation (R2 = 1.00). We also estimated the C-to-T mutation rate at each deoxycytidine position (a total of 128 positions) after 16 h, and we did not find any outlier positions with extraordinary high C-to-T mutation rate. The distribution of mutation rates followed the normal distribution (Shapiro–Wilks normality test, P = 0.91).
Fig. 4.
C-to-T mutations (per 1,000 cytosine sequenced) as a function of incubation time at 90 °C. The regression line fits the equation (C-to-T mutations per thousand positions) = 0.22 + 0.35 (hours of incubation), with R2 = 1.00.
It is of interest to compare the mutation rate per deoxycytidine residue at 90 °C, based on these mutation experiments, with the value that would be expected on the basis of the Arrhenius plots in Fig. 3, which yielded a first order rate constant of 2.2 × 10−7 s−1 for deamination of cytosine at pH 7 and 90 °C. The mutation experiments yielded a first-order rate constant of 0.35 h−1 per thousand deoxycytidine residues, equivalent to 9.7 × 10−8 s−1 per deoxycytidine residue in this ssDNA. Thus, mutation occurred at 44% of the rate expected from the rate of deamination of free cytosine under the same conditions. Rate constants reported by Lindahl and Nyberg for deamination of all of the 14C-labeled deoxycytidine residues in single-stranded E. coli DNA (2.2 × 10−7 s−1 at 95 °C) (22) and by Frederico et al. for reversion of a mutant cytosine residue in the single-stranded DNA of bacteriophage M13 (1.3 × 10−7 s−1 at 90 °C) (23) are equivalent to 65% and 59%, respectively, of the present values for cytosine at the corresponding temperatures. Thus, incorporation of cytosine into single-stranded DNA appears to reduce its rate of deamination by a factor of ∼2. It should be added that the rates of deamination of individual cytosine residues in bacteriophage T4 DNA are not uniform but appear to be site specific in bacteriophage T4 (32).
Discussion
It is of interest to consider the probable time course of deamination-based mutation during Earth’s history, based on the temperature-dependence of cytosine deamination. In the absence of detailed information about the thermal history of the Earth, it seems reasonable to expect a roughly exponential decline of the surface temperature, following Newton's law of cooling, toward a steady state maintained by the balance between the loss of heat to space and the gain of heat from the core and from solar radiation.
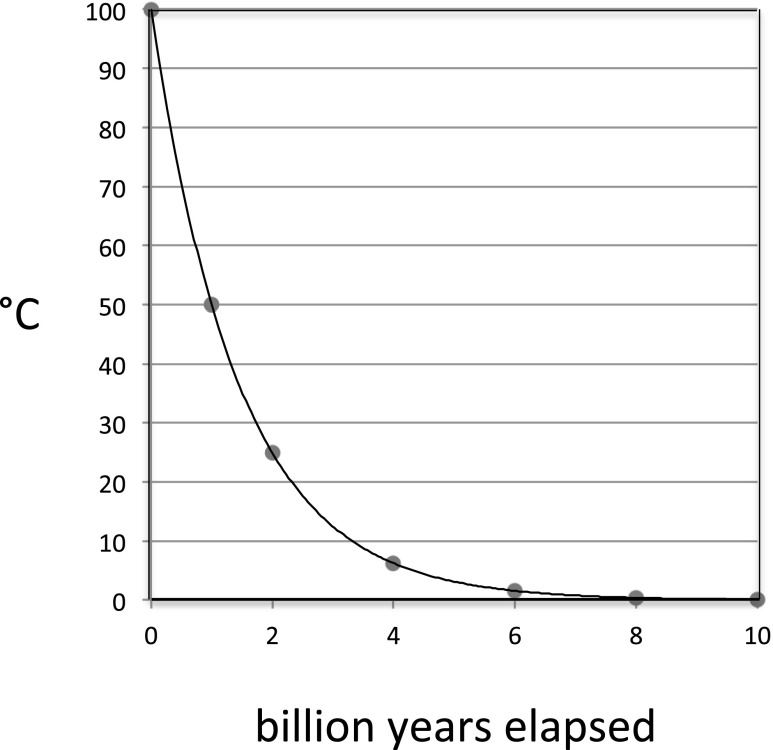
For the sake of argument, let us suppose that the surface temperature declines from T0 = 373° K (100 °C) as a starting point—below which water becomes a liquid at Earth's surface and life becomes possible—toward an ultimate steady state value (T∞) of 273 °K (0 °C). That choice of T∞ is arbitrary, but its actual value is not expected to affect our conclusions in a qualitative sense.
