Abstract
Understanding how geography, oceanography, and climate have ultimately shaped marine biodiversity requires aligning the distributions of genetic diversity across multiple taxa. Here, we examine phylogeographic partitions in the sea against a backdrop of biogeographic provinces defined by taxonomy, endemism, and species composition. The taxonomic identities used to define biogeographic provinces are routinely accompanied by diagnostic genetic differences between sister species, indicating interspecific concordance between biogeography and phylogeography. In cases where individual species are distributed across two or more biogeographic provinces, shifts in genotype frequencies often align with biogeographic boundaries, providing intraspecific concordance between biogeography and phylogeography. Here, we provide examples of comparative phylogeography from (i) tropical seas that host the highest marine biodiversity, (ii) temperate seas with high productivity but volatile coastlines, (iii) migratory marine fauna, and (iv) plankton that are the most abundant eukaryotes on earth. Tropical and temperate zones both show impacts of glacial cycles, the former primarily through changing sea levels, and the latter through coastal habitat disruption. The general concordance between biogeography and phylogeography indicates that the population-level genetic divergences observed between provinces are a starting point for macroevolutionary divergences between species. However, isolation between provinces does not account for all marine biodiversity; the remainder arises through alternative pathways, such as ecological speciation and parapatric (semiisolated) divergences within provinces and biodiversity hotspots.
Keywords: biogeography, coral reefs, evolution, marine biodiversity, speciation
Phylogeography has roots in biogeography, wherein geographic provinces are identified by concordant shifts in species composition. If the partitions defined by taxonomy are regarded as first-order approximations of evolutionary genetic separations, then continuity between biogeography and phylogeography is apparent. Marine biogeography, the study of species’ distributions and evolutionary processes in the sea, began in the mid-19th century based on taxonomic distinctions. Dana (1) divided the surface waters of the world into several temperature zones based on the distributions of corals and crustaceans. Woodward (2) identified a series of marine provinces based on the distributions of mollusks. Forbes (3) made three enduring observations: (i) each biogeographic province is a center of origin for new species, (ii) these new species tend to migrate outward from the center of origin, and (iii) provinces, like species, must be traced back to their historical origins to be understood. These three fundamental contributions appeared in the same decade in which Darwin and Wallace (4) and Darwin (5) identified geography and natural selection as agents of evolutionary change.
It is remarkable that five essential publications in the 1850s (1–5) set the stage for 150 y of biogeographic research. Subsequent effort was devoted to species descriptions, geographic ranges, and relationships. Evolutionary hypotheses were formulated by examining the morphology and distribution of organisms. However, not until the advent of molecular technologies in the 1970s did biogeography transition through another fundamental change (6).
A primary theme emerging from marine biogeography is concordant levels of endemism in very diverse taxa. For example, endemism in Hawai’i is 25% for red algae and fishes (7, 8) and 20% for mollusks (9). The Caribbean Province has 33% endemism for fishes (10), 32% for decapod crustaceans (11), and 37% for corals (12). In the Red Sea, endemism is 13% for fishes and polychaetes, 8% for echinoderms, 17% for ascidians, and 5.5% for corals (13). This concordance across diverse taxonomic groups indicates unifying evolutionary processes.
Here, we demonstrate concordance between biogeographic provinces defined by taxonomy and phylogeographic clusters identified with DNA sequences. At the level of interspecific comparisons, this concordance is obvious; genetic partitions between sister species are expected. However, below this level, at the inception of speciation, it is still unclear how genetic partitions within species (defined by allele-frequency shifts and significant F-statistics) translate into species-level divergences (reciprocal monophyly and morphological distinction). Concordance between taxonomy-based biogeography and genetic-based phylogeography would indicate a continuum from population isolation to morphological divergence to evolutionary innovation. In this review, we examine comparative phylogeography, first across biogeographic provinces and second across taxonomic groups with widely divergent life histories.
A second goal is to summarize aspects of comparative phylogeography that illuminate the origins of marine biodiversity. As in terrestrial and freshwater systems, phylogeographic comparisons among species often reveal a diversity of outcomes, attributed to the idiosyncrasies of individual taxa (14, 15). However, the comparative approach can reveal insights unavailable from any one example (16), as illustrated by the terrestrial biota of Hawai’i (17). Finally, illuminating the origins of new species at biodiversity hotspots and centers of endemism can illustrate conservation priorities for the ocean, the cradle of life on our beleaguered planet.
Biogeographic Provinces
Tropical Oceans.
Tropical oceans are characterized by biodiversity hotspots, including the Caribbean and the Coral Triangle (between the Philippines, Indonesia, and New Guinea) (Fig. 1A) and endemism hotspots, such as Hawai’i and the Red Sea on the periphery of the Indo-Pacific. The evolutionary role of biodiversity hotspots versus endemism hotspots is contentious although biodiversity hotspots are widely recognized as evolutionary incubators producing new species (31, 32).
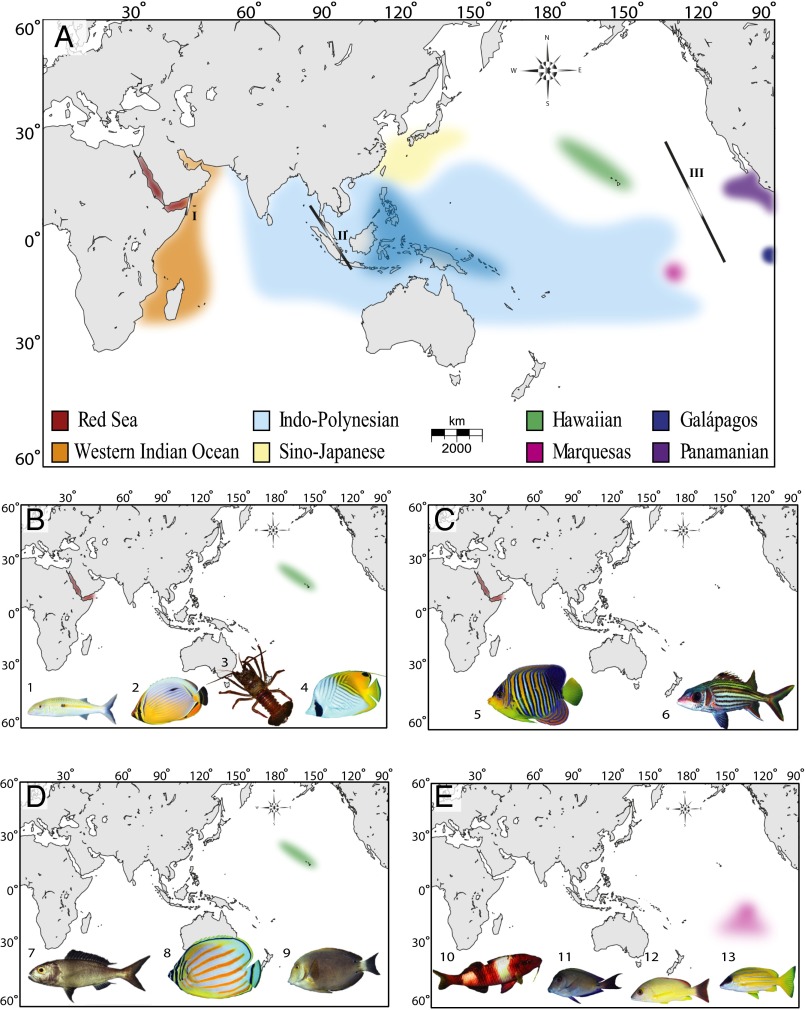
Fig. 1.
(A) Biogeographic provinces of the tropical Indo-Pacific as defined by >10% endemism (18). Coral triangle is indicated in dark blue. Primary barriers include (site I) Red Sea Barrier, (site II) Indo-Pacific Barrier, and (site III) East Pacific Barrier. (B–E) Minimaps illustrating widespread species with phylogeographic separation (strong allele-frequency shifts and significant F-statistics) at peripheral provinces. For each panel, the peripheral region(s) of phylogeographic distinction is highlighted in color, and photos are of the species with genetic evidence for that pattern as follows: (B) Hawai’i and the Red Sea [1, Mulloidichthys flavolineatus (19); 2, Corallochaetodon species complex (20); 3, Panulirus penicillatus (21); 4, Chaetodon auriga (22)]; (C) Red Sea only [5, Pygoplites diacanthus (23); 6, Neoniphon sammara (24)]; (D) Hawai’i only [7, Pristipomoides filamentosus (25); 8, Chaetodon ornatissimus (26); 9, Acanthurus nigroris (27)]; (E) Marquesas/French Polynesia [10, Parupeneus multifasciatus (28); 11, Acanthurus nigrofuscus (29); 12, Lutjanus fulvus (30); 13, Lutjanus kasmira (30)]. Photo credits: J. E. Randall/FishBase (photograph 7); Tane Sinclair-Taylor (all other fish photographs); Matthew Iacchei (photograph of Panulirus penicillatus).
