Significance
Domain of unknown function (DUF) families constitute 3,892 of the 16,295 families in the Pfam database (release 29.0). Given their biological importance, large-scale strategies are required to accomplish their functional assignments. Here, we illustrate an integrated “genomic enzymology” strategy to identify diverse functions within the DUF1537 family (PF07005). We combined high-throughput ligand screening results for transport system solute binding proteins with the synergetic analysis of sequence similarity networks and genome neighborhood networks to establish that the members of the DUF1537 family are novel ATP-dependent four-carbon sugar kinases. This study illustrates the utility of this strategy and enhances our knowledge of bacterial carbohydrate catabolism.
Keywords: DUF1537, kinase, four-carbon acid sugars, conserved genome neighborhoods, genomic enzymology
Abstract
Using a large-scale “genomic enzymology” approach, we (i) assigned novel ATP-dependent four-carbon acid sugar kinase functions to members of the DUF1537 protein family (domain of unknown function; Pfam families PF07005 and PF17042) and (ii) discovered novel catabolic pathways for d-threonate, l-threonate, and d-erythronate. The experimentally determined ligand specificities of several solute binding proteins (SBPs) for TRAP (tripartite ATP-independent permease) transporters for four-carbon acids, including d-erythronate and l-erythronate, were used to constrain the substrates for the catabolic pathways that degrade the SBP ligands to intermediates in central carbon metabolism. Sequence similarity networks and genome neighborhood networks were used to identify the enzyme components of the pathways. Conserved genome neighborhoods encoded SBPs as well as permease components of the TRAP transporters, members of the DUF1537 family, and a member of the 4-hydroxy-l-threonine 4-phosphate dehydrogenase (PdxA) oxidative decarboxylase, class II aldolase, or ribulose 1,5-bisphosphate carboxylase/oxygenase, large subunit (RuBisCO) superfamily. Because the characterized substrates of members of the PdxA, class II aldolase, and RuBisCO superfamilies are phosphorylated, we postulated that the members of the DUF1537 family are novel ATP-dependent kinases that participate in catabolic pathways for four-carbon acid sugars. We determined that (i) the DUF1537/PdxA pair participates in a pathway for the conversion of d-threonate to dihydroxyacetone phosphate and CO2 and (ii) the DUF1537/class II aldolase pair participates in pathways for the conversion of d-erythronate and l-threonate (epimers at carbon-3) to dihydroxyacetone phosphate and CO2. The physiological importance of these pathways was demonstrated in vivo by phenotypic and genetic analyses.
As a result of advances in genome sequencing, the number of sequences in the protein databases is rapidly increasing: for example, >60 million sequences in release 2016_02 of the UniProt database that is increasing in size at the rate of ∼2% per month (1). Approximately two-thirds of the proteins identified in genome projects have annotations based on sequence homology to previously annotated proteins; the remaining proteins are classified as “hypothetical proteins” or “uncharacterized proteins” (2, 3). However, this automated process results in propagation of misleading or incorrect annotations (4–7). Correction of annotations by experimental characterization is essential for realizing the value of genome sequences.
The Pfam database organizes the “protein universe” into homologous families [16,295 protein families in Pfam release 29.0 (8)]. In release 29.0, 3,892 families are annotated as a “domain of unknown function” (DUF) because no member has an experimentally characterized function (9). Given their wide taxonomic distribution, including bacterial pathogens, many DUFs are likely biologically essential (10). Thus, reliable experimental identification of the in vitro enzymatic activities and in vivo physiological functions for the DUF families is important. However, the assignment of functions to uncharacterized proteins is challenging (11–13). The methods now available for discovering the functions of uncharacterized proteins, including DUFs, are inefficient and often depend on inference from the functions of characterized homologs (11). Therefore, new strategies are required to confront and solve the functional assignment challenge.
The DUF1537 family (Pfam families PF07005 and PF17042) contains 4,610 sequences in release 2016_02 of the UniProt database; members of DUF1537 are found in diverse bacterial phyla, including Proteobacteria, Firmicutes, Cyanobacteria, Actinobacteria, and Bacteroidetes. Some organisms contain multiple members of the family, suggesting diverse biological functions. In the Uniprot and National Center for Biotechnology Information databases, DUF1537 proteins often are annotated as “Hrp (hypersensitive reaction and pathogenicity) type III effectors” on the basis of remote functional relationships and low sequence homology. “Hrp” proteins commonly are secreted via a type III secretion system and often are involved in plant tissue infections (14). Although evidence of secretion has not been reported for DUF1537 proteins, one bioinformatic analysis predicted a secretory signal on the N terminus of the Pseudomonas syringae DUF1537 protein HopAN1 (Hrp outer protein effector protein) (Uniprot: Q87V79) (15). The gene encoding VguB (virulence gluconate metabolism), a member of the DUF1537 family, was associated with plant virulence and d-gluconate metabolism in Pectobacterium carotovorum WPP14 (16). In addition, members of the family are elevated in Burkholderia pseudomallei in a human infection model (17). Both the phylogenetic diversity of species with members of the DUF1537 family and the correlation of the proteins with virulence make them productive targets for functional characterization.
As described in this article, we used in vitro and in vivo functional assignment of members of the DUF1537 family to further develop our large-scale genomic enzymology-based strategy for functional assignment of novel enzymes in novel microbial catabolic pathways (18). The strategy takes advantage of the frequently observed genomic colocation of microbial genes that encode the transport systems and enzymes for catabolism of an extracellular solute.
The first step in our strategy is the experimental screening of the ligand specificity for a transport system solute binding protein (SBP) (18). This step identifies the starting metabolite for the pathway and helps locate the genes that encode the enzyme components of the pathway. We focus on the SBPs of bacterial TRAP (tripartite ATP-independent permease) and ATP-binding cassette transport systems; both have an extracellular SBP that binds and delivers its ligand to the integral membrane permease components for transport into the cell (19, 20). We then synergistically use protein family sequence similarity networks (SSNs) and genome neighborhood networks (GNNs) to discover the enzyme components of the pathway and infer their functions (21). A SSN is a readily accessible method (constructed with the Enzyme Function Initiative-Enzyme Similarity Tool web tool) for segregating protein families, including those of SBPs as well as enzymes, into isofunctional groups (22). A GNN (constructed with the Enzyme Function Initiative-Genome Neighborhood Tool web tool) then enables identification of conserved genome neighborhoods for isofunctional groups in the SSN and partitions the enzymes encoded by these neighborhoods into Pfam families, aiding inference of the reactions in the pathway given the expected identity of the substrate for the pathway (the ligand for the SBP).
In this study, we used the experimentally determined specificities of four orthologous TRAP SBPs for four-carbon acid sugars to predict and then experimentally assign kinase functions to members of the DUF1537 family (Pfam families PF07005 and PF17042). With our integrated SSN and GNN analysis, we identified the enzymes and inferred the reactions in three novel catabolic pathways for d-erythronate, d-threonate, and l-threonate; we then biochemically and physiologically verified those predictions. We expect that this strategy will be useful for the discovery of other novel metabolic pathways as well as assigning functions to other DUF families.
Results
Synergistic Analysis of SSNs and GNNs Enables the Prediction That Members of DUF1537 Are Novel Four-Carbon Acid Sugar Kinases.