If, after the elapse of 4 billion years, the surface temperature is now 298 °C (25 °C), then the half-life for cooling is 2 billion years, and the surface temperature decreases with time according to the first-order equation
| [2] |
where To = 100 °C and t = billions of years that have elapsed since 4 billion years ago. Fig. 5 illustrates the expected decline in temperature over the first 10 billion years.
Fig. 5.
Hypothetical decline in temperature (°C) as a function of time elapsed (by) since the earth's surface reached 100 °C (Eq. 1), assuming that T∞ = 0 °C.
Suppose further that the rate constant for mutation (kmut), like the rate constant for cytosine deamination, varies as a logarithmic function of the reciprocal of absolute temperature according to the Arrhenius relationship (Eq. 1 and Fig. 3), with a heat of activation (ΔH‡) of 24 kcal/mol.
Combining the expected rate of mutation as a function of temperature (Eq. 1) with the estimated temperature as a function of time elapsed (Eq. 2), one can estimate the expected rate of mutation as a function of time elapsed. Fig. 6 illustrates the first 4 billion years and shows that the rate of mutation declines much more steeply with time than would be expected for a simple first order process (depicted by the broken black line that has been added as a visual reference). The effect of this steep decline would have been to compress cytosine-deaminating events in ssDNA into the first parts of Earth’s history. Of the ∼4 billion years that we assume to have passed since the origin of life, 95% would have occurred by the end of the first, 99.3% by the end of the second, and 99.9% by the end of the third billion years. Of the total number of mutations per unit biomass that had occurred after 4 billion years, 50% would have occurred after the first 0.2 billion years.
Fig. 6.
Decline in the rate of mutation in single-stranded DNA with time, per unit of biomass, based on an assumed first-order decline in temperature, from 100 °C at 4 billion years ago to 25 °C at present, with T∞ = 0 °C, and ΔH‡ = 24 kcal/mol. The broken line, added solely as a visual reference, shows the behavior that would be expected for a simple first order decline in the rate constant as a function of time.
The steady decline of temperature with time shown in Fig. 6 is obviously an oversimplification. There appear to have been five temporary interruptions, beginning with the Archaic Ice Age ∼2 billion years ago. In other respects, the decline appears to have been fairly steady; only recently, at the outset of the Devonian period 0.4 billion years ago, does the marine environment appear to have arrived at a temperature approaching the temperature at the surface of a modern tropical ocean (33).
