Significance
During chronic infection, bacterial pathogens undergo rapid evolutionary adaptation and extensive genetic diversification affecting patient symptoms and treatment outcomes. Temperate phages are common in pathogen genomes, and phage particles can reach high abundance in human infections, but their role in pathogen evolution is unclear. Using experimental evolution and population genomics, we show that temperate phages found in human infections accelerated pathogen evolution by increasing the supply of beneficial mutations and imposing strong selection on bacterial populations. Notably, phages accelerated the loss of clinically important virulence-related bacterial traits, including motility and quorum sensing. Temperate phages are likely therefore to facilitate rapid evolution of bacterial pathogens and contribute to their adaptation to the host environment and clinical treatments.
Keywords: Pseudomonas aeruginosa, cystic fibrosis, mobile genetic element, experimental evolution, bacteriophage
Abstract
Temperate phages drive genomic diversification in bacterial pathogens. Phage-derived sequences are more common in pathogenic than nonpathogenic taxa and are associated with changes in pathogen virulence. High abundance and mobilization of temperate phages within hosts suggests that temperate phages could promote within-host evolution of bacterial pathogens. However, their role in pathogen evolution has not been experimentally tested. We experimentally evolved replicate populations of Pseudomonas aeruginosa with or without a community of three temperate phages active in cystic fibrosis (CF) lung infections, including the transposable phage, ɸ4, which is closely related to phage D3112. Populations grew as free-floating biofilms in artificial sputum medium, mimicking sputum of CF lungs where P. aeruginosa is an important pathogen and undergoes evolutionary adaptation and diversification during chronic infection. Although bacterial populations adapted to the biofilm environment in both treatments, population genomic analysis revealed that phages altered both the trajectory and mode of evolution. Populations evolving with phages exhibited a greater degree of parallel evolution and faster selective sweeps than populations without phages. Phage ɸ4 integrated randomly into the bacterial chromosome, but integrations into motility-associated genes and regulators of quorum sensing systems essential for virulence were selected in parallel, strongly suggesting that these insertional inactivation mutations were adaptive. Temperate phages, and in particular transposable phages, are therefore likely to facilitate adaptive evolution of bacterial pathogens within hosts.
Comparative genomics suggests that temperate phages play an important role in the evolution and genomic diversification of bacterial pathogens (1). Bacterial genomes often contain a range of intact and remnant prophage elements (1–3), and ecologically important bacterial traits are believed to be phage-derived (e.g., phage-derived bacteriocins) (4). Phage-related sequences are observed more frequently in pathogenic than nonpathogenic strains (5), and prophage acquisition can be associated with changes in pathogen virulence (6, 7). Prophages can directly contribute accessory gene functions (1, 8) or disrupt bacterial genes by insertional inactivation. Of particular note are the transposable class of temperate phages (also known as mutator phages), including D3112 of Pseudomonas aeruginosa (9, 10), which integrate throughout the chromosome disrupting existing genes and increasing the supply of mutations available to selection. Recent reports of high rates of phage mobilization within hosts (11) and high temperate phage abundance in humans (12), including at sites of chronic infection where phage particles have been observed to exceed bacterial host densities by 10- to 100-fold (13), suggests that temperate phages could play an important role in driving within-host evolution of bacterial pathogens. However, experimental tests of the hypothesis that temperate phages contribute to rapid evolutionary adaptation of pathogenic bacteria are lacking.
P. aeruginosa is an important opportunistic pathogen and the major cause of chronic lung infection leading to morbidity and mortality in cystic fibrosis (CF) patients (14). Populations of P. aeruginosa in the CF lung grow as microcolony biofilms suspended within lung sputum and undergo extensive genetic diversification (15–17) and rapid evolutionary adaptation (18, 19) to this host environment. Characteristic bacterial adaptations to life in the CF lung and the transition to chronicity include the evolution of mucoidy, altered metabolism, loss of motility, quorum sensing defects, and resistance to antibiotics (18, 20). Despite detailed knowledge of the targets of selection, we still have only a very limited understanding of the causes of selection driving the evolution of these phenotypes. Phages are known to be present in the CF lung, have been cultured from lung sputa (21, 22), and have been detected at high abundance using culture-independent molecular approaches (13). Moreover, prophages are a common feature of P. aeruginosa sequenced genomes (23), and lysogenic conversion has been linked to the evolution of key clinical phenotypes (e.g., mucoidy) (24, 25). Therefore, it is likely that temperate phages may both impose selection on P. aeruginosa in the CF lung and contribute to pathogen adaptation to this host environment.
We used experimental evolution to directly test how temperate phages affect P. aeruginosa adaptation in artificial sputum medium (ASM), an in vitro environment that recapitulates key physiochemical and biofilm growth properties of CF lung sputum (26). Specifically, we propagated six replicate populations of P. aeruginosa PAO1 in the presence vs. absence of an assemblage of three temperate phages (LESɸ2, ɸ3, and ɸ4) for ∼240 bacterial generations. These temperate phages naturally coexist as prophages in the genome of the P. aeruginosa Liverpool epidemic strain (LESB58) (27), the dominant clone infecting the UK CF population (28), and contribute to its competitiveness in vivo (27, 29–31). Whereas ɸ2 and ɸ3 are insertion site specific, ɸ4 is closely related to D3112, which is known to insert randomly throughout the P. aeruginosa chromosome (9, 10) and may therefore play an important role in facilitating the evolutionary adaptation of P. aeruginosa by increasing mutational supply. All phages display high rates of lytic activity in chronic CF lung infections (13), including being induced into the lytic cycle by clinically relevant antibiotics (21).
Results and Discussion
In the experimental populations, phages had no effect on bacterial densities (Fig. S1A) despite evidence of ongoing phage lysis in all replicate populations of the phage treatment (Fig. S1B). At the end of the experiment, free virions of all phages were detected in four of six populations, whereas in the other two populations, only ɸ3 and ɸ4 virions were detected (Fig. S1C). We observed high rates of lysogeny [i.e., integration of prophage(s) into the bacterial chromosome] in five of six populations, but the phages differed in their propensity to form lysogens: lysogens of the transposable phage ɸ4 approached fixation in five of six populations, whereas lysogenization of bacteria by the other phages was less common, and, where observed, was typically as a polylysogen in combination with ɸ4 (Fig. S2). Thus, lysogeny, and indeed polylysogeny, was rapidly established in our experimental populations; moreover, lysogeny appears to have been essential for the long-term maintenance of phages in populations.
Fig. S1.
Bacterial-free phage dynamics through time when grown in ASM. (A) Colony-forming units per milliliter in each population, separated by treatment. (B) Total free phage (plaque-forming units per milliliter) in phage-treated populations. (C) Relative densities of free virions for each of the LES phages in end point populations from the phage treatment, where relative density is calculated as log10 (copies per microliter + 1), as determined by qPCR. Boxplots represent the median and interquartile range.
Fig. S2.
Lysogen dynamics over time in populations evolving with phages. Each population is represented by a single area plot denoting the frequency of bacterial isolates lysogenic for each specified combination of LES prophages through time.
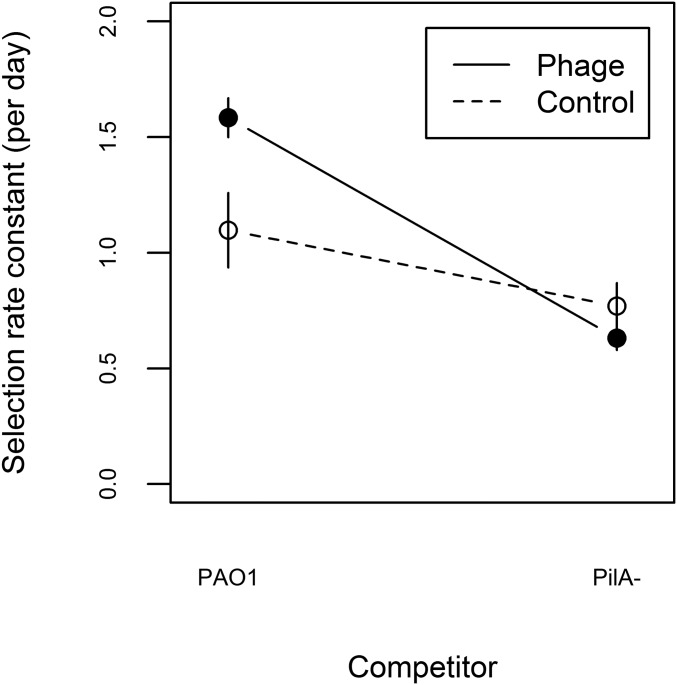
To determine the fitness response to selection, we competed each evolved population against the ancestral PAO1 in ASM, and because lysogens may have higher fitness simply due to phage-mediated killing of susceptible competitors (29, 31, 32), we also performed competitions against a phage-resistant PAO1ΔpilA, an isogenic knockout mutant strain lacking the gene encoding the type IV pilus protein PilA [all of the temperate phages used here infect via the type IV pilus (33)]. We observed that evolved populations from both treatments were fitter relative both to PAO1 and PAO1ΔpilA [Fig. 1; one-sample t test (alt = 0), all significant at an α level of 0.0125]. Populations evolved with phages had higher fitness than populations evolved without phages relative to PAO1, but this fitness advantage of evolving with phages was lost when competing against PAO1ΔpilA (Fig. 1; treatment × competitor interaction: F1,20 = 8.54, P < 0.01; simple effect of treatment against competitor PAO1: F1,10 = 7.12, P < 0.025; and simple effect of treatment against competitor PAO1ΔpilA: F1,10 = 1.53, P = 0.24). Together these data confirm that populations in both treatments adapted to the sputum-like environment and that lysogenised hosts had enhanced competitiveness against phage-susceptible competitors.
Fig. 1.
Fitness response to selection in populations evolving with and without phages. Data points represent the mean ± SE fitness calculated as selection rate for populations evolved with (filled symbols) or without (open symbols) phages in competition against either ancestral PAO1 or an isogenic phage-resistant competitor, PAO1ΔpilA.
To determine the genetic basis of the observed evolutionary adaptation, we performed whole genome sequencing on population samples containing 40 random clones pooled per population from the end of the experiment. All populations contained SNPs and small insertions or deletions (indels), and all replicate populations that had evolved with phages contained integrated prophages. At the genome-wide scale, populations evolved with or without phages did not differ in abundance or frequency of SNPs and indels (excluding insertions caused by prophage integrations) and both groups had high variance of polymorphic sites: between 16 and 173 among the phage-free populations and 17–176 among the phage-containing populations (Table S1).
Table S1.
Total small variants by class in each replicate of each treatment
| Replicate | SNPs | Deletions | Insertions | Insertions and deletions | All small variants* |
| Control 1 | 8 | 5 | 3 | 0 | 16 |
| Control 2 | 49 | 5 | 6 | 0 | 60 |
| Control 3 | 151 | 4 | 14 | 4 | 173 |
| Control 4 | 18 | 6 | 3 | 0 | 27 |
| Control 5 | 13 | 5 | 1 | 0 | 19 |
| Control 6 | 70 | 7 | 10 | 5 | 92 |
| Treatment 1 | 13 | 10 | 2 | 0 | 25 |
| Treatment 2 | 14 | 6 | 0 | 1 | 21 |
| Treatment 3 | 8 | 7 | 1 | 1 | 17 |
| Treatment 4 | 15 | 1 | 1 | 0 | 17 |
| Treatment 5 | 150 | 9 | 13 | 4 | 176 |
| Treatment 6 | 37 | 1 | 1 | 0 | 39 |
Two-sample Wilcoxon test between control and treatment: W = 21, P = 0.7.