The Coral Triangle has been a stable reef habitat for tens of millions of years, and this persistence is believed to be key to the production and export of species (33). Pervasive signals of population structure indicate that novel species are arising by parapatric means within the Coral Triangle, wherein partial isolation between subregions reinforces isolation along ecological gradients (34–37). Based on phylogenies of three reef fish families, Cowman and Bellwood (38) estimate that 60% of Indo-Pacific reef fauna have origins in the Coral Triangle. In contrast, peripheral endemism hotspots were previously regarded as evolutionary dead ends (39, 40), in which rare colonization events can produce endemic species, but with no further evolutionary radiations. This assumption has been challenged in recent years because phylogeographic studies show that both Hawaiian and Red Sea provinces can export novel biodiversity (24, 41).
The dominant feature of tropical marine biogeography is the vast Indo-Polynesian Province (IPP), spanning almost half the planet (Fig. 1A). Concomitant with this large province are unusually large range sizes, averaging 9 million km2 for reef fishes, roughly the size of mainland China (42). Genetic surveys of reef organisms are generally consistent with the boundaries of the IPP, showing little genetic structure across broad areas with a few important exceptions (e.g., Indo-Pacific Barrier) (41). Schultz et al. (43) use bathymetry profiles to demonstrate that dispersal across most of this range (Polynesia to Western Australia) requires no deep-water traverse greater than 800 km. Undoubtedly, this continuity of shallow habitat contributes to the cohesiveness of the IPP.
At the center of this vast province is an intermittent barrier around the Indo-Malay Archipelago, known as the Indo-Pacific Barrier (Fig. 1A). In the mid-Miocene (16–8 Ma), the Australian and Eurasian plates collided and reduced water flow between the Pacific and Indian Oceans (44). During Pleistocene glacial cycles, sea level dropped as much as 130 m below present levels, further constricting connections between these ocean basins. Evidence for interruptions of gene flow can be found in the distributions of sister species, coupled with phylogeographic partitions (as defined by reciprocal monophyly or ΦST > 0.10) in green turtles (45), dugongs (46), and ∼80% of surveyed reef species (Fig. 2) (30, 52). Given the cyclic nature of this barrier, phylogeographic partitions driven by Pleistocene glacial fluctuations are expected to be concordant in terms of geography, but not necessarily concordant in terms of chronology.
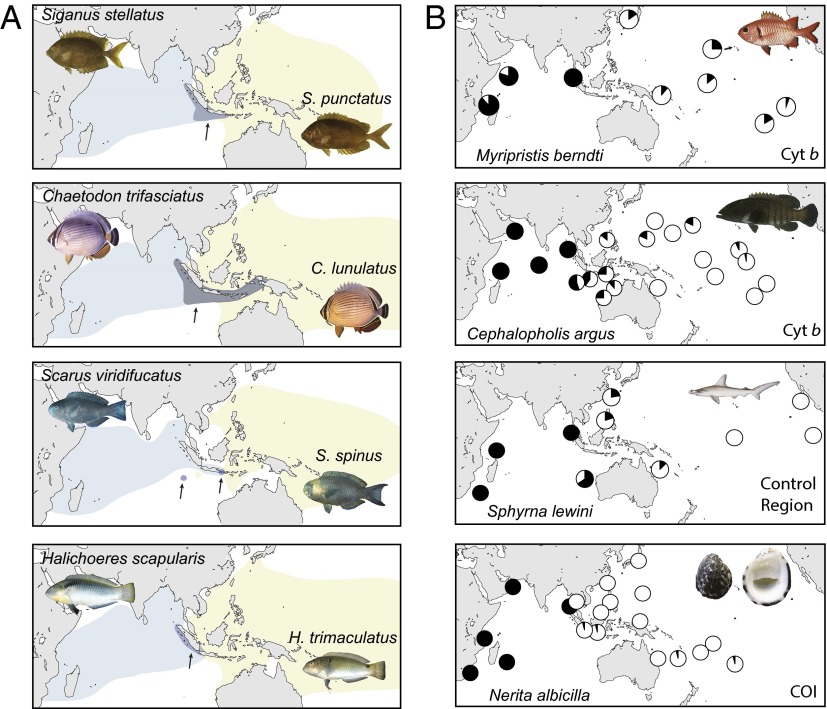
Fig. 2.
Evidence of isolation across the Indo-Pacific Barrier. (A) Distribution patterns of sister species pairs. Distributions shaded in purple (and indicated by arrows) represent areas of species overlap. (B) Phylogeographic studies demonstrating divergent genetic lineages within species. Black and white in pie diagrams indicate distribution of mtDNA phylogroups separated by at least three mutations. In all cases, there is evidence of population expansion with overlap in the Indo Malay-Philippine biodiversity hotspot (Coral Triangle) (47). Myripristis berndti data from Craig et al. (48), Cephalopholis argus data from Gaither et al. (49), Sphyrna lewini data from Duncan et al. (50), and Nerita albicilla data from Crandall et al. (51). COI, cytochrome oxidase subunit 1; Cyt b, cytochrome b. Photo credit: J. E. Randall for fishes, Wikimedia commons/Harry Rose for Nerita albicilla.
On the eastern periphery of the enormous IPP are three isolated provinces with high endemism in reef fishes: (i) the Hawaiian Islands with 25% endemism (8), (ii) the Marquesas Islands with 13.7% endemism (53), and (iii) Easter Island with 21.7% endemism (54). Phylogeographic studies of the first two provinces show strong concordance with biogeographic partitions (Fig. 1B). In Hawai’i, 11 of 16 fishes surveyed are genetically distinct from conspecifics elsewhere in the Pacific (reviewed in ref. 25). At the Marquesas, three of five studies reveal divergences that range from FST ≥ 0.24 at allozyme loci to reciprocal monophyly at mtDNA (28, 30), and a RADSeq study reveals strong divergence between a Marquesan surgeonfish and a widespread sister species (55).
On the western side of the IPP lies the Red Sea biogeographic province, an endemism hotspot characterized by a shallow connection to the Indian Ocean and latitudinal gradients in temperature, salinity, and nutrient load (13, 56). Many Red Sea endemics have sister species in the adjacent Western Indian Ocean (56). This interspecific pattern aligns with mtDNA partitions within species ranging from haplotype frequency shifts to reciprocal monophyly in fishes and invertebrates (table 2 in ref. 56). For example, the Indo-Pacific damselfish (Dascyllus aruanus; 57) and yellowstripe goatfish (Mulloidichthys flavolineatus; 19) both demonstrate similar divisions in mtDNA sequences (ΦST > 0.65) and microsatellite genotypes (FST > 0.03). In some cases, coalescence analyses reveal that Red Sea lineages are older than those in the Indian Ocean, indicating that the former can export biodiversity to adjacent waters (24).
For widely distributed species, genetic divergences at peripheral locations may be the inception of speciation. The pronghorn spiny lobster, Panulirus penicillatus, with a 9-mo pelagic larval duration and a distribution across the entire tropical Indo-Pacific, illustrates genetic diversification at both ends of its range. Iacchei et al. (21) found fixed differences in mtDNA of East Pacific and Red Sea populations (ΦST = 0.74), corroborated by morphological differentiation in the East Pacific (58). Speciation in peripheral provinces is apparent in Thalassoma wrasses (59), Anampses wrasses (60), Acanthurus surgeonfishes (55), Mulloidichthys goatfishes (19), and Montastraea corals (61).
The East Pacific Barrier (EPB) limits the distribution of tropical species (62), with few taxa able to maintain population connectivity across the EPB, as evidenced by the lobster P. penicillatus (21), and the coral Porites lobata (63, 64). However, some fishes (65) and the echinoderm Echinothrix diadema (66) have low or insignificant ΦST values across the EPB.
Atlantic and Indo-Pacific Connections.
Two geological events isolated the tropical Atlantic from the Indo-Pacific: (i) closure of the Tethys Sea ∼13 Ma, brought about by the collision of Africa and Eurasia, and (ii) the rise of the Isthmus of Panama ∼3.5 Ma that separated the Atlantic from the East Pacific Ocean (67). For the latter, some species diverged well before the final closure although the timing of partitions remain controversial (68) (a fruitful topic for genomic studies). Since the closure of the Tethys Sea, natural dispersal between the Atlantic and Indian Oceans has been limited to the hydrographically complex waters around southern Africa (69). A warm-water corridor here was curtailed ∼2.5 Ma by the advent of modern glacial cycles and upwelling in the Benguela Current on the Atlantic side (70). However, the Agulhas Current on the Indian Ocean side occasionally forces warm-water gyres into the Atlantic (71), a potential route of colonization. Phylogeographic studies confirm sporadic dispersal along this route over the last 2.5 My, primarily from the Indian to Atlantic Ocean (72, 73).
Summary.