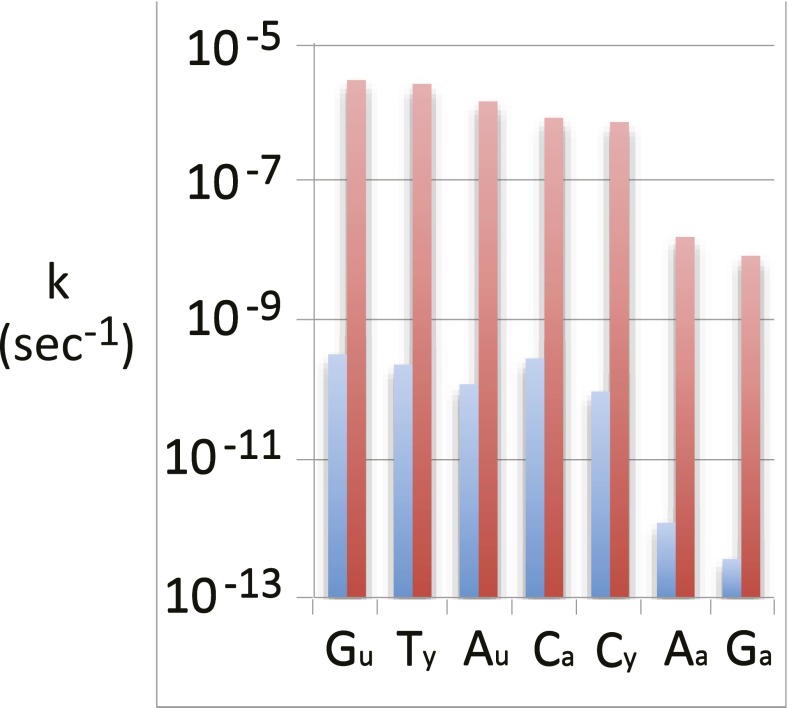
Is cytosine deamination expected to have dominated the rate of mutation at all temperatures? Fig. 7 shows that several other potentially mutagenic processes, including deamination and glycoside cleavage reactions, proceed at comparable rates. Each of these reactions exhibits a larger ΔH‡ value than does cytosine deamination. To the extent that it contributes to spontaneous mutation, the mutation rate would therefore be expected to have declined even more steeply with temperature than would the rate based on cytosine deamination alone.
Fig. 7.
First-order rate constants (s−1) in neutral solution at 25° (blue) and 100 °C (red) for the deamination (a), depurination (u), and depyrimidination (y) of deoxyguanosine, deoxythymidine, deoxycytidine, and deoxyadenosine, from ref. 37. Thus, Ca, Aa, and Ga represent the deamination of deoxycytidine, deoxyadenosine, and deoxyguanosine; Cy and Ty represent cleavage of the cleavage of the linkage between a pyrimidine base and 2-deoxyribose; and Au and Gu represent cleavage of the linkage between a purine base and 2-deoxyribose.
An important factor that we have not discussed up to this point is the influence of base pairing. High temperatures have long been known to favor single-strandedness, as indicated by the melting of double-stranded DNA (34); in a model for the reverse process, the stacking and H-bonding of a terminal AU base pair in double-stranded DNA have been found to release heat (35). In E. coli DNA (22), and at a specific cytosine residue that had been introduced by mutation of the lacZα gene coding sequence of bacteriophage M13mp2 (23), deoxycytidine residues have been found to be deaminated >100-fold more slowly in double-stranded than in ssDNA. In the presence of a complementary strand of DNA, a further retardation in the rate of deamination would be expected to have occurred as the temperature fell below the melting point, accentuating the decline shown for ssDNA in Fig. 6. Organisms adapted to high temperatures may well be equipped with additional mechanisms for reducing DNA damage that remain to be discovered. The detection of phage particles in the water of hot springs, in which the particles are at least partially resistant to extensive exposure to high temperatures (36), may furnish one experimental setting for determining the mechanism and extent to which the constituents of phage particles are able to protect DNA.
Recent work has uncovered the startling extent to which slow reactions are accelerated by elevated temperatures. This acceleration would have collapsed, by many orders of magnitude, the time that would have been required for chemical events on a warm earth before the advent of enzymes (1). It has also been shown that a primitive enzyme, if it enhanced the rate of a reaction by lowering its enthalpy of activation, would have produced a rate enhancement that increased—automatically—as the environment cooled. Simple catalysts have been shown to exhibit that behavior in aqueous solution, as do most present-day enzymes (8). That scenario for enzyme evolution does not take into account improvements arising by natural selection. The present findings confirm and extend earlier evidence that the chemical events that lead to spontaneous mutation are highly sensitive to temperature, furnishing a mechanism for accelerating evolution during Earth’s early history. Later—at lower temperatures—DNA would have become a much more stable repository for genetic information.