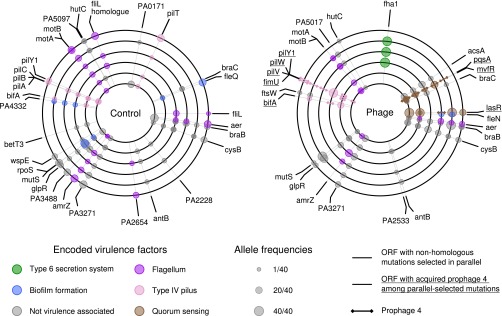
Parallel evolution at a particular locus, where independent mutations are observed more often than expected by chance, is strong evidence for positive selection. For example, in the absence of selection, the probability of observing a mutation in two populations at the conclusion of the experiment is only P = 0.003, and P = 0.0002 if observed in three populations (for an average 1,004-bp protein-coding sequence in the PAO1 genome). Thus, to identify loci likely to have been under selection during experimental evolution, we concentrated our analyses on the subset of genes that had been targeted by mutations in at least two replicate populations per treatment (Fig. 2 and Tables S2 and S3). A greater degree of parallel evolution was observed in the presence of phages (measured as the probability of randomly drawing a pair of mutated genes from different populations, with phages 0.056 ± 0.016 SE and without phages 0.024 ± 0.007 SE, P < 0.05 by bootstrap test). Some parallel targets of selection were shared among treatments, including genes involved in the type IV pilus motility, flagellar motility, biofilm formation, metabolism, and regulation, suggesting that these mutations were beneficial in the sputum-like environment per se. Interestingly, some loci were more likely to evolve in the presence of phages. In particular, mutations affecting the quorum sensing (QS) transcriptional regulators lasR and mvfR were each observed in five of six replicate phage-treated populations compared with only one of six replicate phage-free populations. In addition, three of six populations evolving with phages vs. one of six evolving without phages contained mutations in fha1, which encodes the forkhead-associated (FHA) domain protein that posttranslationally activates type VI secretion (34). A further indication of stronger selection due to phages is that parallel selected loci displayed higher allele frequencies in the phage treatment (mean allele frequency = 34.33 ± 3.2 SEM/40) compared with the control treatment (mean allele frequency = 22.67 ± 3 SEM/40), suggesting that selective sweeps in the phage treatment were closer to fixation (discussed in more detail in SI Materials and Methods and SI Results and Discussion).
Fig. 2.
Genetic loci under positive selection as indicated by parallel genomic evolution in populations evolving with and without phages. Each concentric circle corresponds to a replicate population in either the control (without phages) or treatment group (with phages), as indicated. Positions around each concentric circle, starting at the 12 o'clock position and in a clockwise direction, correspond to positions around the published P. aeruginosa PAO1 single circular chromosome. The smaller circles around each concentric circle indicate the positions of variants in each replicate that were observed in an ORF under positive selection, i.e., mutated in parallel in at least one other replicate. Only variants in fha1 were at precisely the same position and were likely to be homologous. Variants are also listed in Table S6.
Table S2.
Citations for PAO1 genome annotations of features under selection during experiments
| Locus ID | Virulence factor | Citation |
| PA0081 (fha1) | T6SS | PMID:21325275 (66) |
| PA0395 (pilT) | Type IV pilus | PMID:20338182 (67) |
| PA0996 (pqsA) | Quorum sensing | PMID:16735731 (68) |
| PA1003 (mvfR) | Quorum sensing | PMID:16735731 |
| PA1097 (fleQ) | Biofilm formation | PMID:18485075 (69) |
| PA1430 (lasR) | Quorum sensing | PMID:16476569 (70) |
| PA1442 | Flagellum | PMID:10629180 (71) |
| PA1454 (fleN) | Biofilm formation | PMID:18485075 |
| PA1561 (aer) | Flagellum | PMID:16233612 (72) |
| PA2654 | Flagellum | PMID:24291602 (73) |
| PA3385 (amrZ) | Flagellum | PMID:22511872 (74) |
| PA3622 (rpoS) | Biofilm formation | PMID:20735777 (75) |
| PA4332 | Biofilm formation | PMID:23175784 (76) |
| PA4367 (bifA) | Type IV pilus | PMID:17586641 (77) |
| PA4525 (pilA) | Type IV pilus | PMID:19717595 (78) |
| PA4526 (pilB) | Type IV pilus | PMID:12142488 (79) |
| PA4527 (pilC) | Type IV pilus | PMID:12142488 |
| PA4550 (fimU) | Type IV pilus | PMID:8682785 (80) |
| PA4551 (pilV) | Type IV pilus | PMID:7565109 (81) |
| PA4554 (pilY1) | Type IV pilus | PMID:25389296 (82) |
| PA4953 (motB) | Flagellum | PMID:15375113 (83) |
| PA4954 (motA) | Flagellum | PMID:15375113 |
| PA5233 (fliL) | Flagellum | PMID:10629180 |
PMID, PubMed identifier.
Table S3.
Abundance and frequency of mutations (including prophage integrations) in type IV pilus-associated ORFs under selection during the initial experiment
| Treatment | Population | Locus ID | Frequency (n = 40) |
| Phage | 1 | PA4550 (fimU) | 37* |
| Phage | 2 | PA4367 (bifA) | 19 |
| Phage | 2 | PA4550 (fimU) | 1* |
| Phage | 2 | PA4551 (pilV) | 1* |
| Phage | 2 | PA4554 (pilY1) | 4* |
| Phage | 3 | PA4367 (bifA) | 4* |
| Phage | 3 | PA4554 (pilY1) | 35 |
| Phage | 4 | PA4551 (pilV) | 14* |
| Phage | 5 | PA4367 (bifA) | 5 |
| Phage | 5 | PA4551 (pilV) | 36* |
| Phage | 6 | PA4551 (pilV) | 18* |
| Phage | 6 | PA4554 (pilY1) | 2* |
| Phage-free control | 1 | PA0395 (pilT) | 4 |
| Phage-free control | 1 | PA4554 (pilY1) | 5 |
| Phage-free control | 1 | PA4554 (pilY1) | 15 |
| Phage-free control | 2 | PA0395 (pilT) | 2 |
| Phage-free control | 2 | PA4367 (bifA) | 5 |
| Phage-free control | 2 | PA4525 (pilA) | 3 |
| Phage-free control | 2 | PA4526 (pilB) | 7 |
| Phage-free control | 2 | PA4527 (pilC) | 7 |
| Phage-free control | 3 | PA4525 (pilA) | 5 |
| Phage-free control | 3 | PA4526 (pilB) | 4 |
| Phage-free control | 4 | PA0395 (pilT) | 1 |
| Phage-free control | 4 | PA4527 (pilC) | 8 |
| Phage-free control | 5 | PA4367 (bifA) | 5 |
| Phage-free control | 6 | PA0395 (pilT) | 24 |
| Phage-free control | 6 | PA4525 (pilA) | 4 |
| Phage-free control | 6 | PA4554 (pilY1) | 8 |
Prophage integration-mediated mutations.
A key difference between the populations evolving with, vs. without, phages is that a substantial fraction of ORFs under positive selection (8 of 26; Fig. 2) contained mutations caused by prophage integration in the phage-containing treatment. Prophage 2 and 3 were found exclusively at the intergenic sites homologous to their positions in the LESB58 chromosome from where they were isolated (4,629,220 and 4,103,724 bp in the PAO1 chromosome) and were therefore excluded from analyses. In contrast, ɸ4 integrations were observed at 19 different positions across the six replicates, suggesting that ɸ4 integration provided an additional source of genetic diversity for selection to act on. Although ɸ4 is closely related to D3112 and therefore likely to integrate randomly throughout the chromosome (10, 27, 33), we tested for hotspots of integration with an exhaustive search of sequence motifs using the Multiple Em for Motif Elicitation (MEME) and Motif Alignment and Search Tool (MAST) algorithms (35, 36) in the 1,000-bp region around each of the integration sites. Only very weakly conserved motifs were identified (Table S4), suggesting that ɸ4 has the low integration specificity characteristic of transposable phages like D3112. Despite the very low integration specificity of ɸ4, all of the 19 integration sites were located within or adjacent to only seven different operons, which is consistent with positive selection for integrations at those regions (Fig. 2 and Tables S5 and S6). The functions most commonly predicted to be disrupted by positively selected ɸ4 integration events were type IV pilus motility and QS. Thus, transposable temperate phages like ɸ4 may commonly alter the mode of bacterial evolution by increasing the supply of mutations available to natural selection.
Table S4.
Three most specific sequence motifs identified in the 1,000 bp spanning each prophage 4 integration site
| Motif regular expression | E-value | Matches in PAO1 chromosome |
| CTG[GA][TC][CG][GT][CG]CCTG[CT][TA][GC]CTG[AG][TAC]CA[GT]CATCG[CGA][CG]GT[GC][CT]T[GC]G[CG][CG]AT | 1.4 × 10−78 | 67,584 |
| [ACG]AG[GA][AC][AG][AGC]A[GC]AGCG[CT][CG]GT[GA][GC][AGT][AC][CA][AG][TG][CA][TA][GC][AG]CC[TA]TCAC[GC][GC][CA]CA[AT] | 2.1 × 10−70 | 19,256 |
| TT[CG][GA]C[CG][AC][TA]C[TC][GA]CCAG[AG][ACT][CT]GGC[ACT][AT][GC]ACCG[AT][CG][GC]G[GC][GT][GC]C[CG]G[CT][GA]T | 2.1 × 10−62 | 45,395 |
Table S5.