We previously identified four SBPs that bind four-carbon acid sugars, including d-erythronate and l-erythronate (18). When the SSN for the TRAP SBP family (IPR018389) is filtered at an alignment score threshold of 80 (∼45% sequence identity) (SI Appendix, Fig. S1A), the SBPs (Uniprot: Q12HD7, Q12CD8, A1WPV4, and Q1QSK0) are located in two clusters. A GNN generated using the SSN for the TRAP SBP family revealed that many of the genes encoding the members of the SSN clusters are proximal to those encoding members of the DUF1537 family (PF07005) (SI Appendix, Fig. S1B). This colocalization allows the hypothesis that members of DUF1537 are involved in the catabolism of four-carbon acid sugars.
SSNs for the DUF1537 family (PF07005 in Pfam 29.0) were constructed with differing alignment scores (SI Appendix, Fig. S2) and used to generate GNNs (22). Using an SSN filtered at an alignment score of 60 (∼35% sequence identity), the GNN identified proteins encoded by three distinct genome neighborhoods that appear to encode distinct metabolic pathways (Fig. 1); these are distinguished by the presence of genes for members of the 4-hydroxy-l-threonine 4-phosphate dehydrogenase (PdxA) (PF04166), class II aldolase (PF00596), or ribulose 1,5-bisphosphate carboxylase/oxygenase, large subunit (RuBisCO) (PF00016) families (Fig. 1B). In addition, some members of the DUF1537 family are fusion proteins with a member of either the PdxA family (32 sequences from Ralstonia, Arthrobacter, Rhodococcus, and Burkholderia) or the class II aldolase family (five sequences primarily from Clostridium), thereby confirming a functional relationship (23).
Fig. 1.
SSN/GNN analysis of DUF1537 family. (A) SSN for the DUF1537 family (PF07005) displayed at an alignment score of 60 (∼35% sequence identity). The nodes in the network are colored based on conserved genome neighborhoods that encode the members of respective nodes. PdxA2–, aldolase–, and RLP–DUF1537 proteins are denoted by the sea blue, red, and orange nodes, respectively. The UniProt IDs for 22 DUF1537 proteins are also indicated (SI Appendix, Table S1). (B) The cluster-specific GNNs for PdxA2–, aldolase– and RLP–DUF1537 clusters and only the highly conserved [indicated by cluster fraction (CF)], catalytic genome proximal enzyme families specific to these clusters were shown here. (C) Representative PdxA2–DUF1537 and aldolase–DUF1537 gene clusters encoded in R. eutropha H16.
We hypothesized that members of the DUF1537 family catalyze the ATP-dependent phosphorylation of four-carbon acid sugars because: (i) the substrates for members of the PdxA, class II aldolase, and RuBisCO superfamilies are phosphorylated; (ii) the genome neighborhoods lacked known kinase genes; and (iii) the ligands for the SBPs that were used to target the DUF1537 family are four-carbon acid sugars. In the analyses that follow, the DUF1537 proteins, genome neighborhoods, and metabolic pathways are designated as PdxA2–, aldolase–, or RLP–DUF1537 proteins, neighborhoods, and pathways based on the genome neighbors identified in the GNN for the DUF1537 clusters [the RuBisCO homologs belong to the RuBisCO-like protein (RLP) clade of the RuBisCO family; known RLP proteins do not catalyze the carboxylation or oxygenation reactions catalyzed by autotrophic RuBisCOs (24)].
Biochemical Characterization of the PdxA2–DUF1537 Pathway.
High-throughput protein production for members of the DUF1537 family was performed (25). We produced 22 of 122 target proteins (cloned from an in-house collection of ∼400 gDNAs); at least one purified protein was obtained for most of the major clusters in the DUF1537 SSN shown in Fig. 1A. A four-carbon sugar library with 22 potential substrates was assembled, including four-carbon mono- and diacid sugars, aldotetroses, ketotetroses, alditols, and modified sugars/analogs (SI Appendix, Fig. S3A). The purified proteins were screened for ATP-dependent kinase activity using the library. Activity was detected for 20 proteins; the substrates were d-threonate or d-erythronate (SI Appendix, Table S1).
We first focused on members of the DUF1537 family that are encoded genome proximal to members of the PdxA family (Fig. 1 and SI Appendix, Fig. S4; see SI Appendix, Table S11 for a full list of proteins from this study). The SSN for the PdxA family filtered with an alignment score of 80 (∼47% sequence identity) revealed that the members that are encoded genome proximal to DUF1537 proteins (designated a PdxA2 subgroup) are separated from the proteins (PdxA) that catalyze the NAD(P)+-dependent oxidative decarboxylation of 4-hydroxy-l-threonine 4-phosphate (4HT-4P) to form 3-amino-1-hydroxyacetone 1-phosphate in the biosynthetic pathway for pyridoxal 5′-phosphate (PLP) (SI Appendix, Fig. S5) (26, 27), suggesting a distinct function.
Because 4-HT is structurally similar to d-threonate (SI Appendix, Fig. S3B), a hit in our screen, we chemically synthesized 4-HT (28) and compared the in vitro activities of the DUF1537 proteins with 4-HT, d-threonate, and d-erythronate (Table 1 and SI Appendix, Table S2). For five PdxA2–DUF1537 proteins (Uniprot: Q0K4F6, Q8ZRS5, Q6D0N7, B0TBI9, and A0A0H3LX82) (Fig. 1A and SI Appendix, Table S2), d-threonate or d-erythronate was the preferred substrate; although the values of kcat are similar, the values of KM for d-threonate/d-erythronate are ∼100-fold less than those for 4-HT. The PdxA2–DUF1537 enzymes (DtnK, d-threonate kinase and DenK, d-erythronate kinase) catalyze the ATP-dependent phosphorylation of d-threonate or d-erythronate to generate d-threonate 4-phosphate or d-erythronate 4-phosphate.
Table 1.
Selected kinetic parameters for DtnK, PdxA2, LtnD, and DenD
| Uniprot (annotation) | Substrate | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) |
| Q0K4F6 (ReDtnK) | d-Threonate | 45 ± 4 | 0.12 ± 0.03 | 3.8 × 105 |
| 4-Hydroxy-l-threonine | 27 ± 2 | 86 ± 9 | 3.1 × 102 | |
| Q8ZRS5 (SeDtnK) | d-Threonate | 22 ± 1.3 | 0.29 ± 0.05 | 7.5 × 104 |
| 4-Hydroxy-l-threonine | 4.0 ± 0.1 | 3.8 ± 0.5 | 1.1 × 103 | |
| Q0K4F5 (RePdxA2) | d-Threonate 4-phosphate | 3.4 ± 0.09 | 0.12 ± 0.01 | 2.9 × 104 |
| 4-Hydroxy-l-threonine 4-phosphate | 0.35 ± 0.005 | 0.20 ± 0.02 | 1.8 × 103 | |
| P58718 (SePdxA2) | d-Threonate 4-phosphate | 8.9 ± 0.2 | 0.054 ± 0.006 | 1.6 × 105 |
| 4-Hydroxy-l-threonine 4-phosphate | 0.37 ± 0.02 | 0.19 ± 0.04 | 1.9 × 103 | |
| Q0KBC7 (ReLtnD) | l-Threonate, NAD+ | 29 ± 0.7 | 0.35 ± 0.04 | 8.5 × 104 |
| l-Threonate, NADP+ | 2.2 ± 0.07 | 0.30 ± 0.04 | 7.5 × 103 | |
| Q6CZ26 (PaLtnD) | l-Threonate, NAD+ | 54 ± 0.7 | 0.13 ± 0.007 | 4.1 × 105 |
| l-Threonate, NADP+ | 4.7 ± 0.1 | 0.97 ± 0.1 | 4.8 × 103 | |
| Q0KBD2 (ReDenD) | d-Erythronate, NAD+ | 19 ± 0.3 | 0.59 ± 0.04 | 3.3 × 104 |
| d-Erythronate, NADP+ | 3.0 ± 0.08 | 2.4 ± 0.3 | 1.2 × 103 |
Error is the SD.