In conclusion: (i) Biodiversity hotspots and peripheral centers of endemism both produce and export novel evolutionary lineages. (ii) Phylogeographic partitions, as defined by mtDNA monophyly or strong population structure, align well with the biogeographic provinces defined by taxonomy. (iii) Sporadic dispersal around southern Africa is the primary avenue of colonization between Indo-Pacific and Atlantic oceans.
Temperate and Polar Seas.
Northern seas experienced greater extremes in temperature over the Pleistocene than tropical seas, and northern near-shore ecosystems were periodically eradicated by glaciers encroaching onto continental shelves whereas interglacial warming led to colonizations and population expansions. Although phylogeographic structure generally occurs between biogeographic provinces, sub-Arctic shelf fauna have been repeatedly disrupted by glacial cycles (74). Therefore, present-day physical barriers to gene flow may not exert the same influence on phylogeographic patterns as observed in more stable tropical seas. The most notable barriers separating biogeographic domains are the large expanses of ocean waters across the North Pacific and North Atlantic.
North Pacific.
Species in the temperate regions on both sides of the North Pacific show a range of evolutionary divergences that largely depend on dispersal capabilities, temperature tolerances, and climate history. Taxa at higher latitudes tend to have distributions that span the North Pacific (versus taxa at midlatitudes). For example, cold-tolerant cods (Gadus), herring (Clupea), and king crabs (Lithodes, Paralithodes) occur in both the Northwest and Northeast Pacific. Most of these trans-Pacific species show phylogeographic breaks, centered on the Aleutian Archipelago or eastern Bering Sea, that represent secondary contact zones after repeated isolations (75–77). In contrast, temperate fishes, invertebrates, and seaweeds at midlatitudes are generally limited to one side of the North Pacific, with closely related species on the other side. A notable exception are disjunct populations of Pacific sardines (Sardinops) in the Northwest and Northeast Pacific (78).
North Atlantic.
This basin is smaller than the North Pacific and has a U-shaped shoreline with Greenland, Iceland, and Faroe Islands in midocean. Populations of fishes, invertebrates, and seaweeds show a range of genetic divergences across the North Atlantic (79–81). Conspecific populations on either side of the North Atlantic were isolated during glacial episodes, and, in some taxa, the Northwest Atlantic was extirpated and reestablished after the Last Glacial Maximum. Some populations in the Northwest Atlantic show closer genetic affiliations to the North Pacific than to the Northeast Atlantic (seagrass and sea urchins) (82). The Baltic, North Sea, and Mediterranean biogeographic provinces are isolated to some extent from the Atlantic by narrow straits, which often coincide with phylogeographic transitions (83, 84).
Arctic biogeographic province.
The far northern ocean has served as a pathway for dispersal between the North Atlantic and North Pacific (85). Phylogeographic and taxonomic studies reveal sister species in the North Atlantic and North Pacific, including several fishes (86), invertebrates (85), and seaweeds (87). During ∼20% of the Pleistocene, high sea levels breached the 50-m sill across the Bering Strait (88), allowing interocean dispersal as early as 6.4 Ma and again at 3.5 Ma (89). More recent dispersal events have led to the cooccurrence of conspecific populations in both oceans (90).
Antarctic biogeographic province.
The Antarctic is relatively old, ∼25 My, compared with about 2.5 My for the Arctic. The result of this ancient formulation is high endemism: 88% in fishes (91) and 42–56% in four invertebrate classes (92). The high homogeneity of taxa across this vast region is facilitated by the Antarctic Circumpolar Current, which circles the entire continent. Phylogeographic studies are consistent with a highly connected Antarctic Province, showing little (or no) population structure for two decapods (93), one nemertean (94), and four ice fishes (95).
Patterns Within Biogeographic Provinces.
Within the shallow-water provinces, species often share genetic breaks at specific geological features or geographical regions. Examples range from the classic study by Avise (96) on the Carolina Province (Southeast United States), through more recent surveys of the benthic fauna along the coast of New Zealand (97), the northeastern Pacific (98), the Coral Triangle (36), southern Africa (69), and Hawai’i (14). Endemic species confined to a single province tend to show more population structure than widespread species at the same geographic scale (36, 99, 100). Species that lack pelagic development generally show strong genetic structure whereas species with pelagic development are less predictable (101, 102). Regardless of developmental mode, ecological niche, or evolutionary relationships, species showing geographic structuring often have concordant genetic breaks, indicating that shared history or physical factors drive the observed pattern (96). Examination of 47 reef-associated species across the Hawaiian Archipelago reveals that multispecies trends in genetic diversity are driven by a combination of both the dominant physical, historical and ecological features of the seascape, and ecological–genetic feedback within communities (103).
Species that counter these trends may be particularly informative about the process of evolution. For example, Hawaiian limpets of the genus Cellana have diversified within the archipelago along a tidal gradient that indicates ecological speciation (104). Certainly, species sharing population structure at unexpected locations within biogeographic provinces (such as Fiji in the tropical Pacific) (21, 105), or other exceptions to those general trends, will provide evolutionary insights.
Summary.
In conclusion: (i) Species distributions are fundamentally shaped by physiological tolerances to north–south temperature gradients in the North Pacific and North Atlantic. (ii) Glacial cycles impact phylogeography by repeatedly altering species distributions, isolating populations, and creating secondary contact zones. (iii) Shifting interactions between ocean-climate, coastal configuration, and bottom topography produce barriers to dispersal between ocean basins. (iv) Some biogeographic provinces are genetically homogenous, with little opportunity for allopatric divergences, whereas others host heterogeneous habitats that can promote speciation along ecological boundaries.
Taxon-Specific Patterns
Migratory ability and historical dispersal define taxa along a continuum of evolutionary divergence. Clusters of closely related species, each confined to a single biogeographic province, are at one end of the continuum, and highly migratory megafuana are at the other end. Oceanic migrants provide special challenges to both phylogeographic studies and conservation strategies, because both must be conducted on a scale that transcends biogeographic provinces and political jurisdictions (106). Species in the center of the continuum include temperate taxa inhabiting disjunct regions, such as antitropical taxa, sister species separated by the tropics. Comparative phylogeography of these groups provides insights into the roles of dispersal and isolation in contributing to biodiversity.
Antitropical Taxa.
Species with disjunct distributions on both sides of the tropics provide fascinating subjects for phylogeographic study. Equatorial surface waters are lethal to these cold-adapted species, so how do they cross the tropics, and how often can this crossing be accomplished? Sister taxa of fishes on each side of the equator reveal divergences ranging from populations to distinct lineages, but without a clear pattern. For example, a single species of anchovy (Engraulis) occurs in the North Atlantic, southern Africa, and Japan, but three additional species have more restricted ranges (107, 108). In contrast, a single species of sardine (Sardinops) extends from southern Africa to Australia to Chile, California, and Japan (78).
Overall results show that the ability to traverse the tropics is species-specific and that these events have not been limited to particular periods of global cooling. However, one possible point of concordance includes the eastern continental margins of the Atlantic (for anchovies) and the Pacific (for sardines). In both cases, colonizations across the equator have been accomplished recently, as indicated by shared mtDNA haplotypes (Fig. 3).
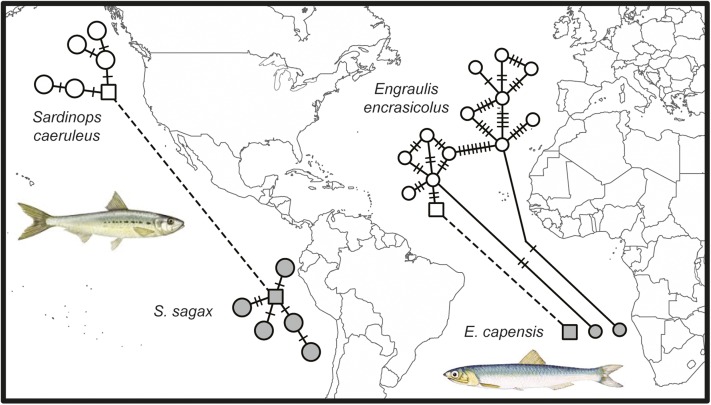
Fig. 3.
Sardines (genus Sardinops) and Anchovies (genus Engraulis) are antitropical species that recently surmounted the warm-water barrier between northern and southern hemispheres, as indicated by mtDNA haplotype networks. For sardines in the East Pacific, transequatorial dispersal is facilitated by a short and steep continental shelf and adjacent deep cold water (78). For anchovies in the East Atlantic, transequatorial dispersal is facilitated by upwelling (cold nutrient-rich water) in low latitudes (107). Light and dark haplotypes indicate northern and southern hemisphere, respectively. Squares connected by a dashed line indicate haplotypes shared between hemispheres. Note that, in the East Pacific sardine, the haplotype shared between northern and southern hemisphere is internal to both networks, indicating an ancient connection. In contrast, the East Atlantic anchovy has connections across the equator that include both interior and peripheral haplotypes in the network.
Cetaceans.