Prophage 4 integration sites
| Replicate population | Frequency* (n = 40) | Position† (bp) | Within ORF | Gene name | Locus ID | Annotation | Positive selection‡ | Within operon | Operon members |
| 1 | 2 | 1086791 | Yes | mvfR | PA1003 | Transcriptional regulator MvfR | Yes | Yes | mvfR |
| 1 | 42 | 5098546 | Yes | fimU | PA4550 | Type 4 fimbrial biogenesis protein FimU | Yes | Yes | fimUpilVWXY1Y2E |
| 2 | 22 | 1079138 | Yes | pqsA | PA0996 | CoA ligase | Yes | Yes | pqsABCDE |
| 2 | 14 | 5098344 | Yes | fimU | PA4550 | Type 4 fimbrial biogenesis protein FimU | Yes | Yes | fimUpilVWXY1Y2E |
| 2 | 4 | 5098931 | Yes | pilV | PA4551 | Type 4 fimbrial biogenesis protein PilV | Yes | Yes | fimUpilVWXY1Y2E |
| 2 | 1 | 5100022 | Yes | pilW | PA4552 | Type 4 fimbrial biogenesis protein PilW | No | Yes | fimUpilVWXY1Y2E |
| 2 | 2 | 5100846 | Yes | pilY1 | PA4554 | Type 4 fimbrial biogenesis protein PilY1 | Yes | Yes | fimUpilVWXY1Y2E |
| 3 | 2 | 1079047 | Yes | pqsA | PA0996 | CoA ligase | Yes | Yes | pqsABCDE |
| 3 | 4 | 4895094 | Yes | bifA | PA4367 | Protein BifA | Yes | Yes | bifA |
| 4 | 13 | 857363 | Yes | — | PA0784 | Transcriptional regulator | No | Yes | — |
| 4 | 10 | 1086270 | Yes | mvfR | PA1003 | Transcriptional regulator MvfR | Yes | Yes | mvfR |
| 4 | 23 | 1558410 | Yes | lasR | PA1430 | Transcriptional regulator LasR | Yes | Yes | lasR |
| 4 | 25 | 5098936 | Yes | pilV | PA4551 | Type 4 fimbrial biogenesis protein PilV | Yes | Yes | fimUpilVWXY1Y2E |
| 5 | 1 | 902610 | Yes | — | PA0827 | Hypothetical protein | No | Yes | — |
| 5 | 4 | 1078619 | Yes | pqsA | PA0996 | CoA ligase | Yes | Yes | pqsABCDE |
| 5 | 44 | 5099014 | Yes | pilV | PA4551 | Type 4 fimbrial biogenesis protein PilV | Yes | Yes | fimUpilVWXY1Y2E |
| 6 | 25 | 5098163 | No | fimT, fimU | PA4549, PA4550 | Type 4 fimbrial biogenesis protein FimT, Type 4 fimbrial biogenesis protein FimU | — | No | fimT, fimUpilVWXY1Y2E |
| 6 | 23 | 5098868 | Yes | pilV | PA4551 | Type 4 fimbrial biogenesis protein PilV | Yes | Yes | fimUpilVWXY1Y2E |
| 6 | 3 | 5101059 | Yes | pilY1 | PA4554 | Type 4 fimbrial biogenesis protein PilY1 | Yes | Yes | fimUpilVWXY1Y2E |
Calculated from depth of paired short reads aligned to P. aeruginosa PAO1 chromosome; values greater than 40 reflect inaccuracies of the method.
Base pair position in P. aeruginosa PAO1 chromosome.
Inferred from parallel selection of mutations in ORFs between replicate populations during experimental evolution.
Table S6.
Variants selected in parallel in each treatment, as plotted in Fig. 2
| Treatment | Replicate | Variant type | Chromosome position (bp) | Frequency (n = 40) | Annotation | Virulence category |
| Control | 1 | Deletion | 194364 | 21 | PA0171 | No virulence known |
| Control | 1 | SNP | 436895 | 4 | pilT (PA0395) | Type IV pilus |
| Control | 1 | SNP | 1733418 | 29 | braB (PA1590) | No virulence known |
| Control | 1 | Insertion | 1894760 | 5 | cysB (PA1754) | No virulence known |
| Control | 1 | SNP | 3662937 | 9 | PA3271 | No virulence known |
| Control | 1 | SNP | 3791349 | 4 | amrZ (PA3385) | Flagellum |
| Control | 1 | Deletion | 3791585 | 1 | amrZ (PA3385) | Flagellum |
| Control | 1 | Insertion | 4015416 | 16 | glpR (PA3583) | No virulence known |
| Control | 1 | Insertion | 5101162 | 5 | pilY1 (PA4554) | Type IV pilus |
| Control | 1 | Deletion | 5101331 | 15 | pilY1 (PA4554) | Type IV pilus |
| Control | 1 | Deletion | 5558959 | 7 | motB (PA4953) | Flagellum |
| Control | 2 | Deletion | 437130 | 2 | pilT (PA0395) | Type IV pilus |
| Control | 2 | SNP | 1162646 | 4 | braC (PA1074) | No virulence known |
| Control | 2 | SNP | 1188091 | 5 | fleQ (PA1097) | Biofilm formation |
| Control | 2 | SNP | 1894307 | 6 | cysB (PA1754) | No virulence known |
| Control | 2 | SNP | 1894501 | 4 | cysB (PA1754) | No virulence known |
| Control | 2 | SNP | 2449784 | 4 | PA2228 | No virulence known |
| Control | 2 | SNP | 2831000 | 7 | antB (PA2513) | No virulence known |
| Control | 2 | SNP | 3662018 | 9 | PA3271 | No virulence known |
| Control | 2 | SNP | 3662937 | 6 | PA3271 | No virulence known |
| Control | 2 | Insertion | 4015416 | 7 | glpR (PA3583) | No virulence known |
| Control | 2 | Deletion | 4015436 | 2 | glpR (PA3583) | No virulence known |
| Control | 2 | SNP | 4058397 | 7 | rpoS (PA3622) | Biofilm formation |
| Control | 2 | SNP | 4409277 | 7 | PA3933 | No virulence known |
| Control | 2 | Deletion | 4895640 | 5 | bifA (PA4367) | Type IV pilus |
| Control | 2 | SNP | 5069342 | 3 | pilA (PA4525) | Type IV pilus |
| Control | 2 | SNP | 5070594 | 7 | pilB (PA4526) | Type IV pilus |
| Control | 2 | SNP | 5072425 | 7 | pilC (PA4527) | Type IV pilus |
| Control | 2 | Deletion | 5558959 | 7 | motB (PA4953) | Flagellum |
| Control | 2 | SNP | 5749291 | 5 | hutC (PA5105) | No virulence known |
| Control | 3 | Insertion | 1572054 | 16 | PA1442 | Flagellum |
| Control | 3 | SNP | 1699673 | 8 | aer (PA1561) | Flagellum |
| Control | 3 | SNP | 1733127 | 13 | braB (PA1590) | No virulence known |
| Control | 3 | SNP | 1733418 | 10 | braB (PA1590) | No virulence known |
| Control | 3 | Insertion | 1733701 | 7 | braB (PA1590) | No virulence known |
| Control | 3 | Indels | 1894433 | 10 | cysB (PA1754) | No virulence known |
| Control | 3 | Deletion | 1894438 | 12 | cysB (PA1754) | No virulence known |
| Control | 3 | Deletion | 1894913 | 1 | cysB (PA1754) | No virulence known |
| Control | 3 | SNP | 2831000 | 5 | antB (PA2513) | No virulence known |
| Control | 3 | SNP | 3003869 | 7 | PA2654 | Flagellum |
| Control | 3 | SNP | 3661850 | 11 | PA3271 | No virulence known |
| Control | 3 | SNP | 3791609 | 13 | amrZ (PA3385) | Flagellum |
| Control | 3 | Indels | 3906126 | 18 | PA3488 | No virulence known |
| Control | 3 | Indels | 4015416 | 32 | glpR (PA3583) | No virulence known |
| Control | 3 | Deletion | 4055899 | 31 | mutS (PA3620) | No virulence known |
| Control | 3 | SNP | 4058528 | 30 | rpoS (PA3622) | Biofilm formation |
| Control | 3 | SNP | 4146829 | 4 | wspE (PA3704) | No virulence known |
| Control | 3 | SNP | 4409968 | 4 | PA3933 | No virulence known |
| Control | 3 | SNP | 4860829 | 7 | PA4332 | Biofilm formation |
| Control | 3 | SNP | 5069342 | 5 | pilA (PA4525) | Type IV pilus |
| Control | 3 | SNP | 5071135 | 4 | pilB (PA4526) | Type IV pilus |
| Control | 3 | Insertion | 5559742 | 12 | motA (PA4954) | Flagellum |
| Control | 3 | SNP | 5891463 | 13 | PA5233 | Flagellum |
| Control | 4 | Deletion | 437130 | 1 | pilT (PA0395) | Type IV pilus |
| Control | 4 | SNP | 1161906 | 5 | braC (PA1074) | No virulence known |
| Control | 4 | SNP | 1162439 | 10 | braC (PA1074) | No virulence known |
| Control | 4 | SNP | 1699709 | 10 | aer (PA1561) | Flagellum |
| Control | 4 | SNP | 1732761 | 19 | braB (PA1590) | No virulence known |
| Control | 4 | Deletion | 1733776 | 2 | braB (PA1590) | No virulence known |
| Control | 4 | SNP | 2449917 | 7 | PA2228 | No virulence known |
| Control | 4 | SNP | 3662246 | 9 | PA3271 | No virulence known |
| Control | 4 | SNP | 3791423 | 8 | amrZ (PA3385) | Flagellum |
| Control | 4 | Insertion | 4015416 | 6 | glpR (PA3583) | No virulence known |
| Control | 4 | SNP | 4860829 | 8 | PA4332 | Biofilm formation |
| Control | 4 | Deletion | 5072606 | 8 | pilC (PA4527) | Type IV pilus |
| Control | 4 | SNP | 5559757 | 8 | motA (PA4954) | Flagellum |
| Control | 4 | Deletion | 5891244 | 8 | PA5233 | Flagellum |
| Control | 5 | Deletion | 194364 | 1 | PA0171 | No virulence known |
| Control | 5 | SNP | 1733418 | 7 | braB (PA1590) | No virulence known |
| Control | 5 | SNP | 1894349 | 8 | cysB (PA1754) | No virulence known |
| Control | 5 | Deletion | 1894397 | 2 | cysB (PA1754) | No virulence known |
| Control | 5 | SNP | 1895161 | 4 | cysB (PA1754) | No virulence known |
| Control | 5 | SNP | 2831000 | 5 | antB (PA2513) | No virulence known |
| Control | 5 | SNP | 3662012 | 17 | PA3271 | No virulence known |
| Control | 5 | SNP | 3791474 | 16 | amrZ (PA3385) | Flagellum |
| Control | 5 | Deletion | 4016098 | 6 | glpR (PA3583) | No virulence known |
| Control | 5 | SNP | 4860766 | 7 | PA4332 | Biofilm formation |
| Control | 5 | SNP | 4860808 | 7 | PA4332 | Biofilm formation |
| Control | 5 | Insertion | 4894768 | 5 | bifA (PA4367) | Type IV pilus |
| Control | 5 | SNP | 5559650 | 17 | motA (PA4954) | Flagellum |
| Control | 5 | Deletion | 5738446 | 5 | PA5097 | No virulence known |
| Control | 5 | SNP | 5749111 | 7 | hutC (PA5105) | No virulence known |
| Control | 6 | SNP | 437147 | 24 | pilT (PA0395) | Type IV pilus |
| Control | 6 | SNP | 1188946 | 24 | fleQ (PA1097) | Biofilm formation |
| Control | 6 | Insertion | 1572373 | 3 | PA1442 | Flagellum |
| Control | 6 | SNP | 1698810 | 23 | aer (PA1561) | Flagellum |
| Control | 6 | SNP | 1732849 | 13 | braB (PA1590) | No virulence known |
| Control | 6 | SNP | 1894270 | 18 | cysB (PA1754) | No virulence known |
| Control | 6 | SNP | 3004487 | 16 | PA2654 | Flagellum |
| Control | 6 | SNP | 3661717 | 26 | PA3271 | No virulence known |
| Control | 6 | Deletion | 3791400 | 25 | amrZ (PA3385) | Flagellum |
| Control | 6 | Deletion | 3906126 | 21 | PA3488 | No virulence known |
| Control | 6 | Insertion | 4015416 | 9 | glpR (PA3583) | No virulence known |
| Control | 6 | Insertion | 4055276 | 2 | mutS (PA3620) | No virulence known |
| Control | 6 | SNP | 4055654 | 24 | mutS (PA3620) | No virulence known |
| Control | 6 | SNP | 4147670 | 14 | wspE (PA3704) | No virulence known |
| Control | 6 | SNP | 5069342 | 4 | pilA (PA4525) | Type IV pilus |
| Control | 6 | SNP | 5103378 | 8 | pilY1 (PA4554) | Type IV pilus |