Acknowledgments
We thank John Drake, Michael Caplow, Howard Fried, Lee Pedersen, Peter Wolfenden, and Günter Wächtershäuser for helpful discussions. These experiments were supported by NIH Grants GM-18325 (to R.W.) and R37 AI44667 (to R.S.), the University of North Carolina (UNC) Center for AIDS Research (NIH Award P30 AI50410), and the UNC Lineberger Comprehensive Cancer Center (NIH Award P30 CA16068).
Footnotes
Conflict of interest statement: University of North Carolina is pursuing intellectual property protection for primer ID, and R.S. is listed as a coinventor. Other authors declare no conflict interest.
Data deposition: The sequence reported in this paper has been deposited in the NIH Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. SRP068325).
References
- 1.Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Acc Chem Res. 2001;34(12):938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Hershberg R, Petrov DA. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6(9):e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wielgoss S, et al. Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3 (Bethesda) 2011;1(3):183–186. doi: 10.1534/g3.111.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drake JW, Baltz RH. The biochemistry of mutagenesis. Annu Rev Biochem. 1976;45:11–37. doi: 10.1146/annurev.bi.45.070176.000303. [DOI] [PubMed] [Google Scholar]
- 6.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276(5313):734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 7.Di Giulio M. The universal ancestor was a thermophile or a hyperthermophile: Tests and further evidence. J Theor Biol. 2003;221(3):425–436. doi: 10.1006/jtbi.2003.3197. [DOI] [PubMed] [Google Scholar]
- 8.Stockbridge RB, Lewis CA, Jr, Yuan Y, Wolfenden R. Impact of temperature on the time required for the establishment of primordial biochemistry, and for the evolution of enzymes. Proc Natl Acad Sci USA. 2010;107(51):22102–22105. doi: 10.1073/pnas.1013647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller SL, Lazcano A. The origin of life--did it occur at high temperatures? J Mol Evol. 1995;41:689–692. doi: 10.1007/BF00173146. [DOI] [PubMed] [Google Scholar]
- 10.Bell EA, Boehnke P, Harrison TM, Mao WL. Potentially biogenic carbon preserved in a 4.1 billion-year-old zircon. Proc Natl Acad Sci USA. 2015;112(47):14518–14521. doi: 10.1073/pnas.1517557112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilde SA, Valley JW, Peck WH, Graham CM. Evidence from detrital zircons for the existence of continental crust and oceans on the Earth 4.4 Gyr ago. Nature. 2001;409(6817):175–178. doi: 10.1038/35051550. [DOI] [PubMed] [Google Scholar]
- 12.Niederberger TD, Götz DK, McDonald IR, Ronimus RS, Morgan HW. Ignisphaera aggregans gen. nov., sp. nov., a novel hyperthermophilic crenarchaeote isolated from hot springs in Rotorua and Tokaanu, New Zealand. Int J Syst Evol Microbiol. 2006;56(Pt 5):965–971. doi: 10.1099/ijs.0.63899-0. [DOI] [PubMed] [Google Scholar]
- 13.González JM, et al. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998;2(2):123–130. doi: 10.1007/s007920050051. [DOI] [PubMed] [Google Scholar]
- 14.Kashefi K, Lovley DR. Extending the upper temperature limit for life. Science. 2003;301(5635):934. doi: 10.1126/science.1086823. [DOI] [PubMed] [Google Scholar]
- 15.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian β-lactamases. J Am Chem Soc. 2013;135(8):2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 16.Akanuma S, et al. Experimental evidence for the thermophilicity of ancestral life. Proc Natl Acad Sci USA. 2013;110(27):11067–11072. doi: 10.1073/pnas.1308215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harcourt AV, Essen WT. On the laws of connexion between the conditions of chemical change and its amount. Philos Trans R Soc Lond. 1866;156:193–221. [Google Scholar]