The NAD+-dependent oxidation activities of members of the PdxA2 group for d-threonate 4-phosphate and d-erythronate 4-phosphate (products of DtnK and DenK, respectively) were evaluated by quantitating the reduction of NAD+. The values of kcat/KM for d-threonate 4-phosphate or d-erythronate 4-phosphate were 10- to 100-fold greater than those for 4-HT-4P (Table 1 and SI Appendix, Table S3). Using both coupled-enzyme spectrophotometric (SI Appendix, Fig. S6) and 1H NMR assays (SI Appendix, Fig. S7), we observed that the PdxA2 proteins catalyze the decarboxylation reaction to generate dihydroxyacetone phosphate (DHAP) and CO2 (analogous to the reaction catalyzed by PdxA). When glycerol 3-phosphate dehydrogenase (G3PDH) was added after the PdxA2-catalyzed reaction was complete, we observed immediate oxidation of NADH (reduction of DHAP) (SI Appendix, Fig. S6), providing further support that PdxA2 catalyzes both the oxidation and subsequent decarboxylation of d-threonate 4-phosphate to produce DHAP. Therefore, the in vitro activities of DtnK or DenK and the PdxA2 proteins suggest catabolic pathways in which DtnK or DenK phosphorylates d-threonate or d-erythronate and the members of the PdxA2 group oxidatively decarboxylate the products to generate DHAP and CO2 (Fig. 2A). The weak promiscuities of DtnK and DenK for 4HT and of PdxA2 for 4-HT-4P provide the potential for evolution of a new catabolic pathway.
Fig. 2.
Pathways for degradation of d/l-threonate and d-erythronate: (A) PdxA2–DUF1537 pathway; (B) aldolase–DUF1537 pathway.
Biological Characterization of the PdxA2–DUF1537 Pathway.
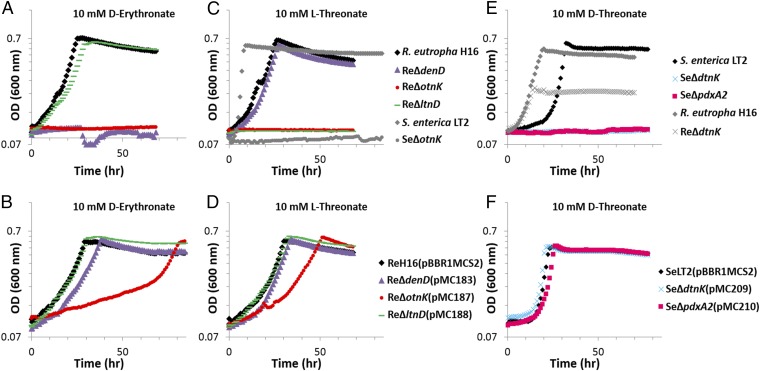
The physiological roles of DtnK and its associated PdxA2 were studied by deleting the gene encoding each protein in Salmonella enterica ser. Typhimurium LT2. In contrast to the wild-type strain, the SeΔdtnK and SeΔpdxA2 strains were unable to use d-threonate as a carbon source (Fig. 3 and SI Appendix, Fig. S13). When the full-length version of the deleted coding region was provided in trans in each mutant strain, the resulting growth mirrored that of the wild-type strain carrying the same empty plasmid (Fig. 3 and SI Appendix, Fig. S13); therefore, both genes are required for d-threonate growth.
Fig. 3.
Growth of selected wild-type, mutant strains (A, C, E), and complemented strains (B, D, F) with 10 mM d-erythronate (A, B), 10 mM l-threonate (C, D), or 10 mM d-threonate (E, F). The same color (similar to SI Appendix, Figs. S4 and S8) and marker represent growth of a strain (A, C, E) and the corresponding complemented strain (B, D, F) for each growth condition. Gray markers represent strains for which no complementation data were collected. Strain identities are presented in the above legends. Plasmid identities are detailed in SI Appendix, Table S13 and strain identities are detailed in SI Appendix, Table S12. For complete growth data, see SI Appendix, Figs. S13–S17.
To investigate whether the physiological role of the dtnK gene cluster is conserved despite its phylogenetic source (different phylogenetic class), dtnK (UniProt: Q0K4F6) was deleted in Ralstonia eutropha H16. The resulting strain also demonstrated impaired d-threonate growth (SI Appendix, Fig. S16).
Biochemical Characterization of the Aldolase–DUF1537 Pathway.
For the members of the DUF1537 family encoded by gene clusters that encode members of the class II aldolase family (PF00596) (Fig. 1 and SI Appendix, Fig. S8), the kinase screen detected activities with d-threonate (SI Appendix, Table S1), although the value of kcat/KM was low (29 M−1 s−1 for A0A0H2VCE6). Therefore, we expected that a structural analog of d-threonate is the physiological substrate. The GNN revealed that this genome context also encodes both an isomerase (PF01261) and a dehydrogenase (PF03446 and PF14833) (Fig. 1B and SI Appendix, Fig. S8). Four dehydrogenases encoded by the aldolase–DUF1537 gene clusters from different organisms (Uniprot: Q6CZ26, A0A0H2VA68, P44979, and Q0KBC7) were screened using a library of 53 sugars (SI Appendix, Table S6). This library included d-gluconate (a possible substrate for a member of the DUF1537 family implicated by studies with P. carotovorum (16); and see Biological Characterization of the Aldolase–DUF1537 Pathway); however, activity was detected only with l-threonate. The dehydrogenases can use either NAD+ or NADP+ as cosubstrate, with a preference for NAD+; the value of kcat/KM for oxidation of l-threonate with NAD+ was ∼105 M−1 s−1 (Table 1 and SI Appendix, Table S4).
Assuming that oxidation of l-threonate is the first reaction in the aldolase–DUF1537 pathway, we conducted a series of coupled-enzyme assays to delineate the sequence of reactions catalyzed by the members of the dehydrogenase, isomerase, and DUF1537 families (SI Appendix, Fig. S9 D–F). l-Threonate is oxidized (LtnD, l-threonate dehydrogenase), isomerized (OtnI, 2-oxo-tetronate isomerase), and phosphorylated (OtnK, 3-oxo-tetronate kinase) before conversion to dihydroxyacetone phosphate and CO2 by the member of the class II aldolase family (OtnC, 3-oxo-tetronate 4-phosphate decarboxylase). The value of kcat/KM for the decarboxylase-catalyzed reaction was estimated (SI Appendix, Fig. S10 and Table S5) and is consistent with the predicted physiological role of the reaction. The same order of reactions was established for the enzymes from the gene clusters in Pectobacterium atrosepticum SCRI 1043, Haemophilus influenza KW20, and R. eutropha H16 (SI Appendix, Fig. S11 A, B, and D).