| Control | 6 | SNP | 5558358 | 3 | motB (PA4953) | Flagellum |
| Control | 6 | Insertion | 5559822 | 24 | motA (PA4954) | Flagellum |
| Control | 6 | Deletion | 5738454 | 3 | PA5097 | No virulence known |
| Control | 6 | Deletion | 5749111 | 8 | hutC (PA5105) | No virulence known |
| Control | 6 | SNP | 5891204 | 23 | PA5233 | Flagellum |
| Phages | 1 | SNP | 969787 | 20 | acsA (PA0887) | No virulence known |
| Phages | 1 | Prophage integration | 1086290 | 18 | mvfR (PA1003) | Quorum sensing |
| Phages | 1 | Prophage integration | 1086648 | 9 | mvfR (PA1003) | Quorum sensing |
| Phages | 1 | Prophage integration | 1086975 | 4 | mvfR (PA1003) | Quorum sensing |
| Phages | 1 | SNP | 1161876 | 5 | braC (PA1074) | No virulence known |
| Phages | 1 | SNP | 1162851 | 5 | braC (PA1074) | No virulence known |
| Phages | 1 | Prophage integration | 1558717 | 38 | lasR (PA1430) | Quorum sensing |
| Phages | 1 | SNP | 1732717 | 3 | braB (PA1590) | No virulence known |
| Phages | 1 | SNP | 1733418 | 9 | braB (PA1590) | No virulence known |
| Phages | 1 | SNP | 1894399 | 4 | cysB (PA1754) | No virulence known |
| Phages | 1 | SNP | 1894630 | 19 | cysB (PA1754) | No virulence known |
| Phages | 1 | SNP | 3662290 | 20 | PA3271 | No virulence known |
| Phages | 1 | Insertion | 3791372 | 10 | amrZ (PA3385) | Flagellum |
| Phages | 1 | Deletion | 4015416 | 6 | glpR (PA3583) | No virulence known |
| Phages | 1 | SNP | 4015517 | 4 | glpR (PA3583) | No virulence known |
| Phages | 1 | Deletion | 5558959 | 1 | motB (PA4953) | Flagellum |
| Phages | 1 | SNP | 5559329 | 17 | motA (PA4954) | Flagellum |
| Phages | 1 | Deletion | 5748779 | 1 | hutC (PA5105) | No virulence known |
| Phages | 2 | SNP | 971285 | 13 | acsA (PA0887) | No virulence known |
| Phages | 2 | Prophage integration | 1086422 | 15 | mvfR (PA1003) | Quorum sensing |
| Phages | 2 | Prophage integration | 1086975 | 3 | mvfR (PA1003) | Quorum sensing |
| Phages | 2 | Prophage integration | 1558717 | 34 | lasR (PA1430) | Quorum sensing |
| Phages | 2 | Prophage integration | 1558834 | 4 | lasR (PA1430) | Quorum sensing |
| Phages | 2 | Deletion | 1699347 | 2 | aer (PA1561) | Flagellum |
| Phages | 2 | SNP | 1733410 | 9 | braB (PA1590) | No virulence known |
| Phages | 2 | SNP | 1733419 | 9 | braB (PA1590) | No virulence known |
| Phages | 2 | SNP | 1894289 | 12 | cysB (PA1754) | No virulence known |
| Phages | 2 | Deletion | 1894665 | 3 | cysB (PA1754) | No virulence known |
| Phages | 2 | SNP | 2831000 | 6 | antB (PA2513) | No virulence known |
| Phages | 2 | SNP | 2860812 | 3 | PA2533 | No virulence known |
| Phages | 2 | SNP | 3661954 | 14 | PA3271 | No virulence known |
| Phages | 2 | Indels | 4015416 | 14 | glpR (PA3583) | No virulence known |
| Phages | 2 | SNP | 4015429 | 4 | glpR (PA3583) | No virulence known |
| Phages | 2 | Prophage integration | 4895587 | 19 | bifA (PA4367) | Type IV pilus |
| Phages | 2 | SNP | 5559650 | 13 | motA (PA4954) | Flagellum |
| Phages | 3 | Deletion | 98901 | 40 | fha1 (PA0081) | T6SS |
| Phages | 3 | Deletion | 971365 | 21 | acsA (PA0887) | No virulence known |
| Phages | 3 | Prophage integration | 1086975 | 1 | mvfR (PA1003) | Quorum sensing |
| Phages | 3 | SNP | 1162203 | 12 | braC (PA1074) | No virulence known |
| Phages | 3 | Deletion | 1733141 | 11 | braB (PA1590) | No virulence known |
| Phages | 3 | SNP | 1894370 | 22 | cysB (PA1754) | No virulence known |
| Phages | 3 | SNP | 3663204 | 21 | PA3271 | No virulence known |
| Phages | 3 | Insertion | 3791425 | 24 | amrZ (PA3385) | Flagellum |
| Phages | 3 | Indels | 4015416 | 14 | glpR (PA3583) | No virulence known |
| Phages | 3 | Prophage integration | 5103656 | 35 | pilY1 (PA4554) | Type IV pilus |
| Phages | 4 | Deletion | 98901 | 38 | fha1 (PA0081) | T6SS |
| Phages | 4 | SNP | 971156 | 8 | acsA (PA0887) | No virulence known |
| Phages | 4 | Prophage integration | 1087007 | 24 | mvfR (PA1003) | Quorum sensing |
| Phages | 4 | SNP | 1584550 | 11 | fleN (PA1454) | Biofilm formation |
| Phages | 4 | SNP | 1698759 | 15 | aer (PA1561) | Flagellum |
| Phages | 4 | SNP | 1733409 | 7 | braB (PA1590) | No virulence known |
| Phages | 4 | SNP | 1894696 | 15 | cysB (PA1754) | No virulence known |
| Phages | 4 | SNP | 2861448 | 4 | PA2533 | No virulence known |
| Phages | 4 | SNP | 3661949 | 11 | PA3271 | No virulence known |
| Phages | 4 | SNP | 3791408 | 11 | amrZ (PA3385) | Flagellum |
| Phages | 4 | SNP | 5558880 | 12 | motB (PA4953) | Flagellum |
| Phages | 4 | SNP | 5559824 | 16 | motA (PA4954) | Flagellum |
| Phages | 4 | SNP | 5641049 | 8 | PA5017 | No virulence known |
| Phages | 5 | Deletion | 98901 | 37 | fha1 (PA0081) | T6SS |
| Phages | 5 | SNP | 1162296 | 5 | braC (PA1074) | No virulence known |
| Phages | 5 | Prophage integration | 1558445 | 36 | lasR (PA1430) | Quorum sensing |
| Phages | 5 | SNP | 1584161 | 11 | fleN (PA1454) | Biofilm formation |
| Phages | 5 | Deletion | 1699874 | 9 | aer (PA1561) | Flagellum |
| Phages | 5 | SNP | 1733418 | 37 | braB (PA1590) | No virulence known |
| Phages | 5 | Insertion | 1894433 | 8 | cysB (PA1754) | No virulence known |
| Phages | 5 | SNP | 1894469 | 4 | cysB (PA1754) | No virulence known |
| Phages | 5 | SNP | 2831000 | 8 | antB (PA2513) | No virulence known |
| Phages | 5 | SNP | 3662861 | 11 | PA3271 | No virulence known |
| Phages | 5 | Insertion | 4015416 | 28 | glpR (PA3583) | No virulence known |
| Phages | 5 | SNP | 4055407 | 37 | mutS (PA3620) | No virulence known |
| Phages | 5 | Prophage integration | 4894991 | 5 | bifA (PA4367) | Type IV pilus |
| Phages | 5 | SNP | 4947128 | 9 | ftsW (PA4413) | No virulence known |
| Phages | 5 | SNP | 5558887 | 11 | motB (PA4953) | Flagellum |
| Phages | 5 | SNP | 5559182 | 11 | motA (PA4954) | Flagellum |
| Phages | 5 | SNP | 5642321 | 11 | PA5017 | No virulence known |
| Phages | 6 | Prophage integration | 1558508 | 19 | lasR (PA1430) | Quorum sensing |
| Phages | 6 | SNP | 1698780 | 12 | aer (PA1561) | Flagellum |
| Phages | 6 | SNP | 1732761 | 14 | braB (PA1590) | No virulence known |
| Phages | 6 | SNP | 1732774 | 4 | braB (PA1590) | No virulence known |
| Phages | 6 | SNP | 1733418 | 4 | braB (PA1590) | No virulence known |
| Phages | 6 | SNP | 1733649 | 3 | braB (PA1590) | No virulence known |
| Phages | 6 | SNP | 1894371 | 12 | cysB (PA1754) | No virulence known |
| Phages | 6 | SNP | 3662438 | 14 | PA3271 | No virulence known |
| Phages | 6 | Deletion | 4016098 | 7 | glpR (PA3583) | No virulence known |
| Phages | 6 | SNP | 4054858 | 5 | mutS (PA3620) | No virulence known |
| Phages | 6 | SNP | 4947128 | 4 | ftsW (PA4413) | No virulence known |
| Phages | 6 | SNP | 5749105 | 3 | hutC (PA5105) | No virulence known |
Because all of the temperate phages used here infect via the type IV pilus (33) and PAO1ΔpilA mutants showed higher fitness compared with PAO1 against lysogenized bacteria evolved in the phage treatment (Fig. 1), we hypothesized that disruption of type IV pilus motility associated genes may have been selected to prevent superinfection and lysis of ɸ4 lysogens. Notably, although ɸ4 lysogens have strong superinfection immunity against ɸ4, they remain susceptible to infection and subsequent lysis by ɸ2 and ɸ3 (33), suggesting that loss of type IV pili could be beneficial in the presence of a diverse phage community. In support of this, we observed higher rates of phage resistance in populations that evolved with vs. without phages (Fig. 3A; Mann–Whitney test; W = 24.0, n1 = n2 = 6, P = 0.02). Correspondingly, type IV pilus-dependent twitching motility was lost more rapidly in phage-containing populations than in phage-free populations, suggesting that the loss of type IV pilus function was more strongly selected in the presence of phages (Fig. 3B). To determine whether loss of type IV pilus twitching motility phenotype was associated with ɸ4 integration, we tracked allele frequency dynamics in two replicate populations. Specific PCR primer sets (Table S7) were used to detect integrated ɸ4 prophage in type IV fimbral biogenesis protein encoding genes fimU and pilV. In both cases, there was a positive association between the allele frequency dynamics and the rise in frequency of twitching motility-deficient mutants (Fig. 3C). We next contrasted the allele frequencies of SNPs, indels, and ɸ4 prophage-integration mutations occurring at type IV pilus associated loci. Across six phage-treated populations, 12 mutations of the type IV pilus-associated genes were detected in parallel, the majority (n = 9) were caused by ɸ4 prophage integration, and half of them (n = 6/12) occurred at high frequencies (≥10/40 clones per population). However, in the absence of phages, although more mutations were detected in these genes in parallel (n = 16), only 13% of these mutations occurred at high frequency (Table S3). Consistent with the phenotypic data (Fig. 3 B and C), this suggests that there was stronger positive selection for mutations disrupting type IV pilus-associated genes in the presence of phages, driving faster selective sweeps, compared with type IV pilus disrupting mutations occurring in the absence of phages. The exception to this pattern is replicate 3 of the phage treatment, where the impairment of twitching motility in 35 out of 40 isolates can be explained by a single frame-shift deletion variant in pilY1 (PA4554; encodes a type IV pili biogenesis protein; Fig. 2). It is notable that a low frequency of lysogeny was observed in this population, unlike all other phage treatment replicates where lysogens approached fixation (Fig. S2).