Patterns of gene flow vary extensively across space and time for cetaceans, driven largely by the wide variety of life history traits (109, 110). Most species exhibit limited gene flow between ocean basins, even in taxa with temperate distributions; but genetic structure within ocean basins varies substantially across species. For Mysticetes (baleen whales), patterns of gene flow are shaped by migratory pathways, with individuals typically exhibiting maternally based site fidelity to tropical breeding and temperate/Arctic feeding areas. This fidelity leads to population genetic separations between ocean basins and among breeding areas, with FST values of 0.05 to 0.1 for right whales (111), blue whales (112), and humpback whales (113).
In contrast, most Odontocetes (toothed whales) do not undertake large-scale migrations and often exhibit genetic structure over relatively short geographic distances due to site fidelity, resource specialization, and social structure. For example, strong fidelity to narrow ranges can result in genetically divergent populations along continuous coastlines or between adjacent islands, as is the case for spinner dolphins (114), Hector’s dolphins (115), and Indo-Pacific humpback dolphins (116). Some Odontocetes have ecologically and behaviorally distinct groups (“ecotypes”), with limited gene flow even in parapatry or sympatry (109). Several dolphin species contain genetically divergent coastal and pelagic ecotypes (117). Killer whales have sympatric ecotypes that differ in prey type, foraging strategy, social structure, and movement (118).
Sea Turtles.
The seven species of sea turtles show patterns of population structure within ocean basins defined by natal homing, the habit of females (and sometimes males) to return to the vicinity of their natal beach, after decades of growth in ocean and coastal habitats. This behavior is the basis for defining regional management units (119). On a global scale, occasional wandering provides connections between nesting populations and ocean basins. Cold-tolerant species, such as the leatherback turtle, pass freely between ocean basins (120). Tropical species, such as the green turtle and the hawksbill turtle, make rarer connections between the Atlantic and Indo-Pacific via southern Africa (121, 122). Bowen and Karl (123) note higher genetic divergences between ocean basins in tropical species, providing a signal that allopatric speciation may predominate in this group.
Pelagic Fishes.
A primary phylogeographic pattern for these oceanic migrants is low to no genetic structure within ocean basins, and strong genetic structure between the Atlantic and Indo-Pacific. Some pelagic species seem to cross the Benguela Barrier (southern Africa) often enough to preclude the development of evolutionary partitions, including albacore tuna (124, 125), wahoo (126), and the common dolphinfish (127). However, these species are likely exceptions, with many large, vagile species demonstrating structured populations across this barrier, including the scalloped hammerhead shark (50), whale shark (128), and blue marlin (129). For tunas in particular, a recurring pattern is two mtDNA lineages: one confined to the Atlantic and an Indo-Pacific lineage that is also found in the Atlantic (table 6 in ref. 126). This pattern indicates extended periods of isolation, punctuated by dispersal around southern Africa.
Plankton.
In the oceanic pelagic zone, where all life stages are planktonic, species' ranges are both extensive and dynamic because adult distributions are not tied to a particular benthic habitat. In turn, biogeographic provinces for the pelagic zone are based on physical and chemical properties (biogeochemical provinces) (130) rather than endemism or species assemblages. Longhurst (131) identified ∼55 biogeochemical provinces (BGCPs), nested within four biomes (Polar, Westerly Winds, Trade Winds, Coastal), across four ocean basins (Atlantic, Pacific, Indian, Southern). Like the species they harbor, the boundaries of the BGCPs fluctuate on both seasonal and annual timescales in accordance with changing environmental conditions (132). Our understanding of pelagic community composition is still nascent, but recent studies have shown concordance between BGCPs and community composition in taxa ranging from viruses (133) to phytoplankton (134) to fishes (135).
Cosmopolitan distributions in the pelagic zone initially prompted the conclusion of little to no population structure in the open ocean, a position that has eroded in recent decades (136, 137). Phylogeographic studies reveal that many cosmopolitan taxa are composed of multiple cryptic species (138, 139), including some that are sympatric over part of their ranges (140). Populations of these cosmopolitan species are subdivided in two ways concordant with the BGCP framework: (i) by continental land masses separating ocean basins, and (ii) by habitat discontinuities in the equatorial region between subtropical gyres in the northern and southern hemispheres (140–142). The few global-scale phylogeographic studies have been restricted to copepods, but evidence from a diversity of other taxa sampled at ocean basin scales indicate that lineages have diverged both in allopatry and sympatry at much smaller geographic distances than anticipated, with examples drawn from chaetognaths (143), euphausiids (144), and mollusks (145).
These combined results indicate that population discontinuities of pelagic species are determined not by the temporal and spatial scales of dispersal, but by habitat characteristics enabling species to maintain viable populations (137, 143). Habitat selection, rather than physical barriers, may be a primary force driving speciation in the pelagic zone (146). Therefore, a biogeographic framework based on water properties is concordant with genetic partitions within species.
Summary.
In conclusion: (i) Several temperate species show disjunct distributions across the tropics, indicating historical dispersals across warm-water barriers. (ii) The deepest phylogeographic separations for oceanic migrants indicate patterns of allopatric isolation between ocean basins, especially for fishes. (iii) Migratory sea turtles and cetaceans show population structure based on reproductive site fidelity. (iv) An ecological component to speciation is indicated by isolation along behavioral barriers in cetaceans, and by the presence of sympatric sister species in the plankton. (v) Planktonic biogeographic provinces are defined by water masses that can change size and position based on oceanographic conditions. (vi) Initial plankton studies indicate concordance between biogeochemical provinces and phylogeographic partitions, particularly at the equatorial break between northern and southern subtropics.
Terrestrial vs. Marine Phylogeography
Life began in the oceans, but the field of phylogeography began with continental biota (6, 15), and many of the insights reviewed here have precedents in terrestrial cases. The biogeographic settings have parallels between land and sea, particularly with latitudinal gradients in biodiversity and concordance between biogeographic provinces and phylogeographic partitions (15, 147). Glacial habitat disruptions in northern seas have a strong parallel in continental faunas (148, 149). Biodiversity hotspots in Indo-Pacific reefs, forests of northern Australia, and Neotropical plant communities are all distinguished by periods of stability, habitat heterogeneity, and the ability to export species (33, 150, 151). A primary difference between marine and terrestrial phylogeography is greater dispersal potential and fewer barriers in the oceans. Although a squirrel in Central Park (New York) cannot deposit progeny in Hyde Park (London), a squirrelfish is capable of dispersing on this scale (48). This difference in evolutionary processes is clear in the Hawaiian Archipelago, where rare terrestrial colonists have proliferated into dozens and hundreds of species (17) whereas marine colonists produce one or a few species (104). Therefore, the evolutionary dramas above and below the waterline have the same ingredients (isolation, selection, adaptation, speciation), but markedly different tempos and outcomes (152).
Conclusion
Marine phylogeography encompasses half-billion year separations and the largest habitat on the planet. Given this diversity, generalizations are few, but some are especially robust. First, phylogeography is the new incarnation of spatial biogeography (153). The alignment of population genetic separations and taxonomic distributions reveals that these are part of a continuum. Evolutionary partitions that could previously be described only with taxonomy are now evaluated with the genomic footprints of isolation, selection, and speciation. Second, the model of allopatric speciation that previously dominated evolutionary thought is an incomplete fit to the dispersive aquatic medium. Phylogeography of oceanic migrants indicates a strong role for allopatric speciation whereas heterogeneous coastal habitats provide more opportunity for sympatric/ecological divergences. Phylogeography in high latitudes is defined by shifting habitats in response to glaciation. Finally, both biodiversity hotspots and endemism hotspots are important in producing novel evolutionary lineages and may work in synergy to enhance biodiversity on the ocean planet.
Acknowledgments
We thank John C. Avise and Francisco J. Ayala, coorganizers of In the Light of Evolution X: Comparative Phylogeography, an Arthur M. Sackler Colloquium of the National Academy of Sciences. For stimulating discussions, advice, and editorial prowess, we thank E. A. Hanni, P. Marko, S. A. Karl, J. Eble, L. Rocha, G. Bernardi, M. Berumen, and the B.B./R.J.T. (ToBo) laboratory. We thank T. Sinclair-Taylor, T. Lilley, and A. Cros for assistance with illustrations and editor John Avise and two anonymous reviewers for comments that improved the manuscript. The authors’ research reported here was funded by the National Science Foundation, the Seaver Institute, the University of Hawaii Sea Grant Program, NOAA, the North Pacific Research Board, and the Saltonstall–Kennedy Grant Program.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution X: Comparative Phylogeography,” held January 8–9, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_X_Comparative_Phylogeography.
This article is a PNAS Direct Submission.