The identities of the pathway intermediates were established by considering that the final products were possible only if the substrate for the decarboxylase (OtnC) was 3-oxo-tetronate 4-phosphate. We propose that the β-ketoacid decarboxylation reaction involves the formation of an enolate anion stabilized by a divalent metal, by virtue of its membership in the class II aldolase family (29). Therefore, OtnK, the member of the DUF1537 family, must have phosphorylated 3-oxo-tetronate, the isomerization product of OtnI. Our results, however, could not distinguish whether the substrate for OtnI/product of LtnD was 2-oxo- or 4-oxo-tetronate until we demonstrated in vivo that hydroxypyruvate isomerase (Hyi, UniProt: Q0K5R4) compensated for the loss of OtnI (UniProt: Q0KBD1) (see Biological Characterization of the Aldolase–DUF1537 Pathway), implying that OtnI catalyzes the isomerization of the carbonyl group between C2 and C3 similar to that catalyzed by Hyi. Therefore, we concluded that the substrate for OtnI was 2-oxo-tetronate, completing the pathway: l-threonate is oxidized to 2-oxo-tetronate (LtnD), 2-oxo-tetronate is isomerized (OtnI) and phosphorylated (OtnK) to 3-oxo-tetronate 4-phosphate, and 3-oxo-tetronate 4-phosphate is decarboxylated (OtnC) to DHAP and CO2 (Fig. 2B).
Approximately 25% of the gene clusters encoding the aldolase–DUF1537 pathway include an additional gene for a protein annotated as “NAD+-dependent epimerase” (PF01370) (Fig. 1 and SI Appendix, Fig. S8). We hypothesized that this protein participates in an alternate version of the pathway, allowing utilization of a different four-carbon acid substrate. Although the protein is annotated as an “epimerase” [homologs catalyze the conversion of UDP-galactose to UDP-glucose in galactose metabolism (30, 31)], the inversion of configuration is accomplished by two successive NAD+-dependent oxidoreductase reactions. We used our sugar library (SI Appendix, Table S6) to screen two orthologous dehydrogenases (Unitprot: P44094 and Q0KBD2) and determined that both catalyze the efficient (kcat/KM > 103 M−1 s−1) oxidation of d-erythronate with either NAD+ or NADP+ as cosubstrate, indicating that they are d-erythronate dehydrogenases (DenD) (Table 1 and SI Appendix, Table S4). Therefore, we hypothesized that DenD participates in a pathway for the utilization of d-erythronate analogous to that for utilization of l-threonate: DenD catalyzes oxidation of d-erythronate to 2-oxo-tetronate that is further catabolized to DHAP and CO2 by OtnI, OtnK, and OtnC in the pathway for l-threonate degradation (Figs. 2B and SI Appendix, Figs. S9 A–C and S11 C and E).
On the basis of these in vitro activities, we conclude that the members of the aldolase (decarboxylase) and DUF1537 (kinase) families participate in convergent pathways for utilization of l-threonate and d-erythronate, with different dehydrogenases generating the common 2-oxo-tetronate (LtnD for l-threonate and DenD for d-erythronate) that is isomerized to 3-oxo-tetronate (OtnI), phosphorylated to 3-oxo-tetronate 4-phosphate (OtnK), and decarboxylated to DHAP and CO2 (OtnC) (Fig. 2B). In this pathway, OtnK catalyzes the ATP-dependent phosphorylation of 3-oxo-tetronate (SI Appendix, Fig. S12), a structural analog of the d-threonate substrate for the PdxA2-DUF1537 protein in the pathway for d-threonate utilization, thereby explaining the low value of kcat/KM observed for d-threonate (3-oxo-tetronate is an unstable β-ketoacid so cannot be added to our screening library).
Biological Characterization of the Aldolase–DUF1537 Pathway.
To investigate whether the aldolase–DUF1537 pathway assembled in vitro is functional in vivo, we determined the effects of deleting the genes in the R. eutropha H16 aldolase–DUF1537 gene cluster that includes denD (Fig. 1C). Wild-type cells grow with either l-threonate or d-erythronate as sole carbon source. With the exception of otnI, deletion of the genes in the neighborhood abolished growth with l-threonate or d-erythronate (Figs. 3 and SI Appendix, Fig. S14). The growth observed for the otnI deletion strain, albeit impaired, is explained by a redundant/promiscuous activity from a hydroxypyruvate isomerase [Hyi, Uniprot: Q0K5R4, 58% identical to and encoded in the same genomic context as the authentic hydroxypyruvate isomerase in Escherichia coli K12 (UniProt: P30147) (32)] that shares 47% sequence identity with OtnI. When hyi was deleted, growth with l-threonate and d-erythronate was unaffected. When hyi and otnI both were deleted in a single strain, growth with l-threonate or d-erythronate was abolished. Because hyi is not essential for optimal l-threonate or d-erythronate growth, OtnI is likely the exclusive isomerase for l-threonate and d-erythronate assimilation in the wild-type strain. Deletion of ltnD did not affect d-erythronate growth, and deletion of denD did not affect l-threonate growth. When each gene was expressed in trans in the corresponding mutant strain, l-threonate or d-erythronate growth was restored, confirming that the product of each gene was required for either l-threonate or d-erythronate catabolism (Fig. 3 and SI Appendix, Fig. S14).
We observed similar phenotypic results when the gene for OtnK (Uniprot: Q8ZMG5) was deleted in S. enterica. Unlike the wild-type strain, SeΔotnK was unable to grow with l-threonate or d-erythronate (SI Appendix, Fig. S17), indicating that the physiological role of the aldolase–DUF1537 protein is conserved among species despite a previous report by Mole et al. that an otnK gene (designated vguB in the study) in P. carotovorum WPP14 was required for virulence in plant leaves and for utilization of d-gluconate as a carbon source (16). With P. carotovorum wild-type, PcΔotnK, and complemented strains provided by Mole et al., we observed no connection between the otnK operon and d-gluconate growth; instead, our results indicate that the otnK operon is required for l-threonate growth (SI Appendix, Fig. S15).
Structural Characterization of the DUF1537 Family.
The structures of several members of the DUF1537 family (∼400 amino acids) were determined by X-ray crystallography. These members included the citrate-bound structure of DtnK from Bordetella bronchiseptica RB50 (BbDtnK, Uniprot: A0A0H3LX82), the unliganded and d-threonate-bound structures of DtnK from P. atrosepticum SCRI 1043 (PaDtnK, Uniprot: Q6D0N7), and the ADP-bound structure of OtnK from R. eutrophia H16 (ReOtnK, Uniprot: Q0KBC8) (SI Appendix, Tables S7–S9). In addition, unliganded structures of two OtnK orthologs (H. influenza KW20, HiOtnK, Uniprot: P44093, PDB ID code 1YZY; and S. enterica ser. Typhimurium LT2, SeOtnK, Uniprot: Q8ZMG5, PDB ID code 3DQQ) had been reported previously (SI Appendix, Fig. S18). Together, these structures represent a diverse set of sequences in the DUF1537 family, with ReOtnK, HiOtnK, and SeOtnK sharing ∼40–50% sequence identity and ReOtnK, BbDtnK, and PaDtnK sharing ∼20% sequence identity (SI Appendix, Table S10).