Fig. 3.
The evolution of resistance to phages and pilus-dependent twitching motility traits. (A) Boxplot of phage resistance in end point populations. The thick horizontal line denotes the median frequency of isolates in a population resistant to one or more LES phages for each treatment. Asterisks denote outliers and narrow horizontal lines denote the upper and lower quartiles. (B) Frequency of bacterial isolates in each population through time displaying impaired twitching motility in the control (gray circles, solid line) and phage treatment (black triangles, dotted line). (C) Allele frequency dynamics of LESφ4 integrated into fimU and pilV for populations P7 and P11, respectively, and loss of twitching motility in these populations. Closed black circles and open white diamonds represent populations P7 and P11, respectively, solid gray lines denote loss of twitching motility data, and dashed black lines denote allele frequency data.
Table S7.
Primers used in the study
| Protocol | Primer name | Sequence | Target | Length of amplicon (bp) | Cycling conditions | Reference |
| Detection of prophage (multiplex PCR assay) | LESp2F | CTCCACTTCTCGGTTGCTTC | LES φ2 | 206 | 95 °C, 4 min then 30 cycles: 95 °C, 30 s; 58 °C, 30 s; 72 °C, 30 s; final extension step: 72 °C, 7 min. | This study |
| LESp2R | ACTAGCCCCGTATCCGAGTT | |||||
| LESp3F | TCAGGAAAACCTTGCCATTC | LES φ3 | 384 | |||
| LESp3R | GTCTTCTGGTGGTCGGTGAT | |||||
| LESp4F | AGTTACGCCTGCTGGTGAGT | LES φ4 | 506 | |||
| LESp4R | CCTCAGTCGTGCCTTCTTTC | |||||
| Determination of LESφ4 integration site in PAO1 chromosome | 7leftF | TTCGAGTTGGATCCGGCC | fimU LJ | 247 | 95 °C, 4 min then 30 cycles: 95 °C, 30 s; 55 °C, 30 s; 72 °C, 30 s; final extension step: 72 °C, 7 min. | This study |
| 7leftR | GTCGTGCTGTGCTGATCTTT | |||||
| 7fimUrF | CGCTCATTCCGTGCCAATTA | fimU RJ | 358 | |||
| 7fimUrR | TCAATGCGATGCTGCAGTAC | |||||
| 7leftF | TTCGAGTTGGATCCGGCC | fimU intact insertion site | 328 | |||
| 7fimUrR | TCAATGCGATGCTGCAGTAC | |||||
| 11leftF | CTTCTTCAAGGCCAAGGGGT | pilV LJ | 458 | |||
| 11leftR | CGATGGCGATACGGTGATGA | |||||
| 11rightF | GACGAGGTGCTTAGACGGAG | pilV RJ | 610 | |||
| 11rightR | ATCATGGACAGGCCCGATTG | |||||
| 11leftF | CTTCTTCAAGGCCAAGGGGT | pilV intact insertion site | 349 | |||
| 11rightR | ATCATGGACAGGCCCGATTG | |||||
| qPCR to detect relative copies of free LES phages | 2totF | AGTAGCCGACCCAGACCTTT | LESφ2 | 141 | 95 °C, 10 min then 40 cycles: 95 °C, 10 s; 60 °C, 15 s; 72 °C, 30s. | (33) |
| 2totR | ATGGAAGCAACCGAGAAGTG | |||||
| 3tot1F | CGCAGGTACCACCAGACTTT | LESφ3 | 122 | |||
| 3tot1R | CATGTCCAGCAGGTTCAAAA | |||||
| 4tot1F | GCTCATGAGTGGCTGACAAC | LESφ4 | 105 | |||
| 4tot1R | TCTTGGGCAGAGAACCATTC | |||||
| Confirmation of complete degradation of bacterial DNA | PS1 | ATGAACAACGTTCTGAAATTCTCTGCT | P. aeruginosa oprI lipoprotein gene | 249 | 94 °C, 5 min then 30 cycles: 94 °C, 1 min; 60 °C, 1 min; 72 °C, 2 min; final extension step: 72 °C, 5 min. | (84) |
| PS2 | CTTGCGGCTGGCTTTTTCCAG |
Our genomic data suggest that temperate phages promoted the loss of QS with positive selection of ɸ4 prophage integrations, SNPs, and indel mutations at the mvfR and lasR loci. Mutations to lasR lead to disruption of the acyl-homoserine-lactone (AHL) signaling system (37), whereas mutations to mvfR lead to disruption of the Pseudomonas quinolone signal (PQS) system (38), suggesting large-scale alterations to QS cell–cell signaling in populations evolving with phages. To test whether QS deficient bacteria have higher fitness in the presence of phages, we competed PAO1 against PAO1ΔlasR in ASM with and without the temperate phages. There was no effect of phages on the fitness of PAO1ΔlasR (Fig. S3; two-sample t test, t10 = −0.7989, P = 0.44), which was substantially fitter relative to PAO1 in both the presence [one-sample t test (alt = 0), t5 = 5.0331, P < 0.01] and absence [one-sample t test (alt = 0), t5 = 6.7065, P = 0.001] of phages. These data suggest that lasR mutations are beneficial in ASM per se. This interpretation is consistent with the observation that single populations in the phage-free treatment also acquired mutations in QS genes, but suggests that the rate of evolution at these loci was higher in the presence of phages. Second, we compared the rates of spontaneous phage lysis of ɸ4 lysogens constructed in both the PAO1 and PAO1ΔlasR backgrounds: there was no significant difference in production of free phages in stationary phase cultures (median free phage density: PAO1, 3.4 × 108 pfu/mL, PAO1ΔlasR, 3.3 × 108 pfu/mL; Mann–Whitney test; W = 92.0, P = 0.345). Thus, although direct interaction between temperate phages and bacterial QS has been reported in other systems, via QS induced lysis by phages (39) or QS mediated alteration of phage receptor expression by bacteria (40), this does not appear to be an important factor in our study. Phages may have simply increased the supply of large effect mutations available to natural selection, notably via ɸ4 prophage integrations into mvfR (Fig. 2). Alternatively, there may have been epistatic interactions between the fitness effects of QS mutations and other positively selected mutations, which strengthened selection for loss of QS in the presence of phages. Mutations in QS regulators are commonly observed to accumulate over time in CF chronic infection (41). Both AHL and PQS signaling are required for full virulence in P. aeruginosa (42), suggesting that temperate phage selection may accelerate the loss of virulence in chronic infections.
Fig. S3.
Fitness of QS-deficient mutants (PAO1ΔlasR) relative to WT PAO1 in the presence and absence of temperate phages in ASM. We find no effect of phage presence on the relative fitness of PAO1ΔlasR (two-sample t test, t10 = −0.7989, P = 0.44), which was fitter than WT in both the presence [one-sample t test (alt = 0), t5 = 5.0331, P < 0.01] and absence [one-sample t test (alt = 0), t5 = 6.7065, P = 0.001] of phages. r = 0, fitness of PAO1ΔlasR = PAO1; r > 0, fitness of PAO1ΔlasR > PAO1. Data are means (n = 6) ± SEM.
In summary, we showed that temperate phages enhanced parallel evolution in P. aeruginosa biofilms in a sputum-like environment. Our data suggest two ways in which this may have occurred: first, the transposable phage ɸ4 mediated adaptive evolution by increasing the supply of positively selected mutations via insertional inactivation of genes caused by prophage integrations, particularly in type IV pilus and QS associated genes. Second, we present evidence that temperate phage strengthened selection, particularly for mutations in type IV pilus associated genes, accelerating the evolutionary loss of type IV dependent pilus motility presumably to avoid superinfection and subsequent lysis by phages which infect via the type IV pilus. A recent transposon sequencing study of P. aeruginosa PA14 shows that mutations in type IV pilus associated genes increase fitness in the murine lung (43). Moreover, loss of both motility (44, 45) and QS (46, 47) functions are known to frequently evolve in P. aeruginosa chronic infections of the CF lung. Temperate phages, including those used here, can be present at very high densities in the CF lung [exceeding bacterial densities by orders of magnitude (13)], which taken together with our findings suggests that temperate phages could play an important role in CF lung infections by driving the evolution of these clinically important traits in P. aeruginosa. In addition, our data suggest that living in a sputum-like environment per se selects for mutations in genes associated with motility, biofilm formation, metabolism and regulation. Similar mutations observed in CF lung isolated P. aeruginosa are therefore likely to be at least partially explained simply as adaptations to selection imposed by the sputum environment, but could have implications for susceptibility to antibiotics (48) or host immune responses (49) as correlated responses (50). Experimental evolution in clinically relevant infection models has the potential to enhance our understanding of the causal links between sources of selection and the evolutionary responses of pathogens in infections (51), advancing our understanding of within host pathogen evolution and our ability to direct this for improved patient health.
SI Materials and Methods
Bacterial Strains, Bacteriophages, and Growth Conditions.
The P. aeruginosa strain PAO1, susceptible to infection by LESφ2–4, was used as a model host to study phage–host dynamics. For the competition experiments, PAO1 was labeled with a gentamicin resistance marker (PAO1 GmR) using a mini-Tn7 transposon as described in ref. 55. The pilA mutant (PAO1 pilA:: TetR) is a mini-Tn5 lx transposon mutant with an insertion in the pilA gene, resulting in the absence of type IV pili (56). Bacteriophages were isolated from P. aeruginosa LES isolate B58 as described previously (33). Before the selection experiment, and in follow-up experiments, all bacterial strains and phages were grown and propagated in standard LB, supplemented with gentamicin (10 µg/mL) or tetracycline (50 µg/mL) as required. Phage suspensions (1 × 108−11 pfu/mL) were stored in LB at 4 °C. ASM was made as described in ref. 57.
Selection Experiment.
Midexponential phase cultures of strain PAO1 (OD600 0.5) were added (1:100) to fresh ASM (12 × 5 mL) and incubated for 24 h [37 °C, 60 rpm (Stuart SI500 16-mm orbital shaking incubator, Bibby Scientific Ltd.)]. Equal numbers of each LES phage (LESφ2–4) were added, once only, to six replicates, to a total multiplicity of infection of 0.1. The remaining six replicates were designated no phage controls. Each population was incubated for a further 72 h before degradation of biofilm structures using Sputasol (Oxoid) as described previously (4). Each Sputasol-treated population was transferred (1:100) into fresh ASM (12 × 5 mL) and incubated for a further 96 h. This process was repeated every 96 h for 120 d (30 transfers and ∼240 bacterial generations). Bacterial density in each population was enumerated every transfer (96 h) by plating a 10-fold dilution series of Sputasol-treated culture onto Columbia agar and calculating the cfu per milliliter. Total free phage density in each population was enumerated every transfer (96 h) by a spot agar overlay assay. Briefly, homogenized cultures were filtered through a 0.2-µm filter (Millipore). Exponentially growing PAO1 was mixed with 5 mL molten 0.4% LB agar and overlaid onto Columbia agar plates. Tenfold dilutions of the filtrate were spotted onto the overlay and incubated overnight at 37 °C before plaques were counted to calculate pfu per milliliter.