References
- 1.Dana JD. On the isothermal cceanic chart: Illustrating the geographical distribution of marine animals. Am J Sci. 1853;16:314–327. [Google Scholar]
- 2.Woodward SP. A Manual of the Mollusca; or, Rudimentary Treatise of Recent and Fossil Shells. John Weale; London: 1851. [Google Scholar]
- 3.Forbes E. The Natural History of the European Seas. John Van Voorst; London: 1859. [Google Scholar]
- 4.Darwin C, Wallace A. On the tendency of species to form varieties; and on the perpetuation of varieties and species by natural means of selection. Proc Linn Soc London. 1858;3(9):45–62. [Google Scholar]
- 5.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. John Murray; London: 1859. [PMC free article] [PubMed] [Google Scholar]
- 6.Avise JC, et al. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 1987;18:489–522. [Google Scholar]
- 7.Abbott IA. Marine Red Algae of the Hawaiian Islands. Bishop Museum; Honolulu: 1999. [Google Scholar]
- 8.Randall JE. Reef and Shore Fishes of the Hawaiian Islands. University of Hawaii Sea Grant College Program; Honolulu: 2007. [Google Scholar]
- 9.Kay EA. Little Worlds of the Pacific: An Essay on Pacific Basin Biogeography (Harold L. Lyon Arboretum Lecture) Lecture No. 9 Lyon Arboretum; Honolulu: 1980. [Google Scholar]
- 10.Floeter SR, et al. Atlantic reef fish biogeography and evolution. J Biogeogr. 2008;35(1):22–47. [Google Scholar]
- 11.Boschi EE. Species of decapod crustaceans and their distribution in the American marine zoogeographic provinces. Revista de Investigación y Desarrollo Pesquero. 2000;13:1–136. [Google Scholar]
- 12.Veron JEN. Corals of the World. Vol 1–3 Australian Institute of Marine Science; Townsville, Australia: 2000. [Google Scholar]
- 13.DiBattista JD, et al. A review of contemporary patterns of endemism for shallow water reef fauna in the Red Sea. J Biogeogr. 2016;43(3):423–439. [Google Scholar]
- 14.Toonen RJ, et al. Defining boundaries for ecosystem-based management: A multispecies case study of marine connectivity across the Hawaiian Archipelago. J Mar Biol. 2011;2011:460173. doi: 10.1155/2011/460173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riddle BR. Comparative phylogeography clarifies the complexity and problems of continental distribution that drove A. R. Wallace to favor islands. Proc Natl Acad Sci USA. 2016;113:7970–7977. doi: 10.1073/pnas.1601072113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bermingham E, Moritz C. Comparative phylogeography: Concepts and applications. Mol Ecol. 1998;7(4):367–369. [Google Scholar]
- 17.Shaw KL, Gillespie RG. Comparative phylogeography of oceanic archipelagos: Hotspots for inferences of evolutionary process. Proc Natl Acad Sci USA. 2016;113:7986–7993. doi: 10.1073/pnas.1601078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briggs JC, Bowen BW. A realignment of marine biogeographic provinces with particular reference to fish distributions. J Biogeogr. 2012;39(1):12–30. [Google Scholar]
- 19.Fernandez-Silva I, et al. Yellow tails in the Red Sea: Phylogeography of the Indo-Pacific goatfish Mulloidichthys flavolineatus reveals isolation in peripheral provinces and cryptic evolutionary lineages. J Biogeogr. 2015;42(12):2402–2413. [Google Scholar]
- 20.Waldrop E, et al. Phylogeography, population structure and evolution of coral‐eating butterflyfishes (Family Chaetodontidae, genus Chaetodon, subgenus Corallochaetodon) J Biogeogr. 2016;43(6):1116–1129. [Google Scholar]
- 21.Iacchei M, Gaither MR, Bowen BW, Toonen RJ. Testing dispersal limits in the sea: Range‐wide phylogeography of the pronghorn spiny lobster Panulirus penicillatus. J Biogeogr. 2016;43(5):1032–1044. [Google Scholar]
- 22.DiBattista JD, et al. Blinded by the bright: A lack of congruence between colour morphs, phylogeography and taxonomy for a cosmopolitan Indo‐Pacific butterflyfish, Chaetodon auriga. J Biogeogr. 2015;42(10):1919–1929. [Google Scholar]
- 23.Coleman RR, et al. Regal phylogeography: Range-wide survey of the marine angelfish Pygoplites diacanthus reveals evolutionary partitions between the Red Sea, Indian Ocean, and Pacific Ocean. Mol Phylogenet Evol. 2016;100:243–253. doi: 10.1016/j.ympev.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 24.DiBattista JD, et al. After continents divide: Comparative phylogeography of reef fishes from the Red Sea and Indian Ocean. J Biogeogr. 2013;40(6):1170–1181. [Google Scholar]
- 25.Gaither MR, et al. High connectivity in the deepwater snapper Pristipomoides filamentosus (Lutjanidae) across the Indo-Pacific with isolation of the Hawaiian archipelago. PLoS One. 2011;6(12):e28913. doi: 10.1371/journal.pone.0028913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiBattista JD, Rocha LA, Craig MT, Feldheim KA, Bowen BW. Phylogeography of two closely related Indo-Pacific butterflyfishes reveals divergent evolutionary histories and discordant results from mtDNA and microsatellites. J Hered. 2012;103(5):617–629. doi: 10.1093/jhered/ess056. [DOI] [PubMed] [Google Scholar]
- 27.DiBattista J, Wilcox C, Craig M, Rocha L, Bowen B. Phylogeography of the Pacific blueline surgeonfish, Acanthurus nigroris, reveals high genetic connectivity and a cryptic endemic species in the Hawaiian Archipelago. J Mar Biol. 2011;2011:839134. [Google Scholar]
- 28.Szabó Z, Snelgrove B, Craig M, Rocha L, Bowen B. Phylogeography of the Manybar Goatfish, Parupeneus multifasciatus reveals moderate structure between the Central and North Pacific and a cryptic endemic species in the Marquesas. Bull Mar Sci. 2014;90(1):493–512. [Google Scholar]
- 29.Eble JA, Rocha LA, Craig MT, Bowen BW. Not all larvae stay close to home: Insights into marine population connectivity with a focus on the brown surgeonfish (Acanthurus nigrofuscus) J Mar Biol. 2011;2011:518516. doi: 10.1155/2011/518516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaither MR, Toonen RJ, Robertson DR, Planes S, Bowen BW. Genetic evaluation of marine biogeographic barriers: perspectives from two widespread Indo-Pacific snappers (Lutjanus spp.) J Biogeogr. 2010;37(1):133–147. [Google Scholar]
- 31.Briggs JC. Marine centres of origin as evolutionary engines. J Biogeogr. 2003;30(1):1–18. [Google Scholar]
- 32.Bowen BW, Rocha LA, Toonen RJ, Karl SA. ToBo Laboratory The origins of tropical marine biodiversity. Trends Ecol Evol. 2013;28(6):359–366. doi: 10.1016/j.tree.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 33.Pellissier L, et al. Quaternary coral reef refugia preserved fish diversity. Science. 2014;344(6187):1016–1019. doi: 10.1126/science.1249853. [DOI] [PubMed] [Google Scholar]
- 34.Timm J, Figiel M, Kochzius M. Contrasting patterns in species boundaries and evolution of anemonefishes (Amphiprioninae, Pomacentridae) in the centre of marine biodiversity. Mol Phylogenet Evol. 2008;49(1):268–276. doi: 10.1016/j.ympev.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 35.Barber PH, Cheng SH, Erdmann MV, Tengardjaja K. Evolution and conservation of marine biodiversity in the Coral Triangle: Insights from stomatopod Crustacea. Crustac Issues. 2011;19:129–156. [Google Scholar]
- 36.Carpenter KE, et al. Comparative phylogeography of the Coral Triangle and implications for marine management. J Mar Biol. 2010;2011:396982. [Google Scholar]
- 37.Tornabene L, Valdez S, Erdmann M, Pezold F. Support for a ‘Center of Origin’ in the Coral Triangle: Cryptic diversity, recent speciation, and local endemism in a diverse lineage of reef fishes (Gobiidae: Eviota) Mol Phylogenet Evol. 2015;82(Pt A):200–210. doi: 10.1016/j.ympev.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 38.Cowman PF, Bellwood DR. The historical biogeography of coral reef fishes: Global patterns of origination and dispersal. J Biogeogr. 2013;40(2):209–224. [Google Scholar]
- 39.Alison Kay E, Palumbi SR. Endemism and evolution in Hawaiian marine invertebrates. Trends Ecol Evol. 1987;2(7):183–186. doi: 10.1016/0169-5347(87)90017-6. [DOI] [PubMed] [Google Scholar]