Members of the DUF1537 family are composed of two domains: an N-terminal domain (residues 1–247, ReOtnK numbering) and a C-terminal domain (residues 266–429) connected by a variable linker sequence (Fig. 3A and SI Appendix, Fig. S19). The N-terminal domain exhibits an α/β-fold composed of an eight-stranded parallel β-sheet (strand order S2N, S3N, S1N, S4N, S11N, S5N, S10N, and S9N) flanked by helices H1N, H5N, H6N, H7N, and H8N on one face and helices H2N, H3N, and H4N on the opposing face of the central β-sheet. The C-terminal domain also exhibits an α/β-fold composed of a seven-stranded mixed β-sheet (strand order S2C, S3C, S1C, S4C, S7C, S6C, and S5C; strands S6C and S7C antiparallel) flanked by helices H1C and H7C on one face and helices H2C, H3C, H4C, H5C, and H6C on the other face of the β-sheet (SI Appendix, Fig. S19). A search for structural homologs for either domain, using PDBeFold (33), yielded low quality hits with RMSDs of >3.0 Å over less than 70% of the structural elements. These distant structural homologs yielded no additional information on the function or the location of active site features.
The central β-sheet of the N-terminal domain abuts the central β-sheet of the C-terminal domain in an antiparallel fashion creating a 15-strand continuous β-sheet. In the case of ReOtnK, HiOtnK, and SeOtnK, the continuous β-sheet is formed within the same subunit (i.e., the protein is monomeric), whereas in BbDtnK and PaDtnK the protein crystallized as a domain swapped dimer with the N-terminal domain of one subunit forming an intermolecular β-sheet with the C-terminal domain of a second subunit (SI Appendix Fig. S18). Regardless of its composition (inter- or intramolecular), every structure has the conserved N- to C-terminal domain β-sheet/β-sheet interaction and, except for the ADP-bound ReOtnK structure (see below), the relative orientation of these two domains is highly conserved (Fig. 4A, open confirmation).
Fig. 4.
Crystallography of DUF1537 proteins. (A) Superposition of DUF1537 structures in the “open” form. Shown are the Cα traces of P44093 (red), Q8ZMG5 (green), Q0KBC8 (blue), Q6D0N7 (magenta), and A0A0H3LX82 (orange). (B) Ribbon diagram of Q6D0N7 in complex with d-threonate (CPK model). The N-terminal domain of subunit A and the C-terminal domain of subunit B are uniquely shown as the potential catalytic unit. (Inset) Fo–Fc omit electron density map for d-threonate contoured at 3σ. (C) LIGPLOT diagram of the interactions of Q6D0N7 with d-threonate. Asp17 is strictly conserved in all DUF1537 proteins, whereas Asp92, conserved among PdxA2–DUF1537 proteins, is a cysteine in Aldolase–DUF1537and RLP–DUF1537 proteins. (D) Ribbon diagram of Q0KBC8 in complex with ADP (CPK model). (Inset) Fo–Fc omit electron density map for ADP contoured at 3σ. (E) LIGPLOT diagram of the interactions of Q0KBC8 with ADP. (F) Molecular model of a ternary d-threonate/ADP complex.
The d-threonate-bound structure of PaDtnK (cocrystallization based on activity screening) demonstrated that the acid-sugar binding site is located in the N-terminal domain adjacent to the domain-domain interface (Fig. 4B). Binding of d-threonate produced no large-scale rearrangements compared with the apo structure (RMSD = 0.23 Å). d-Threonate is bound in a solvent-accessible depression and forms nine direct hydrogen bonds with the protein (Fig. 4C). Two strictly conserved consecutive Asp residues (Asp16 and Asp17 in PaDtnK) and a conserved Arg (Arg60 in PaDtnK) are responsible for coordination of the 4-OH of d-threonate. Asp16 orientates Arg60, which hydrogen bonds to the 4-OH of d-threonate, whereas Asp17 makes a direct hydrogen bond. The position and strict conservation of Asp17 among all DUF1537 family members suggests that Asp17 is the active site base responsible for activation of the alcohol for nucleophilic attack on the γ-phosphate of ATP.
The structure of ReOtnK, determined from crystals formed in the presence of ATP, yielded unambiguous density for ADP (Fig. 4D). ADP is bound only in one of the two molecules (i.e., monomers) per asymmetric unit; the ADP free monomer is in the “open” conformation, whereas in the ADP-bound structure, the C-terminal domain has rotated 30° [as calculated by DynDom (34)] toward the N-terminal domain to form a “closed” structure with a solvent-excluded catalytic site (SI Appendix, Fig. S20). ADP is bound predominately by residues donated by the C-terminal domain, again adjacent to the domain–domain interface. The diphosphate moiety is coordinated by three backbone amides donated from the N-terminal end of H6C, along with a fourth backbone amide from Gly414 (ReOtnK numbering), whereas the adenine moiety is bound within a greasy pocket formed by strand 3, helix 2, helix 4, and helix 5 of the C-terminal domain (Fig. 4E). The only residues from the N-terminal domain that contact ADP are donated by a loop between strand 8 and helix 4, with the side-chains of Pro156 and Leu157 resting against the nonpolar face of adenosine and His155 hydrogen bonding to the β-phosphate. In general, residues of the nucleotide-binding pocket are not strictly conserved, presumably in part due to the large number of mainchain atoms that are used to coordinate the ligand (six of the nine direct hydrogen bonds).
A molecular model of the ternary complex was constructed by superimposing the N-terminal domain of the d-threonate complex with N-terminal domain of the ADP complex (Fig. 4F). The model suggests that a loop between S7C and H7C (i.e., Ser413, Phe416) could additionally be involved in recognition of the sugar substrate when the protein is in the closed ATP bound state. The model highlights the close proximity of the two binding sites when the proteins are in the closed state, where the 4-OH of d-threonate is 5.3 Å from the β-phosphate of ADP and again is consistent with Asp17 being the putative active site base. Structure-based sequence alignments suggest that all family members are kinases with the majority catalyzing the phosphorylation of four-carbon sugars.
Discussion
DUF families are a large set of uncharacterized protein families that are found in the Pfam database, and the number of DUFs is substantially increasing with the rapidly accumulating genome sequencing data (9). Many of the widespread DUFs in bacteria are biologically essential, and the importance of prioritizing these DUFs has been recognized in recent years (10). Before this study, only one DUF family, the DUF849 family of 922 proteins, had been evaluated in large scale by a single report (11), which heavily relied on previously published enzymatic activity and catalytic mechanism/liganded structure of one of its members (35, 36). Recognizing the difficulties associated with DUF functional assignment, we attempt to apply an integrated “genomic enzymology” strategy that had only been used for functional characterization of members of families with known functions (37, 38).
Here, we illustrate the power of the approach to accurately predict and verify functions across the DUF1537 family (PF07005 and PF17042, containing 4,610 sequences). An integrated SSN/GNN analysis of DUF1537 proteins identified three predominant conserved genome neighborhoods and permitted the prediction that DUF1537 proteins catalyze ATP-dependent phosphorylation. We constrained the possible kinase substrates upon recognizing that some of the neighborhoods also encoded SBPs that bound four-carbon monoacid sugars (18). Directed by the prediction, we confirmed functions for DUF1537 proteins from two of the three predominant genome neighborhoods: the PdxA2–DUF1537 and the aldolase–DUF1537 neighborhoods.