Frequency of Prophage Carriage.
Every five transfers (20 d), the frequency of prophage carriage was determined for each phage-treated population, using a multiplex PCR assay of individual colonies (n = 15). Crude DNA was prepared by suspending bacterial colonies in dsH2O (50 µL). Colony suspensions (5 µL) were used as templates in PCR assays. Reactions comprised 10 pmol each primer; 1.5 mM MgCl2; 10 µM each dNTP; 5× GoTaqBuffer, and 1 U GoTaq polymerase (Promega). Details of all primer sequences are shown in Table S7.
qPCR Determination of Relative Free Phage Densities.
Relative free phage densities of each phage were estimated in endpoint population supernatants with a qPCR approach. Filtered ASM supernatant was DNase treated to remove contaminating bacterial/ prophage DNA; 17 µL phage supernatant was mixed with 2 µL incubation buffer and 1 µL DNase I recombinant (Roche) and incubated at 37 °C for 10 min. The reaction was stopped by the addition of EDTA to a final concentration of 8 mM and heating to 75 °C for 10 min. A PCR assay was carried out using primers targeted to the bacterial outer membrane lipoprotein gene (oprI); a negative result confirmed total degradation of bacterial DNA. The phage supernatant was then heated to 100 °C for 5 min to burst the capsid and used as template in the qPCR.
Copy numbers of each phage were estimated by comparison with a concentration gradient of known standards, as described previously (33) using a primer pair targeted to each phage. Each reaction contained 1× SYBR green mastermix (Qiagen), 1 µM each primer, 1 µL DNase-treated phage supernant (or standard), and diethylpyrocarbonate (DEPC)-treated water, to a final volume of 25 µL. Reactions took place in 0.1-mL strip-tubes (Qiagen), which were placed in a Rotor-Gene 6000 (Corbett). Primers and cycling conditions are described in Table S7. Negative controls and standards were included in each run, with two replicates per concentration. Three technical replicates per sample were performed each run and averaged, and each sample was tested on three separate runs.
DNA Extraction and Sequencing.
At the end of the selection experiment, whole populations were plated onto Columbia agar. Forty colonies per population were randomly selected and patched onto fresh media. These isolates were stored in 30% (vol/vol) glycerol-LB at −80 °C. Genomic DNA was extracted from overnight bacterial cultures for each isolate using the Wizard Genomic DNA Purification Kit (Promega). DNA was quantified using the Qubit dsDNA BR assay (Invitrogen) in triplicate and normalized to the isolate with the lowest concentration in each population. DNA was pooled for each population and submitted to Liverpool Centre for Genomic Research for library preparation (500-bp insert size) and sequencing on the Illumina HiSeq 2000 sequencing system (100-bp paired-end reads).
Sequence Data Preparation and Variant Calling.
Preparation of sequence data were performed as described previously (16). Briefly, sequenced read data were trimmed using Cutadapt version 1.2.1 (58) setting option -O 3 and Sickle version 1.2 (https://github.com/najoshi/sickle) setting minimum window quality score to 20 and omitting reads shorter than 10 bp after trimming but retaining single remaining reads from pairs. The Genome Analysis Toolkit (GATK) (59) Indel Realigner module (60) was used to realign raw reads around indels and duplicate reads were identified and removed with Picard (https://github.com/broadinstitute/picard). Single nucleotide polymorphism, insertion, and deletion discovery from the pooled isolate sequence data were performed with GATK's Unified Genotyper module (59) with sample ploidy n = 40, and parameters were set for detection of low-frequency variants in each pooled sample (≤2.5%). Standard conservative filtering parameters were used to provide high-quality variant calls (61).
Genomic Analysis.
Annotations of ORFs from the PAO1 published genome were obtained from National Center for Biotechnology Information (NCBI) GenBank and supplemented from the literature using the text-mining feature of the STRING v10 database website (52) (Table S3). Observation of mutations at an ORF in two or more replicate populations (parallel evolution) was considered evidence of positive selection. To estimate the strength of this evidence, we calculated the probability of each parallel mutation as a result of chance alone
where l is the length of the ORF, s is the number of positions within ORFs in the P. aeruginosa PAO1 genome, p is the total lineages in which an ORF is mutated, and n is the total mutations at the conclusion of the experiment.
We compared the extent of parallelism between treatments by comparing the means of probabilities of two ORFs from different populations being mutated
where pi is the frequency of the ith SNP within an ORF, and m and n are populations within a treatment. This metric was calculated for all ORFs within each treatment for which two populations exhibited mutations and the mean of this metric calculated. Bootstrap samples were drawn within each treatment to estimate SEMs and provide a P value for the difference between the means.
Paired-end read information was used to test for a “multidiverse” signature (Table S8): a prevalence of adjacent polymorphisms not cooccurring in the same cell lineages, i.e., unlinked and not observed on the same sequencing fragments. These polymorphisms were included when assessing allele frequencies as an indicator for selective sweep strength. First, polymorphisms within 1,000 bp were collected (twice the approximate sequencing fragment insert size). Paired sequence reads aligned to the reference genome were then queried for the presence of both members of each variant pair which would indicate linkage on the same genome in the population, not a “multidiverse” signature. The method is implemented in the CheckLinkage option of BAGA (https://github.com/daveuu/baga) (54). It should be noted that only polymorphisms closer than the approximate sequencing fragment length can be tested in this way. However, independent polymorphisms at the same locus are of particular interest for testing parallel selection of mutations because they are likely to affect the same aspect of the phenotype.
Table S8.
Incidence of nearby SNP and indel polymorphisms in reads or read pairs, indicative of a multidiverse signal of variants in separate cell lineages within a population
| Treatment | Population | Variant pair positions* (bp) | Distance (bp) | Linked | Spanning read pairs | Read pairs with variants at both positions | Read pairs with variant at first only | Read pairs with variant at second only | Locus ID | |
| Phage | 1 | 1894399 | 1894630 | 231 | No | 88 | 0 | 15 | 73 | PA1754 (cysB) |
| Phage | 1 | 4015416 | 4015517 | 101 | No | 9 | 0 | 4 | 5 | PA3583 (glpR) |
| Phage | 1 | 1086648 | 1086975 | 327 | No | 12 | 0 | 9 | 3 | PA1003 (mvfR) |
| Phage | 2 | 4015416 | 4015429 | 13 | No | 266 | 0 | 195 | 71 | PA3583 (glpR) |
| Phage | 2 | 1558717 | 1558834 | 117 | No | 41 | 0 | 39 | 2 | PA1430 (lasR) |
| Phage | 2 | 1733410 | 1733419 | 9 | No | 260 | 0 | 132 | 128 | PA1590 (braB) |
| Phage | 3 | 2449914 | 2449916 | 2 | Yes | 19 | 19 | 0 | 0 | PA2228 |
| Phage | 5 | 5558887 | 5559182 | 295 | No | 37 | 1 | 20 | 16 | PA4954 (motA); PA4953 (motB) |
| Phage | 5 | 1894434 | 1894469 | 35 | No | 121 | 0 | 79 | 42 | PA1754 (cysB) |
| Phage | 5 | 2083156 | 2083431 | 275 | No | 27 | 0 | 21 | 6 | PA1910 (femA) |
| Phage | 5 | 1567095 | 1567381 | 286 | Yes | 23 | 21 | 2 | 0 | Inter-ORF; PA1437 |
| Phage | 6 | 1732761 | 1732774 | 13 | No | 406 | 1 | 306 | 99 | PA1590 (braB) |
| Phage | 6 | 1733418 | 1733649 | 231 | No | 62 | 0 | 40 | 22 | PA1590 (braB) |
| Phage | 6 | 4071962 | 4072070 | 108 | No | 57 | 1 | 21 | 35 | PA3637 (pyrG) |
| Phage-free control | 1 | 3791349 | 3791585 | 236 | No | 7 | 0 | 7 | 0 | PA3385 (amrZ) |
| Phage-free control | 1 | 5101163 | 5101331 | 168 | No | 83 | 0 | 19 | 64 | PA4554 (pilY1) |
| Phage-free control | 2 | 1894307 | 1894501 | 194 | No | 54 | 0 | 34 | 20 | PA1754 (cysB) |
| Phage-free control | 2 | 5550250 | 5550276 | 26 | No | 247 | 1 | 130 | 116 | PA4946 (mutL) |
| Phage-free control | 2 | 1183389 | 1183445 | 56 | Yes | 97 | 43 | 3 | 51 | PA1092 (fliC) |
| Phage-free control | 2 | 4015416 | 4015436 | 20 | No | 51 | 0 | 32 | 19 | PA3583 (glpR) |
| Phage-free control | 3 | 1733418 | 1733702 | 284 | No | 38 | 0 | 25 | 13 | PA1590 (braB) |
| Phage-free control | 3 | 1733127 | 1733418 | 291 | No | 53 | 0 | 29 | 24 | PA1590 (braB) |
| Phage-free control | 3 | 1894433 | 1894438 | 5 | No | 72 | 0 | 20 | 52 | PA1754 (cysB) |
| Phage-free control | 3 | 5563126 | 5563198 | 72 | No | 50 | 0 | 29 | 21 | PA4957 (psd) |
| Phage-free control | 4 | 2452199 | 2452205 | 6 | Yes | 31 | 30 | 0 | 1 | PA2229 |
| Phage-free control | 5 | 1894349 | 1894397 | 48 | No | 77 | 0 | 60 | 17 | PA1754 (cysB) |
| Phage-free control | 5 | 4860766 | 4860808 | 42 | No | 188 | 0 | 98 | 90 | PA4332 |
Base pair position in P. aeruginosa PAO1 chromosome
Paired-end read information was also used to identify phage insertion sites (Table S5). Processed BAM files (postduplicate removal and realignment around indels) were scanned, and read pairs where one read mapped to the PAO1 chromosome and one read mapped to the 5′ or 3′ region (500 bp) of the LES prophage 4 sequence were extracted. The mapping positions of the chromosomally mapped reads were pooled and clustered, and the median position of each cluster was used as an approximation for the insertion site. Frequencies of each prophage integration site were estimated as the number of read pairs supporting the integration as a proportion of the mean read pairs at four sites in the region of, but away from, the integration site counted in the same way for a valid comparison. The former were read pairs with one read mapping to the reference chromosome and one read to the prophage chromosome. The latter were read pairs both mapping to the reference chromosome either site of positions 500 and 1,000 bp on each site of the insertion site. Two of these integration sites were validated by subsequent PCR.