- 18.Snider MJ, Wolfenden R. Site-bound water and the shortcomings of a less than perfect transition state analogue. Biochemistry. 2001;40(38):11364–11371. doi: 10.1021/bi011189+. [DOI] [PubMed] [Google Scholar]
- 19.Snider MJ, Reinhardt L, Wolfenden R, Cleland WW. 15N kinetic isotope effects on uncatalyzed and enzymatic deamination of cytidine. Biochemistry. 2002;41(1):415–421. doi: 10.1021/bi011410i. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro R, Klein RS. The deamination of cytidine and cytosine by acidic buffer solutions. Mutagenic implications. Biochemistry. 1966;5(7):2358–2362. doi: 10.1021/bi00871a026. [DOI] [PubMed] [Google Scholar]
- 21.Garrett ER, Tsau J. Solvolyses of cytosine and cytidine. J Pharm Sci. 1972;61(7):1052–1061. doi: 10.1002/jps.2600610703. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 23.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry. 1990;29(10):2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 24.Yamauchi K. Methylation of nucleic acid bases with trimethyl phosphate. J Org Chem. 1976;41(23):3691–3696. [Google Scholar]
- 25.Adachi A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59(2):284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou S, Jones C, Mieczkowski P, Swanstrom R. Primer ID validates template sampling depth and greatly reduces the error rate of next-generation sequencing of HIV-1 genomic RNA populations. J Virol. 2015;89(16):8540–8555. doi: 10.1128/JVI.00522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberg RN, Kishore N, Lennen RM. Thermodynamic quantities for the ionization reactions of buffers. J Phys Chem Ref Data. 2002;31:231–370. [Google Scholar]
- 30.Elguero J, Marzin C, Katritzky AR, Linda P. The Tautomerism of Heterocycles. Advances in Heterocyclic Chemistry, Supplement 1. Academic Press; New York: 1976. pp. 161–162. [Google Scholar]
- 31.Wolfenden R. Degrees of difficulty of water-consuming reactions in the absence of enzymes. Chem Rev. 2006;106(8):3379–3396. doi: 10.1021/cr050311y. [DOI] [PubMed] [Google Scholar]
- 32.Baltz RH, Bingham PM, Drake JW. Heat mutagenesis in bacteriophage T4: The transition pathway. Proc Natl Acad Sci USA. 1976;73(4):1269–1273. doi: 10.1073/pnas.73.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behringer W. A cultural History of Climate. Polity Press; Cambridge, MA: 2010. pp. 20–26. [Google Scholar]
- 34.Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- 35.Petersheim M, Turner DH. Base-stacking and base-pairing contributions to helix stability: Thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983;22(2):256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 36.Breitbart M, Wegley L, Leeds S, Schoenfeld T, Rohwer F. Phage community dynamics in hot springs. Appl Environ Microbiol. 2004;70(3):1633–1640. doi: 10.1128/AEM.70.3.1633-1640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schroeder GK, Wolfenden R. Rates of spontaneous disintegration of DNA and the rate enhancements produced by DNA glycosylases and deaminases. Biochemistry. 2007;46(47):13638–13647. doi: 10.1021/bi701480f. [DOI] [PubMed] [Google Scholar]
- 38.Notari RE. A mechanism for the hydrolytic deamination of cytosine arabinoside in aqueous buffer. J Pharm Sci. 1967;56(7):804–809. doi: 10.1002/jps.2600560703. [DOI] [PubMed] [Google Scholar]
- 39.Snider MJ, Gaunitz S, Ridgway C, Short SA, Wolfenden R. Temperature effects on the catalytic efficiency, rate enhancement, and transition state affinity of cytidine deaminase, and the thermodynamic consequences for catalysis of removing a substrate “anchor”. Biochemistry. 2000;39(32):9746–9753. doi: 10.1021/bi000914y. [DOI] [PubMed] [Google Scholar]
- 40.Callahan BP, Yuan Y, Wolfenden R. The burden borne by urease. J Am Chem Soc. 2005;127(31):10828–10829. doi: 10.1021/ja0525399. [DOI] [PubMed] [Google Scholar]
- 41.Radzicka A, Wolfenden R. Rates of uncatalyzed hydrolysis of peptide bonds in neutral solution, and the transition state affinities of proteases. J Am Chem Soc. 1996;118(26):6105–6109. [Google Scholar]