By characterizing multiple phylogenetically distinct DUF1537 proteins from each neighborhood, we demonstrate that the OtnK and DtnK annotations can be extrapolated to the entire corresponding SSN clusters (1,729 OtnK sequences and 724 DtnK sequences out of 3,673 sequences in the DUF1537 SSN) (Dataset S1). Characterization of the remaining genes and gene products from each neighborhood further permitted identification of novel functions in five additional Pfam families (PF04166, PF03446/PF14833, PF01370, PF01261, and PF00596) (SI Appendix, Table S11) in metabolic pathways for d-threonate, d-erythronate, and l-threonate.
To our knowledge, the pathways provide the first precedence for d-threonate and d-erythronate as biological substrates and offer the first complete catabolic pathway for l-threonate, a degradation product of the abundant plant metabolite l-ascorbate (39–42). The presence of a fusion protein (Uniprot: B3H739) in Arabidopsis thaliana that includes fused domains similar to LtnD, OtnK, and OtnC suggests that the l-threonate catabolic pathway also might be used by plants, potentially answering the 25-y-old question of the biological and environmental fate of l-ascorbate. Together with the previously identified tetritol metabolism in our laboratory (37), we have filled many of the gaps in four-carbon sugar metabolism in the Kyoto Encyclopedia of Genes and Genomes pathway database.
Members of the DUF1537 family with characterized kinase activities were also structurally characterized. Our liganded structures demonstrate that the acid sugar is bound by the N-terminal domain (PF07005) and nucleotide by the C-terminal domain (PF17042). To our knowledge, the studies also yielded the first “closed” DUF1537 structure where a domain rotation of 30° brings the acid-sugar and nucleotide binding site in proximity for catalysis; this structure provides a reliable model for in silico docking studies of kinases with unknown substrate specificities. For example, the functions of the RLP–DUF1537 proteins from the third conserved genome neighborhood remain unknown and are the subject of ongoing studies. From a structure/function comparison of OtnK and DtnK proteins, we observe that the functional groups at the substrate C3 correlate with genome context related conserved residues [Cys for OtnK, Asp for DtnK (Asp92) (Fig. 4C)] that are positioned to coordinate the C3 moiety (carbonyl for OtnK, hydroxyl for DtnK). Because the homologous residue in RLP–DUF1537 proteins is a Cys, we posit that the substrates of RLP–DUF1537 proteins are four-carbon backbone carboxylic acids with a carbonyl group at C3.
Our integrated genomic enzymology strategy, as demonstrated here, is a powerful tool for assigning enzymatic functions across entire protein families and elucidating metabolic pathways, especially when the generic reaction types of families are unknown and other methods, such as computational modeling, are inconclusive. We anticipate we will refine our strategy and experimentally characterize more DUF families.
UniProt Accession IDs.
This manuscript describes functional characterization of proteins with the following UniProt accession IDs: Q6D0N7, A0A0H3LX82, Q8ZRS5, B0TBI9, Q0K4F6, Q6D0N8, A0A0H3LQK8, P58718, B0TBI8, Q0K4F5, Q6CZ26, A0A0H2VA68, P44979, Q0KBC7, P44094, Q0KBD2, Q6CZ24, Q57199, Q0KBC9, A0A0H2VA12, Q0KBD1, Q6CZ23, Q57151, Q0KBC8, Q6CZ25, P44093, Q4KBD3, B1M1V6, A0A0H3KP73, A6VKK5, Q8YB10, A7JVV9, and Q48PB0 (SI Appendix, Table S11).
Materials and Methods
Enzyme assays were performed with a UV-Visible spectrophotometer (Varian CARY 300 Bio). The consumption or formation of NADH was monitored as the decrease or increase in the absorbance at 340 nm with an extinction coefficient (ε) of 6,220 M−1⋅cm−1. Locus tags and Uniprot identifiers for each gene and protein in this study can be found in SI Appendix, Table S11.
ATP-Dependent Kinase Activity Screening with a Focused Sugar Library.
ATP-Dependent kinase activity was determined spectrophotometrically by following consumption of NADH. The formation of ADP was coupled to the oxidation of NADH via pyruvate kinase (PK) and lactate dehydrogenase (LDH) in the presence of ATP, phosphoenolpyruvate (PEP), and NADH. The assay (200 μL) contained 1 μM of purified enzyme, 0.1 M Tris⋅HCl buffer (pH 8.0), 10 mM KCl, 10 mM MgCl2, 2.5 mM ATP, 2.5 mM PEP, 0.3 mM NADH, 5 U PK/LDH from rabbit muscle (Sigma), and 1 mM substrate. The reactions were performed in Corning 96-well clear flat-bottom UV-transparent microplates; the absorbance readings at 340 nm were measured with a TECAN microplate reader after incubating the reaction solution at 25 °C for 10 min, 2 h, and 16 h. The results of different time points allow estimations of relative activities (SI Appendix, Table S1).
Kinetic Assay for PdxA2–DUF1537 Protein Kinase Activity with d-Threonate, d-Erythronate, or 4-HT.
ATP-dependent kinase activities of PdxA2–DUF1537 proteins were assayed by measuring the consumption of NADH (see above for details). The reaction mixture (25 °C) contained variable concentrations of substrate, 100 mM Tris⋅HCl buffer (pH 8.0), 10 mM KCl, 10 mM MgCl2, 2.5 mM ATP, 2.5 mM PEP, 0.16 mM NADH, 5 U PK/LDH from rabbit muscle (Sigma), and enzyme in a final volume of 200 μL. Data were fit to the Michaelis–Menten equation (Table 1 and SI Appendix, Table S2).
Kinetic Assay for PdxA2 Oxidative Activity with d-Threonate 4-Phosphate, d-Erythronate 4-Phosphate, and 4-HT 4-Phosphate.
Oxidation activities of PdxA2 proteins were assayed by measuring the formation of NADH. The reaction mixture (25 °C) contained variable concentrations of substrate, 50 mM Tris⋅HCl buffer (pH 8.0), 1.5 mM NAD+, and enzyme in a final volume of 200 μL. Data were fit to the Michaelis–Menten equation (Table 1 and SI Appendix, Table S3).
Dehydrogenase Activity Screening with a Focused Sugar Library.
Purified dehydrogenases were screened for oxidation activity using a library of 53 sugars (SI Appendix, Table S6). Activity was assayed by measuring the formation of NADH. The assay (200 μL) contained 1.5 μM of purified enzyme, 50 mM Tris⋅HCl buffer (pH 8.0), 1.5 mM NAD+, and 1 mM substrate. The reactions were performed in Corning 96-well clear flat-bottom UV-transparent microplates, and the absorbance readings at 340 nm were measured with a TECAN microplate reader after incubating the reaction solution at 25 °C for 10 min, 2 h, and 16 h.
Kinetic Assay for Dehydrogenase Activity with l-Threonate or d-Erythronate.