- 40.Bellemain E, Ricklefs RE. Are islands the end of the colonization road? Trends Ecol Evol. 2008;23(8):461–468. doi: 10.1016/j.tree.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Eble JA, Bowen BW, Bernardi G. Phylogeography of coral reef fishes. In: Mora C, editor. Ecology of Fishes on Coral Reefs. University of Hawaii Press; Honolulu: 2015. pp. 64–75. [Google Scholar]
- 42.Allen GR. Conservation hotspots of biodiversity and endemism for Indo‐Pacific coral reef fishes. Aquatic Conserv: Mar Freshw Ecosyst. 2008;18(5):541–556. [Google Scholar]
- 43.Schultz JK, et al. Global phylogeography and seascape genetics of the lemon sharks (genus Negaprion) Mol Ecol. 2008;17(24):5336–5348. doi: 10.1111/j.1365-294X.2008.04000.x. [DOI] [PubMed] [Google Scholar]
- 44.Kennett JP, Keller G, Srinivasan M. Miocene planktonic foraminiferal biogeography and paleoceanographic development of the Indo-Pacific region. Geol Soc Am. 1985;163:197–236. [Google Scholar]
- 45.Dethmers KE, et al. The genetic structure of Australasian green turtles (Chelonia mydas): Exploring the geographical scale of genetic exchange. Mol Ecol. 2006;15(13):3931–3946. doi: 10.1111/j.1365-294X.2006.03070.x. [DOI] [PubMed] [Google Scholar]
- 46.Blair D, et al. Pleistocene sea level fluctuations and the phylogeography of the dugong in Australian waters. Mar Mamm Sci. 2014;30(1):104–121. [Google Scholar]
- 47.Crandall ED, Sbrocco EJ, Deboer TS, Barber PH, Carpenter KE. Expansion dating: Calibrating molecular clocks in marine species from expansions onto the Sunda Shelf Following the Last Glacial Maximum. Mol Biol Evol. 2012;29(2):707–719. doi: 10.1093/molbev/msr227. [DOI] [PubMed] [Google Scholar]
- 48.Craig MT, Eble JA, Bowen BW, Robertson DR. High genetic connectivity across the Indian and Pacific Oceans in the reef fish Myripristis berndti (Holocentridae) Mar Ecol Prog Ser. 2007;334(3):245–254. [Google Scholar]
- 49.Gaither MR, et al. Phylogeography of the reef fish Cephalopholis argus (Epinephelidae) indicates Pleistocene isolation across the Indo-Pacific Barrier with contemporary overlap in The Coral Triangle. BMC Evol Biol. 2011;11:189. doi: 10.1186/1471-2148-11-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duncan KM, Martin AP, Bowen BW, De Couet HG. Global phylogeography of the scalloped hammerhead shark (Sphyrna lewini) Mol Ecol. 2006;15(8):2239–2251. doi: 10.1111/j.1365-294X.2006.02933.x. [DOI] [PubMed] [Google Scholar]
- 51.Crandall ED, Frey MA, Grosberg RK, Barber PH. Contrasting demographic history and phylogeographical patterns in two Indo-Pacific gastropods. Mol Ecol. 2008;17(2):611–626. doi: 10.1111/j.1365-294X.2007.03600.x. [DOI] [PubMed] [Google Scholar]
- 52.Ahti PA, et al. Phylogeography of Indo‐Pacific reef fishes: Sister wrasses Coris gaimard and C. cuvieri in the Red Sea, Indian Ocean and Pacific Ocean. J Biogeogr. 2016;43(6):1103–1115. [Google Scholar]
- 53.Delrieu-Trottin E, et al. Shore fishes of the Marquesas Islands: An updated checklist with new records and new percentage of endemic species. Check List. 2015;11(5):1–13. [Google Scholar]
- 54.Randall JE, Cea A. Shore Fishes of Easter Island. Univ of Hawaii Press; Honolulu: 2011. [Google Scholar]
- 55.Gaither MR, et al. Genomic signatures of geographic isolation and natural selection in coral reef fishes. Mol Ecol. 2015;24(7):1543–1557. doi: 10.1111/mec.13129. [DOI] [PubMed] [Google Scholar]
- 56.DiBattista JD, et al. On the origin of endemic species in the Red Sea. J Biogeogr. 2016;43(1):13–30. [Google Scholar]
- 57.Liu SYV, Chang FT, Borsa P, Chen WJ, Dai CF. Phylogeography of the humbug damselfish, Dascyllus aruanus (Linnaeus, 1758): Evidence of Indo‐Pacific vicariance and genetic differentiation of peripheral populations. Biol J Linn Soc Lond. 2014;113(4):931–942. [Google Scholar]
- 58.George R. Tethys sea fragmentation and speciation of Panulirus spiny lobsters. Crustaceana. 2005;78(11):1281–1309. [Google Scholar]
- 59.Bernardi G, Bucciarelli G, Costagliola D, Robertson DR, Heiser J. Evolution of coral reef fish Thalassoma spp. (Labridae). 1. Molecular phylogeny and biogeography. Mar Biol. 2004;144(2):369–375. [Google Scholar]
- 60.Hodge JR, Read CI, van Herwerden L, Bellwood DR. The role of peripheral endemism in species diversification: Evidence from the coral reef fish genus Anampses (Family: Labridae) Mol Phylogenet Evol. 2012;62(2):653–663. doi: 10.1016/j.ympev.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 61.Budd AF, Pandolfi JM. Evolutionary novelty is concentrated at the edge of coral species distributions. Science. 2010;328(5985):1558–1561. doi: 10.1126/science.1188947. [DOI] [PubMed] [Google Scholar]
- 62.Gaither MR, Bowen BW, Rocha LA, Briggs JC. Fishes that rule the world: Circumtropical distributions revisited. Fish Fisheries. 2016 in press. [Google Scholar]
- 63.Baums IB, Boulay JN, Polato NR, Hellberg ME. No gene flow across the Eastern Pacific Barrier in the reef-building coral Porites lobata. Mol Ecol. 2012;21(22):5418–5433. doi: 10.1111/j.1365-294X.2012.05733.x. [DOI] [PubMed] [Google Scholar]
- 64.Forsman Z, Wellington GM, Fox GE, Toonen RJ. Clues to unraveling the coral species problem: Distinguishing species from geographic variation in Porites across the Pacific with molecular markers and microskeletal traits. PeerJ. 2015;3:e751. doi: 10.7717/peerj.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lessios HA, Robertson DR. Crossing the impassable: Genetic connections in 20 reef fishes across the eastern Pacific barrier. Proc Biol Sci. 2006;273(1598):2201–2208. doi: 10.1098/rspb.2006.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lessios HA, Kessing BD, Robertson DR. Massive gene flow across the world’s most potent marine biogeographic barrier. Proc Biol Sci. 1998;265(1396):583–588. [Google Scholar]
- 67.Lessios HA. The great American schism: Divergence of marine organims after the rise of the Central American Isthmus. Annu Rev Ecol Syst. 2008;39:63–91. [Google Scholar]
- 68.Marko PB, Eytan RI, Knowlton N. Do large molecular sequence divergences imply an early closure of the Isthmus of Panama? Proc Natl Acad Sci USA. 2015;112(43):E5766. doi: 10.1073/pnas.1515048112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teske PR, Von Der Heyden S, McQuaid CD, Barker NP. A review of marine phylogeography in southern Africa. S Afr J Sci. 2011;107(5-6):43–53. [Google Scholar]
- 70.Dwyer GS, et al. North Atlantic deepwater temperature change during late Pliocene and late Quaternary climatic cycles. Science. 1995;270(5240):1347–1351. [Google Scholar]
- 71.Hutchings L, et al. The Benguela Current: An ecosystem of four components. Prog Oceanogr. 2009;83(1):15–32. [Google Scholar]
- 72.Reece JS, Bowen BW, Smith DG, Larson A. Molecular phylogenetics of moray eels (Muraenidae) demonstrates multiple origins of a shell-crushing jaw (Gymnomuraena, Echidna) and multiple colonizations of the Atlantic Ocean. Mol Phylogenet Evol. 2010;57(2):829–835. doi: 10.1016/j.ympev.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 73.Gaither MR, et al. Two deep evolutionary lineages in the circumtropical glasseye Heteropriacanthus cruentatus (Teleostei, Priacanthidae) with admixture in the south-western Indian Ocean. J Fish Biol. 2015;87(3):715–727. doi: 10.1111/jfb.12754. [DOI] [PubMed] [Google Scholar]
- 74.Marko PB, et al. The ‘Expansion-Contraction’ model of Pleistocene biogeography: Rocky shores suffer a sea change? Mol Ecol. 2010;19(1):146–169. doi: 10.1111/j.1365-294x.2009.04417.x. [DOI] [PubMed] [Google Scholar]
- 75.Canino MF, Spies IB, Cunningham KM, Hauser L, Grant WS. Multiple ice-age refugia in Pacific cod, Gadus macrocephalus. Mol Ecol. 2010;19(19):4339–4351. doi: 10.1111/j.1365-294X.2010.04815.x. [DOI] [PubMed] [Google Scholar]
- 76.Liu M, et al. What maintains the central North Pacific genetic discontinuity in Pacific herring? PLoS One. 2012;7(12):e50340. doi: 10.1371/journal.pone.0050340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grant WS, Zelenina DA, Mugue NS. Phylogeography of red king crab: Implications for management and stock enhancement. In: Stevens B, editor. The King Crabs. CRC; Boca Raton, FL: 2014. pp. 47–72. [Google Scholar]