To investigate the sequence specificity of the 19 LES phage 4 integrations, we assessed all sequence motifs around ±500 bp of the inferred integration sites. Via the MEME Suite web server (35, 62) at meme-suite.org, the MEME version 4.11.1 algorithm was applied with command line: “meme PP4_insertions_pm500.fna -dna -oc . -nostatus -time 18000 -maxsize 60000 -mod oops -nmotifs 20 -minw 6 -maxw 50 -revcomp” followed by the MAST algorithm with the command line: “mast -oc . -nostatus meme.xml PP4_insertions_pm500.fna.” The search required use of 19 sites because 19 sequences were tested, each with at least one phage 4 integration site. The top three identified motifs were searched for the Find Individual Motif Occurrences (FIMO) algorithm (36) in the complete PAO1 chromosome.
PCR Confirmation of LES Phage 4 Integration Sites.
PCR amplification was conducted to confirm the two most frequent LESφ4 integration sites identified at the end of the selection experiment (fimU gene in P7 and pilV in P11), and to quantify the exact frequency at which the integration occurred in those populations. Dual PCR amplification was used to amplify both the left and right phage-bacterial chromosomal junctions. Positive amplification yielding two products confirmed occupation of the locus by LESφ4, and a single product indicated an intact integration site. PCR setup was as described for multiplex PCR. Primers are described in Table S7.
Twitching Motility Assay.
The 40 sequenced isolates from each end point population were assayed for twitching motility, in addition to 40 randomly selected colonies from each population at transfers 5 and 15. A single colony was stab-inoculated to the bottom of a LB agar plate using a sterile 200-µL pipette tip. Following overnight incubation at 37 °C, the agar was removed with sterile forceps. Each Petri dish was flooded with filtered crystal violet (1% in dsH2O) and incubated at room temperature for 30 min before rinsing. The level of twitching motility was quantified by measuring the diameter of the stained zone. Isolates with a zone diameter <9 mm were considered to have impaired twitching motility. Twitching motility was assayed in duplicate for each isolate, using ancestral PAO1 as a positive control. The frequency of twitching impaired isolates was calculated for each population at each time point.
Competition Experiments Using Evolved Populations.
Fitness of the experimentally evolved populations relative to the ancestor was estimated by competition experiments in ASM used a method similar to those described previously (63). Each end point whole population was competed against PAO1 GmR. To control for the competitive effect of phages, populations were also competed against PAO1 pilA::TetR, which lacks the receptor required by the LES phages. WT PAO1 was competed against both antibiotic marked strains as a control.
Frozen glycerol stocks were defrosted. Approximately 5 × 105 bacterial cells were inoculated into 1 mL ASM in a sterile 24-well tissue culture plate and incubated for 24 h to allow for acclimatization to the competition medium. An equal volume of Sputasol was added, and cultures were diluted fivefold. The OD600 for each competitor was measured and adjusted to 0.4. Competitions were set up with a 1:1 starting ratio of each competitor; 50 µL of each competitor was inoculated into 5 mL ASM and incubated under conditions identical to one cycle of the selection experiment. The exact input cell density was quantified by plating onto Columbia media (∼5 × 107 cfus per competitor). After 4 d, biofilms were homogenized, and diluted cultures were plated onto both nonselective media and the appropriate antibiotic media to quantify the density of each competitor. The Malthusian parameter of each competitor was calculated and the selection rate constant for the evolved population relative to the ancestor was estimated as described previously (64). The selection rate constant (rij) was calculated as
with units of day−1, where mi and mj are the Malthusian parameters of each competitor, estimated by
Five independent biological replicates were performed for each competition.
Competition Experiment Between PAO1LasRΔ and PAO1.
Fitness of the PAO1LasRΔ relative to PAO1 was experimentally determined by competing both strains at a 1:1 starting ratio in ASM in both the presence and absence of phage. Five microliters of sterile ASM was inoculated with ∼5 × 108 bacterial cells of each strain, so that the total cell density was ∼109 cells/mL. Equal numbers of LESφ2–4 were added to phage treatments to a total multiplicity of infection of 0.1. Six replicates were carried out for each treatment. After 96 h, 5 mL sputasol was added to each culture, and vortexed to homogenize biofilms. Diluted cultures were then plated onto both LB agar and LB supplemented with 10 µg/mL gentamicin to quantify the density of each competitor. Malthusian parameters and selection rate constant (r) was calculated as before.
Spontaneous Phage Induction of PAO1 and PAO1LasRΔ LESφ4 Lysogens.
Ten lysogens were constructed in each host background; 100 µL midexponential phase PAO1 or PAO1LasRΔ was mixed with ∼102 pfus of purified LESφ4 and 5 mL molten 0.4% agar, overlaid onto LB agar plates, and incubated overnight at 37 °C. Putative lysogens were isolated from the turbid plaque centers and streaked onto fresh media for single colonies. Colonies were tested for the presence of LESφ4 prophage by standard PCR using the LESp4 primer set (Table S7). Ten lysogens (isolated from separate plaques) were constructed in each host background.
For each construct, 5 mL LB was inoculated with 200 µL overnight culture and incubated for 22 h at 37 °C with shaking at 250 rpm (New Brunswick Innova 44R 25-mm orbital shaking incubator, Eppendorf UK Ltd.). Cultures were diluted and plated onto Columbia for cfus and filtered using a 0.2-µm syringe filter. The pfu per milliliter in the supernatant was quantified using a spot agar overlay assay as described above, using PAO1 as the indicator host.
SI Results and Discussion
Effects of Drift and Hitchhiking for Mutations at Multidiverse Loci and Phage Insertions.
To further assess how temperate phages affected adaptive evolution in these bacteria, we compared the strength of selection between the two treatments: higher allele frequencies indicate stronger positive selection and selective sweeps. For allele frequencies within each replicate, we considered only the highest, because such alleles are the least likely to be neutral, hitch-hiking alleles. Furthermore, to mitigate the effects of genetic drift on allele frequencies, we only included alleles for which we already had evidence for positive selection: those selected in parallel at loci in two or more replicates. Mutations at positions close enough to be sequenced on the same fragment (∼500 bp for our paired end short read sequencing) but that were only observed on separate fragments from the pooled genomic DNA were also included (Table S8). These alleles were also positively selected in parallel, albeit within the same replicate population, and have the “multidiverse” signature, sensu Lieberman et al. (65). The mean for the control was 22.67 ± 3 SEM of 40, whereas for the phage, treatment was 34.33 ± 3.2 SEM, consistent with stronger selective pressure in the presence of phage. In three replicates, a prophage integration allele was within one count of the highest frequency allele. Given that the margin of error in allele frequency measurements is probably greater than one count in 40, the integration allele itself could have been at the highest or equal highest frequency, thus driving the selective sweep during the experiment in those cases.
Materials and Methods
Twelve replicate microcosms (30-mL glass universals containing 5 mL ASM) were inoculated with 5 × 107 cells P. aeruginosa strain PAO1 and grown as a biofilm [37 °C incubation with shaking at 60 rpm (Stuart SI500 16-mm orbital shaking incubator, Bibby Scientific Ltd.)]. LES phages φ2, 3, and 4 were added to six microcosms after 24 h of growth to a total multiplicity of infection of 0.1 (phage treatment), and the remaining six were designated phage-free controls. Phages were added only once, at the beginning of the experiment. After a further 72-h growth, biofilms were homogenized with an equal volume of Sputasol and the homogenate transferred (1:100) into fresh ASM. Transfers were repeated every 4 d, to a total of 30 transfers (∼240 bacterial generations). Every other transfer, bacterial and total free phage densities were enumerated, and every five transfers, the frequency of prophage carriage in the phage treatment was estimated with a multiplex PCR assay using primers targeted to each of the LES phages. At transfers 5, 15, and 30, 40 isolates per population were screened for the type IV pilus-mediated twitching motility phenotype using the agar stab method.
At the end of the experiment, DNA was extracted from 40 isolates per population and pooled, and the pooled DNA sequenced on an Illumina HiSeq 2000. Polymorphisms were called from reads aligned to the published PAO1 genome treating each sample as having ploidy = 40, reflecting the number of pooled isolates. ɸ4 prophage insertion sites were estimated from the mapping locations of reads that mapped to the PAO1 chromosome and whose mate read mapped to the ɸ4 prophage sequence. The MEME software suite (35, 36) was used for motif analysis. First, the 20 most conserved motifs at ɸ4 prophage integration sites were selected using the MEME algorithm. Second, the entire host chromosome was searched for motif occurrences using the MAST algorithm (see SI Materials and Methods for details). Frequencies of each prophage integration site were estimated based on the number of read pairs split between prophage and reference chromosomes as a proportion of read pair depth in that region. ORF annotations from the published sequence were supplemented using the STRING v10 database (52). Counts of ORFs affected by mutations in more than one population per treatment were implemented using the BioPython library ver. 1.65 (53), and chromosome map plots were implemented using the svgwrite library ver. 1.1.3 in Python ver. 2.7.10. Included in the parallel selected loci were those exhibiting the “multidiverse” signature of unlinked polymorphisms in the same ORFs. The method is implemented in the CheckLinkage option of BAGA (https://github.com/daveuu/baga) (54) and discussed in SI Materials and Methods.
Relative free phage abundances in end point populations were estimated separately for each phage using a quantitative PCR (qPCR) assay of DNase-treated supernatants, using primers targeted to each phage and comparison with a set of standards of known concentration. Competitions were performed between end point populations against the ancestral PAO1 (labeled with a gentamicin resistance marker) or an isogenic LES phage-resistant competitor, PAO1ΔpilA (labeled with a tetracycline resistance marker), in conditions identical to one transfer of the selection experiment. The density of each competitor was determined by plating onto antibiotic selective and nonselective media. Fitness was calculated as the selection rate constant. Free phage densities were measured for LESφ4 lysogens in both PAO1 and PAO1ΔlasR. Ten independent lysogens were constructed in each host background (PAO1 and PAO1ΔlasR) and cultured in LB until the stationary phase, and free phage densities in the supernatant were quantified using a plaque assay. Full methods are included in SI Materials and Methods.
Acknowledgments
A. Buckling and V. Friman kindly provided the bacterial strains PAO1ΔpilA and PAO1ΔlasR, respectively, used in this study. This work was funded by The Wellcome Trust project Grants 089215/Z/09/Z (to C.W. and M.A.B.) and 093306/Z/10 (to S.P., C.W., and M.A.B.) and a studentship cofunded by the Medical Research Council and the Institute of Infection and Global Health, University of Liverpool. S.O.B. is funded by The Wellcome Trust Institutional Strategic Support Fund grant supporting the Centre for Chronic Diseases and Disorders at the University of York.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.E.T. is a guest editor invited by the Editorial Board.
Data deposition: The sequence reported in this paper has been deposited in the European Nucleotide Archive, https://www.ebi.ac.uk/ena (accession no. PRJEB9801).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520056113/-/DCSupplemental.