Dehydrogenases were assayed by measuring the formation of formazan by the increase in absorbance at 500 nm. Briefly, NAD(P)H was generated in the oxidation of l-threonate or d-erythronate. This reaction was coupled to diaphorase, which used the NAD(P)H to catalyze reduction of p-iodonitrotetrazolium violet (INT) into formazan (molar extinction coefficient 12,990 M−1⋅cm−1). The reaction mixture (25 °C) contained variable concentrations of substrate, 50 mM Tris⋅HCl buffer (pH 8.5), 1.5 mM NAD(P)+, 0.64 mM INT (Sigma), 2 unit diaphorase from Clostridium kluyveri (Sigma) and enzyme in a final volume of 200 μL. Data were fit to the Michaelis–Menten equation (SI Appendix, Table S4).
Bacterial Strains and Growth Conditions.
R. eutropha H16 (DSM-428) and its derived strains were grown shaking aerobically in nutrient broth (Difco) or in defined media [per liter: 50 mL 20× salts (20 g NH4Cl, 6 g MgSO4·7H2O, 3 g KCl, 0.1 g CaCl2·2H2O, 0.05 g FeSO4·7H2O), 50 mL 20× phosphate buffer pH 7.0 (500 mM KH2PO4 mixed with 500 mM Na2HPO4 until pH 7.0), 1 mL 1,000× trace element solution (per liter: 0.5 g NaEDTA, 0.3 g FeSO4·7H2O, 3 mg MnCl2·4H2O, 5 mg CoCl2·6H2O, 1 mg CuCl2·2H2O, 2 mg NiCl2·6H2O, 3 mg Na2MoO4·2H2O, 5 mg ZnSO4·7H2O, 2 mg H3BO3), 1 mL 1,000× vitamin solution (per liter: 0.1 g cyanocobalamin, 0.3 g pyridoxamine-2 HCL, 0.1 g Ca-d-pantothenate, 0.2 g thiamine dichloride, 0.2 g nicotinic acid, 0.08 g 4-aminobenzoic acid, 0.02 g d-biotin), and 900 mL H2O] with 140 µg/mL kanamycin as necessary. S. enterica serovar Typhimurium LT2 and its derivatives were grown shaking in LB or in defined media at 37 °C with 50 µg/mL kanamycin as necessary. E. coli strains DH5α,S17-1 (43) and BL21(DE3) were grown shaking aerobically in Luria-Bertani (LB) broth at 37 °C with 100 μg/mL ampicillin or 50 μg/mL kanamycin as necessary. P. carotovorum WPP14 and its derived strains (16) were grown at 30 °C in LB or defined media with 25 µg/mL kanamycin and 34 µg/mL chloramphenicol as necessary. Plates were prepared with 1.8% (wt/vol) agar for rich medium and 2.5% (wt/vol) agar for defined medium. Defined medium was supplemented with 10 mM carbon source. Growth studies were performed as 300 µL cultures in a Bioscreen C instrument based on optical density (OD) at 600 nm. The inoculum was derived from mid exponential cultures in rich media; cells were first collected by centrifugation and then rinsed with defined medium once before inoculation.
Isolation and Complementation of Markerless Deletion Strains of R. eutropha and S. enterica.
Deletion strains of R. eutropha were isolated as previously described (44) with modifications described below. Deletions removed in-frame portions of the coding region without introducing any exogenous sequence in the final strain. Briefly, 750–1,000 bp of regions flanking the targeted coding region were amplified by PCR (for primers, see SI Appendix, Table S13). Each fragment contained ≥27 bp of the coding region and preserved the reading frame of the coding region. Using Gibson cloning (New England Biolabs), the fragments were assembled and inserted into pK18mobsacB (45) digested with XbaI/BamHI. The plasmid was transferred into R. eutropha via conjugation with E. coli S17-1 transformed with the plasmid. Single cross-over strains were isolated as kanamycin-resistant on nutrient plates. Colonies were additionally streaked for isolation on selective nutrient plates. Resulting single colonies were incubated in nonselective nutrient broth until OD ∼0.8. A 100-µL aliquot of a 100-fold dilution of the culture was plated on nutrient plates containing 10% sucrose. Double cross-over strains were identified as kanamycin-sensitive after colonies were patched on nutrient plates with and without kanamycin. Among the double crossover strains, wild-type revertants were separated from mutants by colony PCR using primers that directed amplification across the deleted region. Mutant genotypes were confirmed by sequencing the entire region of the genome that was available for recombination with the fragments contained on the corresponding plasmid (see SI Appendix, Table S13 for plasmids).
A modified version of the Datsenko and Wanner (46) protocol was used to delete regions of the S. enterica genome. Genes were inactivated by replacing all but the first and last 27 bp of the coding region with an 85-bp scar sequence. Briefly, each of three PCR products were generated: (i) arm1, ∼1,000 bp upstream of the target gene; (ii) the 1.6-kb kanamycin-resistance cassette from pKD4 (46); and (iii) arm2, ∼1,000 bp downstream of the target gene. Primer sequences are provided in SI Appendix, Table S13. The fragments were assembled by overlapping extension PCR (Ho Gene 1989) based on ∼50 bp of shared sequence between adjacent fragments. Assembly primers for arm1 and the kanamycin-resistance cassette were appropriately chosen among those used to amplify the individual fragments. The final assembled ∼3.6-kb fragment was assembled using appropriate primers and consisted of the kanamycin-resistance cassette flanked by arm1 and arm2. Electrocompetent S. enterica carrying the λ-Red helper plasmid pKD46 was transformed with 100 ng of the final assembled product according to the protocol of Datsenko and Wanner (46). Double cross-over strains were isolated as kanamycin-resistant and confirmed by genomic PCR. The kanamycin-resistance genes were eliminated using helper plasmid pCP20 encoding the FLP recombinase. Final strains were cured of all plasmids as described previously (46).
Complementation of the mutant strains relied on expressing the respective deleted gene from the h16_A1563 promoter (279 bp) for h16_A1557 – h16_A1562 and the stm0161 promoter (234 bp) for stm0162 and stm0163. Promoters and coding regions were amplified separately by PCR (see SI Appendix, Table S13 for primers). The products were fused and ligated into pBBR1MCS2 (47) (digested with EcoRI/XbaI) by Gibson ligation (New England Biolabs). Plasmids were transferred into R. eutropha strains by conjugation with S17-1 transformed with the corresponding plasmid. Plasmids were transferred into S. enterica strains by electroporation.
Supplementary Material
Acknowledgments
This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly Company, which operates the facility. This research was supported by NIH U54GM093342.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4XGJ, 4XFM, 4XFR, 4XG0, and 5DMH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605546113/-/DCSupplemental.