- 78.Bowen B, Grant WS. Phylogeography of the sardines (Sardinops spp.): Assessing biogeographic models and population histories in temperate upwelling zones. Evolution. 1997;51(5):1601–1610. doi: 10.1111/j.1558-5646.1997.tb01483.x. [DOI] [PubMed] [Google Scholar]
- 79.Addison JA, Hart MW. Colonization, dispersal, and hybridization influence phylogeography of North Atlantic sea urchins (Strongylocentrotus droebachiensis) Evolution. 2005;59(3):532–543. [PubMed] [Google Scholar]
- 80.Árnason E. Mitochondrial cytochrome B DNA variation in the high-fecundity Atlantic cod: Trans-Atlantic clines and shallow gene genealogy. Genetics. 2004;166(4):1871–1885. doi: 10.1534/genetics.166.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCusker MR, Bentzen P. Historical influences dominate the population genetic structure of a sedentary marine fish, Atlantic wolffish (Anarhichas lupus), across the North Atlantic Ocean. Mol Ecol. 2010;19(19):4228–4241. doi: 10.1111/j.1365-294X.2010.04806.x. [DOI] [PubMed] [Google Scholar]
- 82.Olsen JL, et al. North Atlantic phylogeography and large-scale population differentiation of the seagrass Zostera marina L. Mol Ecol. 2004;13(7):1923–1941. doi: 10.1111/j.1365-294X.2004.02205.x. [DOI] [PubMed] [Google Scholar]
- 83.Johannesson K, André C. Life on the margin: Genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol Ecol. 2006;15(8):2013–2029. doi: 10.1111/j.1365-294X.2006.02919.x. [DOI] [PubMed] [Google Scholar]
- 84.Patarnello T, Volckaert FA, Castilho R. Pillars of Hercules: Is the Atlantic-Mediterranean transition a phylogeographical break? Mol Ecol. 2007;16(21):4426–4444. doi: 10.1111/j.1365-294X.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- 85.Vermeij GJ. Anatomy of an invasion: The trans-Arctic interchange. Paleobiology. 1991;17(3):281–307. [Google Scholar]
- 86.Grant WS. Genetic divergence between congeneric species of Atlantic and Pacific ocean fishes. In: Ryman N, Utter F, editors. Population Genetics and Fishery Management. Univ of Washington Press; Seattle: 1987. pp. 225–246. [Google Scholar]
- 87.Lindstrom S. The Bering Strait connection: Dispersal and speciation in boreal macroalgae. J Biogeogr. 2001;28(2):243–251. [Google Scholar]
- 88.Miller KG, et al. The Phanerozoic record of global sea-level change. Science. 2005;310(5752):1293–1298. doi: 10.1126/science.1116412. [DOI] [PubMed] [Google Scholar]
- 89.Marincovich L, Gladenkov AY. Evidence for an early opening of the Bering Strait. Nature. 1999;397(6715):149–151. [Google Scholar]
- 90.Carr SM, Kivlichan DS, Pepin P, Crutcher DC. Molecular systematics of gadid fishes: Implications for the biogeographic origins of Pacific species. Can J Zool. 1999;77(1):19–26. [Google Scholar]
- 91.Eastman JT. The nature of the diversity of Antarctic fishes. Polar Biol. 2005;28(2):93–107. [Google Scholar]
- 92.Griffiths HJ, Barnes DK, Linse K. Towards a generalized biogeography of the Southern Ocean benthos. J Biogeogr. 2009;36(1):162–177. [Google Scholar]
- 93.Raupach MJ, et al. Genetic homogeneity and circum-Antarctic distribution of two benthic shrimp species of the Southern Ocean, Chorismus antarcticus and Nematocarcinus lanceopes. Mar Biol. 2010;157(8):1783–1797. [Google Scholar]
- 94.Thornhill DJ, Mahon AR, Norenburg JL, Halanych KM. Open-ocean barriers to dispersal: A test case with the Antarctic Polar Front and the ribbon worm Parborlasia corrugatus (Nemertea: Lineidae) Mol Ecol. 2008;17(23):5104–5117. doi: 10.1111/j.1365-294X.2008.03970.x. [DOI] [PubMed] [Google Scholar]
- 95.Janko K, et al. Did glacial advances during the Pleistocene influence differently the demographic histories of benthic and pelagic Antarctic shelf fishes? Inferences from intraspecific mitochondrial and nuclear DNA sequence diversity. BMC Evol Biol. 2007;7(1):220. doi: 10.1186/1471-2148-7-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Avise JC. Toward a regional conservation genetics perspective: Phylogeography of faunas in the southeastern United States. In: Avise JC, Hamrick J, editors. Conservation Genetics: Case Histories from Nature. Chapman and Hall; New York: 1996. pp. 431–470. [Google Scholar]
- 97.Ross P, Hogg ID, Pilditch CA, Lundquist C. Phylogeography of New Zealand’s coastal benthos. N Z J Mar Freshw Res. 2009;43(5):1009–1027. [Google Scholar]
- 98.Kelly RP, Palumbi SR. Genetic structure among 50 species of the northeastern Pacific rocky intertidal community. PLoS One. 2010;5(1):e8594. doi: 10.1371/journal.pone.0008594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tenggardjaja KA, Bowen BW, Bernardi G. Vertical and horizontal genetic connectivity in Chromis verater, an endemic damselfish found on shallow and mesophotic reefs in the Hawaiian Archipelago and adjacent Johnston Atoll. PLoS One. 2014;9(12):e115493. doi: 10.1371/journal.pone.0115493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tenggardjaja KA, Bowen BW, Bernardi G. Reef fish dispersal in the Hawaiian Archipelago: Comparative phylogeography of three endemic damselfishes. J Mar Biol. 2016;2016:3251814. [Google Scholar]
- 101.Riginos C, Buckley YM, Blomberg SP, Treml EA. Dispersal capacity predicts both population genetic structure and species richness in reef fishes. Am Nat. 2014;184(1):52–64. doi: 10.1086/676505. [DOI] [PubMed] [Google Scholar]
- 102.Liggins L, Treml EA, Possingham HP, Riginos C. Seascape features, rather than dispersal traits, predict spatial genetic patterns in co‐distributed reef fishes. J Biogeogr. 2016;43(2):256–267. [Google Scholar]
- 103.Selkoe KA, et al. Conserving genetic diversity of whole communities: Identifying patterns and landscape drivers. Proc R Soc B. 2016;283:20160354. [Google Scholar]
- 104.Bird CE, Holland BS, Bowen BW, Toonen RJ. Diversification of sympatric broadcast-spawning limpets (Cellana spp.) within the Hawaiian archipelago. Mol Ecol. 2011;20(10):2128–2141. doi: 10.1111/j.1365-294X.2011.05081.x. [DOI] [PubMed] [Google Scholar]
- 105.Drew JA, Barber PH. Comparative phylogeography in Fijian coral reef fishes: A multi-taxa approach towards marine reserve design. PLoS One. 2012;7(10):e47710. doi: 10.1371/journal.pone.0047710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Toonen RJ, et al. Big Ocean Think Tank One size does not fit all: The emerging frontier in large-scale marine conservation. Mar Pollut Bull. 2013;77(1-2):7–10. doi: 10.1016/j.marpolbul.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 107.Grant WS, Leslie RW, Bowen BW. Molecular genetic assessment of bipolarity in the anchovy genus Engraulis. J Fish Biol. 2005;67(5):1242–1265. [Google Scholar]
- 108.Silva G, Horne JB, Castilho R. Anchovies go north and west without losing diversity: Post‐glacial range expansions in a small pelagic fish. J Biogeogr. 2014;41(6):1171–1182. [Google Scholar]
- 109.Hoelzel A. Genetic structure of cetacean populations in sympatry, parapatry, and mixed assemblages: Implications for conservation policy. J Hered. 1998;89(5):451–458. [Google Scholar]
- 110.Andrews KR. Population genetics in the conservation of cetaceans and primates. In: Yamagiwa J, Karczmarski L, editors. Primates and Cetaceans. Springer; New York: 2014. pp. 289–308. [Google Scholar]
- 111.Carroll EL, et al. Cultural traditions across a migratory network shape the genetic structure of southern right whales around Australia and New Zealand. Sci Rep. 2015;5:16182. doi: 10.1038/srep16182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Torres-Florez JP, et al. Blue whale population structure along the eastern South Pacific Ocean: Evidence of more than one population. Mol Ecol. 2014;23(24):5998–6010. doi: 10.1111/mec.12990. [DOI] [PubMed] [Google Scholar]