References
- 1.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68(3):560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bobay LM, Rocha EP, Touchon M. The adaptation of temperate bacteriophages to their host genomes. Mol Biol Evol. 2013;30(4):737–751. doi: 10.1093/molbev/mss279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobay LM, Touchon M, Rocha EP. Pervasive domestication of defective prophages by bacteria. Proc Natl Acad Sci USA. 2014;111(33):12127–12132. doi: 10.1073/pnas.1405336111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hockett KL, Renner T, Baltrus DA. Independent co-option of a tailed bacteriophage into a killing complex in Pseudomonas. MBio. 2015;6(4):e00452. doi: 10.1128/mBio.00452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busby B, Kristensen DM, Koonin EV. Contribution of phage-derived genomic islands to the virulence of facultative bacterial pathogens. Environ Microbiol. 2013;15(2):307–312. doi: 10.1111/j.1462-2920.2012.02886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pallen MJ, Wren BW. Bacterial pathogenomics. Nature. 2007;449(7164):835–842. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- 7.Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999;33(1):167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 8.Figueroa-Bossi N, Uzzau S, Maloriol D, Bossi L. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol Microbiol. 2001;39(2):260–271. doi: 10.1046/j.1365-2958.2001.02234.x. [DOI] [PubMed] [Google Scholar]
- 9.Taylor AL. Bacteriophage-induced mutation in Escherichia coli. Proc Natl Acad Sci USA. 1963;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehmat S, Shapiro JA. Insertion and replication of the Pseudomonas aeruginosa mutator phage D3112. Mol Gen Genet. 1983;192(3):416–423. doi: 10.1007/BF00392184. [DOI] [PubMed] [Google Scholar]
- 11.Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499(7457):219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letarov A, Kulikov E. The bacteriophages in human- and animal body-associated microbial communities. J Appl Microbiol. 2009;107(1):1–13. doi: 10.1111/j.1365-2672.2009.04143.x. [DOI] [PubMed] [Google Scholar]
- 13.James CE, et al. Lytic activity by temperate phages of Pseudomonas aeruginosa in long-term cystic fibrosis chronic lung infections. ISME J. 2015;9(6):1391–1398. doi: 10.1038/ismej.2014.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology. 2007;153(Pt 4):917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 15.Mowat E, et al. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med. 2011;183(12):1674–1679. doi: 10.1164/rccm.201009-1430OC. [DOI] [PubMed] [Google Scholar]
- 16.Williams D, et al. Divergent, coexisting Pseudomonas aeruginosa lineages in chronic cystic fibrosis lung infections. Am J Respir Crit Care Med. 2015;191(7):775–785. doi: 10.1164/rccm.201409-1646OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorth P, et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe. 2015;18(3):307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folkesson A, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat Rev Microbiol. 2012;10(12):841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 19.Diaz Caballero J, et al. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. MBio. 2015;6(5):e00981–e15. doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winstanley C, O’Brien S, Brockhurst MA. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016;24(5):327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fothergill JL, et al. Effect of antibiotic treatment on bacteriophage production by a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2011;55(1):426–428. doi: 10.1128/AAC.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tejedor C, Foulds J, Zasloff M. Bacteriophages in sputum of patients with bronchopulmonary Pseudomonas infections. Infect Immun. 1982;36(1):440–441. doi: 10.1128/iai.36.1.440-441.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol Rev. 2011;35(4):652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 24.Miller RV, Rubero VJ. Mucoid conversion by phages of Pseudomonas aeruginosa strains from patients with cystic fibrosis. J Clin Microbiol. 1984;19(5):717–719. doi: 10.1128/jcm.19.5.717-719.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin DR. Mucoid variation in Pseudomonas aeruginosa induced by the action of phage. J Med Microbiol. 1973;6(1):111–118. doi: 10.1099/00222615-6-1-111. [DOI] [PubMed] [Google Scholar]
- 26.Sriramulu DD, Lünsdorf H, Lam JS, Römling U. Microcolony formation: A novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol. 2005;54(7):667–676. doi: 10.1099/jmm.0.45969-0. [DOI] [PubMed] [Google Scholar]
- 27.Winstanley C, et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res. 2009;19(1):12–23. doi: 10.1101/gr.086082.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J. 2012;40(1):227–238. doi: 10.1183/09031936.00204411. [DOI] [PubMed] [Google Scholar]
- 29.Burns N, James CE, Harrison E. Polylysogeny magnifies competitiveness of a bacterial pathogen in vivo. Evol Appl. 2015;8(4):346–351. doi: 10.1111/eva.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemieux AA, et al. Genes required for free phage production are essential for Pseudomonas aeruginosa chronic lung infections. J Infect Dis. 2016;213(3):395–402. doi: 10.1093/infdis/jiv415. [DOI] [PubMed] [Google Scholar]
- 31.Davies EVJ, et al. April 12, 2016. Temperate phages enhance pathogen fitness in chronic lung infection. ISME J, doi:10.1038/ismej.2016.51.
- 32.Brown SP, Le Chat L, De Paepe M, Taddei F. Ecology of microbial invasions: Amplification allows virus carriers to invade more rapidly when rare. Curr Biol. 2006;16(20):2048–2052. doi: 10.1016/j.cub.2006.08.089. [DOI] [PubMed] [Google Scholar]
- 33.James CE, et al. Differential infection properties of three inducible prophages from an epidemic strain of Pseudomonas aeruginosa. BMC Microbiol. 2012;12:216. doi: 10.1186/1471-2180-12-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9(7):797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 35.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 36.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 38.Diggle SP, Cornelis P, Williams P, Cámara M. 4-quinolone signalling in Pseudomonas aeruginosa: Old molecules, new perspectives. Int J Med Microbiol. 2006;296(2-3):83–91. doi: 10.1016/j.ijmm.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh D, et al. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: An alternative paradigm for prophage induction. Appl Environ Microbiol. 2009;75(22):7142–7152. doi: 10.1128/AEM.00950-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Høyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL. A quorum-sensing-induced bacteriophage defense mechanism. MBio. 2013;4(1):e00362–e12. doi: 10.1128/mBio.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winstanley C, Fothergill JL. The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections. FEMS Microbiol Lett. 2009;290(1):1–9. doi: 10.1111/j.1574-6968.2008.01394.x. [DOI] [PubMed] [Google Scholar]
- 42.Jimenez PN, et al. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76(1):46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roux D, et al. Fitness cost of antibiotic susceptibility during bacterial infection. Sci Transl Med. 2015;7(297):297ra114. doi: 10.1126/scitranslmed.aab1621. [DOI] [PubMed] [Google Scholar]
- 44.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62(2):596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang YS, Klockgether J, Tümmler B. An intragenic deletion in pilQ leads to nonpiliation of a Pseudomonas aeruginosa strain isolated from cystic fibrosis lung. FEMS Microbiol Lett. 2007;270(2):201–206. doi: 10.1111/j.1574-6968.2007.00664.x. [DOI] [PubMed] [Google Scholar]
- 46.Bjarnsholt T, et al. Scandinavian Cystic Fibrosis Study Consortium Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One. 2010;5(4):e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiricny N, et al. Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PLoS One. 2014;9(1):e83124. doi: 10.1371/journal.pone.0083124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011;19(8):419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Alhede M, Bjarnsholt T, Givskov M, Alhede M. Pseudomonas aeruginosa biofilms: Mechanisms of immune evasion. Adv Appl Microbiol. 2014;86:1–40. doi: 10.1016/B978-0-12-800262-9.00001-9. [DOI] [PubMed] [Google Scholar]
- 50.Bell G. Selection: The Mechanism of Evolution. Oxford Univ Press; New York: 2008. [Google Scholar]
- 51.Brockhurst MA. Experimental evolution can unravel the complex causes of natural selection in clinical infections. Microbiology. 2015;161(6):1175–1179. doi: 10.1099/mic.0.000107. [DOI] [PubMed] [Google Scholar]
- 52.Szklarczyk D, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cock PJ, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams D, Paterson S, Brockhurst MA, Winstanley C. Refined analyses suggest that recombination is a minor source of genomic diversity in Pseudomonas aeruginosa chronic cystic fibrosis infections. Microb Genom. 2016;2(3):1–8. doi: 10.1099/mgen.0.000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koch B, Jensen LE, Nybroe O. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J Microbiol Methods. 2001;45(3):187–195. doi: 10.1016/s0167-7012(01)00246-9. [DOI] [PubMed] [Google Scholar]
- 56.Taylor TB, Buckling A. Competition and dispersal in Pseudomonas aeruginosa. Am Nat. 2010;176(1):83–89. doi: 10.1086/652995. [DOI] [PubMed] [Google Scholar]
- 57.Kirchner S, et al. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J Vis Exp. 2012;(64):e3857. doi: 10.3791/3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 59.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinfor. 2013;43:11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucl Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong A, Rodrigue N, Kassen R. Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet. 2012;8(9):e1002928. doi: 10.1371/journal.pgen.1002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138(6):1315–1341. [Google Scholar]
- 65.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43(12):1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hachani A, et al. Type VI secretion system in Pseudomonas aeruginosa: Secretion and multimerization of VgrG proteins. J Biol Chem. 2011;286(14):12317–12327. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giltner CL, Habash M, Burrows LL. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol. 2010;398(3):444–461. doi: 10.1016/j.jmb.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 68.Xiao G, He J, Rahme LG. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology. 2006;152(Pt 6):1679–1686. doi: 10.1099/mic.0.28605-0. [DOI] [PubMed] [Google Scholar]
- 69.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69(2):376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schuster M, Greenberg EP. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296(2-3):73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 71.Dasgupta N, Arora SK, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2000;182(2):357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hong CS, et al. The aerotaxis transducer gene aer, but not aer-2, is transcriptionally regulated by the anaerobic regulator ANR in Pseudomonas aeruginosa. J Biosci Bioeng. 2004;97(3):184–190. doi: 10.1016/S1389-1723(04)70188-7. [DOI] [PubMed] [Google Scholar]
- 73.Kumari H, et al. LTQ-XL mass spectrometry proteome analysis expands the Pseudomonas aeruginosa AmpR regulon to include cyclic di-GMP phosphodiesterases and phosphoproteins, and identifies novel open reading frames. J Proteomics. 2014;96:328–342. doi: 10.1016/j.jprot.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pryor EE, Jr, et al. The transcription factor AmrZ utilizes multiple DNA binding modes to recognize activator and repressor sequences of Pseudomonas aeruginosa virulence genes. PLoS Pathog. 2012;8(4):e1002648. doi: 10.1371/journal.ppat.1002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Irie Y, et al. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78(1):158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Irie Y, et al. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109(50):20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuchma SL, et al. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189(22):8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harvey H, Habash M, Aidoo F, Burrows LL. Single-residue changes in the C-terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility. J Bacteriol. 2009;191(21):6513–6524. doi: 10.1128/JB.00943-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 80.Alm RA, Mattick JS. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178(13):3809–3817. doi: 10.1128/jb.178.13.3809-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alm RA, Mattick JS. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol Microbiol. 1995;16(3):485–496. doi: 10.1111/j.1365-2958.1995.tb02413.x. [DOI] [PubMed] [Google Scholar]
- 82.Nguyen Y, et al. Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J Biol Chem. 2015;290(1):601–611. doi: 10.1074/jbc.M114.616904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doyle TB, Hawkins AC, McCarter LL. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol. 2004;186(19):6341–6350. doi: 10.1128/JB.186.19.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Vos D, et al. Detection of the outer membrane lipoprotein I and its gene in fluorescent and non-fluorescent pseudomonads: Implications for taxonomy and diagnosis. J Gen Microbiol. 1993;139(9):2215–2223. doi: 10.1099/00221287-139-9-2215. [DOI] [PubMed] [Google Scholar]