References
- 1.Galperin MY, Koonin EV. From complete genome sequence to ‘complete’ understanding? Trends Biotechnol. 2010;28(8):398–406. doi: 10.1016/j.tibtech.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galperin MY. Conserved ‘hypothetical’ proteins: New hints and new puzzles. Comp Funct Genomics. 2001;2(1):14–18. doi: 10.1002/cfg.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galperin MY, Koonin EV. ‘Conserved hypothetical’ proteins: Prioritization of targets for experimental study. Nucleic Acids Res. 2004;32(18):5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol. 2003;333(4):863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 5.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLOS Comput Biol. 2009;5(12):e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsiao TL, Revelles O, Chen L, Sauer U, Vitkup D. Automatic policing of biochemical annotations using genomic correlations. Nat Chem Biol. 2010;6(1):34–40. doi: 10.1038/nchembio.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol. 2007;8(12):995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- 8.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman A, Coggill P, Finn RD. DUFs: Families in search of function. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(Pt 10):1148–1152. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodacre NF, Gerloff DL, Uetz P. Protein domains of unknown function are essential in bacteria. MBio. 2013;5(1):e00744-13. doi: 10.1128/mBio.00744-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastard K, et al. Revealing the hidden functional diversity of an enzyme family. Nat Chem Biol. 2014;10(1):42–49. doi: 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, et al. The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat Commun. 2014;5:4339. doi: 10.1038/ncomms5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash A, Yogeeshwari S, Sircar S, Agrawal S. Protein domain of unknown function 3233 is a translocation domain of autotransporter secretory mechanism in gamma proteobacteria. PLoS One. 2011;6(11):e25570. doi: 10.1371/journal.pone.0025570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold R, Jehl A, Rattei T. Targeting effectors: The molecular recognition of type III secreted proteins. Microbes Infect. 2010;12(5):346–358. doi: 10.1016/j.micinf.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson SW. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176(4):1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mole B, Habibi S, Dangl JL, Grant SR. Gluconate metabolism is required for virulence of the soft-rot pathogen Pectobacterium carotovorum. Mol Plant Microbe Interact. 2010;23(10):1335–1344. doi: 10.1094/MPMI-03-10-0067. [DOI] [PubMed] [Google Scholar]
- 17.Su YC, Wan KL, Mohamed R, Nathan S. A genome level survey of Burkholderia pseudomallei immunome expressed during human infection. Microbes Infect. 2008;10(12-13):1335–1345. doi: 10.1016/j.micinf.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 18.Vetting MW, et al. Experimental strategies for functional annotation and metabolism discovery: Targeted screening of solute binding proteins and unbiased panning of metabolomes. Biochemistry. 2015;54(3):909–931. doi: 10.1021/bi501388y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulligan C, Fischer M, Thomas GH. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol Rev. 2011;35(1):68–86. doi: 10.1111/j.1574-6976.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- 20.Higgins CF. ABC transporters: Physiology, structure and mechanism--an overview. Res Microbiol. 2001;152(3-4):205–210. doi: 10.1016/s0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhao S, et al. Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks. eLife. 2014;3:1–32. doi: 10.7554/eLife.03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlt JA, et al. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 2015;1854(8):1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanai I, Derti A, DeLisi C. Genes linked by fusion events are generally of the same functional category: A systematic analysis of 30 microbial genomes. Proc Natl Acad Sci USA. 2001;98(14):7940–7945. doi: 10.1073/pnas.141236298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabita FR, et al. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71(4):576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauder MJ, et al. High throughput protein production and crystallization at NYSGXRC. Methods Mol Biol. 2008;426:561–575. doi: 10.1007/978-1-60327-058-8_37. [DOI] [PubMed] [Google Scholar]
- 26.Cane DE, Hsiung YJ, Cornish JA, Robinson JK, Spenser ID. Biosynthesis of vitamin B-6: The oxidation of 4-(phosphohydroxy)-L-threonine by PdxA. J Am Chem Soc. 1998;120(8):1936–1937. [Google Scholar]
- 27.Sivaraman J, et al. Crystal structure of Escherichia coli PdxA, an enzyme involved in the pyridoxal phosphate biosynthesis pathway. J Biol Chem. 2003;278(44):43682–43690. doi: 10.1074/jbc.M306344200. [DOI] [PubMed] [Google Scholar]
- 28.Wolf E, Spenser ID. [2,3-C-13(2)]-4-hydroxy-L-threonine. J Org Chem. 1995;60(21):6937–6940. [Google Scholar]
- 29.Joerger AC, Gosse C, Fessner WD, Schulz GE. Catalytic action of fuculose 1-phosphate aldolase (class II) as derived from structure-directed mutagenesis. Biochemistry. 2000;39(20):6033–6041. doi: 10.1021/bi9927686. [DOI] [PubMed] [Google Scholar]
- 30.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J Biol Chem. 2001;276(18):15131–15136. doi: 10.1074/jbc.M100220200. [DOI] [PubMed] [Google Scholar]
- 31.Thoden JB, et al. Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36(21):6294–6304. doi: 10.1021/bi970025j. [DOI] [PubMed] [Google Scholar]
- 32.Ashiuchi M, Misono H. Biochemical evidence that Escherichia coli hyi (orf b0508, gip) gene encodes hydroxypyruvate isomerase. Biochim Biophys Acta. 1999;1435(1-2):153–159. doi: 10.1016/s0167-4838(99)00216-2. [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 34.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins. 1998;30(2):144–154. [PubMed] [Google Scholar]
- 35.Kreimeyer A, et al. Identification of the last unknown genes in the fermentation pathway of lysine. J Biol Chem. 2007;282(10):7191–7197. doi: 10.1074/jbc.M609829200. [DOI] [PubMed] [Google Scholar]
- 36.Bellinzoni M, et al. 3-Keto-5-aminohexanoate cleavage enzyme: a common fold for an uncommon Claisen-type condensation. J Biol Chem. 2011;286(31):27399–27405. doi: 10.1074/jbc.M111.253260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, et al. A general strategy for the discovery of metabolic pathways: d-threitol, l-threitol, and erythritol utilization in Mycobacterium smegmatis. J Am Chem Soc. 2015;137(46):14570–14573. doi: 10.1021/jacs.5b08968. [DOI] [PubMed] [Google Scholar]
- 38.Wichelecki DJ, et al. ATP-binding cassette (ABC) transport system solute-binding protein-guided identification of novel d-altritol and galactitol catabolic pathways in Agrobacterium tumefaciens C58. J Biol Chem. 2015;290(48):28963–28976. doi: 10.1074/jbc.M115.686857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smirnoff N. Vitamin C: The metabolism and functions of ascorbic acid in plants. Adv Bot Res. 2011;59:107–177. [Google Scholar]
- 40.Green MA, Fry SC. Vitamin C degradation in plant cells via enzymatic hydrolysis of 4-O-oxalyl-L-threonate. Nature. 2005;433(7021):83–87. doi: 10.1038/nature03172. [DOI] [PubMed] [Google Scholar]
- 41.Loewus FA. Biosynthesis and metabolism of ascorbic acid in plants and of analogs of ascorbic acid in fungi. Phytochemistry. 1999;52(2):193–210. [Google Scholar]
- 42.Williams M, Loewus FA. Biosynthesis of (+)-tartaric acid from l-[4-C]ascorbic acid in grape and geranium. Plant Physiol. 1978;61(4):672–674. doi: 10.1104/pp.61.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Au AC, et al. Study of acute trichinosis in Ghurkas: Specificity and sensitivity of enzyme-linked immunosorbent assays for IgM and IgE antibodies to Trichinella larval antigens in diagnosis. Trans R Soc Trop Med Hyg. 1983;77(3):412–415. doi: 10.1016/0035-9203(83)90175-x. [DOI] [PubMed] [Google Scholar]
- 44.Carter MS, Alber BE. Transcriptional regulation by the short-chain fatty acyl coenzyme A regulator (ScfR) PccR controls propionyl coenzyme A assimilation by Rhodobacter sphaeroides. J Bacteriol. 2015;197(19):3048–3056. doi: 10.1128/JB.00402-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schäfer A, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145(1):69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovach ME, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166(1):175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.