- 113.Jackson JA, et al. Global diversity and oceanic divergence of humpback whales (Megaptera novaeangliae) Proc Biol Sci. 2014;281(1786):20133222. doi: 10.1098/rspb.2013.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Andrews KR, et al. Rolling stones and stable homes: Social structure, habitat diversity and population genetics of the Hawaiian spinner dolphin (Stenella longirostris) Mol Ecol. 2010;19(4):732–748. doi: 10.1111/j.1365-294X.2010.04521.x. [DOI] [PubMed] [Google Scholar]
- 115.Hamner RM, Pichler FB, Heimeier D, Constantine R, Baker CS. Genetic differentiation and limited gene flow among fragmented populations of New Zealand endemic Hector’s and Maui’s dolphins. Conserv Genet. 2012;13(4):987–1002. [Google Scholar]
- 116.Brown AM, et al. Population differentiation and hybridisation of Australian snubfin (Orcaella heinsohni) and Indo-Pacific humpback (Sousa chinensis) dolphins in north-western Australia. PLoS One. 2014;9(7):e101427. doi: 10.1371/journal.pone.0101427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andrews KR, et al. The evolving male: Spinner dolphin (Stenella longirostris) ecotypes are divergent at Y chromosome but not mtDNA or autosomal markers. Mol Ecol. 2013;22(9):2408–2423. doi: 10.1111/mec.12193. [DOI] [PubMed] [Google Scholar]
- 118.Hoelzel AR, Dahlheim M, Stern SJ. Low genetic variation among killer whales (Orcinus orca) in the eastern north Pacific and genetic differentiation between foraging specialists. J Hered. 1998;89(2):121–128. doi: 10.1093/jhered/89.2.121. [DOI] [PubMed] [Google Scholar]
- 119.Wallace BP, et al. Regional management units for marine turtles: A novel framework for prioritizing conservation and research across multiple scales. PLoS One. 2010;5(12):e15465. doi: 10.1371/journal.pone.0015465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dutton PH, Bowen BW, Owens DW, Barragan A, Davis SK. Global phylogeography of the leatherback turtle (Dermochelys coriacea) J Zool (Lond) 1999;248(3):397–409. [Google Scholar]
- 121.Bourjea J, et al. Phylogeography of the green turtle, Chelonia mydas, in the Southwest Indian Ocean. Mol Ecol. 2007;16(1):175–186. doi: 10.1111/j.1365-294X.2006.03122.x. [DOI] [PubMed] [Google Scholar]
- 122.Vargas SM, et al. Phylogeography, genetic diversity, and management units of hawksbill turtles in the Indo-Pacific. J Hered. 2016;107(3):199–213. doi: 10.1093/jhered/esv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bowen BW, Karl SA. Population genetics and phylogeography of sea turtles. Mol Ecol. 2007;16(23):4886–4907. doi: 10.1111/j.1365-294X.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- 124.Vinas J, Alvarado Bremer J, Pla C. Inter-oceanic genetic differentiation among Albacore (Thunnus alalunga) populations. Mar Biol. 2004;145(2):225–232. [Google Scholar]
- 125.Montes I, et al. Worldwide genetic structure of albacore Thunnus alalunga revealed by microsatellite DNA markers. Mar Ecol Prog Ser. 2012;471:183–191. [Google Scholar]
- 126.Theisen TC, Bowen BW, Lanier W, Baldwin JD. High connectivity on a global scale in the pelagic wahoo, Acanthocybium solandri (tuna family Scombridae) Mol Ecol. 2008;17(19):4233–4247. doi: 10.1111/j.1365-294x.2008.03913.x. [DOI] [PubMed] [Google Scholar]
- 127.Díaz-Jaimes P, et al. Global phylogeography of the dolphinfish (Coryphaena hippurus): The influence of large effective population size and recent dispersal on the divergence of a marine pelagic cosmopolitan species. Mol Phylogenet Evol. 2010;57(3):1209–1218. doi: 10.1016/j.ympev.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 128.Castro ALF, et al. Population genetic structure of Earth’s largest fish, the whale shark (Rhincodon typus) Mol Ecol. 2007;16(24):5183–5192. doi: 10.1111/j.1365-294X.2007.03597.x. [DOI] [PubMed] [Google Scholar]
- 129.Buonaccorsi VP, McDowell JR, Graves JE. Reconciling patterns of inter-ocean molecular variance from four classes of molecular markers in blue marlin (Makaira nigricans) Mol Ecol. 2001;10(5):1179–1196. doi: 10.1046/j.1365-294x.2001.01270.x. [DOI] [PubMed] [Google Scholar]
- 130.Longhurst A. Seasonal cycles of pelagic production and consumption. Prog Oceanogr. 1995;36(2):77–167. [Google Scholar]
- 131.Longhurst A. Ecological Geography of the Sea. 2nd Ed Academic; San Diego: 2007. [Google Scholar]
- 132.Reygondeau G, et al. Dynamic biogeochemical provinces in the global ocean. Global Biogeochem Cycles. 2013;27(4):1046–1058. [Google Scholar]
- 133.Brum JR, et al. Patterns and ecological drivers of ocean viral communities. Science. 2015;348(6237):1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 134.Alvain S, Moulin C, Dandonneau Y, Bréon F-M. Remote sensing of phytoplankton groups in case 1 waters from global SeaWiFS imagery. Deep-Sea Res. 2005;52(11):1989–2004. [Google Scholar]
- 135.Reygondeau G, et al. Biogeography of tuna and billfish communities. J Biogeogr. 2012;39(1):114–129. [Google Scholar]
- 136.Miya M, Nishida M. Speciation in the open ocean. Nature. 1997;389(6653):803–804. [Google Scholar]
- 137.Norris RD. Pelagic species diversity, biogeography, and evolution. Paleobiology. 2000;26(sp4):236–258. [Google Scholar]
- 138.Miyamoto H, Machida RJ, Nishida S. Global phylogeography of the deep-sea pelagic chaetognath Eukrohnia hamata. Prog Oceanogr. 2012;104:99–109. [Google Scholar]
- 139.Hirai J, Tsuda A, Goetze E. Extensive genetic diversity and endemism across the global range of the oceanic copepod Pleuromamma abdominalis. Prog Oceanogr. 2015;138:77–90. [Google Scholar]
- 140.Andrews KR, Norton EL, Fernandez-Silva I, Portner E, Goetze E. Multilocus evidence for globally distributed cryptic species and distinct populations across ocean gyres in a mesopelagic copepod. Mol Ecol. 2014;23(22):5462–5479. doi: 10.1111/mec.12950. [DOI] [PubMed] [Google Scholar]
- 141.Goetze E. Population differentiation in the open sea: Insights from the pelagic copepod Pleuromamma xiphias. Integr Comp Biol. 2011;51(4):580–597. doi: 10.1093/icb/icr104. [DOI] [PubMed] [Google Scholar]
- 142.Norton EL, Goetze E. Equatorial dispersal barriers and limited population connectivity among oceans in a planktonic copepod. Limnol Oceanogr. 2013;58(5):1581–1596. [Google Scholar]
- 143.Peijnenburg KT, Fauvelot C, Breeuwer JA, Menken SB. Spatial and temporal genetic structure of the planktonic Sagitta setosa (Chaetognatha) in European seas as revealed by mitochondrial and nuclear DNA markers. Mol Ecol. 2006;15(11):3319–3338. doi: 10.1111/j.1365-294X.2006.03002.x. [DOI] [PubMed] [Google Scholar]
- 144.Bucklin A, et al. DNA barcodes for species identification of euphausiids (Euphausiacea, Crustacea) J Plankton Res. 2007;29(6):483–493. [Google Scholar]
- 145.Burridge AK, Goetze E, Raes N, Huisman J, Peijnenburg KT. Global biogeography and evolution of Cuvierina pteropods. BMC Evol Biol. 2015;15(1):39. doi: 10.1186/s12862-015-0310-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Peijnenburg KT, Goetze E. High evolutionary potential of marine zooplankton. Ecol Evol. 2013;3(8):2765–2781. doi: 10.1002/ece3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schluter D. Speciation, ecological opportunity, and latitude (American Society of Naturalists Address) Am Nat. 2016;187(1):1–18. doi: 10.1086/684193. [DOI] [PubMed] [Google Scholar]
- 148.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc Lond. 1996;58(3):247–276. [Google Scholar]
- 149.Bernatchez L, Wilson CC. Comparative phylogeography of Nearctic and Palearctic fishes. Mol Ecol. 1998;7(4):431–452. [Google Scholar]
- 150.Moritz C, Ens E-J, Potter S, Catullo R. The Australian monsoonal tropics: An opportunity to protect unique biodiversity and secure benefits for Aboriginal communities. Pac Conserv Biol. 2013;19(4):343–355. [Google Scholar]
- 151.Antonelli A, et al. An engine for global plant diversity: Highest evolutionary turnover and emigration in the American tropics. Front Genet. 2015;6:130. doi: 10.3389/fgene.2015.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bowen BW. 2016. The three domains of conservation genetics: Case histories from Hawaiian waters. J Hered 107(4):309–317.
- 153.Arbogast BS, Kenagy G. Comparative phylogeography as an integrative approach to historical biogeography. J Biogeogr. 2001;28(7):819–825. [Google Scholar]